Abstract
Gelsolin is a calcium-, pH- and lipid-dependent actin filament severing/capping protein whose main function is to regulate the assembly state of the actin cytoskeleton. Gelsolin is associated with membranes in cells, and it is generally assumed that this interaction is mediated by PPIs (polyphosphoinositides), since an interaction with these lipids has been characterized in vitro. We demonstrate that non-PPI lipids also bind gelsolin, especially at low pH. The data suggest further that gelsolin becomes partially buried in the lipid bilayer under mildly acidic conditions, in a manner that is not dependent of the presence of PPIs. Our data also suggest that lipid binding involves a number of sites that are spread throughout the gelsolin molecule. Linker regions between gelsolin domains have been implicated by other work, notably the linker between G1 and G2 (gelsolin domains 1 and 2 respectively), and we postulate that the linker region between the N-terminal and C-terminal halves of gelsolin (between G3 and G4) is also involved in the interaction with lipids. This region is compatible with other studies in which additional binding sites have been located within G4–6. The lipid–gelsolin interactions reported in the present paper are not calcium-dependent, and are likely to involve significant conformational changes to the gelsolin molecule, as the chymotryptic digest pattern is altered by the presence of lipids under our conditions. We also report that vesicle-bound gelsolin is capable of binding to actin filaments, presumably through barbed end capping. Gelsolin bound to vesicles can nucleate actin assembly, but is less active in severing microfilaments.
Keywords: actin cytoskeleton, gelsolin, lipid, polyphosphoinositides
Abbreviations: Br-PC, 1,2-dibromostearoyl-sn-glycero-3-phosphocholine; DPH, 1,6-diphenyl-1,3,5-hexatriene; FRET, fluorescence resonance energy transfer; G1–6, The six repeated domains of gelsolin; IPTG, isopropyl β-D-thiogalactoside; KD, Stem–Volmer constant; NBD-PC, 1-palmitoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphocholine; PC, phosphatidylcholine (1,2-dioleyl-sn-glycero-3-phosphocholine); PG, phosphatidylglycerol (1,2-diacyl-sn-glycero-3-phosphoglycerol); PPI, polyphosphoinositide; PS, phosphatidylserine (1,2-diacyl-sn-glycero-3-phospho-L-serine); SUV, small unilamellar vesicle
INTRODUCTION
The actin cytoskeleton is a major factor that determines and maintains the shape of typical eukaryotic cells. It also confers the ability of cells to crawl and is responsible for other motile phenomena, such as the movement of vesicles and cytokinesis. Actin microfilaments are regulated by approx. 100 actin-binding proteins [1] that arrange and organize the microfilaments. The functions of the actin-binding proteins are in turn regulated by various signalling molecules, including a variety of lipids [2]. The PPIs (polyphosphoinositides) have been particularly well characterized in this regard.
The PPIs are a group of lipids with phosphates attached to carbons in the inositol ring that endow the head groups with an overall negative charge and the ability to be specifically bound by a large number of proteins that are involved in signal transduction. A huge variety of different signalling lipids are generated from the PPIs, and these relay messages downstream by their binding proteins [3]. Signalling proteins contain various motifs such as PH (pleckstrin homology) [4], PX (phox homology) [5] and FYVE (Fab-1, YOTB, Vac1p and EEA1) [6] through which these proteins bind specific PPIs. Profilin was the first actin-binding protein to be discovered to also be a PtdIns(4,5)P2-binding protein [7]; however, profilin does not contain a recognized motif for binding PPIs that has homologous regions in other proteins. Subsequently, a large number of other actin-binding proteins have been found to bind PPIs (reviewed in [8]), and genetic evidence suggests a role for these lipids in actin polymerization in vivo [9].
One of these proteins is gelsolin, an actin-binding protein that is regulated by calcium [10], pH [11,12] and PPIs [13]. Gelsolin consists of six similar repeated domains (G1–6) that have four distinct actin-binding sites, within G1, G2, G4 and G6 (reviewed in [14]). Through these, gelsolin is able to nucleate actin polymerization from the pointed end, sever actin filaments and cap the barbed end of filaments. In resting cells, the barbed ends of filaments are capped by proteins such as gelsolin, and these are removed in some regions of the cell upon stimulation with chemotactic stimuli or growth factors. The presence of PPIs removes gelsolin from the pivotal barbed end of actin filaments, thereby controlling actin polymerization in vitro [13], and is reported to do so in vivo [15]. These early in vitro studies [13] used PtdIns(4,5)P2 micelles, whose surfaces do not accurately reflect those encountered on the inner membrane because they have a high curvature, being only 6 nm in diameter [16]. Subsequent studies using more physiological PPI-containing vesicles also demonstrated that these lipids dissociated the gelsolin–actin complexes and uncapped the barbed end of gelsolin bound filaments [17]. PPI-binding regions within gelsolin have been localized to two regions within the N-terminal half of gelsolin (G1–3), with additional contributions being made from parts of the C-terminal half (G4–6). Similar sites are reported within the N-terminus of villin, with an additional site in the C-terminal ‘head domain’ [18].
In the present paper, we report that at low pH values within the physiologically normal range, gelsolin binds lipids in the absence of PPI, and that it becomes partially embedded in vesicles. We confirm the contribution of the C-terminal half of gelsolin (G4–6) in this interaction. Gelsolin may therefore provide a stable membrane anchor for actin filaments in regions of low PPI concentration that may then be released upon local increases of PPI production.
In addition to binding to actin and regulating the actin cytoskeleton, gelsolin has numerous ancillary functions, such as an involvement in apoptosis [19], and functions that do not have any clear connection to the cytoskeleton. For example, gelsolin doubles as a crystallin in the fish eye [20], is reported to act as a morphogen in development [21] and may function to carry certain lipids in serum [22]. Lipid binding may play an important part in all of these other functions that gelsolin performs. In the present paper, we report that gelsolin binds to lipid vesicles by partial insertion, and that the vesicle-bound gelsolin is capable of binding to actin filaments, presumably through barbed end capping.
MATERIALS AND METHODS
Proteins and peptides
All recombinant proteins in this study were expressed in Escherichia coli BL21 derivatives using the pMW172 vector [23]. A gelsolin amino-acid numbering system based on the human plasma gelsolin [24] is used in the present paper as has become standard [25–27]. For example, G1 is defined as extending from Pro39 to Tyr133, and G2 as being Gly137 to Leu247. G1–3 (residues 1–407) was expressed in BL21(de3) cells and was purified from the soluble fraction of the bacteria as described previously [26]. G4–6 (residues 407–755) [26] was produced in BL21(de3) cells and was purified from inclusion bodies. Whole gelsolin was produced in E. coli BL21(de3). Soluble protein was dialysed against 10 mM Tris/HCl, pH 8.0, 1 mM EGTA, 1 mM sodium azide and 50 mM NaCl, and was added to a DE52 column equilibrated with the same buffer. Pure gelsolin was eluted from the column with 10 mM Tris/HCl, pH 8.0, 2 mM CaCl2, 1 mM sodium azide and 50 mM NaCl [28]. Rhodamine-labelled synthetic peptide derived from gelsolin sequences, RhoB–QRLFQVKGRR was obtained as described previously [29]. Biotinylation of gelsolin by BACNHS (biotinamidocaproate N-hydroxysuccinimide ester) was performed as previously reported [30]. Excess reagents were eliminated by chromatography on a PD10 column (Amersham Biosciences) in 0.1 M NaHCO3 buffer, pH 8.6. Rabbit skeletal muscle actin was produced from acetone powder [31], and was labelled at Cys374 by pyrenyl iodoacetamide [32].
Gelsolin chymotryptic fragments were electroblotted on to a PVDF membrane and subjected to N-terminal sequencing using a PerkinElmer Procise 492 sequencer operated according to the supplied pulsed liquid program.
Preparation of liposomes: SUVs (small unilamellar vesicles)
PC (1,2-dioleyl-sn-glycero-3-phosphocholine or phosphatidylcholine), Br-PC [1,2-dibromostearoyl-sn-glycero-3-phosphocholine; catalogue number 850366] and NBD-PC (1-palmitoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphocholine; catalogue number 810130) were purchased from Avanti Polar Lipids. PG (1,2-diacyl-sn-glycero-3-phosphoglycerol or phosphatidylglycerol), PS (1,2-diacyl-sn-glycero-3-phospho-L-serine or phosphatidylserine) and PtdIns(4,5)P2 [1,2-diacyl-sn-glycero-3-phospho-(1-o-myo-inositol 4,5-bisphosphate] were purchased from Sigma.
