Abstract
Liver-specific inactivation of Trsp, the gene for selenocysteine tRNA, removes SePP (selenoprotein P) from plasma, causing serum selenium levels to fall from 298 μg/l to 50 μg/l and kidney selenium to decrease to 36% of wild-type levels. Likewise, glutathione peroxidase activities decreased in plasma and kidney to 43% and 18% respectively of wild-type levels. This agrees nicely with data from SePP knockout mice, supporting a selenium transport role for hepatically expressed SePP. However, brain selenium levels remain unaffected and neurological defects do not occur in the liver-specific Trsp knockout mice, while SePP knockout mice suffer from neurological defects. This indicates that a transport function in plasma is exerted by hepatically derived SePP, while in brain SePP fulfils a second, hitherto unexpected, essential role.
Keywords: brain, glutathione peroxidase, selenium, selenocysteine, selenoprotein P, tRNA[Ser]Sec
Abbreviations: Alb-Cre, Cre recombinase transgene under control of the rat albumin promoter; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GPx, glutathione peroxidase; cGPx, cellular GPx; eGPx, extracellular GPx; KO, knockout; Sec, selenocysteine; SePP, selenoprotein P; Trsp, tRNA[Ser]Sec gene
INTRODUCTION
Se (selenium) is an essential micronutrient in the diet of humans and other mammals, and has numerous health effects [1]. It occurs in proteins in the form of the unusual amino acid Sec (selenocysteine). Sec-containing proteins, designated selenoproteins, are likely to account for many of the health benefits derived from Se. The human genome contains 25 genes encoding selenoproteins [2]; among these, the GPxs (glutathione peroxidases) [3,4], thioredoxin reductases [5] and thyroid hormone deiodinases [6] are the best characterized. Sec is co-translationally incorporated into the growing polypeptide chain in response to an in-frame UGA codon. In eukaryotes, the presence of a specific RNA stem-loop structure in the 3′-untranslated region of the mRNA recodes the opal STOP codon to dictate Sec incorporation (see [7] for a review).
SePP (selenoprotein P) is a unique selenoprotein, in that it is the only such protein known that contains multiple Sec residues; it may carry between 10 and 17 Sec moieties depending on the species. More than 50% of plasma Se exists in SePP, suggesting a role for this protein in Se transport [8]. Gene targeting of SePP leads to a substantial decrease in Se levels in plasma, kidney, testis, heart and brain which is associated with subsequent decreases in selenoenzyme activities in these organs [9,10]. Supplementation of SePP KO (knockout) mice with Se-containing compounds restored tissue Se levels and selenoenzyme activities [11,12], further supporting a transport role for SePP. SePP mRNA expression is highest in the liver, but most other organs, including brain, also express SePP mRNA. The brain is a privileged organ with respect to Se supply, suggesting a vital role for selenoproteins in brain function [13]. Accordingly, dietary Se depletion and gene targeting of individual selenoproteins have provided evidence for specific roles of selenoproteins in brain function [14]. For example, SePP KO mice display a neurological phenotype with a movement disorder and epileptic seizures associated with greatly decreased brain Se content [9,11,12]. Maintaining rodents on a Se-deficient diet for several generations has not achieved the same decrease in brain Se content, nor has it led to spontaneous neurological phenotypes like those observed in SePP KO mice [15].
Therefore we wondered whether the liver is the sole source of the plasma SePP pool distributing Se within the body, or whether SePP may also have specific local functions in other organs. We took advantage of our SePP KO mice and compared them with mice lacking the Trsp (tRNA[Ser]Sec) gene only in the liver [16]. Without a functional Trsp gene in hepatocytes, these mice are unable to synthesize and secrete hepatic selenoproteins into the circulatory system. We show that liver-specific gene targeting of Trsp greatly decreases plasma SePP and thereby plasma Se levels, leading to similar impairments of renal GPx expression as described in SePP KO mice. However, the neurological dysfunction present in SePP KO mice was not replicated in liver-specific Trsp KO mice, a finding that strongly suggests an additional, hitherto unrecognized, local role of SePP in the brain.
MATERIALS AND METHODS
Animals
Mice that were homozygous for the floxed tRNA[Ser]Sec gene (Trspfl/fl) and heterozygous for Alb-Cre (an albumin–Cre transgene), wherein the Cre recombinase is driven by the hepatocyte-specific albumin promoter and Trspfl/fl is targeted for removal in the liver, were described recently [16]. Animals were maintained in a 12 h/12 h light/dark cycle with free access to food and water as described [9] and fed with standard lab chow (Altromin, Lage, Germany). Blood was taken after killing by cardiac puncture and treated or not with heparin, and plasma and serum were prepared after centrifugation at 10000 g at 4 °C for 10 min and frozen at −80 °C.
