Significance
Since colonizing the Mackenzie Delta in northwestern Canada ~1200 CE, Inuvialuit have been heavily reliant on belugas for their livelihoods and cultural heritage. However, little is known of the impact of centuries of sustained Inuvialuit subsistence hunting on the beluga population inhabiting the Mackenzie Delta. Using paleogenomic and stable isotope analysis of zooarchaeological remains, and comparing the findings with contemporary data, we investigate temporal changes in beluga diversity, structuring, and foraging ecology. We show Inuvialuit harvests had a negligible impact on the genetic diversity of contemporary Mackenzie belugas, and highlight the applicability of combining genomic sexing and isotope analysis of zooarchaeological remains for advancing our understanding of past hunting practices and faunal ecologies.
Keywords: Arctic, paleogenomics, stable isotopes, subsistence hunting, zooarchaeology
Abstract
Beluga whales play a critical role in the subsistence economies and cultural heritage of Indigenous communities across the Arctic, yet the effects of Indigenous hunting on beluga whales remain unknown. Here, we integrate paleogenomics, genetic simulations, and stable δ13C and δ15N isotope analysis to investigate 700 y of beluga subsistence hunting in the Mackenzie Delta area of northwestern Canada. Genetic identification of the zooarchaeological remains, which is based on radiocarbon dating, span three time periods (1290 to 1440 CE; 1450 to 1650 CE; 1800 to 1870 CE), indicates shifts across time in the sex ratio of the harvested belugas. The equal number of females and males harvested in 1450 to 1650 CE versus more males harvested in the two other time periods may reflect changes in hunting practices or temporal shifts in beluga availability. We find temporal shifts and sex-based differences in δ13C of the harvested belugas across time, suggesting historical adaptability in the foraging ecology of the whales. We uncovered distinct mitochondrial diversity unique to the Mackenzie Delta belugas, but found no changes in nuclear genomic diversity nor any substructuring across time. Our findings indicate the genomic stability and continuity of the Mackenzie Delta beluga population across the 700 y surveyed, indicating the impact of Inuvialuit subsistence harvests on the genetic diversity of contemporary beluga individuals has been negligible.
Human settlement of the Arctic has been heavily reliant on the availability of marine resources, including cetaceans (1). Zooarchaeological remains and ethnohistoric records indicate beluga whales (white whales, Delphinapterus leucas) were fundamental to the survival of communities occupying the coastline in several regions of Alaska, northern Canada, and Greenland (2). The East Channel of the Mackenzie River near Tuktoyaktuk, Northwest Territories, Canada (Fig. 1 A and B), is home to the Inuvialuit, who have hunted beluga whales in the area for at least the past 700 y (3). Before the Inuvialuit occupation of the Mackenzie Delta, a series of Paleo-Inuit occupations began ca. 3000 y ago. However, due to the large-scale erosion occurring across the region, few archaeological sites from these earlier periods remain (4).
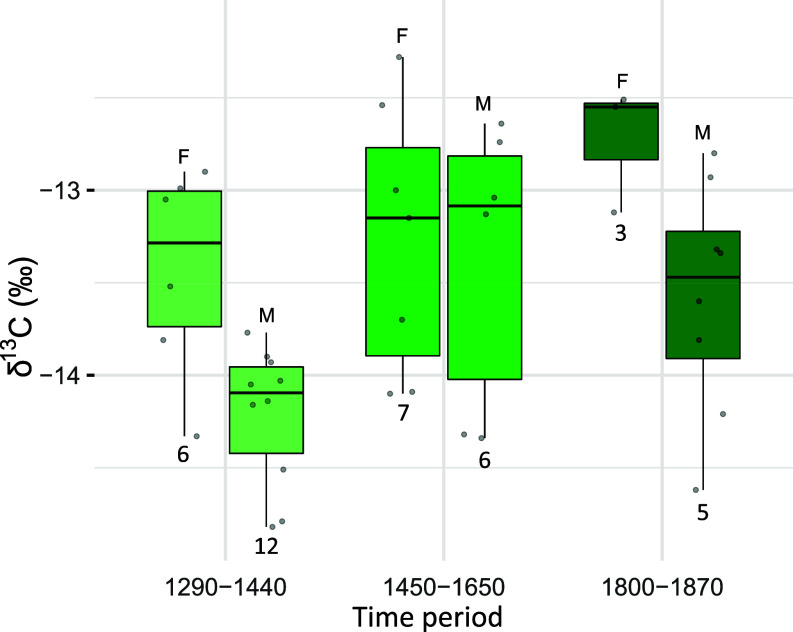
Fig. 1.
