Abstract
The biosynthesis of the antifungal agent pimaricin by Streptomyces natalensis has been proposed to involve a cytochrome P450 encoded by the gene pimD. Pimaricin is derived from its immediate precursor de-epoxypimaricin by epoxidation of the C-4–C-5 double bond on the macrolactone ring. We have overproduced PimD with a N-terminal His6 affinity tag in Escherichia coli and purified the enzyme for kinetic analysis. The protein showed a reduced CO-difference spectrum with a Soret maximum at 450 nm, indicating that it is a cytochrome P450. Purified PimD was shown to catalyse the in vitro C-4–C-5 epoxidation of 4,5-de-epoxypimaricin to pimaricin. The enzyme was dependent on NADPH for activity with optimal pH at 7.5, and the temperature optimum was 30 °C. The kcat value for the epoxidation of de-epoxypimaricin was similar to the values reported for other macrolide oxidases. Enzyme activity was inhibited at high substrate concentration. This is the first time that a polyene macrolide P450 mono-oxygenase has been expressed heterologously and studied. The unique specificity of this epoxidase should be useful for the oxidative modification of novel polyene macrolide antibiotics.
Keywords: cytochrome P450 mono-oxygenase, epoxidase, pimaricin, polyene, polyketide synthase
Abbreviations: BCIP, 5-bromo-4-chloroindol-3-yl phosphate; IPTG, isopropyl β-D-thiogalactoside; LC/MS, liquid chromatography/mass spectrometry; Ni-NTA, Ni2+-nitrilotriacetate; TBS, Tris-buffered saline
INTRODUCTION
The biosynthesis of macrolides often requires the tailoring action of specific oxidases in order to construct the final bioactive compound. Such oxidases belong to a family of enzymes that is one of the most widely distributed in nature, the cytochromes P450. These are b-type cytochromes that carry out oxygenation reactions on an enormous array of substrates [1], among them the polyketide precursors of macrolide antibiotics. Examples include reactions involved in the biosynthesis of erythromycin [2,3], tylosin [4], oleandomycin [5], narbomycin [6,7], epothilone [8,9], amphotericin [10], nystatin [11], pimaricin [12,13] and rapamycin [14], among others. These reactions typically occur during the late stages of the biosynthesis, once the macrolide ring has been constructed by the polyketide synthase, and constitute an important contribution for biological activity [6,15]. However, although cytochromes P450 using polyketide structures as substrates are common, there are considerable variations in the substrates that are utilized, and the positions of the resulting hydroxy and/or epoxide substituents to such compounds introduced by these enzymes. This diversity may be exploited to obtain novel products by bioconversions. It is, therefore, of great importance to characterize additional macrolide mono-oxygenases in order to target the increasing number of novel polyketides obtained by combinatorial techniques.
Thus far, the erythromycin hydroxylases EryF and EryK from Saccharopolyspora erythraea [16,17], the methymycin/picromycin hydroxylase PicK (PikC) from Streptomyces venezuelae [6,7] and the epothilone epoxidase EpoK [8], are the only enzymes from this family to be expressed, purified and characterized. The first three enzymes catalyse hydroxylation of 12- or/and 14-membered macrolactones. While EryF and EryK exhibit strict substrate specificity and are responsible for the hydroxylation at single positions of the 14-membered erythromycin aglycone [16,17], PicK has broader substrate specificity, accepting both 12- and 14-membered ring macrolides as substrates, as well as catalysing the addition of a hydroxy group at two different positions in the case of the 12-membered macrolactone [7,18]. EpoK specifically converts deoxyepothilone into epothilone, a 16-membered macrolactone [8].
Pimaricin is a 26-membered polyene macrolide antifungal antibiotic [19], which is widely used for the treatment of fungal keratitis, and also in the food industry in order to prevent mould contamination of cheese and other non-sterile foods (i.e. cured meat, sausages, ham etc.). The biosynthetic gene cluster for pimaricin has been characterized [12,13,15,20]. Insertional inactivation of one of the genes of the pimaricin cluster, pimD, generated a mutant that accumulated a glycosylated pimaricin precursor, 4,5-de-epoxypimaricin [15], thus suggesting that this gene codes for a mono-oxygenase that is responsible for the introduction of the epoxy group at C-4–C-5 of the pimaricin molecule. In the present paper, we report the heterologous expression of PimD in Escherichia coli and the in vitro characterization of this enzyme as the epoxidase that transforms de-epoxypimaricin into pimaricin. The availability of the purified PimD enabled measurement of kinetic parameters of the reaction catalysed by the enzyme and comparison with the oxidations performed by the 12- and 14-membered macrolactone hydroxylases. The present study constitutes the first for a polyene macrolide, and for any other large-size ring macrolactone.