Testing of motor co-ordination on the rotarod was performed as described in [11]. For footprint analysis, stamp pad ink was applied to front (red) and back (black) paws and the animal was placed on a paper-covered path leading to a dark box. Stride length for front and back paws as well as the base width (i.e. the distance between the two hindpaw footprints) were determined for each animal at least three times. Results comparing base widths showed the same pattern and significance as those for stride length, and are available upon request. Animal studies were conducted according to legal regulations and approved by the local authorities in Berlin, Germany.
Se determinations
Se measurements were performed by a fluorimetric assay as described [11]. A commercial Se standard (Aldrich #247928; Sigma, Munich, Germany) and an independent seronorm standard (Sero AS, Billingstad, Norway) were used as reference samples to verify the assay.
Enzymatic assay
Enzymatic activity of eGPx (extracellular GPx) was determined using mouse plasma samples as described in [11]. cGPx (cellular GPx) activity was determined in tissue homogenates prepared as described in [9]. In brief, NADPH consumption by glutathione reductase was monitored at 340 nm as a measure of the GSSG formation catalysed by GPx. Enzymatic activity was recorded at 37 °C in a buffer containing 0.02 M potassium phosphate, pH 7.0, 0.6 mM EDTA, 0.15 mM NADPH, 2 mM GSH and 4 units of glutathione reductase (Calbiochem, Darmstadt, Germany). The reaction was started by addition of t-butylhydroperoxide (0.1 mM final concentration) as substrate. Background NADPH consumption was determined in the presence of 100 mM mercaptosuccinate and subtracted. Protein concentration was determined by the method of Bradford using IgG as a standard.
Northern blot analysis
cDNA fragments encompassing the complete reading frames of murine SePP, cGPx and eGPx were amplified by PCR, subcloned into pGEM-T vectors (Promega, Mannheim, Germany) and verified by DNA sequencing (BigDye Termination Sequencing Kit; Applied Biosystems, Warrington, U.K.). The inserts were prepared by restriction digestion and preparative gel electrophoresis, and radioactively labelled with [32P]dCTP (Hartmann Analytic, Braunschweig, Germany) using a random priming kit (High Prime; Roche Diagnostics GmbH, Mannheim, Germany). Hybridizations were conducted essentially as described in [9], and specific signals were quantified employing a phosphoimager (Cyclon; Perkin Elmer, Köln, Germany). Signals for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were subsequently recorded and used to calculate relative changes in transcript levels.
Western blot analysis
Aliquots of 50 μg of liver homogenates were separated by SDS/PAGE (10% polyacrylamide gel), and polyclonal antibodies against mouse SePP were used as described [11].
RESULTS
Reduced plasma Se levels in liver-specific selenoprotein KO mice
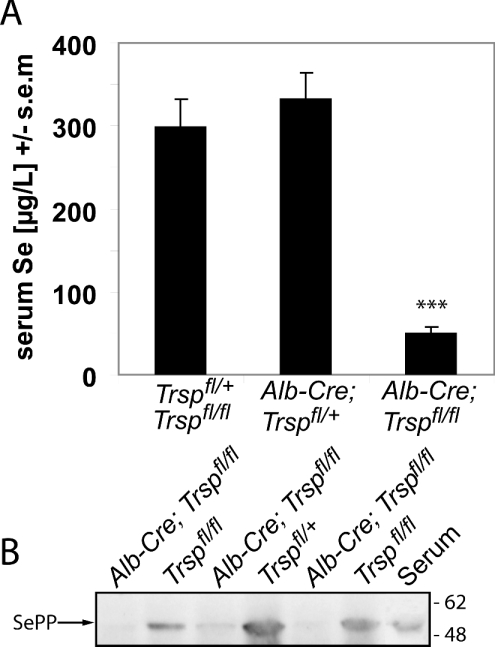
Trsp was selectively removed from the hepatocytes of mice (Alb-Cre; Trspfl/fl; referred to herein as liver-specific selenoprotein KO mice) using Cre-loxP technology as described in the Materials and methods section. These mice were analysed at the age of 5 weeks, i.e. the age at which SePP KO mice were analysed previously in our lab [9,11]. At this age and under our housing conditions, liver-specific selenoprotein KO mice did not show any signs of illness, and we used them, in first approximation, as a model for liver-specific SePP KO mice. In order to determine the contribution of the liver to plasma Se content, we compared liver-specific selenoprotein KO mice with appropriate littermate controls that encoded Trsp as either Trspfl/fl or Trspfl/+. As shown in Figure 1(A), there was a marked decrease in plasma Se concentration in the liver-specific selenoprotein KO mice. Synthesis of SePP, the main plasma Se carrier, is almost absent in the liver of these mice (Figure 1B). The remaining plasma Se content was the same as in SePP KO mice [9], suggesting that SePP secreted by the liver is the main determinant of plasma Se levels. Although most organs are known to express SePP mRNA, they obviously do not contribute significantly to plasma SePP levels.
