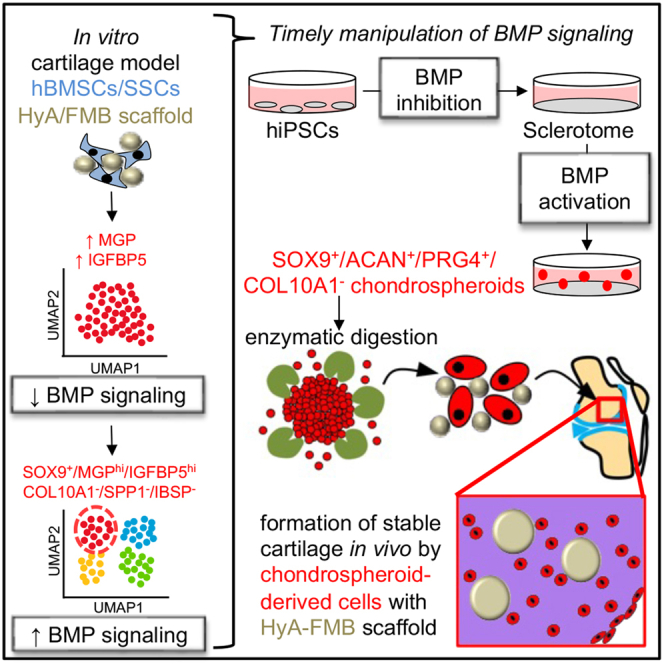
Summary
Stem cell therapies for degenerative cartilage disease are limited by an incomplete understanding of hyaline cartilage formation and maintenance. Human bone marrow stromal cells/skeletal stem cells (hBMSCs/SSCs) produce stable hyaline cartilage when attached to hyaluronic acid-coated fibrin microbeads (HyA-FMBs), yet the mechanism remains unclear. In vitro, hBMSC/SSC/HyA-FMB organoids exhibited reduced BMP signaling early in chondrogenic differentiation, followed by restoration of BMP signaling in chondrogenic IGFBP5+/MGP+ cells. Subsequently, human-induced pluripotent stem cell (hiPSC)-derived sclerotome cells were established (BMP inhibition) and then treated with transforming growth factor β (TGF-β) −/+ BMP2 and growth differentiation factor 5 (GDF5) (BMP signaling activation). TGF-β alone elicited a weak chondrogenic response, but TGF-β/BMP2/GDF5 led to delamination of SOX9+ aggregates (chondrospheroids) with high expression of COL2A1, ACAN, and PRG4 and minimal expression of COL10A1 and ALP in vitro. While transplanted hBMSCs/SSCs/HyA-FMBs did not heal articular cartilage defects in immunocompromised rodents, chondrospheroid-derived cells/HyA-FMBs formed non-hypertrophic cartilage that persisted until at least 5 months in vivo.
Subject areas: cell biology, molecular biology, Physiology, stem cells research
Graphical abstract
Highlights
-
•
A bone marrow stromal cell subset bound to fibrin beads do not hypertrophy in vivo
-
•
BMP signaling is initially reduced, then activated in stable chondroprogenitors
-
•
Temporal manipulation of BMP signaling in iPSC cultures generates chondrospheroids
-
•
Chondrospheroid-derived cells transplanted with fibrin beads form stable cartilage
Cell biology; Molecular biology; Physiology; Stem cells research
Introduction
Osteoarthritis (OA), a disease characterized by the permanent loss of articular cartilage, is among the most prevalent disabling diseases, affecting over 300 million people worldwide.1 For end-stage OA, surgical joint replacements provide excellent outcomes for pain relief and restoration of quality of life, but limitations in longevity and the risk of revision limit use in early OA and younger patients.2 Cell therapy has the potential to effectively treat OA, yet current strategies are largely ineffective. For instance, microfracture—a technique by which small holes are drilled into the subchondral bone to liberate bone marrow (which contains bone marrow stromal cells [BMSCs]/skeletal stem cells [SSCs]) to the affected area—is limited by the overproduction of fibrocartilage, enriched in type I collagen (COL1A1) and less suited to reduce friction with joint movement.3 Similarly, direct injection of human bone marrow stromal cells (hBMSCs)/SSCs into arthritic joints has led to inconsistent results in restoring the architecture of hyaline cartilage and relieving pain.4 Autologous chondrocyte implantation (ACI)—a two-step procedure by which healthy chondrocytes from less-weight-bearing areas are removed, expanded ex vivo, and transplanted into the affected joint—has been recommended as a cost-effective procedure5 for chondral defects in patients with no previous cartilage repair surgery and in knees with minimal OA. However, ACI is limited by the dedifferentiation of chondrocytes during ex vivo expansion6 and issues surrounding manufacture and supply. Alternative single-step procedures, utilizing the autologous harvesting of cartilage and bone (osteochondral) plugs and repositioning them in a suitably prepared area of cartilage damage (e.g., mosaicplasty), are limited by donor site morbidity and incomplete coverage of the damaged area, leading to suboptimal healing.7 Therefore, cartilage repair remains an unmet clinical need, where regenerative cell therapies are limited by an inadequate understanding of the development and maintenance of articular cartilage.
Following conception and the formation of three germ layers during gastrulation, embryonic mesenchyme expressing COL1A1 condenses in an area that will develop into a long bone.8 The induction of the sex-determining region Y-box (SOX) family of transcription factors—SOX5, SOX6, SOX8, and SOX9—promotes chondrogenic differentiation, leading to the formation of a cartilage template rich in type II collagen (COL2A1) and Aggrecan (ACAN). As development progresses, the majority of chondroprogenitors express hypertrophic markers (e.g., Type X Collagen [COL10A1]) and either go through apoptosis or become osteogenic, resulting in the replacement of cartilage by bone through the process of endochondral ossification.9 However, a condensation of flattened cells in the presumptive joint area—the interzone—begins to regulate the development of articular chondrocytes that resist hypertrophy.10 The interzone is rich in cells that secrete transforming growth factor β (TGF-β), growth differentiation factor 5 (GDF5), and wingless-type MMTV integration site (WNT) ligands, whereas a nearby area contains cells that are exposed to BMP inhibitors (e.g., Noggin) and may progressively migrate to the interzone region.11,12,13,14 During joint cavitation, interzone cells synthesize high amounts of hyaluronic acid, which binds to CD44 and Aggrecan, forming aggregates with anionic glycosaminoglycans that attract water into the developing matrix.8,10 A layered architecture begins to appear in articular cartilage. Lubricin (PRG4) becomes preferentially expressed at the surface layer of articular cartilage and interacts with cartilage oligomeric matrix protein (COMP) and COL2A1 expressed in surface and deeper layers.15,16 The surface layer is also marked by the expression of bone morphogenetic protein receptor type-1B (BMPR1B), a receptor for BMP2 and GDF5.17,18 The surface layer is typically the most damaged by OA, displaying fibrillations and fissures with joint aging and OA, leading to the exposure of deeper layers and subchondral bone that elicit pain with joint movement.17 Since BMP signaling plays key roles in interzone development and surface articular cartilage homeostasis, these findings suggest that this pathway can be fine-tuned in stem cell differentiation strategies to enhance regeneration.
One stem cell population that has been extensively tested for OA regenerative therapies is hBMSCs/SSCs, a subset of which are pericytes, derived from perichondral cells during endochondral ossification that attach to invading blood vessels, where they preserve their primitive state in adult marrow.19 Adult hBMSCs/SSCs are identified by their ability to generate bone, hematopoiesis-supporting stroma, and marrow adipocytes in transplantation studies and their ability to self-renew across serial transplantation.20,21,22 hBMSCs/SSCs do not directly form cartilage during development or in steady state, but maintain a chondrogenic memory, as their chondrogenic capacity is revealed upon in vitro pellet culture conditions with TGF-β supplementation.19 However, current chondrogenic induction strategies of hBMSCs/SSCs seem to follow a default pathway of endochondral ossification, evidenced by the expression of hypertrophic markers in vitro and matrix calcification in vivo,23,24 questioning whether hBMSCs/SSCs have the potential to form surface hyaline cartilage.25 The attachment of hBMSCs/SSCs to a three-dimensional (3D) scaffold, such as dense solid fibrin microbeads (FMBs),26,27,28 may promote a switch from an endochondral ossification program to that which promotes and maintains stable cartilage. Modification of the FMB scaffold by surface coating with hyaluronic acid (HyA-FMBs) was shown to promote the differentiation of hBMSCs/SSCs to stable hyaline-like cartilage in an ectopic site, which could be maintained for up to 28 weeks after in vivo subcutaneous implantation into immunocompromised mice.29 Thus, hBMSCs/SSCs attached to the HyA-FMB scaffold presents an attractive model to study mechanisms that promote stable cartilage development.
An alternative stem cell population that can be harnessed for OA regenerative therapies are human-induced pluripotent stem cells (hiPSCs). Since these cells can be reprogrammed from a patient’s somatic cells and resist replicative senescence, hiPSCs offer advantages in preventing graft rejection and generating expandable and scalable tissues and lack ethical restraints associated with the use of human embryonic stem cells (hESCs). Several differentiation strategies have been used to generate articular chondrocyte-like cells, including coculture with chondrocytes,30,31 formation of a BMSC/SSC-like intermediate prior to chondrogenic differentiation,32,33 and the stepwise differentiation that mimics embryonic gastrulation and formation of neural crest or mesoderm intermediates prior to chondrogenic induction.34,35,36,37 Few chondrogenic differentiation strategies have yielded chondrocyte-like cells that are able to resist hypertrophy in functional transplantation studies38,39,40; however, these studies employed undefined, animal-derived factors (e.g., serum) in differentiation, hampering clinical translation. Though some serum-free strategies have shown effective chondrogenic differentiation in vitro,41,42 follow-up transplantation studies have focused on hESC-derived chondrocyte-like cells.43,44 Therefore, the derivation of articular chondrocyte-like cells in serum-free and defined conditions that resist hypertrophy in long-term transplantation studies has been an ongoing challenge in hiPSC translational research.
Herein, we show that HyA-FMBs combined with hBMSCs/SSCs in vitro promote early expression of non-collagenous proteins (i.e., cartilage oligomeric matrix protein [COMP], dermatopontin [DPT], insulin-like growth factor-binding protein 5 [IGFBP5], matrix Gla protein [MGP]) with reduced BMP signaling marked by expression and protein analyses. However, by days 5 and 10 of chondrogenic differentiation, an early chondrogenic subpopulation with high expression levels of IGFBP5 and MGP shows restored BMP signaling and low expression of hypertrophic and osteogenic markers, confirmed in transplanted hBMSC/SSC-derived stable chondrocytes. Applying this mechanistic knowledge to chondrogenic differentiation of hiPSCs, we inhibited BMP signaling and other pathways during an initial sclerotome induction phase and activated BMP signaling using BMP2 and GDF5 during a subsequent chondrogenic induction phase. This biphasic BMP signaling produced chondrocyte-like cells derived from a 3D structure (herein termed a “chondrospheroid”) with a stable phenotype in vitro when applied to a purified SOX9-expressing subpopulation of sclerotome cells but produced a hypertrophic phenotype when applied to all sclerotome cells, indicating a time- and cell-specific nature of BMP signaling in stable chondrogenic differentiation. Further, chondral transplantation of cells liberated from day 35 chondrospheroids, which mimicked the transcriptional identity of a fetal chondrocyte, produced stable hyaline-like cartilage for up to 5 months in NSG mice and SRG rats when attached to HyA-FMBs, offering an articular-like cell source from a serum-free and defined protocol that can be employed in cartilage regenerative therapies.
Results
HyA-FMBs promote early expression of non-collagenous proteins
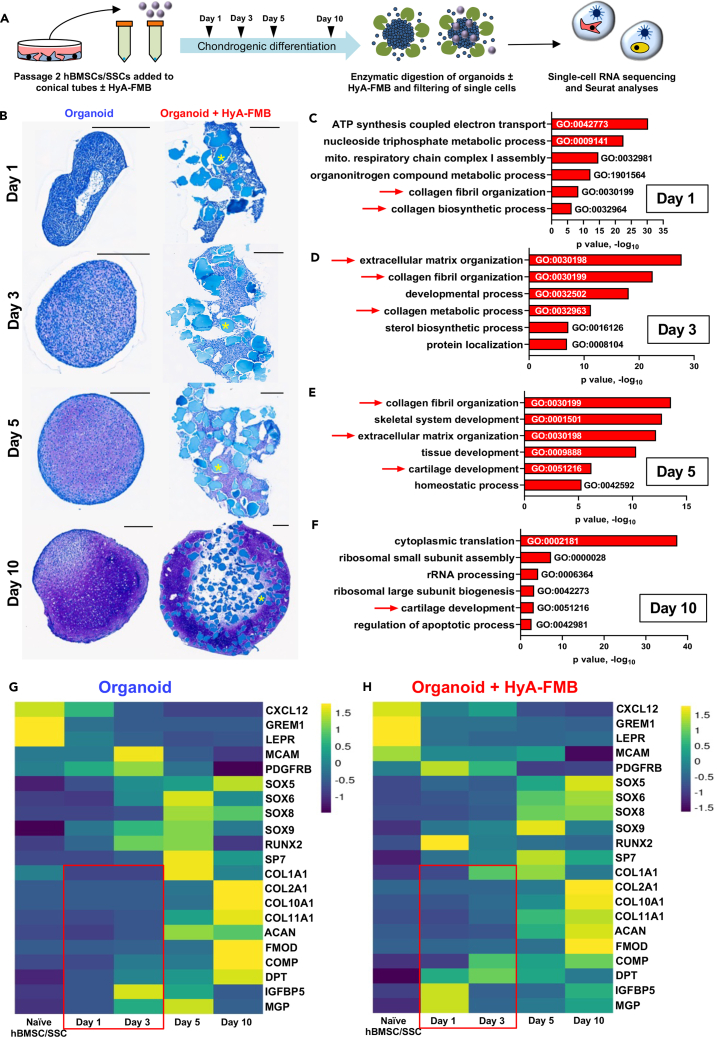
hBMSCs/SSCs differentiate into chondrocytes that form stable hyaline-like cartilage for up to 28 weeks when transplanted on HyA-FMBs subcutaneously in mice.29 Because of the stability of this cartilage, we sought to determine what influence the HyA-FMBs have on hBMSCs/SSCs that would direct them into a non-hypertrophic chondrogenic fate. To address this issue, we used these constructs as a model system to examine transcriptomic mechanisms that guide stable cartilage development and maintenance. Specifically, we cultured hBMSCs/SSCs attached to HyA-FMBs in chondrogenic differentiation medium and performed single-cell RNA sequencing (scRNA-seq) from digested organoids at days 1, 3, 5, and 10 (Figure 1A) in comparison with organoids that did not have HyA-FMBs. Over time, organoids formed a glycosaminoglycan-rich cartilage matrix indicated by toluidine blue staining (Figure 1B) and required TGF-β1 supplementation for proper chondrogenic differentiation (Figures S1A and S1B).
Figure 1.
HyA-FMBs promote early expression of non-collagenous proteins
(A) hBMSCs/SSCs were attached to HyA-FMBs and differentiated in chondrogenic medium, forming HyA-FMB organoids, which were digested at days 1, 3, 5, and 10 for scRNA-seq analyses in (C)–(H).
(B) Toluidine blue staining of control and HyA-FMB organoids at days 1, 3, 5, and 10 of chondrogenic differentiation. Scale bars, 300 μm. Asterisks (∗) indicate HyA-FMBs.
(C–F) Gene ontology analysis showing enriched pathways in HyA-FMB organoids compared with controls at days 1 (C), 3 (D), 5 (E), and 10 (F) of chondrogenic differentiation. Analyses performed using gProfiler’s driver GO pathway analysis from global differential gene expression (logfc>0.25, p < 0.05) between control and HyA-FMB organoids. Arrows indicate pathways of interest.
(G and H) Gene expression from days 0, 1, 3, 5, and 10 of chondrogenic differentiation. These data represent integrated data within control (G) and HyA-FMB (H) datasets for an overview of gene expression over time in each condition. Boxes indicate key trends in genes of interest (see also Figure S1).
A total of 37,792 cells from control and 41,545 cells from HyA-FMB organoids were analyzed by scRNA-seq across all time points following ambient RNA and doublet removal (along with other quality control measures; see STAR methods), and Seurat integration45 was performed within control and HyA-FMB datasets. Gene ontology analysis of differentially expressed genes in HyA-FMB organoids showed an enrichment of collagen fibril organization and extracellular matrix organization at early timepoints, especially at day 3 (Figures 1C–1F). Global expression analysis across 10 days of chondrogenic differentiation revealed early induction of COMP, DPT, IGFBP5, and MGP in HyA-FMB organoids (Figures 1G and 1H). Thus, HyA-FMBs promote early induction of non-collagenous and secreted proteins in hBMSCs/SSCs during chondrogenic differentiation. Trends for COMP, DPT, and MGP were confirmed at the protein level (see Figures 2G–2L).