EXPERIMENTAL
Bacterial strains and cultivation
Streptomyces natalensis A.T.C.C. 27448 was routinely grown in YEME (yeast extract–malt extract) medium [21] without sucrose. Sporulation was achieved in TBO (tomato paste–oat flakes) medium [12] at 30 °C. For pimaricin production, the strain was grown in YEME without sucrose. The same media were supplemented with thiostrepton when used for St. natalensis 6D4 growth and/or de-epoxypimaricin production [15]. E. coli strain DH5α was used as a host for DNA manipulation. E. coli BL21 (DE3) was used for expression studies.
Plasmids and DNA manipulation procedures
PBluescript was used as the routine cloning vector, and pQE-30 (Qiagen) was the vector used to construct the PimD expression plasmid, pMVM5. Plasmid, cosmid and genomic DNA preparation, DNA digestion, fragment isolation, Southern hybridization, and transformation of E. coli were performed by standard procedures [22]. PCRs were carried out using Pfu DNA polymerase as described by the enzyme supplier (Stratagene). DNA sequencing was accomplished by the dideoxynucleotide chain-termination method using the PerkinElmer Amplitaq Gold Big Dye-terminator sequencing system with an Applied Biosystems model 310 sequencer (Foster City, CA, U.S.A.).
Construction of PimD E. coli expression plasmid
The pimD gene was amplified for insertion into the His6-tag expression vector pQE-30 using PCR. The forward primer used (5′-CCCATGACCGCCGGATCCCACGACCTGCCC-3′) introduced a unique BamHI site at the 5′ end of the gene, while the reverse primer (5′-GTCCTCCCCGCGCTGCAGCCCCCAG-3′) carries a PstI site 36 nucleotides downstream from the TGA translational stop codon. The amplified DNA fragment was digested with BamHI and PstI, and was cloned into the same sites of pQE-30 to generate pMVM5. The amplified DNA fragment was sequenced from the expression vector in order to discard any mistakes introduced by the DNA polymerase.
Expression and purification of His6–PimD
E. coli cells were grown at 30 °C in 100 ml of LB (Luria–Bertani) medium containing 1% (w/v) glycerol and 100 μg/ml ampicillin until a D600 of between 0.7 and 0.9 was reached, and then induced by adding IPTG (isopropyl β-D-thiogalactoside) to a final concentration of 0.5 mM, and grown for an additional 3 h. Cells were harvested, washed with 0.9% (w/v) NaCl, resuspended in binding buffer (20 mM Tris/HCl, pH 8.0, 500 mM NaCl and 20 mM imidazole) and lysed by sonication using an ultrasonic processor XL apparatus (Misonix, Farmingdale, NY, U.S.A.). The insoluble material was separated by centrifugation at 40000 g for 30 min, and the soluble fraction was applied to a FPLC Ni-NTA (Ni2+-nitrilotriacetate)–agarose (Qiagen) HR5/5 column equilibrated with binding buffer. The column was washed with 25 ml of binding buffer, and the protein was eluted with a 15 ml linear gradient (20–500 mM imidazole in binding buffer). Protein elution was monitored at 280 nm, and the presence of PimD was assessed by SDS/PAGE (12% gels). Fractions containing the PimD protein were pooled and dialysed against storage buffer (45 mM Tris/HCl, pH 7.5, 0.1 mM EDTA, 0.2 mM dithiothreitol and 10% glycerol). Enzyme concentration and yield were determined by the Bradford method [23] using BSA as a standard.
Immunoblot analysis
Cell-free extracts were run on SDS/12% polyacrylamide gels and transferred on to nylon membranes using standard techniques. Membranes were then blocked by incubation at room temperature (22 °C) in 5% (w/v) skimmed dried milk in TBS (Tris-buffered saline solution) for 1 h. After washing, blots were successively incubated with mouse monoclonal anti-His-tag antibodies (Qiagen), goat anti-mouse immunoglobulin–alkaline phosphatase conjugate (Sigma) and BCIP (5-bromo-4-chloroindol-3-yl phosphate)–Nitro Blue Tetrazolium substrate in alkaline phosphatase buffer (100 mM NaCl, 5 mM MgCl2 and 20 mM Tris/HCl, pH 9.5). A triple wash with TBS containing 2 g/l Tween 20 was performed between incubations, except for the last one in which we used alkaline phosphatase buffer just before the addition of the substrate solution. Colour development was stopped by washing with tap water.