A mixture containing phospholipids in chloroform were dried under a stream of N2 with a TurboVap Evaporator (Zymark, Hopkinton, MA, U.S.A.). They were dried further under high vacuum for 1 h. The dried lipids were resuspended in water at 10 mg/ml and then sonicated with a micro-tip sonicator (Branson Digital Sonifier) at 10% amplitude until nearly optically clear.
Immunological techniques
The ELISA technique, described previously in detail in [33], was used to monitor the interaction of biotinylated gelsolin with phospholipids that were immobilized on plastic microtitre wells. Lipids were coated at 40 μg/ml by the ELISA methods that we have characterized previously and have proven to be specific [33]. We have rigorously tested the binding assay used in the present study [34]. Peptides derived from CTP:phosphocholine-cytidylyltransferase were used to test specific SUV binding [34], and we found that only those peptides from the well characterized lipid–protein interface interacted with coated SUVs (M. Larvor, R. Cerdan and C. Roustan, unpublished work). Interaction was monitored using alkaline-phosphatase-labelled streptavidin (diluted 1/1000). Control assays were carried out in wells saturated with gelatin and with gelatin hydrolysate alone. Each assay was conducted in triplicate, and the mean values were plotted after subtraction of non-specific absorption. The binding parameters (apparent dissociation constant, Kd, and the maximal binding, Amax) were determined by non-linear fitting:
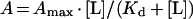 |
(1) |
where A is the absorbance at 405 nm and [L] is the ligand concentration, by using the CurveFit software developed by K. Raner (Mt Waverley, Vic, Australia). Additional details on the different experimental conditions are given in the Figure legends.
Fluorescence measurements
Fluorescence experiments were conducted with a LS 50 PerkinElmer luminescence spectrometer. Emission spectra for the intrinsic fluorescence of tryptophan were obtained with a wavelength of excitation at 280 nm. The fluorescent reagent DPH (1,6-diphenyl-1,3,5-hexatriene) was purchased from Sigma–Aldrich.
Collisional quenching of a fluorophore, such as tryptophan in the present study, is described by the Stern–Volmer equation, F°/F=1+KD·[Q] where F and F° are the fluorescence intensities in the presence and in the absence of the quencher (Q) respectively, and KD is the Stern–Volmer constant [35]. The constant KD depends upon the lifetime of fluorescence without quencher and the bimolecular rate constant for the quencher. In the present study, acrylamide was chosen as the quencher.
Actin polymerization
Actin polymerization and depolymerization were monitored by fluorescence measurements. Fluorescence increase of pyrenyl-actin [32,36] was used as an indicator of actin polymerization. Excitation and emission wavelengths for pyrenyl-actin were set at 355 and 386 nm respectively. Actin polymerization was induced by the addition of 2 mM MgCl2 and 0.1 M KCl. Nucleation enhanced by gelsolin was determined by measuring the rate of actin polymerization [37,38]. G-actin (2.3 μM) was added to 2 mM MgCl2 and 0.1 M KCl, 20 μM ATP, 0.5 mM CaCl2 and 20 mM Tris/HCl buffer at pH 7.5 or 2 mM MgCl2 and 0.1 M KCl, 20 μM ATP and 20 mM Mes buffer at pH 5.8 in the presence or absence of 26 nM gelsolin. The severing activity of gelsolin is directly related to the depolymerization rate [26] observed when 25 μM fluorescent F-actin (pre-capped by 1:1000 molar ratio of gelsolin) is diluted to 480 nM in the two buffers described above in the presence of 26 or 52 nM gelsolin. Results obtained with or without SUVs (0.2 mg/ml) (25% PG+75% PC) were compared.
Confocal microscopy observations
Liposomes and actin filaments were observed at 25 °C using confocal microscopy (Leica TCS 4D microscope). To observe liposomes by fluorescence microscopy, the SUVs were prepared by adding 2% NBD-PC. To visualize F-actin, rhodamine–phalloidin (R+415; Molecular Probes) was added. The final concentration of rhodamine–phalloidin in microscope specimens was about 20 nM. The preparations were mounted in Mowiol.
Analytical methods
Protein concentrations were determined by UV absorbance using a Varian MS 100 spectrophotometer. Gelsolin domain concentrations were determined spectrophotometrically using values of A280 (1 cm−1)=15.5 μM for G4–6, 21.0 μM for G1–3 and 8.93 μM for whole gelsolin. These extinction coefficients were calculated by tryptophan, tyrosine and cysteine content [39]. Electrophoresis was carried out on 15% (w/v) polyacrylamide slab gels, according to the method of Laemmli [40], and stained with Coomassie Blue.
RESULTS
Interaction of gelsolin with various phospholipid mixtures: evidence by intrinsic tryptophan fluorescence
It has been reported that a critical concentration of PtdIns(4,5)P2 is necessary in order to inhibit gelsolin activity in a background of mixed lipids [41]; no effect was observed when PtdIns(4,5)P2 was present at 3% compared with the change observed in the presence of 10% PtdIns(4,5)P2. Interestingly, it was observed that addition of 20% PS to the vesicles dropped this critical concentration of gelsolin activity to 3% PtdIns(4,5)P2 (data not shown in [41]). We therefore tested the possibility that gelsolin would interact with mixed lipids (PC+PG or PS) in vesicular form (SUV), even in the absence of PPI.
In a first set of experiments, we analysed the ability of gelsolin to interact with liposomes at neutral pH by monitoring fluorescence changes due to the modification of the gelsolin tryptophan environment. Table 1 shows that the changes in tryptophan fluorescence depend on the composition of the liposomes. No significant change was observed with 100% PC, a small, but significant, fluorescence enhancement (1.04) occurs in mixed lipids (75% PC+25% PS or PG). This interaction is not calcium-dependent. In contrast, the presence of 30% PtdIns(4,5)P2 with PC and PG, induced a specific change (fluorescence quenching) in the environment of the gelsolin tryptophan residues. As shown in Table 1, a 10% fluorescence decrease was observed. In addition, while 3% PtdIns(4,5)P2 in pure PC liposomes does not induce a significant fluorescence change, the presence of PG in the same micelles induces a fluorescence quenching which indicates that gelsolin binds to PtdIns(4,5)P2. In a control experiment, we observed the expected fluorescence quenching (up to 36% in our experimental conditions; Table 1) induced by the binding of gelsolin to pure PtdIns(4,5)P2 micelles. These last results are in agreement with previous reports [41,42].
Table 1. Interaction of gelsolin (20 μg/ml) with PG/PC SUVs (0.4 mg/ml) was monitored by fluorescence.
Enhancement (F/F°) in the intensity of the fluorescence emission spectra of gelsolin tryptophan residues were recorded at pH 7.0 in 0.15 M NaCl and 1 mM EGTA or 1 mM CaCl2 and 0.05 M Tris/HCl buffers. Excitation was fixed at 280 nm and emission was recorded at 340 nm.
| Lipid mixture | Changes in tryptophan fluorescence (F/F°) |
|---|---|
| PC (100%) | 1.00±0.01 |
| PC (75%)+PG (25%)+EGTA | 1.04±0.01 |
| PC (75%)+PG (25%)+Ca2+ | 1.04±0.02 |
| PC (75%)+PS (25%)+EGTA | 1.04±0.02 |
| PC (70%)+PtdIns(4,5)P2 (30%) | 0.90±0.02 |
| PC (97%)+PtdIns(4,5)P2 (3%) | 1.00±0.01 |
| PC (75%)+PG (22%)+PtdIns(4,5)P2 (3%) | 0.96±0.02 |
| PtdIns(4,5)P2 (100%) | 0.64±0.04 |
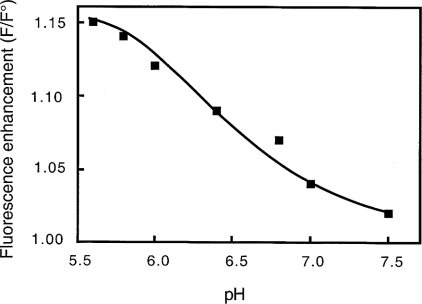
In a previous study [12], we demonstrated that the change in gelsolin conformation during activation resulting from a drop in pH is different from that induced by calcium. In the present study, we tested the effect of pH on the interaction of gelsolin to liposomes consisting of PC/PG or PC/PS. In the presence of PC/PG liposomes, an increase in tryptophan fluorescence induced by the interaction with SUVs following a pH decrease was observed (Figure 1). Similar results were obtained with PC/PS liposomes (results not shown). A maximum fluorescence enhancement of 16±2% (mean of five experiments) is observed at pH below 6. The data reported in Figure 1 are consistent with an assumed pKa of approx. 6.5. These results suggest a direct pH-dependent interaction with mixed phospholipids, resulting in an alteration of the environment of intrinsic tryptophan in gelsolin. This fluorescence change, as shown in Figure 2, is time-dependent, with an apparent rate constant of 0.60±0.06 min−1.
