Abstract
The mammalian neprilysin (NEP) family members are typically type II membrane endopeptidases responsible for the activation/inactivation of neuropeptides and peptide hormones. Differences in substrate specificity and subcellular localization of the seven mammalian NEPs contribute to their functional diversity. The sequencing of the Drosophila melanogaster genome has revealed a large expansion of this gene family, resulting in over 20 fly NEP-like genes, suggesting even greater diversity in structure and function than seen in mammals. We now report that one of these genes (Nep2) codes for a secreted endopeptidase with a highly restricted pattern of expression. D. melanogaster NEP2 is expressed in the specialized stellate cells of the renal tubules and in the cyst cells that surround the elongating spermatid bundles in adult testis, suggesting roles for the peptidase in renal function and in spermatogenesis. D. melanogaster NEP2 was found in vesicle-like structures in the syncytial cytoplasm of the spermatid bundles, suggesting that the protein was acquired by endocytosis of protein secreted from the cyst cells. Expression of NEP2 cDNA in D. melanogaster S2 cells confirmed that the peptidase is secreted and is only weakly inhibited by thiorphan, a potent inhibitor of human NEP. D. melanogaster NEP2 also differs from human NEP in the manner in which the peptidase cleaves the tachykinin, GPSGFYGVR-amide. Molecular modelling suggests that there are important structural differences between D. melanogaster NEP2 and human NEP in the S1′ and S2′ ligand-binding subsites, which might explain the observed differences in inhibitor and substrate specificities. A soluble isoform of a mouse NEP-like peptidase is strongly expressed in spermatids, suggesting an evolutionarily conserved role for a soluble endopeptidase in spermatogenesis.
Keywords: Drosophila melanogaster, Malpighian tubules, neprilysin, neutral endopeptidase, spermatogenesis, testes
Abbreviations: ECE, endothelin-converting enzyme; EST, expressed sequence tag; LomTK, locustatachykinin; MMEL2, membrane metalloendopeptidase-like 2; NEP, neprilysin; NL1, neprilysin-like 1; PTr, 0.5% Triton X-100 in PBS; PTrN, 5% normal goat serum in 0.5% Triton X-100/PBS; PTw, 0.1% Tween 20 in PBS; ECE-i, 4-chloro-N-[(4-cyano-3-methyl-1-phenyl-1H-pyrazol-5-yl)amino]carbonyl benzene sulphonamide, monosodium salt; SEP, secreted endopeptidase
INTRODUCTION
The mammalian neprilysin (NEP), or M13 family of zinc metallopeptidases, comprises seven members in the human genome, although physiological functions have only been ascribed to NEP itself and to the endothelin-converting enzymes (ECE-1 and ECE-2) [1]. NEP family members are typically type II integral membrane glycoproteins of approx. 700 amino acids, with a single transmembrane domain connecting the N-terminal cytoplasmic segment with the much larger C-terminal luminal or extracellular domain containing the active site. This region contains the HEXXH zinc-binding motif found in many zinc peptidases. The histidines in this motif (His-583 and His-587 in the human NEP sequence) constitute two of the zinc ligands, with a glutamate residue [Glu-646 in the conserved sequence EXX(A/G)D] acting as the third zinc ligand and classifying NEP as a gluzincin. Glu-584 in the HEXXH motif is essential for catalysis, and other key residues in substrate binding and catalysis have been identified by site-directed mutagenesis [2]. Many of these established features have been confirmed by X-ray crystallographic studies of the extracellular domain of human NEP complexed with its inhibitor, phosphoramidon [3].
NEP has diverse functions in humans, being principally involved in turning off regulatory peptide signals in the brain and periphery [2]. Key substrates in vivo include the enkephalins, substance P, atrial natriuretic peptides and endothelins, implicating NEP in the regulation of pain, inflammatory responses and the cardiovascular system. NEP is identical with the common acute lymphoblastic leukaemia antigen (CALLA or CD10), and is known to be down-regulated in a number of cancers, especially of the prostate [4]. Most recently, NEP has been shown to be involved in metabolism and removal of the neurotoxic amyloid β-peptide, identifying the enzyme as a potential target in the treatment of Alzheimer's disease [5]. The NEP homologues, ECE-1 and ECE-2, appear to have a more restricted role in the proteolytic processing of some prohormones, particularly the endothelin precursor ‘big endothelin’ [1]. Previously, it has been shown that one of the NEP family members, known as either SEP (secreted endopeptidase) or NL1 (neprilysin-like 1) in mouse [6,7], NEP2 in rat [8] and MMEL2 (membrane metallo-endopeptidase-like 2) in human [9], can exist both as a membrane-bound form and as a soluble secreted protein. SEP and its homologues are strongly expressed in mammalian testes [6–9]. The fact that all other mammalian NEP/ECE proteins are strictly membrane-associated has focused special attention on the mechanism by which the secreted SEP/NL1 is generated and the physiological significance of the alternative trafficking of the enzyme. The two mouse isoforms (SEP/NL1 and SEP/NL1Δ) arise by differential mRNA splicing, which leads to the insertion of a 23-amino-acid peptide, containing a proteolytic cleavage site, into the ectodomain of SEP/NL1 [6–8,10]. This larger isoform undergoes proteolysis to generate a soluble enzyme destined for secretion, whereas the integral membrane form (SEP/NL1Δ) is retained in the endoplasmic reticulum. SEP/NL1 mRNA is primarily restricted to round and elongated spermatids, suggesting an important role for the enzyme in reproduction [6]. This has recently been confirmed in a study of male SEP/NL1-deficient mice, which sire smaller litters compared with wild-type animals [11]. Physiological substrates for mammalian SEP/NL1 have not as yet been identified, however, in vitro studies indicate relatively minor differences between the substrate specificities of SEP/NL1 and NEP.
NEP-like proteins have been identified in bacteria, archaea and the animal kingdom, and are more diverse in invertebrates (see entries under M13 family in the MEROPS database; http://merops.sanger.ac.uk) [12]. NEP-like activity which is inhibited by phosphoramidon, a potent inhibitor of human NEP, has been identified in molluscs [13,14], a nematode [15], an annelid [16] and several insect species, including the house fly (Musca domestica) [17], fruit fly (Drosophila melanogaster) [18,19], the gypsy moth (Lymantria dispar) [20], the tomato moth (Lacanobia oleracea) [19], two locust species (Schistocerca gregaria [21] and Locusta migratoria [19]) and a cockroach (Leucopheae maderae) [19,22]. NEP-like activity is enriched in synaptic membranes prepared from the locust central nervous system [21], and has been shown histochemically to be present in neuropil regions of the cockroach and locust brain [22]. These data suggest an evolutionarily conserved role for NEPs in the metabolic inactivation of neuropeptides in the central nervous system. Recently, an ECE-like endopeptidase has been characterized from L. migratoria and shown to have a broad tissue distribution, including strong expression in male and female reproductive tissues and the brain [23].
Expansion of the NEP gene family has led to functional diversification in mammals through selection based on substrate specificity, linked in some instances to differences in cellular and subcellular expression patterns. Comparative genomics revealed that NEP gene expansions have occurred independently of the chordates in insects and nematodes [24], which may have resulted in adaptive evolution of peptidases with novel substrate specificities and functions. The sequencing of the entire genome of D. melanogaster has revealed more than 20 NEP/ECE-like genes, which provides a valuable model system with powerful genetic tools to explore the biochemical and functional diversity of the M13 family. In the present study, we have analysed the NEP family in D. melanogaster. Of the 24 NEP/ECE-like protein sequences identified by BLAST analysis using human NEP and ECE-1 as queries, five (Nep1, FlyBase number CG5894; Nep2, CG9761; Nep3, CG9565; Nep4, CG4058; Nep5, CG6265) show a particularly high degree of similarity to mammalian NEP and ECEs. We now report that one of these genes, Nep2, codes for a secreted endopeptidase with a limited pattern of expression in stellate cells of adult renal tubules and in elongating cysts in the testes. We show that recombinant D. melanogaster NEP2 displayed phosphoramidon-sensitive endopeptidase activity, but differs from mammalian NEP with respect to peptide-bond specificity and sensitivity to NEP inhibitors, which can be attributed to differences in the molecular structure of the S1′ and S2′ subsites. We propose that this new secreted member of the NEP/ECE family may have a role in the metabolism of peptides involved in regulating renal tubule function and in reproduction.
