Abstract
The high-affinity IgE receptor FcεRI plays a key role in triggering allergic reactions. We recently reported that human FcεRI β-chain gene expression was down-regulated by a transcription factor, MZF-1, through an element in the fourth intron. In the present study, we found that this transcriptional repression by MZF-1 required FHL3 (four and a half LIM domain protein 3) as a cofactor. Yeast two-hybrid and immunoprecipitation assays demonstrated that FHL3 bound MZF-1 in vitro and in vivo. Overexpression of FHL3 in KU812 cells suppressed the β-chain promoter activity through the element in the fourth intron in an MZF-1-dependent manner. Furthermore, results from pull-down assays and gel-filtration chromatography employing nuclear extracts indicated that MZF-1 and FHL3 formed a complex of high molecular mass with some additional proteins in the nucleus. Granulocyte–macrophage colony-stimulating factor, which was reported to decrease FcεRI expression, induced the accumulation of FHL3 in the nucleus, in accordance with the repressive role of FHL3 in β-chain gene expression.
Keywords: allergy, cofactor, FcεRIβ, four and a half LIM domain protein 3 (FHL3), gene expression, MZF-1
Abbreviations: AD, activation domain; Ade, adenine; BD, DNA-binding domain; CREB, cAMP-response-element-binding protein; EMSA, electrophoretic mobility-shift assay; FHL, four and a half LIM domain protein; GM-CSF, granulocyte-macrophage colony-stimulating factor; HA, haemagglutinin; 6HN, 6×His-Asn; IL-3, interleukin 3; RT, reverse transcriptase; SD medium, minimal synthetic dropout medium
INTRODUCTION
The high-affinity receptor for IgE, FcεRI, plays a central role in triggering the IgE-mediated allergic reaction. FcεRI is expressed in limited types of cells such as mast cells, basophils, eosinophils [1], monocytes [2], Langerhans cells [3,4], platelets [5,6] and neutrophils [7]. Cross-linking of FcεRI on effector cells by antigen (allergen)–IgE complexes induces not only the release of chemical mediators in the early-phase reaction but also cytokine gene expressions, leading to the late-phase reaction.
FcεRI is composed of three different subunits, α, β and γ, of which the α-chain directly binds IgE, while the β- and γ-chains are responsible for mediating intracellular signals. Although a functional receptor is expressed both as a tetramer (αβγ2) and a trimer (αγ2) in humans [8], intracellular signals [9,10] in addition to cell-surface expression [11] have been reported to be amplified significantly by the β-chain, indicating that the β-chain increases the sensitivity of cell activation to stimulation by allergens. Therefore functional inhibition of the β-chain is a reasonable strategy to suppress allergic reaction by raising the threshold of FcεRI-mediated cell activation by decreasing both the intracellular signals and the cell-surface FcεRI expression. Among many methods of inhibiting the function of the β-chain, repression of the β-chain gene transcription would be an effective solution, because the β-chain expression is limited to specific types of cells expressing FcεRI.
The genomic structure of the human FcεRI β-chain gene has already been determined [12]; however, only a few analyses were performed on regulatory mechanisms of the β-chain gene transcription. At first, our group reported that two Oct-1-binding sites in the 5′-untranslated region were essential for the activation of the β-chain gene promoter [13]. Recently, we also reported that a transcription factor, MZF-1, repressed the β-chain gene expression through an element in the fourth intron [14]. Since it was suggested that cofactors were required for the transcriptional suppression by MZF-1 [14], we searched a human cDNA library for them by the yeast two-hybrid assay. Out of the identified proteins interacting with MZF-1 in the assay, FHL3 (four and a half LIM domain protein 3), was found to act as a repressive cofactor in the MZF-1-mediated transcriptional regulation of the β-chain gene.
EXPERIMENTAL
Plasmid construction
Human MZF-1 cDNA, obtained as described previously [14], was inserted into pGBKT7 (BD Biosciences Clontech, Palo Alto, CA, U.S.A.) harbouring a GAL4 BD (DNA-binding domain) and a c-Myc tag at the SmaI site to produce pGBKT7-MZF1. Deletion mutants of MZF-1 and FHL3 were constructed with restriction endonuclease digestion and PCR technique. A DNA fragment encoding the full-length human FHL3 obtained by PCR using pACT2-FHL3 as a template was inserted into pCR3.1 (Invitrogen, Carlsbad, CA, U.S.A.) to yield pCR3.1-FHL3. Human FHL1 and FHL2 cDNAs were obtained by RT (reverse transcriptase)–PCR and cloned into pCR3.1 to yield pCR3.1-FHL1 and pCR3.1-FHL2 respectively. The nucleotide sequences of synthetic oligonucleotides used as PCR primers are as follows [15,16]: FHL1, 5′-CATGGCGGAGAAGTTTGACTGCCAC-3′ and 5′-GTTTACAGCTTTTTGGCACAGTCGG-3′; FHL2, 5′-AATGACTGAGCGCTTTGACTGCCACC-3′ and 5′-GTGTTGAATTCAGATGTCTTTCCCAC-3′.
A double-stranded synthetic oligonucleotide prepared by the annealing of 5′-CTAGCATGAGACATAATCATAATCATAATCATAATCATAATCACA-3′ and 5′-AGCTTGTGATTATGATTATGATTATGATTATGATTATGTCTCATG-3′ was introduced into pCR3.1-FHL3 at the NheI–HindIII site to construct pCR3.1-6HN-FHL3 (where 6HN stands for 6×His-Asn). A DNA fragment encoding BD-c-Myc-MZF1 prepared from pGBKT7-MZF1 was inserted into pCR3.1 and the resulting plasmid was named pCR3.1-BD-c-Myc-MZF1.
Yeast two-hybrid assay
A Matchmaker Gal4 two-hybrid system 3 (Clontech) was used according to the manufacturer's instructions. Yeast AH109 cells were sequentially transformed with pGBKT7-MZF1 and with a human lymph node cDNA library in a pACT2 vector (Clontech) containing a GAL4 AD (activation domain) and an HA (haemagglutinin) tag. Co-transformed cells grew on an SD −Trp/−Leu medium (minimal synthetic dropout medium lacking tryptophan and leucine), because pGBKT7 carried a TRP1 gene allowing the cells to grow without Trp and pACT2 carried an LEU2 gene allowing the cells to grow without Leu. Protein interaction in yeast AH109 cells was detected by the expression of three reporter genes (HIS3, ADE2 and MEL1) under the control of distinct GAL4 upstream activating sequences and TATA boxes to decrease false positives. After selecting the clones expressing HIS3 on SD −Trp/−Leu/−His plates, growing colonies were transferred on to SD −Trp/−Leu/−His/−Ade (adenine)/+X-α-Gal plates (where X-α-Gal stands for 5-bromo-4-chloro-3-indoyl-α-D-galactopyranoside) to select those expressing the other reporter genes of ADE2 and MEL1. pACT2 plasmids carrying cDNA inserts were rescued from the positive clones expressing all three reporter genes of HIS3, ADE2 and MEL1. After sorting the cDNA inserts by AluI digestion, their nucleotide sequences were determined. The candidate interaction was re-tested by co-transformation of AH109 cells with pGBKT7-MZF1 and pACT2-FHL3. Controls containing bait or prey alone, respectively, were also employed for the assay.