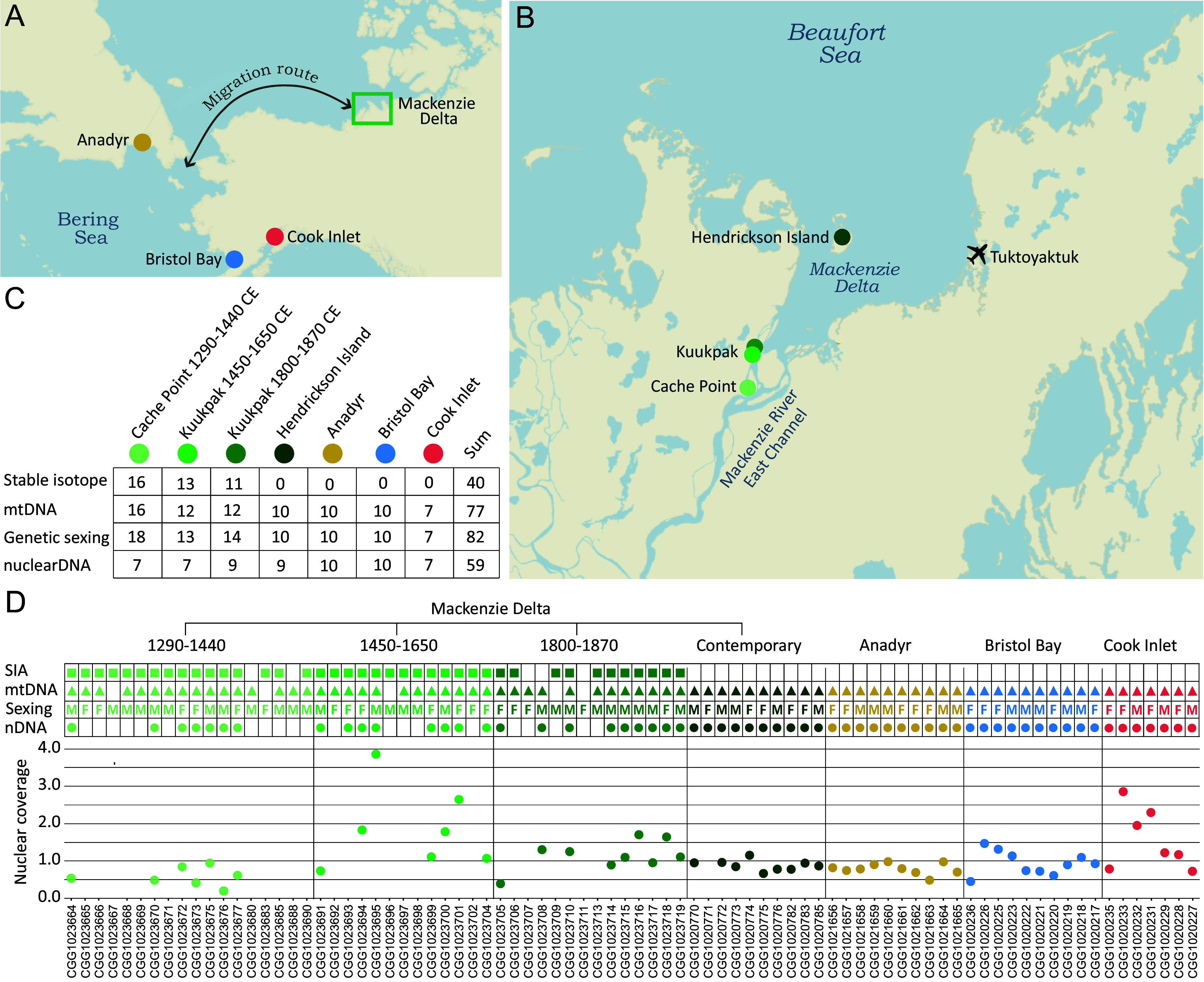
Sample localities and data overview. (A) Regional map of the Pacific Arctic showing the location of the Mackenzie Delta, and the three adjacent beluga whale populations included in this study. The annual migration route of the Mackenzie beluga whales is indicated. (B) Locality of the archaeological and contemporary (Hendrickson Island) beluga whale sample sites within the Mackenzie Delta. (C) Sample sizes of the stable isotopes, mitochondrial (mtDNA), genetic sexing, and nuclear DNA datasets. (D) Overview of the data available for each specimen. Colored symbols (squares, triangles, circles) indicate sufficient data for analysis. Nuclear coverage indicates the genome-wide read depth of each specimen; only specimens with > 0.2 × coverage are shown. Sample size of the nuclear DNA data is smaller in (C) than in (D), as two pairs of related individuals were identified genetically; one pair among the 1450 to 1650 CE samples and one pair in the contemporary Mackenzie Delta samples. For each pair, the individual with lowest coverage was excluded from further nuclear analyses.
The Mackenzie Delta contains numerous archaeological sites along the river banks, where the Inuvialuit relocated their settlements farther and farther downstream (northward), as more and more sediments were deposited over time. The archaeological sites indicate continuous occupation from ca. 1300 CE to the present (5, 6). The early part of this period, from ca. 1300 to 1400 CE, is referred to by archaeologists as the “Thule period,” and after ca. 1400 CE as the Inuvialuit (or Mackenzie Inuit) period, due to changes in the form of houses and artifacts. However, all sites form an unbroken cultural continuum, which we refer to in aggregate as Inuvialuit (7).
