Abstract
To elucidate the specific biological role of the yeast homologues of PTPA (phosphatase 2A phosphatase activator), Ypa1 and Ypa2 (where Ypa stands for yeast phosphatase activator), in the regulation of PP2A (protein phosphatase 2A), we investigated the physical interaction of both Ypa proteins with the catalytic subunit of the different yeast PP2A-like phosphatases. Ypa1 interacts specifically with Pph3, Sit4 and Ppg1, whereas Ypa2 binds to Pph21 and Pph22. The Ypa1 and Ypa2 proteins do not compete with Tap42 (PP2A associating protein) for binding to PP2A family members. The interaction of the Ypa proteins with the catalytic subunit of PP2A-like phosphatases is direct and independent of other regulatory subunits, implicating a specific function for the different PP2A–Ypa complexes. Strikingly, the interaction of Ypa2 with yeast PP2A is promoted by the presence of Ypa1, suggesting a positive role of Ypa1 in the regulation of PP2A association with other interacting proteins. As in the mammalian system, all yeast PP2A-like enzymes associate as an inactive complex with Yme (yeast methyl esterase). Ypa1 as well as Ypa2 can reactivate all these inactive complexes, except Pph22-Yme. Ypa1 is the most potent activator of PP2A activity, suggesting that there is no direct correlation between activation potential and binding capacity.
Keywords: phosphatase 2A phosphatase activator (PTPA), protein phosphatase 2A (PP2A), PP2A methyl esterase 1 (PME-1), Saccharomyces cerevisiae, Tap42, target of rapamycin (TOR)
Abbreviations: GST, glutathione S-transferase; HA, haemagglutinin; IVTT, in vitro transcribed and translated; LCMT-1, leucine carboxyl methyl transferase 1; PP2A, protein phosphatase 2A; PME-1, PP2A methyl esterase 1; PP2Ac, catalytic subunit of PP2A; PP2Ai, inactive form of PP2A; PTPA, phosphatase 2A phosphatase activator; Sap, Sit4 associating protein; Tap42, PP2A associating protein; Tip41, Tap42 interacting protein; TOR, target of rapamycin; Yme, yeast methyl esterase; Ypa, yeast phosphatase activator
INTRODUCTION
PP2A (protein phosphatase 2A) represents a major fraction of the cellular Ser/Thr phosphatase activity, responsible for the regulation of many different cellular events (see [1] for a review). PP2A is a highly conserved enzyme during evolution. In Saccharomyces cerevisiae, two related genes, PPH21 and PPH22, encode the catalytic subunit of PP2A (PP2AC). Deletion of both genes confers a severe growth defect and in the absence of a third gene, encoding the yeast PP2A-like phosphatase Pph3, this double deletion strain loses viability. Two other PP2A-like phosphatases are described in yeast: Ppg1, possibly involved in glycogen metabolism [2] and Sit4, an essential protein for proper progression through the G1/S transition [3].
The catalytic subunit of PP2A is part of a core enzyme together with the A/PR65 subunit, encoded by TPD3 in yeast, which is a scaffold protein targeting the third regulatory B subunits to the PP2A core structure. There are three families of mutually exclusive regulatory B subunits (B/PR55, B′/PR61 and B″/PR72). No sequence similarity exists between these families, except for two conserved ASBD (A subunit binding domains) [4]. All B subunit families consist of different members, resulting in a multitude of theoretically possible holoenzyme formations. Association of the B subunit to the PP2A core structure is considered to be important for regulation of the Ser/Thr phosphatase activity of the different PP2A holoenzymes by conferring substrate specificity or determining the subcellular localization [1]. In yeast, only one member of the B/PR55 subunit family (Cdc55) and one member of the B′/PR61 subunit family (Rts1) exist, whereas no homologue of the B″/PR72 subunit family can be found in the yeast genome.
The Sit4 PP2A-like phosphatase catalytic subunit interacts with four different Saps (Sit4 associating proteins), which are positive regulators of the biological function of Sit4 [5]. No regulatory subunits for Ppg1 or Pph3 are known.
The catalytic subunit of PP2A as well as Sit4 can interact with Tap42 (PP2A associating protein) known as a component of the TOR (target of rapamycin) pathway [6] and as the orthologue of the mammalian B cell receptor associated α4 protein [7]. TOR is a phosphatidylinositol kinase-related Ser/Thr protein kinase, implicated in cell growth control in response to nutrients by regulating several processes such as transcription, translation, protein stability and ribosome biogenesis [8]. Dominant TOR mutations as well as a temperature-sensitive TAP42 mutant render a cell resistant to rapamycin. The complex formation of Tap42 with all PP2A-like catalytic subunits is conserved from yeast [9] to human [10], is independent of other regulatory subunits and might be abolished by rapamycin or nutrient deprivation ([9], but see [10–12] for another view). PP2A-Tap42 as well as Sit4-Tap42 holoenzymes are negative regulators of the TOR pathway implicated in the control of the nutrient-dependent growth control [13].
PP2A can also be regulated by post-translational modifications such as threonine as well as tyrosine phosphorylation and methylation of PP2AC. At least two of these post-translational modifications occur in the C-terminal part of the PP2AC protein, which is highly conserved among species and different PP2A-like family members [1]. Reversible methylation occurs at the end-standing leucine residue by specific methyltransferase and methylesterase enzymes. These proteins are found from human [LCMT-1 (leucine carboxyl methyl transferase 1), LCMT-2 and PME-1 (PP2A methyl esterase 1)] to yeast (Ymt1, Ymt2 and Yme, where Yme stands for yeast methyl esterase). Biochemical analysis in mammalian systems and genetic analysis in yeast show that only the LCMT-1/Ymt1 and not the LCMT-2/Ymt2 isoform displays methyltransferase activity towards PP2AC. Methylation does not alter PP2A activity, but is required for the association of the third regulatory subunit to the PP2A core enzyme [14,15].
PP2A is also regulated by PTPA (phosphatase 2A phosphatase activator), initially known as the phosphotyrosyl phosphatase activator, since PTPA activates the phosphotyrosyl phosphatase activity of the PP2A core enzyme in vitro [16]. PTPA was found to be a highly conserved protein during evolution [17,18], suggesting an important biological function for this protein. In S. cerevisiae, two genes encode PTPA, YPA1 and YPA2. Ypa1 and Ypa2 (where Ypa stands for yeast phosphatase activator) have C-terminal extensions relative to vertebrate PTPA, and these extensions do not show any similarity to each other. Phenotypic analysis of single and double deletion mutants revealed a specific role for Ypa1 in the G1 phase of the cell cycle [19], whereas Ypa2 functions in the M phase [20]. Ypa1 might regulate the G1/S transition through the TOR pathway, since deletion of YPA1 confers rapamycin resistance [19,21]. Deletion of both genes is lethal [20,21], indicating that PTPA activity, displayed by both partially redundant proteins is essential for viability. Furthermore, we [20] and others [21,22] have already shown genetic evidence for a functional interaction between PTPA and the yeast PP2A homologues or Sit4. Recently, we showed that PTPA also affects the Ser/Thr phosphatase activity of PP2Ai (inactive form of PP2A), a special inactive form that was isolated as a complex with PME-1 [23]. Therefore the meaning of PTPA was changed into PP2A phosphatase activator.
To elucidate further the biological role of PTPA as PP2A regulator, we investigated the physical interaction of Ypa1 and Ypa2 with the different PP2A-like phosphatases in yeast (Pph21, Pph22, Pph3, Sit4 and Ppg1) by using the yeast two-hybrid system as well as GST (glutathione S-transferase) pull-down assays. We show that Ypa1 preferentially binds to Ppg1, Sit4 and Pph3, whereas Ypa2 favours interaction with Pph21 and Pph22 and this latter interaction is promoted by the presence of Ypa1. In addition, we investigated the potential link between the Ypa proteins and Tap42 in the regulation of the TOR pathway as suggested by the rapamycin-resistant phenotype of ypaΔ [19,21] and tap42ts [6] mutants. We provide genetic evidence for the functional interaction between Tap42 and Ypa1. We observed no significant reciprocal influence at the level of interaction with the different PP2A-like phosphatases, but provide evidence for the existence of a trimeric Tap42–Ypa1 phosphatase complex. Furthermore, we found no direct correlation between the specificity of either of the two Ypa proteins to bind to different PP2A-like enzymes and their possibility to activate the different PP2Ai-like phosphatase complexes. Therefore activation and binding are two distinct Ypa functions that might operate independently.
