Abstract
With glossy, wax-coated leaves, Rubus leucanthus is one of the few heat-tolerant wild raspberry trees. To ascertain the underlying mechanism of heat tolerance, we generated a high-quality genome assembly with a genome size of 230.9 Mb and 24,918 protein-coding genes. Significantly expanded gene families were enriched in the flavonoid biosynthesis pathway and the circadian rhythm-plant pathway, enabling survival in subtropical areas by accumulating protective flavonoids and modifying photoperiodic responses. In contrast, plant–pathogen interaction and MAPK signaling involved in response to pathogens were significantly contracted. The well-known heat response elements (HSP70, HSP90, and HSFs) were reduced in R. leucanthus compared to two other heat-intolerant species, R. chingii and R. occidentalis, with transcriptome profiles further demonstrating their dispensable roles in heat stress response. At the same time, three significantly positively selected genes in the pathway of cuticular wax biosynthesis were identified, and may contribute to the glossy, wax-coated leaves of R. leucanthus. The thick, leathery, waxy leaves protect R. leucanthus against pathogens and herbivores, supported by the reduced R gene repertoire in R. leucanthus (355) compared to R. chingii (376) and R. occidentalis (449). Our study provides some insights into adaptive divergence between R. leucanthus and other raspberry species on heat tolerance.
Keywords: Raspberry trees, thermotolerance, HSP70, cuticle wax, Rubus leucanthus
1. Introduction
Temperature directly influences plants by affecting the rates of biochemical reactions, as well as their growth, reproduction, and survival. Plants are limited to environments that fall within their physiological tolerances; therefore, thermotolerance is a key determinant of a species’ geographic range and distribution.1 Natural variation in thermotolerance, both between species (interspecific) and within species (intraspecific), is often associated with adaptations to various thermal environments.2 Species and populations living in warmer climates tend to possess higher thermotolerance limits compared to those in cooler regions. Greater thermotolerance offers a fitness advantage in hotter environments by enabling organisms to maintain their performance at elevated temperatures. The current trend of global warming poses a significant threat to crop productivity worldwide, making the development of thermotolerant cultivars urgent. Fundamental to this effort is the understanding of the genetic architecture underlying thermotolerance. The ‘cost of domestication’ hypothesis suggests that the long-term focus on achieving high yields and quality in crop cultivars has led to a depletion of genetic diversity, particularly in terms of biotic and abiotic stress tolerances.3 In contrast, crop wild relatives (CWRs) harbor a substantial amount of genetic diversity for such tolerances. CWRs are utilized not only as donor parents in hybridization efforts but also provide opportunities to elucidate the mechanisms underlying these traits.
The term ‘thermomorphogenesis’ refers to the effects of temperature on plant growth and morphology.4 Typical responses to thermomorphogenesis include elongation of hypocotyls and petioles, reduced stomatal density, and the development of smaller, thinner leaves.5 Thermotolerances can be categorized based on its sources into two types. The first is basal thermotolerance, which denotes a plant’s inherent capacity to endure heat stress. The second is acquired thermotolerance or heat stress priming, which is an induced tolerance to severe heat stress conditions that would typically be lethal, activated by prior exposure to mild heat stress.6 The induction of heat shock proteins (HSPs) is the most well-documented aspect of acquired thermotolerance. During the acclimation process, massively increase the transcription and translation of HSPs. These proteins are believed to act as molecular chaperones, protecting cellular proteins from irreversible denaturation due to heat and aiding in the refolding of proteins that have been damaged by heat.7 Without heat acclimation, plants rely on certain regulatory and acclimation proteins to achieve basal thermotolerance. These include the transcriptional regulator MBF1c (multiprotein bridging factor 1c) and the reactive oxygen species (ROS) detoxifying enzyme, catalase.8 Basal and acquired thermotolerances are often interlinked and play distinct roles at various stages of development and in different plant tissues.9 To date, the molecular and genetic mechanisms regulating plant thermomorphogenesis have remained largely elusive, with the bulk of research conducted on model species such as Arabidopsis and rice. In Arabidopsis, morphological adaptations that facilitate acclimation to elevated temperatures are predominantly orchestrated by the transcription factor PHYTOCHROME-INTERACTING FACTOR 4 (PIF4), PIF4 is regulated by various thermosensing mechanisms that empower plants to adjust to rising temperatures.10 In rice, the naturally occurring quantitative trait locus THERMOTOLERANCE 2 (TT2) confers thermotolerance across both vegetative and reproductive growth stages without compromising yield.11 This advantage arises from the plant’s ability to maintain a higher content of protective wax on its surface under high temperatures. Additionally, intrinsic mechanisms for thermotolerance include heat shock proteins, such as Hsp70 and Hsp90, which offer formidable defense against heat stress. While model species have provided initial insights, a broader examination involving a wide spectrum of plant species in crucial to fully comprehend the molecular and genetic controls of plant thermomorphogenesis.
The genus Rubus, belonging to the family Roseaceae, is estimated to contain between 600 and 800 species. This includes two commercially significant fruits: the red raspberry (R. idaeus L.) and black raspberry (R. occidentalis L.), both in the subgenus Idaeobatus.12 Raspberry cultivation commenced in Europe around 450 years ago. The optimal growing conditions for raspberries are regions with mild winters and prolonged, temperate summers, which are prevalent in areas such as Russia, Europe, and the Pacific coast of North America. Raspberries are one of the most extensively cultivated and cherished berries around the world, flouring in temperate climates across the globe. The boast an estimated annual production of 822,493 metric tonnes (https://www.atlasbig.com/en-us/countries-raspberry-production). China harbors approximately 200 wild species of the Rubus species, with the highest diversity and the most primitive taxa predominantly found in the Southwest. This area is recognized as the origin and diversity hub for the Rubus genus in East Asia.13 Although there is a rich diversity of wild Rubus species concentrated in Southwest China, commercial raspberry cultivation and production started only a few decades ago and has primarily been limited to the northern regions of the country. With the rise of increasingly extreme temperatures in China, there is a pressing necessity to breed raspberry varieties that are tolerant to heat. Some wild Rubus are found in tropical regions of China, having adapted over time to flourish in such warm climates. Consequently, these tropical Rubus species from China could offer valuable germplasm for breeding programs aimed at developing cultivars with enhanced heat tolerance. To utilize these wild Rubus species, it is essential to capture the genetic diversity and genomic resources of these thermotolerant wild species. As of 12 January 2023, genome assemblies are available for five Rubus species from the subgenus Idaeobatus, including R. occidentalis, R. ideaus, R. argutus, R. chingii, R. corchorifoliu (Subsect. Corchorifolii) (tracked from https://www.plabipd.de/plant_genomes_pa.ep). The last three species are widely distributed in East Asia and exhibit a great deal of local adaptation. Nonetheless, they are generally found in subtropical China and display only a modest degree of thermotolerance. Here, we present the genome assembly of a common Rubus species in tropical/subtropical China, R. leucanthus (Subsect. Leucanthi, subgenus Idaeobatus). This shrub is identifiable among other Rubus species by its glossy and leathery compound leaves14 (Fig. 1A). Notably adapted to tropical climates, R. leucanthus also produces abundant red berries, making it an attractive prospect for the development of new tropical raspberry cultivars. Our study delivers a high-quality genome assembly for R. leucanthus, and conduct comparative genomic analysis to shed light on evolutionary mechanisms underlying thermotolerance.
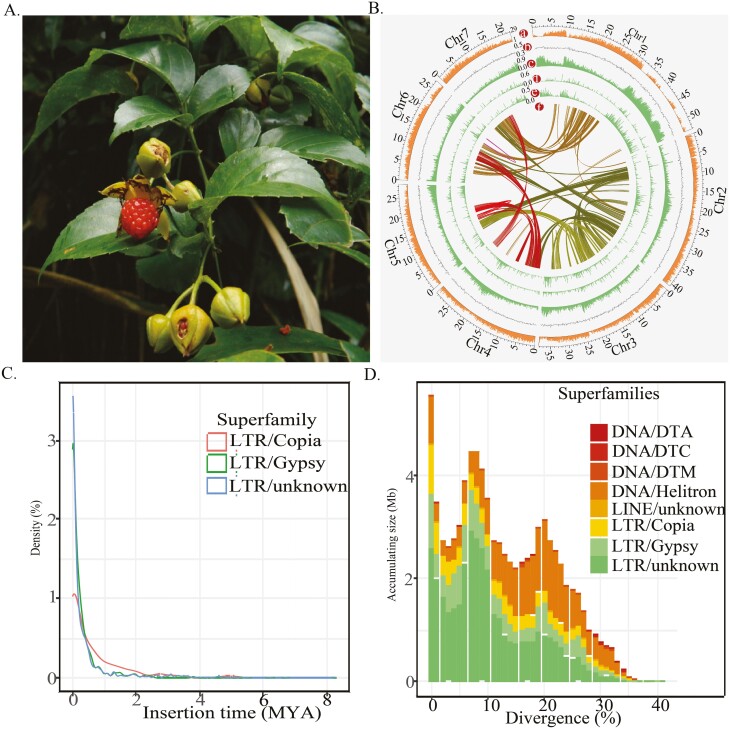
Figure 1.
(A) Photograph of Rubus leucanthus shown for its leaves and fruits. (B) Density distributions of insertion time for superfamilies LTR/Copia, LTR/Gypsy, and LTR/unknown. (C) Accumulating sizes for different age groups witin transposable elements superfamilies. (D) Genomic features of R. leucanthus. The concentric circles, from outermost to innermost were gene density (a), GC content (b), overall transposable element contents (c), LTR/Copia content (d), LTR/Gypsy content (e), and intragenomic collinearity blocks, respectively.