Figure 1. Reduced SePP expression in liver-specific selenoprotein KO mice.
(A) Serum Se levels in male mice at 5 weeks of age. The following genotypes were analysed: Trspfl/fl and Trspfl/+ (n=14), Alb-Cre; Trspfl/+ (n=10), and Alb-Cre; Trspfl/fl (n=8). Significant difference compared with wild type: ***P<0.001 (one-way ANOVA with Dunnett's post hoc test). (B) Western blot reveals a marked decrease in SePP expression in livers from liver-specific selenoprotein KO mice. Samples from individual mice of the indicated genotypes are shown. Serum served as a positive control for SePP. Positions of marker proteins in kDa are indicated.
Impaired expression of GPx in kidney
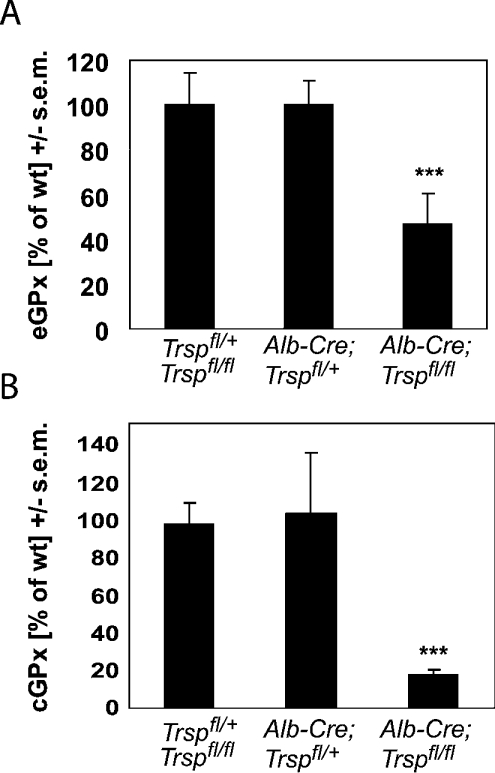
The other major selenoprotein in plasma is eGPx (plasma GPx). We determined eGPx activity in plasma from liver-specific selenoprotein KO mice and littermate controls (Figure 2A). Again, eGPx activity was decreased by approx. 50% in liver-specific selenoprotein KO mice, as in SePP-deficient mice [11]. The main site of eGPx secretion into plasma is the kidney [17]. We have shown previously that the reduced renal GPx activity observed in SePP KO mice can be rescued by Se supplementation, suggesting that kidney selenoprotein and Se levels depend on Se provided by the circulatory system [11]. Accordingly, renal GPx activity, as a measure for Se bioavailability in kidney, was drastically decreased in liver-specific selenoprotein KO mice (Figure 2B). This nicely parallels the reduced kidney Se content (Table 1). Since we found almost exactly the same decrease in renal GPx activity in liver-specific selenoprotein KO and SePP KO mice [9], the impairment of renal Se availability is likely to be caused by the lack of hepatically derived SePP.
Figure 2. Kidney selenoprotein expression depends on liver-derived SePP.
(A) eGPx activity was measured in the plasma of 5-week-old male mice. The following genotypes were analysed: Trspfl/fl and Trspfl/+ (n=8), Alb-Cre; Trspfl/+ (n=10), and Alb-Cre; Trspfl/fl (n=7). (B) cGPx activity was determined in kidney homogenates with t-butylhydroperoxide as a substrate (male mice; n=6–9). Wild-type (Trspfl/fl and Trspfl/+) GPx activity was 516±60 nmol of NADPH/min per mg of protein. Significant differences compared with wild type: ***P<0.001 (one-way ANOVA with Dunnett's post hoc test).
Table 1. Se content in liver-specific selenoprotein KO mice.
Measurements were performed using 5-week-old male mice. Values are means±S.E.M. (n=6–9). *P<0.05, **P<0.01 compared with control (Trspfl/fl and Trspfl/+) (one-way ANOVA with Dunnett's test).