Figure 1. Effects of pH on the interaction of gelsolin with PG/PC SUVs.
Interaction of gelsolin with SUVs (0.4 mg/ml) (25% PG+75% PC) was monitored by fluorescence. Enhancement (F/F°) in the intensity of the fluorescence emission spectra of gelsolin tryptophan residues were recorded at various pH between 5.6 and 7.5 in 0.15 M NaCl, 1 mM EGTA and 0.05 M Mes or Tris/HCl buffers. Excitation was fixed at 280 nm and emission was recorded at 340 nm.
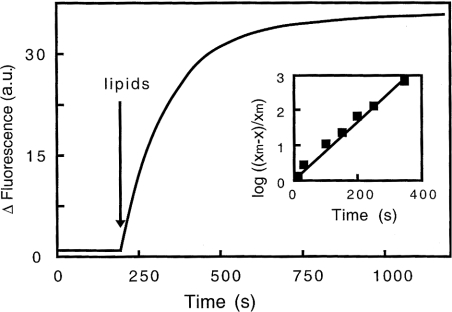
Figure 2. Kinetics of the interaction of gelsolin with PG/PC SUVs at pH 5.6 monitored by fluorescence.
SUVs (25% PG+75% PC, 0.4 mg/ml) were added to gelsolin in 0.15 M NaCl, 5 mM EGTA and 0.05 M acetate buffer, pH 5.6, and changes in fluorescence intensity of gelsolin tryptophan residues at 340 nm were recorded over time. Inset, graphical determination of the constant rate related to the fluorescence enhancement induced by gelsolin interaction with PG/PC SUVs, log [(xm−x)/xm] is plotted against time, where xm is the maximum fluorescence change observed and x is the fluorescence change at time t. a.u., arbitrary units.
Adsorption of gelsolin to PC/PG liposomes
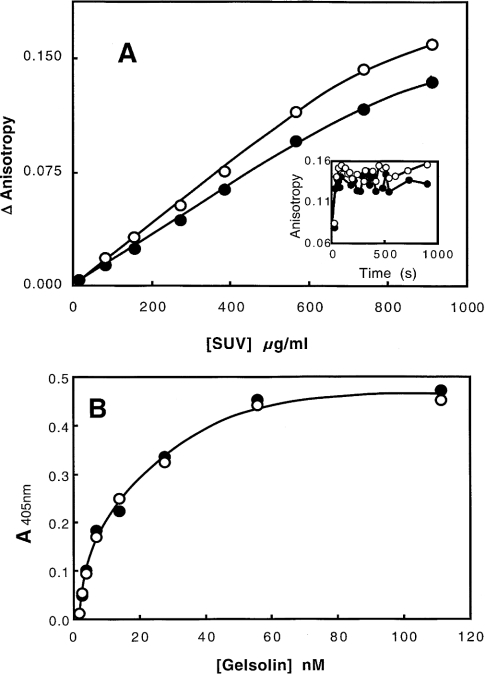
The ability of gelsolin to bind to liposomes was analysed by fluorescence anisotropy changes and ELISA. Figure 3(A) shows that the fluorescence anisotropy of gelsolin tryptophan increases with the concentration of phospholipids varied between 0 and 0.9 mg/ml, thus demonstrating a direct association between the protein and liposomes. A similar change is observed for two pH values (7.0 and 5.6). However, as the size distribution of the liposomes is likely to depend in part on the lipid concentration, a quantitative analysis of the gelsolin binding was not possible. In addition, the increase in tryptophan fluorescence shown in Figure 2 is time-dependent, whereas the increase in fluorescence anisotropy is instantaneous (Figure 3A, inset). The binding corresponds to a direct interaction with PG/PC SUVs, followed by a reorganization of the gelsolin–phospholipid complex.
Figure 3. Binding of gelsolin to PG/PC SUVs.
(A) Interaction of gelsolin monitored by tryptophan fluorescence anisotropy. Experiments were carried out in 0.15 M NaCl, 5 mM EGTA and 0.05 M acetate buffer, pH 5.6 (●), or 0.15 M NaCl, 1 mM EGTA and 0.05 M Tris/HCl buffer, pH 7.0 (○). Anisotropy change is plotted against PG/PC SUV concentration. Inset, PG/PC SUVs (0.4 mg/ml) were added to gelsolin at zero time, and anisotropy was recorded over time at pH 5.6 (●) or pH 7.0 (○). (B) Interaction of gelsolin with coated phospholipids (0.04 mg/ml) (25% PG+75% PC). Biotinylated gelsolin was added at various concentrations (0–110 nM) at pH 5.6 (●) or pH 7.0 (○) in the same buffers, and the binding was monitored at 405 nm.
In order to circumvent these problems, we studied the interaction further using an ELISA approach. In these experiments, lipids were coated to the wells of plastic microtitre plates, and the interaction of biotinylated gelsolin was monitored using alkaline-phosphatase-labelled streptavidin. A similar saturation curve was obtained at the two pH values tested (pH 6.0 and 7.2), and an apparent Kd of 20 nM was estimated (Figure 3B). The orientation of the gelsolin molecule with respect to the phospholipid surface has yet to be determined.
Does gelsolin insert into phospholipid liposomes?
To determine if the gelsolin molecule intercalates into the hydrophobic part of the lipids in liposomes, three approaches were tested:
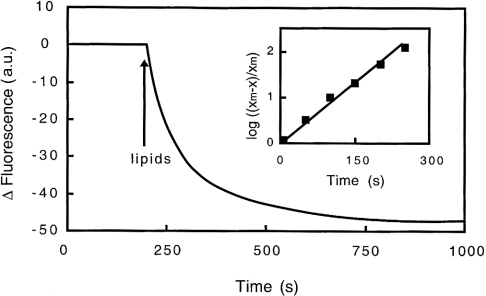
(i) When gelsolin is added to brominated vesicles containing 25% PG, 37.5% Br-PC and 37.5% PC, the emission spectrum of tryptophan is quenched up to 19% at pH 5.6 (Figure 4). The spectral effect is time-dependent, with a constant rate similar to that reported for unlabelled SUVs (0.6 min−1). The fluorescence decrease is also pH-dependent, the effect being of only 10% at pH 6.4 and less than 3% at pH 7.0. At neutral pH, the small variation observed is not enhanced in the presence of calcium (Table 2). The fluorescence quenching observed at acidic pH suggests proximity between gelsolin and the quencher (Br) located into the hydrophobic core of the vesicle.
Figure 4. Kinetics of the interaction of gelsolin with Br-PC/PG SUVs at pH 5.6 monitored by fluorescence.
SUVs (25% PG, 37.5% Br-PC and 37.5% PC) at 0.4 mg/ml were added to gelsolin in 0.15 M NaCl, 5 mM EGTA and 0.05 M acetate buffer, pH 5.6, and fluorescence quenching of gelsolin tryptophan residues was recorded over time. Inset, graphical determination of the constant rate related to the fluorescence quenching induced by gelsolin interaction with Br-PC/PG SUVs, log [(xm−x)/xm] is plotted against time, where xm is the maximum fluorescence change observed and x is the fluorescence change at time t. a.u., arbitrary units.
Table 2. BrPC-PG SUVs (0.4 mg/ml) were added to gelsolin (20 μg/ml) in 0.15 NaCl, 5 mM EGTA and 0.05 M acetate or Mes for pH 5.6, 0.05 M Mes for pH 6.4 and 0.05 M Tris/HCl for pH 7.0.
| Lipid mixture | pH | Quenching (%) |
|---|---|---|
| 37.5% PC+37.5% Br-PC+25% PG+EGTA | 5.6 | 19±2 |
| 37.5% PC+37.5% Br-PC+25% PG+EGTA | 6.4 | 10±1 |
| 37.5% PC+37.5% Br-PC+25% PG+EGTA | 7.0 | 1–3 |
| 37.5% PC+37.5% Br-PC+25% PG+Ca2+ | 7.0 | 1–3 |
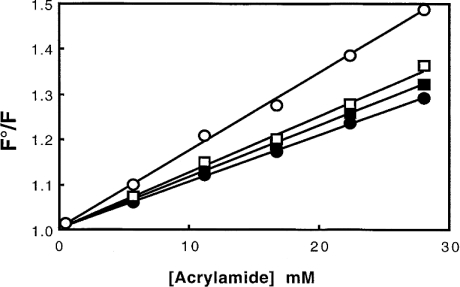
(ii) Experiments were also performed to test the accessibility of the gelsolin tryptophan residues in the presence or the absence of liposomes (25% PG and 75% PC) at pH 5.6 and 7.0. The results obtained using acrylamide as a quenching reagent (Figure 5) show that the tryptophan residues of gelsolin are, at pH 5.6, somewhat less shielded from the solvent than at pH 7.0, since the apparent KD is less for the last condition (KD=13.2±0.5 M−1) than for the pH 5.6 (KD=18.0±1 M−1), according to the opening of the molecule induced by a pH decrease [11,12]. In contrast, at neutral pH, the addition of lipids induced only a small change in the tryptophan accessibility (KD=12.0±0.5 M−1, instead of KD=13.2±0.5 M−1) and a significant insertion of gelsolin into the PG/PC SUV surface is indicated at pH 5.6 (KD=11.0±1 M−1, instead of KD=18.0±1 M−1).