In conclusion, Ypa1 and Ypa2 both interact genetically and physically with different PP2A-like phosphatases, indicating a different and specific function for both proteins in the in vivo regulation of PP2A-like enzymes.
MATERIALS AND METHODS
Plasmids, strains and materials
Plasmids used for the expression of GST or His-tagged fusion proteins in Escherichia coli were made by subcloning the appropriate coding region in the pGEX-4T-3 (Amersham Biosciences) or pET15b (Novagen) bacterial expression vectors respectively.
All strains used have the same W303-1A genetic background. The wild-type, ypa1Δ and ypa2Δ strains used are described in [19]. The HA-Pph21 (where HA stands for haemagglutinin) and HA-Pph22 constructs were introduced in the yeast genome in single copy and were kindly provided by Dr K. Arndt, Wyeth-Ayerst Research, U.S.A. (strain CY1624) and Dr M. Stark, University of Dundee, U.K. (strain DEY103HA) respectively. Sit4 and Pph3 were produced by the HA-tagged YCpIF17 expression vector with a Gal-inducible promoter that was kindly provided by Dr J. Arino (Universitat Autonoma Barcelona, Spain). Ppg1 was N-terminally HA-tagged by subcloning in the pMV-HA tag vector (a gift from Dr M. Voorhoeve, The Netherlands Cancer Institute, Amsterdam, The Netherlands) and subsequently subcloned in the Gal-inducible pYES2 yeast expression vector (Invitrogen). The HA-Pph21 and HA-Pph22 strains were grown in YPD [10 g of yeast extract, 20 g of peptone, 2% (w/v) glucose in 1 litre of water] medium, whereas strains transformed with all other constructs were grown in synthetic medium lacking the appropriate amino acid and supplemented with 2% (w/v) galactose and 1% raffinose.
Full-length coding sequences for all different proteins used in the present study were obtained by PCR on yeast genomic DNA with the appropriate forward and reverse primers containing the start and stop codons respectively and subcloned in the pBluescript plasmid.
Yeast two-hybrid system
Ypa1 was subcloned in the pAS2 vector, containing the GAL4 DNA-binding domain, and used as a bait to screen an S. cerevisiae expression library subcloned in a modified version of the pGAD424 vector [24]. The PJ69-4A strain was subsequently transformed with both bait and library and assayed for growth on plates containing synthetic medium without adenine (Ade−) and for expression of the β-galactosidase reporter gene [24]. Fragments encoding Ypa1-interacting proteins were isolated from yeast and subcloned in bacterial plasmids to determine the DNA sequence as described in the Matchmaker GAL4 two-hybrid user manual of ClonTech. Full-length fragments of PP2A-like yeast phosphatases were obtained by PCR (see above) and subcloned in frame with the GAL4 activation and DNA-binding domain of the pGAD424 and pGBT9 vectors respectively. Ade− growth and α-galactosidase assays were performed by co-transforming the AH109 strain (ClonTech) with these plasmids together with Ypa1 or Ypa2 in the pGBT9 or pGAD424 vectors.
GST pull-down assays
[35S]Methionine-labelled yeast proteins were obtained by using the TNT-coupled rabbit reticulocyte lysate system (Promega) with the appropriate RNA polymerase (T3 or T7) on pBluescript plasmids containing the coding region of Ppg1, Pph3, Sit4, Pph21, Pph22, Tpd3, Cdc55, Rts1, Tap42, Sap185, Sap155 and Tip41 (Tap42 interacting protein) according to the manufacturer's instructions. Otherwise, cell-free yeast extracts were made from HA-tagged PP2A-like phosphatases expressing yeast strains as described in [19]. GST fusion proteins (GST, GST-Ypa1, GST-Ypa2, GST-Tap42 and GST-Yme) and His-tagged Ypa1, Ypa2, Yme and Tap42 were expressed in E. coli BL21-pLys bacteria and purified on glutathione–Sepharose (Amersham Biosciences) and Ni2+-PDC Agarose (Affiland, Liège, Belgium) respectively according to the manufacturer's instructions. IVTT (in vitro transcribed and translated, 20 μl) yeast PP2A protein or 100 μl of yeast extracts with equal protein concentrations were added to equal amounts (5 μg) of GST fusion proteins bound to glutathione–Sepharose beads in 500 μl NENT-100 [20 mM Tris/HCl, pH 7.4, 1 mM EDTA, 0.1% Nonidet P40, 25% (v/v) glycerol and 100 mM NaCl] buffer supplemented with the protease inhibitors pepstatin (2 μg/ml), leupeptin (2 μg/ml) and PMSF (1 mM) and BSA (1 mg/ml). For competition experiments, equal amounts (5 μg) of the His-tagged protein of interest were added. The GST pull-down binding reactions were incubated for at least 4 h on a rotating wheel at 4 °C. The beads were subsequently washed three times with NENT-100 buffer with 1 mg/ml BSA and twice with NENT-100 buffer without BSA. Bound proteins were eluted by the addition of 20 μl of SDS sample buffer and boiling. The eluted proteins were analysed by loading 10 μl of this mixture on a SDS/PAGE and the [35S]methionine-labelled PP2A-like proteins were visualized by the Phosphoimager (Amersham Biosciences). To estimate the binding capacity of the IVTT protein to the GST fusion protein, 3 μl of total IVTT PP2A-like products were loaded on the same gel as input. SDS/PAGE of GST pull-down assays with yeast extracts was analysed by Western blotting. Alternatively, a fraction of the washed glutathione–Sepharose beads were immediately assayed in 20 mM Tris/HCl (pH 7.4), 1 mM dithiothreitol for phosphorylase a phosphatase activity after an additional wash in this buffer.
Immunoprecipitation and Western-blot analysis
Yeast extracts were first precleared with 20 μl of Protein G–Sepharose (Amersham Biosciences) for 1 h at 4 °C. The precleared supernatant was incubated overnight with 10 μl of the appropriate undiluted antibody (Sigma) in 0.5 ml of TBS (25 mM Tris/HCl, pH 7.4, 0.1% Nonidet P40, 150 mM NaCl) buffer supplemented with the protease inhibitors pepstatin (2 μg/ml), leupeptin (2 μg/ml) and PMSF (1 mM) at 4 °C on a rotating wheel. Afterwards, 20 μl of Protein G–Sepharose was added for another 2 h at 4 °C. Subsequently, the immunoprecipitate was washed five times with TBS where 0.1 M LiCl was optionally added.
After separation on SDS/PAGE, the proteins were transferred on to a PVDF membrane. The Western blots were preincubated in PBS, supplemented with 0.1% Tween 20 and 5% (w/v) skimmed milk powder. Subsequently, Western blots were incubated with the mouse monoclonal anti-HA tag (Sigma; 1/10.000) or rabbit polyclonal anti-Tap42 (a gift from K. Arndt; 1/200) primary antibodies in the same buffer for at least 2 h. Rabbit polyclonal anti-Ypa1 primary antibodies were made towards bacterially expressed Ypa1, purified as described in [18] and used in a dilution of 1/500. Rabbit anti-mouse or swine anti-rabbit immunoglobulins coupled with horseradish peroxidase (Dako Cytomation, Glostrup, Denmark) were used as secondary antibody at a dilution of 1/5000. After extensive washing, Western blots were visualized using ECL® (enhanced chemiluminescence; Amersham Biosciences).