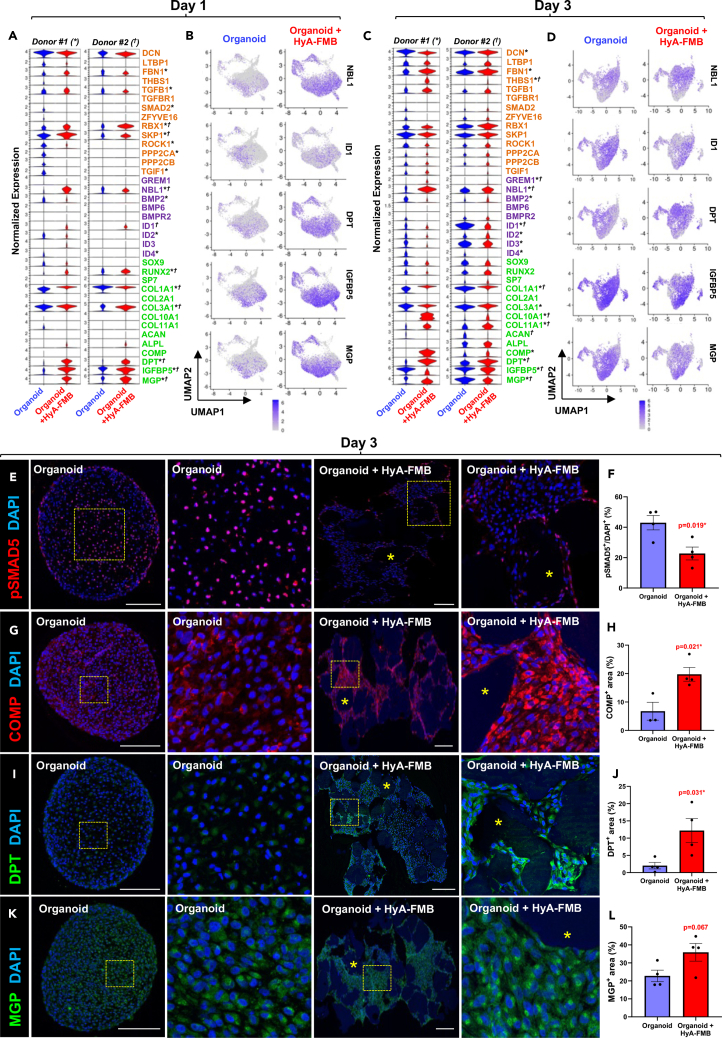
Figure 2.
HyA-FMBs suppress BMP signaling early in chondrogenic differentiation
(A–D) Global expression analyses from scRNA-seq of control (blue) and HyA-FMB (red) organoids at day 1 (A and B) and day 3 (C and D) of chondrogenic differentiation. Control and HyA-FMB datasets at each time point were normalized and integrated using Seurat. Cluster analyses at each time point are provided in Figures S2A–S2F.
(A, C) Violin plots depict expression of genes associated with TGFβ signaling (orange), BMP signaling (purple), and chondro-osteogenesis (green). Statistical significance (logfc>0.25, p < 0.05) is shown with an asterisk (∗) for donor #1 and dagger (†) for donor #2.
(B and D) Feature plots show genes of interest.
(E, G, I, and K) Immunofluorescence analyses in day 3 control and HyA-FMB organoids. High-magnification insets shown to the right of their corresponding images. Nuclei counterstained with DAPI. Scale bars, 200 μm. Asterisks (∗) indicate HyA-FMBs.
(F, H, J, and L) Area quantification of protein expression in day 3 control and HyA-FMB organoids. Each dot represents a biological replicate. Data are mean ± SEM; ∗p < 0.05, unpaired two-tailed t test (see also Figures S1 and S2).
HyA-FMBs suppress BMP signaling early in chondrogenic differentiation
Since many differences associated with HyA-FMB organoids were observed early, we jointly analyzed control and HyA-FMB datasets from two hBMSC/SSC donors at days 1 and 3 of chondrogenic differentiation. We processed 26,307 control and 30,890 HyA-FMB single-cell transcriptomes following quality control and performed Seurat integration between control and HyA-FMB datasets for direct comparison. Enrichment analysis of differentially expressed genes against the KEGG Pathway Database revealed a higher number of genes from the TGF-β signaling pathway in control organoids than in HyA-FMB organoids (Figures S1C and S1D). At day 1, expression of TGF-β and BMP inhibitors (Ring-Box1 [RBX1], S-phase kinase-associated protein 1 [SKP1], and DAN Family BMP Antagonist [NBL1]) were elevated in HyA-FMB organoids (Figures 2A and 2B). At day 3, BMP inhibitor expression (GREM1 and NBL1) remained elevated with a corresponding reduction in BMP-inducible genes, ID1-4 in HyA-FMB organoids (Figures 2C and 2D), confirmed by pathway analysis (Figures S1E and S1F). HyA-FMB organoids showed global reduction in phospho-SMAD5 (pSMAD5) protein expression, suggesting a reduction in BMP signaling46 (Figures 2E and 2F). Confocal microscopy also confirmed early induction of COMP and DPT expression in HyA-FMB organoids (Figures 2G–2L).
To address heterogeneity of these early organoid cultures, we performed cluster analyses. The majority of the cells did not display a discernible phenotype at day 1 (Cluster #4, Figures S2A–S2C), but a subgroup of COL2A1+ chondrogenic cells enriched in IGFBP5 and MGP emerged by day 3 of differentiation (Cluster #1, Figures S2D–S2F, solid black box in F). This chondrogenic population exhibited lower levels of NBL1 and higher levels of ID1 compared with an early osteogenic population (RUNX2+/IBSP+) (Cluster #4, Figure S2F, dashed black box). These differences between a chondrogenic cluster and an osteogenic cluster prompted us to further examine BMP signaling in MGP/IGFBP5-enriched cells during chondrogenic differentiation.
HyA-FMBs restore BMP signaling in MGP/IGFBP5-enriched chondrogenic cells
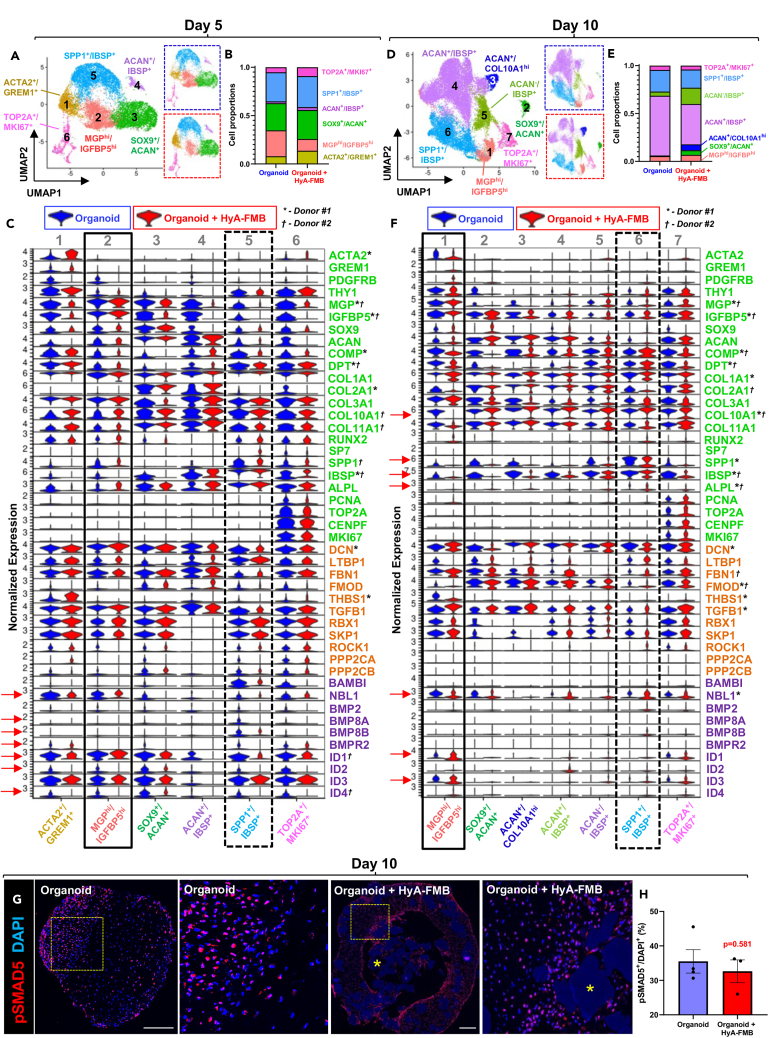
At day 5 of chondrogenic differentiation, more identifiable chondrogenic and osteogenic populations emerged in both control and HyA-FMB organoids. Specifically, cluster analysis of integrated day 5 datasets identified six populations: primitive actin alpha2 (ACTA2)+/GREM1+ cells, MGPhi/IGFBP5hi early chondrogenic cells, SOX9+/ACAN+ chondrogenic cells, ACAN+/bone sialoprotein (IBSP)+ chondro-osteogenic cells, secreted phosphoprotein 1(SPP1)+/IBSP+ osteogenic cells, and DNA topoisomerase II alpha (TOP2A)+/marker of proliferation Ki-67 (MKI67)+ cycling chondro-osteoprogenitor cells (Figure 3A). Cluster proportions were similar between control and HyA-FMB datasets, with the exception of the MGPhi/IGFBP5hi early chondrogenic cluster, which comprised 12% of HyA-FMB organoids compared with 27% of controls (Figure 3B). Expression in BMP signaling genes were similar between organoids in MGPhi/IGFBP5hi early chondrogenic cells (Figure 3C, solid black box, arrows); however, reduced levels of BMP8A, BMP8B, BMPR2, ID1, ID2, and ID4 were observed in SPP1+/IBSP+ osteogenic cells (Figure 3C, dashed black box, arrows) cultured with HyA-FMBs. These data suggest that HyA-FMBs reduce BMP signaling in SPP1+/IBSP+ osteogenic-committed cells but restore BMP signaling in MGPhi/IGFBP5hi early chondrogenic cells by day 5 of chondrogenic differentiation.
Figure 3.
HyA-FMBs restore BMP signaling in MGP/IGFBP5-enriched chondrogenic cells
(A–F) Cluster analyses from scRNA-seq of control (blue) and HyA-FMB (red) organoids at day 5 (A–C) and day 10 (D–F) of chondrogenic differentiation. Control and HyA-FMB datasets at each time point were normalized and integrated using Seurat. Marker genes for each cluster are listed in the corresponding color. Data are shown from donor #1.
(A, D) UMAP representation of integrated control and HyA-FMB datasets with annotated clusters. Split UMAPs are shown on the right for control organoids (blue dashed outline) and HyA-FMB organoids (red dashed outline).
(B and E) Bar chart depicting proportion of cell clusters in control and HyA-FMB datasets.
(C and F) Violin plots depicting gene expression split by experimental condition (blue violin plots are control organoids, red violin plots are HyA-FMB organoids) from integrated datasets at day 5 (C) and day 10 (F) of chondrogenic differentiation. Genes associated with TGF-β signaling (orange), BMP signaling (purple), and chondro-osteogenesis (green) are shown. Arrows depict notable gene expression differences (logfc>0.25, p < 0.05) between MGPhi/IGFBP5hi cluster (solid black box) and SPP1+/IBSP+ cluster (dashed black box). Global significance (logfc>0.25, p < 0.05) between control and HyA-FMB organoids is shown with an asterisk (∗) for donor #1 and dagger (†) for donor #2.
(G) Immunofluorescence of phospho-Smad5 (pSMAD5) in day 10 organoids. High-magnification insets shown to the right of their corresponding images. Nuclei counterstained with DAPI. Scale bars, 300 μm. Asterisks (∗) indicate HyA-FMBs.
(H) Quantification of pSMAD5+ nuclei in day 10 organoids. Each dot represents a biological replicate. Data are mean ± SEM (see also Figure S2).
At day 10, IBSP+ cell populations comprised 84% ± 6% of control organoids compared with 67% ± 7% of HyA-FMB organoids, suggesting that the default osteogenic program in hBMSCs/SSCs was reduced in HyA-FMB organoids (Figures 3D and 3E). Importantly, MGP/IGFBP5-enriched cells from HyA-FMB organoids exhibited little to no expression of hypertrophic and osteogenic markers (COL10A1, Alkaline Phosphatase [ALPL], IBSP, and SPP1) compared with control organoids (Figure 3F, black box, arrows). Expression of NBL1, ID1, and ID3 were almost exclusive to MGPhi/IGFBP5hi early chondrogenic cells and TOP2A+/MKI67+ cycling chondro-osteoprogenitor cells. However, NBL1 expression in MGP/IGFBP5-enriched cells from HyA-FMB organoids was reduced in comparison with control organoids, whereas BMP target genes, ID1 and ID3, were increased, indicative of restoration of BMP signaling (Figure 3F, black box, arrows), and confirmed by pathway analysis (Figure S2G). Confocal microscopy showed no differences in pSMAD5 expression between control and HyA-FMB organoids at day 10 of chondrogenic differentiation (Figures 3G and 3H). Overall, these data suggest that HyA-FMBs restore BMP signaling (as measured by the phosphorylation status of Smad5) in MGPhi/IGFBP5hi early chondrogenic cells, which exhibit low expression of hypertrophic and osteogenic markers.
BMP signaling is increased in stable, hyaline-like cartilage upon ectopic transplantation
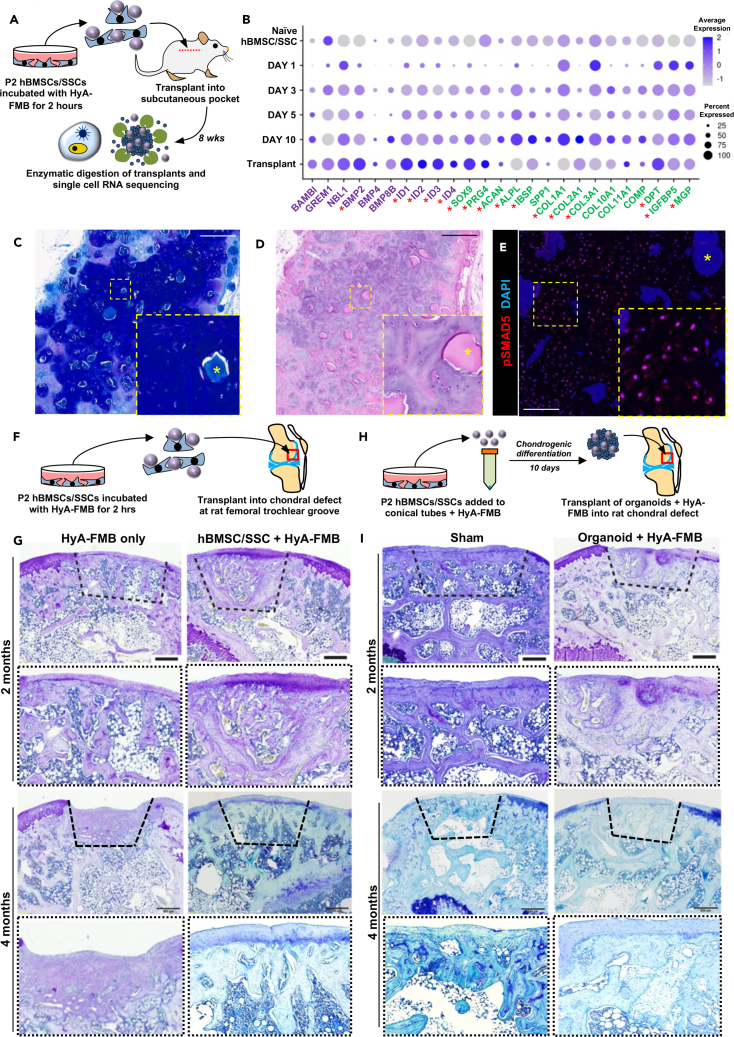
We next examined BMP signaling in stable, transplanted cartilage. Specifically, we transplanted hBMSCs/SSCs attached to HyA-FMBs subcutaneously into immunocompromised NSG mice and performed scRNA-seq of digested cartilage tissues 8 weeks post-transplant (Figure 4A), at which point, chondrogenesis is well underway. Integration of in vitro and in vivo HyA-FMB datasets was performed for cross-comparison. Transplanted tissues exhibited increased levels of PRG4 and decreased levels of ALPL, IBSP, SPP1, COL1A1, and COL10A1 compared with in vitro culture, confirming a more hyaline-like phenotype in the in vivo model (Figure 4B). Moreover, BMP2, ID1, ID2, ID3, and ID4 were increased in 8-week transplants, which was confirmed by pathway analysis, suggesting a potential role of BMP activation in maintaining stable cartilage (Figures 4B and S3A). Ectopic HyA-FMB transplants formed a hyaline-like cartilage matrix by 8 weeks, shown by toluidine blue and H&E staining (Figures 4C and 4D) with nearly half (48.8% ± 2.7%) of transplanted cells exhibiting pSMAD5 immunoreactivity (Figure 4E), similar to what was found in 10-day HyA-FMB organoids in vitro (Figure 2F). These data confirm the activation of BMP signaling genes in the formation of stable, mature cartilage.