Figure 5. Quenching of tryptophan fluorescence of gelsolin by acrylamide.
Stern–Volmer plot for the quenching of gelsolin by acrylamide in 0.15 M NaCl, 5 mM EGTA and 0.05 M acetate buffer, pH 5.6 (○) or 0.15 M NaCl, 1 mM EGTA and 0.05 M Tris/HCl buffer, pH 7.0 (□). The same experiments were also performed in the presence of PG/PC SUVs (0.4 mg/ml) (25% PG+75% PC) at pH 5.6 (●) or pH 7.0 (■). F°/F was determined as described in the Materials and methods section. The excitation wavelength was set at 280 nm.
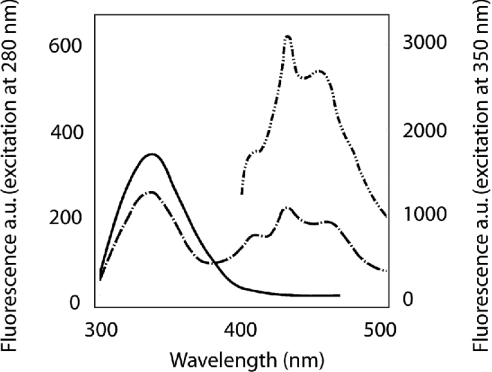
(iii) In a third series of experiments, the interaction of gelsolin was investigated by FRET (fluorescence resonance energy transfer). The fluorescent reagent DPH was incorporated into the hydrophobic layer of the phospholipids mixture, and fluorescence of gelsolin tryptophan was monitored at pH 5.6. The excitation spectrum of the reagent overlaps with the fluorescence emission of tryptophan. As shown in Figure 6, we observed that quenching of the tryptophan fluorescence upon gelsolin binding to labelled liposomes correlated with the appearance of an emission spectrum corresponding to DPH. These data corresponding to FRET are indicative of proximity between the two lots of chromophore. However, a more quantitative approach is not possible as the observed FRET is the integration of several of the 15 tryptophan residues present in gelsolin.
Figure 6. Interaction of gelsolin with phospholipids measured by FRET.
Tryptophan emission spectra of gelsolin–PG/PC SUV complexes were performed in 5 mM EGTA and 0.05 M acetate buffer, pH 5.6, in the absence (—) and in the presence of 4 μM DPH (-·-·-). Gelsolin was used at 0.4 μM and PG/PC SUVs (25% PG+75% PC) were at 0.4 mg/ml. Excitation was at 280 nm and emission was recorded between 300 and 500 nm. The emission spectrum between 400 and 500 nm of the tertiary complex resulting from the excitation of DPH fluorophore at 350 nm is also included (-··-··-··-, secondary y-axis). a.u., arbitrary units.
Taken together, these data indicate that the interaction of gelsolin with lipids is superficial at neutral pH and that gelsolin partially intercalates into the hydrophobic core of the vesicles at more acidic pH within the physiological range.
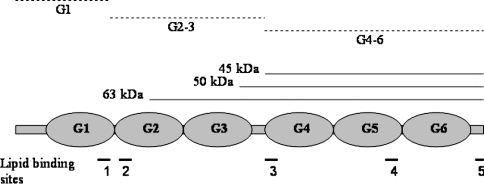
Cleavage of gelsolin by chymotrypsin
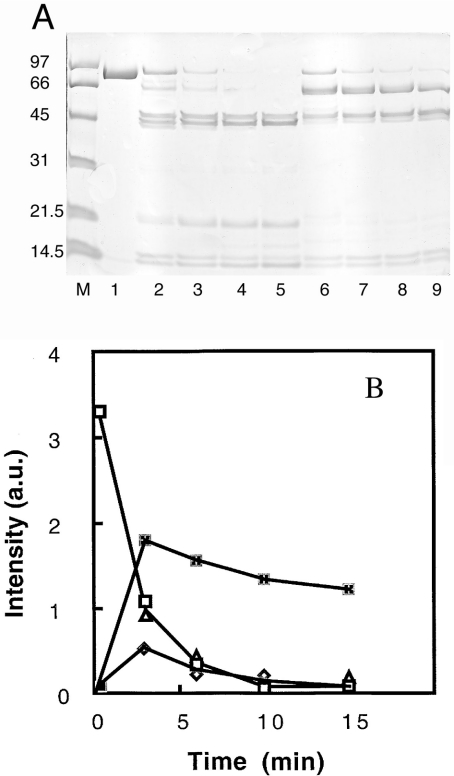
The chymotryptic digestion of gelsolin carried out at neutral pH produces three major fragments G1, G2–3 and G4–6 [43,44]. A fine transitory band of approx. 75–80 kDa is also present [45]. As shown in Figure 7, neither the cleavage pattern of gelsolin nor the rate of proteolysis is significantly different in the presence of lipids at pH 7.0. The same digestion conducted at pH 6.0, however, is quite different. A 63 kDa fragment appears first and is quickly degraded (Figure 8). Two major fragments with molecular masses of 50 kDa and 45 kDa appear also in the beginning of the proteolysis. We have determined that the N-terminal sequence of the 63 kDa fragment is ERLK (Glu-Arg-Leu-Lys), corresponding to a cleavage at position 209. The N-terminal sequences of the two other fragments are LSSH (Leu-Ser-Ser-His) and AAQH (Ala-Ala-Gln-His), corresponding to positions 383 and 407 respectively. Many uncharacterized bands below 20 kDa were also observed. These data suggest that, at pH 6.0, a cleavage takes place within the G2 domain (not observed at neutral pH) in addition to the production of the G4–6 fragment. However, we did not observe a 30 kDa peptide corresponding to the degradation of the G1–3 domains into G1 and G2–3. More interestingly, this proteolysis is significantly slowed by the presence of phospholipids (Figure 8). Since the vanishing rate of the entire gelsolin band seems unaffected in the presence of PG/PC SUVs (Figure 8), the degradation of the 63 kDa fragment into the G4–6 fragment is drastically and specifically impeded. This result suggests the participation of the link between the N- and C- terminal domains in the pH-dependent phospholipid interaction.
Figure 7. Effect of PG/PC SUVs on susceptibility of gelsolin to chymotryptic digestion at pH 7.0.
Digestion of gelsolin in 0.15 M NaCl, 1 mM CaCl2 and 0.05 M Tris/HCl buffer, pH 7.0, by chymotrypsin (chymotrypsin/gelsolin ratio, 1:300, w/w) for various times (3, 6, 10 and 15 min) in the absence (lanes 2–5 respectively) or in the presence of 0.4 mg/ml PG/PC SUVs (25% PG+75% PC), (lanes 6–9 respectively). Gelsolin at zero time is in lane 1. Molecular-mass markers are in lane M; standards are phosphorylase B (97.4 kDa), BSA (66.2 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), soybean trypsin inhibitor (21.5 kDa) and lysozyme (14.5 kDa). Samples were separated by SDS/15% (w/v) PAGE.
Figure 8. Effects of PG/PC SUVs on the susceptibility of gelsolin to chymotryptic digestion at pH 6.
(A) Digestion of gelsolin in 0.1 M Mes buffer, pH 6.0, by chymotrypsin (chymotrypsin/gelsolin ratio, 1:200, w/w) for various times (3, 6, 10 and 15 min) in the absence (lanes 2–5 respectively) or in the presence of 0.4 mg/ml PG/PC SUVs (25% PG+75% PC) (lanes 6–9 respectively). Gelsolin at zero time is in lane 1. Molecular-mass markers are in lane M and are as in Figure 7. Samples were separated by SDS/12.5% PAGE. (B) Kinetics of chymotryptic digestion. Bands in the electrophoretic gel presented above were integrated and the intensity plotted against time. Results are for 90 kDa (gelsolin) compound with (△) or without PG/PC SUVs (□), and 63 kDa fragment with (×) or without PG/PC SUVs (◇). a.u., absorbance units.