2. Materials and methods
2.1. Sample collection and genome sequencing
Genomic DNA was extracted from the fresh leaves of R. leucanthus, transplanted from Huadu, Guangzhou, China, using the DNAsecure Plant Kit (Tiangen Biotech, Beijing, China). Before library preparation, sample purity and concentration were evaluated using a Qubit fluorometer (ThermoFisher Scientific, MA, USA) and 1% agarose gel electrophoresis. A 350 bp insert fragment library was sequenced on the Illumina HiSeq XTM Ten platform (Illumina Inc., San Diego, CA, USA) to generate paired-end reads for genome surveying. The genome size and heterozygosity of R. leucanthus were estimated with GenomeScope 2.015 by analyzing the 21-mer distribution using about 50-fold Illumina sequencing coverage. A 20 Kbp insert SMRTbell library was constructed for long-read sequencing following PacBio’s standard protocol and sequenced on the PacBio Sequel platform using the DNA Sequencing Kit 4.0 V2. For the Hi-C library, young leaves from the same R. leucanthus accession were fixed with 1% formaldehyde. Subsequent steps included nuclei extraction, permeabilization, chromatin digestion with DpnII enzyme, and proximity ligation, as previously described.16 The resulting Hi-C library was sequenced on the Illumina HiSeq XTM Ten platform (Illumina Inc., San Diego, CA, USA) to generate 150 bp paired-end reads.
2.2. Genome assembly and scaffolding
The heterozygous R. leucanthus genome was assembled using Single Molecule Real-Time sequencing via the pb-assembly pipeline (https://github.com/PacificBiosciences/pb-assembly). This pipeline, which uses the Falcon and Falcon-unzip algorithms,17 was applied to 100-fold raw subreads. The process involved raw subread overlapping and consensus calling, error-corrected subread overlapping, contig assembly, diploid assembly of primary contigs and haplotigs based on read alignment, SNP calling, read phasing, and polishing of contigs using BLASR.18 The estimated haploid genome size for R. leucanthus was used to identify potential allelic contig pairs among the polished primary assemblies. These allelic pairs were then removed to produce haplotype-fused assemblies using Purge Haplotigs (version 1.1.0) with the parameters ‘-a 70’.19
To scaffold, order, and orient the purged R. leucanthus assemblies, capture chromatin contact information between physically proximate DNA regions were captured from the generated Hi-C libraries. Hi-C data generated from these libraries were evaluated using HiC-Pro v2.7.1.20 Raw reads from qualified Hi-C libraries were then processed into a non-redundant list of paired-end alignments using Juicer v1.6.21 The 3D-DNA pipeline was applied to order and orient the raw assemblies based on valid chromatin interaction data. Finally, mis-assemblies identified in the scaffolded assemblies were visually inspected and manually corrected using Juicebox Assembly Tools.
The final R. leucanthus genome assembly quality was evaluated using four methods: First, QUAST v5.2.022 calculated assembly continuity metrics (N50, N90) for contiguity assessment. Second, assembly completeness and redundancy were evaluated by aligning two BUSCO gene sets to the assemblies using BUSCO v5.4,23 including 2326 eudicot genes and 255 core eukaryotic genes. Third, PacBio subreads, Illumina resequencing reads and RNA-seq reads generated in this study (details provided Materials and Methods section 2.6) were mapped back to the assembled genomes and mapping rates were summarized. Finally, the LTR assembly index (LAI) was calculated to assess completeness of assembled long terminal repeat (LTR) retrotransposon sequences. In summary, continuity, completeness, accuracy, and LTR content were quantified to evaluate the R. leucanthus genome assembly.
2.3. Gene predictions and functional annotation
For identify and mask repetitive elements in the R. leucanthus genome assembly, we first constructed a de novo species-specific repeat library using RepeatModeler v1.0.11.24 This library was combined with existing repeat databases Dfam 3.0 and Repbase (20181026) to generate a comprehensive custom library. The R. leucanthus genome assembly was then masked using RepeatMasker v4.0.925 against the custom repeat library to identify interspersed and tandem repeat sequences. Using Infernal v1.1.226 and tRNAscan-SE,27 non-coding RNAs in the genome assemblies were identified.
For protein-coding gene prediction, we used the MAKER v3.01.02 pipeline,28 implementing both ab initio and homology-based approaches. Augustus v3.3.329 and SNAP (2013.11.29)30 were used for ab initio prediction. Prior to running MAKER, we conducted at least two rounds of iterative training for the hidden Markov models (HMMs) in Augustus and SNAP to optimize gene prediction parameters. These trained HMMs were then utilized alongside protein and transcript evidence from related Roseaceae species, including R. leucanthus (transcripts only), Fragaria vesca, Prunus persica, Pyrus communis, Rubus occidentalis, and Malus domestica, to predict gene structures. In the final step, MAKER combined ab initio predictions, homology-based predictions, and transcript alignments into a weighted consensus gene set using EvidenceModeler v1.1.1.31 To functionally annotate predicted proteins, we searched against InterPro v5.53.87.0, RefSeq (release99, 2020-05-11), and Swiss-Prot (release-2022-05) databases using InterProScan v532 or BLASTP. gene ontology terms and KEGG pathways were assigned by mapping to databases in KOBAS v3.0333 with an E-value cutoff of 1e−3. Transposable elements in the R. leucanthus genome assembly were identified and classified using the Extensive de-novo TE Annotator (EDTA) program,34 transposable elements in the genome assembly of R. leucanthus were identified and classified.
Using MCScanX with default parameters,35 intra-/inter-species collinear blocks among R. leucanthus and other species grape, R. chingii and R. occidentalis were determined, syntenic depth between R. leucanthus and other species were used to infer the whole genome duplication events.
2.4. Gene family cluster, phylogenetic tree construction, and divergence time estimation
To explore the genome evolution of R. leucanthus, we retrieved genomes of 11 other related species from online databases (Supplementary Table 1). These included Rubus occidentalis, Rubus chingii, Dryas drummondii, Rosa chinensis, Fragaria vesca, Potentilla micrantha, Eriobotrya japonica, Pyrus communis, Malus domestica, Prunus persica from the Roseaceae family, and the outgroup species Morus notabilis. Orthogroups between R. leucanthus and the other 11 species were inferred using OrthoFinder v2.3.1436 with Diamond Blast mode and Markov Cluster Algorithm (MCL) with an inflation parameter of 1.5.
Using single-copy orthogroups, we estimated the species tree via the multi-species coalescent model implemented in ASTRAL II.37 For each single-copy nuclear gene, protein sequences were aligned using MAFFT v7.0 with L-INS-I strategy.38 The corresponding coding sequences were then aligned to the protein alignments with no gaps or mismatches using PAL2NAL v14.39 The aligned coding sequences were concatenated into a supermatrix for substitution model testing using jModelTest 2 with the Akaike Information Criterion.40 Using the best-fit model and Morus notabilis as the outgroup, the phylogenetic tree relating R. leucanthus and the other 11 species was constructed using RAxML v8.2.1041 with 1000 bootstrap replicates. All these achieved with custom perl scripts (https://github.com/altingia/phylogenomics_pipeline).
Using the single-copy nuclear genes, we estimated Bayesian molecular dating between R. leucanthus and R. chingii as well as other nodes across the phylogenetic tree using the MCMCTREE program in PAML v4.9e.42 Following recommended procedures for divergence time estimation using genome-scale data (https://github.com/mariodosreis/divtime), we conducted an approximate likelihood estimation of branch lengths and divergence times using Markov chain Monte Carlo (MCMC) methods under a relaxed clock model with independent rates constraints. Two calibration points were used: one for the root node age between M. notabilis and E. japonica (62–118 million years ago, MYA), and another for the node between D. drummondii and R. chinensis (46–74 MYA).43
2.5. Gene family evolution and positive selection in R. leucanthus
To identify rapidly evolving gene families, we used the program CAFE v544 to analyze gene family size evolution across the ultrametric species phylogeny. CAFE was run with 1 to 4 gamma rate categories, with at least five iterations per category to ensure convergence. The best-fitting number of birth and death rate parameters was determined through likelihood ratio tests. This analysis enabled detection of gene families that have expanded or contracted significantly faster than expected under a random birth-death model along particular lineage like R. leucanthus.
To identify genes under positive selection in R. leucanthus, we utilized GWideCodeML v1.1,45 implementing branch-site models in PAML v4.9e.42 Using the 12-species phylogeny with R. leucanthus as the foreground branch, GWideCodeML estimated the nonsynonymous to synonymous substitutions ratio (ω) and generated dN/dS summaries for each orthogroup. We extracted key results from GWideCodeML outputs, including site class proportions, model likelihoods, and per-site posterior probabilities, using a custom Perl script. Positively selected genes met these criteria: ω>1, alignment length >=150 amino acid sites, significant likelihood ratio test difference between alternative and null models (P < 0.05), and individual sites with >95% posterior probability for site classes 2a or 2b. For accuracy, only orthogroups with all 12 species were analyzed, and duplicates were randomly reduced to one member. R. leucanthus was the ‘foreground’ branch, others were ‘background’. Enrichment analysis of gene ontology terms and KEGG pathways for positively selected and rapidly evolving genes was performed using KOBAS v3.03 at FDR < 0.05. Using online tool AlphaFold server 3,46 protein structures for the consensus sequences of the 12 species and sequences in R. leucanthus were predicted, and demonstrated and visualized with pyMOLv3.0.47
2.6. Heat shock proteins (HSPs) and heat shock transcription factors (HSFs) identification and transcriptome profiling in R. leucanthus
We searched the protein databases of R. leucanthus, R. chingii, R. occidentalis, and Arabidopsis using HMM profiles for Hsp70 (PF00012), Hsp90 (PF00183), and heat stress transcription factors (HSFs, PF00447) from Pfam. Hit proteins were aligned and used to build species-specific HMM profiles in an iterative search procedure. Candidate Hsp70s were screened for the presence of at least one intact nucleotide-binding domain (NBD) using InterProScan. Hsp90 candidates were verified to contain the key domains-N-terminal ATPase domain (NTD), middle substrate-binding domain (MD), and C-terminal dimerization domain (CTD). HSF candidates were confirmed to have intact DNA-binding domain (DBD) and HR-A/B regions. This domain/region screening ensured that only canonical proteins were included in further analyses. Using the pacakge NLGenomeSweeper,48 NBS-LRR (NLR) disease resistance genes in the genome assembly of R. leucanthus, R. chingii, R. occidentalis were identified with NB-ARC domain (PF00931) respectively.