Figure 4.
BMP signaling is increased in stable, hyaline-like cartilage upon ectopic transplantation
(A) hBMSCs/SSCs were incubated with HyA-FMBs for 2 h and transplanted ectopically into NSG mice, followed by digestion of transplanted tissue 8 weeks post-transplant and scRNA-seq analysis shown in (B).
(B) Dot plot demonstrating global gene expression of integrated in vitro HyA-FMB datasets (from days 0, 1, 3, 5, and 10 of chondrogenic differentiation) and in vivo transplant of hBMSCs/SSCs attached to HyA-FMBs analyzed 8 weeks post-transplant. Genes associated with BMP signaling (purple) and chondro-osteogenesis (green) are shown. Asterisks indicate differentially expressed genes (logfc>0.35) in transplanted hBMSCs/SSCs attached to HyA-FMBs, compared with all in vitro HyA-FMB datasets.
(C and D) Toluidine blue (C) and H&E (D) staining of ectopic transplant of hBMSCs/SSCs attached to HyA-FMBs at 8 weeks post-transplant with high-magnification insets. Scale bars, 500 μm. Asterisks (∗) indicate HyA-FMBs.
(E) Immunofluorescence analysis of pSMAD5 (red) at 8 weeks post-transplant with high-magnification insets. Nuclei counterstained with DAPI (blue). Scale bar, 100 μm. Asterisks (∗) indicate HyA-FMBs.
(F) hBMSCs/SSCs were incubated with HyA-FMBs for 2 h and transplanted into a 2-mm defect at the femoral trochlear groove in SRG rats, followed by histology analyses in (G).
(G) Toluidine blue staining of defect areas (dashed lines) from chondral transplants at 2 and 4 months post-transplant in SRG rats. Scale bars, 500 μm. High-magnification images shown below each image.
(H) hBMSCs/SSCs were cultured with HyA-FMBs in chondrogenic medium for 10 days and transplanted into a 2-mm defect at the femoral trochlear groove in SRG rats, followed by histology analyses in (I).
(I) Toluidine blue staining of defect areas (dashed lines) from organoid chondral transplants at 2 and 4 months post-transplant in SRG rats. Scale bars, 500 μm. High-magnification images shown below each image (see also Figure S3; Table S1).
Rat chondral transplantation of hBMSC/SSC/HyA-FMB constructs yields suboptimal chondrogenesis
To test the stability of newly formed cartilage by hBMSCs/SSCs attached to HyA-FMBs in a chondral defect model, we transplanted hBMSCs/SSCs attached to HyA-FMBs into a 2-mm defect at the femoral trochlear groove in immunodeficient SRG rats, followed by histological analyses of the defect site at 1, 2, and 4 months post-transplantation (Figure 4F). Unexpectedly, transplanted hBMSC/SSC/HyA-FMBs were degraded within 1–2 months at the defect site, as identified by h-vimentin staining of transplanted human cells (Figures S3B and S3C). Toluidine blue-stained cartilage matrix was evident on the joint surface in the defect site at 2 months post-transplant, yet surface toluidine blue staining was less evident by 4 months, with organization of subchondral bone and marrow in the defect site (Figures 4G and S3D). We examined whether a 10-day chondrogenic induction period in vitro would enhance the stability of transplanted hBMSC/SSC/HyA-FMBs when placed into the defect site of SRG rats (Figure 4H). Toluidine blue and H&E staining revealed a similar degradation of HyA-FMBs by 1–2 months, vascular invasion by 2 months, and replacement by bone by 4 months (Figures 4I, S3E, and S3F). Cartilage and bone from defect areas were graded according to an established scoring system,29 indicating low cartilage formation and abundant bone formation by hBMSC/SSC/HyA-FMB transplants (Table S1). Thus, upon degradation of HyA-FMBs, hBMSC/SSCs regenerate subchondral bone following injury and may be relevant for osteochondral regeneration (e.g., osteochondral plug surgery). Given the degradation of HyA-FMBs and propensity of hBMSCs/SSCs to undergo endochondral ossification in the absence of HyA-FMBs, we directed our attention to another potential source of chondrogenic cells, hiPSCs, which may escape this default program upon directed differentiation.
BMP activation in SOX9+-purified early chondrogenic cells promotes stable chondrogenesis in vitro
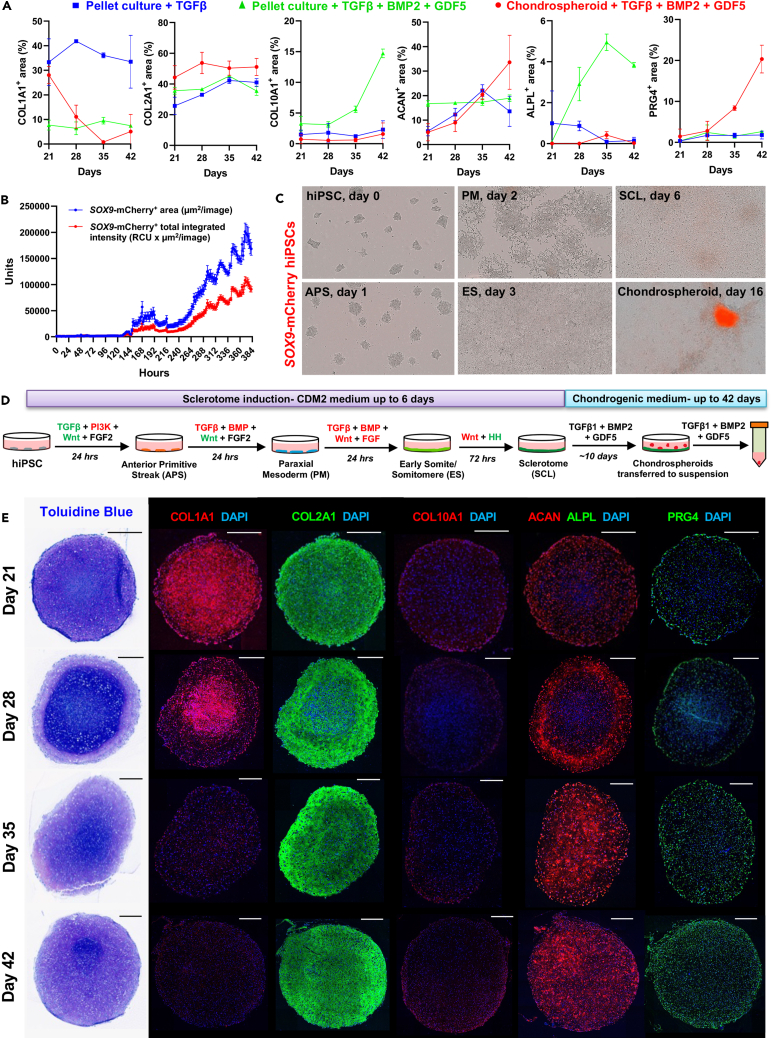
Based on the mechanistic knowledge that we gained from the HyA-FMB model system in vitro and in vivo transplants in immunocompromised mice, we sought to establish a serum-free hiPSC differentiation protocol that employs BMP inhibition during initial differentiation, followed by BMP activation in early chondrogenic cells, with the goal of obtaining a stable chondrocyte-like cell for clinical translation. First, we employed an initial 6-day sclerotome induction period in hiPSCs in which BMP signaling was inhibited, as previously proposed in human ESCs,36 and confirmed transcriptional identity of resulting anterior primitive streak, paraxial mesoderm, early somite/somitomere, and sclerotome cells (Figures S4A–S4D). Since sclerotome cells are enriched for IGFBP5 (as shown below), mimicking its expression in early organoids with HyA-FMBs in vitro when BMP signaling was low, our next approach was to activate BMP signaling in a chondrogenic induction protocol, based on the fact that we saw BMP2 expression in the transcriptome analyses, and we observed pSMAD5 in maturing cartilage in vitro and in vivo. We also included GDF5 because it is known that it signals through the same receptors as BMP2, but it is also known that GDF5 prefers BMPR1B over BMPR1A, which is preferred by BMP2. Consequently, we included it because it has been reported to be important for condensation of chondrogenic progenitors and enhances chondrogenic differentiation of hBMSCs/SSCs. As a negative control, supplementation of sclerotome-derived pellet cultures with TGF-β1 alone led to stable COL1A1 and COL2A1 expression across 42 days, with sporadic ACAN and minimal PRG4 expression (Figures S4E and 5A, blue lines); these data confirm incomplete chondrogenesis in cultures treated with only TGF-β8 and no HyA-FMBs (see Figure 3). We then formed pellet cultures from sclerotome cells and supplemented them with TGF-β1 and the BMP signaling activators, BMP2 and GDF5, which produced strong COL2A1 and ACAN expression, but a progressive increase in COL10A1 and ALPL across 42 days (Figures S5A and 5A, green lines), suggesting improved chondrogenic differentiation but increased hypertrophy of hiPSC-derived chondroprogenitors. Since BMP signaling is restored in chondrogenic-enriched cell populations in the development of hypertrophic-resistant chondrocytes (see Figure 3), we hypothesized that targeted BMP activation in a purified, early chondrogenic subset of cells would produce a more homogeneous and stable chondrogenic phenotype (compared with broad BMP activation of the entire culture). To select for an early chondrogenic subset of cells, we treated monolayer sclerotome cultures derived from SOX9-mCherry hiPSCs with chondrogenic medium supplemented with TGF-β1, BMP2, and GDF5. Incucyte imaging revealed a progressive increase in SOX9-mCherry area and intensity in sclerotome cultures across 10 days of chondrogenic induction (Figure 5B). Interestingly, groups of SOX9-mCherry+ cells began to condense after 6 days of chondrogenic induction in sclerotome cultures, similar to the condensation observed during in vivo limb development, and formed SOX9-mCherrybright aggregates that detached around 10 days, whereas surrounding adherent cells showed little to no SOX9-mCherry expression (Figure 5C and Video S1). Therefore, we transferred these SOX9-mCherrybright aggregates—hereafter termed chondrospheroids—to suspension cultures and continued BMP signaling activation selectively in this subgroup of early chondrogenic cells (see full chondrospheroid protocol in Figure 5D). Chondrospheroids transiently expressed COL1A1, followed by expression of strong COL2A1, ACAN, and PRG4 expression, starting from the exterior and moving inward, with little to no COL10A1 expression (Figure 5E); chondrospheroid formation and expression patterns were confirmed in an additional hiPSC line and quantified over time (Figure 5A, red lines). Overall, non-selective BMP activation in all sclerotome cells induced hypertrophic chondrogenesis, yet selective BMP activation in a SOX9+ chondrogenic subset induced stable chondrogenesis in vitro.
Figure 5.
BMP activation in SOX9+-purified early chondrogenic cells promotes stable chondrogenesis in vitro
(A) Quantification of protein expression in tissues derived from three chondrogenic differentiation strategies: iPSC-derived sclerotome cells were pelleted in chondrogenic medium supplemented with TGF-β (blue) or TGF-β, BMP2, and GDF5 (green) or formed chondrospheroids that were cultured in chondrogenic medium supplemented with TGF-β, BMP2, and GDF5 (red). n = 2–4 replicates. Data are mean ± SD. Confocal images associated with these area quantifications are shown in Figures 5E, S4E, and S5A.
(B) SOX9-mCherry culture area (blue) and fluorescence intensity (red) of SOX9-mCherry hiPSCs across 16 days of differentiation on monolayer cultures from the protocol depicted in Figure 5D, measured by Incucyte analysis software. Sum of three independent experiments.
(C) Representative brightfield and mCherry fluorescence images depicting cell morphology across differentiation, and appearance of SOX9-mCherrybright chondrospheroid by 16 days of adherent culture. Abbreviation names are shown in Figure 5D.
(D) Schematic of sclerotome and chondrogenic differentiation strategy from hiPSCs: pathway activators (green), inhibitors (red), and recombinant growth factors (black) were added across 6 days of adherent culture, followed by chondrogenic induction to produce chondrospheroids.
(E) Toluidine blue staining and immunofluorescence analyses of chondrospheroids across time. Nuclei counterstained with DAPI. Scale bars, 300 μm (see also Figures S4 and S5).
A serum-free chondrogenic differentiation strategy beginning with sclerotome induction. Brightfield and mCherry live cell Incucyte imaging of SOX9-mCherry hiPSC differentiation from day 0 to day 16 of chondrospheroid protocol outlined in Figure 5D.
BMP2 and GDF5 supplementation enhance chondrogenesis in iPSC-derived sclerotome
To confirm the role of BMP activation in strengthening the chondrogenic phenotype, we differentiated hiPSCs to sclerotome and treated sclerotome cells with either TGF-β alone or in combination with BMP2 and GDF5, using the chondrospheroid protocol, followed by scRNA-seq. At 42 days of differentiation, cluster analyses of integrated datasets showed three chondrogenic populations, 1 cycling population, and 5 off-target populations with tendon and neuronal expression markers (Figures S5B and S5C). Seventy-six percent of cultures treated with TGF-β along with BMP2 and GDF5 were composed of chondrogenic and cycling populations compared with 54% of cultures treated with TGF-β alone (Figure S5D). Further, supplementation of TGF-β along with BMP2 and GDF5 promoted elevated expression of chondrogenic markers (i.e., ACAN, MGP, COMP, decorin (DCN), MATN3, COL2A1; Figure S5E), indicating enhanced chondrogenic differentiation compared with cultures treated with TGF-β alone.
Chondrospheroid transcriptomes show efficient chondrogenic differentiation
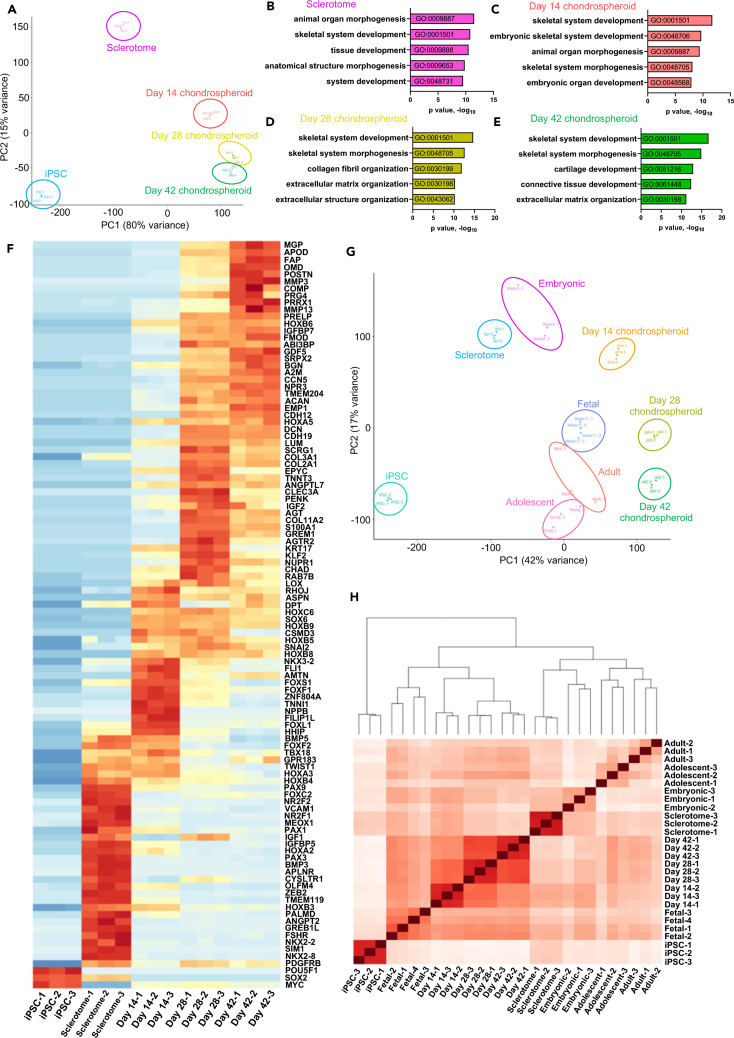
We next performed bulk RNA sequencing of sclerotome cells and sclerotome-derived chondrospheroids at days 14, 28, and 42 of differentiation. Principal-component analysis (PCA) (Figure 6A) and hierarchical clustering (Figure S6A) showed similarities among chondrospheroids on days 14, 28, and 42, with clear differences between chondrospheroids and undifferentiated iPSCs and sclerotome cells. Gene ontology (GO) analysis of sclerotome cells enriched for genes associated with tissue development and organ morphogenesis (Figure 6B), with high expression of FOXC2, IGFBP5, mesenchyme homeobox 1 (MEOX1), PAX1, and PAX9 (Figure 6F). Additionally, many primitive hBMSC/SSC markers were highly expressed in sclerotome cells—such as C-X-C motif chemokine ligand 12 (CXCL12), leptin receptor (LEPR), and platelet-derived growth factor B (PDGFRB)—whereas others peaked later in differentiation, including CD146 (melanoma cell adhesion molecule/MCAM) and GREM1 (Figures 6F and S6B). Several homeobox (HOX) family genes were differentially expressed early in chondrogenic differentiation, as day 14 chondrospheroids were enriched for genes associated with embryonic skeletal system and organ development (Figures 6C and 6F). Day 28 and 42 chondrospheroids, which were the most similar, were enriched for genes involved in skeletal system development and morphogenesis; however, day 28 chondrospheroid expression showed an enrichment for collagen and extracellular matrix organization (Figure 6D), evidenced by transiently high expression of COL2A1, COL3A1, COL9A1, COL11A1, and COL11A2 (Figures 6F and S6B). In contrast, day 42 chondrospheroids showed enrichment for genes involved in cartilage and connective tissue development (Figure 6E), showing higher expression of COMP, GDF5, MGP, and PRG4 and sustained expression of ACAN, DCN, DPT, fibromodulin (FMOD), and IGFBP7 (Figure 6F). Notably, these transcriptional profiles matched protein expression (see Figures 5A and 5E) with early, transient COL1A1 expression; progressive increased expression of ACAN and PRG4; uniformly low expression of COL10A1 (with the exception of one replicate); and absence of ALPL expression (Figures 6F and S6B). BMP activation in chondrospheroids led to the induction of ID genes with low expression of osteogenic genes (COL1A1, SPP1, IBSP, ALPL) similar to hBMSC/SSC-derived chondrocytes in stable hyaline-like cartilage formed subcutaneously (see Figures 4A–4E). Taken together, these results provide transcriptional confirmation of sclerotome and chondrogenic identity using a serum-free chondrospheroid differentiation protocol with selective, timed BMP activation.