Orientation of the gelsolin molecule towards the vesicle surface
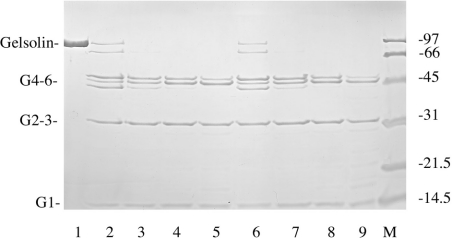
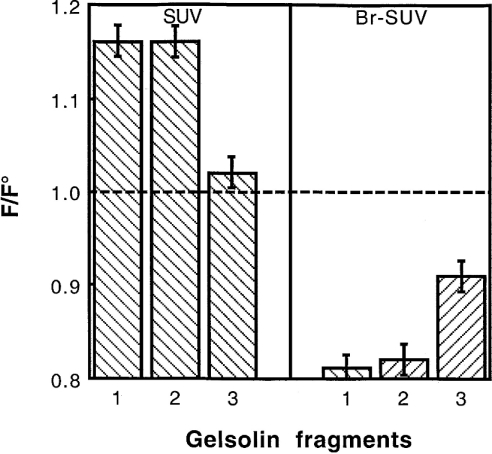
The ability of gelsolin fragments, G1–3 and G4–6 to interact with the phospholipid mixture was then tested at pH 5.6 (Figure 9). In a first experiment, we observed a 16% enhancement of the tryptophan emission spectrum induced by G1–3 incorporation and only 2% enhancement for the G4–6 domains. By using brominated phospholipids (25% PG, 37.5% Br-PC and 37.5% PC), a more significant quenching is observed with the G1–3 domains (18%) and with the G4–6 domains (9%). The data obtained with the G1–3 domains used alone are similar to those observed with the entire gelsolin molecule, which might suggest that the G1–3 domains alone are inserted in the lipid core; however, other explanations are possible. Finally, we have found that the N-terminal sequence of the G2 domain (RhoB–QRLFQVKGRR) that is known to interact with PtdIns(4,5)P2 [29] can also insert into PG/PC SUVs in the absence of PtdIns(4,5)P2. We observed that, after incubation of this rhodamine-labelled peptide with phospholipids, a complete quenching of the fluorophore was observed together with an increase in the rhodamine fluorescence anisotropy (results not shown). Similar results were observed at either pH 5.6 or pH 7.5.
Figure 9. Tryptophan fluorescence of gelsolin (1), G1–3 (2) and G4–6 (3) in the presence of PG/PC SUVs and Br-PC/PG SUVs at pH 5.6.
Effect of PG/PC SUV on gelsolin activity
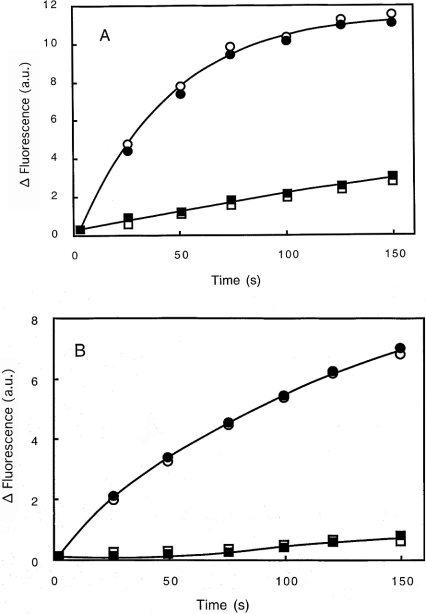
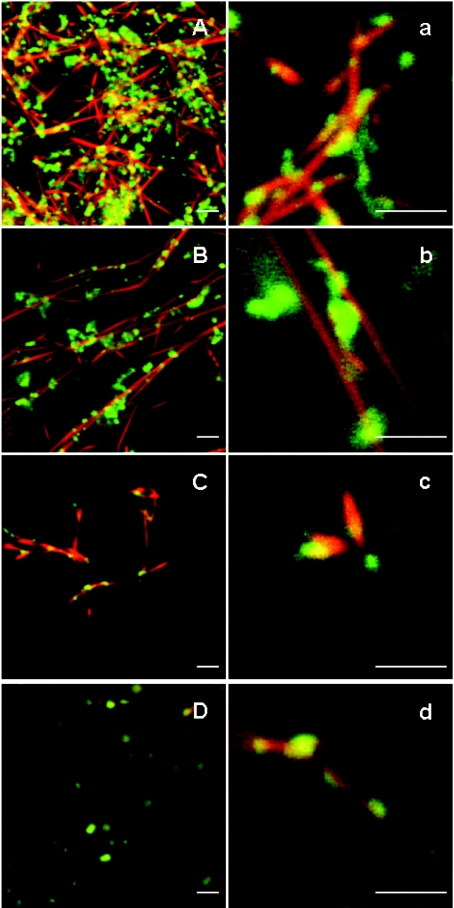
Two sets of experiments were performed to test gelsolin activity. Gelsolin greatly enhanced the rate of actin polymerization whatever the pH used (pH 7.5 or 5.8), as shown in Figure 10, and accordingly to the well-established properties of gelsolin [11,26,38,42]. More interestingly, the presence of PG/PC SUVs (25% PG+75% PC) does not significantly alter the gelsolin activity (Figures 10A and 10B), indicating that interaction of gelsolin with PG/PC SUVs does not impede G-actin–gelsolin association leading to nuclei formation. This proposition was tested by confocal microscopy. Polymerization of G-actin (3 μM) in the presence of gelsolin (52 nM) and PG/PC SUVs (0.2 mg/ml) was performed as for nucleation experiments, except that the actin was stained with rhodamine-phalloidin, and the PG/PC SUVs were prepared with 2% NBD-PC. Actin and PG/PC SUVs in the absence of gelsolin are used as references. We observed the formation of short actin filaments in the presence of gelsolin–PG/PC SUV complex at the two pH values tested (pH 5.8 and 7.5) (Figure 11). This was in contrast with the reference experiments showing very long actin filaments. Figure 11 also shows localization of PG/PC SUVs along actin filaments in all cases, but in the absence of gelsolin, more PG/PC SUVs seem free from F-actin. More interestingly, in the presence of gelsolin, we observe a co-localization (Figures 11A, 11a, 11C and 11c) of PG/PC SUVs (yellow/orange) with short filaments (red) all around liposomes. These results clearly suggest that gelsolin associated with liposomes allows actin polymerization from these loci of nucleation.
Figure 10. Effect of PG/PC SUVs on the activation of gelsolin nucleating activity.
Labelled 2.3 μM G-actin was added to 2 mM MgCl2 and 0.1 M KCl, 20 μM ATP and 20 mM Mes buffer at pH 5.8 (A) or 2 mM MgCl2 and 0.1 M KCl, 20 μM ATP, 0.5 mM CaCl2 and 20 mM Tris/HCl buffer at pH 7.5 (B) in the presence (circles) or in the absence (squares) of 26 nM gelsolin, as described in the Materials and methods section. The increase in fluorescence in the presence (open symbols) or in the absence (closed symbols) of PG/PC SUVs (25% PG+75% PC) at 0.2 mg/ml was plotted against time. a.u., arbitrary units.
Figure 11. Confocal image of actin filaments in association with PG/PC SUVs and gelsolin.
G-actin (3 μM) was added to 2 mM MgCl2 and 0.1 M KCl, 20 μM ATP and 20 mM Mes buffer at pH 5.8 or 2 mM MgCl2 and 0.1 M KCl, 20 μM ATP, 0.5 mM CaCl2 and 20 mM Tris/HCl buffer at pH 7.5 in the presence of PG/PC SUVs at 0.2 mg/ml (25% PG+75% PC) containing 2% NBD-labelled PC. Actin filaments were stained with rhodamine-labelled phalloidin before visualization. (A), (a), (B) and (b) show experiments performed at pH 5.8 in the presence or absence of 52 nM gelsolin respectively. (C), (c), (D) and (d) show experiments performed at pH 7.5 in the presence or absence of 52 nM gelsolin respectively. Scale bar, 2 μm.