EXPERIMENTAL
Insect culture
D. melanogaster strains were maintained on a standard diet [4.16% (w/v) porridge oats; 4.16% (w/v) sugar, 10% (w/v) yeast, 1.7% (w/v) agar, 0.16% (w/v) methyl p-hydroxybenzoate] at 25 °C with a 12 h–12 h light–dark cycle. tud1 flies were obtained from the Bloomington Stock Centre (http://fly.bio.indiana.edu/). Male survivors from tud1/tud1 females are sterile with testes that lack germ cells; these flies are hereinafter referred to as tudor flies.
Immunocytochemistry
Two staining protocols were employed depending on whether the secondary antibody was conjugated to peroxidase or Alexa 488.
Antibody conjugated to peroxidase
Tissues from wandering third instar larvae (L3) and newly eclosed male Oregon R or tudor flies, were dissected in PBS (137 mM NaCl, 2.68 mM KCl, 4.02 mM Na2HPO4, 1.76 mM KH2PO4, pH 7.4) and treated with 2% (v/v) H2O2 in methanol for 5 min. The tissues were subjected to three 5 min washes in PBS before being fixed for 20 min in 4% (w/v) paraformaldehyde in PBS. Fixation was terminated by three 5 min washes in PBS/0.5% (v/v) Triton-X 100 (PTr). Tissues were then placed in blocking solution containing 10% (w/v) BSA and 10% (v/v) goat serum in PTr for 1 h before incubation for 18 h at 4 °C with the primary antibody (D. melanogaster anti-NEP2 antibody; 1:1000) in 3% (w/v) BSA in PTr. The primary antibody was removed by washing in PTr containing 3% (w/v) BSA for 30 min at room temperature (24 °C). This washing procedure was repeated twice. Tissues were then incubated with biotinylated secondary antibody (Vectastain® ABC Kit; Vector Laboratories, Peterborough, U.K.) diluted 1:5000 in PBS with 1.5% (v/v) normal goat serum. After 1 h the tissues were placed in PBS for 10 min, a procedure that was repeated a further three times prior to incubation with the Vectastain® ABC reagent prepared according to the manufacturer's instructions. After 30 min, the tissues were washed three times for 5 min with PBS before staining with Sigma Fast DAB (Sigma–Aldrich, Poole, Dorset, U.K) for 5 min. Colour development was terminated by three 5 min washes with PBS and the tissues were mounted on glass slides in glycerol/gelatine (Sigma–Aldrich).
Antibody conjugated to Alexa 488
Testes of adult flies were dissected in PBS and fixed with 2% (v/v) methanol in PBS for 5 min. Following this, the tissue was washed three times with PBS, each of 5 min duration, and fixed with 4% (w/v) paraformaldehyde in PBS on ice for 30 min. For antibody visualization, samples were washed in PTr several times for a total time of 1.5 h and were then blocked with 5% (v/v) normal goat serum in PTr (PTrN) for 30 min and incubated with primary antibody (D. melanogaster NEP2 antibody, 1:1000) at 4 °C overnight. The samples were subjected to a series of washes: two 15 min washes with PTrN; two 45 min washes with PTr; two 45 min washes with PTrN. Tissues were then incubated with secondary antibody [goat anti-(rabbit Alexa-Fluor 488); Molecular Probes] at a dilution of 1:500 in PTr at 4 °C for 17 h. After frequent washes with PTr over 2 h, the samples were incubated in PBS at 4 °C for 17 h before being mounted on to glass slides in Vectashield (Vector Laboratories). Images were collected by confocal microscopy using a Leica SP2 DMIRE2 and processed using Leica SP2 AOBS software.
In situ hybridization
Tissues were dissected from adult D. melanogaster in PBS and transferred without delay to a microcentrifuge tube containing 5% (v/v) formaldehyde in PTr. After 20 min, tissues were washed 3 times in PTr. In situ hybridization using sense and antisense riboprobes generated from a Nep2 EST (expressed sequence tag) cDNA clone (LD45441, BDGP) was carried out according to the method described previously [25]. LD45441 was linearized with SmaI to transcribe the antisense probe with Sp6 RNA polymerase (Promega, Southampton, U.K.) and XhoI to transcribe the sense probe with T7 RNA polymerase (Promega).
Preparation of membrane and soluble fractions from testes and Malpighian tubules of Oregon R and tudor flies
Malpighian tubules and testes were dissected into 100 μl of PBS and homogenized in a glass homogenizer. The resulting homogenate was then centrifuged at 4 °C in a microcentrifuge for 5 min at 1000 g. The supernatant was removed and placed into a 1.5-ml ultracentrifuge tube (Beckman Coulter, High Wycombe, Bucks., U.K.) while the pellet was re-homogenized in 100 μl of PBS and spun for 5 min at 1000 g at 4 °C. The combined supernatants were centrifuged (Optima™ MAX, Beckman Coulter, TLA 110 rotor) at 50000 g at 4 °C for 1 h to separate the membrane and soluble fractions. The resulting supernatant (soluble fraction) was collected and the pellet re-suspended in 100 mM Tris/HCl, 500 mM NaCl, pH 7.5, buffer. The washed membranes were pelleted by centrifugation at 50000 g for 1 h and re-suspended in 200 μl of buffer (100 mM Tris/HCl, 150 mM NaCl, pH 7.5) to form the membrane fraction.
Antibody production and purification
Antibodies recognizing an internal sequence of the D. melanogaster NEP2 were generated by Pepceuticals Ltd (Leicester, U.K.) by immunizing rabbits with the peptide CTAERPETEPKHFRLPN conjugated to BSA via an N-terminal cysteine residue using standard procedures [26]. Antibody purification was undertaken using the Sulpholink™ column (Perbio Science UK Ltd, Tattenhall, Cheshire, U.K.) according to manufacturer's instructions. Antibody fractions were stored in PBS containing 0.05% (w/v) sodium azide at 4 °C.
Immunoelectrophoresis
Gel electrophoresis was carried out using a Mini Protean® II Electrophoresis Cell (Bio-Rad, Hemel Hempstead, Herts., U.K.) on a SDS/PAGE gel (10%). Proteins were transferred on to Hybond P® PVDF or Hybond-ECL® nitrocellulose membrane (Amersham Pharmacia, Little Chalfont, Bucks., U.K.) in transfer buffer [25 mM Tris/HCl, 190 mM glycine, 2% (v/v) methanol] using a Mini Trans-Blot® Cell (Bio-Rad) at 100 V for 1 h. The membrane was washed once in PBS with 0.1% Tween 20 (PTw) and then blocked in a 5% (w/v) non-fat dried milk solution in PTw for 1 h. The membrane was then washed twice in PTw before incubation with the purified D. melanogaster NEP2 antibody, diluted to 1:5000 in 5% (w/v) non-fat dried milk solution in PTw and incubated overnight at 4 °C. Bound antibody was then labelled with an anti-rabbit horseradish-peroxidase-conjugated secondary antibody diluted to 1:5000 in 5% (w/v) non-fat dried milk solution in PTw. A SuperSignal® (Pierce) chemiluminescent detection kit was used to detect the presence of the secondary antibody, according to the manufacturer's instructions. Immunoblot membranes were exposed to Hyperfilm® (Amersham) and developed in a Xograph Compact X4 automatic X-ray film processor (Xograph Imaging Systems, Tetbury, U.K.). All procedures were performed at room temperature unless otherwise stated.
Assay for peptidase activity in testes and Malpighian tubules
A 2-μl aliquot from the soluble fractions of Oregon R testes and Malpighian tubules was incubated with 100 μM peptide substrate, locustatachykinin 1 (LomTK-1; GPSGFYGVR-amide from Bachem Ltd, St. Helens, Merseyside, U.K.) in 100 mM Tris/HCl, pH 7.5, in a total volume of 20 μl at 37 °C. Enzyme activity was stopped by decreasing the pH to 2 by the addition of 5 μl of 8% (v/v) trifluoroacetic acid, and the final volume was made up to 260 μl with 0.1% (v/v) trifluoroacetic acid. Peptide fragments generated by peptidase activity were resolved and quantified by reverse-phase HPLC using a Jupiter 5μ column [C18, 250 mm×4.5 mm (internal diameter); Phenomenex, Macclesfield, U.K.] and detection at 214 nm, as described previously [19]. The effects of inhibitors on enzyme activity were investigated by preincubating the inhibitor with enzyme for 10 min at room temperature prior to starting the reaction by the addition of peptide substrate. IC50 values were calculated using a curve fitting computer programme (FigP, Biosoft, Cambridge, U.K.). Phosphoramidon and thiorphan were obtained from Sigma–Aldrich and ECE-i {4-chloro-N-[(4-cyano-3-methyl-1-phenyl-1H-pyrazol-5-yl)amino]-carbonyl benzene sulphonamide, monosodium salt} was given by Professor R. A. Grigg (School of Chemistry, University of Leeds, Leeds, U.K.). The identity of GPSGFYG was confirmed by MS, and the endopeptidase activity is expressed as pmol of GPSGFYG formed/h per insect [19].