Figure 6.
Chondrospheroid transcriptomes reveal a fetal-like chondrogenic identity
(A) PCA of hiPSC, sclerotome, and chondrospheroid datasets. Each dot represents a technical replicate.
(B–E) Gene ontology analysis representing top five pathways from sclerotome (B), day 14 (C), day 28 (D), and day 42 (E) chondrospheroids.
(F) Heatmap depicting differential gene expression: top 100 differentially expressed genes from each dataset were assessed against GO pathways listed in (B)–(E), reducing the list to 103 relevant genes.
(G) PCA of hiPSC, sclerotome, and chondrospheroid datasets, which were batch-corrected and normalized using ComBat-seq to previously published datasets,42 which included 5- to 6-week-old embryonic, 17-week-old fetal, adolescent, and adult primary human chondrocytes.
(H) Hierarchical clustering analysis of sclerotome, chondrospheroid, and primary human chondrocyte42 populations (see also Figure S6; Table S2. Excel file containing additional data too large to fit in a PDF, related to Figure 6G, H, Table S3. Excel file containing additional data too large to fit in a PDF, related to Figure 6G, H, Table S4. Excel file containing additional data too large to fit in a PDF, related to Figure 6G, H).
Chondrospheroid transcriptomes reveal a fetal-like chondrogenic identity
To determine the chondrogenic maturity of cultured chondrospheroids, we normalized and compared our datasets with primary human embryonic (5–6 weeks old), fetal (17 weeks old), adolescent, and adult chondrocytes from a recent study42 using ComBat-Seq (see STAR methods). Both PCA and hierarchical clustering confirmed the transcriptional similarity among adolescent and adult chondrocytes reported in the initial study42 (Figures 6G and 6H). PCA placed embryonic chondrocytes close to the 14 chondrospheroids; fetal chondrocytes were near day 28 and 42 chondrospheroids, with day 42 chondrospheroids approaching adult chondrocyte populations (Figure 6G). Hierarchical clustering grouped sclerotome cells with embryonic chondrocytes and all chondrospheroid datasets with fetal chondrocytes (Figure 6H). Differentially expressed genes among chondrospheroid and human chondrocyte populations are provided in Table S2. Excel file containing additional data too large to fit in a PDF, related to Figure 6G, H, Table S3. Excel file containing additional data too large to fit in a PDF, related to Figure 6G, H, Table S4. Excel file containing additional data too large to fit in a PDF, related to Figure 6G, H. Therefore, the initial chondrogenic differentiation of sclerotome cells mimicked an embryonic chondrogenic state, progressing to a fetal-like state in day 28 and 42 chondrospheroids.
We also sought to compare our chondrospheroids with hESC-derived chondrocytes42 that have recently been shown to repair damaged articular cartilage in pigs with stability for up to 6 months.44 Interestingly, PCA and hierarchical clustering showed transcriptional similarity between chondrospheroids (day 28 and day 42) and hESC-derived chondrocytes (day 60) that have shown stable chondrogenesis in preclinical studies (Figures S6C and S6D).
Chondral transplantation of chondrospheroid cells attached to HyA-FMBs yields stable chondrogenesis
Our final goal was to examine stable chondrogenesis in transplanted chondrospheroids, and we first used subcutaneous transplantation to compare three cell delivery methods (Figure S7A). The first method involved the transplantation of undigested day 35 chondrospheroids, which led to enlarged lacunae and widespread hypertrophy at 2 months (Figures S7B and S7C). The latter two methods utilized enzymatic digestion of day 35 chondrospheroids and transplantation of isolated cells either in Matrigel or with HyA-FMB scaffolds. Chondrospheroid cells transplanted in Matrigel formed a patchy cartilage matrix by 2 months with minimal evidence of hypertrophy (Figures S7D and S7E). In contrast, HyA-FMB transplants with chondrospheroid-derived cells formed a uniform metachromatic stain of cartilage matrix as shown by toluidine blue and COL2A1 staining at 2 months (Figures S7F and S7G), suggesting improved chondrogenesis stability in vivo. Similar to stable ectopic transplants of hBMSC/SSC/HyA-FMB constructs (see Figure 4E), about half (51.71% ± 2.13%) of transplanted chondrospheroids with HyA-FMBs exhibited pSMAD5 immunoreactivity (Figures S7H and S7I), confirming active BMP signaling in stable cartilage.
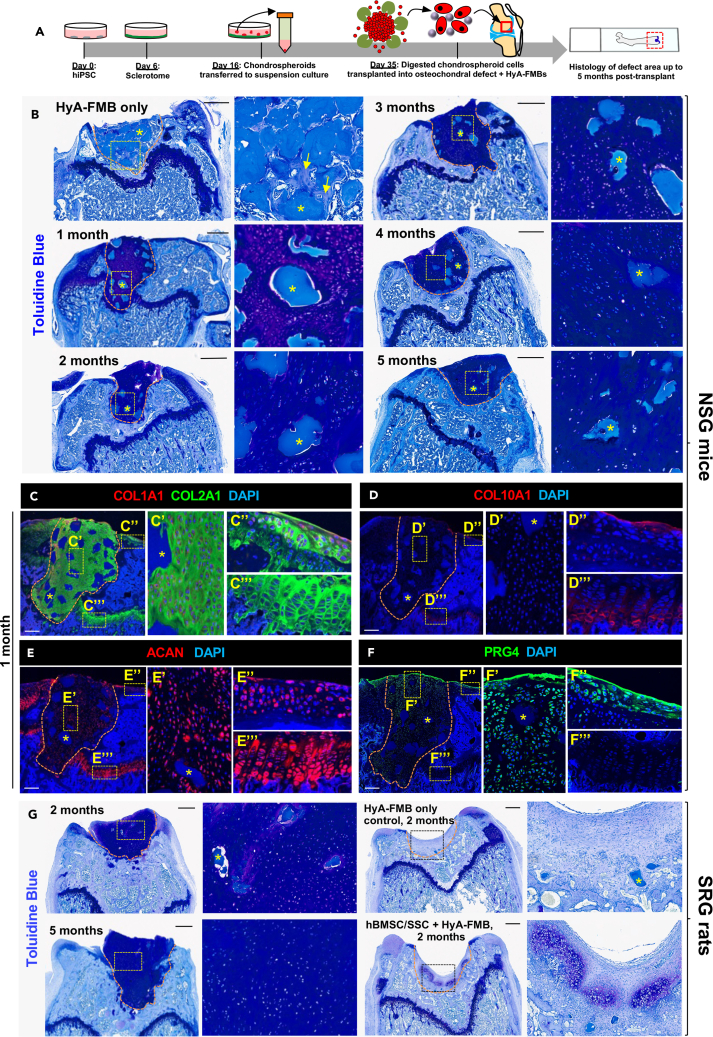
We then transplanted digested chondrospheroid cells attached to HyA-FMBs into articular cartilage defects in vivo (Figure 7A). Day 35 chondrospheroid-derived cells attached to HyA-FMBs formed stable, hyaline-like cartilage for up to 5 months upon chondral transplantation in NSG mice compared with undigested chondrospheroids without Hy-A-FMBs and cell-free controls, which formed bone at the defect site (Figures 7B and S8A–S8C). Of note, the animals were fully ambulatory during the entire period from transplantation to euthanasia. No major differences were noted in the integrity of the transplants from early and late time points. These transplanted human cells, which were confirmed by h-vimentin expression (Figure S8D), exhibited similar protein expression to surface articular chondrocytes and disparate expression to growth plate chondrocytes; i.e., immunofluorescence analyses showed uniform COL2A1 expression, no COL10A1 expression, COL1A1 expression near subchondral bone, widespread ACAN expression, and surface PRG4 expression in transplanted chondrospheroid cells. Although PRG4 may be absorbed to the surface of cartilage from the synovial fluid, cells well below the surface were also PRG4 positive, but to a lesser extent. All in all, these results confirm an articular-like phenotype (Figures 7C–7F and S8E). Similarly, chondral transplantation of day 35 chondrospheroid-derived cells attached to HyA-FMBs in SRG rats produced dense metachromatic staining with toluidine blue at the defect site after 2 and 5 months, which proved superior to the patchy, hypertrophic chondrogenesis and fibrocartilage formation seen with transplantation of hBMSC/SSC/HyA-FMB constructs and the absent chondrogenesis in controls (Figures 7G and S8F; Table S1). Interestingly, unlike the transplantation of hBMSC/SSC/HyA-FMB constructs, the HyA-FMBs were not degraded with time in chondrospheroid-derived cell constructs and were still observed at 5 months in vivo. Overall, these data confirm stable chondrogenesis in two animal models with a defined, serum-free, articular-like cell source obtained from timely and selective BMP activation.
Figure 7.
Chondral transplantation of chondrospheroid cells attached to HyA-FMBs yields stable chondrogenesis
(A) hiPSC-derived sclerotome was differentiated to chondrospheroids, which were digested at day 35, incubated with HyA-FMBs for 2 h, and transplanted into a femoral trochlear groove defect in NSG mice (B–F) and SRG rats (G).
(B) Toluidine blue staining of defect areas from transplanted day 35 chondrospheroids attached to HyA-FMBs at 1–5 months post-transplant in NSG mice. High-magnification insets shown to the right of their corresponding images. Arrows depicting the formation of bone in control transplants at 1 month. Scale bars, 500 μm.
(C–F) Immunofluorescence analyses of femoral defects from NSG mice 1 month post-transplant. High-magnification insets depicting transplanted cells, surface articular cartilage, and growth plate cartilage shown to the right of their corresponding images. Nuclei counterstained with DAPI. Scale bars, 200 μm.
(G) Toluidine blue staining of defect areas from transplanted day 35 chondrospheroids attached to HyA-FMBs at 2 and 5 months post-transplant in SRG rats (left). Right images depict control transplants, including HyA-FMBs only and hBMSCs/SSCs attached to HyA-FMBs at 2 months. High-magnification insets shown to the right of their corresponding images. Scale bars, 500 μm.
(B–G) Asterisks (∗) indicate HyA-FMBs. Orange dashed lines indicate area of transplanted tissue (see also Figures S7 and S8).
Discussion
In this study, we have confirmed our previous data showing that attachment of hBMSCs/SSCs to HyA-FMBs supports the formation of stable articular-like cartilage in subcutaneous transplants. We then utilized hBMSC/SSC cartilage organoids without and with HyA-FMBs in vitro to determine pathways by which HyA-FMBs support stable cartilage. In day 1 and 3 HyA-FMB organoids, increased levels of non-collagenous proteins, including COMP, DPT, IGFBP5, and MGP, and decreased BMP signaling (assessed by pSMAD5 levels) were noted in comparison with control organoids. At day 5 and 10, BMP signaling was apparent in MGP+/IGFBP5+ early chondrogenic cells devoid of hypertrophic and osteogenic markers in HyA-FMB organoids, as well as in 8-week hBMSC/SSC/HyA-FMB transplants in vivo. Using this information, we developed a serum-free method for chondrogenic differentiation of hiPSCs that incorporated inhibition of BMP signaling for the differentiation of sclerotome-like cells, followed by treatment with BMP2/GDF5 in addition to TGFβ, which stimulated BMP signaling and resulted in delamination of Sox9+ chondrospheroids that exhibited high expression of COL2A1, ACAN, and PRG4 with little to no COL10A1, similar to fetal chondrocytes. When chondrospheroid-derived cells were transplanted along with HyA-FMBs into articular defects, the defects were filled with stable cartilage up to at least 5 months in immunocompromised mice and rats, whereas hBMSCs/SSCS/HyA-FMBS only temporarily formed cartilage that went on to undergo hypertrophy and bone formation. Overall, these studies highlight the biphasic nature of BMP signaling in the formation and maintenance of stable cartilage and provide an articular cartilage-like cell source for regenerative OA and cartilage repair therapies.
It is possible that the HyA-FMBs absorbed a particular subset of hBMSCs/SSCs that elicited early changes in the HyA-FMB organoids. However, we hypothesize that the nature of the initial interaction is mediated, in part, by the highly condensed and cross-linked low-heat denatured fibrin, by cell attachment to specific highly conserved C-termini on different fibrinogen chains, termed “Haptides.”47,48 Additionally, hyaluronic acid linked to HyA-FMBs may further enhance cell interaction through the hyaluronic acid receptor, CD44. Our previous study has shown that covalent linkage of hyaluronic acid on HyA-FMBs improves stable chondrogenesis,29 either by binding to CD44, which is abundant on hBMSCs/SSCs, or by an indirect effect whereby physical composition (e.g., stiffness) associated with covalent linkage is altered, as it is known that hBMSC/SSC cell fate is influenced by extracellular matrix composition and substrate stiffness.19,49
HyA-FMBs organoids express MGP, IGFBP5, and DPT after only 1–3 days of TGF-β treatment. MGP prevents extracellular calcium deposition, as Mgp-deficient mice exhibit arterial and cartilage calcification. MGP nonsense mutations in humans with Keutel syndrome results in diffuse cartilage calcification, leading to facial abnormalities and cardiovascular defects.50,51 IGFBP5 is known to regulate chondro-osteogenic cell fate, as high IGFBP5 levels inhibit osteoblast differentiation in C3H10T1/2 cells and promote early chondrocyte differentiation in RCJ cells.52,53 Our studies show that IGFBP5 is induced early in chondrogenic differentiation, labeling an early chondrogenic population, confirming results from other studies,38 with consistent coexpression of MGP. The function of DPT is not well understood in chondrogenesis, though studies in epithelial cells indicate a TGF-β binding site on DPT, which in conjunction with decorin, can modulate TGF-β signaling on target cells.54 Overall, early changes in extracellular matrix composition may influence early hBMSC/SSC cell fate decisions and intracellular signaling.
The best demonstration of hBMSC/SSC chondrogenic capacity in vitro is in pellet cultures supplemented with TGF-β.19 During fetal development, TGF-βs are expressed at the developing joint interzone, stimulate proliferation, inhibit osteogenesis and adipogenesis in hBMSCs/SSCs, and promote chondrogenic cell fate in hBMSCs/SSCs after one day of exposure in vitro.11,55,56,57 In our study, TGF-β alone was insufficient in promoting uniform and stable chondrogenic differentiation of hBMSCs/SSCs and hiPSCs, consistent with previous reports.8 For this reason, many studies have targeted the BMP signaling pathway8,38,40,41; however, there is uncertainty whether BMP signaling must be activated or repressed. Some studies have shown that BMP2 is induced after one day of TGF-β exposure in hBMSCs/SSCs micropellet cultures, suggesting that it may be skewing differentiation to hypertrophy and osteogenesis.55 Other studies have shown that BMP4 promotes hypertrophic differentiation, leading some to conclude that BMP activation should be avoided in chondrogenic differentiation.33,41 While blocking BMP signaling hampers mineralization in osteogenic and chondrogenic cultures treated with β-glycerophosphate,58 chronic BMP blockade using LDN 193189 does not prevent hypertrophic expression in hBMSC/SSC micropellet cultures.59 Our study suggests that BMP inhibition may be time-sensitive, as HyA-FMB organoids show overall BMP inhibition during the first 3 days of chondrogenic induction, followed by an expression signature indicating restored BMP signaling in early chondrogenic cells at days 5 and 10. Consistent with these results, transient and early BMP inhibition using an engineered compound that targets ALK2 and ALK3 receptors and downstream Smad1/5/8 signaling reduces hypertrophic expression in vitro and bone formation in vivo.60
GDF5 has become a primary marker in identifying the joint interzone61 and has been shown to induce chondrogenesis. GDF5 is less potent in inducing hypertrophy compared with BMP2 based on its preferential binding to BMPR1B, a marker for articular chondrocyte progenitors.17,40,62 However, while BMP2 signaling through BMPR1A can promote hypertrophy and endochondral ossification, loss of BMPR1A leads to severe articular cartilage degeneration and fibrillation, suggesting a potential role of BMP2 in articular cartilage maintenance.63,64 Our studies show increased levels of BMP2 and downstream ID genes in engineered stable, hyaline-like cartilage, suggesting the importance of BMP signaling in the later stages of articular cartilage maturation and maintenance.