Measurement of phosphatase activity
Purified PP2Ai from porcine brain [22] was initially used to measure the reactivation capacity of Ypa1 and Ypa2 compared with rPTPA (purified His-tagged recombinant Ypa1, Ypa2 and mammalian PTPA bacterially expressed proteins). The reactivation assay was performed with equal amounts of PTPA proteins in the presence or absence of protamine using 32P-labelled phosphorylase a as substrate. Glutathione–Sepharose beads of GST-Yme pull-down assays were preincubated in the absence (control) or presence of Ypa1, Ypa2 or rPTPA and 0.33 mM ATP and 1.67 mM Mg2+ at 30 °C for 10 min. 32P-labelled phosphorylase a (1 mg/ml) was added in the absence (basal activity) or presence of protamine [33 μg/ml in 16 μM (NH4)2SO4] for another 10 min at 30 °C. The reaction was stopped by trichloric acid (20%) precipitation and the liberated 32P was measured in the scintillation counter. Percentage of maximal reactivation is the percentage of PP2A activity relative to the maximal reactivation after the control (no PTPA or Mn2+) is subtracted. Inhibition of Ypa1 activity by Ypa2 was measured by the addition of different concentrations of Ypa2 to 125 pM Ypa1 in the preincubation with mammalian PP2Ai.
RESULTS
In vivo interaction of Ypa proteins with yeast PP2A-like enzymes in the yeast two-hybrid system
Genetic analysis indicates that Ypa1 is implicated in different cellular pathways [19,21,22]. To localize the role of Ypa1 in these pathways, we made use of the yeast two-hybrid system to identify in vivo Ypa1-interacting proteins that may act as regulators or targets of Ypa1. A yeast genomic library was screened with Ypa1 as a bait. Four different positive clones were isolated, based on their ability to grow on agar plates where adenine is omitted. DNA sequence analysis revealed partial open reading frames corresponding to four different proteins, including a member of the PP2A-like phosphatase family, Ppg1, possibly involved in glycogen metabolism [2].
In the present study, the Ppg1–Ypa1 interaction was further examined, since a physical interaction between Sit4, another PP2A-like phosphatase, and both Ypa1 and Ypa2 proteins were already described in vitro (MBP-Sit4 pull-down assay, where MBP stands for maltose-binding protein) as well as in vivo (HA-Sit4 immunoprecipitation assay) [22]. Hence Ypa1 and Ypa2 seem to interact physically with different PP2A-like phosphatases in yeast. Therefore we examined the interaction of Ypa1 and Ypa2 not only with Ppg1 but with all PP2A-like enzymes in yeast. Pph21, Pph22, Pph3, Sit4 and Ppg1 encoding sequences were generated by PCR on yeast genomic DNA. These PCR fragments were subcloned in both the GAL4 activation and GAL4 DNA-binding domain vectors and co-transformed with either of the two Ypa proteins encoding plasmids in an AH109 yeast strain. The resulting transformants were subjected to α-galactosidase and Ade− growth assays. Under these conditions, Ypa1 specifically interacted with Ppg1, Sit4 and Pph3, whereas Ypa2 bound only to the yeast PP2A homologues Pph21 and Pph22. This suggests that the two yeast homologues of PTPA specifically regulate different PP2A-like phosphatases by a physical interaction with a specific partner.
In vitro interaction of yeast PP2A-like enzymes with Ypa proteins in GST-Ypa pull-down assays
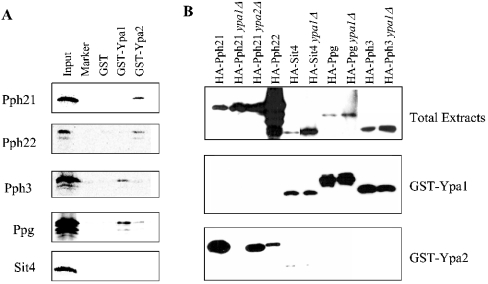
To confirm the yeast two-hybrid results, GST-Ypa1 and GST-Ypa2 pull-down assays were performed in two different methods to visualize the associated PP2A-like proteins. PP2A-like enzymes were either produced in a heterologous system as IVTT [35S]methionine-labelled proteins and detected by autoradiography (Figure 1A), or HA-tagged versions were expressed in different yeast strains and after pull-downs detected by Western blotting (Figure 1B). Both methods gave qualitatively the same results as the dual hybrid experiments: Ypa2 preferentially interacted with Pph21 and Pph22, whereas Sit4, Pph3 and Ppg1 associated with Ypa1 (the reason why the IVTT Sit4 did not interact with Ypa1 is argued in the Discussion section). Moreover, it was observed that Ypa2 has a higher affinity for Pph21 than for Pph22 (Figure 1B); although HA-Pph22 was expressed at a much higher level than HA-Pph21, much more HA-Pph21 was bound to GST-Ypa2 than HA-Pph22.
Figure 1. Specific interaction of Ypa1 or Ypa2 with different PP2A-like phosphatases.
(A) Different PP2A-like phosphatases were IVTT as [35S]methionine-labelled proteins and used in GST-Ypa1 and GST-Ypa2 pull-down assays as described in the Materials and methods section. (B) Different PP2A-like phosphatases were overexpressed in different yeast strains as HA-tagged proteins and analysed by Western blot with anti-HA tag antibodies. The top panel shows the expressing level of the HA-PP2A-like phosphatases in 5 μl of cell-free extracts. In this case, Western blot of HA-Sit4 and HA-Ppg1 needs a longer exposure time (2 min versus 15 s) to detect the expressed HA-tagged proteins. The middle and the lower panels show the GST-Ypa1 and GST-Ypa2 pull-downs respectively of these yeast extracts (exposure time 15 s).
To investigate the reciprocal influence of the presence of Ypa1 or Ypa2 on their interaction with the PP2A-like enzymes, the experiments with the expressed HA-PP2A-like enzymes were also performed in strains missing one of the two Ypas (Figure 1B). Contrary to our expectation, the presence of Ypa1 was a prerequisite for binding of Pph21 to Ypa2. No such relationship was observed for binding of Sit4, Ppg1 or Pph3 to Ypa1. Although the mechanism of this interference is not yet known, it might be the underlying reason for the lethality of the Ypa double deletion (see further in the Discussion section).
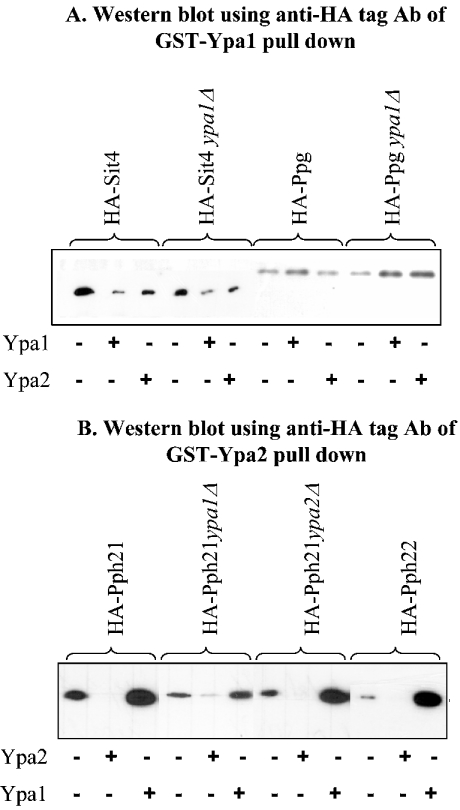
We sought further independent evidence for specific interactions and potential reciprocal influence of Ypa1 and Ypa2 by examining the potential competition between Ypa1 and Ypa2. We investigated the consequence of the absence of endogenous Ypa (ypaΔ strains) or the presence of an excess of exogenous Ypa (addition of purified His-Ypa) on GST-Ypa pull-downs of their specific PP2A-like interaction partners. As shown in Figure 2(A), the presence of endogenous Ypa1 did not significantly compete for binding of Ypa1 to Ppg1 or Sit4. The addition of an excess of purified recombinant His-Ypa1 during the GST-Ypa1 pull-down assays decreased the binding of Sit4 to GST-Ypa1, whereas almost no difference in binding was observed for Ppg1 with or without His-Ypa1 in the GST-Ypa1 pull-down assay. Maybe, GST-Ypa1 has a higher affinity or a more accessible interaction domain for Ppg1 than the recombinant His-Ypa1 or endogenous Ypa1 protein. The addition of His-Ypa2 in the GST-Ypa1 pull-down of HA-Sit4 or HA-Ppg1 expressing extracts hardly changed the binding of Sit4 or Ppg1 to GST-Ypa1 (Figure 2A).