Immunoprecipitation of D. melanogaster NEP2
A 25 μl aliquot of a soluble fraction (50000 g supernatant) prepared from homogenates of testes and Malpighian tubules of adult D. melanogaster, in 475 μl of 50 mM Tris/HCl (pH 7.5)/150 mM NaCl, was incubated with 100 μl of 6 mg/ml Protein A–Sepharose beads (Sigma–Aldrich) with rotation at 4 °C. After 2 h, the beads were removed by centrifugation for 5 min at 5000 rev./min in an MSE microcentrifuge and the supernatant collected. Affinity-purified NEP2 antibody or control non-immune rabbit IgG (Sigma–Aldrich) were equalized for protein concentration (0.68 mg/ml) and used at a dilution of 1:10. The antibodies were incubated with the treated supernatant fraction at 4 °C for 18 h before the addition of 100 μl of 6 mg/ml of fresh Protein A–Sepharose. After 3 h rotating at 4 °C, the beads and the supernatant fraction were separated by centrifugation as described above. The beads were washed extensively in 50 mM Tris/HCl (pH 7.5)/150 mM NaCl containing 1% Triton X-100, before Western blot analysis was carried out. The supernatant fractions were saved for measuring any remaining NEP-like activity as described above.
Expression of recombinant D. melanogaster NEP2
cDNA clone LD45441 was used to generate full-length D. melanogaster Nep2 with dA incorporated at the 3′ ends by using a Taq/Pwo enzyme mix (Expand High Fidelity System, Lewes, East Sussex, U.K.). PCR was carried out under standard conditions according to the manufacturer's instructions using an annealing temperature of 58 °C and 5 s increments to the extension time from cycle 11 of 25 cycles. The primer sequences were: primer 1, 5′-TAACCCGGGATGCAGACGGTAATC-3′; primer 2, 5′-GTTTTCACGCTCCAAACCATCGAGCTCTTTA-3′. The 3′ A at either end of the PCR product facilitated subcloning into the pMT/V5-His-TOPO® vector using the DES® TOPO® TA expression kit (Invitrogen) according to the manufacturer's instructions. The construct sequence was verified by primer walking (DBS Genomics, Department of Biological Sciences, University of Durham, Durham, U.K.). DNA for transfection of S2 cells was prepared using the Qiagen HiSpeed Plasmid Maxi Kit (Qiagen Ltd., Dorking, U.K.) according to the manufacturer's instructions.
D. melanogaster S2 cells were grown at 25 °C in complete Schneider's medium supplemented with 10% (v/v) heat-inactivated fetal calf serum and 50 units/ml penicillin G and 50 μg/ml streptomycin (Invitrogen). Cells were transfected using reagents supplied with the inducible D. melanogaster Expression System® from Invitrogen. The protocol supplied was followed until induction of expression, when cells were transferred to serum-free media for 24 h, after which cells and media were harvested to assess expression levels. The media from transfected cells from 4-ml cultures were dialysed against 2×1 litre of 20 mM Tris/HCl, pH 8, and concentrated using a Microsep Centrifugal Concentrator (Pall Filtron, VWR International, Poole, Dorset, U.K.). The concentrate (0.4 ml) was used undiluted for Western blots, but was diluted 1:2 with de-ionized water to assay for NEP-like activity. Cells were washed four times in PBS before being resusupended in 0.2 ml of PBS. Cells were disrupted either by heating in SDS/PAGE sample buffer for blotting or by incubating in 1% (w/v) Triton X-100 in PBS by rotating for 1 h at 4 °C for use in enzyme assays.
Molecular modelling
The model of D. melanogaster NEP2 was based on the crystal structure of human neprilysin (Protein DataBank accession code, 1DMT). An initial crude model was generated from the primary sequence of D. melanogaster NEP2 by the threading program 3D-PSSM [27]. Main chain breaks were limited to the loop regions on the surface of the protein and were corrected using the REFI function in the molecular visualization program O [28]. Zinc was manually positioned co-ordinated by His-598, Glu-660 and His-602, which were deduced to be the co-ordinating residues by sequence alignment. This model was improved by energy minimization and molecular dynamics simulations using the DS Modeling software (Accelrys, San Diego, CA, U.S.A.). All energy calculations were performed using the CHARM22 force field. The distance dependent dielectric was 4 and the non-bonded cut-off was set to 12 Å. Initial optimization was performed by two stages of energy minimization, firstly 500 steps of a steepest descent algorithm, followed by 1000 steps using the adopted basis Newton–Raphson algorithm. This was followed by heating to and equilibrium at 300 K before a 1000-step molecular dynamics simulation with time steps of 0.001 ps.
The co-ordinates of LomTK-1 with an amidated C-terminus were generated in PyMOL [29] (http://www.pymol.org). The peptide was manually positioned into the proposed active site of D. melanogaster NEP2 using the molecular visualization program O. The peptide was aligned such that the carboxyl group of the scissile peptide bond was orientated towards the zinc according to the proposed catalytic mechanism [3]. A large cavity allowed positioning of the seven N-terminal residues, whereas two smaller cavities allowed positioning of the C-terminal Val–Arg residues. The protein–ligand simulation function of the DS Modelling package was then used to improve this model. This function applies the same optimization procedure as described above, except the simulated system is restricted to within 20 Å of the ligand's centre of mass, and the distance dependent dielectric was set to 1. The quality of the model was checked using PROCHECK [30].
RESULTS
Comparative structure of the Nep2 (FlyBase number CG9761) protein
A search for D. melanogaster genes encoding NEP-like proteins was undertaken using the BLASTP algorithm. BLASTP hits obtained using human NEP, MMEL2 and ECE-1 protein sequences as queries included the predicted CG9761 protein with scores of 451, 501 and 479 respectively. CG9761, annotated as Nep2, is located on the right arm of chromosome 3 (cytologic location 82D2), and it covers 12077 nt of genomic sequence comprising nine exons [31]. D. melanogaster NEP2 is a 763-amino-acid preprotein with a predicted molecular mass of 86838 Da. The SignalP V3.0 programme predicts the presence of a signal peptide and cleavage between Gly-41 and Lys-42, with a mean S score of 0.575 (prediction cut-off value, 0.47; [32]). Comparison with the protein sequence of human NEP, MMEL2 and ECE-1 in a ClustalW alignment shows that key active-site residues, including those involved in co-ordination with zinc and those required for catalysis, have been conserved (Figure 1). In addition, all ten cysteines used in the formation of disulphide bridges in human NEP are conserved in D. melanogaster NEP2 [3]. There is no equivalent to Cys-428 of human ECE-1, which forms the covalent linkage between ECE-1 dimers [33]. There are eight potential N-glycosylation sites in D. melanogaster NEP2. The position of two of these sites (Asn-173 and Asn-642) are conserved in several members of the human NEP family (NEP, MMEL2 and ECE-1) (see Figure 1).
Figure 1. ClustalW alignment of the amino acid sequences of D. melanogaster NEP2 (DmNEP2), human NEP (HuNEP), human MMEL2 (HuMMEL2) and human ECE1 (HuECE1).
—, signal peptide; |, predicted cleavage site for a signal peptidase; *, potential N-glycosylation sites conserved in all four proteins; =, conserved active site residues; ↑, conserved cysteines; □, cysteine involved in the formation of the disulphide-linked ECE-1 dimer; open up arrow, amino acids that contribute side-chains to the S1′ subsite of human NEP.
Expression of Nep2 in adult Malpighian tubules
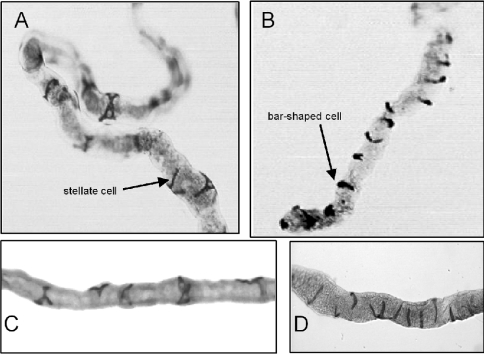
The cellular localization of NEP2 mRNA was investigated using digoxygenin-labelled antisense riboprobes to detect mRNA transcripts. Strong expression of Nep2 was restricted to the stellate cells in the main segment and to the bar-shaped cells in the initial segment of Malpighian tubules from both male and female adult D. melanogaster (Figures 2A and 2B). Affinity-purified antibodies, raised against an internal peptide sequence of D. melanogaster NEP2, stained the same stellate and bar-shaped cells, which intercalate the principal cells of the renal tubules (Figures 2C and 2D).