A zooarchaeological study of the Kuukpak site in the Mackenzie Delta, which was inhabited from ca. 1400 to 1870 CE, indicated beluga whales as the most commonly occurring species, as measured by number of specimens (8). When bone numbers were converted to meat weights, beluga whales were estimated to provide ~66% of all consumed meat and fat at the site. The Mackenzie Delta beluga whale harvests provided enough resources to make the settlements on the river banks among the largest 19th-century communities in the entire Canadian Arctic (6). For example, the Kuukpak site held a minimum of 29 multifamily houses, though not all would have been occupied simultaneously (9). Beluga whales [qilalugaq in the Siglit Inuvialuit dialect (10)] to this day remain of central importance to the region’s culture and economy (11).
Inuvialuit historians such as Nuligak (12), as well as 19th-century European observers (13, 14), describe the late summer beluga whale hunt as it occurred before the introduction of European technologies in the late 19th century. These intensive hunts were performed by dozens of men in kayaks forming a line and driving beluga whales into the shallows, where they could be harpooned and lanced. It is difficult to determine with certainty how far back in time this beluga whale hunting method goes. Previous zooarchaeological analysis of beluga whale age distributions from the Kuukpak site indicates that drive hunting has been practiced well into the precontact past (2). However, we do not know when these drive methods were first developed, or whether the earliest Thule people hunted with this method.
While precise numbers harvested during these hunts are unclear, one estimate based on notes taken by Isaac Stringer, an Anglican missionary, indicated that in 1893 this method yielded approximately 155 beluga whales at Kitigaaryuit (15). This is equivalent to nearly one beluga whale for each woman, man, and child at the settlement (159 total in 1893). In earlier periods, before the tragic impacts of epidemic diseases, Inuvialuit populations at the two major beluga hunting sites of Kuukpak and Kitigaaryuit were much higher, with most estimates indicating a combined summer population of at least 1,000 people (6, 16). Thus, if hunting success rates per capita were even half those recorded in 1893, an average summer would see hundreds of beluga whales harvested, perhaps 500 or more.
Subsistence harvests of this beluga whale population continued throughout the 20th and 21st centuries in the six communities of the Inuvialuit Settlement Region, to a total of ca. 100 individuals taken each year (17). The majority of these beluga whales were and continue to be landed in the Mackenzie Delta, suggesting variable but relatively high harvest levels from the 14th century up to the present day.
Contemporary beluga whales in the Mackenzie Delta belong to the Eastern Beaufort Sea population, which numbers ~40,000 individuals (18) and is among the largest beluga whale populations globally (19). The population annually migrates several thousand kilometers (Fig. 1A). We assume this migratory behavior has been continuous throughout the 700 y investigated in this study, as beluga whales have strong site fidelity to their summering grounds, which can be maintained over millennia (20). The Beaufort Sea beluga whales form large summering aggregations in the Mackenzie Delta estuary (21, 22), and then travel further offshore in the eastern Beaufort Sea and the adjacent waters of the Canada Basin, Amundsen Gulf, and Viscount Melville Sound, when the areas become ice-free during the summer (23–25). As sea ice forms in autumn, individuals migrate west through the Bering Strait to the Bering Sea, where several other beluga whale populations overwinter (26). As sea ice starts to break up and the Bering Strait opens in spring, they migrate back, thus completing their annual migration.
Time-series faunal data from archaeological deposits provide insights into past human resource use and the ecology of past faunal populations. Ancient biomolecular approaches that integrate radiocarbon dating, paleogenomics, and stable carbon (δ13C) and nitrogen (δ15N) isotope analysis, provide the toolbox necessary to further elucidate past Inuvialuit hunting practices and their possible impacts on the beluga whale population. Genome-wide ancient DNA can be used for genetic sexing to i) address possible sex bias in the beluga whale harvests, ii) estimate levels of genetic diversity, and iii) assess patterns of population subdivision and continuity through time. Bone collagen stable carbon (δ13C) and nitrogen (δ15N) isotope records provide information on the long-term foraging ecology and habitat use of the harvested individuals, and can be used to uncover temporal ecological shifts, which may have affected the beluga whale populations and the Inuvialuit communities reliant on them for survival.