Determination of the interacting domains of MZF-1 and FHL3
Yeast AH109 cells were co-transformed with pACT2-FHL3 and various deletion mutants of MZF-1 in pGBKT7. The cells were selected on SD −Trp/−Leu plates and then examined for their ability to grow on SD −Trp/−Leu/−His/−Ade (adenine) plates. Similar analyses were performed with the cells co-transformed by pGBKT7-MZF1 (full-length or amino acids 1–217) and various deletion mutants of FHL3 in pGADT7 (Clontech).
Immunoprecipitation of in vitro translated MZF-1 and FHL3
c-Myc-tagged MZF-1 (amino acids 1–217) and HA-tagged FHL3 were produced by the in vitro transcription/translation method with TNT T7 Quick Coupled Transcription/Translation System (Promega, Madison, WI, U.S.A.) employing pGBKT7-MZF1-(amino acids 1–217) and pGADT7-FHL3. pGBKT7-53 and pGADT7-T (Clontech), encoding murine p53 and SV40 (simian virus 40) large T antigen respectively, were employed as controls. The products were mixed for immunoprecipitation with anti-c-Myc monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.) or anti-HA polyclonal antibody (Santa Cruz Biotechnology) followed by immunoblotting with anti-c-Myc and anti-HA (Roche, Basel, Switzerland) monoclonal antibodies.
Cell culture
KU812 cells (human basophillic leukaemia cell line) and Jurkat cells (human T cell line) were cultured in RPMI 1640 (Sigma, St. Louis, MO, U.S.A.) at 37 °C in a humidified incubator with 5% CO2. Similarly, HMC-1 cells (human mast cell line) and HeLa cells (human epithelial cell line) were cultured in Iscove's modified Dulbecco's medium (Invitrogen) and Dulbecco's modified Eagle's medium (Sigma) respectively. All media contained 10% (v/v) fetal bovine serum (JRH Bioscience, Lenexa, KS, U.S.A.), 100 units/ml penicillin (Banyu Pharmaceutical, Tokyo, Japan) and 100 μg/ml streptomycin (Meiji Seika, Tokyo, Japan).
Reporter assay with luciferase activity
Transfection of the cells and measurement of the luciferase activities were performed as described before [14]. Cells were transfected with 5 μg of a reporter plasmid pGLβ(−95/+102) [13] or pGβp-4180/4260 [14] with 2 or 5 μg of FHL expression plasmids. An empty plasmid pCR3.1-self [14] was used as a control. For MZF-1 antisense experiments, 10 μg of pCR3.1-hMZF1antisense [14] or an equivalent amount of a scrambled oligonucleotide of 20-mers as a control was introduced into the cells. To verify the protein expression of FHL1, FHL2 and FHL3 in the cells transfected with each FHL expression plasmid, cells were collected 24 h after the transfection and lysed in SDS/PAGE loading buffer before Western-blot analysis with anti-FHL1, -FHL2 and -FHL3 antibodies.
RT–PCR
To detect the FHL2 variant by RT–PCR, total RNA was prepared from each cell line with TRIzol® (Invitrogen). After RT reaction using 1 μg of the total RNA as a template and an oligo(dT)12–18 primer (Invitrogen), PCR was performed with two primer sets. Nucleotide sequences of the primers used for the PCR are as follows. Set 1: 1F, 5′-ATGACTGAGCGCTTTGACTGCCACC-3′, and 1R, 5′-GTGTTGAATTCAGATGTCTTTCCCAC-3′; set 2: 2F, 5′-CTGCGTGGTGTGCTTTGAGACCCTG-3′, and 2R, 5′-GAAGCGCTGCCCAGACAGCTGCTTC-3′. A thermal cycle of 95 °C for 30 s, 55 °C for 1 min and 72 °C for 1.5 min was repeated 32 times.
To quantify the mRNAs for MZF-1 and FHL3, PCR was performed with oligonucleotide primers whose sequences are represented below, after RT reaction using a random hexamer primer. For MZF-1 [17]: 5′-CTTCAGCCGCAGCTCGCACCTGCT-3′ and 5′-CTACTCGGCGCTGTGGACGCGCTGGT-3′; for FHL3 [18]: 5′-CATGGCATGAGCACTGCTTCCTG-3′ and 5′-GCTTAGGGCCCTGCCTGGCTACAGC-3′; for glyceraldehyde-3-phosphate dehydrogenase [19]: 5′-CCACCCATGGCAAATTCCATGGCA-3′ and 5′-TCTAGACGGCAGGTCAGGTCCACC-3′. A thermal cycle of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 1 min was repeated 28 times for MZF-1, 26 times for FHL3 and 20 times for glyceraldehyde-3-phosphate dehydrogenase.
Preparation of cell extracts
Nuclear extracts of various KU812 transfectants were prepared as follows. Cells were collected 24 h after the transfection and washed with ice-cold PBS and resuspended in ice-cold buffer A [10 mM Hepes (pH 7.9), 10 mM potassium chloride, 10 mM 2-mercaptoethanol, 1 mM PMSF, 1 μg/ml leupeptin and 1 μg/ml aprotinin]. Then, the cells were incubated on ice for 10 min and solubilized with 0.5% (v/v) Nonidet P40 for an additional 15 min. After centrifugation at 9000 g for 1 min, the pellets were resuspended in the extracting buffer [20 mM Hepes (pH 7.9), 400 mM potassium chloride, 4.5 mM magnesium chloride, 10 mM 2-mercaptoethanol, 1 mM PMSF, 1 μg/ml leupeptin and 1 μg/ml aprotinin] and incubated on ice for 1 h. The cell lysates were centrifuged at 10000 g for 10 min to collect the supernatants.
Cytoplasmic and nuclear fractions of KU812 cells treated with or without GM-CSF (granulocyte-macrophage colony-stimulating factor) were prepared using NE-PER nuclear and cytoplasmic extraction reagents (Pierce Biotechnology, Rockford, IL, U.S.A.).
EMSA (electrophoretic mobility-shift assay)
EMSA was performed as described previously [14] using a double-stranded oligonucleotide of 5′-AGTTAGTGGGGACGTT-3′ as the probe. Nuclear extracts (10 μg) from KU812 cells transfected with 10 μg of MZF-1 antisense or an equivalent amount of a scrambled oligonucleotide of 20-mers were used for the assay.