These mechanistic studies corroborate current understanding of articular cartilage formation during joint development. Cells located in the distal proliferative zone beneath the joint interzone13 are highly exposed to the BMP inhibitor, Noggin, before migrating to the presumptive joint surface where they are primed as early chondrocytes and exposed to GDF5, promoting the maturation and maintenance of early articular chondrocytes.10 This is similar to our process for differentiating iPSCs into stable cartilage. After initial inhibition, we determined that selective BMP activation in SOX9+ cells led to cell aggregation, as seen during limb development, confirming our data generated by the HyA-FMB organoids. The early chondrogenic populations are enriched for IGFBP5 and SOX9,65 which have been shown to prevent BMP2-mediated osteogenic transcription.53,66,67 This pattern is followed by expression of ACAN, COL2A1, COMP, DCN, FMOD, GDF5, MGP, matrix metalloproteinase 3 (MMP3), and PRG4 in day 28 and 42 chondrospheroids. Thus, it would be interesting to explore whether an altered response to BMP signaling could precede articular cartilage degeneration in OA, since SOX9 and IGFBP5 expression in articular cartilage decrease with advanced age and OA.68,69
Previously we showed that subcutaneous transplantation of naive hBMSCs/SSCs attached to HyA-FMBs produced stable, hyaline-like cartilage.29 However, transplantation of hBMSC/SSC/HyA-FMB organoids into injured cartilage led to the formation of bone. It was also noted that in this scenario, the HyA-FMBs rapidly disappeared. Likewise, transplantation of undigested hiPSC-derived chondrospheroids led to progressive hypertrophy by 2 months but adding freshly digested chondrospheroid-derived cells to HyA-FMBs prior to transplantation produced stable articular chondrocyte-like cells for up to 5 months. These results emphasize the role of the HyA-FMBs in stable chondrogenesis in vivo and demonstrate superior cartilage formation when single cells freshly attached to HyA-FMBs are transplanted rather than organoids incubated in vitro. Further, larger organoids often contain diffusion gradients, leading to differences in signaling, extracellular matrix composition, and cell phenotype across the tissue.70,71 For this reason, digested cells or smaller microtissues with more homogeneity, similar to the microwell-mesh system previously described,71 may provide superior clinical efficacy and reproducibility. Finally, transplantation of hiPSC-derived chondrospheroid cells proved superior to hBMSC/hSSC-derived cells, which failed to form stable cartilage in the articular defect. This suggests that the predisposition of hBMSCs/SSCs toward an osteogenic fate is not present in hiPSC-derived cells. Additional studies are needed to determine the cellular and molecular events that are associated with priming of hBMSCs/SSCs toward ossification and how they can be overcome to achieve stable cartilage.
A successful pre-clinical hiPSC-cartilage regenerative therapy would rely on using defined factors and resisting hypertrophy in long-term transplantation studies in vivo. One group has performed extensive transcriptional screening of an articular protocol based on knowledge of human development.17,42 They have demonstrated stable chondrogenesis from ESC-derived chondrocytes,44 which show transcriptional similarity to our chondrospheroids; thus, our studies provide a hiPSC alternative approach. A serum-free protocol using both hiPSCs and hESCs has been reported, generating stable hESC-derived cartilage; however, the hiPSC-derived cartilage showed signs of hypertrophy,41 and their subsequent transplantation studies have focused only on hESCs.43 Other groups have shown resistance of their cells to hypertrophy in vivo but are limited by the use of serum and short follow-up periods.38,39,40 Thus, our data show that TGFβ/BMP2/GDF5 promote efficient chondrogenesis in SOX9+ chondrospheroid-derived cells in serum-free conditions, which promote stable cartilage at the joint surface up to 5 months upon attachment to HyA-FMBs. These data support the potential for clinical utility in cartilage repair and regeneration within the context of OA as a cell therapy.
Limitations of the study
While the results presented here are encouraging, there are a number of limitations to this study. We established an in vitro system to examine the mechanism by which HyA-FMBs facilitate formation of stable cartilage, since transcriptomic studies of in vivo HyA-FMB transplants are limited by the lack of appropriate controls; i.e., scaffold-free cells do not survive, and Matrigel would add confounding factors (due to presence of growth factors and soft mechanical influence) for sequencing analyses. However, the in vitro approach does not perfectly mimic their in vivo properties, as demonstrated by expression of COL10A1 in vitro but not in vivo. However, the disappearance of COL10A1 in MGPhi/IGFBP5hi early chondrogenic cells by day 10 of chondrogenic differentiation suggests potential transient hypertrophic expression in vitro. To best address this limitation, we performed transcriptomic measurements from hBMSC/SSC/HyA-FMB transplants and compared results with in vitro studies. An additional limitation includes the lack of stability of HyA-FMBs in chondral transplants with hBMSCs/SSCs. There are a number of potential reasons that could be addressed such as increasing the stability of the HyA-FMBs in the injured joint, modifying the hostile environment, or modifying the hBMSCs/SSCs. By comparing the transcriptomes of the less than successful hBMSCs/SSCs with the successful iPSC-derived chondrospheroid cells, more clues as to which gene regulatory networks are at play in the formation of stable cartilage may shed light on how hBMSCs/SSCs can be altered. Finally, the chondral defect model was designed to assess chondrocyte stability at the joint site, yet future studies using more superficial defects (without potential damage to subchondral bone) in large animal models could be used to assess surface chondrocyte integration and stability with greater weight-bearing forces. In spite of these limitations, our study provides a process by which hiPSCs can be used to generate stable cartilage in cartilage defects and suggests that modifications of hBMSC/SSC processing and/or HyA-FMBs may provide another source of cells for the regeneration of cartilage lost due to trauma and disease.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-MGP | Abcam | Cat. No. ab224367 |
| Rabbit anti-COMP | Abcam | Cat. No. ab231977 |
| Rabbit anti-DPT | ThermoFisher | Cat. No. PA514396; RRID: AB_2094317 |
| Rabbit anti-COL1A1 | Abcam | Cat. No. ab34710; RRID: AB_731684 |
| Mouse anti-COL2A1 | Dev. Studies Hybridoma Bank | Cat. No. II-II6B3; RRID: AB_528165 |
| Rabbit anti-COL10A1 | Kuznetsov et al., 201929 | Made by SBS/NIDCR/NIH |
| Rabbit anti-ACAN | Millipore Sigma | Cat. No. AB1031; RRID: AB_90460 |
| Goat anti-ALPL | ThermoFisher | Cat. No. PA547419; RRID: AB_2609590 |
| Rabbit anti-PRG4 | Abcam | Cat. No. ab28484; RRID: AB_776089 |
| Alexa 488 Goat anti-hVIMENTIN | R&D Systems | Cat. No. IC8104G; RRID: AB_2827831 |
| Alexa Flour 488 Donkey anti-Goat | ThermoFisher | Cat. No. A11055; RRID: AB_2534102 |
| Alexa 488 Donkey anti-Mouse | ThermoFisher | Cat. No. R37114; RRID: AB_2556542 |
| Alexa Flour 546 Donkey anti-Rabbit | ThermoFisher | Cat. No. A10040; RRID: AB_2534016 |
| Biological samples | ||
| Human bone marrow stromal cells/skeletal stem cells | Human bone specimens | see Table S1 |
| NCRM NL5 human induced pluripotent stem cell line | Baghbaderani et al., 201572 | NIH-Center for Regenerative Medicine |
| MIXL1-GFP human induced pluripotent stem cell line | This paper | N/A |
| SOX9-mCherry human induced pluripotent stem cell line | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Fetal Bovine Serum | GeminiBio | Cat. No. 100-50 |
| Dexamethasone | Sigma | Cat. No. D4902 |
| L-ascorbic acid phosphate magnesium salt n-hydrate | Wako | Cat. No. 013-19641 |
| Vitronectin XF | Stemcell Technologies | Cat. No. 07180 |
| Insulin-transferrin-selenium | ThermoFisher | Cat. No. 51500056 |
| Recombinant TGFβ1 | Peprotech | Cat. No. 100-21 |
| Collagenase II | ThermoFisher | Cat. No. 17101015 |
| Hyaluronidase | Merck | Cat. No. H3506 |
| Dispase | Sigma | Cat. No. D4693 |
| Bovine serum albumin | Miltenyi Biotec | Cat. No. 130-091-376 |
| Pepsin | Millipore Sigma | Cat. No. R2283 |
| Citrate buffer | Millipore Sigma | Cat. No. C9999 |
| Matrigel | Corning | Cat. No. 356237 |
| Polyvinyl alcohol | Sigma | Cat. No. P8136 |
| Lipid concentrate | ThermoFisher | Cat. No. 11905031 |
| Monothioglycerol | Sigma | Cat. No. M6145 |
| Recombinant Activin A | R&D systems | Cat. No. 338-AC-050/CF |
| CHIR99021 (Wnt agonist) | R&D Systems | Cat. No. 4423 |
| Recombinant FGF-2 | R&D Systems | Cat. No. 233-FB |
| PIK90 (PI3K inhibitor) | Millipore Sigma | Cat. No. 528117 |
| SB-431542 (TGFβ inhibitor) | Millipore Sigma | Cat. No. S4317 |
| LDN-193189 (BMP inhibitor) | Sigma | Cat. No. SML0559 |
| C59 (Wnt inhibitor) | Cellagen Technology | Cat. No. C7641-2s |
| PD173074 (FGF inhibitor) | R&D Systems | Cat. No. 3044 |
| Purmorphamine (Hedgehog agonist) | ReproCell | Cat. No. 04-0009 |
| Recombinant BMP-4 | R&D systems | Cat. No. 314-BP-050 |
| Recombinant BMP-2 | Peprotech | Cat. No. AF-120 |
| Recombinant GDF-5 | Peprotech | Cat. No. 120-01 |
| Critical commercial assays | ||
| EndoFree Plasmid Maxi Kit | Qiagen | Cat. No/12362 |
| RNeasy Micro Kit with DNase treatment | Qiagen | Cat. No. 74004 |
| SuperScript III First-Strand Synthesis SuperMix | ThermoFisher | Cat. No. 11752050 |
| Deposited data | ||
| hiPSC RNA sequencing data | GEO | NCBI GEO accession number: GSE242526 |
| Code | Github | GitHub: https://github.com/gadomskisj/Gadomski_et_al_iScience_2024 |
| Experimental models: organisms/strains | ||
| Mouse: NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ | The Jackson Laboratory | Strain No. 005557 |
| Rat: Sprague Dawley-Rag2em2heraIl2rgem1hera/HblCrl | Charles River | Strain No. 400 |
| Oligonucleotides | ||
| Primers for qRT-PCR | U.S. Food and Drug Administration | see Table S2 |
| Recombinant DNA | ||
| Plasmid: pUC57 | Genscript Biotech Corp. | Cat. No. SD1176 |
| Plasmid: LentiCRISPR v2 | Addgene | Plasmid #52961 |
| Software and algorithms | ||
| Cell Ranger version 7.0 | 10x Genomics | RRID:SCR_017344 |
| SoupX (ambient RNA removal) | Young and Behjati, 202073 | RRID:SCR_019193 |
| scDblFinder (doublet removal) | Germain et al., 202274 | RRID:SCR_022700 |
| Seurat (integration and cluster analysis) | Stuart et al., 201945 | RRID:SCR_016341 |
| Clustree (determining cluster resolution) | Zappia & Oshlack, 201875 | RRID:SCR_016293 |
| clusterProfiler (pathway analysis) | Wu et al., 202176 | RRID:SCR_016884 |
| Bioconductor’s edgeR package (RNA sequencing analysis) | Robinson et al., 200977 | RRID:SCR_012802 |
| gProfiler (gene ontology) | Raudvere et al., 201978 | version e107_e.g.,54_p17_bf42210 |
| ComBat-Seq (adjusting batch effects with cross-comparison of published datasets) | Zhang et al., 202079 | RRID:SCR_010974 |
| Incucyte Image Analysis Software | Sartorius, UK | RRID:SCR_017316 |
| ImageJ/Fiji Software | National Institutes of Health | RRID:SCR_002285 |
| Flowjo 10.6 Software | FLOWJO, LLC | RRID:SCR_008520 |
| GraphPad Prism 8 Software | GraphPad Software | RRID:SCR_002798 |
| Other | ||
| HyA-FMBs | Gorodetsky et al., 199926; Kuznetsov et al., 201929 | N/A |
| 0.9 mm and 2.1 mm drill bits | FineScienceTools | Cat. No. 19007 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Pamela G. Robey (probey@dir.nidcr.nih.gov).
Materials availability
-
•
HyA-FMBs were constructed as a modification of the original cell binding condensed fibrin-based microbeads at the laboratory of Prof. Raphael Gorodetsky,26 for better adhesion of hBMSCs/SSCs for chondrogenesis.27 The FMBs were further chemically cross-linked with hyaluronic acid coating to form HyA-FMBs, as previously described.29 The fully detailed HyA-FMBs production protocols are available. HyA-FMBs have a density of 1.2–1.3, and the mean molecular weight of the HyA-FMB polymer is 20,000.
-
•
MIXL1-GFP and SOX9-mCherry hiPSC lines will be made available upon request.
Data and code availability
-
•
Single-cell and bulk RNA sequencing have been deposited at NCBI’s GEO Database and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
All original code has been deposited at Github and is publicly available as of the date of publication. The link is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Animals
Immunocompromised female 3–4-month-old NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, the Jackson Laboratory, Strain#005557) and male and female 20–25-week-old SRG rats (Sprague Dawley-Rag2em2heraIl2rgem1hera/HblCrl, Charles River) were used as recipients for transplant studies. All animal experiments were approved by the National Institutes of Health (NIH)/National Institute of Dental and Craniofacial Research (NIDCR) Animal Care and Use Committee (protocol #19–888 and 19–889) in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Human BMSC/SSC cultures
Human bone specimens were obtained from surgical waste in accordance with NIH regulations from four donors under the OHSR exemption #373 and #170 NIDCR-00183 (Table S5). Primary hBMSCs/SSCs were isolated and cultured, as previously described.29 Briefly, human bone marrow fragments were removed and placed in α-modified Minimum Essential Medium (αMEM, ThermoFisher, Cat. No. 12571-063), followed by repeated pipetting and mechanical digestion through 16 and 19 G needles (Becton Dickinson). Cell suspensions were filtered through 70 μm cell strainers (Biologix, Cat. No. 15–1070) and plated at a density of 2-6.7 × 104 cells/cm2 in αMEM supplemented with 20% non-heat-inactivated fetal bovine serum (FBS, pre-selected lot, GeminiBio, Cat. No. 100-50), 10 nM dexamethasone (Sigma, Cat. No. D4902), 25 μg/mL L-ascorbic acid phosphate magnesium salt n-hydrate (Wako, Cat. No. 013–19641), and 1% penicillin/streptomycin (ThermoFisher, Cat. No. 15070-063). hBMSC/SSC cultures were incubated at 37°C, 5% CO2 with biweekly medium replacements, and passaged using 0.05% Trypsin-EDTA (ThermoFisher, Cat. No. 25300-054).
Human iPSC cultures
hiPSCs were derived from CD34+ peripheral blood cells using episomal reprogramming (NL5, NCRM-5, fetal male), NIH Center for Regenerative Medicine, Bethesda, MD).72 A SOX9 mCherry reporter hiPSC line (described below) was also used for differentiation experiments. hiPSCs were cultured on 6-well plates treated with 10 μg/mL Vitronectin XF (Stemcell Technologies, Cat. No. 07180) overnight at 37°C and maintained feeder-free in Essential 8 medium (ThermoFisher, Cat. No. A1517001). Spontaneous differentiation was prevented by timely passaging and the addition of 50 μm ROCK Inhibitor (Y-27632, Sigma, Cat. No. SCM075) to E8 medium during passage. hiPSC cultures were incubated at 37°C, 5% CO2 with daily medium replacements, and passaged as aggregates using 0.5 mM EDTA/PBS solution.