In this study, we used a combined biomolecular approach of radiocarbon dating, paleogenomics, and stable δ13C and δ15N isotope analysis, to investigate whether and to what degree human harvesting over the past 700 y impacted the beluga whale population that continues to aggregate in the Mackenzie Delta. Specifically, we investigated i) patterns of sex-biased hunting, and whether practices changed through time; ii) population diversity and continuity over time based on patterns of genetic variation and structuring; iii) the long-term foraging ecology of the harvested beluga whales, using stable isotope data as a proxy; and iv) the spatiotemporal relationship of the Mackenzie belugas with adjacent beluga populations. We analyzed 45 zooarchaeological specimens from the Cache Point and Kuukpak archaeological sites, which covered three distinct time periods: 1290 to 1440 CE; 1450 to 1650 CE; and 1800 to 1870 CE (Fig. 1 and SI Appendix, Tables S1 and S2). For comparison with the zooarchaeological remains, we included ten contemporary Mackenzie Delta beluga whales from recent Inuvialuit subsistence hunts near Hendrickson Island, and adjacent populations in Anadyr (Russia), Bristol Bay, and Cook Inlet (Alaska, US).
Results
Radiocarbon Dating.
The sixteen radiocarbon dates obtained for this study, in combination with one available date (3), allowed us to refine the temporal phases of the Cache Point and Kuukpak sites. The sites were split into three distinct and successive time periods in a temporal sequence: 1290 to 1440 CE (Cache Point site houses 6 and 8); 1450 to 1650 CE (Kuukpak site Area 3 House 5); and 1800 to 1870 CE (Kuukpak site Area 5 House 1) (Fig. 1 and SI Appendix, Table S1). We further discuss the dating evidence in the Materials and Methods section.
Stable Isotope Analysis.
Stable isotope analysis was carried out on the zooarchaeological material only, as we had no bone material available from contemporary Mackenzie Delta beluga whales, or the adjacent populations in Anadyr, Bristol Bay, and Cook Inlet. In total, 40 samples passed the stable isotope analysis quality control with an atomic C:N ratio of 2.9 to 3.6 (27), which indicates that the collagen’s isotopic composition was not altered in the burial environment (SI Appendix, Table S2).
The collagen yields were relatively low (average of 4.5%, SI Appendix, Table S2) but this reflects the low collagen content of the skeletal element (the extremely dense petrous portion of the temporal bone) rather than low levels of organic preservation. Accordingly, collagen quality control indicators based on elemental compositions were prioritized over those based on collagen yield. Isotopic data were excluded from the analysis if they had atomic C:N ratios greater than 3.6 or less than 2.9 (27) and if they had <13% C or 4.8% N by weight (28). The stable isotope dataset included 16 samples in the early (1290 to 1440 CE), 13 samples in the middle (1450 to 1650 CE), and 11 samples in the late (1800 to 1870 CE) time period (Fig. 1C and SI Appendix, Tables S2 and S3).
Taking only time into account, we compared data across the three archaeological time periods. Bone collagen δ13C differed significantly among the three time periods [ANOVA, F(2,36) = 4.8, P = 0.01], Fig. 2 and SI Appendix, Fig. S1 and Table S3). Values were lower during 1290 to 1440 CE than during 1450 to 1650 CE and 1800 to 1870 CE (Tukey HSD, P = 0.03 and P = 0.05 for each comparison respectively), but did not significantly differ between 1450 to 1650 CE and 1800 to 1870 CE (Tukey HSD, P = 0.99).
Fig. 2.
Bone collagen δ13C for female (F) and male (M) Mackenzie Delta beluga whales for the three time periods analyzed. Sample sizes are indicated below each bar plot.
Taking only sex into account and ignoring time periods, we compared females and males from the combined zooarchaeological samples (16 females and 24 males). Bone collagen δ13C differed significantly between sexes [ANOVA, F(1,36) = 8.2, P < 0.01]; females had higher δ13C than males. When we took time into account, bone collagen δ13C was significantly higher in females than in males for the early and late time periods [1290 to 1440 CE; t = 3.01, df = 8, P = 0.02), (1800 to 1870 CE; t = 2.89, df = 7, P = 0.02, Fig. 2 and SI Appendix, Fig. S1). In the middle time period, δ13C did not differ between sexes (1450 to 1650 CE; t = 0.25, df = 10, P = 0.81). We acknowledge there are only three data points for females in 1800 to 1870 CE, and thus the results need to be interpreted with caution.
Bone collagen δ15N did not differ significantly among time periods, or between sexes pooled across time (Kruskal–Wallis X2 = 0.21, df = 1, P = 0.65 and Kruskal–Wallis X2 = 4.11, df = 2, P = 0.13 respectively) (SI Appendix, Fig. S1). Thus, we did not compare bone collagen δ15N between sexes within each time period, given we found no differences overall.
Comparing niche size, we find females (SEAB = 1.18, SEAC = 1.20) in 1290 to 1440 CE had a larger isotopic niche than males (SEAB = 0.35, SEAC = 0.35, proportion (p) of simulated ellipses that are larger in females than males = 0.99, SI Appendix, Fig. S1). In 1450 to 1650 CE, we observed no significant differences in isotopic niche size between females (SEAB = 1.20, SEAC = 1.20) and males (SEAB = 2.00, SEAC = 0.35, P = 0.82). We did not compare isotopic niches between males (SEAB = 1.34, SEAC = 1.35) and females in 1800 to 1870 CE, as females had too few data points (n = 3) to accurately estimate a SEA. Ellipse overlap between the isotopic niches of females and males was 4% in 1290 to 1440 CE and 50% in 1450 to 1650 CE (SI Appendix, Fig. S1). We could not estimate overlap in 1800 to 1870 CE due to the limited data for females.