Stem cuttings from young plants were cultivated for 3 months before transferred to a growth chamber. The chamber conditions were set to 70% humidity and a light intensity of 222 µmol.m-2. s-1 at 28°C for 2 hours. Leaves from three biological replications were then collected for ecophysiological assays and RNA sequencing. Following this, another three biological replications were exposed to a temperature of 46°C for an additional 2 hours, after which sampling was conducted in the same manner. Posttreatment, leaves for RNA sequencing were preserved with Liquid- Nitrogen, and stored at −80°C. Free malondialdehyde (MDA) content, soluble protein concentration, and SOD activity were measured using commercial kits. Total RNA was extracted using a Trizol reagent kit, and its quality was assessed using an Agilent 2100 Bioanalyzer and agarose gel electrophoresis. The total RNA was enriched for mRNA using Oligo (dT) beads, fragmented, and reverse transcribed into cDNA. The cDNA fragments were processed and sequenced on an Illumina Novaseq6000 platform. Clean reads were mapped to the reference genome using HISAT2. 2.4 with default parameters.49 FPKM (fragment per kilobase of transcript per million mapped reads) values were calculated for each transcription region using RSEM software.50 Differential expression analysis was performed by DESeq2.51 The genes/transcripts with false discovery rate (FDR) below 0.05 and absolute fold change not less than 2 were considered differentially expressed genes (DEGs). The STRING v12.0 database (https://string-db.org/) predicted protein-protein interactions among DEGs. The DEG list was input into STRING, and a PPI network was built with a confidence interaction score threshold of 0.4. The resulting STRING PPI network was then visualized and analyzed using Cytoscape software v3.10.0 and the cytoHubba plugin v0.1.52 cytoHubba identified hub nodes in the network using various topological parameters. The maximal clique centrality (MCC) metric was used to predict hub genes.
3. Results and discussion
3.1 Genome assembly and annotation
The genome size of R. leucanthus was estimated to be 239.08 Mb with a heterozygosity of 0.91% (Supplementary Fig. 1), well in according with the genome size of R. chingii, which is 239.4 Mb with a heterozygosity of 0.80%.53 In the sequencing and assembly of R. leucanthus genome, we progressed through several critical steps, each contributing to the refinement of the genome’s quality and completeness as detailed in Supplementary Table 2. Initially, we obtained 25.04 Gb of raw reads with 105.17-fold coverage of the genome. Through the hierarchical genome assembly process (HGAP), these were corrected to produce 21.11 Gb of preassembled reads (preads), achieving a pre-assembly rate 0.88, indicating superior quality beyond the threshold of 0.50. Further rounds of HGAP and haplotype phasing, polishing and deduplications, yielded a final haploid genome assembly of 239.67 Mb, consisting of 188 contigs with an N50 of 2.40 Mb (Supplementary Table 2). Using the 3.98 million valid Hi-C interactions (Supplementary Table 3), we successfully anchored and oriented 96.3% (230.9 Mb) of the draft assemblies onto seven pseudo-chromosomes, achieving an N50 of 40.2 Mb (Supplementary Fig. 2). Additionally, the quality of the genome assembly was also assessed by mapping rates of the PacBio and Illumina sequences. Mapping results showed that 96.0% of the PacBio genomic reads were mapped, while Illumina genomic and transcriptomic reads had mapping rates of 97.9% and 93.4%, respectively (Supplementary Table 4). The assembled genome of R. leucanthus achieved an LAI score of 20.92, satisfying the gold quality assembly criteria.54 BUSCO analysis revealed 97.6% and 97.0% completeness for the eukaryotic (Eukaryotes_odb10) and eudicot (Eudicots_odb10) gene sets, respectively (Supplementary Fig. 3). Based on the metrics of contiguity, completeness, and accuracy, we have obtained a high-quality genome assembly for R. leucanthus.
The analysis of repeat sequences revealed that 95.3 Mb, which constitutes 39.8% of the genome, was made up of repetitive elements in the R. leucanthus genome assembly. This is comparable to the 36.5% found in the R. chingii genome.53 The vast majority of which were transposable elements (TEs), accounting for 36.7% of the assembly. The composition of these TEs included 21.6% long terminal repeat (LTR) elements, 0.2% non-LTR elements, 10.5% terminal inverted repeat (TIR) elements, and 4.4% Helitrons (Supplementary Table 5). Retrotransposable elements comprised 21.8% of the genome assembly, with 63.3% of these retroelements being highly fragmented and not classified at the superfamily level. Of the classified retroelements, the content of Gypsy superfamily (5.0% of the assembly) was nearly double that of the Copia superfamily (2.8%). DNA transposons accounted for 15.0% of the genome assembly, with the most abundant DNA transposon superfamily being the DTM, comprising 4.8% of the assembly. The second most abundant were subclass 2 Helitrons, accounting for 4.4% of the genome assembly (Supplementary Table 5). No obvious LTR bursts were observed (Fig. 1C). However, two periods of increased transposable element (TE) activity were identified, with accumulation peaks at genetic distances of 0.08 and 0.21, suggesting historical bursts TE activity. Furthermore, there was a notable presence of older transposable element within the genome (Fig. 1D). Intriguingly, as the age of these elements increased, the genome retained a higher number of DNA transposons in comparison to retrotransposons. These dynamics of repetitive elements offer valuable insights into the compact nature of the R. leucanthus genome.
Using the maker pipeline, a total of 24,918 protein-encoding genes were predicted (Table 1, Supplementary Table 6, Supplementary Fig. 4). Over 95% of these genes were corroborated by supporting evidence lines, with an Annotation Edit Distance (ADE) score < 0.5 as recommend55 (Supplementary Fig. 5). With a threshold E-value of 0.001, the proportions of genes in the R. leucanthus genome that could be annotated were 81.4%, 70.0%, 86.8%, 16.8%, and 67.1% according to the InterPro, Swiss-Prot, RefSeq, KEGG, and GO databases, respectively (Supplementary Table 7). In addition to protein-encoding genes, other non-coding RNAs including 112 miRNA, 109 rRNA, 431 tRNA, and 336 snoRNA were also identified using the Hidden Markov model of RNA families as provide in the RFAM v13.0 databases56 (Supplementary Table 8).
Table 1.
Summary of genome assembly of Rubus leucanthus
| Assemble feature | Statistics | |
|---|---|---|
| Estimated genome size | 238094177 | |
| Heterzygosity rate | 0.91% | |
| Repeat proportion | 32.80% | |
| PacBio sequencing assembly | ||
| Number of contigs | 188 | |
| Contig N50 | 2404816 | |
| Contig L50 | 32 | |
| Contig N90 | 878157 | |
| Contig L90 | 94 | |
| Longest contig | 9418599 | |
| GC content | 36.14% | |
| Total contig length | 239668171 | |
| Hi-C scaffolding assembly | ||
| Number of scaffolds | ||
| Scaffold N50 | 40220118 | |
| Scaffold L50 | 3 | |
| Scaffold N90 | 19636704 | |
| Scaffold L90 | 7 | |
| Longest scaffold | 44099573 | |
| GC content | 36.14% | |
| Total scaffold length | 239911671 |
3.2. Lineage unique gene families, gene family expansions, and contractions in R. leucanthus genome
In a comparative analysis, a total of 70, 445 orthologous groups among the 409, 829 genes from R. leucanthus and 11 other species were identified (Supplementary Table 9). Within these groups, 2,427 genes were found to be unique to R. leucanthus, distributed across 2, 211 orthologous groups. These unique gene families were significantly enriched in 10 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, which were mainly involved in the metabolism of ribosomes, amino acids, fatty acids (Supplementary Table 10). Enrichment analysis pinpointed two amino acid degradation pathways associated with the response to abiotic stress: the degradation of valine, leucine, and isoleucine (KO00280, FDR = 3.08E−4) and lysine degradation (KO00640, FDR = 4.06E−4).57 Moreover, six genes unique to R. leucanthus were identified as members of transcription factor families, including bHLH (Rle11390), GRAS (Rle21912, Rle21913), NAC (Rle22254), FAR1 (Rle9875), and HD-ZIP (Rle21888), respectively (Supplementary Table 11). The complex roles of these genes in diverse aspects of plant development and their responses to biotic/abiotic stresses have been the subject of extensive studies.58–60
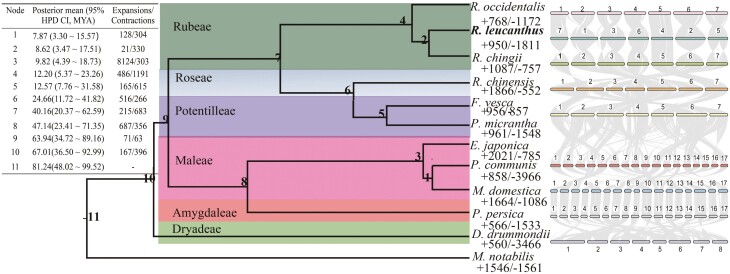
Phylogenetic analyses were conducted using 628 single-copy genes identified within those orthogroups. Both concatenation or coalescent methods resulted in phylogenetic trees with identical topologies, albeit with minor differences in bootstrap support values (Supplementary Fig. 6). The estimated divergence time between R. leucanthus and R. chingii was about 8.62 million years ago (Mya), with a 95% Highest Posterior Density Credible Interval (95% HPD CI) ranging from 3.47 to 17.51 Mya (Fig. 2, Supplementary Fig. 7). This divergence occurred after the separation of their common ancestor from the eastern North American native species R. occidentalis, which took place around 12.20 Mya, with a 95% HPD CI of 5.37 to 23.36 Mya. Additionally, the syntenic depth ratio of 1:1 between R. leucanthus and grape (Supplementary Fig. 8), suggested that R. leucanthus retains the ancient whole-genome triplication event that is shared by core eudicots.61
Figure 2.