Figure 2. Distribution of NEP2 mRNA and protein in adult Malpighian tubules.
(A and B) In situ hybridization was carried out using a digoxygenin-labelled Nep2 anti-sense riboprobe. (C and D) Immunocytochemistry was performed using a Vectastain® ABC Kit and NEP2 antibodies. High levels of NEP2 mRNA and protein were detected in the stellate cells and the related bar-shaped cells, but not the principal cells of the Malpighian tubules. Control experiments performed with either a sense-strand riboprobe or non-immune IgG gave no specific staining of the Malpighian tubules (results not shown).
Expression of Nep2 in adult testes
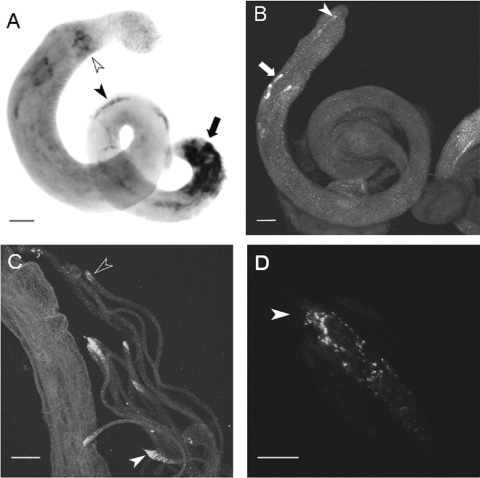
NEP2 cDNAs are represented in the D. melanogaster testis EST database, indicating that the gene is expressed in the male gonad. In situ hybridization revealed high levels of Nep2 transcripts in a group of cells towards the base of the testes (Figure 3A). The position and morphology of these cells is consistent with them being somatic cyst cells [34]. NEP2 mRNA was also detected in elongated cells, surrounding spermatid bundles and waste bags, confirming these cells as tail cyst cells (Figure 3A). Immunofluorescence showed that NEP2 was concentrated at the tail end of elongating cysts. Strongest fluorescence was observed in cysts which had elongated to three quarters the length of the testis (Figures 3B and 3C). Weaker fluorescence was present at the tail end of fully elongated cysts, but was undetectable in cysts earlier than the half-elongated stage. Confocal optical sections revealed a punctate pattern of fluorescence which was distributed throughout the spermatid syncytial cytoplasm, strongest at the tail end and extending for approx. 50 μm towards the head end (Figure 3D). There was no specific immunostaining for NEP2 above background levels in tudor testes lacking germ cells. In addition, D. melanogaster NEP2 was not detected in male accessory glands of either wild-type or tudor males (results not shown).
Figure 3. Distribution of NEP2 mRNA and protein in the adult testes.
(A) In situ hybridization. Strong staining is present in cyst cells at the base of the testis (arrow). Staining is also apparent in tail cyst cells of elongating cysts (solid arrowhead) and in tail cyst cells surrounding the waste bags (open arrowhead). (B–D) Immunofluorescence studies. (B) Intact testis. Strong fluorescence is observed at the tail end of elongating cysts at approx. 75% of the length of the testis (arrow) and weaker fluorescence is observed at the tail end of fully elongated cysts (arrowhead). (C) Disrupted testis. Fluorescence is associated with the tail end of elongated cysts (e.g. closed arrowhead), but is weaker in more fully elongated cysts (open arrowhead). (D) Longitudinal optical section through the tail end of an elongating cyst. Punctate fluorescence is apparent within the spermatid bundle and is strongest at the tail end (arrowhead). Scale bars: (A–C) 50 μm; (D) 10 μm.
Malpighian tubule and testicular NEP2 is not an integral membrane protein
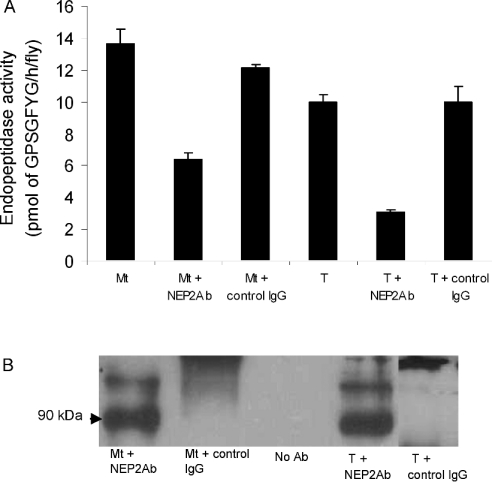
Affinity-purified antibodies were used to probe for D. melanogaster NEP2 on a Western blot of membrane and soluble fractions obtained from Malpighian tubules and testes after high-speed centrifugation of tissue homogenates (Figure 4). The D. melanogaster NEP2 antibodies recognized a protein band with an estimated molecular mass of approx. 90000 in the soluble protein fraction from both Malpighian tubules and testes, but, importantly, no immunoreactive protein was detected in the membrane fraction from either of these tissues. Western blot analysis confirmed the absence of D. melanogaster NEP2 from tudor testes.
Figure 4. Demonstration of a soluble NEP2 in the Malpighian tubules and testes of adult flies.
(A) Endopeptidase activity was measured in soluble extracts of Malpighian tubules (Mt) and testes by determining the rate of cleavage of the Gly–Val bond of the GPSGFYGVR-amide peptide using HPLC to quantify the reaction product GPSGFYG. Reactions were carried out in 100 mM Tris/HCl, pH 7.5, in either the absence or presence of 100 μM phosphoramidon (P), as described in the Experimental section. The results are expressed as the means±S.E.M. (n=3). (B, C) Western blot analysis of the distribution of NEP2 in membrane (P) and soluble (S) fractions prepared from Malpighian tubules from 10 adult wild-type flies and testes (5 pairs per lane) from wild-type and tudor flies. Equivalent amount of membrane and soluble fractions were loaded on to the SDS/PAGE gel. Blotted proteins were probed with purified NEP2 antibody as the primary antibody to detect a protein band of approx. 90 kDa.
To demonstrate the existence of a soluble NEP-like activity, the Malpighian tubule and testis soluble protein fractions were incubated with an insect peptide (GPSGFYGVR-amide, LomTK1), known to be a good substrate for both insect and mammalian endopeptidases (Figure 4) [19]. The major peptide fragment generated by the soluble endopeptidase from both tissues was GPSGFYG, indicating that cleavage occurred at the Gly–Val peptide bond. The formation of this fragment was completely blocked in the presence of the NEP inhibitor phosphoramidon (IC50 values of 5.3 μM for the tubule enzyme and 1.7 μM for the testicular enzyme). The endopeptidase activity from both tissues was weakly inhibited by 100 μM thiorphan (18% and 33% inhibition of the tubule and testis enzymes respectively).
Confirmatory evidence that a soluble NEP2 was responsible for the aforementioned endopeptidase cleavages was sought using the affinity-purified antibody to immunoprecipitate the enzyme activity. The Malpighian tubule and testicular soluble fractions were assayed for endopeptidase activity before and after incubation with D. melanogaster NEP2 antibodies and separation of the IgG–NEP2 complex from unbound protein using Protein A–Sepharose beads. This resulted in a decrease in endopeptidase activity of between 50% and 70% compared with a loss of approx. 10% in experiments where non-specific IgG replaced the NEP2 antibodies (Figure 5A). A Western blot of the immunoprecipitated material showed that D. melanogaster NEP2 was bound to the Protein A–Sepharose beads, only when the soluble fractions were incubated with NEP2 antibodies (Figure 5B).
Figure 5. Immunoprecipitation of the soluble endopeptidase activity of Malpighian tubules and testes from adult flies.
(A) Endopeptidase activity was measured in soluble fractions from Malpighian tubules and testes by determining the rate of hydrolysis of the Gly–Val peptide bond of LomTK-1 before and after immunoprecipitation with purified D. melanogaster NEP2 antibodies/Protein A–Sepharose. In control experiments, the D. melanogaster NEP2 antibody was replaced with non-immune rabbit IgG at the same protein concentration. The results are expressed as the mean of two independent experiments and were within a range ±5%. (B) The Protein A–Sepharose beads were analysed by Western blotting using the D. melanogaster NEP2 antibody (NEP2Ab) to probe for immunoprecipitated NEP2 in the soluble fraction prepared from Malpighian tubules (Mt) and testes (T). Specificity was controlled using IgG from non-immunized rabbits (Sigma–Aldrich).