Genomic Analysis.
The genome-wide nuclear dataset included 45 zooarchaeological specimens (Fig. 1 and SI Appendix, Table S2). Twenty-one of these specimens had a coverage, estimated as genome-wide average read depth, below 0.2× and were only used for genetic sexing. The genome-wide coverage of the remaining 24 zooarchaeological specimens ranged from 0.2× to 3.9×, with the majority of samples (19 out of 24) having between 0.5× and 1.8× coverage. For the 37 samples from the four contemporary beluga whale populations, coverage ranged from 0.4× to 2.9× (Dataset S1).
Genetic sexing of the zooarchaeological dataset identified 18 females and 27 males, but with differences in the ratio of females and males in each time period (Fig. 1 C and D and SI Appendix, Table S2). In the earliest time period (1290 to 1440 CE), we identified 6 females and 12 males. In the middle time period (1450 to 1650 CE) we identified 7 females and 6 males. In the most recent period (1800 to 1870 CE) we identified 5 females and 9 males. For the four contemporary populations, an equal proportion of females and males were targeted for DNA sequencing, and this was confirmed by genetic sexing (SI Appendix, Table S2).
The NGSrelate analysis showed that our nuclear dataset of 24 zooarchaeological and 37 contemporary samples included two pairs of closely related individuals, determined by relatedness coefficients (r) > 0.3 (SI Appendix, Fig. S2). The relatedness coefficients cannot be used to identify the degree of relatedness, such as parent–offspring or siblings, but rather can be used as an indicator of shared minor alleles. In each pair, we excluded the individual with lowest nuclear coverage from further nuclear analysis.
The first individual excluded from the nuclear analyses was CGG1023693 from 1450 to 1650 CE, which was related to CGG1023691, also from 1450 to 1650 CE (r = 0.68). Both were sampled from house A3H5 and were genetically identified as females, with autosome to sex-linked coverage ratios of 0.96 and 1.03, respectively. They also had the same mitochondrial haplotype. However, both samples were petrous bones from the left side, thus ruling out that they could be the same individual. This is further supported by r = 0.68, which would lie very close to 1 if both samples were from the same individual; duplicate samples would be expected to always share the minor allele.
The second individual excluded from the nuclear analyses was CGG1020771, a contemporary sample from Hendrickson Island, which was related to CGG1020772 from the same locality (r = 0.46). Both individuals were genetically identified as male, with identical autosome to sex-linked coverages of 0.55 (SI Appendix, Table S2), and shared mitochondrial haplotypes. They were collected on the same date, July 3rd 2004. However, the low r value makes it unlikely that the samples were from the same individual. Related belugas are often part of the same pod (29) and thus it makes sense that two related males may have been harvested on the same day.
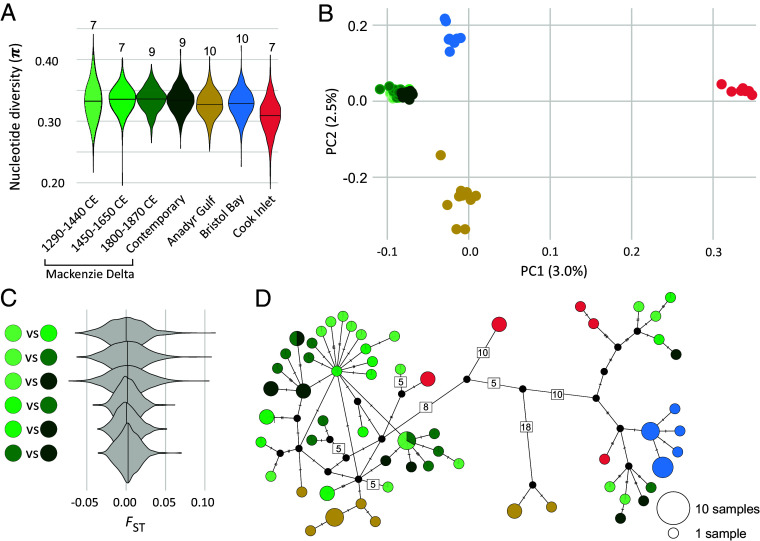
After removal of the two related individuals, our nuclear analysis included 23 zooarchaeological specimens: seven from 1290 to 1440 CE; seven from 1450 to 1650 CE; nine samples 1800 to 1870 CE, and 36 contemporary samples from four sites in Canada, Russia, and Alaska (Fig. 1 C and D). The filtered genomic dataset of the 59 samples used for analysis of diversity and differentiation included 467,920 variable sites. The mean nuclear nucleotide diversity of the Mackenzie Delta remained stable across time (Fig. 3A): 0.332 (1290 to 1440 CE), 0.335 (1450 to 1650 CE), 0.335 (1800 to 1870 CE), and 0.334 (Hendrickson Island). The three adjacent populations had nucleotide diversities of 0.327 (Anadyr), 0.328 (Bristol Bay), and 0.308 (Cook Inlet).