Phylogenetic relationships among Rubus leucanthus and related species. The central diagram represents the maximum likelihood species tree, constructed using single copy nuclear genes. Adjacent to each tip taxon, significant gene family expansions and contractions are indicated. In the upper left, summaries of divergence times and significant gene family expansions and contractions for each node across the tree are provided. On the right, macro-syntenic blocks between closely related species are displayed.
In the genome of R leucanthus, compared to its ancestor, a total of 243 rapidly evolved gene families, encompassing 513 genes, were identified with a significance level of 0.05 (Fig. 2). Among these, 19 gene families, which include 202 genes, were significantly expanded, while 224 gene families, comprising 311 genes, underwent significant contraction. The expanded gene families showed significant enrichment in the flavonoid biosynthesis pathway (KO00941, FDR = 1.2E−6) and the plant circadian rhythm pathway (KO04712, FDR = 3.0E−6) (Supplementary Fig. 9). Flavonoids, a diverse group of plant secondary metabolites, play crucial roles in plant growth, development, and defense against environmental stresses.62 Additionally, studies have shown that flavonoid biosynthesis is upregulated in response to high temperature stress in some plant species.63 The circadian rhythm is vital for optimizing an organism’s adaptation to its environment, with the photoperiodic response being key to ensuring plants flower during favorable seasons.64 In areas experiencing high summer, plant species in lower latitudes tend to flower in response to shorter days to evade extreme heat.65 The expansion of these two KEGG pathways in R. leucanthus sheds light on its adaptive strategies for survival in subtropical regions, suggesting an evolution towards accumulating protective flavonoids and adjusting its photoperiodic response for enhanced fitness. For the contracted families, enrichment analysis highlighted pathways including plant–pathogen interaction (KO04626, FDR = 5.9E−5) and MAPK signaling (KO04016, FDR = 0.03), both crucial in defense mechanisms against pathogens.66 Interestingly, despite inhabiting warmer subtropical/tropical regions where biotic stresses are typically more intense, R. leucanthus has presented a contraction in these defense-related pathways compared to its ancestor. A plausible explanation could be that R. leucanthus may rely more on physical and chemical defenses, such as thorns, trichomes, and an increased production of flavonoids, rather than traditional pathogen-defense signaling pathways. This hypothesis aligns with the observed upregulation of flavonoid biosynthesis, suggesting a strategic shift in defense mechanisms to adapt to its environment.
3.3. Positively selected genes (PSGs) in R. leucanthus and associated species
In this study, we narrowed our focus on two species: R. leucanthus and R. chingii, both of which are native to subtropical/tropical China and considered to be more heat-tolerant compared to the deciduous temperate species R. occidentalis. By setting the common ancestor of the two thermophilic species as the foreground, a total of 40 genes were identified as being under positive selection (Supplementary Table 12). Although no significant enriched functional terms were found (Supplementary Table 13), some genes have been associated with adaptive traits pertinent to these heat-tolerant species. For instance, one positively selected gene, Rle1148, is homologous to the thioredoxin superfamily proteins found in Arabidopsis. These small proteins are ubiquitous across various organisms and are crucial for signal transduction during both pathogenic and abiotic stresses by mediating oxidative reactions.67 Plants in tropical climates encounter a broader range of challenges compared to those in temperate zones, including a diversity of biotic stresses, such as various pathogens, and abiotic stresses, like heat. Another gene exhibiting signs of positive selection is Rle8963, akin to photo-assimilate-responsive protein (PAR -1), which is part of a disease resistance-responsive protein family found in tomato and tabaco plants.68 Additionally, Rle17273, which is homologous to the gene encoding for Fatty acid/sphingolipid desaturase (AT3G61580), was under positive selection. These desaturases are key in multiple biological processes within plants, encompassing responses to environmental stress (high temperature, drought, and salt stress), growth and development regulation, and lipid metabolism.68 A number of PSGs in the common ancestor of R. leucanthus and R. chingii are associated with various transcription factors known for their critical roles in plant stress responses. This includes Rle2698, which encodes for the bHLH transcription factor PIF1 involved in heat stress response; Rle1705 and Rle1567, corresponding to genes for the transcription factors ATH1 and MYB44, respectively, both mediators in drought stress response; and Rle3214, coding for the NAC domain-containing protein NAC73, implicated in salt stress signaling. The positive selection observed on these transcription factor genes suggests active adaptation by the ancestor of R. leucanthus and R. chingii to novel environmental stresses, necessitating alterations in the regulatory networks that manage biotic and abiotic stress responses.
Out of the 4106 filtered gene families, the branch-site model test identified 486 positively selected genes (PSGs) in the genome of R. leucanthus, derived from a comparative analysis involving R. leucanthus and an additional 11 species (Supplementary Table 14). No significant KEGG or Gene ontology (GO) terms were identified after false discovery rate correction (Supplementary Table 15). Among the PSGs, 28 genes were homologous to 17 transcription factors in Arabidopsis, spanning a variety of families critical for regulatory pathways that are essential for adapting to tropical environments (Supplementary Table 16). These include TFs controlling plant growth and development such as ARF,69ERF,70MYB.71 Other among the positively selected TFs are implicated in hormone and light signaling pathways, such as GRAS,72GATA.73 Additionally, certain TFs that have specialized functions for tropical plants, like HD-ZIP58 and Trihelix,74 were also under positive selection. Stress-responsive TF families, such as WRKY75 and SBP,76 were targets of selection as well.
The positive selection on these diverse transcription factor families likely enabled the fine-tuning of key pathways to tailor R. leucanthus’s growth, physiology, and stress responses for adaptation to tropical environments. The extensive range of transcription factor families under selection in R. leucanthus underscores that adaptation involved incremental modifications across multiple regulatory pathways. For instance, alterations in transcription factors related to auxin, gibberellic acid, or circadian signaling might have adjusted developmental timing to suit tropical day lengths.77 Modifications in transcription factors associated with stomatal or root patterning could enhance growth processes tailored for tropical conditions.78,79 Lastly, advancements in defense signaling transcription factors provide a buffer against tropical pests and pathogens.80 The positively selected transcription factor families emphasize the role of regulatory pathway adjustments in fine-tuning R. leucanthus’s growth, reproduction, reproductive strategies, and overall survival in tropical climates. The collection of these positively selected transcription factors demonstrates a significant adaptation of R. leucanthus’s regulatory systems throughout its evolutionary history in tropical settings.
3.4. Identification of key genes involved in wax and cuticle biosynthesis in the R. leucanthus
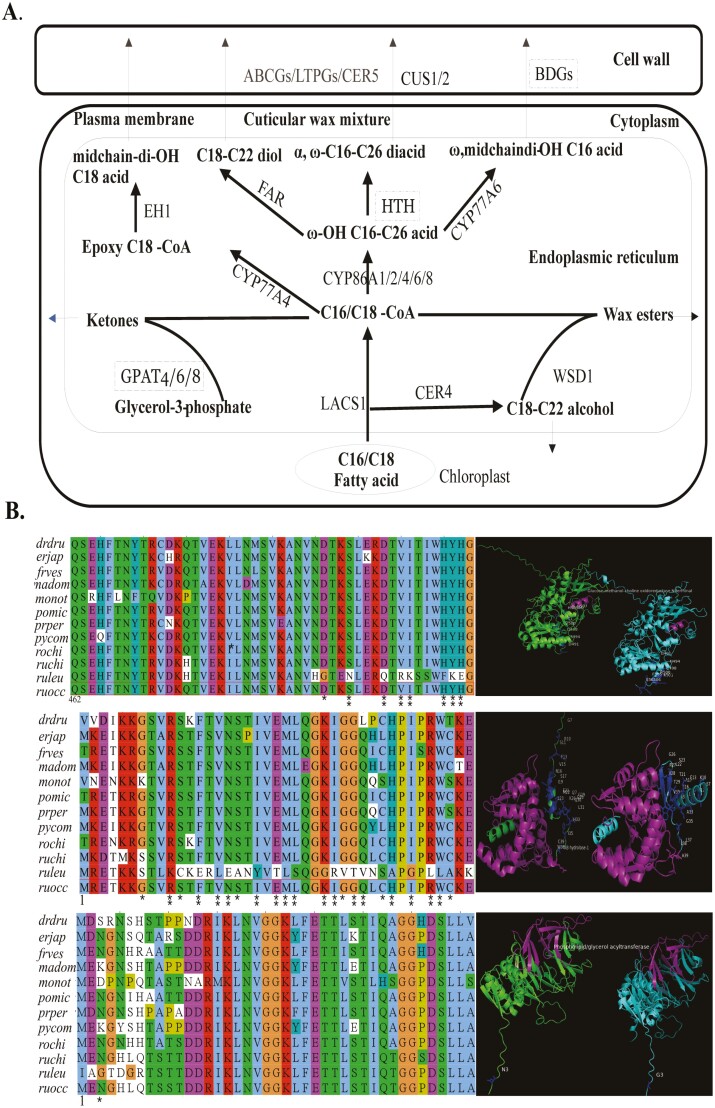
R. leucanthus is characterized by its leathery and glossy leaves, which often confer greater heat tolerance compared to its counterparts, R. chingii and R. occidentalis. The waxy cuticle on the leaves of R. leucanthus’s is likely a critical barrier, serving to protect against water loss and overheating. We analyzed 47 genes known to be involved in cutin and wax biosynthesis in model species such as Arabidopsis, rice, and tomato (Fig. 3, Supplementary Table 17).81,82 Through this analysis, we identified three PSGs in R. leucanthus: Rle10928, Rle12018, and Rle20301 (Table 2, Fig. 3, Supplementary Table 17). Rle10928 is homologous to BDG/BDG3 in Arabidopsis, which encode extracellular enzymes that synthesize cutin monomers (Fig. 3A, Fig. 3B).83 Overexpression of BDG increases total cutin contents by nearly four times in Arabidopsis.84Rle12018 and Rle6573 are orthologs to GPAT4/6/8 in Arabidopsis, known for their bifunctional acyl- transferase/phosphatase activity, which is essential for cutin biosynthesis (Fig. 3A and B).85,86 Although HOTHEAD (HTH) in Arabidopsis is assumed to catalyze cutin biosynthesis, its function in other species remains uncertain since dicarboxylic acids, which it uses as substrates, are less prevalent outside of Arabidopsis.81,83,87 The tertiary structure analysis showed that all mutations subjected to selection were located outside the respective domains (Glucose-methanol-choline oxidoreductase N-terminal for HOTHEAD, AB hydrolase-1 for BDG/BDG3, and Phospholipid/glycerol acyltransferase for GPAT4/6/8). This suggests conservation of function within these domains, despite significant changes in the 3D structure being observed (see Fig. 3B). Such structural alterations could contribute to their functional divergence. However, further experimental validation will be necessary to elucidate the detailed mechanisms underlying these observations.