Functional expression of soluble NEP2 in D. melanogaster S2 cells
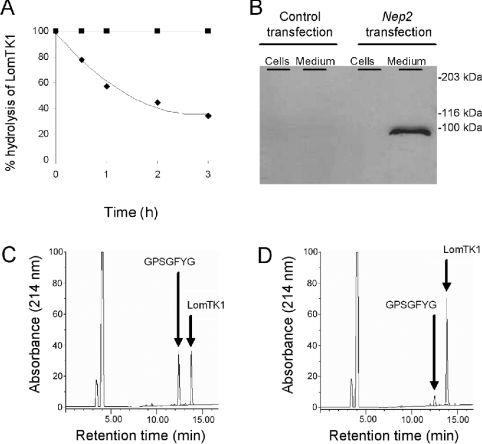
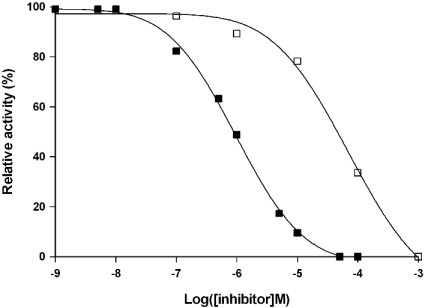
S2 cells were transiently transfected with either a full-length NEP2 cDNA or an empty vector DNA as a control experiment. Expression was induced by addition of 500 μM CuSO4 to the cultures and at 24 h post-induction, cells and medium were analysed for endopeptidase activity and for D. melanogaster NEP2 immunoreactivity using Western blotting. Strong endopeptidase activity was found in medium from Nep2 transfected, but not control cell cultures (Figure 6A). This activity cleaved LomTK-1 to generate GPSGFYG as the only detectable peptide fragment (Figure 6C) and was totally inhibited by 100 μM phosphoramidon. The rate of hydrolysis occurring with the conditioned medium was around 5-fold faster than that observed in the cell extract (Figures 6C and 6D), establishing that recombinant NEP2 is mainly secreted from the transfected S2 cells. This was confirmed by Western blotting which showed that D. melanogaster NEP2 immunoreactivity was in the medium and not the cells (Figure 6B). The endopeptidase activity found in the medium was strongly inhibited by phosphoramidon (IC50, 0.96 μM), but was less affected by thiorphan (IC50, 69 μM) (Figure 7). The endopeptidase activity was not inhibited at all by ECE-i, a selective inhibitor of ECE-1, at a final concentration of 100 μM.
Figure 6. Characterization of soluble recombinant D. melanogaster NEP2 expressed in S2 cells.
(A) Time-course for the hydrolysis of LomTK-I by the conditioned medium from a culture of S2 cells over-expressing NEP2 (◆) and control S2 cells transformed with an empty vector (■). In this experiment aliquots (5 μl) of the unconcentrated media were incubated with LomTK-1 (100 μM, final concentration) in 100 mM Tris/HCl, pH 7.5, for 3 h before termination of the reaction and HPLC analysis, as described in the Experimental section. The extent of hydrolysis was determined by quantifying the decline in the parent peptide with time. No hydrolysis was observed when the peptide was incubated with medium from cells transfected with empty vector. (B) Western blot of recombinant NEP2 expressed in S2 cells. S2 cells transfected with either empty vector or Nep2 cDNA were grown to confluency before being maintained for 24 h in serum-free medium. Medium was harvested, dialysed and concentrated before analysis as described in the Experimental section. Collected cells were lysed with 1% (w/v) Triton X-100 in PBS and an equivalent amount of the media and cell extract were loaded on to SDS/PAGE gels for direct comparison of the distribution of NEP2. (C and D) HPLC chromatograms showing the cleavage of LomTK-1 by the medium (C) and detergent-lysed cells (D) from a culture of S2 cells expressing NEP2 under the control of the inducible metallothionein promoter. The only hydrolysis product detected by HPLC was identified as GPSGFYG by co-chromatography with authentic material. The predicted second cleavage product, VR-amide, was not detectable with our chromatography system. Equivalent amounts of medium and cells were incubated with the peptide substrate in 100 mM Tris/HCl, pH 7.5, for 1h.
Figure 7. Inhibition of recombinant D. melanogaster NEP2 activity by inhibitors of human NEP/ECE.
Culture medium (2 μl) from S2 cells transiently expressing Nep2 was incubated with LomTK-1 (100 μM) in 100 mM Tris/HCl, pH 7.5, in the absence and presence of different concentrations of phosphoramidon (■) and thiorphan (□). The rate of cleavage of the Gly–Val peptide bond of LomTK-1 was determined by HPLC quantification of the reaction product (GPSGFYG) at 214 nm. The results are expressed as a percentage of the uninhibited activity.
Molecular modelling of the S1′ and S2′ subsites of D. melanogaster NEP2
The high-resolution crystal structure of human NEP shows that the S1′ subsite comprises a large hydrophobic pocket lined by the side-chains Phe-106, Ile-558, Phe-563, Met-579, Val-580, Val-692 and Trp-693 [3,35]. In D. melanogaster NEP2, five of these seven residues are conserved, but the equivalent of human NEP Met-579 and Val-692 are a tyrosine and a threonine residue respectively (Table 1 and Figure 1). The S2′ subsites of the two enzymes are also predicted to be markedly different. The human NEP S2′ subsite is formed from the side-chains of Arg-102, Phe-106, Asp-107 and Arg-110. Apart from Phe-106, all the equivalent amino acids in D. melanogaster NEP2 are serine residues which, because of their smaller and polar side-chains, are predicted to form a more open uncharged pocket (Table 1).
Table 1. Comparison of the amino acids that contribute side-chains to the S1′ and S2′ subsites of human NEP and D. melanogaster NEP2.
For the identification of the human NEP subsite amino acids, see [3].
| Human NEP | D. melanogaster NEP2 | ||
|---|---|---|---|
| S1′ | S2′ | S1′ | S2′ |
| Phe-106 | Arg-102 | Phe-134 | Ser-130 |
| Ile-558 | Phe-106 | Ile-573 | Phe-134 |
| Phe-563 | Asp-107 | Phe-578 | Ser-135 |
| Met-579 | Arg-110 | Tyr-594 | Ser-138 |
| Val-580 | Val-595 | ||
| Val-692 | Thr-706 | ||
| Trp-693 | Trp-707 | ||
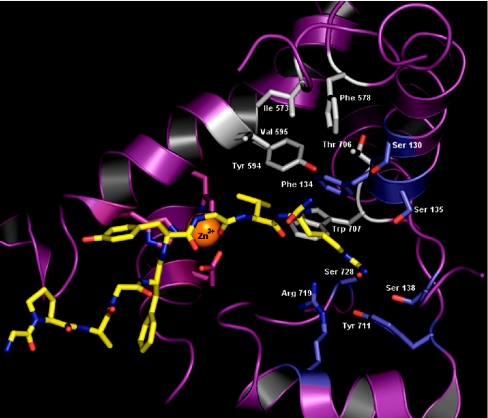
A homology model of D. melanogaster NEP2 suggests that the S1′ pocket of the insect enzyme is smaller than the human S1′ subsite and less hydrophobic, mainly due to the substitutions Met→Tyr and Val→Thr (Figure 8). The model of LomTK-1 bound to D. melanogaster NEP2 shows that the valine residue of LomTK-1 can make favourable van der Waals interactions with residues Val-595, Phe-134 and Trp-707 (Figure 8). Cleavage at the Gly–Val bond of LomTK-1 would require accommodation of the basic side-chain of the C-terminal arginine in the S2′ subsite. The model of D. melanogaster NEP2 suggests that this pocket is now larger compared with that in the human enzyme and is lined by polar residues, at least two (Ser-728 and Tyr-711) of which are predicted to make favourable hydrogen bonds to the C-terminal arginine of LomTK-1.
Figure 8. Model of D. melanogaster NEP2 with LomTK-1 docked at the active site.
A ribbon diagram showing the active-site space with substrate and metal ion co-ordinating residues. The peptide substrate LomTK-1 is shown in yellow and the ligand-binding subsites S1′ and S2′ are shown with their side-chains in grey and light blue respectively. Associated helices and zinc co-ordinating ligands are shown in magenta.