Fig. 3.
Genomic analysis of belugas sampled across four time periods from the Mackenzie Delta, and three adjacent populations. (A) Genome-wide nucleotide diversity for each group, with sample sizes indicated. (B) Principal component analysis, indicating the percentage of variation described by each axis. (C) Fixation index FST of all pairwise comparisons of the four Mackenzie Delta time-periods. (D) Network of the 60 mitochondrial haplotypes found among 77 beluga individuals. Relative circle size indicates the number of individuals sharing a haplotype. The number of substitutions among haplotypes are indicated by hashes or by numbers for >4. Of note, length of branches between haplotypes is not drawn to scale.
To gain an intuition of the expected trajectory of nucleotide diversity across the time frame of the population history of belugas investigated in this study, we conducted population genetic simulations. To capture the uncertainties of the demographic history of belugas, we performed simulations across a grid of parameters which reflect the effective population size and reasonable rates of hunting (SI Appendix, Fig. S3). We found that—regardless of the parameter combination in each simulation scenario—the distributions of simulated nucleotide diversities across time do not indicate any significant changes in nucleotide diversity across the time frame studied here (SI Appendix, Fig. S4 and Table S4), thus supporting the evidence from our empirical data (Fig. 3A).
In the principal component analysis, the Cook Inlet beluga whales separated from the remaining samples on PC1, which captured 3.0% of the variation in the dataset, while the Anadyr and Bristol Bay beluga whales separated on PC2, which captured 2.5% of the variation (Fig. 3B). The archaeological and contemporary Mackenzie Delta beluga whales clustered together, suggesting population continuity through time. The pairwise FST comparisons among the Mackenzie Delta time periods, both zooarchaeological and contemporary, were close to zero (0.0005 to 0.0032), indicating a lack of temporal structuring and supporting population continuity across the 700 y covered by our study (Fig. 3C). Our D-Stat analysis of D[H1-Zooarchaeological, H2-Contemporary, H3-Adjacent, Outgroup] combinations of samples yielded low D values between −0.07 and 0.04 (SI Appendix, Fig. S5) and associated z scores between −4.03 and 3.20 (SI Appendix, Fig. S6). Only 26 and 2 of the 5589 z scores were below −3 and above 3, respectively. These results suggest no significant gene flow events between the four Mackenzie Delta time periods and the three adjacent populations.
The mitochondrial dataset had 40 archaeological and 37 contemporary samples (Fig. 1 C and D and 3D), including the four related individuals identified genetically. When calling sites with less than five reads in a sample as missing data, the mitochondrial alignment had data for all 77 sampled individuals in 16,361 of the 16,386 base pairs, which constitutes 99.9% of the mitochondrial genome. Missing data were only found in the ends of the mitochondrial genome, where no variation has been reported based on a range-wide dataset of 206 beluga whales sampled across 18 of the 21 recognized management units (20). This was most likely due to poor mapping related to the circular mitochondrial genome being treated as a linear sequence in the genome aligner used [BWA; (30)].
The mitochondrial alignment included 156 segregating sites and 60 haplotypes, 35 of which were exclusively found in the zooarchaeological samples (Fig. 3D). Mean estimates of mitochondrial nucleotide diversity for each time period in Mackenzie Delta were 0.0011, (1290 to 1440 CE), 0.0009 (1450 to 1650 CE), 0.0007 (1800 to 1870 CE), and 0.0011 (Contemporary) (SI Appendix, Table S5) and were not significantly different with P values between 0.209 and 0.926. For each of the contemporary adjacent populations, the nucleotide diversity was estimated to be 0.0011 (Anadyr), 0.0001 (Bristol Bay), and 0.0015 (Cook Inlet).
Mitochondrial haplotype diversity of the four Mackenzie Delta time periods ranged between 0.96 and 1, while haplotype diversities of the adjacent populations ranged between 0.80 and 0.91 (SI Appendix, Table S5). One haplotype was shared between the earliest (1290 to 1440 CE) and latest (1800 to 1870 CE) time period, and one haplotype was shared between 1800 and 1870 CE and the contemporary Mackenzie Delta samples (Fig. 3D). Neither the zooarchaeological nor the contemporary Mackenzie Delta samples shared any haplotypes with the adjacent populations. When comparing the Mackenzie Delta mitochondrial haplotypes with a global dataset (20), we saw a similar pattern; none were shared with other localities (SI Appendix, Fig. S7). Hence, all but one haplotype in the archaeological Mackenzie sequences are new to the species.