Figure 3.
Putative biosynthesis, regulation, and key candidate genes for leaf cuticular wax in the species Rubus leucanthus. (A) Illustrations of the proposed biosynthesis of cuticular wax, with candidate regulatory genes under positive selection highlighted in dashed rectangle. (B) Left panel shown for alignments for the three genes under positive selection (top: Rle20301; middle: Rle10928; bottom: Rle12018) in R. leucanthus, with positively selected sites marked by single or double asterisks to indicate a Bayes Empirical Bayes test with posterior probabilities exceeding 95% and 98%, respectively. Species abbreviated as: ruleu: R. leucanthus; ruchi: Rubus chingii; ruocc: Rubus occidentalis; frves: Fragaria vesca; prper: Prunus persica; pycom: Pyrus communis; madom: Malus domestica; rochi: Rosa chinensis, drdru: Dryas drummondii; monot: Morus notabilis; pomic: Potentilla micrantha. The right panel illustrates the tertiary structure of the protein derived from consensus sequences and sequences under positive selection for R. leucanthus. Domains that have been identified are highlighted in purple, and residues under positive selection are marked accordingly.
Table 2.
Summaries of three positive selection genes involved in the wax-cuticle biosynthesis in Rubus leucanthus by branch-site model test
| Positive selection gene | Null model | Alternative model | lnLnull | lnLalt | 2ΔLnL | df | P -value | Positive selection sites |
|---|---|---|---|---|---|---|---|---|
| Rle10928 (BDG/BDG3) | p0 = 0.67, p1 = 0.08, p2a = 0.23, p2b = 0.02; b: ω0 = 0.06, ω1 = 1.00, ω2a = 0.06, ω2b = 1.00; f: ω0 = 0.06, ω1 = 1.00, ω2a = 1.00, ω2b = 1.00 |
p0 = 0.82, p1 = 0.10, p2a = 0.07, p2b = 0.01; b: ω0 = 0.06, ω1 = 1.00, ω2a = 0.06, ω2b = 1.00; f: ω0 = 0.06, ω1 = 1.00, ω2a = 261.64, ω2b = 261.64 |
−4528.38 | −4461.37 | 134.02 | 1 | 0 | 7S*, 10K**, 11C*, 13E**, 15L**, 16E**, 17A*, 19Y**, 21T**, 22L**, 23S**, 26G**, 27R**, 28V*, 29T**, 30V**, 31N*, 33A**, 35G**, 37L**, 38L**, 39A* |
| Rle12018(GPAT4/6/8) | p0 = 0.84, p1 = 0.16, p2a = 0.00, p2b = 0.00; b: ω0 = 0.10, ω1 = 1.00, ω2a = 0.10, ω2b = 1.00; f: ω0 = 0.10, ω1 = 1.00, ω2a = 0.10, ω2b = 1.00 |
p0 = 0.83, p1 = 0.15, p2a = 0.01, p2b = 0.01; b: ω0 = 0.10, ω1 = 1.00, ω2a = 0.10, ω2b = 1.00; f: ω0 = 0.10, ω1 = 1.00, ω2a = 672.50, ω2b = 672.50 |
−7146.67 | −7133.74 | 25.86 | 1 | 3.68E−07 | 3G* |
| Rle20301(HOTHEAD) | p0 = 0.57, p1 = 0.14, p2a = 0.23, p2b = 0.06; b: ω0 = 0.07, ω1 = 1.00, ω2a = 0.07, ω2b = 1.00; f: ω0 = 0.07, ω1 = 1.00, ω2a = 1.00, ω2b = 1.00 |
p0 = 0.78, p1 = 0.19, p2a = 0.02, p2b = 0.01; b: ω0 = 0.07, ω1 = 1.00, ω2a = 0.07, ω2b = 1.00; f: ω0 = 0.07, ω1 = 1.00, ω2a = 999.00, ω2b = 999.00 |
−6616.17 | −6590.67 | 51 | 1 | 9.24E−13 | 491G*, 494N**, 498Q**, 500R**, 501K**, 505E**, 506K**, 507E** |
f: foreground branch, b: background branches; site classes: 0—purifying selection in f & b 1—neutral evolution in f & b, 2a - purifying in b, positive selection in f, 2b - neutral in b, positive selection in f; p0, p1, p2a, p2b: proportions of sites in classes 0, 1, 2a, 2b; 2ΔlnL: likelihood ratio test statistic; *P > 95%, **P > 99%.
The distinct glossy and leathery appearance of R. leucanthus leaves is likely due to a combination of reduced epicuticular wax crystals and an increased cutin content in the cuticle.88 The identification of three PSGs involved in cuticle biosynthesis in R. leucanthus may indicate alterations in the regulatory networks that control wax and cutin production. These changes could be key in differentiating this species from R. chingii and R. occidentalis. Nonetheless, the exact functional implications of these genetic variations are not yet fully understood. To gain deeper insight into the possible regulatory roles of these genes, comprehensive chemical profiling of the cuticular lipids and cutin monomers in R. leucanthus is warranted. Furthermore, combining these with functional genetic studies would be invaluable in shedding light on how these potential regulators fine-tune wax and cutin metabolism. Such research could uncover the genetic and biochemical strategies that equip R. leucanthus for survival in high-temperature environments through adaptive modifications to the composition and structure of the leaf cuticle. In summary, the PSGs suggest that changes in cuticle biosynthesis contribute to the heat-adapted phenotype of R. leucanthus. However, to fully understand their role in adaptation, further chemical and genetic analysis is essential.
3.5. Gene repertoire comparison of HSP70/90 and heat shock transcription factor gene families among three Rubus species
Genome wide analyses identified 8, 41, and 33 canonical HSP70 genes in R. leucanthus, R. chingii, and R. occidentalis, respectively. Together with the 18 HSP70 genes in the Arabidopsis thaliana,89 a maximum likelihood phylogenetic tree was constructed for all identified HSP70 genes (Supplementary Fig. 10). Based on homology with Arabidopsis, the subcellular localizations for these proteins were identified as the cytoplasm, endoplasmic reticulum (ER), mitochondria, and plastids. The phylogenetic analysis highlighted that R. leucanthus is missing the largest subgroup of cytoplasmic HSP70s found in R. chingii and R. occidentalis. A collinearity analysis indicated that HSP70s in R. leucanthus were either singletons or dispersed, contrasting with the tandem duplications observed in R. chingii (Supplementary Table 18). Additionally, fewer canonical HSP90 and HSF genes were identified in R. leucanthus (6 HSP90s, 3 HSFs) and R. chingii (4 HSP90s, 5 HSFs), as compared to R. occidentalis (4 HSP90s, 16 HSFs). Phylogenetic classification revealed differences in the distribution of HSF classes among the species, documenting there were 2/0/1, 4/1/0, 13/3/0 members of classes A/B/C in the genomes of R. leucantus, R. chingii and R. occidentalis, respectively (Supplementary Fig. 11). Overall, R. leucanthus possessed fewer canonical heat response genes, especially among the cytoplasmic HSP70s. This suggests a reliance on alternative mechanisms for heat tolerance, highlighting how gene family sizes and compositions can vary dramatically across plant taxa in response to adaptive requirements.
Heat shock proteins (HSP70s/90s) function as molecular chaperones that stabilize proteins during heat stress and facilitate the release of HSFs, which in turn induce the expression of HSPs.90 The diversification of heat response elements in plants tends to reflect their adaptation to various thermal environments.91 For instance, Camelina saliva, which has a wide geographical distribution, possesses 108 HSFs, which may be linked to its capacity to adapt to diverse climates (https://planttfdb.gao-lab.org/family.php?fam=HSF). In contrast, our findings show that R. leucanthus has a more modest complement of canonical heat response genes (17 in total) compared to R. chingii (50) and R. occidentalis (53). This disparity suggests that R. leucanthus may have evolved alternative mechanisms to withstand heat stress. Notably, While R. leucanthus is native to subtropical China, R. chingii and R. occidentalis have more extensive distributions, inhabiting East China and Eastern North America, respectively. The larger heat gene repertories of these two species may be indicative of their broader adaptation to thermal variations.