DISCUSSION
In the present study we have shown that Nep2, a member of a relatively large family of D. melanogaster NEP/ECE-like genes, encodes a secreted endopeptidase that has a very restricted pattern of expression in the adult fly. D. melanogaster NEP2 is predicted to be a 763-amino-acid preprotein with a N-terminal signal peptide, which is cleaved after directing the newly synthesized protein into the secretory pathway. Western blotting was used to estimate the size of secreted NEP2 in D. melanogaster tissues to be approx. 90 kDa, which is slightly larger than the molecular mass of 82083 predicted for the secreted protein. A likely explanation for this difference is the possible addition of carbohydrate at one or more of the eight potential N-glycosylation sites. We have confirmed that NEP2 is indeed a soluble secreted enzyme in adult fly tissues, and that expression of the Nep2 gene in S2 cells generates a secreted enzyme and not a membrane-associated form. All mammalian members of the NEP family are synthesized as type II integral membrane proteins. Even the soluble form of SEP/NL1 is first synthesized as a membrane-bound proprotein that becomes soluble upon proteolytic cleavage in the secretory pathway [10]. Whether an endopeptidase is tethered or fully secreted will have important implications, such as determining the field of activity, as well as the concentration of enzyme at the membrane surface.
We have identified the Malpighian tubules and the testes as the principal sites of expression of D. melanogaster Nep2 in adult insects by detecting RNA transcripts and the expressed protein. Malpighian tubules are the insect's renal organs responsible for water and ion balance, and excretion of waste metabolites and toxic chemicals [36]. D. melanogaster tubules consist of two main cell types, principal cells and stellate cells, which are distinguished by their morphology and by their transporting properties [36]. Peptides are known to be powerful regulators of diuresis in insects by selective action on principal and stellate cells [36,37]. The drosokinin peptide is a potent stimulator of chloride conductance in stellate cells [38], whereas the D. melanogaster CRF (corticotropin-releasing factor)-like diuretic hormone, calcitonin-like diuretic hormone and the CAP2b peptides stimulate fluid secretion by the principal cells through stimulation of V-ATPase activity and cation transport [39,40]. Insect tachykinins are also potent stimulators of fluid secretion by Malpighian tubules [41]. Although it has not been shown directly that tachykinin peptides stimulate D. melanogaster tubules, the demonstration that a D. melanogaster tachykinin receptor gene (Takr, CG7887) is expressed in Malpighian tubules suggests that these peptides do have a role to play in the renal physiology of this insect [42]. Peptidases that are exclusive to one cell type, as is the case for D. melanogaster NEP2, might satisfy a need for peptide-degrading enzymes with different substrate specificities to regulate the quality and duration of the signalling activity of peptide hormones in a cell-specific manner. A recent comparative study of the transcriptomes of Malpighian tubules and whole adult D. melanogaster showed that a second neprilysin, Nep3, in addition to Nep2, is up-regulated in renal tubules by approx. 3-fold [42]. D. melanogaster NEP3, unlike NEP2, is predicted to be a typical type II membrane protein, anchored by a hydrophobic transmembrane peptide and a cytoplasmic domain comprising the first 52 amino acids. Interestingly, Nep3 is expressed in the principal cells, but not the stellate cells, of D. melanogaster tubules (J. E. Thomas, A. D. Shirras, A. J. Turner and R. E. Isaac, unpublished work). These differences in cell expression and final destination of the enzymes suggest very distinct roles for NEP2 and NEP3 in D. melanogaster renal physiology. In mammals, a variety of peptidase activities are enriched in membranes of the kidney brush border with NEP constituting around 4% of the total membrane protein in rabbit kidney [43]. This renal NEP is primarily responsible for inactivating circulating atrial natriuretic peptide [44], which regulates sodium and water balance, blood volume and arterial pressure by direct effects on the mammalian kidney. We speculate that the NEP2 and NEP3 might have equally important roles in homoeostasis through the metabolism of endocrine peptides that regulate Malpighian tubule physiology in D. melanogaster.
Strong Nep2 expression also occurred in the cyst cells of the testes. In D. melanogaster two somatic cyst cells are associated with each developing cyst which, prior to meiosis, contains sixteen germ cells (for a review of spermatogenesis in D. melanogaster, see [45]). During cyst elongation the head cyst cell remains small, but the tail cyst cell elongates to become extremely long and thin along with the spermatid tails, which it encases. In situ hybridization detected NEP2 transcripts in tail cyst cells just prior to and during elongation, and in the fully elongated cysts. NEP2 on the other hand was located within the syncytial cytoplasm of spermatid bundles during later stages of elongation. It is concentrated towards the tail end of the spermatids, the punctate pattern of fluorescence suggesting that the protein is localized in vesicles. Our interpretation of these results is that the Nep2 gene is expressed in the tail cyst cell, from where the protein is secreted and endocytosed by the spermatid bundle – the punctate staining we observed is presumably endocytic vesicles. Although there are no other reports of endocytosis by spermatids of proteins secreted by cyst cells in D. melanogaster, mammalian spermatids are known to be active in endocytosis [46] and endocytose androgen-binding protein [47] and mannose 6-phosphate-containing glycoproteins [48] secreted by somatic Sertoli cells. Another point to note is that whereas NEP2 mRNA is detectable in non-elongated cyst cells right through to the fully elongated stage, the protein is only detectable in cysts which are substantially elongated, with protein staining being strongest in a small developmental window at approx. three quarters elongation. This temporal difference between mRNA and protein levels may result from control at the level of translation or endocytosis/vesicle trafficking. The absence of NEP2 in tudor testes is presumably because, in the absence of germ cells, cyst cells are prevented from reaching the developmental stage at which the Nep2 gene is activated. The function of D. melanogaster NEP2 in spermato-genesis is unclear, but its strong localization at the tail end of elongating cysts suggests a role in the elongation process.
The identity of the natural substrates for NEP2 and its precise role in reproductive physiology is open to speculation. D. melanogaster NEP2, unlike most of the other members of the NEP family, is not an integral membrane protein, which appears to be a special feature of the enzyme allowing movement between the cyst cells and the spermatid bundles. It is also possible that NEP2 is secreted into the extracellular space in the testes and is diluted to a level undetected by our antibody. D. melanogaster NEP2 is similar to mammalian SEP/NL1 in that both are highly expressed in the testes as secreted proteins. The reduced fertility of male mice that do not express a functional SEP/NL1 appears to be due to a lower success rate of egg fertilization and early embryonic development [11]. Other peptidases that have been implicated in the metabolism of peptide signalling molecules are also known to be important for male fertility in mammals. Male mice lacking germinal ACE (angiotensin-converting enzyme) [49] and PC4, a proprotein convertase of the subtilisin family, also have impaired fertility [50]. Although peptide and protein substrates for these enzymes are not known, the clear implication is that peptide processing and, possibly, inactivation are crucial aspects of the biology of mammalian reproduction. Several previous studies suggest that peptidases are also important for reproduction in D. melanogaster. Male-derived peptidases have been implicated in the processing and activation of hormone-like peptides of the male accessory glands [51], and a recent report provides evidence that male germinal Ance, which codes for a D. melanogaster dipeptidyl carboxypeptidase (angiotensin-converting enzyme or ACE) is important for spermatogenesis [52]. The demonstration of Nep2 expression in the cyst cells of D. melanogaster testes and the presence of NEP2 protein in spermatid bundles adds to the accumulating evidence that peptidases have important and, possibly, evolutionarily conserved roles in spermatogenesis and fertilization.
Mammalian NEP is by far the best-characterized member of the M13 family, in terms of substrate specificity [2] and molecular structure [35]. The similarity of the primary structure of D. melanogaster NEP2 and human NEP, and the conservation of not only important active-site residues, but also all the cysteines involved in the formation of intramolecular disulphide bonds and some of the glycosylation sites, suggests similarity in the overall architecture of the two proteins. The crystal structure of human NEP, complexed with either phosphoramidon or other potent inhibitors, has revealed the molecular interactions between enzyme and inhibitor, and identified the amino acids whose side-chains form the S1′ and S2′ subsites [3,35]. The S1′ subsite is a deep hydrophobic pocket, which explains the preference for amino acids with bulky neutral aliphatic and aromatic side-chains in the P1′ position of peptide substrates (e.g. LomTK-1) and the inhibitors phosphoramidon and thiorphan. The S2′ subsite is relatively large and accommodates the indole side-chain of phosphoramidon.