| Se content (ng/mg of protein) | |||
|---|---|---|---|
| Organ | Trspfl/fl and Trspfl/+ | Alb-Cre; Trspfl/+ | Alb-Cre; Trspfl/fl |
| Kidney | 2.5±0.4 | 2.1±0.3 | 0.9±0.2* |
| Liver | 2.5±0.4 | 2.4±0.3 | 0.9±0.3** |
| Brain | 1.1±0.2 | 0.9±0.3 | 0.8±0.3 |
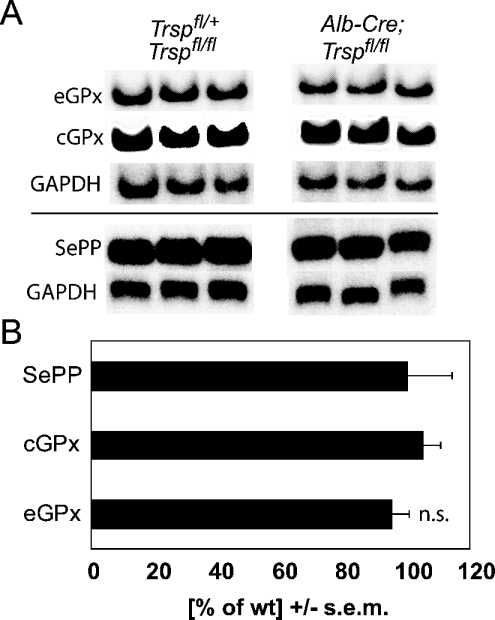
In order to test whether GPx down-regulation in liver-specific selenoprotein KO mice occurs at the transcript level, we performed Northern blot experiments on kidneys from these mice. As in SePP-deficient mice, there was no difference in the amounts of renal eGPx transcripts between liver-specific selenoprotein KO mice and controls (Figure 3). This implies that renal eGPx expression is subject to translational control, possibly depending on the supply of Sec-tRNA[Ser]Sec. In summary, liver-specific selenoprotein KO mice resemble SePP KO mice with respect to decreased plasma and kidney Se levels and diminished renal GPx expression.
Figure 3. Selenoprotein transcript levels are not altered in kidneys from liver-specific selenoprotein KO mice.
(A) Northern blot analysis of eGPx, cGPx and SePP mRNAs. GAPDH was used as a standard. One representative Northern blot from two independent experiments is shown. Levels were measured in kidneys from 5-week-old male mice (n=5–6). (B) Quantification of selenoprotein mRNA expression. Phosphorimager signals were normalized using GAPDH. Results for liver-specific selenoprotein KO mice are expressed as a fraction of those for wild-type (wt) littermate controls. n.s., not significant (Student's t test).
Brain-specific function of SePP
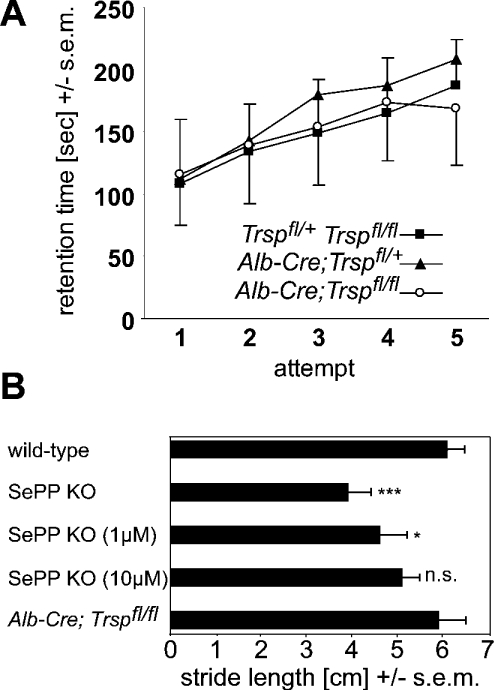
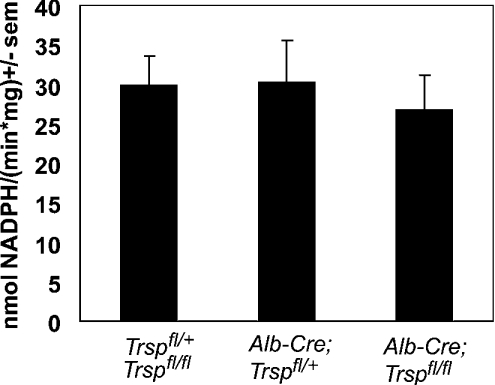
We and others have reported that SePP KO mice display a neurological phenotype involving seizures and a movement disorder [9,10,12]. According to a transport hypothesis for the physiological role of SePP, it appeared conceivable that the neurological dysfunction as well as decreased selenoenzyme activities in the brains of SePP KO mice resulted from impaired Se transport from the liver to the brain. This was believed to be mediated by the lack of plasma SePP, resulting in a drastically reduced Se concentration in plasma. However, liver-specific selenoprotein KO mice do not suffer from seizures, and nor could we observe any abnormal behaviour, despite markedly decreased plasma Se levels. Unlike SePP KO mice, their rotarod performance was indistinguishable from that of littermate controls (Figure 4A). In addition, footprint analyses did not reveal an abnormal gait of these mice, although SePP KO mice had a reduced stride length as assessed by this assay (Figure 4B). Se content in brains from liver-specific selenoprotein KO mice was unaltered (Table 1). In these mice, cGPx activity, a sensitive marker of tissue Se levels, was also unchanged (Figure 5). We conclude from these findings that the brain can maintain its Se and selenoenzyme levels in the absence of hepatic SePP production, and thus is largely independent of plasma Se levels as long as local SePP expression in brain is preserved. Our results thus provide the first direct evidence for a physiological function for local SePP expression in brain, distinct from its role as a plasma Se carrier.