Method details
Generation of chondrogenic organoids
To generate control organoids, 2.5 × 105 hBMSCs/SSCs (passage 2) were suspended in 14 mL polyethylene conical tubes with 1 mL chondrogenic medium, which consisted of high glucose Dulbecco’s Modified Eagle Medium (DMEM) with sodium pyruvate (ThermoFisher, Cat. No. 11995073), 1% insulin-transferrin-selenium (ThermoFisher, Cat. No. 51500056), 100 nM dexamethasone, 50 μg/mL L-ascorbic acid phosphate magnesium salt n-hydrate (Wako, Cat. No. 013–19641), 40 μg/mL L-Proline (Sigma, Cat. No. P5607), 1% penicillin/streptomycin, and 10 ng/mL TGFβ1 (Peprotech, Cat. No. 100-21). The same process was employed to generate HyA-FMB organoids, except each conical tube contained 3 mg of pre-sterilized HyA-FMBs (70% ethanol washes overnight), which were constructed according to established protocols.29 The ratio of cells to scaffold was approximately the same as our previous in vivo study.29 For both control and HyA-FMB organoids, each tube was centrifuged at 193xg for 6 min, then incubated with loosened caps at 37°C, 5% CO2 with medium replacements 2-3x/week.
Tissue processing and histological staining
Tissue processing
hBMSC/SSC and hiPSC-derived tissues from in vitro pellet culture studies were fixed in 4% formaldehyde at room temperature with mixing for 2 h, then embedded in 3% agarose disks and placed in 70% ethanol for paraffin embedding. In vivo mouse subcutaneous and chondral transplants were harvested and fixed overnight in 4% formaldehyde at 4°C with mixing and demineralized with 250 mM EDTA/dH2O solution (Quality Biological, Cat. No. 351-027-101) for 1.5–2 weeks at 4°C with mixing. Rat chondral transplants were fixed for 2 days and demineralized for 4 weeks in the same conditions. Transplants were stored in 70% ethanol at 4°C and embedded in paraffin within one week.
Toluidine blue staining
Tissues were sectioned at 6 μm and rehydrated in washes of xylene (2×) for 5 min, 100% ethanol (2×) for 2 min, 95% ethanol (2×) for 2 min, and tap water (1×) for 2–3 min. Then, tissues were stained with 0.1% w/v Toluidine Blue Stain Solution (Mercedes Scientific, Cat No. EKI14949250ML, pH 4.148) for 4 min, rinsed with tap water (1×) for 1 min, rinsed with distilled water (1×) for 1 min, and allowed to air dry before placing in xylene for mounting with Optic Mount I (Mercedes Scientific, Cat. No. MER7722).
H&E staining
Rehydrated tissues were treated with the following reagents (all from Fischer Scientific): Hematoxylin 2 (Cat. No. 22050113)(1 × 3 min), tap water (2 × 2 min), Clarifier 2 (Cat. No. 22050117)(1 × 1 min), tap water (2 × 1 min), Bluing reagent (Cat. No. 22050114)(1 × 1 min), tap water (1 × 1 min), 95% ethanol (1 × 1 min), Eosin Y with Phloxine (Cat. No. 22050198)(1 × 40 s), 95% ethanol (2 × 10 dips), 100% ethanol (1 × 2 min), xylene (2 × 3 min), and mounting with Optic Mount I.
Single-cell RNA sequencing and analysis
Passage 2 hBMSCs/SSCs from donors 1–2 (see Table S5) were used to generate organoids for single cell RNA sequencing studies, and were shown to form bone, hematopoietic supporting-stroma, and adipocytes upon ectopic transplantation. Control and HyA-FMB organoids from days 1–10 and ectopic HyA-FMB transplant were digested in high-glucose DMEM containing 0.2% collagenase II (ThermoFisher, Cat. No. 17101015), 0.1% hyaluronidase (Merck, Cat. No. H3506), and 0.1% dispase (Sigma, Cat. No. D4693) with gentle pipetting every 20–30 min. Enzymatic digestion was observed in 6 well-plates, and digestion was terminated when aggregates were completely dissolved, for up to 2 h. Enzyme was inhibited with 20% FBS, and cells were filtered through 70 μm strainers and re-suspended in PBS supplemented with 0.04% high-quality bovine serum albumin (BSA, Miltenyi Biotec, Cat. No. 130-091-376) at 1000 cells/μL for single-cell capture. Only samples with >95% viability were submitted for single-cell capture, as measured by Vi-CELL BLU Cell Viability Analyzer (Beckman Coulter). Naive hBMSC/SSC controls (from donor 3) required a 10-min incubation with collagenase II on monolayer cultures prior to cell detachment and cell capture. Single-cell RNA sequencing was performed using 10× Genomics Chromium instruments. Reads were mapped using GRCh38-2020-A reference transcriptome, and libraries were processed using Cell Ranger version 7.0. The following quality control measurements were performed: Ambient RNA removal using SoupX,73 cell cycle regression using Seurat’s Cell Cycle Scoring (https://satijalab.org/seurat/articles/cell_cycle_vignette.html), doublet identification and removal using scDblFinder,74 and removal of low-quality or dead cells that expressed fewer than 150–200 genes and >10–15% mitochondrial reads. Captures from the HyA-FMB ectopic transplant contained an abnormally high number of doublets, so only global transcriptional measurements were described from this dataset. The merging of datasets using anchoring techniques (https://satijalab.org/seurat/articles/integration_introduction.html), dimensionality reduction, cluster analysis and UMAP projection was performed using Seurat’s R package with cluster resolution determined by Clustree.75 Differentially expressed genes, which represented 25% of each dataset (logfc>0.25, p < 0.05), were analyzed using gProfiler (https://biit.cs.ut.ee/gprofiler/gost), and pathway analysis was performed using clusterProfiler and pathview, with focused analysis on KEGG signal transduction pathways (FDR<0.20).76 Raw data are deposited in the Gene Expression Omnibus (GEO).
Immunofluorescence and confocal imaging
Staining procedure
Paraffin-embedded tissues were used for immunofluorescence. Tissues were sectioned at 6 μm and heated for 1 h at 60°C, then rehydrated in 2 washes of xylene (5 min), 100% ethanol (5 min), 95% ethanol (5 min), 70% ethanol (5 min), and distilled water (5 min). Two different antigen retrieval methods were used: (1) for staining with COL1A1, COL2A1, COL10A1, PRG4, and hVIMENTIN, tissues were treated with 2 mg/mL hyaluronidase/PBS solution for 1 h at 37°C, followed by a PBS wash and treatment with Pepsin (Millipore Sigma, Cat. No. R2283) for 5–10 min at 37°C; (2) for staining with MGP, COMP, DPT, ACAN, ALPL, and pSMAD5, tissues were incubated in citrate buffer (Millipore Sigma, Cat. No. C9999) for 1 h at 70°C, followed by a 15-s wash with distilled water. Cytofix/Cytoperm buffer (BD, Cat. No. 554722) was used in some cases to enhance intracellular staining. Tissues were blocked in staining solution for 30 min in PBS supplemented with 5% donkey serum (Sigma, Cat. No. D9663) and 0.5% IgePal (Sigma, Cat. No. I3021), then stained with primary antibodies overnight at 4°C. On the following day, tissues were washed in PBS (4x for 5 min) and incubated with secondary antibodies for 1 h at room temperature, followed by a PBS wash and mounting with Prolong Gold Antifade Mountant with DAPI (ThermoFisher, Cat. No. P36931). For negative controls, non-immune antibodies of the same isotype and host species as primary antibodies were incubated overnight at 4°C, followed by secondary staining. The following primary antibodies were used: Rabbit anti-MGP (Abcam, Cat. No. ab224367), Rabbit anti-COMP (Abcam, Cat. No. ab231977), Rabbit anti-DPT (ThermoFisher, Cat. No. PA514396), Rabbit anti-COL1A1 (Abcam, Cat. No. ab34710), Mouse anti-COL2A1 (Developmental Studies Hybridoma Bank, Cat. No. II-II6B3), Rabbit anti-COL10A1 (synthesized in the Skeletal Biology Section in consultation with Dr. Larry W. Fisher, according to Kuznetsov et al., 2019), Rabbit anti-ACAN (Millipore Sigma, Cat. No. AB1031), Goat anti-ALPL (ThermoFisher, Cat. No. PA547419), Rabbit anti-PRG4 (Abcam, Cat. No. ab28484), Rabbit anti-Phospho-Smad5 (Ser463, Ser465; Bioss, Cat. No. BSM-52206R), and Alexa 488 Goat anti-hVIMENTIN (R&D Systems, Cat. No. IC8104G). The following secondary antibodies were used: Alexa 546 Donkey anti-Rabbit (ThermoFisher, Cat. No. A10040), Alexa 488 Donkey anti-Mouse (ThermoFisher, Cat. No. R37114), and Alexa 488 Donkey anti-Goat (Abcam, Cat. No. ab150129).
Confocal imaging
Tiled z-stacks were acquired using the Zeiss LSM 880 confocal microscope with Airyscan using the 20× objective and 20% overlap. Maximum intensity projections were merged in Fiji/ImageJ (National Institutes of Health) for image analysis and quantifications. Second harmonic generation images were acquired using a Nikon A1R + MP, two photon, resonant scanner with the 40× lens water objective. Live-cell imaging of SOX9-mCherry hiPSCs was performed using Essen Incucyte Zoom S3 with 2018B software.
qRT-PCR and RNA sequencing
qRT-PCR
Lysates were processed according to Qiagen’s RNeasy Mini Kit with DNAse treatment (Cat. No. 74004), and RNA quality and concentration was measured using a NanoDrop. cDNA was constructed using SuperScript III First-Strand Synthesis SuperMix (ThermoFisher, Cat. No. 11752050) with no Reverse Transcriptase added to negative controls. Samples were measured in triplicate using the Quantstudio 6 Flex system (Applied Biosystems), and 2−ΔCT values were calculated using GAPDH as the housekeeping gene. Primer sets used for qRT-PCR are listed in Table S6.
RNA sequencing
RNA from hiPSCs, sclerotome cells, and chondrospheroids (days 14, 28, and 42) was isolated using Qiagen’s RNeasy Mini Kit with DNAse treatment. mRNA libraries were prepared using the polyA TruSeq method (Illumina) and sequenced on an Illumina NextSeq500 configured for 100 paired-end reads. FASTQ files were pre-processed using the snakemake/5.6.0 utility and aligned using the STAR v2.7.3a aligner with mapping parameters derived from the GENCODE project. All samples had >50% alignment with <1% reads mapping to ribosomal transcripts. Sequencing depth at 50 million reads was shown to be sufficient in saturation curves. Quantification, normalization, PCA, hierarchical clustering, and differential expression analysis were performed using Bioconductor’s edgeR package.77 Top 100 differentially expressed genes from each dataset were assessed against top 5 gene ontology pathways in gProfiler,78 which reduced the list to 103 relevant genes for plotting using the heatmap.2 function (https://cran.r-project.org/web/packages/gplots/index.html). The ComBat-Seq method79 was employed to generate batch-corrected expression matrices for normalization of previously published datasets42 with chondrospheroid datasets. Briefly, a combined gene expression matrix was assembled from ours and previously published datasets and then submitted to the ComBat-seq method after specifying one batch per study. This allows the independent modeling the variance of gene expression at the study level and then the creation of a batch-corrected raw counts expression matrix that was submitted to downstream (PCA) analysis. Of note, a comparison of the single-end sequenced human chondrocyte populations with paired-end sequenced chondrospheroids could create some technical variation. Raw data are deposited in the Gene Expression Omnibus (GEO).
In vivo transplantation
Ectopic transplantation
Subcutaneous transplantation in male NSG rats were performed according to an established method19,29 under an NIDCR ACUC-approved protocol. Briefly, the dorsal skin of the back was shaved and sterilized using alternating Betadyne and ethanol scrubs. Then, a 1.5 cm incision was made on the back, and hBMSC/SSC- and hiPSC-derived cells were placed in a subcutaneous pocket using a sterile spatula, with up to 4 transplants per mouse. Three methods of cell delivery were used for day 35 chondrospheroids. The first method involved the placement of undigested chondrospheroids directly into the subcutaneous pocket. The latter two methods involved the digestion of chondrospheroids to liberate cells using 0.2% collagenase II and 0.1% dispase in high glucose DMEM for up to 2 h with gentle pipetting every 30 min, after which cells were washed and filtered through 70 μm strainers. The second approach involved the re-suspension of ∼1.7 × 106 cells in 100 μL Matrigel (Corning, Cat. No. 356237). The final approach involved incubation of ∼1.7 × 106 cells with 10 mg pre-sterilized HyA-FMBs for 1.5 h at 37°C with gentle rocking. In both cases, transplants were placed on ice for ∼1–2 h during transfer and preparation of surgical site, then placed into a subcutaneous pocket. Incisions were closed using resorbable sutures (Ethicon, 5-0 Monocryl, Cat. No. Y303H) in a horizontal mattress pattern and dissected 1–2 months later.
Chondral transplantation
Passage 2 hBMSCs/SSCs from donors 1 and 4 (see Table S5) were detached using Trypsin/EDTA, and 3x106 hBMSCs/SSCs were incubated with 30 mg pre-sterilized HyA-FMBs, according to previous studies.29 In some cases, hBMSCs/SSCs were subjected to chondrogenic differentiation for 10 days, as described above, prior to transplantation of pre-formed organoids. For hiPSC transplants, ∼1.7 × 106 cells were incubated with 10 mg HyA-FMBs, as noted above. Both immunocompromised NSG mice and SRG rats were used for chondral transplantation under a NIDCR ACUC-approved animal protocol, and both left and right knees were used, often with one knee serving as a HyA-FMB only control (no cells) or sham control (no cells or HyA-FMBs). Surgical draping (Vitality Medical, Cat. No. 005714) and autoclaved instruments, as well as alternating Betadyne and alcohol scrubs of the shaved surgical site, were employed to maintain sterility. An incision was made at the medial aspect of the knee through the overlying skin and joint capsule with the mouse in a supine position. Next, the patella was gently dislocated laterally using a 25 G needle, exposing the trochlear groove of the femur, which is then stabilized with large, serrated tweezers clamped to a ring-stand. The Ideal micro-drill (CellPoint Scientific) was used to create a chondral defect using a 0.9 mm drill bit for mice and 2.1 mm drill bit for rats (FineScienceTools, Cat. No. 19007) on the trochlear surface, and a 0.5 mm micro-curette was used to introduce and pressurize hBMSCs/SSCs/HyA-FMB or hiPSC/HyA-FMB constructs into the defect. Finally, the patella was gently relocated; two vertical sutures (Ethicon, 5-0 Vicryl, Cat. No. J303H) were tied at the joint capsule and 2–3 sutures (Ethicon, 5-0 Monocryl, Cat. No. Y303H) were placed in a horizontal mattress pattern in the skin. Heat pads were placed underneath the surgical area and surgical recovery area to maintain core temperature, and animals were administered Buprenorphine (ZooPharm LLC) subcutaneously before surgery and as needed afterward. Femurs were gently dissected at several timepoints and processed, as noted above.
Grading of cartilage and bone formation
Cartilage and bone formation was quantitatively assessed using an established grading system.29 Briefly, cartilage was graded from 0 to 4: 0 indicates no cartilage formation by Toluidine Blue staining, 1 indicates minimal cartilage formation (a single or few small cartilaginous areas), 2 indicates low cartilage formation (cartilaginous areas occupy only a small area), 3 indicates moderate cartilage formation (cartilaginous areas occupy a significant area but less than one-half of transplant), and 4 indicates abundant cartilage formation (mature cartilaginous areas in more than one-half of transplant). Levels of metachromasia were also considered in cartilage grading, as previously described.29 Bone was also graded from 0 to 4: 0 indicates no bone formation by H&E staining, 1 indicates minimal bone formation (a single or a few bone trabeculae), 2 indicates low bone formation (multiple bone trabeculae present in several areas but the new osseous tissue occupies only a small portion), 3 indicates moderate bone formation (bone occupies a significant area, but did not exceed one-half of the transplant), and 4 indicates abundant bone formation (osseous tissue occupies greater than one-half of the transplant).
Generation of reporter hiPSC lines
MIXL1-GFP
The donor plasmid (based on GRCh38.p11, 2017) was synthesized and cloned into Puc57-Kan plasmid by Genscript Biotech Corp. (Piscataway, NJ). The donor contains the MIXL1 left homology arm, exon 2 with TGA stop codon removed and fused to neonGreen, rBGpA, CMVPuro, BGHpA and right homology arm. The donor has 5 bases changed to the 3′UTR to prevent cutting by CRISPR/Cas9 guides as follows with changes underlined: AGTGGATTCTGGGAGAATTCGAGATAAGCTCTGAGAAGCCATGACTGACAGCCTGAGAGA. LentiCRISPR v2 was a gift from Feng Zhang (Addgene plasmid #5296180) and was used as the backbone to create the CRISPR-Cas9 plasmids. The following primers were used to clone the guides targeting the 3′UTR into the lentiCRISPR v2 according to Sanjana et al. (2014): 5′-caccgTGAGGATTCTGGGAGAATTC-3’ (1a-MIXL-CRISPR-3UTR) with 5′-aaacGAATTCTCCCAGAATCCTCAc-3’ (1b-MIXL-CRISPR-3UTR); 5′-caccgAGAATTCGGGATAAGCTCTG-3’ (2a-MIXL-CRISPR-3UTR) with 5′-aaacCAGAGCTTATCCCGAATTCTc-3’ (2b-MIXL-CRISPR-3UTR).