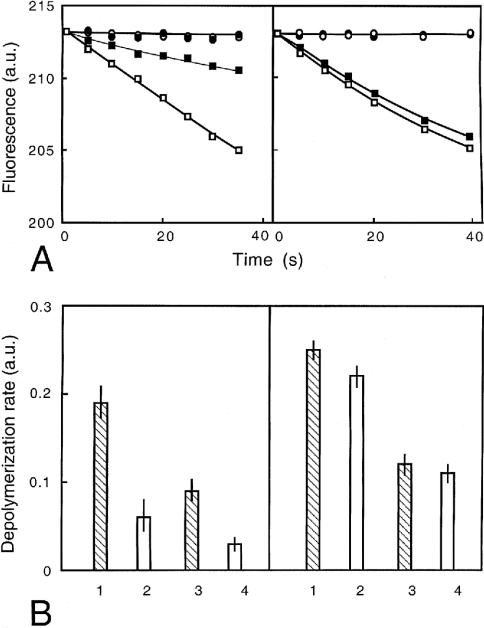
In severing assays, the rate of fluorescence decrease was measured after dilution (1/52) of gelsolin-capped F-actin in the presence of several gelsolin concentrations (0, 26 and 52 nM) (Figure 12). In these experiments, the effect of PG/PC SUVs on gelsolin activity was larger at acidic pH (70% inhibition) than at pH 7.5 (10% inhibition). The same relative extent of inhibition was observed at the two gelsolin concentrations used.
Figure 12. Effects of PG/PC SUVs on the severing activity of gelsolin.
(A) Labelled 25 μM F-actin (pre-capped by 1:1000 molar ratio of gelsolin) was diluted to 480 nM in either 2 mM MgCl2 and 0.1 M KCl, 20 μM ATP and 20 mM Mes buffer at pH 5.8 (left-hand panel) or 2 mM MgCl2 and 0.1 M KCl, 20 μM ATP, 0.5 mM CaCl2, 20 mM Tris/HCl buffer at pH 7.5 (right-hand panel) in the presence of 52 nM gelsolin (squares) or in the absence of gelsolin (circles). Fluorescence at 386 nm (a.u., arbitrary units) was reported over time in the absence (open symbols) and in the presence (closed symbols) of PG/PC SUVs (25% PG+75% PC) at 0.2 mg/ml. (B) Rates of depolymerization were measured from experiments conducted as in (A). Left-hand panel, pH 5.8 buffer; right-hand panel, pH 7.5 buffer, with 52 nM (lanes 1 and 2) or 26 nM (lanes 3 and 4) gelsolin. The rate of actin depolymerization (a.u.) was reported in the absence (lanes 1 and 3) and in the presence (lanes 2 and 4) of PG/PC SUVs (25% PG+75% PC) at 0.2 mg/ml.
DISCUSSION
When presented with high lipid concentrations, gelsolin can bind as many as ten PtdIns(4,5)P2 molecules [41]. The value of the ratio between gelsolin and PtdIns(4,5)P2 has been contentious, and has been complicated by differences in the state or presentation of the lipid between studies. When presented as a minor component with other lipids, one PtdIns(4,5)P2 binds one gelsolin [41]. This is probably close to the physiological situation, as PPIs are relatively scarce. For example, PtdIns(4,5)P2 accounts for 0.3–1.5% of the plasma membrane of a variety of mammalian cells [46]. However, it is known that PPIs forms aggregates within the bilayer under the influence of certain proteins [46] and so there may be many possible modes of PPI and other lipid binding. The finding that several sites within gelsolin can be cross-linked to PPI analogues would seem to support this view [47]. Together with our present data, indicating that gelsolin binds non-PPI lipids with high affinity, it seems likely that gelsolin may bind up to four PtdIns(4,5)P2 if they are available, through direct interactions with the sites (Figure 13) on gelsolin, but, at lower PtdIns(4,5)P2 concentrations, these sites may be occupied by other lipids. This is in agreement with others who conclude that gelsolin requires a net negative charge created by lipids other than PPIs, a hydrophobic interface or PPI for membrane interaction [41].
Figure 13. Domain structure of gelsolin showing proteolytic products at pH 7.0 (broken lines) and pH 6.0 (solid lines, 45, 50 and 63 kDa products) in the presence of PG/PC SUVs.
Lipid-binding regions are also shown in the lower part of the diagram: 1 and 2, the well-documented PtdIns(4,5)P2-binding sites 135–149 and 150–169; 3, a possible lipid-binding site suggested by the present study; 4 and 5, other sites in G4–6.
Gelsolin has been found to be associated with both membranes and actin filaments in activated macrophages and platelets [15,49]. This is a surprise, since PtdIns(4,5)P2 has been assumed to be the binding site and yet this lipid dissociates the gelsolin–actin complex [13,17]. It is possible that the binding sites for the gelsolin–actin complexes in these macrophages and platelet membranes are lipids other than PPIs and that these do not dissociate the complex. We have found that binding of gelsolin to the PG/PC SUVs does not inhibit the nucleation of actin polymerization, but reduces the severing rate caused by gelsolin at low pH. We have also shown that gelsolin increases the amount of actin microfilaments associated with PG/PC SUVs by microscopy. These observations are consistent with the barbed end capping of actin microfilament by gelsolin, with the simultaneous binding to the PG/PC SUVs, and that gelsolin bound by PG/PC SUVs has a reduced capacity for actin filament severing. Thus, unlike PPIs, these non-PPI lipids do not lead to dissociation of the gelsolin–actin complexes. Our microscopic observation also reveals that there is a weak interaction between the PG/PC SUVs and actin in the absence of gelsolin in agreement with previous studies [48]. Another non-exclusive possibility is that barbed-end-associated gelsolin binds the membrane through other protein intermediaries as a number of possible partners exist. For example, gelsolin binds directly to phospholipase D [50], is localized to the focal adhesion through an interaction with proline-rich tyrosine kinase-2 [51], and binds to phosphoinositide 3-kinase [52].
Tryptophan fluorescence quenching by acrylamide is pH-dependent, in agreement with our previous study with G4–6 [12]. Lower pH activated gelsolin in a similar, but distinct, manner observed in the presence of calcium. Lower pH ‘opened up’ the gelsolin molecule and this is reflected by greater acrylamide quenching and enhanced tryptophan fluorescence. The presence of the PG/PC SUVs did not markedly affect quenching by acrylamide at pH 7.0, whereas at pH 5.6, a large difference was found. We hypothesize that PG/PC SUV binding causes large conformational changes in gelsolin that are distinct from that caused by low pH alone, as tryptophan fluorescence is quenched in the presence of PG/PC SUVs at low pH. The tryptophan quenching data indicates an intercalation into the lipid bilayer and corresponds to a pKa of approx. 6.5. This is in contrast with the pH effect on the gelsolin activity with regard to microfilament severing, which shows a maximum effect below pH 6 [11]. This intercalation is also supported by our FRET experiments.
Although the primary PtdIns(4,5)P2-binding sites are reported to be within G1–3, several studies conclude that G4–6 is also involved [47,53]. G1–3 bound to PtdIns(4,5)P2 with approx. 7-fold lower Kd than G4–6 under a variety of conditions [53]. The present study has raised the possibility that not only does gelsolin bind the surface of lipids, but also that it becomes partially embedded within the lipid core. Previous studies have indicated that various peptides derived from PPI-binding regions of gelsolin have this capacity in isolation. The rhodamine-labelled peptide can not only embed within membranes, but also passes through them, and, in so doing, may exert microbial toxicity [54]. We have also found that this peptide can incorporate into our PG/PC SUVs in the absence of PPIs. The importance of hydrophobic interactions between gelsolin and PPIs has been suggested by a molecular dynamic study in which the PPIs are pulled out to some extent from the bilayer [55].
Our work indicates that G4–6 also contains a lipid-binding region as shown in Figure 9. Tryptophan fluorescence is quenched by Br-PC/PG SUVs, and we suspect that Trp423 may be one of these residues, since it is partially exposed on the surface of closed gelsolin [27] and is part of the connection between the two halves of the molecule towards the N-terminus of G4–6. This site becomes protected in the presence of lipids possibly by being buried in or stabilized by the lipid bilayer. Others have shown that this region is also protected against caspase 3 when bound by either micelles or vesicles containing PtdIns(4,5)P2 or PtdIns(3,4)P2 lipids [56].
The PtdIns(4,5)P2-binding domain P2 has a propensity to form a helix in the presence of lipid [57,58], whereas it is otherwise a random coil [27]. The effector domain of the MARCKS (myristoylated alanine-rich C-kinase substrate) protein is similar to P2 in that it is basic, being lysine-rich, and, like P2, has phenylalanine residues within the lysine-rich regions (Figure 13). The effector domain, like P2, also passes through vesicles, and it is proposed that the five phenylalanine residues penetrate the hydrophobic core of the membrane [46].