Spectral analysis
Spectra of the purified PimD in storage buffer were recorded from 300 to 600 nm. The ferric haem was then reduced by adding Na2S2O4 to the protein solution, and the spectrum of the reduced enzyme was then recorded. Treatment of the Na2S2O4-reduced sample with CO and comparison with the Na2S2O4-reduced sample gave the CO-difference spectrum.
De-epoxypimaricin extraction and analysis
De-epoxypimaricin was extracted from the ΔPimD mutant St. natalensis 6D4 culture broths following published procedures [15]. Quantitative determination of pimaricin or de-epoxypimaricin was performed with a Waters 600 HPLC with a diode array ultraviolet detector set at 304 nm, fitted with a μ-Bondapack RP-C18 column (10 μm; 3.9 mm×300 mm). Elution was with a gradient (1 ml/min) of 100% methanol (methanol concentration: 50%, 0–3 min; up to 90%, 3–12 min, 90%, 12–20 min; down to 50%, 20–25 min; 50%, 25–30 min). Retention time for pimaricin was 14.7 min, and for 4,5-de-epoxypimaricin it was 16.5 min.
Conversion of de-epoxypimaricin into pimaricin in vitro
A standard conversion of 4,5-de-epoxypimaricin into pimaricin was accomplished by combining 10–150 nM PimD, 10–250 μM de-epoxypimaricin, 100 μg/ml spinach ferredoxin, 0.2 unit/ml spinach ferredoxin–NADP+ reductase, 1.4 mM NADPH, 10 mM glucose 6-phosphate and 8 units/ml glucose-6-phosphate dehydrogenase in 50 mM Tris/HCl, pH 7.5. The reactions were carried out for different time periods at 30 °C and terminated by the addition of 8 vol. of butanol. The resulting organic extracts were dried and resuspended in methanol for HPLC analysis. The identities of de-epoxypimaricin and pimaricin were determined by LC/MS (liquid chromatography/mass spectrometry) using an ES–TOF (electrospray-time of flight) apparatus (Micromass, Manchester, U.K.). A pimaricin solution (5 mg/ml methanol) was used for tuning. Analyses were run using the positive-ionization mode. Chromatography and column characteristics were as indicated above.
RESULTS
Heterologous expression of pimD
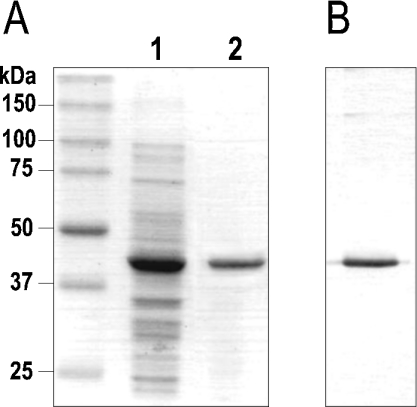
The involvement of the product of the gene pimD in the formation of the epoxide group of the pimaricin molecule has been suggested on the basis of gene inactivation experiments [15], but has not been proven in vitro. To analyse the epoxidation reaction catalysed by PimD, the pimD gene was overexpressed in E. coli, and the corresponding product was purified and functionally characterized. For expression, the pimD gene was placed under the control of a T5 promoter including a His6-tag introduced at its 3′ terminus (plasmid pMVM5; see the Experimental section). Induction of E. coli BL21(DE3)/pMVM5 with IPTG resulted in the production of a soluble 44 kDa protein as determined by SDS/PAGE (12% gels) analysis, consistent with the predicted molecular mass of PimD (43533 Da) (Figure 1). This band was absent from a control culture of BL21(DE3) containing the vector plasmid pQE30 with no insert. Purification of His6–PimD was achieved with a single chromatographic step on a Ni-NTA–agarose column. The purified His6–PimD fraction contained a single detectable band on a Coomassie-Blue-stained SDS/12%-polyacrylamide gel, and reacted strongly with anti-His-tag antibodies in Western blots (Figure 1).