Figure 2. Competition between Ypa1 and Ypa2 for binding to their specific PP2A-like partners.
Western blot incubated with anti-HA tag antibodies of (A) GST-Ypa1 and (B) GST-Ypa2 pull-downs from extracts of different yeast strains expressing HA-PP2A-like phosphatases in the absence or presence of an excess bacterially expressed and purified His-tagged Ypa1 or Ypa2. The exposure time of the Western blots after ECL® was ten times longer (2 min versus 15 s) for GST-Ypa pull-downs of extracts prepared from HA-Ppg (A), HA-Pph21 ypa1Δ and HA-Pph22-expressing strains HA-Pph21 (B).
The presence of endogenous Ypa2 did not seem to drastically influence the interaction of GST-Ypa2 with Pph21, probably due to excess of GST-Ypa2 in the pull-down assay (Figure 2B). As expected, an excess of His-Ypa2 competes for binding of Pph21 or Pph22 with GST-Ypa2. In contrast, the absence of endogenous Ypa1 significantly decreased the interaction of Pph21 with GST-Ypa2 (Figure 2B, strain HA-Pph21 ypa1Δ), as previously shown in Figure 1(B). Moreover, an excess of His-Ypa1 ameliorates the binding of Pph21 or Pph22 to GST-Ypa2 (Figure 2B). This further substantiates a positive function of Ypa1 in the specific binding of Ypa2 to Pph21 or Pph22.
Ypa directly interacts with the catalytic subunit of PP2A-like enzymes
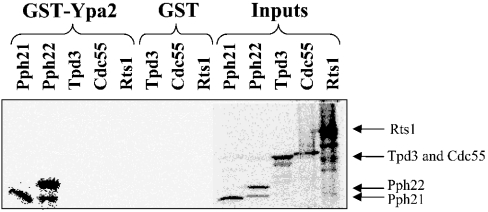
The catalytic subunits of the different PP2A-like phosphatases associate with distinct regulatory subunits. Pph21 and Pph22 interact with Tpd3 and Cdc55 or Rts1, whereas Sit4 forms complexes with Saps [5]. The interaction of Ypa1 and Ypa2 with the catalytic subunit of these phosphatases is probably direct and not through the associated regulatory subunits. This was demonstrated by a direct PP2AC–Ypa interaction in both the yeast two-hybrid system and the heterologous rabbit reticulocyte lysate. However, both systems contain highly conserved endogenous regulatory subunits. Hence an interaction of Ypa with one of the regulatory subunits present in these systems could not be excluded. Consequently, an interaction of IVTT [35S]methionine-labelled Pph21, Pph22, Tpd3, Cdc55 and Rts1 proteins was examined by GST-Ypa2 pull-down (Figure 3). It is clear that Ypa2 interacted directly only with the catalytic subunits, Pph21 and Pph22, and did not interact with the Tpd3 scaffold protein, or Cdc55 or Rts1 regulatory subunits. Moreover, GST-Ypa2 pull-downs of yeast extracts from HA-PP2A-like phosphatases expressing strains did not contain Tpd3, Cdc55 or Rts1 as observed by Western-blot analysis (results not shown). Also no interaction between Ypa1 or Ypa2 and the Saps (Sap155 and Sap185) or other Sit4 regulatory proteins (Tap42 and Tip41) could be demonstrated in similar GST-Ypa pull-down experiments with IVTT [35S]methionine-labelled Sap155, Sap185, Tap42 or Tip41 (results not shown).
Figure 3. Ypa2 binds directly to Pph21 and Pph22 independent of the A/Tpd3 or B/Cdc55 and B′/Rts1 subunits.
IVTT [35S]methionine-labelled yeast PP2A subunits were pulled down as described in the Materials and methods section.
PP2A–Ypa association is independent of Tap42
Besides the Ypa proteins, Tap42 is another common, highly conserved regulatory subunit of different PP2A-like enzymes, directly associating with the catalytic subunit in the absence of other regulatory subunits [6]. Furthermore, a temperature-sensitive TAP42 mutation [6] as well as deletion of YPA1 and to a lesser extent a deletion of YPA2 confers rapamycin resistance [19,21]. Therefore we wondered whether the function of the Ypas and Tap42 might be linked in the TOR pathway. First, we found that overexpression of Tap42 suppresses the rapamycin resistance of the ypa1Δ mutant (Table 1). These genetic data provide evidence for a functional interaction between both proteins. Both Tap42 and Ypa might be involved in the TOR pathway by direct physical interaction with PP2A-like enzymes. They could modulate phosphatase activity towards specific targets in this signalling pathway. For instance, Tap42 might inhibit or promote the association of the Ypa proteins with the different PP2A-like catalytic subunits, modulating the dephosphorylation of specific TOR substrates. In GST-Ypa pull downs of extracts prepared from cells expressing HA-PP2A-like phosphatases, the addition of an excess of bacterially expressed and purified His-Tap42 did not inhibit the Ypa1 or Ypa2 interaction with the different PP2A-like enzymes tested (results not shown). This suggests that Ypa has a higher affinity for these PP2A-like phosphatases compared with Tap42. Otherwise, Ypa proteins and Tap42 might bind to different domains of the PP2A catalytic subunit and form trimeric Ypa–PP2A–Tap42 complexes. Hence Tap42 does not seem to affect the PP2A–Ypa association.
Table 1. Rapamycin resistance of different yeast strains.
Rapamycin sensitivity of the different strains (all have a W303-1A genetic background) was measured on solid YPD medium supplemented with 0.2 or 1 μg/μl rapamycin. All strains were grown as liquid YPD cultures to A600 of unity at the appropriate temperature. Subsequently, 10 μl of a serial diluton (four times 1/10) were spotted on YPD plates in the presence or absence of rapamycin. Growth was scored after 2 days incubation at the appropriate temperature, ranging from no growth (−) to growth similar to that in the absence of rapamycin (++). Temperature sensitive strains indicated as ‘ts’ were incubated at the permissive temperature of 24 °C.
| Rapamycin | ||
|---|---|---|
| Strain | 0.2 μg/ml | 1 μg/ml |
| Wild-type [19] | − | − |
| ypa1Δ [19] | ++ | ++ |
| ypa2Δ [19] | − | − |
| HA-PPH21 [6] | − | − |
| HA-PPH21 ypa1Δ (this study) | ++ | ++ |
| pph22ts [30] | +/− | − |
| pph22tspph3Δ [30] | − | − |
| ypa1Δ TAP42 on PYX042 (this study) | − | − |
| tap42ts [6] | +/− | +/− |
| ypa1Δ tap42ts (this study) | + | + |
| tap42tsYPA1 on pXL2 (this study) | +/− | − |
PP2A–Tap42 association is independent of Ypa
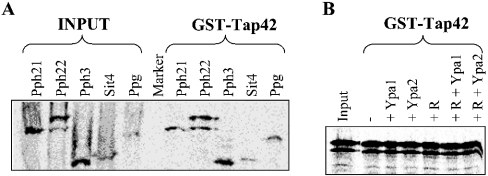
On the other hand, Ypa might regulate the rapamycin-sensitive association of Tap42 with PP2A-like catalytic subunits. To investigate this, GST-Tap42 pull-down assays of all IVTT [35S]methionine-labelled PP2A-like phosphatases were performed. We demonstrated a Tap42 interaction with all different PP2A-like enzymes (Figure 4A). The Pph22–Tap42 complex was used to examine the competition of Tap42 with Ypa proteins. No difference in Pph22–Tap42 association was observed in a GST-Tap42 pull down with IVTT [35S]methionine-labelled Pph22 in the absence or presence of bacterially expressed and purified His-Ypa1 or His-Ypa2 (Figure 4B). Hence Ypa does not seem to inhibit the PP2A–Tap42 association. Furthermore, the sensitivity of this complex formation to rapamycin as reported in [6] was also investigated. As shown in Figure 4(B), the addition of 20 μM rapamycin during the GST pull-down assays did not alter the Pph22–Tap42 interaction, as previously reported by other researchers [10–12]. More recently [25], an indirect effect of rapamycin on PP2A–Tap42 association is shown, caused by the inhibition of TOR kinase phosphorylation of Tip41, resulting in an increased binding of Tip41 to Tap42. Therefore the direct addition of rapamycin to the in vitro GST-Tap42 pull-down of PP2A might not disrupt the PP2A–Tap42 interaction.