However, adaptability to heat stress involves multiple facets and is not solely dictated by the number of heat response genes. R. leucanthus likely compensates for its fewer heat shock genes with other strategies, such as its glossy and waxy leaves, which could reduce heat stress and the risk of pathogen attacks. Intriguingly, our research also indicates that the tropical R. leucanthus has a smaller assortment of disease resistance genes (355), in comparison to the subtropical R. chingii (376) and the temperate R. occidentalis (449) (Supplementary Table 19). This finding supports the idea that R. leucanthus may rely on different mechanisms to manage the challenges posed by pathogens in subtropical environments, unlike R. chingii and R. occidentalis, which possess more extensive disease resistance gene repertoires.
3.6. Physiological response and transcriptome profile upon heat stress
Upon exposure to heat stress at 46°C, R. leucanthus demonstrated physiological responses indicative of stress activation. There was a notable increase in malondialdehyde (MDA) content, a marker of oxidative damage, from 60.61 ± 1.17 nmol/g under normal temperature to 63.32 ± 1.46 nmol/g at 46°C (t = 15.72, df = 2, P = 0.004). Concurrently, the activity of the antioxidant enzyme superoxide dismutase (SOD) showed a significant rise from 208.70 ± 2.21 U/g to 217.31 ± 4.92 U/g under heat stress conditions (t = 5.34, df = 2, P = 0.03). Furthermore, the level of proline, an amino acid associated with stress response, increased from 236.90 ± 2.85 mg/kg at ambient temperatures to 306.21 ± 4.03 mg/kg at the elevated temperature of 46°C (t = 35.84, df = 2, P = 0.001). These marked increases in MDA content, SOD activity, and proline levels in R. leucanthus upon exposure to heat stress suggest the activation of various stress responses, including oxidative, antioxidant, and osmotic stress mechanisms.
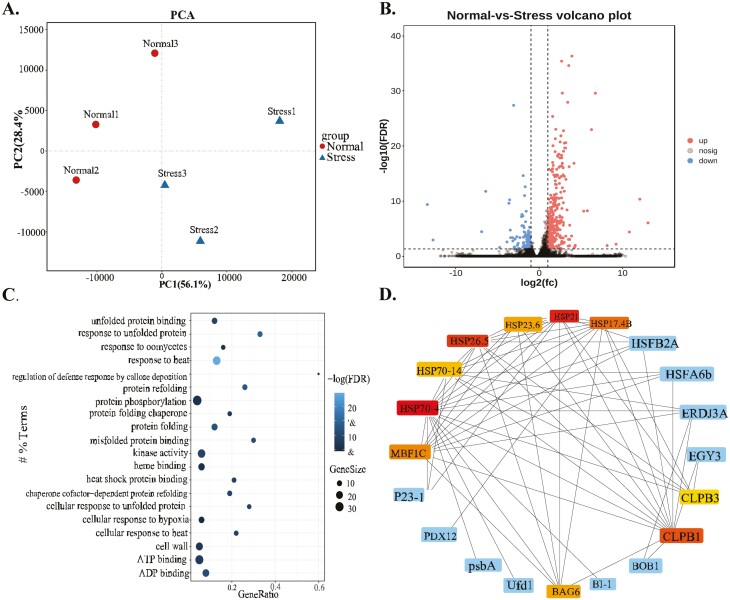
Transcriptomic analysis revealed 432 differentially expressed genes (DEGs) in R. leucanthus when comparing conditions of 28°C to 46°C. Of these, 312 genes were up-regulated, and 120 were down- regulated (Fig. 4a and b). GO terms enrichment analysis identified 32 significantly enriched terms among the DEGs after false discovery rate (FDR) correction (Fig. 4c). The enriched GO terms highlight transcriptional changes in response to elevated temperatures, including activation of heat shock pathways (‘response to heat’ [GO:0009408], ‘cellular response to heat’[GO:0034605]), protein stability (‘response to unfolded protein’ [GO:0006986], ‘protein refolding’ [GO:0042026]), defense against pathogens (‘response to oomycetes’[GO:0002239], ‘defense response to bacterium’ [GO:0009816], ‘defense response to fungus’ [GO:0050832]), redox regulation (‘monooxygenase activity’ [GO:0004497], ‘oxidoreductase activity’ [GO:0016705]), signal transduction (‘ATP binding’ [GO:0005524], ‘protein phosphorylation’ [GO:0006468], ‘kinase activity’ [GO:0016301]), and reactions to cadmium signaling (‘response to cadmium ion’ [GO:0046686]). In essence, R. leucanthus exhibits widespread transcriptional modulation in response to acute heat stress, with a notable upregulation of genes associated with heat shock responses, protein maintenance, pathogen resistance, and redox balance. Further investigations are needed to clarity the precise functions and interactions of these DEGs in the enriched pathways as they relate to temperature adaptation.
Figure 4.
Transcriptome profile upon heat stress for Rubus leucanthus. A samples group clustering by principal component analysis. (B) Vocanol plot shown for magnitutude and significance of the differential expression between normal and stress conditions. (C) Top 20 significantly enriched gene ontology terms for differentially expressed genes between normal and stress conditions, and the size and heatmap intensity of each entry representing the number of genes and false discovery rate-adjusted P-value, respectively. (D) Identification of hub genes using protein–protein interaction network among differential expression genes upon heat stress for R. leucanthus.
Gene expression analysis identified 5 intact HSP70 genes (Rle21724, Rle23870, Rle11632, Rle24327, Rle23760) and 4 HSP90 genes (Rle20121, Rle9016, Rle18054, Rle8017) that were significantly upregulated in R. leucanthus under heat stress conditions, along with the heat shock transcription factor HSF1 (Rle18137) (Supplementary Table 20). To delve into the interactions and significance of these heat shock proteins, a protein–protein interaction (PPI) network was constructed using the STRING database. Subsequent topological analysis of this network through the cytoHubba plugin within Cytoscape identified several heat shock proteins as central hub genes, based on their maximal clique centrality scores. Remarkably, the truncated HSP70-4 gene (Rle23898) was identified as the principal hub gene, underscoring its pivotal role in managing the heat shock response network. In contrast, the intact HSP70 and HSP90 genes, despite being upregulated, were not identified as hub genes. Additional notable genes in the network include small heat shock proteins such as HSP21 (MSTRG.21029), HSP26.5 (Rle12860), HSP101 (Rle6051), HSP17.4B (Rle13354), along with various co-chaperones, anti-apoptotic genes, and protein degradation factors (Fig. 4d). The network analysis highlights HSP70-4 as a critically important hub gene connecting the broader heat shock response, while the intact HSP70/90 genes may have more peripheral, specialized roles despite their induction. These results demonstrated the dispensable roles for the HSP70/90s in heat stress response, also implicating other alternative strategy for R. leucanthus.
4. Conclusion
In this study, we provided a high-quality genome assembly and annotation for R. leucanthus. Through comparative genomic analysis, we explored evolutionary adaptations that might enable the species to withstand hot climates. Contrary to expectations-where one might predict an enlargement of gene families related to heat shock responses, like the HSP70/90s and HSFs-our research reveals a different scenario. We identified three genes linked to cuticle and wax biosynthesis that exhibit signs of positive selection. These genes could contribute to the development of glossy and wax leaves, offering a distinctive adaptation to environment stressors.
Plants have evolved various strategies to cope with heat stress. For instance, they developed thermotolerance through heat shock proteins and antioxidant enzymes.92 They utilize osmo-protectants like proline and trehalose to preserve cellular hydration and integrity. Additionally, they can modulate photosynthesis to reduce the production of heat-sensitive compounds. Tropical plants often display an array of morphological features to handle high temperatures and humidity, such as succulent stems for enduring heat93; thick bark for heat protection,94 and large leaves for optimal photosynthesis while minimizing heat stress.95
Our study found that R. leucanthus developed thick, waxy leaves which reduce water loss and reflect excess solar radiation, contrasting with the thinner, duller leaves of R. occidentalis and R. chingii. This dense waxy coating could also protect against pathogens and herbivores,96 indicated by the fewer resistance (R) genes compared to temperate relatives. This suggests that R. leucanthus relies more on physical defenses like cuticle wax rather than on chemical defenses mediated by R proteins. The leathery, waxy leaves provided dual advantage of conserving water and protecting against biotic stress, crucial for adaptation to hot environments and could offer insights into R. leucanthus’ survival in hot, moist habitats with a compact genome.
Three PSGs identified in this study, associated with cuticle wax biosynthesis, may play a role in heat tolerance in R. leucanthus. Future experiments to functionally validate these genes are crucial to confirm their adaptive roles and to evaluate their potential in breeding heat-tolerant Rubus raspberry cultivars. Specifically, transgenic techniques that overexpressing these wax biosynthetic genes in cold-tolerant Rubus species may determine whether they enhance leaf integrity and water retention under high temperatures. Understanding the genetics behind the glossy, wax leaves of R. leucanthus provides potential targets for introducing beneficial traits into crop breeding programs aimed at hot, arid regions.
Supplementary Material
Acknowledgements
We appreciate Mr Yubing Zhou for sample preparation and sequencing, and we also thank Dr Shaohua Xu for the use of pyMOL package. Special thanks are also given to the reviewers and editors for their efforts in improving our manuscript.
Contributor Information
Wei Wu, College of Horticulture and Landscape Architecture, Zhongkai University of Agriculture and Engineering, Guangzhou, 510225, Guangdong, China.
Longyuan Wang, College of Horticulture and Landscape Architecture, Zhongkai University of Agriculture and Engineering, Guangzhou, 510225, Guangdong, China.
Weicheng Huang, Plant Science Center, South China Botanical Garden, Chinese Academy of Science, , Guangzhou, 510650, Guangzhou, China.
Xianzhi Zhang, College of Horticulture and Landscape Architecture, Zhongkai University of Agriculture and Engineering, Guangzhou, 510225, Guangdong, China.