Discussion
We present a combined biomolecular analysis of beluga whale faunal remains from archaeological contexts and contemporary populations in the Mackenzie Delta, Northwest Territories, Canada, spanning the past seven centuries. Our simultaneous investigation of DNA and stable isotopes across the time series offers a unique opportunity to elucidate the impact of premodern subsistence harvests of beluga whales and to assess temporal patterns in their ecology, diversity, and structuring.
Shifting Hunting Practices.
We found differences in the sex ratio of beluga whales harvested across the three archaeological time periods analyzed, although we acknowledge our sample sizes are limited (Fig. 1). In the early (1290 to 1440 CE) and late (1800 to 1870 CE) time periods, we observed a ca. 1:2 female to male ratio, but in the intermediate time period (1450 to 1650 CE) the sex ratio was close to equal. The earliest time period in our dataset (1290 to 1440 CE) coincides with the first colonization of Thule Inuit from Alaska (7), which differed in several significant ways from later Inuvialuit culture. In particular, newly arrived Thule Inuit may have had no prior experience hunting beluga whales regularly and in large numbers, as indicated by the small proportion of beluga whales identified in archaeological findings in Alaska (31). As a result, it is possible that the later Inuvialuit methods of mass drive hunting had not yet been developed, and early Thule may have instead targeted a single individual whale in each hunt. If larger individuals were selected, harvests may have been biased toward males, as males are around 30% larger than females (32).
Ethnographic records of the 19th century describe the later Inuvialuit hunting practices as drive hunts (2). The beluga whales were guided into shallow waters, where they could more easily be harvested. Mandible growth layer distributions from previous studies of the Kuukpak site indicate that premodern Inuvialuit drive hunts were most likely indiscriminate regarding ages (2), targeting all individuals in the pod, which usually comprises all ages and both sexes (33). This suggests the individuals from the intermediate (1450 to 1650 CE) and late (1800 to 1870 CE) time periods may represent a random sample of beluga whales available to the Inuvialuit in the Mackenzie Delta. Thus, the observed difference in sex ratio between these two later time periods may indicate either a change in the sex ratio of the available beluga whales, or perhaps a shift in hunting practices, with a preference for males in the later time period (1800 to 1870 CE).
The present-day beluga whale hunt in the Mackenzie Delta by Inuvialuit is strongly male-biased; between 1973 and 1999, 2.3 males were harvested for every female (n = 3,687) (34). As a result of a local hunting practice aimed at the conservation of reproductive females, the male bias in beluga whale harvests increased to four males for every female between 2005 and 2016 (n = 1,200) (17). Thus, the male bias in beluga whale harvests in the Mackenzie Delta is not a recent phenomenon but has increased from an equal ratio (1450 to 1650 CE), to 2:1 (1800 to 1870 CE), to 2.3:1 (1973 to 1999) and finally to 4:1 (2005 to 2016). Male-biased harvests may play a positive demographic role in the conservation of beluga whale populations, as it may minimize the effect of the harvests on female reproductive rates and on the survival of nursing calves, which would likely die if their mothers were taken during the hunt.
Temporal Patterns in Beluga Whale Foraging Ecology.
Our data suggest the dietary niches of female and male beluga whales have changed over time, and may reflect either temporal changes in prey preference or habitat use, or underlying shifts in the nutrient composition in the area. While females and males had similar δ13C in the middle time period (1450 to 1650 CE), females had significantly higher δ13C in both the early (1290 to 1440 CE) and late (1800 to 1870 CE) time periods (Fig. 2). Comparable bone collagen stable isotope compositions from contemporary beluga whales in the Canadian Arctic and western Greenland did not show differences in δ13C between sexes (35, 36), which in combination with our findings suggest dietary niches of beluga whales are variable in time and space. Size-based differences in δ13C have been reported in soft tissues from contemporary Mackenzie Delta beluga whales, with higher δ13C values in females than in large and medium-sized males, but no difference was found between females and small males (37). Although differences in turnover rate between soft tissue (months) and bone collagen (years) mean the data are not directly comparable with ours, these previous findings, in combination with our results, suggest both sex and size influence δ13C levels in beluga whales, as has been shown in other toothed whales (36, 38). We were unable to estimate the size of the beluga whale individuals in our analysis based on the available zooarchaeological material (petrous bones), as no conversion ratio is available, but this would be a valuable variable to include in future studies.
Quantitative analyses of fatty acids have identified three species of fish of importance for beluga whales in the Beaufort Sea: Arctic cod (Boreogadus saida), capelin (Mallotus villosus), and Canadian eelpout (Lycodes polaris) (39). Medium and large males rely primarily on Arctic cod, while females and small males also rely on capelin and Canadian eelpout. Arctic cod and capelin have similar δ13C values, but Canadian eelpout, which make up more than 25% of the diets of females and small males, have significantly higher δ13C (39). This dietary differentiation between sex and size may explain the higher values of δ13C in female beluga whales found in the early (1290 to 1440 CE) and late (1800 to 1870 CE) Mackenzie Delta time period, as well as in tissue samples from contemporary beluga whales from the Beaufort Sea (37). Dietary differentiation related to sex and size may reflect differential habitat use and energetic needs (40); females prefer to stay near shore with their calves, where they would focus feeding on coastal fish species, such as capelin and Canadian eelpout, while large males may search for Arctic cod further north under the sea ice (37).