Figure 1. Purification of PimD in E. coli BL21.
(A) SDS/PAGE (12% gels) of crude E. coli lysate expressing PimD (lane 1) and after purification by Ni2+ chromatography (lane 2). (B) Immunoblot detection of His6–PimD with anti-His-tag antibodies.
PimD is an authentic cytochrome P450
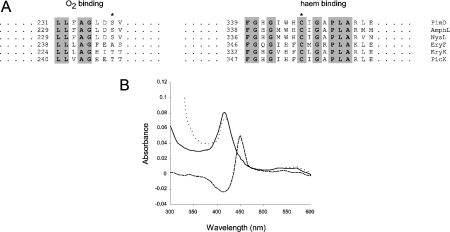
The translated amino acid sequence of PimD was compared with those of other macrolide P450 enzymes. As expected, PimD contains the conserved signature sequences associated with cytochrome P450 enzymes, namely an O2-binding site and a C-terminal haem-binding domain containing the invariant cysteine residue that co-ordinates the haem (Figure 2A).
Figure 2. Identification of PimD as an authentic cytochrome P450.
(A) Comparison of O2-binding pockets and haem-binding sequences from some macrolide P450 mono-oxygenases. The asterisk on the left alignment shows the conserved threonine/serine (alanine in EryF) residues that are believed to be involved in O2 scission, whereas the asterisk on the right indicates the haem-binding cysteine residue. Conserved amino acids are shown in bold and are highlighted. Numbers indicate amino acid residues from the N-terminus of the protein. AmphL, St. nodosus P450 involved in amphotericin biosynthesis (GenBank® accession number AAK73504); NysL, St. noursei P450 involved in nystatin biosynthesis (GenBank® accession number AAF71769); EryF (GenBank® accession number Q00441) and EryK (GenBank® accession number P48635), P450s from Sa. erythraea involved in erythromycin biosynthesis; PicK, methymycin/picromycin hydroxylase from St. venezuelae (GenBank® accession number AAC64105). (B) CO difference spectrum of purified PimD. The continuous line indicates the absorbance spectrum of the pure protein in storage buffer. The dotted line denotes the spectrum of the protein reduced with Na2S2O4. The reduced CO-difference spectrum is indicated by a broken line.
The UV–visible absorption spectrum of the purified protein contained the characteristic 390–450 nm cytochrome P450 peak with a maximum at 415.5 nm. Reduction with Na2S2O4 and exposure to CO gave the distinctive CO complex spectrum characteristic of cytochromes P450, with a maximum at 450 nm (Figure 2B). This spectral characterization provided the first physical evidence that PimD was in fact a cytochrome P450.
Conversion of de-epoxypimaricin into pimaricin in vitro
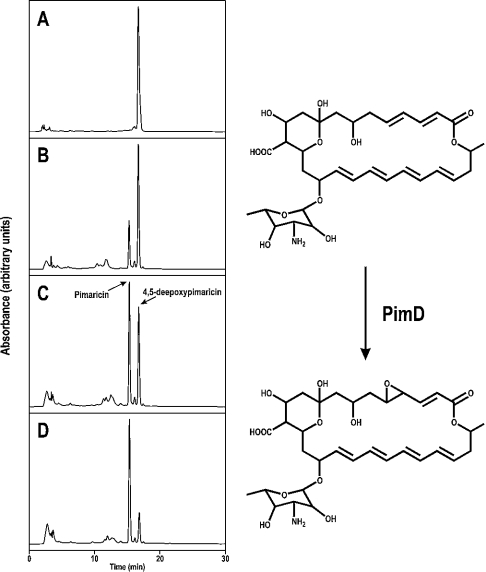
The enzymic activity of PimD was measured by converting its natural substrate 4,5-de-epoxypimaricin (isolated from the ΔPimD mutant St. natalensis 6D4, which accumulates de-epoxypimaricin; [15]) into pimaricin. Purified PimD protein was incubated at 30 °C with de-epoxypimaricin in the presence of spinach ferredoxin, spinach ferredoxin–NADP+ reductase and NADPH in vitro. An NADPH-regenerating system based on glucose 6-phosphate and glucose-6-phosphate dehydrogenase was also included in the reaction mixture. In the presence of such an electron-donating system, the enzyme readily converted de-epoxypimaricin into pimaricin. Figure 3 shows an enzyme-dependent conversion of de-epoxypimaricin into pimaricin, as measured by HPLC. The identity of the pimaricin peak was assessed by LC/MS. In the absence of the NADPH-regenerating system, no conversion was observed at all. Furthermore, in its presence, removal of any of the three remaining protein components of the reaction, namely the spinach ferredoxin, the ferredoxin–NADP+ reductase, or the PimD enzyme, resulted in the complete loss of catalytic activity (from a rate of 30.95 nmol of pimaricin formed/nmol of PimD per min when all components were present).