Affinity purification
Nuclear extracts prepared from KU812 cells co-transfected with pCR3.1-6HN-FHL3/pCR3.1-BD-c-Myc-MZF1, pCR3.1-6HN-FHL3/pCR3.1-BD-c-Myc or pCR3.1-6HN/pCR3.1-BD-c-Myc-MZF1 were affinity-purified using BD™ TALON metal affinity Co2+ resin (Clontech) according to the manufacturer's instructions. For equilibration and washing, 50 mM sodium phosphate buffer (pH7.0) containing 400 mM sodium chloride and 5% (v/v) glycerol was employed. The eluates were analysed by Western blotting with anti-c-Myc and anti-FHL3 antibodies (Santa Cruz Biotechnology). Similarly, nuclear extracts from the cells co-transfected with pCR3.1-6HN-FHL3/pCR3.1-BD-c-Myc-MZF1 or pCR3.1-6HN/pCR3.1-BD-c-Myc (as a control) were purified with an affinity column of BD™ TALON CellThru resin (Clontech) and subsequently immunoprecipitated with an anti-c-Myc antibody. The immunoprecipitates were analysed by SDS/PAGE followed by silver staining.
Gel-filtration chromatography
Nuclear extracts were applied on to a Superdex-200 column (Amersham Bioscience, Piscataway, NJ, U.S.A.) equilibrated with PBS and run at a flow rate of 0.25 ml/min. The eluate was collected in 1 ml fractions. Each fraction was analysed by Western blotting with anti-FHL3 and anti-c-Myc antibodies (Santa Cruz Biotechnology). Protein standards [thyroglobulin, ferritin and ovalbumin (Amersham Biosciences)] were applied on to the column separately.
RESULTS
Screening for MZF-1-interacting proteins by the yeast two-hybrid assay
We screened a human cDNA library fused to a GAL4 AD and an HA tag to determine MZF-1-interacting proteins using the yeast two-hybrid assay. Since some transcription factors independently activate the reporter to yield false positives when used as bait, first we examined whether full-length and deletion mutants of MZF-1 could be used as bait without autonomous activation. When introduced into a yeast strain AH109, only a deletion mutant containing amino acids 185–285 activated the reporter, whereas the other proteins, whose expressions in the yeast cells were detected by Western blotting, did not (Figure 1 and Table 1). Therefore we employed full-length MZF-1 fused to a GAL4 BD and a c-Myc tag as the bait. We screened 4.62×106 clones and obtained 141 positive clones expressing HIS3, ADE2 and MEL1 reporter genes. The AD-library plasmids were rescued from the positive clones to determine the nucleotide sequences of their cDNA inserts. It was revealed that nine of the clones did not have cDNA inserts in frame. The remaining 132 clones encoded 32 different proteins in frame, of which 14 proteins were encoded by multiple positive clones, whereas 18 proteins were solely encoded by a single clone. Among the proteins encoded by multiple kinds of positive clones, which probably reflected the true interaction, FHL3 was revealed to repress human FcεRI β-chain gene transcription and we further analysed this interaction in detail.
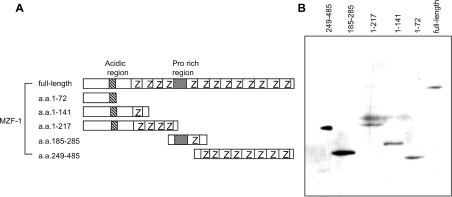
Figure 1. MZF-1 deletion mutants and their expression in yeast cells.
(A) Schematic drawing of full-length MZF-1 and deletion mutants (amino acids 1–27, 1–141, 1–217, 185–285 and 249–485). Number of amino acid residues counted from the N-terminus is represented; Z, zinc finger motif. (B) Yeast AH109 cells were transformed with expression plasmids for c-Myc-tagged full-length and deletion mutants of MZF-1 fused to BD. To verify the expression in yeast, the cell lysate prepared from each transformant was analysed by Western blotting with anti-c-Myc antibody.
Table 1. Examination of autonomous activation.
Full-length and deletion mutants (containing amino acids 1–72, 1–141, 1–217, 185–285 or 249–485) of human MZF-1 as a fusion to GAL4 BD were expressed in yeast AH109. The activity of α-galactosidase, a secreted enzyme encoded by the MEL1 gene under the control of a GAL4-responsive promoter, was detected on SD−Trp/+X-α-Gal medium. pCL-1 (Clontech) encoding full-length GAL4 and an empty vector pGBK were used as positive and negative controls respectively.
| α-Galactosidase activity | |
|---|---|
| Full-length MZF-1 | − |
| MZF-1-(1–72) | − |
| MZF-1-(1–141) | − |
| MZF-1-(1–217) | − |
| MZF-1-(185–285) | + |
| MZF-1-(249–485) | − |
| pCL-1 | ++ |
| pGBK | − |
FHL3 binds MZF-1
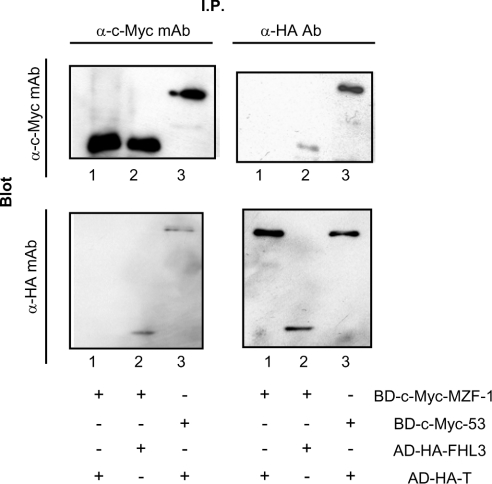
To verify the interaction between MZF-1 and FHL3, yeast AH109 cells were co-transformed with the constructs carrying BD–MZF-1 and AD–FHL3. The yeast cells were examined for growth on −His/−Ade medium to test the expression of two reporter genes of HIS3 and ADE2. The cells expressing BD–MZF-1/AD–FHL3 grew on the −His/−Ade medium, whereas those expressing BD alone/AD–FHL3 or BD–MZF-1/AD alone did not grow on it (results not shown). These results revealed that AD–FHL3 bound BD–MZF-1 not through the AD or BD region but through the direct interaction between FHL3 and MZF-1. To confirm further the interaction of MZF-1 and FHL3, c-Myc-tagged BD–MZF-1 and HA-tagged AD–FHL3 were produced by the in vitro transcription/translation method. A mixture of them was immunoprecipitated with anti-c-Myc-tag or anti-HA-tag antibody. Since the full-length MZF-1 fused to BD was not expressed efficiently by in vitro transcription/translation, the amino acids 1–217 region (see Figure 3) of MZF-1 was employed. AD–FHL3 but not AD-T-antigen was co-immunoprecipitated with BD–MZF-1 both by anti-c-Myc-tag and anti-HA-tag antibodies (Figure 2), also indicating that FHL3 directly bound MZF-1.
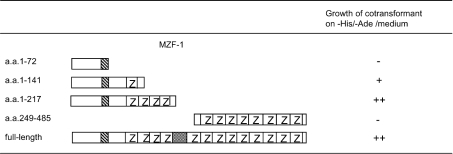
Figure 3. Determination of the FHL3-interacting region on MZF-1.