Our model of D. melanogaster NEP2 predicts that the S1′ subsite forms a smaller and less hydrophobic pocket than that found in human NEP, which would explain why the Gly–Phe bond of LomTK-1 is not cleaved by the insect enzyme. The S2′ subsite might also contribute towards this difference in peptide bond specificity. The S2′ side-chains of NEP2 are predicted to have favourable interactions with the C-terminal α-amidated arginine of the substrate LomTK-1, whereas the presence of the positively charged side-chains of Arg-102 and Arg-110 in the vicinity of the S2′ subsite of human NEP might discourage binding of the C-terminus of LomTK-1 in this pocket. The reduced potency of phosphoramidon and thiorphan as inhibitors of D. melanogaster NEP2, when compared with mammalian NEP [53], may also be explained by structural differences of the S1′ and S2′ subsites. In human NEP, the tryptophan side-chain of phosphoramidon forms Van der Waals' interactions with the side-chain of Phe-106 and hydrogen bonds with several other amino acids. The marked changes in the shape of the S2′ pocket predicted for D. melanogaster NEP2 is likely to have significant effects on these interactions and hence on the binding affinity of phosphoramidon. Thiorphan has even lower potency as an inhibitor of NEP2, probably because of a poor fit of the P1′ phenylalanine side-chain in the S1′ pocket combined with weak interactions of the C-terminal glycine residue in the S2′ subsite.
The enlarged family of NEP genes in D. melanogaster provides us with an opportunity to study the extent to which NEP-like endopeptidases have diversified in terms of their structure, enzymatic properties and function during the course of evolution. More detailed structure–function studies are likely to provide additional information about the protein architecture that defines substrate specificity in this family.
Acknowledgments
We thank the Drosophila Genomics Resource Center for cDNAs, and Helen White-Cooper for helpful comments on the testes expression of Nep2. We are also grateful to Sue Matthews for technical support during the course of this work. This work was supported by a grant (24/S12813) from the Biotechnology and Biological Sciences Research Council, U.K.
References
- 1.Turner A. J., Isaac R. E., Coates D. The neprilysin (NEP) family of zinc metalloendopeptidases: genomics and function. Bioessays. 2001;23:261–269. doi: 10.1002/1521-1878(200103)23:3<261::AID-BIES1036>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 2.Roques B. P., Noble F., Dauge V., Fournie-Zaluski M. C., Beaumont A. Neutral endopeptidase 24.11: structure, inhibition, and experimental and clinical pharmacology. Pharmacol. Rev. 1993;45:87–146. [PubMed] [Google Scholar]
- 3.Oefner C., D'Arcy A., Hennig M., Winkler F. K., Dale G. E. Structure of human neutral endopeptidase (Neprilysin) complexed with phosphoramidon. J. Mol. Biol. 2000;296:341–349. doi: 10.1006/jmbi.1999.3492. [DOI] [PubMed] [Google Scholar]
- 4.Papandreou C. N., Usmani B., Geng Y., Bogenrieder T., Freeman R., Wilk S., Finstad C. L., Reuter V. E., Powell C. T., Scheinberg D., et al. Neutral endopeptidase 24.11 loss in metastatic human prostate cancer contributes to androgen-independent progression. Nat. Med. 1998;4:50–57. doi: 10.1038/nm0198-050. [DOI] [PubMed] [Google Scholar]
- 5.Marr R. A., Guan H., Rockenstein E., Kindy M., Gage F. H., Verma I., Masliah E., Hersh L. B. Neprilysin regulates amyloid beta peptide levels. J. Mol. Neurosci. 2004;22:5–11. doi: 10.1385/JMN:22:1-2:5. [DOI] [PubMed] [Google Scholar]
- 6.Ghaddar G., Ruchon A. F., Carpentier M., Marcinkiewicz M., Seidah N. G., Crine P., Desgroseillers L., Boileau G. Molecular cloning and biochemical characterization of a new mouse testis soluble-zinc-metallopeptidase of the neprilysin family. Biochem. J. 2000;347:419–429. doi: 10.1042/0264-6021:3470419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda K., Emoto N., Raharjo S. B., Nurhantari Y., Saiki K., Yokoyama M., Matsuo M. Molecular identification and characterization of novel membrane-bound metalloprotease, the soluble secreted form of which hydrolyzes a variety of vasoactive peptides. J. Biol. Chem. 1999;274:32469–32477. doi: 10.1074/jbc.274.45.32469. [DOI] [PubMed] [Google Scholar]
- 8.Tanja O., Facchinetti P., Rose C., Bonhomme M. C., Gros C., Schwartz J. C. Neprilysin II: A putative novel metalloprotease and its isoforms in CNS and testis. Biochem. Biophys. Res. Commun. 2000;271:565–570. doi: 10.1006/bbrc.2000.2664. [DOI] [PubMed] [Google Scholar]
- 9.Bonvouloir N., Lemieux N., Crine P., Boileau G., DesGroseillers L. Molecular cloning, tissue distribution, and chromosomal localization of MMEL2, a gene coding for a novel human member of the neutral endopeptidase-24.11 family. DNA Cell Biol. 2001;20:493–498. doi: 10.1089/104454901316976127. [DOI] [PubMed] [Google Scholar]
- 10.Raharjo S. B., Emoto N., Ikeda K., Sato R., Yokoyama M., Matsuo M. Alternative splicing regulates the endoplasmic reticulum localization or secretion of soluble secreted endopeptidase. J. Biol. Chem. 2001;276:25612–25620. doi: 10.1074/jbc.M101703200. [DOI] [PubMed] [Google Scholar]
- 11.Carpentier M., Guillemette C., Bailey J. L., Boileau G., Jeannotte L., DesGroseillers L., Charron J. Reduced fertility in male mice deficient in the zinc metallopeptidase NL1. Mol. Cell. Biol. 2004;24:4428–4437. doi: 10.1128/MCB.24.10.4428-4437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rawlings N. D., Tolle D. P., Barrett A. J. MEROPS: the peptidase database. Nucleic Acids Res. 2004;32:D160–D164. doi: 10.1093/nar/gkh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shipp M. A., Stefano G. B., D'Adamio L., Switzer S. N., Howard F. D., Sinisterra J., Scharrer B., Reinherz E. L. Downregulation of enkephalin-mediated inflammatory responses by CD10/neutral endopeptidase 24.11. Nature (London) 1990;347:394–396. doi: 10.1038/347394a0. [DOI] [PubMed] [Google Scholar]
- 14.Bawab W., Aloyz R. S., Crine P., Roques B. P., DesGroseillers L. Identification and characterization of a neutral endopeptidase activity in Aplysia californica. Biochem. J. 1993;296:459–465. doi: 10.1042/bj2960459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sajid M., Isaac R. E. Identification and properties of a neuropeptide-degrading endopeptidase (neprilysin) of Ascaris suum muscle. Parasitol. 1995;111:599–608. doi: 10.1017/s0031182000077088. [DOI] [PubMed] [Google Scholar]
- 16.Laurent V., Salzet M. Metabolism of angiotensins by head membranes of the leech Theromyzon tessulatum. FEBS Lett. 1996;384:123–127. doi: 10.1016/0014-5793(96)00293-1. [DOI] [PubMed] [Google Scholar]
- 17.Lamango N. S., Isaac R. E. Metabolism of insect neuropeptides: properties of a membrane-bound endopeptidase from heads of Musca domestica. Insect Biochem. Mol. Biol. 1993;23:801–808. doi: 10.1016/0965-1748(93)90068-4. [DOI] [PubMed] [Google Scholar]
- 18.Isaac R. E., Priestly R. M. Neuropeptide metabolism: properties of an endopeptidase from heads of Drosophila melanogaster. In: Borkovec A. B., Masler E. P., editors. Insect Neurochemistry and Neurophysiology. Clifton, NJ: Humana Press; 1990. pp. 267–269. [Google Scholar]
- 19.Isaac R. E., Parkin E. T., Keen J. N., Nassel D. R., Siviter R. J., Shirras A. D. Inactivation of a tachykinin-related peptide: identification of four neuropeptide-degrading enzymes in neuronal membranes of insects from four different orders. Peptides. 2002;23:725–733. doi: 10.1016/s0196-9781(01)00653-2. [DOI] [PubMed] [Google Scholar]