A number of sites in gelsolin have been proposed to bind PtdIns(4,5)P2, and these tend to be located in linker regions between the discrete domains of gelsolin. The main sites appear to be in the G1–2 linker region 135–149 and 150–169; secondary sites have been identified within the C-terminal half of gelsolin in the G5–6 linker region 620–634 and within the C-terminal 23 amino acids 732–755 [47]. If these primary regions are inserted into the equivalent positions in G4–6 by mutagenesis, PPI binding results [59], suggesting that these linker regions are also close, or can be brought close to the lipid membrane. We hypothesize that in addition to these linker regions, the extended linker region between G3 and G4, which is the region that connects the two halves of gelsolin, constitutes a further lipid-binding site. Evidence for the involvement of this region comes from the lipid-dependent inhibition of proteolytic cleavage of this site (Figure 8), and the fact that G4–6 intrinsic fluorescence is also quenched by Br-Pc lipids (Figure 9). We suggest further that our data indicate that this region becomes inserted into the core of the lipid bilayer in a PPI-independent manner at low pH.
Acknowledgments
This research was supported by grants from AFM (Association Française contre les Myopathies) and the British Council (S.K.M. and C.R.). We thank J. L. Derancourt for kindly performing the structural analyses of chymotrypsin gelsolin fragments.
References
- 1.dos Remedios C. G., Chhabra D., Kekic M., Dedova I. V., Tsubakihara M., Berry D. A., Noseworthy N. J. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 2003;83:433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 2.Isenberg G., Goldmann W. H. Actin-binding protein–lipid interaction. In: Pryme H., editor. The Cytoskeleton, vol. 1. Greenwich, CT/London: JAI Press Inc.; 1995. pp. 169–204. [Google Scholar]
- 3.Irvine R. F., Schell M. J. Back in the water: the return of the inositol phosphates. Nat. Rev. Mol. Cell Biol. 2001;2:327–338. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- 4.Dowler S., Currie R. A., Campbell D. G., Deak M., Kular G., Downes C. P., Alessi D. R. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellson C. D., Andrews S., Stephens L. R., Hawkins P. T. The PX domain: a new phosphoinositide-binding module. J. Cell Sci. 2002;115:1099–1105. doi: 10.1242/jcs.115.6.1099. [DOI] [PubMed] [Google Scholar]
- 6.Misra S., Miller G. J., Hurley J. H. Recognizing phosphatidylinositol 3-phosphate. Cell. 2001;107:559–562. doi: 10.1016/s0092-8674(01)00594-3. [DOI] [PubMed] [Google Scholar]
- 7.Lassing I., Lindberg U. Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature (London) 1985;314:472–474. doi: 10.1038/314472a0. [DOI] [PubMed] [Google Scholar]
- 8.Niggli V. Structural properties of lipid-binding sites in cytoskeletal proteins. Trends Biochem. Sci. 2001;26:604–611. doi: 10.1016/s0968-0004(01)01927-2. [DOI] [PubMed] [Google Scholar]
- 9.Suchy S. F., Nussbaum R. L. The deficiency of PIP2 5-phosphatase in Lowe syndrome affects actin polymerization. Am. J. Hum. Genet. 2002;71:1420–1427. doi: 10.1086/344517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin H. L., Stossel T. P. Control of cytoplasmic actin gel–sol transformation by gelsolin, a calcium dependent regulatory protein. Nature (London) 1979;281:583–586. doi: 10.1038/281583a0. [DOI] [PubMed] [Google Scholar]
- 11.Lamb J. A., Allen P. G., Tuan B. Y., Janmey P. A. Modulation of gelsolin function: activation at low pH overrides Ca2+ requirement. J. Biol. Chem. 1993;268:8999–9004. [PubMed] [Google Scholar]
- 12.Lagarrigue E., Ternent D., Maciver S. K., Fattoum A., Benyamin Y., Roustan C. The activation of gelsolin by low pH: the calcium latch is sensitive to calcium but not pH. Eur. J. Biochem. 2003;270:4105–4112. doi: 10.1046/j.1432-1033.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- 13.Janmey P. A., Stossel T. P. Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature (London) 1987;325:362–364. doi: 10.1038/325362a0. [DOI] [PubMed] [Google Scholar]
- 14.McGough A., Staiger C. J., Min J.-K., Simonetti K. D. The gelsolin family of actin regulatory proteins: modular structures, versatile functions. FEBS Lett. 2003;552:75–81. doi: 10.1016/s0014-5793(03)00932-3. [DOI] [PubMed] [Google Scholar]
- 15.Hartwig J. H., Bokoch G. M., Carpenter C. L., Janmey P. A., Taylor L. A., Toker A., Stossel T. P. Thrombin receptor ligation and activated Rac uncap actin filament barbed ends through phosphoinositide synthesis in permeabilized human platelets. Cell. 1995;82:643–653. doi: 10.1016/0092-8674(95)90036-5. [DOI] [PubMed] [Google Scholar]
- 16.Sugiura Y. Structure of molecular aggregates of 1-(3-sn-phosphatidyl)-L-myo-inositol 3,4-bis(phosphate) in water. Biochim. Biophys. Acta. 1981;641:148–159. doi: 10.1016/0005-2736(81)90578-2. [DOI] [PubMed] [Google Scholar]
- 17.Janmey P. A., Iida K., Yin H. L., Stossel T. P. Polyphosphoinositide micelles and polyphosphoinositide-containing vesicles dissociate endogenous gelsolin–actin complexes and promote actin assembly from the fast-growing end of actin filaments blocked by gelsolin. J. Biol. Chem. 1987;262:12228–12236. [PubMed] [Google Scholar]
- 18.Kumar N., Zhao P., Tomar A., Galea C. A., Khurana S. Association of villin with phosphatidylinositol 4,5-bisphosphate regulates the actin cytoskeleton. J. Biol. Chem. 2004;279:3096–3110. doi: 10.1074/jbc.M308878200. [DOI] [PubMed] [Google Scholar]
- 19.Ohtsu M., Sakai N., Fujita H., Kashiwagi M., Gasa S., Shimizu S., Eguchi Y., Tsujimoto Y., Sakiyama Y., Kobayashi K., Kuzumaki N. Inhibition of apoptosis by the actin-regulatory protein gelsolin. EMBO J. 1997;16:4650–4656. doi: 10.1093/emboj/16.15.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y.-S., Kantorow M., Davis J., Piatigorsky J. Evidence for gelsolin as a corneal crystallin in zebrafish. J. Biol. Chem. 2000;275:24645–24652. doi: 10.1074/jbc.M001159200. [DOI] [PubMed] [Google Scholar]
- 21.Kanungo J., Kozmik Z., Swamynathan S. K., Piatigorsky J. Gelsolin is a dorsalizing factor in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3287–3292. doi: 10.1073/pnas.0634473100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goetzl E. J., Lee H., Azuma T., Stossel T. P., Turck C. W., Karliner J. S. Gelsolin binding and cellular presentation of lysophosphatidic acid. J. Biol. Chem. 2000;275:14573–14578. doi: 10.1074/jbc.275.19.14573. [DOI] [PubMed] [Google Scholar]
- 23.Way M., Pope B., Gooch J., Hawkins M., Weeds A. G. Identification of a region of segment 1 of gelsolin critical for actin binding. EMBO J. 1990;9:4103–4109. doi: 10.1002/j.1460-2075.1990.tb07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwiatkowski D. J., Stossel T. P., Orkin S. H., Mole J. E., Colten H. R., Yin H. L. Plasma and cytoplasmic gelsolins are encoded by a single gene and contain a duplicated actin-binding domain. Nature (London) 1986;323:455–458. doi: 10.1038/323455a0. [DOI] [PubMed] [Google Scholar]
- 25.Way M., Weeds A. G. Nucleotide sequence of pig plasma gelsolin: comparison of protein sequence with human gelsolin and other actin-severing proteins shows strong homologies and evidence for large internal repeats. J. Mol. Biol. 1988;203:1127–1133. doi: 10.1016/0022-2836(88)90132-5. [DOI] [PubMed] [Google Scholar]
- 26.Way M., Gooch J., Pope B., Weeds A. G. Expression of human plasma gelsolin in E. coli and dissection of actin binding sites by segmental deletion mutagenesis. J. Cell Biol. 1989;109:593–605. doi: 10.1083/jcb.109.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burtnick L. D., Koepf E. K., Grimes J., Jones E. Y., Stuart D. I., McLaughlin P. J., Robinson R. C. The crystal structure of plasma gelsolin: implications for actin severing, capping and nucleation. Cell. 1997;90:661–670. doi: 10.1016/s0092-8674(00)80527-9. [DOI] [PubMed] [Google Scholar]