Figure 4. Tap42 interacts with all PP2A-like phosphatases, independent of Ypa and rapamycin.
GST-Tap42 pull-downs of (A) IVTT [35S]methionine-labelled PP2A-like phosphatases or (B) IVTT [35S]methionine-labelled Pph22 with (+) or without (−) bacterially expressed and purified His-Ypa1 or His-Ypa2 in the presence or absence of 20 μM rapamycin (R) as described in the Materials and methods section.
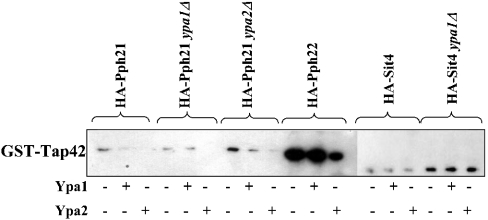
We further investigated the competition of the Ypa proteins for Tap42 binding to the different PP2A-like phosphatases. We performed a GST-Tap42 pull-down assay with extracts from cells expressing HA-PP2A-like phosphatases with or without the addition of an excess of bacterially expressed His-Ypa1 or His-Ypa2 (Figure 5). Ypa2, as specific Pph21/Pph22-interacting protein, and slightly also Ypa1, competed with Tap42 for interaction with Pph21 and somewhat with Pph22, but not for interaction with Sit4. This suggests a higher affinity of Sit4 for Tap42 in comparison with Pph21/Pph22. Moreover, the absence of the specific interacting Ypa protein (HA-Pph21 ypa2Δ and HA-Sit4 ypa1Δ) increases the association of Tap42 with the PP2A-like phosphatases (Figure 5). Ypa2 preferentially binds Pph21 (Figure 1B), whereas Tap42 prefers interaction with Pph22 (Figure 5), suggesting a specific but distinct cellular function for both Pph21–Ypa2 and Pph22–Tap42 complexes.
Figure 5. PP2A–Tap42 association is independent of Ypa1 and Ypa2.
GST-Tap42 pull-downs of extracts from different yeast strains expressing HA-PP2A-like phosphatases were performed in the presence (+) or absence (−) of bacterially expressed and purified His-Ypa1 or His-Ypa2 and analysed by Western blotting with anti-HA tagged antibodies as described in the Materials and methods section. Note that the experiments with HA-Sit4-expressing strains need a longer exposure time (2 min versus 15 s) to detect the expressed HA-tagged proteins.
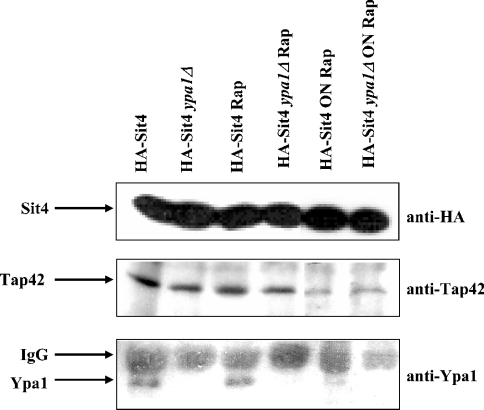
A potential interplay between Tap42 and Ypa was further examined in vivo by co-immunoprecipitation experiments. Extracts from HA-Sit4 expressing strains whether or not deleted for Ypa1, were incubated with anti-HA tag antibodies and co-immunoprecipitated Tap42 or Ypa1 was visualized by Western-blot analysis with anti-Tap42 or anti-Ypa1 antibodies. In these experimental conditions, Sit4, Tap42 and Ypa1 co-immunoprecipitated in the same complex (Figure 6), suggesting the existence of a trimeric Ypa1–Sit4–Tap42 complex in vivo, although this result can also be explained by the presence of separate Sit4–Tap42 and Sit4–Ypa1 complexes. Deletion of Ypa1 does not alter the Sit4–Tap42 association and also does not alter the rapamycin sensitivity of the Sit4–Tap42 interaction (Figure 6). Moreover, the rapamycin-dependent dissociation of the Sit4–Tap42 complex, as reported in [6] is only partial after a prolonged overnight rapamycin treatment of the cultures (Figure 6). Interestingly, the Sit4–Ypa1 association also seems to be diminished by the overnight rapamycin treatment (Figure 6). Thus the trimeric Tap42–Sit4–Ypa1 complex might be sensitive to rapamycin. Also, no difference in Pph21–Tap42 association was observed in wild-type versus ypa2Δ strains treated with or without rapamycin (results not shown). All these results suggest that there are no drastic reciprocal effects of Tap42 and Ypa at the level of binding to PP2A-like phosphatases. PP2A–Tap42 and PP2A–Ypa complexes might exist separately or as trimeric complexes acting on their respective substrates. Some of these substrates might be common for all complexes, which might explain why overexpression of Tap42 can compensate for the absence of Ypa1 for rapamycin sensitivity as shown in Table 1.
Figure 6. Rapamycin sensitive co-immunoprecipitation of Tap42 and Ypa1 with Sit4.
Exponentionally growing wild-type and ypa1Δ strains expressing HA-Sit4 were incubated with 0.2 μg/ml rapamycin (Rap) for 4 h or overnight (ON). Cell-free extracts were immunoprecipitated with anti-HA tag antibodies and analysed by Western blotting with anti-HA tag (exposure time 10 s), anti-Tap42 or anti-Ypa1 antibodies (exposure times 2 min) as described in the Materials and methods section.
Effect of Ypa on PP2A phosphatase activity
To elucidate the role of the physical interaction of Ypa with yeast PP2A-like phosphatases, we analysed the effect of Ypa on the activity of PP2A and made use of the PP2A reactivation assay by PTPA as described in [23]. Briefly, we isolated an inactive PP2AD–PME-1 complex, PP2Ai, from porcine brain. PP2Ai can be reactivated by recombinant mammalian PTPA, also with phosphorylase a as substrate. We made use of this feature to measure the activity of the yeast PTPA homologues, Ypa1 and Ypa2.
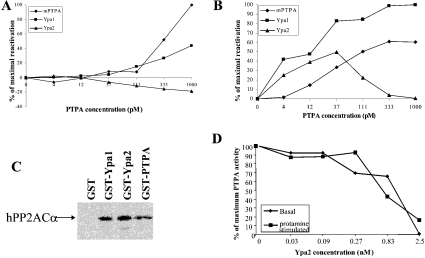
In yeast, only Ypa2 and not Ypa1 interacted with both PP2A homologues, Pph21 and Pph22. Therefore we expected that Ypa2 would reactivate the inactive mammalian PP2A. However, as shown in Figure 7(A), Ypa2 could not reactivate the basal activity of PP2Ai, and was even inhibitory at higher concentrations. However, Ypa1 reactivated PP2Ai almost as efficiently as mammalian PTPA. Also, the protamine-stimulated PP2A activity was reactivated much better by Ypa1 than by Ypa2 (Figure 7B). Although Ypa2 specifically bound to the yeast PP2A homologues, whereas Ypa1 was not a Pph21/Pph22 interacting protein, both Ypa1 and Ypa2, as well as the only mammalian PTPA protein (mPTPA) all associated with the IVTT α-isoform of the catalytic subunit of human PP2A (hPP2ACα) (Figure 7C). Therefore stable binding of Ypa to PP2A is not required for PP2Ai reactivation. Previously, it was shown that Ypa2 is not capable of activating the phosphotyrosyl phosphatase activity of PP2A [18]. Hence Ypa2 might be a competitive inhibitor of Ypa1 by binding but not activating the PP2Ai. The biphasic activation curve of PP2Ai by Ypa2 (Figure 7B) is in line with this interpretation: at higher concentrations, Ypa2 might bind to a secondary site that would prevent activation. To explore this possibility, we added different concentrations of Ypa2 to Ypa1 in the PP2A reactivation assay. As shown in Figure 7(D), Ypa2 inhibited the PTPA-mediated reactivation of the basal as well as the protamine stimulated PP2A activity of Ypa1 in a dose-dependent manner. Also Ypa2 inhibits mPTPA activity (results not shown).