- 20.Masler E. P., Wagner R. M., Kovaleva E. S. In vitro metabolism of an insect neuropeptide by neural membrane preparations from Lymantria dispar. Peptides. 1996;17:321–326. doi: 10.1016/0196-9781(95)02098-5. [DOI] [PubMed] [Google Scholar]
- 21.Isaac R. E. Neuropeptide-degrading endopeptidase activity of locust (Schistocerca gregaria) synaptic membranes. Biochem. J. 1988;255:843–847. doi: 10.1042/bj2550843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isaac R. E., Nassel D. R. Identification and localization of a neprilysin-like activity that degrades tachykinin-related peptides in the brain of the cockroach, Leucophaea maderae, and locust, Locusta migratoria. J. Comp. Neurol. 2003;457:57–66. doi: 10.1002/cne.10561. [DOI] [PubMed] [Google Scholar]
- 23.Macours N., Poels J., Hens K., Luciani N., De Loof A., Huybrechts R. An endothelin-converting enzyme homologue in the locust, Locusta migratoria: functional activity, molecular cloning and tissue distribution. Insect Mol. Biol. 2003;12:233–240. doi: 10.1046/j.1365-2583.2003.00406.x. [DOI] [PubMed] [Google Scholar]
- 24.Coates D., Siviter R., Isaac R. E. Exploring the Caenorhabditis elegans and Drosophila melanogaster genomes to understand neuropeptide and peptidase function. Biochem. Soc. Trans. 2000;28:464–469. [PubMed] [Google Scholar]
- 25.Siviter R. J., Coast G. M., Winther A. M., Nachman R. J., Taylor C. A., Shirras A. D., Coates D., Isaac R. E., Nassel D. R. Expression and functional characterization of a Drosophila neuropeptide precursor with homology to mammalian preprotachykinin A. J. Biol. Chem. 2000;275:23273–23280. doi: 10.1074/jbc.M002875200. [DOI] [PubMed] [Google Scholar]
- 26.Harlow J. M., Lane D. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. Antibodies: A Laboratory Manual. [Google Scholar]
- 27.Kelley L. A., MacCallum R. M., Sternberg M. J. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 28.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr., Sect. A: Found. Crystallogr. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.DeLano W. L. San Carlos, CA, U.S.A.: DeLano Scientific; 2002. The PyMOL User's Manual. [Google Scholar]
- 30.Laskowski R. A., Moss D. S., Thornton J. M. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 31.The FlyBase Consortium. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 2003;31:172–175. doi: 10.1093/nar/gkg094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 33.Hoang V. M., Sansom C. E., Turner A. J. Mutagenesis and modeling of endothelin converting enzyme. Biochem. Soc. Trans. 1996;24:471S. doi: 10.1042/bst024471s. [DOI] [PubMed] [Google Scholar]
- 34.Gonczy P., Viswanathan S., DiNardo S. Probing spermatogenesis in Drosophila with P-element enhancer detectors. Development. 1992;114:89–98. doi: 10.1242/dev.114.1.89. [DOI] [PubMed] [Google Scholar]
- 35.Oefner C., Roques B. P., Fournie-Zaluski M. C., Dale G. E. Structural analysis of neprilysin with various specific and potent inhibitors. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:392–396. doi: 10.1107/S0907444903027410. [DOI] [PubMed] [Google Scholar]
- 36.Dow J. T., Davies S. A. Integrative physiology and functional genomics of epithelial function in a genetic model organism. Physiol. Rev. 2003;83:687–729. doi: 10.1152/physrev.00035.2002. [DOI] [PubMed] [Google Scholar]
- 37.Gade G. Regulation of intermediary metabolism and water balance of insects by neuropeptides. Annu. Rev. Entomol. 2004;49:93–113. doi: 10.1146/annurev.ento.49.061802.123354. [DOI] [PubMed] [Google Scholar]
- 38.Radford J. C., Davies S. A., Dow J. A. Systematic G-protein-coupled receptor analysis in Drosophila melanogaster identifies a leucokinin receptor with novel roles. J. Biol. Chem. 2002;277:38810–38817. doi: 10.1074/jbc.M203694200. [DOI] [PubMed] [Google Scholar]
- 39.Cabrero P., Radford J. C., Broderick K. E., Costes L., Veenstra J. A., Spana E. P., Davies S. A., Dow J. A. The Dh gene of Drosophila melanogaster encodes a diuretic peptide that acts through cyclic AMP. J. Exp. Biol. 2002;205:3799–3807. doi: 10.1242/jeb.205.24.3799. [DOI] [PubMed] [Google Scholar]
- 40.Coast G. M., Webster S. G., Schegg K. M., Tobe S. S., Schooley D. A. The Drosophila melanogaster homologue of an insect calcitonin-like diuretic peptide stimulates V-ATPase activity in fruit fly Malpighian tubules. J. Exp. Biol. 2001;204:1795–1804. doi: 10.1242/jeb.204.10.1795. [DOI] [PubMed] [Google Scholar]
- 41.Johard H. A., Coast G. M., Mordue W., Nassel D. R. Diuretic action of the peptide locustatachykinin I: cellular localisation and effects on fluid secretion in Malpighian tubules of locusts. Peptides. 2003;24:1571–1579. doi: 10.1016/j.peptides.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 42.Wang J., Kean L., Yang J. Y., Allan A. K., Davies S. A., Herzyk P., Dow J. A. T. Function-informed transcriptome analysis of Drosophila renal tubule. Genome Biol. 2004;5:R69. doi: 10.1186/gb-2004-5-9-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kerr M. A., Kenny A. J. The purification and specificity of a neutral endopeptidase from rabbit kidney brush border. Biochem. J. 1974;137:477–488. doi: 10.1042/bj1370477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kenny A. J., Stephenson S. L. Role of endopeptidase-24.11 in the inactivation of atrial natriuretic peptide. FEBS Lett. 1988;232:1–8. doi: 10.1016/0014-5793(88)80375-2. [DOI] [PubMed] [Google Scholar]
- 45.Fuller M. T. Spermatogenesis in Drosophila. In: Bate M., Martinez-Arias A., editors. The Development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 71–147. [Google Scholar]
- 46.Segretain D., Egloff M., Gerard N., Pineau C., Jegou B. Receptor-mediated and absorptive endocytosis by male germ cells of different mammalian species. Cell Tissue Res. 1992;268:471–478. doi: 10.1007/BF00319154. [DOI] [PubMed] [Google Scholar]
- 47.Gerard A. Endocytosis of androgen-binding protein (ABP) by spermatogenic cells. J. Steroid Biochem. Mol. Biol. 1995;53:533–542. doi: 10.1016/0960-0760(95)00109-d. [DOI] [PubMed] [Google Scholar]
- 48.O'Brien D. A., Gabel C. A., Eddy E. M. Mouse Sertoli cells secrete mannose 6-phosphate containing glycoproteins that are endocytosed by spermatogenic cells. Biol. Reprod. 1993;49:1055–1065. doi: 10.1095/biolreprod49.5.1055. [DOI] [PubMed] [Google Scholar]
- 49.Krege J. H., John S. W. M., Langenbach L. L., Hodgin J. B., Hagaman J. R., Bachman E. S., Jennette J. C., O'Brien D. A., Smithies O. Male–female differences in fertility and blood-pressure in ACE-deficient mice. Nature (London) 1995;375:146–148. doi: 10.1038/375146a0. [DOI] [PubMed] [Google Scholar]
- 50.Mbikay M., Tadros H., Ishida N., Lerner C. P., De Lamirande E., Chen A., El-Alfy M., Clermont Y., Seidah N. G., Chretien M., et al. Impaired fertility in mice deficient for the testicular germ-cell protease PC4. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6842–6846. doi: 10.1073/pnas.94.13.6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park M., Wolfner M. F. Male and female cooperate in the prohormone-like processing of a Drosophila melanogaster seminal fluid protein. Dev. Biol. 1995;171:694–702. doi: 10.1006/dbio.1995.1315. [DOI] [PubMed] [Google Scholar]
- 52.Hurst D., Rylett C. M., Isaac R. E., Shirras A. D. The Drosophila angiotensin-converting enzyme homologue Ance is required for spermiogenesis. Dev. Biol. 2003;254:238–247. doi: 10.1016/s0012-1606(02)00082-9. [DOI] [PubMed] [Google Scholar]
- 53.Jeng A. Y., Ansell J., Erion M. D. pH- and time-dependent inhibition of rat kidney neutral endoprotease 24.11 by thiorphan and phosphoramidon. Life Sci. 1989;45:2109–2114. doi: 10.1016/0024-3205(89)90076-3. [DOI] [PubMed] [Google Scholar]