SOX9-mCherry
The SOX9-mCherry donor plasmid (based on GRCh38.p13, 2019) was inserted into Puc57 plasmid by GenScript Biotech Corp. (Piscataway, NJ). The TGA stop codon in exon 3 of the SOX9 coding region was replaced with GGC, followed by mCherry in frame. The synthesized product was inserted into Xba1 to HindIII sites of Puc57. The CMV-puro-BGHpA, flanked by loxP sites (also synthesized by GenScript), was cloned into the Pac1 to Spe1 sites of the SOX9-mCherry donor to confer selection capability and excision if necessary. The donor was modified to prevent cutting by CRISPR/Cas9 guides as follows with changes underlined: TATACGAAGTTATACTAGTGGAGGCCTCCCACGA. LentiCRISPR v2 was a gift from Feng Zhang (Addgene plasmid #5296180). The following primers were used to clone the guides into the LentiCRISPRv2 according to Sanjana et al. (2014): 5′-caccgCAGCTCACTCGACCTTGAGG-3’ (FWD) with 5′-aaacCCTCAAGGTCGAGTGAGCTGc-3’ (REV)[CRISPR Set 1], and 5′-caccgCTTGAGGAGGCCTCCCACGA-3’ (FWD) with 5′-aaacTCGTGGGAGGCCTCCTCAAGc-3’ (REV)[CRISPR Set 2].
Transfection and screening
NL5 hiPSCs were grown in Nutristem (Stemgent) for 1–2 weeks prior to transfection with AMAXA mouse ES transfection kit (A-023), 5 μg donor and 5 μg CRISPR/Cas9 (2.5 μg each). Five to six days after transfection, the colonies were treated with 0.25–0.5 μg/mL puromycin (Thermofisher Scientific) for 2 to 3 days. Colonies were expanded and retreated with 0.25–0.5 μg/mL puromycin for 3 days for additional selection. Individual colonies were picked and expanded in E8 medium. During passaging, some cells from each clone were used to test for the correct insertion of the donor plasmid. The following PCR primers were used to screen for correct insertion: 5′-AAAAGGGGGCTGTCCAGTGTGT-3’ (FWD-SOX9scrn-Ex3-2330, outside SOX9 donor region) with 5′-AGCCCTCCATGTGCACCTTGAA-3’ (REV-mCherryscrn-1045, inside SOX9 donor region); 5′-GAAATTGCATCGCATTGTCTGAGTAGG-3’ (FWD BGHpA, inside MIXL1 donor region) and 5′-5537 TTGCATAGCTGTCCTGCAGG-3’ (3Rev-MX-RHA-scrn, outside MIXL1 donor region).
hiPSC differentiation
Sclerotome differentiation
hiPSCs were passaged at 1:12-1:20 as evenly distributed small aggregates onto Vitronectin-coated 6-well plates. After one day, Essential 8 medium was replaced with CDM2 medium36 which consisted of 50% IMDM medium (ThermoFisher, Cat. No. 31980030), 50% F12 medium (ThermoFisher, Cat. No. 31765035), 1 mg/mL pre-dissolved polyvinyl alcohol (Sigma, Cat. No. P8136), 1% lipid concentrate (ThermoFisher, Cat. No. 11905031), 450 μM monothioglycerol (Sigma, Cat. No. M6145), 1% insulin-transferrin-selenium, and 1% penicillin/streptomycin. Differentiation to sclerotome was achieved using selective pathway activators and inhibitors supplemented in CDM2 medium, according to previous reports.36 At day 1, hiPSC cultures were treated with 30 ng/mL Activin A (R&D systems, Cat. No. 338-AC-050/CF), 4 μM CHIR99021 (R&D Systems, Cat. No. 4423), 20 ng/mL FGF2 (R&D Systems, Cat. No. 233-FB), and 100 nM PIK90 (Millipore Sigma, Cat. No. 528117) for 24 h to induce anterior primitive streak. At day 2, cultures were treated with 4 μM SB-431542 (Millipore Sigma, Cat. No. S4317), 3 μM CHIR99021, 250 nM LDN-193189 (Sigma, Cat. No. SML0559), and 20 ng/mL FGF2 for 24 h to induce paraxial mesoderm. At day 3, cultures were treated with 4 μM SB-431542, 250 nM LDN-193189, 1 μM C59 (Cellagen Technology, Cat. No. C7641-2s), and 500 nM PD173074 (R&D Systems, Cat. No. 3044) for 24 h to induce early somite/somitomere. At days 4–6, cultures were treated with 2 μM purmorphamine (ReproCell, Cat. No. 04–009) and 1 μM C59 for 72 h to induce sclerotome; purmorphamine was shown to be an effective substitute for the commercially synthesized Hedgehog agonist 21K, according to previous reports.38 Each day, cultures were washed with CDM2 medium prior to adding fresh differentiation medium.
Chondrogenic differentiation
At day 7, three chondrogenic differentiation strategies were employed from sclerotome cultures derived from NL5 hiPSC and SOX9-mCherry hiPSC cell lines. The first two strategies involved cell detachment using Accutase, and the pelleting of 5 × 105 sclerotome cells at 193xg in 14 mL conical tubes with 1 mL chondrogenic medium, consisting of high glucose Dulbecco’s Modified Eagle Medium (DMEM) with sodium pyruvate (ThermoFisher, Cat. No. 11995073), 1% insulin-transferrin-selenium (ThermoFisher, Cat. No. 51500056), 100 nM dexamethasone, 50 μg/mL L-ascorbic acid phosphate magnesium salt n-hydrate (Wako, Cat. No. 013–19641), 40 μg/mL L-Proline (Sigma, Cat. No. P5607), and 1% penicillin/streptomycin. In the first approach, pellet cultures were supplemented with 10 ng/mL TGFβ1, and in the second approach cultures were treated with 10 ng/mL TGFβ1, 10 ng/mL BMP2 (Peprotech, Cat. No. AF-120), and 10 ng/mL GDF5 (Peprotech, Cat. No. 120-01); in both cases, medium was changed 2-3x/week. The final approach, which is referred to as the formation of chondrospheroids, involved treatment of monolayer sclerotome cultures with chondrogenic medium supplemented with 10 ng/mL TGFβ1, 10 ng/mL BMP2, and 10 ng/mL GDF5, with daily medium replacements. Following 8–11 days of chondrogenic treatment, developing nodules were loosely adhered or completely detached from monolayer cultures; these chondrospheroids were either gently detached or collected via pipetting and transferred to 14 mL conical tubes with 1 mL chondrogenic medium supplemented with 10 ng/mL TGFβ1, 10 ng/mL BMP2, and 10 ng/mL GDF5, with medium replacements 2-3x/week. We found that reproducibility of this protocol is especially dependent on the initial plating density. Plating of hiPSCs at 1:12 at day 0 led to the formation of more developed chondrospheroids, but risks delamination of the entire cell layer during chondrospheroid formation. Timely collection of chondrospheroids is essential in this case (prior to delamination); chondrospheroids can also be collected and separated from delaminated cultures by gentle pipetting. Plating of hiPSCs at 1:20 at day 0 leads to smaller chondrospheroids but reduces the chance of cell layer delamination during chondrogenic differentiation. Tissues were collected at several timepoints and processed for histology, sequencing, and transplantation, as noted above.
Flow cytometry
For MIXL1-GFP measurements, day 1 anterior primitive streak cells were detached using Accutase and resuspended in PBS supplemented with 0.2% BSA. Cells were acquired using BD LSR Fortessa1 with analysis using FlowJo (Tree Star).
Quantification and statistical analysis
Area measurements from maximum intensity projections were performed as previously shown.81 Following confocal acquisition, the channel of interest was isolated using “Split Channel” and quantified using “Color Threshold” in Fiji/ImageJ Software (National Institutes of Health). Measurements of both positively stained area and total tissue section area were obtained. Positive staining area was calculated as a percentage of total tissue area, which did not include HyA-FMB particles. Data are shown as mean ± standard error of the mean (SEM) and are representative of at least two independent experiments, unless otherwise noted, with N values representing biological or technical replicates, which are outlined in the figure legends. Unpaired two-tailed t tests were used for two-group comparisons with α = 0.05. Significant differences between groups were indicated as: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Statistical analyses and graphics were performed with Microsoft Excel and GraphPad Prism 8 software.
Acknowledgments
We would like to acknowledge the NIDCD/NIDCR Genomics and Computational Biology Core (National Institute on Deafness and Other Communication Disorders), which operates with funds from the NIDCD Division of Intramural Research/NIH (ZIC DC000086 to the GCBC). This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). This research was made possible by the NIH OXCAM Program and the NIH Medical Research Scholars Program, a public-private partnership supported jointly by the NIH and contributions to the Foundation for the NIH from the Doris Duke Charitable Foundation, Genentech, the American Association for Dental Research, the Colgate-Palmolive Company, Hadassah Hospital Research fund, Versus Arthritis (Grant 21156), and NIHR (National Institute for Health and Care Research) Cambridge Biomedical Research Centre, and the National Institute of Dental and Craniofacial Research (ZIA DE000380), a part of the Intramural Research Program of the National Institutes of Health, Department of Health and Human Services.
Author contributions
S.J.G. designed, performed, and analyzed most experiments, prepared figures, and wrote the manuscript. B.W.H.M, S.S.P, J.F., L.L., A.H., J.-C.K., S.A.K., K.F., and R.K.M. performed experiments and analyses. R.G. led the development of HyA-FMBs and supplied them for the project. A/L.-H., D.M., N.G.C.B.C., and A.W.M. provided critical advice and resources for this project. P.G.R. designed the overall study, supervised experiments, and wrote the manuscript. All authors edited the manuscript.
Declaration of interests
P.G.R., S.A.K., R.G., A.H.-L., and J.F. have a patent on the hyaluronic-acid-coated fibrin microbeads (US Patent #1094021).
Published: July 18, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110537.
Supplemental information
Transcriptomic comparison of Day 42 chondrospheroids and fetal chondrocytes.
Transcriptomic comparison of Day 28 chondrospheroids and fetal chondrocytes.
Transcriptomic comparison of Day 14 chondrospheroids and fetal chondrocytes.
References
- 1.Boer C.G., Hatzikotoulas K., Southam L., Stefánsdóttir L., Zhang Y., Coutinho de Almeida R., Wu T.T., Zheng J., Hartley A., Teder-Laving M., et al. Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations. Cell. 2021;184:4784–4818.e17. doi: 10.1016/j.cell.2021.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayliss L.E., Culliford D., Monk A.P., Glyn-Jones S., Prieto-Alhambra D., Judge A., Cooper C., Carr A.J., Arden N.K., Beard D.J., Price A.J. The effect of patient age at intervention on risk of implant revision after total replacement of the hip or knee: a population-based cohort study. Lancet. 2017;389:1424–1430. doi: 10.1016/S0140-6736(17)30059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carey J.L. Fibrocartilage following microfracture is not as robust as native articular cartilage. J. Bone Joint Surg. 2012;94:10–11. doi: 10.2106/JBJS.L.00319. [DOI] [PubMed] [Google Scholar]
- 4.Ha C.W., Park Y.B., Kim S.H., Lee H.J. Intra-articular Mesenchymal Stem Cells in Osteoarthritis of the Knee: A Systematic Review of Clinical Outcomes and Evidence of Cartilage Repair. Arthroscopy. 2019;35:277–288.e2. doi: 10.1016/j.arthro.2018.07.028. [DOI] [PubMed] [Google Scholar]
- 5.NICE Autologous chondrocyte implantation for repairing symptomatic articular cartilage defects of the knee. 2017. https://www.nice.org.uk/guidance/ta477/chapter/1-Recommendations
- 6.Tsumaki N., Okada M., Yamashita A. iPS cell technologies and cartilage regeneration. Bone. 2015;70:48–54. doi: 10.1016/j.bone.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Andrade R., Vasta S., Pereira R., Pereira H., Papalia R., Karahan M., Oliveira J.M., Reis R.L., Espregueira-Mendes J. Knee donor-site morbidity after mosaicplasty – a systematic review. J. Exp. Orthop. 2016;3:31. doi: 10.1186/s40634-016-0066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Kinderen P., Meester J., Loeys B., Peeters S., Gouze E., Woods S., Mortier G., Verstraeten A. Differentiation of Induced Pluripotent Stem Cells Into Chondrocytes: Methods and Applications for Disease Modeling and Drug Discovery. J. Bone Miner. Res. 2022;37:397–410. doi: 10.1002/jbmr.4524. [DOI] [PubMed] [Google Scholar]
- 9.Yang L., Tsang K.Y., Tang H.C., Chan D., Cheah K.S.E. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc. Natl. Acad. Sci. USA. 2014;111:12097–12102. doi: 10.1073/pnas.1302703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Archer C.W., Dowthwaite G.P., Francis-West P. Development of synovial joints. Birth Defects Res. C Embryo Today. 2003;69:144–155. doi: 10.1002/bdrc.10015. [DOI] [PubMed] [Google Scholar]
- 11.Spagnoli A., O’Rear L., Chandler R.L., Granero-Molto F., Mortlock D.P., Gorska A.E., Weis J.A., Longobardi L., Chytil A., Shimer K., Moses H.L. TGF-β signaling is essential for joint morphogenesis. J. Cell Biol. 2007;177:1105–1117. doi: 10.1083/jcb.200611031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pacifici M., Koyama E., Shibukawa Y., Wu C., Tamamura Y., Enomoto-Iwamoto M., Iwamoto M. Cellular and molecular mechanisms of synovial joint and articular cartilage formation. Ann. N. Y. Acad. Sci. 2006;1068:74–86. doi: 10.1196/annals.1346.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray A., Singh P.N.P., Sohaskey M.L., Harland R.M., Bandyopadhyay A. Precise spatial restriction of BMP signaling is essential for articular cartilage differentiation. Development. 2015;142:1169–1179. doi: 10.1242/dev.110940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shwartz Y., Viukov S., Krief S., Zelzer E. Joint Development Involves a Continuous Influx of Gdf5-Positive Cells. Cell Rep. 2016;15:2577–2587. doi: 10.1016/j.celrep.2016.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flowers S.A., Zieba A., Örnros J., Jin C., Rolfson O., Björkman L.I., Eisler T., Kalamajski S., Kamali-Moghaddam M., Karlsson N.G. Lubricin binds cartilage proteins, cartilage oligomeric matrix protein, fibronectin and collagen II at the cartilage surface. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-13558-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maly K., Sastre E.A., Farrell E., Meurer A., Zaucke F. Comp and tsp-4: Functional roles in articular cartilage and relevance in osteoarthritis. Int. J. Mol. Sci. 2021;22:1–23. doi: 10.3390/ijms22052242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu L., Bluguermann C., Kyupelyan L., Latour B., Gonzalez S., Shah S., Galic Z., Ge S., Zhu Y., Petrigliano F.A., et al. Human developmental chondrogenesis as a basis for engineering chondrocytes from pluripotent stem cells. Stem Cell Rep. 2013;1:575–589. doi: 10.1016/j.stemcr.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nohe A., Hassel S., Ehrlich M., Neubauer F., Sebald W., Henis Y.I., Knaus P. The mode of bone morphogenetic protein (BMP) receptor oligomerization determines different BMP-2 signaling pathways. J. Biol. Chem. 2002;277:5330–5338. doi: 10.1074/jbc.M102750200. [DOI] [PubMed] [Google Scholar]
- 19.Robey P.G., Riminucci M. Principles of Bone Biology. Academic Press; 2020. Skeletal stem cells: tissue-specific stem/progenitor cells of cartilage, bone, stroma, and marrow adipocytes; pp. 45–71. [Google Scholar]
- 20.Bianco P., Cao X., Frenette P.S., Mao J.J., Robey P.G., Simmons P.J., Wang C.Y. The meaning, the sense and the significance: Translating the science of mesenchymal stem cells into medicine. Nat. Med. 2013;19:35–42. doi: 10.1038/nm.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Méndez-Ferrer S., Michurina T.V., Ferraro F., Mazloom A.R., MacArthur B.D., Lira S.A., Scadden D.T., Mag’Ayan A., Enikolopov G.N., Frenette P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou B.O., Yue R., Murphy M.M., Peyer J.G., Morrison S.J. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelttari K., Winter A., Steck E., Goetzke K., Hennig T., Ochs B.G., Aigner T., Richter W. Premature induction of hypertrophy during in vitro chondrogenesis of human mesenchymal stem cells correlates with calcification and vascular invasion after ectopic transplantation in SCID mice. Arthritis Rheum. 2006;54:3254–3266. doi: 10.1002/art.22136. [DOI] [PubMed] [Google Scholar]