Figure 7. Reactivation of mammalian PP2Ai by yeast as well as mammalian PTPA.
Purified PP2Ai (1 nM) was incubated with the indicated concentrations of purified recombinant mammalian PTPA (◆), Ypa1 (■) and Ypa2 (▲) with ATP, Mg2+ and assayed for (A) basal and (B) protamine-stimulated phosphorylase a phosphatase activity. (C) GST pull-down assay of IVTT [35S]methionine-labelled human PP2ACα. (D) Ypa2 inhibits Ypa1 activity. Mammalian PP2Ai is reactivated by 125 pM Ypa1 as in (A, B) in the presence of the indicated concentrations of Ypa2. Subsequently, basal and protamine-stimulated phosphorylase a phosphatase activity was measured.
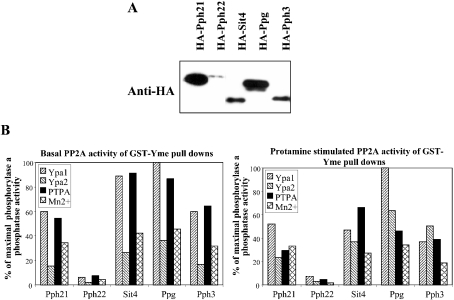
We examined whether this PTPA reactivation mechanism of PP2Ai also exists in yeast. Yeast PP2A-like phosphatases were expressed as HA-tagged proteins in yeast and cell-free extracts were prepared and precipitated by GST-Yme, a GST fusion protein of the Yme homologue. As shown in Figure 8(A), all yeast PP2A-like phosphatases interacted with Yme. Similar to the mammalian system, the yeast PP2A-like phosphatase–Yme complexes were inactive and except for the weak GST–Yme interacting phosphatase, Pph22, they could be reactivated by PTPA. We compared the potency of Ypa1 and Ypa2 to reactivate these inactive yeast PP2A members with that of PTPA. As shown in Figure 8(B), Ypa1 was the best PP2A-reactivating enzyme in yeast. Again, Ypa2 was less active and had a lower reactivation potency than mammalian PTPA in this yeast system. However, when yeast phosphatase activity was stimulated by protamine, the difference in PTPA activity between both Ypa proteins was less clear. Furthermore, the reactivation of inactive Sit4 and Ppg1 was most pronounced, whereas less Sit4 was associated with GST-Yme in comparison with Pph21, indicating that once activated, Sit4 might have a higher specific activity than Pph21.
Figure 8. Inactive yeast PP2A-like phosphatase–Yme complexes are reactivated by yeast and mammalian PTPA and also partially by Mn2+.
(A) Western-blot analysis with anti-HA tag antibody of GST-Yme pull-downs of extracts from different yeast strains expressing HA-PP2A-like phosphatases. (B) Reactivation of GST-Yme precipitated HA-PP2A-like phosphatases by the addition of bacterially expressed and purified mammalian and yeast His-tagged PTPA, in comparison with stimulation by 1 mM Mn2+.
DISCUSSION
To identify cellular targets of Ypa involved in different signalling pathways as suggested by the multiple phenotypic traits of the ypa1Δ mutant [19], we used the yeast two-hybrid system to screen an expressed yeast genomic library with Ypa1 as bait. Four different positive clones were isolated and DNA sequence analysis revealed partial open reading frames encoding Ppg1, a PP2A-like phosphatase [2], and three other proteins with known functions. Consulting the Comprehensive Yeast Genome Database (http://mips.gsf.de/genre/proj/yeast/index.jsp) Pex15, a protein required for peroxisome assembly, is reported to be another Ypa1 interaction partner. Although this interaction was found by a comprehensive two-hybrid analysis [26], Pex15 was not isolated as a Ypa1 interaction partner in our screen. In addition, we only isolated Ppg1 and no other PP2A-like phosphatases in this yeast two-hybrid screen. This might be due to a lower representation of some clones after multiple rounds of library amplification. However, we previously implied by in vitro [18] and genetic [19,20] data that Ypa1 and also Ypa2 are in vivo regulators of PP2A. Sit4, another phosphatase of the PP2A-like family, is also already identified as a Ypa1 and Ypa2 interacting protein [22]. Ypa1 and Ypa2 display different functions [20], suggesting that they might interact with different cellular proteins. Therefore we investigated the physical interaction of both Ypa proteins with all the different PP2A-like phosphatases.
By yeast two-hybrid and GST pull-down assays, we demonstrated a specific interaction of Ypa1 with Sit4, Ppg1 and Pph3, whereas Ypa2 specifically binds to the yeast PP2A homologues, Pph21 and Pph22. Although the specificity of these interactions is beyond any doubt, this specificity is not absolute. For instance, a weak Sit4–Ypa2 interaction could also be observed (Figure 1B). Therefore the interaction between Sit4 and Ypa2 as reported in [22] is not necessarily contradictory to our results. Also, the previously reported Sit4–Ypa1 interaction [22] could be confirmed in the yeast two-hybrid system as well as by GST-Ypa1 pull-downs with yeast extracts overexpressing HA-tagged Sit4 (Figure 1B). Nevertheless, no interaction of Sit4 with either Ypa1 or Ypa2 could be demonstrated by GST-Ypa pull-down assays of IVTT Sit4 in the rabbit reticulocyte system (Figure 1A). This apparent discrepancy might be explained by the presence of a hypothetical third partner in yeast possibly essential for the Sit4–Ypa1 interaction. Tap42 [6] as well as the Saps [5] is known as regulatory subunits of Sit4 and, therefore, they might be responsible for indirect binding of Ypa1 to Sit4. However, none of these proteins or the Tap42 interacting protein, Tip41 [25], interacts directly with Ypa1 (results not shown). Alternatively, a post-translational modification of Sit4 in the rabbit reticulocyte lysate, not occurring in yeast, might inhibit interaction of Sit4 with Ypa1. More probably, interaction between Ypa1 and Sit4 can only be demonstrated when Sit4 is overexpressed. Under normal conditions, all Sit4 might be associated with either Tap42 or one of the Saps leaving no free Sit4 to interact with Ypa1. This might be the situation in rabbit reticulocyte lysates, since mammalian homologues of Sit4, Tap42 as well of the Saps are known. Therefore, Ypa1 might interact directly with Sit4, but with much lower affinity than Tap42 or the Saps.
Ypa2 directly interacted with the catalytic subunits of yeast PP2A. No regulatory PP2A subunits such as Tpd3, Cdc55 or Rts1 could be detected in these PP2A–Ypa2 complexes. Interestingly, this result implies that Ypa2 might interact with a free catalytic subunit or could recruit the catalytic subunit from other PP2A holoenzymes in living yeast cells. Up to now, such a direct interaction with a free catalytic subunit, independent of the regulatory A or B subunits is only described for Tap42 and Pph21 or Pph22 [6]. Interaction of Tap42 with PP2A alters the activity of PP2A towards specific substrates, determining the function of PP2A in TOR-mediated signalling pathways [12,27,28]. Similarly, the Pph21/Pph22–Ypa2 complex can be considered as a novel PP2A holoenzyme with an altered activity towards specific substrates, conferring specific cellular functions to PP2A, which are mediated by the Ypa2 regulatory subunit. One of these functions might be the regulation of PP2A in the spindle assembly checkpoint, since Ypa2 is shown to be specifically involved in this cell-cycle checkpoint [20]. Although Ypa1 did not directly interact with the yeast PP2A homologues, the presence of Ypa1 positively regulated the Ypa2 interaction with yeast PP2A. This might explain the lethality of a ypa1Δ ypa2Δ double deletion mutant despite the different cellular functions of Ypa1 and Ypa2, as suggested by the different phenotypes of ypaΔ single deletion mutants and different PP2A-like interaction partners.