FHL3 as a fusion to AD and various deletion mutants of MZF-1 fused to BD were co-expressed in yeast AH109 cells. The co-transformed cells were examined for their ability to grow on −His/−Ade medium. When both of the two reporter genes, HIS3 and ADE2, are expressed, the cells can grow on the medium, which represents the interaction between FHL3 and the deletion mutant of MZF-1; −, no growth was observed; +, growth was observed on −His/−Ade medium; ++, significantly more growth was observed than +.
Figure 2. Detection of MZF-1–FHL3 interaction by immunoprecipitation.
c-Myc-tagged MZF-1 (amino acids 1–217) and HA-tagged FHL3 were produced by the in vitro transcription/translation method. A mixture of them was subjected to immunoprecipitation with anti-c-Myc or anti-HA antibody followed by blotting with anti-c-Myc and anti-HA antibodies. c-Myc-tagged p53 and HA-tagged SV40 large T-antigen were used as controls. Lane 1, BD-c-Myc-MZF-1 (amino acids 1–217)/AD-HA-T; lane 2, BD-c-Myc-MZF-1 (amino acids 1–217)/AD-HA-FHL3; lane 3, BD-c-Myc-53/AD-HA-T.
The N-terminal cluster of zinc finger motifs on MZF-1 interacts with the second and third LIM domains of FHL3
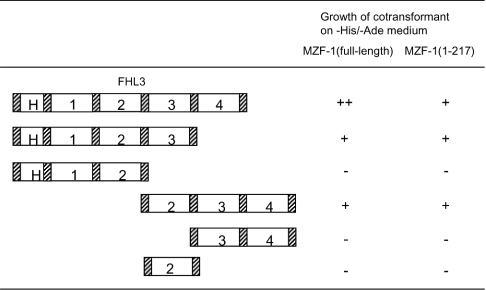
Next, we analysed which domains of MZF-1 and FHL3 interacted with each other by employing the deletion mutants of both proteins. At first, yeast AH109 cells were co-transformed with the expression plasmids of full-length FHL3 as a fusion to AD and various MZF-1 deletion mutants fused to BD to test the expression of HIS3 and ADE2 reporter genes. As shown in Figure 3, the cells expressing the region (amino acids 1–217) of MZF-1 lacking the C-terminal cluster of zinc finger motifs grew on −His/−Ade medium, but the cells expressing the amino acids 1–72 or 249–485 of MZF-1 (both of which lacked the N-terminal zinc finger cluster) did not. The cells expressing the amino acids 1–141 region of MZF-1, including a part of the N-terminal zinc finger cluster, grew slowly. These results indicated that the N-terminal cluster of zinc finger motifs on MZF-1 was required for the interaction with FHL3. On the other hand, when yeast cells were co-transformed with full-length MZF-1 and various deletion mutants of FHL3, the cells expressing FHL3 without either the second or third LIM domain did not grow on −His/−Ade medium (Figure 4). Similar results were obtained when amino acids 1–217 of MZF-1, which was shown to bind FHL3 in Figure 3, was expressed instead of the full-length MZF-1 (Figure 4). Collectively, it was revealed that the N-terminal cluster of zinc finger motifs on MZF-1 interacted with the second and third LIM domains of FHL3.
Figure 4. Determination of the MZF-1-interacting region on FHL3.
MZF-1 (full-length or amino acids 1–217) as a fusion to BD and various deletion mutants of FHL3 fused to AD were co-expressed in yeast AH109 cells. The co-transformed cells were examined for their ability to grow on −His/−Ade medium; H, half LIM domain. The numbers 1–4 represent the first, second, third and fourth LIM domains respectively.
FHL3 acts as a repressive cofactor of MZF-1 in human FcεRI β-chain gene transcription
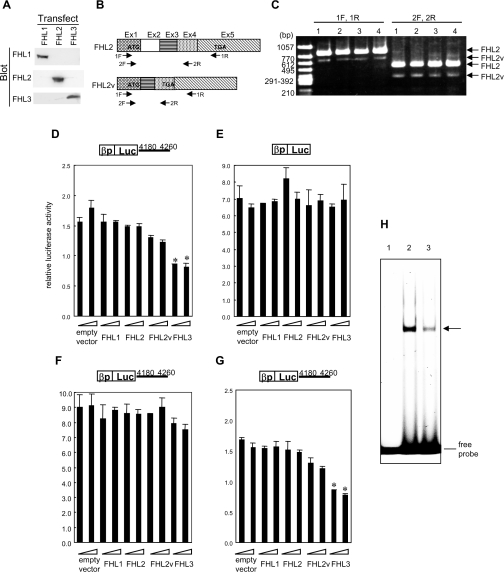
An expression plasmid of FHL3 was introduced into a human basophillic leukaemia cell line, KU812, known to express mast cell-specific molecules, for a transient expression assay with a reporter plasmid carrying a luciferase gene under the control of the human FcεRI β-chain promoter in the presence or absence of the β-chain gene 4180–4260 nt region, which mediated the transcriptional repression by MZF-1. In addition to FHL3, expression plasmids of other members of the FHL family (FHL1 and FHL2) were introduced, except for FHL4 and ACT that are only expressed in testis. The expression of FHL1, FHL2 and FHL3 in the cells transfected with each expression plasmid was verified by Western blotting (Figure 5A). Although the amount of each protein could not be compared strictly, because of potential differences in the affinity of the antibodies, all exogenous expression of FHL1, FHL2 and FHL3 was significantly abundant compared with their endogenous expression represented by remarkably weak or almost undetectable bands. Since an isoform of FHL2 was obtained when FHL2 cDNA was cloned from KU812 cells by RT–PCR, a variant named FHL2v was also used for the assay. The nucleotide sequence of FHL2v was determined (DDBJ/EMBL/GenBank® accession no. AB158503) and it revealed that FHL2v lacked the second exon and encoded a protein containing only the N-terminal region of FHL2 due to a frame shift yielding a stop codon in the fourth exon (Figure 5B). It was confirmed that FHL2v was expressed in various types of cells by RT–PCR using two different primer sets (Figure 5C). Since the antibody for FHL2 recognized the C-terminus of FHL2, the expression of FHL2v at the protein level could not be detected. In Figure 5(D), overexpression of FHL3 decreased the luciferase activity in the presence of 4180–4260 nt and the reduced luciferase activity was much lower than the basal level. (The relative activity 1.0 is determined without the promoter.) In contrast, overexpression of FHL3 had no effect in the absence of 4180–4260 nt (Figure 5E). FHL2v also decreased the luciferase activity slightly in the presence of nt 4180–4260, while FHL1 and FHL2 had no effect (Figure 5D). Furthermore, MZF-1 antisense inhibited the nt 4180–4260-dependent suppressive activity of FHL3 (Figure 5F), whereas a scrambled oligonucleotide did not inhibit it (Figure 5G). Since the antibody for MZF-1 was not available, the effects of MZF-1 antisense on MZF-1 expression levels were verified by EMSA. A double-stranded oligonucleotide probe carrying a consensus MZF-1-binding sequence and nuclear extracts from the cells transfected with MZF-1 antisense or an equivalent amount of a scrambled oligonucleotide was employed for the assay (Figure 5H). The intensity of the shifted band was decreased on introducing the antisense compared with the case of the scrambled oligonucleotide introduction, suggesting that the MZF-1 antisense actually decreased the MZF-1 expression level. All these results indicated that FHL3 repressed the β-chain promoter activity through the 4180–4260 nt element in an MZF-1-dependent manner.