Figure 4. Unlike SePP KO mice, liver-specific selenoprotein KO mice do not display a movement disorder.
(A) Rotarod analysis. No differences in motor co-ordination were observed between genotypes Trspfl/fl and Trspfl/+ (n=21), Alb-Cre; Trspfl/+ (n=12), and Alb-Cre; Trspfl/fl (n=15). (B) Footprint analysis. Stride length is compared between SePP KO mice, SePP KO mice rescued with selenite from birth (1 μM or 10 μM in drinking water) and liver-specific selenoprotein KO mice. Selenite supplementation dose-dependently leads to amelioration of the movement disorder of SePP KO mice, while liver-specific selenoprotein KO mice are indistinguishable from wild-type mice. Results from wild-type littermate controls of each group were similar. (n=4–6). Significance of differences: n.s. not significant; *P<0.05; ***P<0.001 (one-way ANOVA with Dunnett's post hoc test).
Figure 5. Brain GPx activity is unaltered in liver-specific selenoprotein KO mice.
GPx activity was determined in brain homogenates from 5-week-old mice with t-butylhydroperoxide as a substrate. The following genotypes were analysed: Trspfl/fl and Trspfl/+ (n=16), Alb-Cre; Trspfl/+ (n=12), and Alb-Cre; Trspfl/fl (n=16).
DISCUSSION
The current model of Se distribution within the body states that dietary Se after intestinal absorption is taken up by the liver, where it is transformed and incorporated into selenoproteins, including SePP [18]. SePP in turn is secreted into plasma to serve as a Se transport protein supplying Se to the other organs (Figure 6). Several lines of evidence support this model: (1) mammalian SePP contains the unusual number of 10–12 Sec residues, none of which (with one exception) have been implicated in any enzymatic mechanism; (2) plasma SePP is a marker for dietary Se status [19]; (3) for cultured cells, SePP is a bioavailable source of Se [20,21], and its Sec-rich C-terminus can be liberated specifically by proteolytic cleavage [22]; (4) 75Se-labelled SePP injected into rats is taken up readily by other tissues, including the brain, in contrast with 75Se-labelled cGPx [23]; (5) genetic inactivation of SePP in mice results in greatly diminished plasma Se levels and decreased Se content in kidney, testis, heart and brain [9,10]; and (6) supraphysiological Se supplementation of SePP KO mice rescues their biochemical and neurological phenotype [11,12]. Here we report that liver-specific inactivation of selenoprotein biosynthesis leads to a loss of SePP in plasma and a significant decrease in plasma Se content that resembles the situation in SePP KO mice. Similarly, kidney GPx activity, a sensitive measure of Se bioavailability, is greatly diminished in liver-specific selenoprotein KO mice, as is renal secretion of eGPx. In addition, our data suggest that, despite SePP mRNA expression in other organs, these organs do not contribute significantly to plasma SePP or Se levels. Thus our data seemingly support the current model of Se transport from the liver via SePP to other organs.
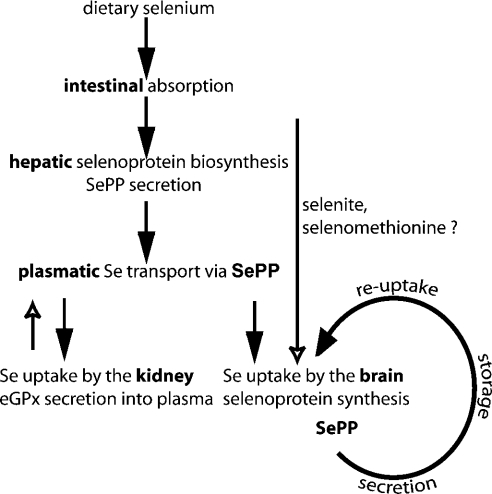
Figure 6. Summary of proposed function for SePP in Se transport and storage.