Yongquan Li, College of Horticulture and Landscape Architecture, Zhongkai University of Agriculture and Engineering, Guangzhou, 510225, Guangdong, China.
Wei Guo, College of Horticulture and Landscape Architecture, Zhongkai University of Agriculture and Engineering, Guangzhou, 510225, Guangdong, China.
Conflict of interest
The authors declare no conflict of interest.
Funding
This study was supported by Science and Technology Program from Forestry Administration of Guangdong Province (2021-KJCX015), Guangdong Province University Innovative Team Project: Innovation and Development Application of Ornamental Plant Germplasm with Lingnan Characteristics (2023KCXTD017).
Data availability
The raw data from the whole-genome sequencing and RNA sequencing have been deposited in the Sequence Read Archive (SRA) with SRA accession numbers: SRR29481249 to SRR29481260, under the project PRJNA1125881 at the National Center for Biotechnology Information (NCBI). The genome assemblies and annotations have been deposited at figshare with DOI: 10.6084/m9.figshare.24882360.
References
- 1. Grace, J. 1987, Climatic tolerance and the distribution of plants, New Phytol., 106, 113–30. [Google Scholar]
- 2. Driedonks, N., Wolters-Arts, M., Huber, H., et al. 2018, Exploring the natural variation for reproductive thermotolerance in wild tomato species, Euphytica., 214, 67. [Google Scholar]
- 3. Moyers, B.T., Morrell, P.L., and McKay, J.K.. 2017, Genetic costs of domestication and improvement, J. Hered., 109, 103–16. [DOI] [PubMed] [Google Scholar]
- 4. Casal, J.J. and Balasubramanian, S.. 2019, Thermomorphogenesis, Annu. Rev. Plant Biol., 70, 321–46. [DOI] [PubMed] [Google Scholar]
- 5. Ludwig, W., Hayes, S., Trenner, J., Delker, C., and Quint, M.. 2021, On the evolution of plant thermomorphogenesis, J. Exp. Bot., 72, 7345–58. [DOI] [PubMed] [Google Scholar]
- 6. Perrella, G., Bäurle, I., and van Zanten, M.. 2022, Epigenetic regulation of thermomorphogenesis and heat stress tolerance, New Phytol., 234, 1144–60. [DOI] [PubMed] [Google Scholar]
- 7. Larkindale, J., Hall, J.D., Knight, M.R., and Vierling, E.J.P.P.. 2005, Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance, Plant Physiol., 138, 882–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bokszczanin, K., Fragkostefanakis, S., Bostan, H., et al. 2013, Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance, Front. Plant Sci., 4, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yeh, C.-H., Kaplinsky, N.J., Hu, C., and Charng, Y.-Y.J.P.S.. 2012, Some like it hot, some like it warm: phenotyping to explore thermotolerance diversity, Plant Sci., 195, 10–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou, D., Wang, X., Wang, X., and Mao, T.J.T.P.C.. 2023, PHYTOCHROME INTERACTING FACTOR 4 regulates microtubule organization to mediate high temperature-induced hypocotyl elongation in Arabidopsis, Plant Cell, 35, 2044–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kan, Y., Mu, X.-R., Zhang, H., et al. 2022, TT2 controls rice thermotolerance through SCT1-dependent alteration of wax biosynthesis, Nat. Plants, 8, 53–67. [DOI] [PubMed] [Google Scholar]
- 12. Thompson, M.M. 1995, Chromosome numbers of Rubus species at the national clonal germplasm, HortScience, 30, 1447–52. [Google Scholar]
- 13. Wang, Y., Chen, Q., Chen, T., Tang, H., Liu, L., and Wang, X.. 2016, Phylogenetic insights into Chinese Rubus (Rosaceae) from multiple chloroplast and nuclear DNAs, Front. Plant Sci., 7, 1– 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu, L. and Boufford, D.. 2003, Rosaceae. In: Wu, Z. and Raven, P. H. (eds), Flora of China, Science Press \& Missouri Botanical Garden Press: Beijing & St. Louis, pp. 195–285. [Google Scholar]
- 15. Vurture, G.W., Sedlazeck, F.J., Nattestad, M., et al. 2017, GenomeScope: fast reference-free genome profiling from short reads, Bioinformatics, 33, 2202–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Louwers, M., Splinter, E., van Driel, R., de Laat, W., and Stam, M.. 2009, Studying physical chromatin interactions in plants using Chromosome Conformation Capture (3C), Nat. Protoc., 4, 1216–29. [DOI] [PubMed] [Google Scholar]
- 17. Chin, C.-S., Peluso, P., Sedlazeck, F.J., et al. 2016, Phased diploid genome assembly with single-molecule real-time sequencing, Nat. Methods., 13, 1050–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chaisson, M.J. and Tesler, G.. 2012, Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory, BMC Bioinf., 13, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roach, M.J., Schmidt, S.A., and Borneman, A.R.. 2018, Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies, BMC Bioinf., 19, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Servant, N., Varoquaux, N., Lajoie, B.R., et al. 2015, HiC-Pro: an optimized and flexible pipeline for Hi-C data processing, Genome Biol., 16, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Durand, N.C., Robinson, J.T., Shamim, M.S., et al. 2016, Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom, Cell Syst., 3, 99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G.. 2013, QUAST: quality assessment tool for genome assemblies, Bioinformatics, 29, 1072–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Manni, M., Berkeley, M.R., Seppey, M., Simão, F.A., and Zdobnov, E.M.. 2021, BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral Genomes, Mol. Biol. Evol., 38, 4647–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smit, A.F.A. and Hubley, R.. 2015, RepeatModeler Open-1.0. RepeatModeler Open-1.0. https://www.repeatmasker.org/RepeatModeler [Google Scholar]
- 25. Smit, A.F.A., Hubley, R. and Green, P.. 2013, RepeatMasker Open-4.0. https://repeatmasker.org [Google Scholar]
- 26. Nawrocki, E.P. and Eddy, S.R.. 2013, Infernal 1.1: 100-fold faster RNA homology searches, Bioinformatics, 29, 2933–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chan, P. P. and Lowe, T. M.. 2019, tRNAscan-SE: searching for tRNA genes in genomic sequences. In: Kollmar, M. (ed), Gene Prediction: Methods and Protocols, Springer New York: NY, pp. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Campbell, M.S., Holt, C., Moore, B., and Yandell, M.. 2014, genome annotation and curation using MAKER and MAKER-P, Curr. Protoc.Bioinformatics, 48, 4.11.11–14.11.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stanke, M., Diekhans, M., Baertsch, R., and Haussler, D.. 2008, Using native and syntenically mapped cDNA alignments to improve de novo gene finding, Bioinformatics, 24, 637–44. [DOI] [PubMed] [Google Scholar]
- 30. Korf, I. 2004, Gene finding in novel genomes, BMC Bioinf., 5, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haas, B.J., Salzberg, S.L., Zhu, W., et al. 2008, Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments, Genome Biol., 9, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jones, P., Binns, D., Chang, H.-Y., et al. 2014, InterProScan 5: genome-scale protein function classification, Bioinformatics, 30, 1236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xie, C., Mao, X., Huang, J., et al. 2011, KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases, Nucleic Acids Res., 39, W316–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ou, S., Su, W., Liao, Y., et al. 2019, Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline, Genome Biol., 20, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang, Y., Tang, H., DeBarry, J.D., et al. 2012, MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity, Nucleic Acids Res., 40, e49–e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emms, D.M. and Kelly, S.. 2015, OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy, Genome Biol., 16, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mirarab, S. and Warnow, T.. 2015, ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes, Bioinformatics, 31, i44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Katoh, K. and Standley, D.M.. 2013, MAFFT multiple sequence alignment software Version 7: improvements in performance and usability, Mol. Biol. Evol., 30, 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suyama, M., Torrents, D., and Bork, P.. 2006, PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments, Nucleic Acids Res., 34, W609–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Darriba, D., Taboada, G.L., Doallo, R., and Posada, D.. 2012, jModelTest 2: more models, new heuristics and parallel computing, Nat. Methods., 9, 772–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stamatakis, A. 2014, RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies, Bioinformatics, 30, 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang, Z. 2007, PAML 4: Phylogenetic analysis by maximum likelihood, Mol. Biol. Evol., 24, 1586–91. [DOI] [PubMed] [Google Scholar]
- 43. Kumar, S., Stecher, G., Suleski, M., and Hedges, S.B.. 2017, TimeTree: a resource for timelines, timetrees, and divergence Times, Mol. Biol. Evol., 34, 1812–9. [DOI] [PubMed] [Google Scholar]
- 44. Mendes, F.K., Vanderpool, D., Fulton, B., and Hahn, M.W.. 2021, CAFE 5 models variation in evolutionary rates among gene families, Bioinformatics, 36, 5516–8. [DOI] [PubMed] [Google Scholar]
- 45. Macías, L.G., Barrio, E., and Toft, C.. 2020, GWideCodeML: a Python package for testing evolutionary hypotheses at the genome-wide level, G3 (Bethesda, Md.), 10, 4369–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Abramson, J., Adler, J., Dunger, J., et al. 2024, Accurate structure prediction of biomolecular interactions with AlphaFold 3, Nature, 630, 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schrodinger, LLC 2015, The PyMOL Molecular Graphics System, Version 1.8.