Figure 3. In vitro conversion of de-epoxypimaricin into pimaricin by PimD.
The results of analytical HPLC after reaction with different enzyme concentrations are shown as follows: (A), no enzyme; (B) 25 nM PimD; (C) 50 nM PimD; (D) 100 nM PimD. Reactions were carried out for 30 min. Chromatographic peaks corresponding to 4,5-de-epoxypimaricin and pimaricin are indicated by arrows.
Optimal conditions for activity
In a standard 45 min reaction, the optimal temperature was found to be in the range 28–32 °C. Outside this range, the enzyme activity was lost rapidly, especially at higher temperatures. This activity loss was particularly evident at 37 °C, where only 30% activity remains after 15 min of incubation. Within the optimal temperature range, the linear phase of reaction lasted for 20 min, and the enzyme remained active for at least 2 h.
The pH-dependence of enzyme activity was measured in 50 mM Mes/NaOH, pH 5.5–7.5, 50 mM Tris/HCl, pH 7.0–8.5, and 50 mM glycine/NaOH, pH 8.5–9.5 buffers. The enzyme activity became apparent throughout the pH range 6.5–9.0, with a peak at pH 7.5 (results not shown).
Kinetic parameters of the reaction catalysed by His6–PimD
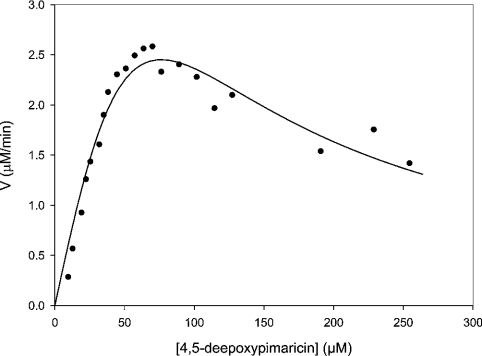
Initial velocities for the epoxidation of 4,5-de-epoxypimaricin were determined by directly measuring the formation of pimaricin by HPLC. At a substrate concentration of 1 mM, the initial rate of epoxidation was found to correlate linearly with enzyme concentrations in the range 10–100 nM His6–PimD. An enzyme concentration of 75 nM was selected to obtain steady-state kinetic parameters. Initial velocities were obtained with de-epoxypimaricin concentrations between 10 and 250 μM. Product formation was linear for the period monitored (0–20 min). A double-reciprocal plot of the data obtained indicated an asymptotic rise in 1/V at low values of 1/[S]. This behaviour is typical of substrate inhibition. For this reason, we fitted the initial velocity data to the equation v=Vmax/{1+(Km/[S])+([S]/Ki)}, which takes substrate inhibition into account (Figure 4). In this case, the steady-state parameters were a Km of 93±29 μM and a Vmax of 6.8±1.5 μM/min, corresponding to a kcat value of 1.5±0.34 s−1. Alternatively, at low substrate concentrations, where inhibition is negligible, the data could be fitted to the Michaelis–Menten equation, resulting in values for Km of 33±5.5 μM and Vmax of 3.5±0.2 μM/min. Assuming that all the enzyme is active, this would correspond to a kcat value of 0.78±0.04 s−1 and a kcat/Km of 0.024±0.003 μM−1·s−1 (Table 1).
Figure 4. Least-squares fit of initial rate data.
Initial rates were determined by applying a quadratic fit to the linear region of the time trace. The data were then fitted to the equation v=Vmax/{1+(Km/[S])+([S]/Ki)}.