- 24.Scotti C., Piccinini E., Takizawa H., Todorov A., Bourgine P., Papadimitropoulos A., Barbero A., Manz M.G., Martin I. Engineering of a functional bone organ through endochondral ossification. Proc. Natl. Acad. Sci. USA. 2013;110:3997–4002. doi: 10.1073/pnas.1220108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Somoza R.A., Welter J.F., Correa D., Caplan A.I. Chondrogenic differentiation of mesenchymal stem cells: Challenges and unfulfilled expectations. Tissue Eng. Part B Rev. 2014;20:596–608. doi: 10.1089/ten.teb.2013.0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorodetsky R., Clark R.A.F., An J., Gailit J., Levdansky L., Vexler A., Berman E., Marx G. Fibrin microbeads (FMB) as biodegradable carriers for culturing cells and for accelerating wound healing. J. Invest. Dermatol. 1999;112:866–872. doi: 10.1046/j.1523-1747.1999.00600.x. [DOI] [PubMed] [Google Scholar]
- 27.Gurevich O., Vexler A., Marx G., Prigozhina T., Levdansky L., Slavin S., Shimeliovich I., Gorodetsky R. Fibrin microbeads for isolating and growing bone marrow-derived progenitor cells capable of forming bone tissue. Tissue Eng. 2002;8:661–672. doi: 10.1089/107632702760240571. [DOI] [PubMed] [Google Scholar]
- 28.Ben-Ari A., Rivkin R., Frishman M., Gaberman E., Levdansky L., Gorodetsky R. Isolation and implantation of bone marrow-derived mesenchymal stem cells with fibrin micro beads to repair a critical-size bone defect in mice. Tissue Eng. Part A. 2009;15:2537–2546. doi: 10.1089/ten.tea.2008.0567. [DOI] [PubMed] [Google Scholar]
- 29.Kuznetsov S.A., Burks S., Hailu-Lazmi A., Cherman N., Fernandez de Castro L., Robey P.G., Gorodetsky R. In vivo formation of stable hyaline cartilage by transplantation of naive human bone marrow stromal cells. Stem Cells Transl Med. 2019;26:586–592. doi: 10.1016/j.joca.2018.02.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang N.S., Varghese S., Elisseeff J. Derivation of chondrogenically-committed cells from human embryonic cells for cartilage tissue regeneration. PLoS One. 2008;3:e2498. doi: 10.1371/journal.pone.0002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bigdeli N., Karlsson C., Strehl R., Concaro S., Hyllner J., Lindahl A. Coculture of human embryonic stem cells and human articular chondrocytes results in significantly altered phenotype and improved chondrogenic differentiation. Stem Cell. 2009;27:1812–1821. doi: 10.1002/stem.114. [DOI] [PubMed] [Google Scholar]
- 32.Chang Y.H., Wu K.C., Ding D.C. Induced Pluripotent Stem Cell-Differentiated Chondrocytes Repair Cartilage Defect in a Rabbit Osteoarthritis Model. Stem Cells Int. 2020;2020:8867349. doi: 10.1155/2020/8867349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diederichs S., Klampfleuthner F.A.M., Moradi B., Richter W. Chondral Differentiation of Induced Pluripotent Stem Cells Without Progression Into the Endochondral Pathway. Front. Cell Dev. Biol. 2019;7:270–310. doi: 10.3389/fcell.2019.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oldershaw R.A., Baxter M.A., Lowe E.T., Bates N., Grady L.M., Soncin F., Brison D.R., Hardingham T.E., Kimber S.J. Directed differentiation of human embryonic stem cells toward chondrocytes. Nat. Biotechnol. 2010;28:1187–1194. doi: 10.1038/nbt.1683. [DOI] [PubMed] [Google Scholar]
- 35.Chijimatsu R., Ikeya M., Yasui Y., Ikeda Y., Ebina K., Moriguchi Y., Shimomura K., Hart D.A., Hideki Y., Norimasa N. Characterization of Mesenchymal Stem Cell-Like Cells Derived from Human iPSCs via Neural Crest Development and Their Application for Osteochondral Repair. Stem Cells Int. 2017;2017:1960965. doi: 10.1155/2017/1960965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loh K.M.M., Chen A., Koh P.W.W., Deng T.Z.Z., Sinha R., Tsai J.M.M., Barkal A.A.A., Shen K.Y.Y., Jain R., Morganti R.M.M., et al. Mapping the Pairwise Choices Leading from Pluripotency to Human Bone, Heart, and Other Mesoderm Cell Types. Cell. 2016;166:451–467. doi: 10.1016/j.cell.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith C.A., Humphreys P.A., Naven M.A., Woods S., Mancini F.E., O’Flaherty J., Meng Q.-J., Kimber S.J. Directed differentiation of hPSCs through a simplified lateral plate mesoderm protocol for generation of articular cartilage progenitors. PLoS One. 2023;18:e0280024. doi: 10.1371/journal.pone.0280024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu C.L., Dicks A., Steward N., Tang R., Katz D.B., Choi Y.R., Guilak F. Single cell transcriptomic analysis of human pluripotent stem cell chondrogenesis. Nat. Commun. 2021;12:362. doi: 10.1038/s41467-020-20598-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J., Taylor S.E.B., Smeriglio P., Lai J., Maloney W.J., Yang F., Bhutani N. Early induction of a prechondrogenic population allows efficient generation of stable chondrocytes from human induced pluripotent stem cells. FASEB J. 2015;29:3399–3410. doi: 10.1096/fj.14-269720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamashita A., Morioka M., Yahara Y., Okada M., Kobayashi T., Kuriyama S., Matsuda S., Tsumaki N. Generation of scaffoldless hyaline cartilaginous tissue from human iPSCs. Stem Cell Rep. 2015;4:404–418. doi: 10.1016/j.stemcr.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Craft A.M., Rockel J.S., Nartiss Y., Kandel R.A., Alman B.A., Keller G.M. Generation of articular chondrocytes from human pluripotent stem cells. Nat. Biotechnol. 2015;33:638–645. doi: 10.1038/nbt.3210. [DOI] [PubMed] [Google Scholar]
- 42.Ferguson G.B., Van Handel B., Bay M., Fiziev P., Org T., Lee S., Shkhyan R., Banks N.W., Scheinberg M., Wu L., et al. Mapping molecular landmarks of human skeletal ontogeny and pluripotent stem cell-derived articular chondrocytes. Nat. Commun. 2018;9:3634. doi: 10.1038/s41467-018-05573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gardner O.F.W., Juneja S.C., Whetstone H., Nartiss Y., Sieker J.T., Veillette C., Keller G.M., Craft A.M. Effective repair of articular cartilage using human pluripotent stem cell-derived tissue. Eur. Cell. Mater. 2019;38:215–227. doi: 10.22203/eCM.v038a15. [DOI] [PubMed] [Google Scholar]
- 44.Petrigliano F.A., Liu N.Q., Lee S., Tassey J., Sarkar A., Lin Y., Li L., Yu Y., Geng D., Zhang J., et al. Long-term repair of porcine articular cartilage using cryopreservable, clinically compatible human embryonic stem cell-derived chondrocytes. NPJ Regen. Med. 2021;6:77. doi: 10.1038/s41536-021-00187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lowery J.W., Rosen V. The BMP pathway and its inhibitors in the skeleton. Physiol. Rev. 2018;98:2431–2452. doi: 10.1152/physrev.00028.2017. [DOI] [PubMed] [Google Scholar]
- 47.Gorodetsky R., Vexler A., Shamir M., An J., Levdansky L., Shimeliovich I., Marx G. New cell attachment peptide sequences from conserved epitopes in the carboxy termini of fibrinogen. Exp. Cell Res. 2003;287:116–129. doi: 10.1016/S0014-4827(03)00120-4. [DOI] [PubMed] [Google Scholar]
- 48.Levy-Beladev L., Levdansky L., Gaberman E., Friedler A., Gorodetsky R. A family of cell-adhering peptides homologous to fibrinogen C-termini. Biochem. Biophys. Res. Commun. 2010;401:124–130. doi: 10.1016/j.bbrc.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 49.Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 50.Guangbin L., Patricia D., McKee M.D., Pinero G.J., Loyer E., Behringer R.R., Evelyne L., Gerard K. Spontaneous Calcification of Arteries and Cartilage in Mice Lacking Matrix GLA Protein. Nature. 1997;386:78–81. doi: 10.1038/386078a0. [DOI] [PubMed] [Google Scholar]
- 51.Cancela M.L., Laizé V., Conceição N., Kempf H., Murshed M. Keutel Syndrome, a Review of 50 Years of Literature. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.642136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kiepe D., Ciarmatori S., Haarmann A., Tönshoff B. Differential expression of IGF system components in proliferating vs. differentiating growth plate chondrocytes: The functional role of IGFBP-5. Am. J. Physiol. Endocrinol. Metab. 2006;290:363–371. doi: 10.1152/ajpendo.00363.2005. [DOI] [PubMed] [Google Scholar]
- 53.Mukherjee A., Rotwein P. Insulin-like growth factor-binding protein-5 inhibits osteoblast differentiation and skeletal growth by blocking insulin-like growth factor actions. Mol. Endocrinol. 2008;22:1238–1250. doi: 10.1210/me.2008-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okamoto O., Fujiwara S. Dermatopontin, a novel player in the biology of the extracellular matrix. Connect. Tissue Res. 2006;47:177–189. doi: 10.1080/03008200600846564. [DOI] [PubMed] [Google Scholar]
- 55.Futrega K., Robey P.G., Klein T.J., Crawford R.W., Doran M.R. A single day of TGF-β1 exposure activates chondrogenic and hypertrophic differentiation pathways in bone marrow-derived stromal cells. Commun. Biol. 2021;4:29. doi: 10.1038/s42003-020-01520-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alliston T., Choy L., Ducy P., Karsenty G., Derynck R. TGF-β-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 2001;20:2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar A., Ruan M., Clifton K., Syed F., Khosla S., Oursler M.J. TGF-β mediates suppression of adipogenesis by estradiol through connective tissue growth factor induction. Endocrinology. 2012;153:254–263. doi: 10.1210/en.2011-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hellingman C.A., Davidson E.N.B., Koevoet W., Vitters E.L., Van Den Berg W.B., Van Osch G.J.V.M., Van Der Kraan P.M. Smad signaling determines chondrogenic differentiation of bone-marrow-derived mesenchymal stem cells: Inhibition of Smad1/5/8P prevents terminal differentiation and calcification. Tissue Eng. Part A. 2011;17:1157–1167. doi: 10.1089/ten.tea.2010.0043. [DOI] [PubMed] [Google Scholar]
- 59.Franco R.A.G., McKenna E., Robey P.G., Shajib M.S., Crawford R.W., Doran M.R., Futrega K. Inhibition of BMP signaling with LDN 193189 can influence bone marrow stromal cell fate but does not prevent hypertrophy during chondrogenesis. Stem Cell Rep. 2022;17:616–632. doi: 10.1016/j.stemcr.2022.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Occhetta P., Pigeot S., Rasponi M., Dasen B., Mehrkens A., Ullrich T., Kramer I., Guth-Gundel S., Barbero A., Martin I. Developmentally inspired programming of adult human mesenchymal stromal cells toward stable chondrogenesis. Proc. Natl. Acad. Sci. USA. 2018;115:4625–4630. doi: 10.1073/pnas.1720658115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koyama E., Shibukawa Y., Nagayama M., Sugito H., Young B., Yuasa T., Okabe T., Ochiai T., Kamiya N., Rountree R.B., et al. A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev. Biol. 2008;316:62–73. doi: 10.1016/j.ydbio.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mang T., Kleinschmidt-Doerr K., Ploeger F., Schoenemann A., Lindemann S., Gigout A. BMPR1A is necessary for chondrogenesis and osteogenesis, whereas BMPR1B prevents hypertrophic differentiation. J. Cell Sci. 2020;133:jcs246934. doi: 10.1242/jcs.246934. [DOI] [PubMed] [Google Scholar]
- 63.Rountree R.B., Schoor M., Chen H., Marks M.E., Harley V., Mishina Y., Kingsley D.M. BMP receptor signaling is required for postnatal maintenance of articular cartilage. PLoS Biol. 2004;2:e355. doi: 10.1371/journal.pbio.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kobayashi T., Lyons K.M., McMahon A.P., Kronenberg H.M. BMP signaling stimulates cellular differentiation at multiple steps during cartilage development. Proc. Natl. Acad. Sci. USA. 2005;102:18023–18027. doi: 10.1073/pnas.0503617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bian Q., Cheng Y.H., Wilson J.P., Su E.Y., Kim D.W., Wang H., Yoo S., Blackshaw S., Cahan P. A single cell transcriptional atlas of early synovial joint development. Development. 2020;147:dev185777. doi: 10.1242/dev.185777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao C., Jiang W., Zhou N., Liao J., Yang M., Hu N., Liang X., Xu W., Chen H., Liu W., et al. Sox9 augments BMP2-induced chondrogenic differentiation by downregulating Smad7 in mesenchymal stem cells (MSCs) Genes Dis. 2017;4:229–239. doi: 10.1016/j.gendis.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liao J., Hu N., Zhou N., Lin L., Zhao C., Yi S., Fan T., Bao W., Liang X., Chen H., et al. Sox9 potentiates BMP2-induced chondrogenic differentiation and inhibits BMP2-induced osteogenic differentiation. PLoS One. 2014;9:e89025. doi: 10.1371/journal.pone.0089025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haseeb A., Kc R., Angelozzi M., de Charleroy C., Rux D., Tower R.J., Yao L., da Silva R.P., Pacifici M., Qin L., Lefebvre V. SOX9 keeps growth plates and articular cartilage healthy by inhibiting chondrocyte dedifferentiation/osteoblastic redifferentiation. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2019152118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tardif G., Hum D., Pelletier J.P., Duval N., Martel-Pelletier J. Regulation of the IGFBP-5 and MMP-13 genes by the microRNAs miR-140 and miR-27a in human osteoarthritic chondrocytes. BMC Musculoskelet. Disord. 2009;10:148–211. doi: 10.1186/1471-2474-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toda S., Blauch L.R., Tang S.K.Y., Morsut L., Lim W.A. Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science. 2018;361:156–162. doi: 10.1126/science.aat0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Futrega K., Palmer J.S., Kinney M., Lott W.B., Ungrin M.D., Zandstra P.W., Doran M.R. The microwell-mesh: A novel device and protocol for the high throughput manufacturing of cartilage microtissues. Biomaterials. 2015;62:1–12. doi: 10.1016/j.biomaterials.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 72.Baghbaderani B.A., Tian X., Neo B.H., Burkall A., Dimezzo T., Sierra G., Zeng X., Warren K., Kovarcik D.P., Fellner T., Rao M.S. CGMP-manufactured human induced pluripotent stem cells are available for pre-clinical and clinical applications. Stem Cell Rep. 2015;5:647–659. doi: 10.1016/j.stemcr.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Young M.D., Behjati S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. GigaScience. 2020;9:giaa151. doi: 10.1093/gigascience/giaa151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Germain P.L., Robinson M.D., Lun A., Garcia Meixide C., Macnair W. Doublet identification in single-cell sequencing data using scDblFinder. F1000Res. 2022;10:979–1026. doi: 10.12688/f1000research.73600.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zappia L., Oshlack A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. GigaScience. 2018;7:giy083. doi: 10.1093/gigascience/giy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J. G:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y., Parmigiani G., Johnson W.E. ComBat-seq: Batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2020;2 doi: 10.1093/nargab/lqaa078. lqaa078–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gadomski S., Fielding C., García-García A., Korn C., Kapeni C., Ashraf S., Villadiego J., Toro R. del, Domingues O., Skepper J.N., et al. A cholinergic neuroskeletal interface promotes bone formation during postnatal growth and exercise. Cell Stem Cell. 2022;29:528–544.e9. doi: 10.1016/j.stem.2022.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A serum-free chondrogenic differentiation strategy beginning with sclerotome induction. Brightfield and mCherry live cell Incucyte imaging of SOX9-mCherry hiPSC differentiation from day 0 to day 16 of chondrospheroid protocol outlined in Figure 5D.
Transcriptomic comparison of Day 42 chondrospheroids and fetal chondrocytes.
Transcriptomic comparison of Day 28 chondrospheroids and fetal chondrocytes.
Transcriptomic comparison of Day 14 chondrospheroids and fetal chondrocytes.
Data Availability Statement
-
•
Single-cell and bulk RNA sequencing have been deposited at NCBI’s GEO Database and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
All original code has been deposited at Github and is publicly available as of the date of publication. The link is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.