- 48. Toda, N., Rustenholz, C., Baud, A., et al. 2020, NLGenomeSweeper: a tool for genome-wide NBS-LRR resistance gene edentification, Genes, 11, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim, D., Langmead, B., and Salzberg, S.L.. 2015, HISAT: a fast spliced aligner with low memory requirements, Nat. Methods, 12, 357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li, B. and Dewey, C.N.. 2011, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome, BMC Bioinf., 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Love, M.I., Huber, W., and Anders, S.. 2014, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chin, C.-H., Chen, S.-H., Wu, H.-H., Ho, C.-W., Ko, M.-T., and Lin, C.-Y.. 2014, cytoHubba: identifying hub objects and sub-networks from complex interactome, BMC Syst. Biol., 8, S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang, L., Lei, T., Han, G., et al. 2021, The chromosome-scale reference genome of Rubus chingii Hu provides insight into the biosynthetic pathway of hydrolyzable tannins, Plant J., 107, 1466–77. [DOI] [PubMed] [Google Scholar]
- 54. Ou, S., Chen, J., and Jiang, N.. 2018, Assessing genome assembly quality using the LTR assembly endex (LAI), Nucleic Acids Res., 46, e126–e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yandell, M. and Ence, D.. 2012, A beginner’s guide to eukaryotic genome annotation, Nat. Rev. Genet., 13, 329–42. [DOI] [PubMed] [Google Scholar]
- 56. Kalvari, I., Argasinska, J., Quinones-Olvera, N., et al. 2017, Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families, Nucleic Acids Res., 46, D335–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Trovato, M., Funck, D., Forlani, G., Okumoto, S., and Amir, R.. 2021, Editorial: amino acids in plants: regulation and functions in development and stress defense, Front. Plant Sci., 12, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ariel, F.D., Manavella, P.A., Dezar, C.A., and Chan, R.L.. 2007, The true story of the HD-Zip family, Trends Plant Sci., 12, 419–26. [DOI] [PubMed] [Google Scholar]
- 59. Nuruzzaman, M., Sharoni, A.M., and Kikuchi, S.. 2013, Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants, Front. Microbiol., 4, 1– 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Waseem, M., Nkurikiyimfura, O., Niyitanga, S., Jakada, B.H., Shaheen, I., and Aslam, M.M.. 2022, GRAS transcription factors emerging regulator in plants growth, development, and multiple stresses, Mol. Biol. Rep., 49, 9673–85. [DOI] [PubMed] [Google Scholar]
- 61. Jiao, Y., Leebens-Mack, J., Ayyampalayam, S., et al. 2012, A genome triplication associated with early diversification of the core eudicots, Genome Biol., 13, R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Khalid, M., Bilal, M., and Huang, D.-F.J.J.O.I.A.. 2019, Role of flavonoids in plant interactions with the environment and against human pathogens—a review, J Integr. Agric., 18, 211–30. [Google Scholar]
- 63. Jan, R., Kim, N., Lee, S.-H., et al. 2021, Enhanced flavonoid accumulation reduces combined salt and heat stress through regulation of transcriptional and hormonal mechanisms, Front. Plant Sci., 12, 796956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pruneda-Paz, J.L. and Kay, S.A.. 2010, An expanding universe of circadian networks in higher plants, Trends Plant Sci., 15, 259–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gil, K.-E. and Park, C.-M.. 2019, Thermal adaptation and plasticity of the plant circadian clock, New Phytol., 221, 1215–29. [DOI] [PubMed] [Google Scholar]
- 66. Dodds, P.N. and Rathjen, J.P.. 2010, Plant immunity: towards an integrated view of plant–pathogen interactions, Nat. Rev. Genet., 11, 539–48. [DOI] [PubMed] [Google Scholar]
- 67. Kneeshaw, S., Gelineau, S., Tada, Y., Loake, G.J., and Spoel, S.H.. 2014, Selective protein denitrosylation activity of thioredoxin-h5 modulates plant immunity, Mol. Cell, 56, 153–62. [DOI] [PubMed] [Google Scholar]
- 68. Etalo, D.W., Stulemeijer, I.J.E., Peter van Esse, H., de Vos, R.C.H., Bouwmeester, H.J., and Joosten, M.H.A.J.. 2013, System-wide hypersensitive response-associated Ttranscriptome and metabolome reprogramming in tomato, Plant Physiol., 162, 1599–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Finet, C., Berne-Dedieu, A., Scutt, C.P., and Marlétaz, F.. 2012, Evolution of the ARF gene family in land plants: old domains, new tricks, Mol. Biol. Evol., 30, 45–56. [DOI] [PubMed] [Google Scholar]
- 70. Xie, Z., Nolan, T.M., Jiang, H., and Yin, Y.. 2019, AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis, Front. Plant Sci., 10, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L.. 2010, MYB transcription factors in Arabidopsis, Trends Plant Sci., 15, 573–81. [DOI] [PubMed] [Google Scholar]
- 72. Jaiswal, V., Kakkar, M., Kumari, P., Zinta, G., Gahlaut, V., and Kumar, S.. 2022, Multifaceted roles of GRAS transcription factors in growth and stress responses in plants, iScience, 25, 105026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. An, Y., Zhou, Y., Han, X., et al. 2019, The GATA transcription factor GNC plays an important role in photosynthesis and growth in poplar, J. Exp. Bot., 71, 1969–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kaplan-Levy, R.N., Brewer, P.B., Quon, T., and Smyth, D.R.. 2012, The trihelix family of transcription factors --light, stress and development, Trends Plant Sci., 17, 163–71. [DOI] [PubMed] [Google Scholar]
- 75. Rushton, P.J., Somssich, I.E., Ringler, P., and Shen, Q.J.. 2010, WRKY transcription factors, Trends Plant Sci., 15, 247–58. [DOI] [PubMed] [Google Scholar]
- 76. Zhang, D., Han, Z., Li, J., et al. 2020, Genome-wide analysis of the SBP-box gene family transcription factors and their responses to abiotic stresses in tea (Camellia sinensis), Genomics, 112, 2194–202. [DOI] [PubMed] [Google Scholar]
- 77. Claeys, H., De Bodt, S., and Inzé, D.. 2014, Gibberellins and DELLAs: central nodes in growth regulatory networks, Trends Plant Sci., 19, 231–9. [DOI] [PubMed] [Google Scholar]
- 78. Robbins, N.E., II and Dinneny, J.R.. 2015, The divining root: moisture-driven responses of roots at the micro- and macro-scale, J. Exp. Bot., 66, 2145–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pillitteri, L.J. and Torii, K.U.. 2012, Mechanisms of stomatal development, Annu. Rev. Plant Biol., 63, 591–614. [DOI] [PubMed] [Google Scholar]
- 80. Birkenbihl, R.P., Liu, S., and Somssich, I.E.. 2017, Transcriptional events defining plant immune responses, Curr. Opin Plant Biol., 38, 1–9. [DOI] [PubMed] [Google Scholar]
- 81. Yeats, T.H. and Rose, J.K.C.. 2013, The formation and function of plant cuticles, Plant Physiol., 163, 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fich, E.A., Segerson, N.A., and Rose, J.K.C.. 2016, The plant polyester cutin: biosynthesis, structure, and biological roles, Annu. Rev. Plant Biol., 67, 207–33. [DOI] [PubMed] [Google Scholar]
- 83. Kurdyukov, S., Faust, A., Nawrath, C., et al. 2006, The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis, Plant Cell, 18, 321–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jakobson, L., Lindgren, L.O., Verdier, G., et al. 2016, BODYGUARD is required for the biosynthesis of cutin in Arabidopsis, New Phytol., 211, 614–26. [DOI] [PubMed] [Google Scholar]
- 85. Li-Beisson, Y., Pollard, M., Sauveplane, V., Pinot, F., Ohlrogge, J., and Beisson, F.. 2009, Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester, Proc. Natl. Acad. Sci. U.S.A., 106, 22008–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Li, Y., Beisson, F., Koo, A.J.K., Molina, I., Pollard, M., and Ohlrogge, J.. 2007, Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers, Proc. Natl. Acad. Sci. USA, 104, 18339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Serra, O. and Geldner, N.. 2022, The making of suberin, New Phytol., 235, 848–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Koch, K. and Ensikat, H.-J.. 2008, The hydrophobic coatings of plant surfaces: Epicuticular wax crystals and their morphologies, crystallinity and molecular self-assembly, Micron, 39, 759–72. [DOI] [PubMed] [Google Scholar]
- 89. Lin, B.-L., Wang, J.-S., Liu, H.-C., et al. 2001, Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana, Cell Stress Chaperones, 6, 201–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jacob, P., Hirt, H., and Bendahmane, A.. 2017, The heat-shock protein/chaperone network and multiple stress resistance, Plant Biotechnol. J., 15, 405–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Baniwal, S.K., Bharti, K., Chan, K.Y., et al. 2004, Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors, J. Biosci., 29, 471–87. [DOI] [PubMed] [Google Scholar]
- 92. Mittler, R. 2002, Oxidative stress, antioxidants and stress tolerance, Trends Plant Sci., 7, 405–10. [DOI] [PubMed] [Google Scholar]
- 93. Ogburn, R.M. and Edwards, E.J.. 2010, The ecological water-use strategies of succulent plants, Adv. Bot. Res., 55, 179–225. [Google Scholar]
- 94. Rosell, J.A. 2016, Bark thickness across the angiosperms: more than just fire, New Phytol., 211, 90–102. [DOI] [PubMed] [Google Scholar]
- 95. Valladares, F., Wright, S.J., Lasso, E., Kitajima, K., and Pearcy, R.W.. 2000, Plastic phenotypic response to light of 16 congeneric shrubs from a panamanian rainforest, Ecology, 81, 1925–36. [Google Scholar]
- 96. Lewandowska, M., Keyl, A., and Feussner, I.. 2020, Wax biosynthesis in response to danger: its regulation upon abiotic and biotic stress, New Phytol., 227, 698–713. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data from the whole-genome sequencing and RNA sequencing have been deposited in the Sequence Read Archive (SRA) with SRA accession numbers: SRR29481249 to SRR29481260, under the project PRJNA1125881 at the National Center for Biotechnology Information (NCBI). The genome assemblies and annotations have been deposited at figshare with DOI: 10.6084/m9.figshare.24882360.