Table 1. Steady-state kinetic parameters for His6–PimD and other macrolide oxidases.
| Enzyme | Substrate | kcat (s−1) | Km (μM) | Kcat/Km (μM−1·s−1) | Source |
|---|---|---|---|---|---|
| PimD | 4,5-De-epoxypimaricin | 1.5±0.34 | 93±29 | 0.016±0.002 | This work |
| PimD* | 4,5-De-epoxypimaricin | 0.78±0.04 | 33±5.5 | 0.024±0.003 | This work |
| EryF | 6-Deoxyerythronolide B | 1.7 | 1.98±0.26 | 0.85±0.11 | [16] |
| EryK | Erythromycin D | 6.2±1.2 | 44±10 | 0.14±0.042 | [17] |
| EryK* | Erythromycin D | 1.8±0.18 | 8±2.3 | 0.22±0.068 | [17] |
| PicK | YC-17 | 1.1±0.08 | 130±20 | 0.008±0.004 | [18] |
| PicK | Narbomycin | 2.0±0.3 | 88±24 | 0.023±0.01 | [18] |
| PicK* | YC-17 | 0.6±0.05 | 20.4±5.6 | 0.029±0.008 | [7] |
| PicK* | Narbomycin | 1.0±0.11 | 43.7±5.9 | 0.023±0.018 | [7] |
* Apparent values obtained from fitting the Michaelis–Menten equation to the low-concentration data in the linear region of the saturation curve.
DISCUSSION
PimD [CYP161A2, according to the standard nomenclature (see http://drnelson.utmem.edu/CytochromeP450.html)] represents the first polyene macrolide P450 mono-oxygenase to be expressed heterologously and studied, and constitutes the first oxidase tailoring a large-size macrolactone ring to be characterized in some detail. Thus far, only three macrolide hydroxylases have been studied regarding their kinetics in vitro. These are EryF and EryK, both involved in the hydroxylation of erythromycin A precursors, and PicK, involved in the oxidation of methymycin, neomethymycin and picromycin precursors (namely YC-17 and narbomycin). All of them catalyse hydroxylation of 12- and 14-membered macrolactones [7,16–18], while PimD uses a 26-membered aglycone as substrate.
PimD tailors 4,5-de-epoxypimaricin, the immediate precursor of pimaricin [15], while it seems unable to use 4,5-de-epoxypimaricinolide as substrate. Mutants in the mycosaminyl transferase gene, pimK, present in the pimaricin gene cluster in St. natalensis, accumulate 4,5-de-epoxypimaricinolide as a major product (M. V. Mendes, J. F. Martín and J. F. Aparicio, unpublished work), indicating that epoxidation is impaired in the absence of glycosylation and suggesting that the oxidase shows strict molecular recognition of the glycosylated polyene substrate. Similar results have been observed upon inactivation of the amphDIII gene, which encodes a GDP-D-mannose 4,6-dehydratase that is involved in the biosynthesis of mycosamine, from the amphotericin gene cluster in Streptomyces nodosus [24]. In this case, the mutant accumulated 8-deoxyamphoteronolides, suggesting that glycosylation precedes C-8 hydroxylation, and that the AmphL mono-oxygenase that is responsible for the hydroxylation of 8-deoxyamphotericins at C-8 shows strict substrate specificity [24]. PimD and AmphL, and also their counterpart NysL from nystatin-producing Streptomyces noursei [11], belong to the same phylogenetic group, and are thought to be responsible of specific oxidations in the polyol region of polyene macrolide precursors [19].
It is unknown if the His6-tag addition to the N-terminus of the protein affects the measured kinetic values of PimD. In any case, the kcat value measured for His6–PimD (0.8–1.5 s−1, depending on the method used to calculate the kinetic parameters; see the Results section) is similar to the values reported for EryF towards its substrate 6-deoxyerythronolide B, EryK towards erythromycin D, and PicK towards both YC-17 and narbomycin (Table 1); while the Km value (33–93 μM) is higher than those reported for EryF and EryK, and similar to the parameters for PicK (Table 1). This represents a substantially lower substrate specificity of PimD compared with that of EryF (kcat/Km=0.85 μM−1·s−1) and EryK (kcat/Km=0.14–0.22 μM−1·s−1), almost all of which can be attributed to the lower affinity of PimD towards its substrate, and similar to that of PicK towards its substrates, narbomycin and YC-17.
Interestingly, PimD was inhibited by a high concentration of its natural substrate de-epoxypimaricin. Substrate inhibition has previously been observed in the hydroxylations catalysed by EryK (towards erythromycin D) [17] and PicK (towards both YC-17 and narbomycin) [18].