Figure 5. FHL3 down-regulated the FcεRI β-chain gene promoter activity through the element in the fourth intron in an MZF-1-dependent manner.
(A) Expression of FHL1, FHL2 and FHL3 in KU812 cells transfected with each expression plasmid was detected by Western blotting with anti-FHL1, anti-FHL2 and anti-FHL3 antibodies. (B) cDNA structures of full-length FHL2 (FHL2) and a splice variant (FHL2v) are schematically represented. (C) Expression of mRNAs for FHL2 and FHL2v in various cell lines was detected by RT–PCR employing two primer sets (1F/1R and 2F/2R). Lane 1, KU812 cells; lane 2, HMC-1 cells; lane 3, Jurkat cells; lane 4, HeLa cells. The primers employed for PCR are indicated in (B). (D–G) For a transient expression assay, expression plasmids of FHL3 and other members of the FHL family (FHL1, FHL2 and FHL2v) were introduced into KU812 cells with a reporter plasmid carrying a luciferase gene under the control of the human FcεRI β-chain promoter in the presence (D, F, G) or absence (E) of the β-chain gene region 4180–4260 nt, which mediates the transcriptional repression by MZF-1. In addition, MZF-1 antisense (F) or the same amount of a scrambled oligonucleotide as a control (G) was introduced into the cells. Luciferase activities relative to that from a reporter construct solely containing a luciferase gene without a promoter are shown. Results are expressed as the means±S.D. for three independent experiments. *P<0.05, significantly different from the cells transfected with an equivalent amount of an empty vector. (H) A double-stranded oligonucleotide probe carrying a consensus sequence of MZF-1-binding motif and the nuclear extracts from the cells transfected with MZF-1 antisense or an equivalent amount of a scrambled oligonucleotide were employed for EMSA. Lane 1, no nuclear extract; lane 2, nuclear extracts of the cells in which a scrambled oligonucleotide was introduced; lane 3, nuclear extracts of MZF-1 antisense-introduced cells.
GM-CSF facilitates the accumulation of FHL3 in the nucleus
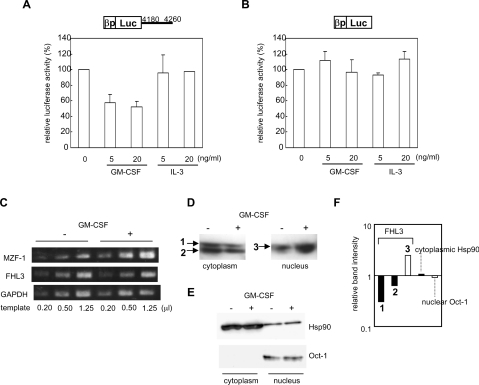
MZF-1 was known to be up-regulated by GM-CSF [20]. On the other hand, GM-CSF was reported to decrease the FcεRI expression [21,22]. These observations raise a possibility that the repressive activity of GM-CSF on FcεRI expression is partly due to suppression of β-chain gene expression by the up-regulated MZF-1, in addition to the decrease of α-chain gene expression reported by Welker et al. [22]. However, overexpression of MZF-1 alone did not decrease the β-chain gene transcription in our previous experiments [14]. We therefore examined the behaviour of FHL3 in the presence of GM-CSF, in addition to that in the presence of IL-3 (interleukin 3), which was reported to increase β-chain gene expression [23]. GM-CSF decreased the β-chain promoter activity in a transient expression assay using a reporter plasmid including nt 4180–4260, but IL-3 had no effect on it (Figure 6A). On the other hand, neither GM-CSF nor IL-3 modulated the β-chain promoter activity when a reporter without nt 4180–4260 was used (Figure 6B). These results revealed that GM-CSF increased the down-regulating activity of the MZF-1-binding motif (4180–4260 nt), but had no effect on the β-chain promoter itself. GM-CSF up-regulated MZF-1 expression as reported by Hui et al. [20], whereas the expression of FHL3 mRNA was not affected (Figure 6C). However, accumulation of FHL3 in the nucleus was observed on the addition of GM-CSF, which was revealed through Western blotting of the cytoplasmic and nuclear fractions of the cells (Figure 6D). FHL3 was increased in the nucleus and decreased in the cytoplasm by GM-CSF (Figure 6D), whereas subcellular localization of the control proteins, Hsp90 (heat-shock protein 90), mainly existing in the cytoplasm, and Oct-1, mainly existing in the nucleus, was not affected by GM-CSF (Figure 6E). As shown in Figure 6(F), the intensity of the band of nuclear FHL3 was increased 2.48-fold by GM-CSF and that of cytoplasmic FHL3 was decreased 0.304-fold for the upper band that was commonly observed in the cytoplasmic and nuclear fractions (‘1’ in Figure 6D) and 0.632-fold for the lower band only observed in the cytoplasmic fraction (‘2’ in Figure 6D). In contrast, the intensity of the bands for Hsp90 in the cytoplasmic fraction and for Oct-1 in the nuclear fraction was not changed by GM-CSF.
Figure 6. GM-CSF facilitated the accumulation of FHL3 in the nucleus.
(A, B) KU812 cells were transfected with a reporter plasmid including the human FcεRI β-chain gene promoter and a luciferase gene with (A) or without (B) the 4180–4260 nt region of the β-chain gene for a transient expression assay. GM-CSF or IL-3 was added to the medium at the indicated concentrations. The luciferase activities relative to that without the addition of GM-CSF or IL-3 are shown. Results are expressed as the means±S.D. for three independent experiments. (C) Expression levels of mRNAs for MZF-1 and FHL3 in KU812 cells cultured in the presence or absence of 10 ng/ml GM-CSF were semi-quantified by RT–PCR. (D) Cytoplasmic and nuclear fractions from KU812 cells treated with or without 10 ng/ml GM-CSF were prepared. Total protein (50 μg) from each fraction was analysed by Western blotting with anti-FHL3 antibody. (E) As a control, total proteins (50 μg) from each cytoplasmic and nuclear fraction were analysed by Western blotting with anti-Oct-1 antibody (Santa Cruz Biotechnology). Similarly, 5 μg of total protein from each fraction was employed for immunoblotting with anti-Hsp90 antibody (Santa Cruz Biotechnology). (F) Changes in the amount of cytoplasmic (black bars) and nuclear (white bars) FHL3 with GM-CSF treatment are expressed by calculating the ratio of the band intensity in the presence of GM-CSF to that in the absence of GM-CSF. Similarly, changes in the amount of Hsp90 in the cytoplasmic fraction and that of Oct-1 in the nuclear fraction are presented as controls.