The current model states that dietary Se compounds are taken up by the liver, where selenoproteins, including SePP, are synthesized. SePP is secreted into plasma and provides Se to the rest of the body. The kidney takes up liver-derived SePP from plasma for biosynthesis of selenoproteins, including eGPx. A similar scenario was expected for brain. However, in the absence of hepatic selenoprotein biosynthesis, plasma SePP and plasma Se levels are severely reduced, but brain selenoprotein metabolism remains unaffected, indicating an alternative route for Se uptake available to the brain. We propose SePP to also serve as a local Se storage and recycling protein. All available Se not needed to maintain selenoenzyme activities is incorporated into SePP to serve as an extracellular Se store. After proteolytic cleavage, the Se-rich C-terminus is taken up by the neuron and re-used for selenoprotein biosynthesis. Thus genetic inactivation of SePP compromises brain selenoprotein metabolism and Se retention, while liver-specific inactivation of SePP does not affect brain Se status, selenoprotein biosynthesis or Se retention.
However, brain selenoprotein biosynthesis is not impaired in liver-specific selenoprotein KO mice, and no neurological phenotype is observed, in contrast with SePP KO mice. This finding hints at a hitherto less appreciated role of local SePP expression in the brain. Obviously, the brain can maintain its Se status even in the absence of liver-derived plasma SePP and reduced Se levels in plasma, provided that SePP expression is preserved inside the brain. Likewise, in rats bred and maintained on a Se-deficient diet for generations, plasma and liver Se levels decrease drastically, while normal brain Se levels are maintained [24]. The lack of SePP expression in the brain is apparently more deleterious than its lack in the liver or plasma (or both). Interestingly, SePP has been purified as a neurotrophic activity from bovine serum [20]. Several investigators have found SePP mRNA in human [25], mouse [9] and bovine [26] brain, and in cultured brain cells [27]. Thus Se could be organified and concentrated within the brain via local SePP biosynthesis. However, the pertinent question is what role SePP plays for neurons. It may harbour a neuroprotective activity, e.g. it may act as a peroxynitrite scavenger [28], as a Hg- and Cd-detoxifying protein [29] or as a phospholipid hydroperoxide peroxidase [30]. These hypotheses, however, have so far not been supported by in vivo experiments. For example, Se supplementation of SePP KO mice normalizes the neurological condition – this would not be possible if the SePP KO phenotype is caused by lack of a specific activity related to SePP protein other than Se transport or storage. In the light of our new findings, we propose a novel role for SePP as a local Se organifier and an extracellular Se storage protein. Owing to their inherent reactivity, Se compounds are potentially hazardous thiol-modifying agents. Therefore Sec is not a storage form for Se, and nor is it directly charged on to tRNA[Ser]Sec. We hypothesize that available Se is incorporated safely into SePP, which can be exported into the extracellular space for storage. Hence SePP could be taken up by cells at times of need in order to recycle its Se for selenoprotein synthesis (Figure 6). The only missing component would then be a receptor for SePP. The existence of such a receptor has been proposed based on biochemical experiments [23,31], but thus far it has not been cloned. Our new model is consistent with SePP being a survival-promoting factor and a form of Se that is more bioavailable than selenite for neurons [20]. This model could also explain why the brain does not lose its Se during times of dietary Se restriction [24], because it implies that Se is trapped within a ‘selenoprotein cycle’ within the brain [32]. Accordingly, SePP KO mice, initially rescued with selenite, lose their serum Se within 10 days after discontinuation of supraphysiological Se supplementation and start to develop the neurological phenotype after a few weeks. Hence Se loss occurs from the brain in the absence of SePP, even if a Se-adequate diet is provided (U. Schweizer, J. Köhrle and L. Schomburg, unpublished work). Selenocysteine β-lyase (Scly) could close this cycle, as it specifically liberates Se from Sec [33]. Such an enzyme would not be necessary if the Se liberated was not destined for selenoprotein re-synthesis, because Sec is also a substrate for common cysteine desulphurases. Finally, our hypothesis does not exclude any of the proposed ‘moonlighting’ activities of SePP that may be of importance when aging beyond the life span of a rodent is considered.
A functional dichotomy between plasma and brain transport protein expression is reminiscent of the iron transport protein transferrin, which is expressed by the liver (representing a source of plasma transferrin), but is also expressed in the brain [34].
Our finding that brain Se levels can be maintained in liver-specific selenoprotein KO mice implies that brain Se levels and plasma Se levels are not necessarily a function of each other – in other words, plasma Se measurements cannot be taken as a measure of brain Se status, but rather represent a measure of hepatic Se status. Thus clinical studies where plasma Se is used as a surrogate marker for brain selenoprotein activity in neurodegenerative disease should be interpreted with due caution (see [35] for a review). The evaluation of SePP content in cerebrospinal fluid should represent a superior measure of brain Se status, because it is likely to reflect the brain's ability to accumulate Se and synthesize SePP and other essential selenoproteins in sufficient amounts for proper brain functioning.
Acknowledgments
We thank Silke Kappler, Vartitér Seher, and Anita Kinne for technical assistance. Funding was provided by the Deutsche Forschungsgemeinschaft and Deutsche Krebshilfe.