Combinatorial biosynthesis is leading to a growing number of novel macrolide analogues. In many cases, however, the modified compounds lack biological activity, due to their deficiency in post-polyketide synthase oxidations, possibly due to the apparent strict substrate specificities of many of these oxidizing enzymes. The identification of novel macrolide mono-oxygenases with activities towards alternative substrates is therefore a crucial aspect for the oxidative tailoring of novel polyketides, and should facilitate the search for new bioactive macrolide compounds. It is conceivable that PimD could also accept other substrates and prove to be valuable for the introduction of epoxy substituents into designer macrolides, including pimaricin, amphotericin or nystatin derivatives. The ease of its purification can greatly facilitate its use for the in vitro epoxidation of non-natural substrates that may require high concentrations of enzyme.
Acknowledgments
This work was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (CICYT) to J.F.A. (BIO2001-0040) and J.F.M. (BIO2003-01489). M.V.M. received a fellowship of the Fundação para a Ciência e a Tecnologia (PRAXIS XXI/BD/15850/98). N.A. was the recipient of a F.P.U. fellowship from Ministerio de Educación, Cultura y Deporte (AP2002-1446). We thank E. Recio and L.L. Enríquez for helpful discussions.
References
- 1.Munro A. W., Lindsay J. G. Bacterial cytochromes P-450. Mol. Microbiol. 1996;20:1115–1125. doi: 10.1111/j.1365-2958.1996.tb02632.x. [DOI] [PubMed] [Google Scholar]
- 2.Haydock S. F., Dowson J. A., Dhillon N., Roberts G. A., Cortés J., Leadlay P. F. Cloning and sequencing analysis of genes involved in erythromycin biosynthesis in Saccharopolyspora erythraea: sequence similarities between EryG and a family of S-adenosylmethionine-dependent methyltransferases. Mol. Gen. Genet. 1991;230:120–128. doi: 10.1007/BF00290659. [DOI] [PubMed] [Google Scholar]
- 3.Stassi D., Donadio S., Staver M. J., Katz L. Identification of a Saccharopolyspora erythraea gene required for the final hydroxylation step in erythromycin biosynthesis. J. Bacteriol. 1993;175:182–189. doi: 10.1128/jb.175.1.182-189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merson-Davies L. A., Cundliffe E. Analysis of five tylosin biosynthetic genes from the tylIBA region of Streptomyces fradiae genome. Mol. Microbiol. 1994;13:349–355. doi: 10.1111/j.1365-2958.1994.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 5.Rodríguez A. M., Olano C., Méndez C., Hutchinson C. R., Salas J. A. A cytochrome P450-like gene possibly involved in oleandomycin biosynthesis by Streptomyces antibioticus. FEMS Microbiol. Lett. 1995;127:117–120. doi: 10.1111/j.1574-6968.1995.tb07459.x. [DOI] [PubMed] [Google Scholar]
- 6.Betlach M. C., Kealey J. T., Betlach M. C., Ashley G. W., McDaniel R. Characterisation of the macrolide P-450 hydroxylase from Streptomyces venezuelae which converts narbomycin to picromycin. Biochemistry. 1998;37:14937–14942. doi: 10.1021/bi981699c. [DOI] [PubMed] [Google Scholar]
- 7.Xue Y., Wilson D., Zhao L., Liu H., Sherman D. H. Hydroxylation of macrolactones YC-17 and narbomycin is mediated by the pikC-encoded cytochrome P450 in Streptomyces venezuelae. Chem. Biol. 1998;5:661–667. doi: 10.1016/s1074-5521(98)90293-9. [DOI] [PubMed] [Google Scholar]
- 8.Tang L., Shah S., Chung L., Carney J., Katz L., Khosla C., Julien B. Cloning and heterologous expression of the epothilone gene cluster. Science. 2000;287:640–642. doi: 10.1126/science.287.5453.640. [DOI] [PubMed] [Google Scholar]
- 9.Molnár I., Schupp T., Ono M., Zirkle R. E., Milnamow M., Nowak-Thompson B., Engel N., Toupet C., Stratmann A., Cyr D. D., et al. The biosynthetic gene cluster for the microtubule-stabilising agents epothilones A and B from Sorangium cellulosum So ce90. Chem. Biol. 2000;7:97–109. doi: 10.1016/s1074-5521(00)00075-2. [DOI] [PubMed] [Google Scholar]