FHL3 and MZF-1 form a complex of high molecular mass in the nucleus
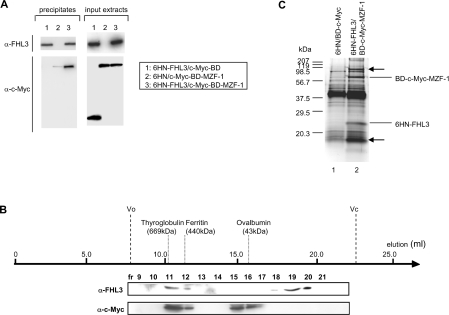
KU812 cells were co-transfected with the expression plasmids for 6HN-tagged FHL3 and c-Myc-tagged BD–MZF-1. Nuclear extracts of the co-transfected cells were subjected to a pull-down assay with metal affinity Co2+ resin against the 6HN tag, followed by blotting with anti-c-Myc and anti-FHL3 antibodies. As shown in Figure 7(A), c-Myc-tagged BD–MZF-1 was precipitated together with 6HN-tagged FHL3, representing their actual interaction in the nuclei of mammalian cells (lane 3). c-Myc-tagged BD was not precipitated with 6HN-tagged FHL3 (lane 1). Similarly, a very little amount of c-Myc-tagged BD–MZF-1 was precipitated with 6HN tag (lane 2), indicating that c-Myc-tagged BD–MZF-1 hardly binds the Co2+ resin non-specifically and that MZF-1 bound FHL3 directly, not through c-Myc-BD or 6HN tag. The nuclear extracts were next separated by gel-filtration chromatography. FHL3 was detected in fractions 11–12 and 18–20 by Western blotting using anti-FHL3 antibody. Similarly, by Western blotting with anti-c-Myc antibody, MZF-1 was detected in fractions 11–12 and 15–16 (Figure 7B). Fractions solely containing FHL3 (fractions 18–20) and those of MZF-1 (fractions 15–16) seemed to contain a monomeric or oligomeric form of each protein. FHL3 and MZF-1 were co-eluted in the same fractions (fractions 11–12) of high molecular mass (400–700 kDa), indicating that these proteins formed a complex probably containing some additional proteins judging from the molecular mass. To confirm this possibility, nuclear extracts from the transfected KU812 cells were purified with metal affinity chromatography for the 6HN tag using Co2+ resin, and subsequently immunoprecipitated with anti-c-Myc antibody. Figure 7(C) represents the results of SDS/PAGE analysis of the immunoprecipitated proteins. The bands indicated by arrows were observed in the immunoprecipitants from the co-transfected cells (lane 2) but not observed in those from the cells transfected with empty vectors (lane 1), suggesting that these proteins formed a complex with MZF-1 and FHL3.
Figure 7. MZF-1 and FHL3 formed a complex of high molecular mass in the nucleus.
KU812 cells were co-transfected with expression plasmids for c-Myc-tagged BD–MZF-1 and 6HN-tagged FHL3. (A) Nuclear extracts were prepared from the cells and employed for a pull-down assay with metal affinity Co2+ resin against the 6HN tag, followed by blotting with anti-c-Myc and anti-FHL3 antibodies. Nuclear extracts from cells co-transfected with 6HN-tagged FHL3/c-Myc-tagged BD or 6HN-tag/c-Myc-tagged BD–MZF-1 were used as controls. Expression of 6HN-tagged FHL3, c-Myc-tagged MZF-1 and c-Myc-tagged BD in a volume 1/50th of the input extracts was detected by Western blotting with anti-c-Myc and anti-FHL3 antibodies. (B) The nuclear extracts prepared from the co-transfected cells were separated by gel-filtration chromatography. Each fraction was analysed by Western blotting using anti-c-Myc and anti-FHL3 antibodies. (C) Nuclear extracts from the co-transfected cells were purified with a Co2+ affinity column for 6HN tag and subsequently immunoprecipitated with anti-c-Myc antibody. The immunoprecipitates were analysed by SDS/PAGE and the resolved protein species were visualized by silver staining. Nuclear extracts from the cells transfected with empty vectors were used as a control.
DISCUSSION
In the present study, we found that FHL3 acted as a co-repressor of MZF-1 in the transcriptional regulation of the human FcεRI β-chain gene. Although the presence of cofactors interacting with MZF-1 was predicted from the observation that MZF-1 both activated and suppressed transcription depending on cell types [24], no factors binding with MZF-1 or functioning as co-activators or co-repressors have been identified so far. This is the first example to identify an MZF-1-binding protein as a transcriptional cofactor.
FHL proteins (FHL1, KyoT2, FHL2, FHL3, FHL4 and ACT) [15,16,18,25–27] contain only a single N-terminal half-LIM domain followed by four sequential LIM domains, belonging to a subset of the LIM protein superfamily, which possesses cysteine-rich double zinc finger motifs called LIM. No interaction between FHL1 and MZF-1 and poor interaction between FHL2 and MZF-1 were detected in the yeast two-hybrid assay (results not shown). Therefore FHL3 was believed to bind specifically MZF-1 among the FHL family members, which would lead to the results in Figure 5(D) where FHL3 but not FHL1 and FHL2 inhibited the β-chain promoter activity. Although LIM proteins regulate transcription in the nucleus [28–30], the LIM zinc finger does not interact with DNA but with proteins [31,32]. This coincides with our results that FHL3 itself did not modulate the FcεRI β-chain gene transcription in the absence of MZF-1 (Figure 5F). Among the FHL members, ACT was first reported to act as a tissue-specific, CBP [CREB (cAMP-response-element-binding protein)-binding protein]-independent transcriptional co-activator for the transcription factor CREB [28] and, subsequently, FHL2 and FHL3 were also revealed to provide an activating function to CREB in the co-expression experiments [29]. Recently, it was reported that FHL3 binds BKLF (basic Kruppel-like factor)/KLF3 (Kruppel-like factor 3), a transcription factor harbouring zinc finger motifs such as MZF-1, and its co-repressor, CtBP, to repress transcription [33]. From these results and ours, FHL3 was revealed to behave both as a co-activator and a co-repressor, suggesting that FHL3 functions as an adapter molecule linking other factors with a transcription factor–DNA complex and stabilizing the assembled multi-protein complex. There will be complicated mechanisms by which either an activation- or repression-type of regulatory complex is formed depending on the transcription factor–DNA context, cofactor availability etc. In fact, FHL3 formed a complex of high molecular mass in the nucleus (Figure 7), similar to the case of Turner et al. [33], and we are now analysing the factors constituting the complex. This will enable us to find specific molecules or specific interactions modulating the β-chain gene expression, although FHL3 itself is broadly expressed in many types of human cell lines (results not shown).