References
- 1.Rayman M. P. The importance of selenium to human health. Lancet. 2000;356:233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 2.Kryukov G. V., Castellano S., Novoselov S. V., Lobanov A. V., Zehtab O., Guigo R., Gladyshev V. N. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 3.Brigelius-Flohé R. Tissue-specific functions of individual glutathione peroxidases. Free Radical Biol. Med. 1999;27:951–965. doi: 10.1016/s0891-5849(99)00173-2. [DOI] [PubMed] [Google Scholar]
- 4.Arthur J. R. The glutathione peroxidases. Cell. Mol. Life Sci. 2000;57:1825–1835. doi: 10.1007/PL00000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rundlof A. K., Arner E. S. Regulation of the mammalian selenoprotein thioredoxin reductase 1 in relation to cellular phenotype, growth, and signaling events. Antioxid. Redox Signalling. 2004;6:41–52. doi: 10.1089/152308604771978336. [DOI] [PubMed] [Google Scholar]
- 6.Köhrle J. The deiodinase family: selenoenzymes regulating thyroid hormone availability and action. Cell. Mol. Life Sci. 2000;57:1853–1863. doi: 10.1007/PL00000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatfield D. L., Gladyshev V. N. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motsenbocker M. A., Tappel A. L. A selenocysteine-containing selenium-transport protein in rat plasma. Biochim. Biophys. Acta. 1982;719:147–153. doi: 10.1016/0304-4165(82)90318-x. [DOI] [PubMed] [Google Scholar]
- 9.Schomburg L., Schweizer U., Holtmann B., Flohé L., Sendtner M., Köhrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem. J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill K. E., Zhou J., McMahan W. J., Motley A. K., Atkins J. F., Gesteland R. F., Burk R. F. Deletion of selenoprotein P alters distribution of selenium in the mouse. J. Biol. Chem. 2003;278:13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 11.Schweizer U., Michaelis M., Köhrle J., Schomburg L. Efficient selenium transfer from mother to offspring in selenoprotein-P-deficient mice enables dose-dependent rescue of phenotypes associated with selenium deficiency. Biochem. J. 2004;378:21–26. doi: 10.1042/BJ20031795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill K. E., Zhou J., McMahan W. J., Motley A. K., Burk R. F. Neurological dysfunction occurs in mice with targeted deletion of the selenoprotein P gene. J. Nutr. 2004;134:157–161. doi: 10.1093/jn/134.1.157. [DOI] [PubMed] [Google Scholar]
- 13.Schweizer U., Bräuer A. U., Köhrle J., Nitsch R., Savaskan N. E. Selenium and brain function: a poorly recognized liaison. Brain Res. Rev. 2004;45:164–178. doi: 10.1016/j.brainresrev.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Schweizer U., Schomburg L., Savaskan N. E. The neurobiology of selenium: lessons from transgenic mice. J. Nutr. 2004;134:707–710. doi: 10.1093/jn/134.4.707. [DOI] [PubMed] [Google Scholar]
- 15.Savaskan N. E., Bräuer A. U., Kühbacher M., Eyupoglu I. Y., Kyriakopoulos A., Ninnemann O., Behne D., Nitsch R. Selenium deficiency increases susceptibility to glutamate-induced excitotoxicity. FASEB J. 2003;17:112–114. doi: 10.1096/fj.02-0067fje. [DOI] [PubMed] [Google Scholar]
- 16.Carlson B. A., Novoselov S. V., Kumaraswamy E., Lee B. J., Anver M. R., Gladyshev V. N., Hatfield D. L. Specific excision of the selenocysteine tRNA[Ser]Sec (Trsp) gene in mouse liver demonstrates an essential role of selenoproteins in liver function. J. Biol. Chem. 2004;279:8011–8017. doi: 10.1074/jbc.M310470200. [DOI] [PubMed] [Google Scholar]
- 17.Maser R. L., Magenheimer B. S., Calvet J. P. Mouse plasma glutathione peroxidase. cDNA sequence analysis and renal proximal tubular expression and secretion. J. Biol. Chem. 1994;269:27066–27073. [PubMed] [Google Scholar]
- 18.Burk R. F., Hill K. E., Motley A. K. Selenoprotein metabolism and function: evidence for more than one function for selenoprotein P. J. Nutr. 2003;133:1517S–1520S. doi: 10.1093/jn/133.5.1517S. [DOI] [PubMed] [Google Scholar]