- 10.Caffrey P., Lynch S., Flood E., Finnan S., Oliynyk M. Amphotericin biosynthesis in Streptomyces nodosus: deductions from analysis of polyketide synthase and late genes. Chem. Biol. 2001;8:713–723. doi: 10.1016/s1074-5521(01)00046-1. [DOI] [PubMed] [Google Scholar]
- 11.Brautaset T., Sekurova O. N., Sletta H., Ellingsen T. E., Strom A. R., Valla S., Zotchev S. B. Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem. Biol. 2000;7:395–403. doi: 10.1016/s1074-5521(00)00120-4. [DOI] [PubMed] [Google Scholar]
- 12.Aparicio J. F., Colina A. J., Ceballos E., Martín J. F. The biosynthetic gene cluster for the 26-membered ring polyene macrolide pimaricin. J. Biol. Chem. 1999;274:10133–10139. doi: 10.1074/jbc.274.15.10133. [DOI] [PubMed] [Google Scholar]
- 13.Aparicio J. F., Fouces R., Mendes M. V., Olivera N., Martín J. F. A complex multienzyme system encoded by five polyketide synthase genes is involved in the biosynthesis of the 26-membered polyene macrolide pimaricin in Streptomyces natalensis. Chem. Biol. 2000;7:895–905. doi: 10.1016/s1074-5521(00)00038-7. [DOI] [PubMed] [Google Scholar]
- 14.Molnár I., Aparicio J. F., Haydock S. F., Khaw L. E., Schwecke T., König A., Staunton J., Leadlay P. F. Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of genes flanking the polyketide synthase. Gene. 1996;169:1–7. doi: 10.1016/0378-1119(95)00799-7. [DOI] [PubMed] [Google Scholar]
- 15.Mendes M. V., Recio E., Fouces R., Luiten R., Martin J. F., Aparicio J. F. Engineered biosynthesis of novel polyenes: a pimaricin derivative produced by targeted gene disruption in Streptomyces natalensis. Chem. Biol. 2001;8:635–644. doi: 10.1016/s1074-5521(01)00033-3. [DOI] [PubMed] [Google Scholar]
- 16.Andersen J. F., Tatsuta K., Gunji H., Ishiyama T., Hutchinson C. R. Substrate specificity of 6-deoxyerythronolide B hydroxylase, a bacterial cytochrome P450 of erythromycin biosynthesis. Biochemistry. 1993;32:1905–1913. doi: 10.1021/bi00059a004. [DOI] [PubMed] [Google Scholar]
- 17.Lambalot R. H., Cane D. E., Aparicio J. J., Katz L. Overproduction and characterization of the erythromycin C-12 hydroxylase, EryK. Biochemistry. 1995;34:1858–1866. doi: 10.1021/bi00006a006. [DOI] [PubMed] [Google Scholar]
- 18.Graziani E. I., Cane D. E., Betlach M. C., Kealey J. T., McDaniel R. Macrolide biosynthesis: a single cytochrome P450, PicK, is responsible for the hydroxylations that generate methymycin, neomethymycin, and picromycin in Streptomyces venezuelae. Bioorg. Med. Chem. Lett. 1998;8:3117–3120. doi: 10.1016/s0960-894x(98)00553-8. [DOI] [PubMed] [Google Scholar]
- 19.Aparicio J. F., Caffrey P., Gil J. A., Zotchev S. B. Polyene antibiotic biosynthesis gene clusters. Appl. Microbiol. Biotechnol. 2003;61:179–188. doi: 10.1007/s00253-002-1183-5. [DOI] [PubMed] [Google Scholar]
- 20.Antón N., Mendes M. V., Martín J. F., Aparicio J. F. Identification of PimR as a positive regulator of pimaricin biosynthesis in Streptomyces natalensis. J. Bacteriol. 2004;186:2567–2575. doi: 10.1128/JB.186.9.2567-2575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. Norwich: John Innes Foundation; 2000. Practical Streptomyces Genetics. [Google Scholar]
- 22.Sambrook J., Russell D. W. 3rd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning: a Laboratory Manual. [Google Scholar]
- 23.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–252. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 24.Byrne B., Carmody M., Gibson E., Rawlings B., Caffrey P. Biosynthesis of deoxyamphotericins and deoxyamphoterololides by engineered strains of Streptomyces nodosus. Chem. Biol. 2003;10:1215–1224. doi: 10.1016/j.chembiol.2003.12.001. [DOI] [PubMed] [Google Scholar]