FHL3 was accumulated in the nucleus when GM-CSF was added to the cell culture. In Figure 6(D), double bands were detected in the cytoplasmic fraction, but it could not be determined whether the lower band was from an unmodified or partially digested form of FHL3. Since the cytoplasmic FHL3 was decreased by GM-CSF, translocation of FHL3 from the cytoplasm to the nucleus rather than the stabilization of FHL3 in the nucleus by forming a multi-protein complex might contribute to the accumulation of the nuclear FHL3. In addition to the accumulation of FHL3 in the nucleus, GM-CSF up-regulated the expression of MZF-1 (Figure 6C), suggesting that GM-CSF decreased the β-chain expression by promoting MZF-1–FHL3 complex formation in the nucleus, which mediated transcriptional repression of the β-chain gene through the element in the fourth intron.
In the present study, we found a novel variant of FHL2, probably yielded by alternative splicing, which lacked a sequence corresponding to the second exon (Figures 5B and 5C). As for FHL1, an alternative splicing variant called KyoT2 was reported to negatively regulate RBP-J (J recombination signal sequence binding protein)-mediated transcription by competing with EBNA2 (Epstein–Barr virus nuclear antigen 2) and Notch 1 for binding to RBP-J [25]; however, the mechanisms by which the FHL2 variant slightly decreased the FcεRI β-chain promoter activity in Figure 5(D) is not clear at present.
In our two-hybrid screen, several other proteins in addition to FHL3 were revealed to interact with MZF-1 (K. Takahashi, C. Matsumoto and C. Ra, unpublished work). Many of them are reported to be involved in the regulation of apoptosis and cell-cycle progression, confirming the reported roles of MZF-1 in the regulation of cell differentiation and proliferation [34–36].
The N-terminal cluster of zinc fingers containing four motifs and the C-terminal cluster of nine motifs of MZF-1 were described to bind DNA independently [37], but functions of other domains remain to be analysed. The proline-rich region of MZF-1 was suggested to be a transcriptional AD by our analyses, because it independently activated transcription when fused to GAL4-BD (Table 1). In the analyses using deletion mutants, the forward cluster of zinc finger motifs of MZF-1 interacted with FHL3. This suggests that one of the two DNA-binding domains of MZF-1 actually interacts with DNA depending on the binding of a specific cofactor, which will result in a specific interaction of the MZF-1–cofactor complex with the target DNA. Therefore it will be interesting to investigate regulatory mechanisms of the transcriptional suppression by MZF-1 in our case by analysing the manner of MZF-1–cofactor interaction.
In the present study, we found FHL3 to be a novel factor that modulates the FcεRI β-chain gene expression. A more detailed study will identify the entire regulatory mechanism of FcεRI β-chain expression and provide new molecular targets for the development of therapeutic or prophylactic drugs for allergy.
Acknowledgments
This work was partially supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to C. R.) and a grant from Nihon University School of Medicine on the occasion of its 50th anniversary of founding.
References
- 1.Gounni A. S., Lamkhioued B., Ochiai K., Tanaka Y., Delaporte E., Capron A., Kinet J. P., Capron M. High-affinity IgE receptor on eosinophils is involved in defense against parasites. Nature (London) 1994;367:183–186. doi: 10.1038/367183a0. [DOI] [PubMed] [Google Scholar]
- 2.Maurer D., Fiebiger E., Reininger B., Woff-Winski B., Jouvin M. H., Kilgus O., Kinet J. P., Stingl G. Expression of functional high affinity immunoglobulin E receptors (FcεRI) on monocytes of atopic individuals. J. Exp. Med. 1994;179:745–750. doi: 10.1084/jem.179.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieber T., de la Salle H., Wollenberg A., Hakimi J., Chizzonite R., Ring J., Hanau D., de la Salle C. Human epidermal Langerhans cells express the high affinity receptor for immunoglobulin E (FcεRI) J. Exp. Med. 1992;175:1285–1290. doi: 10.1084/jem.175.5.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang B., Rieger A., Kilgus O., Ochiai K., Maurer D., Fodinger D., Kinet J. P., Stingl G. Epidermal Langerhans cells from normal human skin bind monomeric IgE via FcεRI. J. Exp. Med. 1992;175:1353–1365. doi: 10.1084/jem.175.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joseph M., Gounni A. S., Kusnierz J. P., Vorng H., Sarfati M., Kinet J. P., Tonnel A. B., Capron A., Capron M. Expression and functions of the high-affinity IgE receptor on human platelets and megakaryocyte precursors. Eur. J. Immunol. 1997;27:2212–2218. doi: 10.1002/eji.1830270914. [DOI] [PubMed] [Google Scholar]
- 6.Hasegawa S., Pawankar R., Suzuki K., Nakahata T., Furukawa S., Okumura K., Ra C. Functional expression of the high affinity receptor for IgE (FcεRI) in human platelets and its intracellular expression in human megakaryocytes. Blood. 1999;93:2543–2551. [PubMed] [Google Scholar]
- 7.Gounni A. S., Lamkhioued B., Koussih L., Ra C., Renzi P. M., Hamid Q. Human neutrophils express the high-affinity receptor for immunoglobulin E (Fc epsilon RI): role in asthma. FASEB J. 2001;15:940–949. doi: 10.1096/fj.00-0378com. [DOI] [PubMed] [Google Scholar]
- 8.Miller L., Blank U., Metzer H., Kinet J. P. Expression of high-affinity binding of human immunoglobulin E by transfected cells. Science. 1989;244:334–337. doi: 10.1126/science.2523561. [DOI] [PubMed] [Google Scholar]
- 9.Lin S., Cicala C., Scharenberg A. M., Kinet J. P. The FcεRI β subunit functions as an amplifier of FcεRI γ-mediated cell activation signals. Cell (Cambridge, Mass.) 1996;85:985–995. doi: 10.1016/s0092-8674(00)81300-8. [DOI] [PubMed] [Google Scholar]
- 10.Dombrowicz D., Lin S., Flamand V., Brini A. T., Koller B. H., Kinet J. P. Allergy-associated FcεRβ is a molecular amplifier of IgE- and IgG-mediated in vivo responses. Immunity. 1998;8:517–529. doi: 10.1016/s1074-7613(00)80556-7. [DOI] [PubMed] [Google Scholar]
- 11.Donnadieu E., Jouvin M. H., Kinet J. P. A second amplifier function for the allergy-associated FcεRI β subunit. Immunity. 2000;12:515–523. doi: 10.1016/s1074-7613(00)80203-4. [DOI] [PubMed] [Google Scholar]