- 28.Kurokawa H., Fujii W., Ohmi K., Sakurai T., Nonomura Y. Simple and rapid purification of brevin. Biochem. Biophys. Res. Commun. 1990;168:451–457. doi: 10.1016/0006-291x(90)92342-w. [DOI] [PubMed] [Google Scholar]
- 29.Cunningham C. C., Vegners R., Bucki R., Funaki M., Korde N., Hartwig J. H., Stossel T. P., Janmey P. A. Cell permeant polyphosphoinositide-binding peptides that block cell motility and actin assembly. J. Biol. Chem. 2001;276:43390–43399. doi: 10.1074/jbc.M105289200. [DOI] [PubMed] [Google Scholar]
- 30.Papa I., Astier C., Kwiatek O., Lebart M.-C., Raynaud F., Benyamin Y., Roustan C. Use of a chaotropic anion iodide in the purification of Z-line proteins: isolation of CapZ from fish white muscle. Protein Expression Purif. 1999;17:1–7. doi: 10.1006/prep.1999.1124. [DOI] [PubMed] [Google Scholar]
- 31.Spudich J. A., Watt S. The regulation of rabbit skeletal muscle contraction: biochemical studies of the interaction of the tropomyosin–troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 32.Kouyama T., Mihashi K. Fluorimetry study of N-(1-pyrenyl) iodoacetamide-labelled F-actin. Eur. J. Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- 33.Méjean C., Lebart M. C., Poyer M., Roustan C., Benyamin Y. Localization and identification of actin structures involved in the filamin–actin interaction. Eur. J. Biochem. 1992;209:555–562. doi: 10.1111/j.1432-1033.1992.tb17320.x. [DOI] [PubMed] [Google Scholar]
- 34.Larvor M. P., Cerdan R., Gumila C., Maurin L., Seta P., Roustan C., Vial H. Characterization of the lipid-binding domain of the Plasmodium falciparum CTP:phosphocholine cytidylyltransferase through synthetic-peptide studies. Biochem. J. 2003;375:653–661. doi: 10.1042/BJ20031011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakowicz J. R. New York: Plenum Publishing Corp.; 1983. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 36.Frieden C. Polymerization of actin: mechanism of the Mg2+-induced process at pH 8 and 20 °C. Proc. Natl. Acad. Sci. U.S.A. 1983;80:6513–6517. doi: 10.1073/pnas.80.21.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janmey P. A., Stossel T. P. Kinetics of actin monomer exchange at the slow growing ends of actin filaments and their relation to the elongation of filaments shortened by gelsolin. J. Muscle Res. Cell Motil. 1986;7:446–454. doi: 10.1007/BF01753587. [DOI] [PubMed] [Google Scholar]
- 38.Janmey P. A., Chaponnier C., Lind S. E., Zaner K. S., Stossel T. P., Yin H. L. Interactions of gelsolin and gelsolin–actin complexes with actin: effects of calcium on actin nucleation, filament severing and end blocking. Biochemstry. 1985;24:3714–3723. doi: 10.1021/bi00335a046. [DOI] [PubMed] [Google Scholar]
- 39.Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 40.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 41.Tuominen E. K. J., Holopainen J. M., Chen J., Prestwich G. D., Bachiller P. R., Kinnunen P. K. J., Janmey P. J. Fluorescent phosphoinositide derivatives reveal specific binding of gelsolin and other actin regulator proteins to mixed lipid bilayers. Eur. J. Biochem. 1999;263:85–92. doi: 10.1046/j.1432-1327.1999.00464.x. [DOI] [PubMed] [Google Scholar]
- 42.Janmey P. A., Stossel T. P. Gelsolin–polyphosphoinositide interaction. J. Biol. Chem. 1989;264:4825–4831. [PubMed] [Google Scholar]
- 43.Chaponnier C., Janmey P. A., Yin H. L. The actin filament-severing domain of plasma gelsolin. J. Cell Biol. 1986;103:1473–1481. doi: 10.1083/jcb.103.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soua Z., Porte F., Harricane M., Feinberg J., Capony J. Bovine serum brevin: purification by hydrophobic chromatography and properties. Eur. J. Biochem. 1985;153:275–287. doi: 10.1111/j.1432-1033.1985.tb09298.x. [DOI] [PubMed] [Google Scholar]
- 45.Kwiatkowski D. J., Janmey P. A., Mole J. E., Yin H. L. Isolation and properties of two actin-binding domains in gelsolin. J. Biol. Chem. 1985;260:15232–15238. [PubMed] [Google Scholar]
- 46.Gambhir A., Hangyas-Mihalyne G., Zaitseva I., Cafiso D. S., Wang J., Murray D., Pentyala S. N., Smith S. O., McLaughlin S. Electrostatic sequestration of PIP2 on phospholipid membranes by basic/aromatic regions of proteins. Biophys. J. 2004;86:2188–2207. doi: 10.1016/S0006-3495(04)74278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng L., Mejillano M., Yin H. L., Chen J., Prestwich G. D. Full-contact domain labelling: identification of a novel phosphoinositide binding site on gelsolin that requires the complete protein. Biochemistry. 2001;40:904–913. doi: 10.1021/bi000996q. [DOI] [PubMed] [Google Scholar]
- 48.St-Onge D., Gicquaud C. Evidence of direct interaction between actin and membrane lipids. Biochem. Cell Biol. 1989;67:297–300. doi: 10.1139/o89-045. [DOI] [PubMed] [Google Scholar]
- 49.Hartwig J. H., Chambers K. A., Stossel T. P. Association of gelsolin with actin and cell membranes of macrophages and platelets. J. Cell Biol. 1989;108:467–479. doi: 10.1083/jcb.108.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steed P. M., Nagar S., Wennogle L. P. Phospholipase D regulation by a physical interaction with the actin-binding protein gelsolin. Biochemistry. 1996;35:5229–5237. doi: 10.1021/bi952370j. [DOI] [PubMed] [Google Scholar]
- 51.Wang Q., Xie Y., Du Q.-S., Wu X.-J., Feng X., Mei L., McDonald J. M., Xiong W.-C. Regulation of the formation of osteoclastic actin rings by proline-rich tyrosine kinase 2 interacting with gelsolin. J. Cell Biol. 2003;160:565–575. doi: 10.1083/jcb.200207036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chellaiah M., Fitzgerald C., Alvarez U., Hruska K. c-Src is required for stimulation of gelsolin-associated phosphatidylinositol 3-kinase. J. Biol. Chem. 1998;273:11908–11916. doi: 10.1074/jbc.273.19.11908. [DOI] [PubMed] [Google Scholar]
- 53.Lin K.-M., Wenegieme E., Lu P.-J., Chen C.-S., Yin H. L. Gelsolin binding to phosphatidylinositol 4,5-bisphosphate is modulated by calcium and pH. J. Biol. Chem. 1997;272:20443–20450. doi: 10.1074/jbc.272.33.20443. [DOI] [PubMed] [Google Scholar]
- 54.Bucki R., Pastore J. J., Randhawa P., Vegners R., Weiner D. J., Janmey P. A. Antibacterial activities of rhodamine B-conjugated gelsolin-derived peptides compared to those of the antimicrobial peptides cathelicidin LL37, magainin II, and melittin. Antimicrob. Agents Chemother. 2004;48:1526–1533. doi: 10.1128/AAC.48.5.1526-1533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liepina I., Czaplewski C., Janmey P., Liwo A. Molecular dynamics study of a gelsolin-derived peptide binding to a lipid bilayer containing phosphatidylinositol 4,5-bisphosphate. Biopolymers. 2003;71:49–70. doi: 10.1002/bip.10375. [DOI] [PubMed] [Google Scholar]
- 56.Azuma T., Koths K., Flanagan L., Kwiatkowski D. Gelsolin in complex with phosphatidylinositol 4,5-bisphosphate inhibits caspase-3 and -9 to retard apoptotic progression. J. Biol. Chem. 2000;275:3761–3766. doi: 10.1074/jbc.275.6.3761. [DOI] [PubMed] [Google Scholar]
- 57.Xian W., Garver T. M., Braunlin W. H., Janmey P. A. NMR and CD conformational studies of a 20 amino-acid PIP2-binding site on gelsolin. FASEB J. 1992;6:A87. [Google Scholar]
- 58.Xian W., Vegners R., Janmey P., Braunlin W. Spectroscopic studies of a phosphoinositide-binding peptide from gelsolin: behavior in solutions of mixed solvent and anionic micelles. Biophys. J. 1995;69:2695–2702. doi: 10.1016/S0006-3495(95)80140-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xian W., Janmey P. A. Dissecting the gelsolin–polyphosphoinositide interaction and engineering of a polyphosphoinositide-sensitive gelsolin C-terminal half protein. J. Mol. Biol. 2002;322:755–771. doi: 10.1016/s0022-2836(02)00841-0. [DOI] [PubMed] [Google Scholar]