- 19.Persson-Moschos M., Alfthan G., Akesson B. Plasma selenoprotein P levels of healthy males in different selenium status after oral supplementation with different forms of selenium. Eur. J. Clin. Nutr. 1998;52:363–367. doi: 10.1038/sj.ejcn.1600565. [DOI] [PubMed] [Google Scholar]
- 20.Yan J., Barrett J. N. Purification from bovine serum of a survival-promoting factor for cultured central neurons and its identification as selenoprotein-P. J. Neurosci. 1998;18:8682–8691. doi: 10.1523/JNEUROSCI.18-21-08682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito Y., Takahashi K. Characterization of selenoprotein P as a selenium supply protein. Eur. J. Biochem. 2002;269:5746–5751. doi: 10.1046/j.1432-1033.2002.03298.x. [DOI] [PubMed] [Google Scholar]
- 22.Saito Y., Sato N., Hirashima M., Takebe G., Nagasawa S., Takahashi K. Domain structure of bi-functional selenoprotein P. Biochem. J. 2004;381:841–846. doi: 10.1042/BJ20040328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burk R. F., Hill K. E., Read R., Bellew T. Response of rat selenoprotein P to selenium administration and fate of its selenium. Am. J. Physiol. 1991;261:E26–E30. doi: 10.1152/ajpendo.1991.261.1.E26. [DOI] [PubMed] [Google Scholar]
- 24.Behne D., Hilmert H., Scheid S., Gessner H., Elger W. Evidence for specific selenium target tissues and new biologically important selenoproteins. Biochim. Biophys. Acta. 1988;966:12–21. doi: 10.1016/0304-4165(88)90123-7. [DOI] [PubMed] [Google Scholar]
- 25.Dreher I., Schmutzler C., Jakob F., Köhrle J. Expression of selenoproteins in various rat and human tissues and cell lines. J. Trace Elem. Med. Biol. 1997;11:83–91. doi: 10.1016/S0946-672X(97)80031-4. [DOI] [PubMed] [Google Scholar]
- 26.Saijoh K., Saito N., Lee M. J., Fujii M., Kobayashi T., Sumino K. Molecular cloning of cDNA encoding a bovine selenoprotein P-like protein containing 12 selenocysteines and a (His-Pro) rich domain insertion, and its regional expression. Mol. Brain Res. 1995;30:301–311. doi: 10.1016/0169-328x(94)00007-2. [DOI] [PubMed] [Google Scholar]
- 27.Yang X., Hill K. E., Maguire M. J., Burk R. F. Synthesis and secretion of selenoprotein P by cultured rat astrocytes. Biochim. Biophys. Acta. 2000;1474:390–396. doi: 10.1016/s0304-4165(00)00035-0. [DOI] [PubMed] [Google Scholar]
- 28.Arteel G. E., Mostert V., Oubrahim H., Briviba K., Abel J., Sies H. Protection by selenoprotein P in human plasma against peroxynitrite-mediated oxidation and nitration. Biol. Chem. 1998;379:1201–1205. [PubMed] [Google Scholar]
- 29.Yoneda S., Suzuki K. T. Equimolar Hg-Se complex binds to selenoprotein P. Biochem. Biophys. Res. Commun. 1997;231:7–11. doi: 10.1006/bbrc.1996.6036. [DOI] [PubMed] [Google Scholar]
- 30.Saito Y., Hayashi T., Tanaka A., Watanabe Y., Suzuki M., Saito E., Takahashi K. Selenoprotein P in human plasma as an extracellular phospholipid hydroperoxide glutathione peroxidase. Isolation and enzymatic characterization of human selenoprotein P. J. Biol. Chem. 1999;274:2866–2871. doi: 10.1074/jbc.274.5.2866. [DOI] [PubMed] [Google Scholar]
- 31.Wilson D. S., Tappel A. L. Binding of plasma selenoprotein P to cell membranes. J. Inorg. Biochem. 1993;51:707–714. doi: 10.1016/0162-0134(93)85003-q. [DOI] [PubMed] [Google Scholar]
- 32.Schomburg L., Schweizer U., Köhrle J. Selenium and selenoproteins in mammals: extraordinary, essential, enigmatic. Cell. Mol. Life Sci. 2004;61:1988–1995. doi: 10.1007/s00018-004-4114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mihara H., Kurihara T., Watanabe T., Yoshimura T., Esaki N. cDNA cloning, purification, and characterization of mouse liver selenocysteine lyase. Candidate for selenium delivery protein in selenoprotein synthesis. J. Biol. Chem. 2000;275:6195–6200. doi: 10.1074/jbc.275.9.6195. [DOI] [PubMed] [Google Scholar]
- 34.Burdo J. R., Connor J. R. Brain iron uptake and homeostatic mechanisms: an overview. Biometals. 2003;16:63–75. doi: 10.1023/a:1020718718550. [DOI] [PubMed] [Google Scholar]
- 35.Chen J., Berry M. J. Selenium and selenoproteins in the brain and brain diseases. J. Neurochem. 2003;86:1–12. doi: 10.1046/j.1471-4159.2003.01854.x. [DOI] [PubMed] [Google Scholar]