- 12.Kuster H., Zhang L., Brini A. T., MacGlashan D. W., Kinet J. P. The gene and cDNA for the human high affinity immunoglobulin E receptor beta chain and expression of the complete human receptor. J. Biol. Chem. 1992;267:12782–12787. [PubMed] [Google Scholar]
- 13.Akizawa Y., Nishiyama C., Hasegawa M., Maeda K., Nakahata T., Okumura K., Ra C., Ogawa H. Regulation of human FcεRI β chain gene expression by Oct-1. Int. Immunol. 2003;15:549–556. doi: 10.1093/intimm/dxg055. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi K., Nishiyama C., Hasegawa M., Akizawa Y., Ra C. Regulation of the human high affinity IgE receptor β-chain gene expression via an intronic element. J. Immunol. 2003;171:2478–2484. doi: 10.4049/jimmunol.171.5.2478. [DOI] [PubMed] [Google Scholar]
- 15.Greene W. K., Baker E., Rabbitts T. H., Kees U. R. Genomic structure, tissue expression and chromosomal location of the LIM-only gene, SLIM1. Gene. 1999;232:203–207. doi: 10.1016/s0378-1119(99)00125-0. [DOI] [PubMed] [Google Scholar]
- 16.Chan K. K., Tsui S. K., Lee S. M., Luk S. C., Liew C. C., Fung K. P., Waye M. M., Lee C. Y. Molecular cloning and characterization of FHL2, a novel LIM domain protein preferentially expressed in human. Gene. 1998;210:345–350. doi: 10.1016/s0378-1119(97)00644-6. [DOI] [PubMed] [Google Scholar]
- 17.Hromas R., Collins S. J., Hickstein D., Raskind W., Deaven L. L., O'Hara P., Hagen F. S., Kaushansky K. A retinoic acid-responsive human zinc finger gene, MZF-1, preferentially expressed in myeloid cells. J. Biol. Chem. 1991;266:14183–14187. [PubMed] [Google Scholar]
- 18.Morgan M. J., Madgwick A. J. Slim defines a novel family of LIM-proteins expressed in skeletal muscle. Biochem. Biophys. Res. Commun. 1996;225:632–638. doi: 10.1006/bbrc.1996.1222. [DOI] [PubMed] [Google Scholar]
- 19.Tokunaga K., Nakamura Y., Sakata K., Fujimori K., Ohkubo M., Sawada K., Sakiyama S. Enhanced expression of glyceraldehyde-3-phosphate dehydrogenase gene in human lung cancers. Cancer Res. 1987;47:5616–5619. [PubMed] [Google Scholar]
- 20.Hui P., Guo X., Bradford P. G. Isolation and functional characterization of the human gene encoding the myeloid zinc finger protein MZF-1. Biochemistry. 1995;34:16493–16502. doi: 10.1021/bi00050a033. [DOI] [PubMed] [Google Scholar]
- 21.Welker P., Grabbe J., Zuberbier T., Henz B. M. GM-CSF downregulates expression of tryptase, FcεRI and histamine in HMC-1 mast cells. Int. Arch. Allergy Immunol. 1997;113:284–286. doi: 10.1159/000237574. [DOI] [PubMed] [Google Scholar]
- 22.Welker P., Grabbe J., Zuberbier T., Grutzkau A., Henz B. M. GM-CSF downmodulates c-kit, FcεRIα and GM-CSF receptor expression as well as histamine and tryptase levels in cultured human mast cells. Arch. Dermatol. Res. 2001;293:249–258. doi: 10.1007/s004030100225. [DOI] [PubMed] [Google Scholar]
- 23.Saini S. S., Richardson J. J., Wofsy C., Lavens-Phillips S., Bochner B. S., Macglashan D. W., Jr Expression and modulation of FcεRIα and FcεRIβ in human blood basophils. Int. Arch. Allergy Immunol. 2001;107:832–841. doi: 10.1067/mai.2001.114653. [DOI] [PubMed] [Google Scholar]
- 24.Hromas R., David B., Rauscher F. J., III, Klemsz M., Tene D., Hoffman S., Xu D., Morris J. F. Hematopoietic transcriptional regulation by the myeloid zinc finger gene, MZF-1. Curr. Top. Microbiol. Immunol. 1996;211:159–164. doi: 10.1007/978-3-642-85232-9_16. [DOI] [PubMed] [Google Scholar]
- 25.Taniguchi Y., Furukawa T., Tun T., Han H., Honjo T. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol. Cell. Biol. 1998;18:644–654. doi: 10.1128/mcb.18.1.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan M. J., Madgwick A. J. The fourth member of the FHL family of LIM proteins is expressed exclusively in the testis. Biochem. Biophys. Res. Commun. 1999;255:251–255. doi: 10.1006/bbrc.1999.0180. [DOI] [PubMed] [Google Scholar]
- 27.Morgan M. J., Madgwick A. J., Charleston B., Pell J. M., Loughna P. T. The developmental regulation of a novel muscle LIM-protein. Biochem. Biophys. Res. Commun. 1995;212:840–846. doi: 10.1006/bbrc.1995.2045. [DOI] [PubMed] [Google Scholar]
- 28.Fimia G. M., De Cesare D., Sassone-Corsi P. CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature (London) 1999;398:165–169. doi: 10.1038/18237. [DOI] [PubMed] [Google Scholar]
- 29.Fimia G. M., De Cesare D., Sassone-Corsi P. A family of LIM-only transcriptional coactivators: tissue-specific expression and selective activation of CREB and CREM. Mol. Cell. Biol. 2000;20:8613–8622. doi: 10.1128/mcb.20.22.8613-8622.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller J. M., Isele U., Metzger E., Rempel A., Moser M., Pscherer A., Breyer T., Holubarsch C., Buettner R., Schule R. FHL2, a novel tissue-specific coactivator of the androgen receptor. EMBO J. 2000;19:359–369. doi: 10.1093/emboj/19.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dawid I. B., Breen J. J., Toyama R. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 32.Bach I. The LIM domain: regulation by association. Mech. Dev. 2000;91:5–17. doi: 10.1016/s0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- 33.Turner J., Nicholas H., Bishop D., Matthews J. M., Crossley M. The LIM protein FHL3 binds Kruppel-like factor/Kruppel-like factor 3 and its co-repressor C-terminal-binding protein 2. J. Biol. Chem. 2003;278:12786–12795. doi: 10.1074/jbc.M300587200. [DOI] [PubMed] [Google Scholar]
- 34.Lenny N., Westendorf J. J., Hiebert S. W. Transcriptional regulation during myelopoiesis. Mol. Biol. Rep. 1997;24:157–168. doi: 10.1023/a:1006859700409. [DOI] [PubMed] [Google Scholar]
- 35.Robertson K. A., Hill D. P., Kelley M. R., Tritt R., Crum B., Van Epps S., Srour E., Rice S., Hromas R. The myeloid zinc finger gene (MZF-1) delays retinoic acid-induced apoptosis and differentiation in myeloid leukemia cells. Leukemia. 1998;12:690–698. doi: 10.1038/sj.leu.2401005. [DOI] [PubMed] [Google Scholar]
- 36.Nagamura-Inoue T., Tamura T., Ozato K. Transcription factors that regulate growth and differentiation of myeloid cells. Int. Rev. Immunol. 2001;20:83–105. doi: 10.3109/08830180109056724. [DOI] [PubMed] [Google Scholar]
- 37.Morris J. F., Hromas R., Rauscher F. J. Characterization of the DNA-binding properties of the myeloid zinc finger protein MZF1: two independent DNA-binding domains recognize two DNA consensus sequence with a common G-rich core. Mol. Cell. Biol. 1994;14:1786–1795. doi: 10.1128/mcb.14.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]