Both Tap42 and Ypa can directly interact with PP2A-like phosphatases, although with different affinities. Tap42 interacted with all PP2A-like phosphatases (Figure 4A). This result corroborates the recent characterization of a conserved Tap42-binding motif in the N-terminal domain of all PP2A-like enzymes, responsible for their interaction with Tap42 [9]. Interestingly, we observed that Ypa2 preferentially interacts with Pph21, whereas Tap42 prefers interaction with Pph22. This suggests a specific and different function of Pph21–Ypa2 and Pph22–Tap42 complexes, probably by altered PP2A activity towards specific cellular substrates or by directing the complexes to distinct cellular locations. PP2A-like phosphatases associated with Tap42 function in the TOR pathway [6] and a role for Ypa in this pathway is suggested by the similar rapamycin-resistant phenotype of the ypa1Δ and the tap42ts strains ([6,19], this study). Moreover, a partially overlapping role for Ypa1- and Tap42-associated PP2A-like phosphatases could be predicted from the genetic data that Tap42 overexpression suppresses the rapamycin resistance of the ypa1Δ strain (Table 1). Therefore we investigated whether the physical interaction between PP2A-like phosphatases and Ypa proteins as demonstrated in the present study might regulate the association of PP2A-like phosphatases with Tap42. Only a minor effect of Ypa on the PP2A–Tap42 interaction was observed. In addition, no direct effect of rapamycin on the PP2A–Tap42 association was observed in our experiments (Figure 4B), as was previously reported by others [10–12]. However, disruption of the TOR kinase phosphorylation-dependent PP2A–Tap42 complex by rapamycin is also notified [6]. More recently [25], an indirect effect of rapamycin on PP2A–Tap42 association was described, caused by the inhibition of TOR kinase phosphorylation of Tip41, resulting in an increased binding of Tip41 to Tap42. This might explain the rapamycin effect in vivo on the Sit4–Tap42 interaction by prolonged incubation (Figure 6). Moreover, similar to Tap42, Ypa1 was also co-immunoprecipitated with Sit4 and also this complex formation was rapamycin-sensitive under these experimental conditions. These results indicate that although Ypa1 is not required for Sit4–Tap42 interaction, since this complex is also present in a ypa1Δ strain, trimeric Ypa1–Sit4–Tap42 complexes most probably also exist. Therefore Ypa's function in the TOR pathway might be to direct the phosphatase activity of the different Sit4–Tap42 complexes towards specific substrates.
We examined the functional consequence of the physical interaction of Ypa1 and Ypa2 with specific yeast PP2A-like phosphatases. So far, no phosphatase activity could be measured in GST-Ypa1 or GST-Ypa2 pull-downs of yeast PP2A-like phosphatases, but this might be due to the fact that the physiological substrates are not yet known. When Ypa1 and Ypa2 were used in the reactivation assay of the inactive mammalian PP2A–PME-1 complex [23], Ypa1 increased PP2A activity almost to the same level as that of mammalian PTPA. In contrast, Ypa2 was rather a dose-dependent inhibitor of the PP2A activation and inhibited Ypa1 activity towards PP2A. In contrast with the specific binding of Ypa2 to the yeast PP2A homologues, both Ypa1 and Ypa2 bound to PP2AC and Ypa2 hampers Ypa1 in its function. On the other hand, Ypa1 is essential for Ypa2 binding to Pph21 and Pph22. Two different interacting sites for Ypa1 and Ypa2 each with a different functional consequence might be the physical basis for both the potentiating effect of Ypa1 on Ypa2 binding and for the inhibiting effect of Ypa2 on Ypa1 activity. By inducing a conformational change in PP2Ai and activating the phosphatase activity, Ypa1 might also induce the right conformation in Pph21 and Pph22 to allow Ypa2 binding. Binding of Ypa2 (on a second site) would then impair Ypa1 in its function. Since the activation reaction is enzymic, stable binding to site 1 might not be essential for full activation. Ypa2 can partially replace Ypa1 in this reactivation reaction (Figure 8B) and at higher concentrations, it inhibits this reaction (Figures 7A, 7B and 7D). These functional differences of Ypa1 and Ypa2 might be the explanation for the different specific phenotypes of single ypa1Δ and ypa2Δ mutants [20,21].
Subsequently, we examined whether this mechanism of PP2A reactivation by Ypa proteins also exists in yeast. All PP2A-like phosphatases interact with Yme (Figure 8A). All yeast PP2A-like phosphatases, except Pph22, are reactivated by both Ypa proteins, with a higher reactivation potential for Ypa1 on the basal phosphorylase a phosphatase activity. Also, Mn2+ reactivates the inactive PP2A–Yme complex up to a certain level. Such Mn2+ stimulation has previously been reported for a Ypa1/Ypa2-deficient strain [29], but as discussed in [23], this phenomenon probably represents the reactivation of a PP2Ai form that has lost its essential metal ions. Since reactivation by Ypa1 is much more pronounced than by Mn2+, most of the Yme associated PP2Ai probably represents a PP2Ai form inactivated by an unknown mechanism and stabilized by Yme, as already demonstrated in rabbit skeletal muscles and porcine brain [23].
This is the first report that demonstrated in vitro phosphorylase a phosphatase activity displayed by Ppg1 and Sit4. It might be that Sit4 and Ppg1 are present as inactive enzymes in the cells that can be activated by Ypa1. This is consistent with the fact that partially purified PP6, the mammalian Sit4 homologue, also displays only a very low phosphatase activity towards several in vitro substrates relative to PP2AC [11]. This might imply a spatial or temporal control of Sit4 (and its mammalian homologue PP6) and Ppg1 activity by the presence of PTPA. Although some Pph22 was associated with GST-Yme, no reactivation of this PP2A homologue could be observed. Therefore, as also suggested by the different preference for Ypa2 and Tap42 association, Pph21 and Pph22 might be differentially regulated in yeast, resulting in different cellular functions of both PP2A homologues.
In conclusion, in yeast, Ypa1 and to a lesser extent Ypa2 are functional PTPA homologues. Reactivation of inactive Yme–PP2A-like phosphatase complexes by Ypa is independent of the specific affinity of both Ypa proteins for the different PP2A-like phosphatases. This implies that the tight association of Ypa with PP2A-like enzymes is not required for the up-regulation of the phosphatase activity. This is not so surprising, since the reactivation is an enzymic reaction, probably inducing a conformational change of PP2AC from an inactive to an active structure by PTPA's peptidyl-prolyl cis/trans isomerase activity (J. Jordens, V. Janssens, I. Stevens, E. Martens, S. Longin, G. Bultynck, Y. Engelborghs, E. Waelkens, J. Goris and C. Van Hoof, unpublished work). Therefore the catalytic function of PTPA does not require stoichiometric association with PP2A. Physical interaction between PTPA and PP2A might be functionally important for the substrate specificity or subcellular localization of PP2A by directing PP2A towards specific substrates or cellular compartments.
Acknowledgments
This work was supported by the ‘Geconcerteerde Onderzoeksacties van de Vlaamse Gemeenschap’, the F.W.O-Vlaanderen and the ‘Interuniversity Attraction Poles’. C.V.H. and V.J. are post-doctoral fellows of the F.W.O.-Vlaanderen. We highly appreciate technical assistance of M. Veeckmans and M.-R. Verbiest.
References
- 1.Janssens V., Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Posas F., Clotet J., Muns M. T., Corominas A., Arino J. The gene PPG encodes a novel yeast protein phosphatase involved in glycogen accumulation. J. Biol. Chem. 1993;268:1349–1354. [PubMed] [Google Scholar]
- 3.Sutton A., Immanuel D., Arndt K. T. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell. Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X., Virshup D. M. Two conserved domains in regulatory B subunits mediate binding to the A subunit of protein phosphatase 2A. Eur. J. Biochem. 2002;269:546–552. doi: 10.1046/j.0014-2956.2001.02680.x. [DOI] [PubMed] [Google Scholar]
- 5.Luke M., Della Seta F., Di Como C. J., Sugimoto H., Kobayashi R., Arndt K. T. The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell. Biol. 1996;16:2744–2755. doi: 10.1128/mcb.16.6.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Como C. J., Arndt K. T. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 7.Murata K., Wu J., Brautigan D. L. B cell receptor-associated protein alpha4 displays rapamycin-sensitive binding directly to the catalytic subunit of protein phosphatase 2A. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10624–10629. doi: 10.1073/pnas.94.20.10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacinto E., Hall M. N. TOR signaling in bugs, brain and brawn. Nat. Rev. Mol. Cell Biol. 2003;4:117–126. doi: 10.1038/nrm1018. [DOI] [PubMed] [Google Scholar]
- 9.Wang H., Wang X., Jiang Y. Interaction with Tap42 is required for the essential function of Sit4 and type 2A phosphatases. Mol. Biol. Cell. 2003;14:4342–4351. doi: 10.1091/mbc.E03-02-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J., Peterson R. T., Schreiber S. L. Alpha 4 associates with protein phosphatases 2A, 4, and 6. Biochem. Biophys. Res. Commun. 1998;247:827–832. doi: 10.1006/bbrc.1998.8792. [DOI] [PubMed] [Google Scholar]
- 11.Kloeker S., Reed R., McConnell J. L., Chang D., Tran K., Westphal R. S., Law B. K., Colbran R. J., Kamoun M., Campbell K. S., et al. Parallel purification of three catalytic subunits of the protein serine/threonine phosphatase 2A family (PP2A(C), PP4(C), and PP6(C)) and analysis of the interaction of PP2A(C) with alpha4 protein. Protein Expr. Purif. 2003;31:19–33. doi: 10.1016/s1046-5928(03)00141-4. [DOI] [PubMed] [Google Scholar]
- 12.Nanahoshi M., Nishiuma T., Tsujishita Y., Hara K., Inui S., Sakaguchi N., Yonezawa K. Regulation of protein phosphatase 2A catalytic activity by alpha4 protein and its yeast homolog Tap42. Biochem. Biophys. Res. Commun. 1998;251:520–526. doi: 10.1006/bbrc.1998.9493. [DOI] [PubMed] [Google Scholar]
- 13.Zabrocki P., Van Hoof C., Goris J., Thevelein J. M., Winderickx J., Wera S. Protein phosphatase 2A on track for nutrient-induced signalling in yeast. Mol. Microbiol. 2002;43:835–842. doi: 10.1046/j.1365-2958.2002.02786.x. [DOI] [PubMed] [Google Scholar]
- 14.Bryant J. C., Westphal R. S., Wadzinski B. E. Methylated C-terminal leucine residue of PP2A catalytic subunit is important for binding of regulatory Balpha subunit. Biochem. J. 1999;339:241–246. [PMC free article] [PubMed] [Google Scholar]
- 15.Wei H., Ashby D. G., Moreno C. S., Ogris E., Yeong F. M., Corbett A. H., Pallas D. C. Carboxymethylation of the PP2A catalytic subunit in Saccharomyces cerevisiae is required for efficient interaction with the B-type subunits Cdc55p and Rts1p. J. Biol. Chem. 2001;276:1570–1577. doi: 10.1074/jbc.M008694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cayla X., Goris J., Hermann J., Hendrix P., Ozon R., Merlevede W. Isolation and characterization of a tyrosyl phosphatase activator from rabbit skeletal muscle and Xenopus laevis oocytes. Biochemistry. 1990;29:658–667. doi: 10.1021/bi00455a010. [DOI] [PubMed] [Google Scholar]
- 17.Cayla X., Van Hoof C., Bosch M., Waelkens E., Vandekerckhove J., Peeters B., Merlevede W., Goris J. Molecular cloning, expression and characterization of PTPA, a protein that activates the tyrosyl phosphatase activity of protein phosphatase 2A. J. Biol. Chem. 1994;269:15668–15675. [PubMed] [Google Scholar]
- 18.Van Hoof C., Janssens V., Dinishiotu A., Merlevede W., Goris J. Functional analysis of conserved domains in the phosphotyrosyl phosphatase activator: molecular cloning of the homologues from Drosophila melanogaster and Saccharomyces cerevisiae. Biochemistry. 1998;37:12899–12908. doi: 10.1021/bi980496l. [DOI] [PubMed] [Google Scholar]
- 19.Van Hoof C., Janssens V., De Baere I., de Winde H., Winderickx J., Dumortier F., Thevelein J., Merlevede W., Goris J. The Saccharomyces cerevisiae homologue YPA1 of the mammalian phosphotyrosyl phosphatase activator of protein phosphatase 2A controls progression through the G1 phase of the yeast cell cycle. J. Mol. Biol. 2000;302:103–119. doi: 10.1006/jmbi.2000.4062. [DOI] [PubMed] [Google Scholar]
- 20.Van Hoof C., Janssens V., De Baere I., Stark M. J. R., de Winde H., Winderickx J., Thevelein J. M., Merlevede W., Goris J. The Saccharomyces cerevisiae phosphotyrosyl phosphatase activator proteins are required for a subset of the functions disrupted by protein phosphatase 2A mutations. Exp. Cell Res. 2001;264:372–387. doi: 10.1006/excr.2000.5144. [DOI] [PubMed] [Google Scholar]
- 21.Rempola B., Kaniak A., Migdalski A., Rytka J., Slonimski P. P., di Rago J.-P. Functional analysis of RRD1 (YIL153w) and RRD2 (YPL152w), which encode two putative activators of the tyrosyl phosphatase activity of PP2A in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000;262:1081–1092. doi: 10.1007/pl00008651. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell D. A., Sprague G. F., Jr The phosphotyrosyl phosphatase activator, Ncs1p (Rrd1p), functions with Cla4p to regulate the G(2)/M transition in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:488–500. doi: 10.1128/MCB.21.2.488-500.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longin S., Jordens J., Stevens I., Janssens V., Martens E., Rondelez E., De Baere I., Derua R., Waelkens E., Goris J., et al. An inactive protein phosphatase 2A population is associated with methylesterase and can be reactivated with the phosphotyrosyl phosphatase activator, PTPA. Biochem. J. 2004;380:111–119. doi: 10.1042/BJ20031643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacinto E., Guo B., Arndt K. T., Schmelzle T., Hall M. N. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell. 2001;8:1017–1026. doi: 10.1016/s1097-2765(01)00386-0. [DOI] [PubMed] [Google Scholar]
- 26.Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duvel K., Santhanam A., Garrett S., Schneper L., Broach J. R. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell. 2003;11:1467–1478. doi: 10.1016/s1097-2765(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 28.Wang H., Jiang Y. The Tap42-protein phosphatase type 2A catalytic subunit complex is required for cell cycle-dependent distribution of actin in yeast. Mol. Cell. Biol. 2003;23:3116–3125. doi: 10.1128/MCB.23.9.3116-3125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fellner T., Lackner D. H., Hombauer H., Piribauer P., Mudrak I., Zaragoza K., Juno C., Ogris E. A novel and essential mechanism determining specificity and activity of protein phosphatase 2A (PP2A) in vivo. Genes Dev. 2003;17:2138–2150. doi: 10.1101/gad.259903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans D., Stark M. Mutations in the Saccharomyces cerevisiae type 2A protein phosphatase catalytic subunit reveal roles in cell wall integrity, actin cytoskeleton organization and mitosis. Genetics. 1997;145:227–241. doi: 10.1093/genetics/145.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]