We observed a notable pattern when we combined the dietary niche of females and males with the sex ratio of the harvested beluga whales within a given time period. In the early (1290 to 1440 CE) and late (1800 to 1870 CE) time periods, females had significantly higher δ13C values than males and were harvested at a 1:2 ratio. In the intermediate time period (1450 to 1650 CE), we observed no differences in δ13C between sexes, and an equal ratio of harvested females and males. While this relationship may call for a unified explanation, we were unable to further identify any biological, environmental, or cultural driver that may offer a combined explanation.
Impact of Subsistence Harvests.
We observed similar levels of genomic diversity across time periods in the Mackenzie Delta (Fig. 3A). This is consistent with population genetic simulations of the expected trajectories of nucleotide diversity across a range of reductions in effective population size over time, which also do not indicate any significant changes in nucleotide diversity within the time frame studied here (SI Appendix, Fig. S4). Of course, average genome-wide nucleotide diversity is but one measure of population diversity. When permitted by sufficient data quality, future studies should consider complementing this statistic with other metrics, such as those quantifying extended runs of homozygosity. We also observe a lack of population differentiation across time periods in the Mackenzie Delta (Fig. 3C), suggesting the belugas analyzed were part of the same, continuous population.
At a global scale, beluga whales have high mitochondrial diversity, with few shared haplotypes among individuals (20). The Mackenzie Delta samples do not share any mitochondrial haplotypes with the adjacent populations (Fig. 3D) or with beluga populations elsewhere (SI Appendix, Fig. S7). During the annual migration to their wintering grounds in the Bering sea (Fig. 1A), the Beaufort Sea beluga whales come close to other beluga whale populations that also winter in the area (26). Beluga whales are believed to mate during winter, but our data do not indicate gene flow between Mackenzie delta beluga whales and adjacent populations (SI Appendix, Figs. S5 and S6), which are recognized and managed as separate stocks (19).
Beluga whales have a generation time of 32 y (41); after reaching sexual maturity at 8 to 13 y, females produce a single calf every 3 y (42). The considerable number of beluga whales harvested each year by premodern Inuvialuit, which based on ethnographic records is estimated to number in the hundreds (15), translates into thousands of individuals removed per generation from the Beaufort Sea population, to which the Mackenzie Delta beluga whales belong. Aerial surveys of the Beaufort Sea population, carried out in 1992 and in 2019, estimated a census size of ~40,000 individuals (18, 21). This makes it one of the largest beluga whale populations globally, second only to the population in Hudson Bay, Canada (19), which may in part have buffered any impact of the Inuvialuit harvests.
We investigate Indigenous subsistence whaling using time-series, population-level biomolecular data. Other studies comparing (primarily mitochondrial) DNA from historic samples and contemporary individuals have focused on large marine mammal species (baleen whales, sperm whale [Physeter macrocephalus], and walrus [Odobenus rosmarus]), which were targeted by commercial overexploitation between the late-16th and mid-20th centuries (43). Comparisons of the genetic diversity of individuals pre- and postcommercial harvests in humpback whales (Megaptera novaeangliae), gray whales (Eschrichtius robustus), North Atlantic right whales (Eubalaena glacialis), and Pacific walrusses (Odobenus rosmarus divergens), have not found any significant difference when applying mitochondrial or microsatellite markers (44–47). However, analyses of high-coverage genomes of contemporary bowhead whales (Balaena mysticetus) and fin whales (Balaenoptera physalus) have indicated a genetic impact of commercial whaling, identified as modeled changes in recent effective population sizes (48–50).
To our knowledge, only one other study has utilized time-series paleogenomes to estimate changes in diversity across a time frame that included prehistoric Indigenous subsistence harvests. Westbury et al. (51) investigated bowhead whale fossils spanning the past 11,000 y in age, including contemporary individuals, from the Canadian Arctic Archipelago, and found no genetic impact of Thule subsistence harvests on the population. Rather, they found a loss of genetic diversity in contemporary populations associated with commercial whaling that is still ongoing, and will continue many centuries into the future.
While belugas were not targeted by large-scale commercial whaling due to their size, local overharvests led to the extirpation of a beluga population in southwest Greenland a century ago. Between 1872 and 1930, 8,000 to 10,000 beluga individuals were harvested, primarily from Nuuk and Maniitsoq in west Greenland (52, 53). Indeed, south of 66°N, more than 7,600 individuals were harvested between 1919 and 1926, with 1500 beluga individuals taken in a single year (1926). Ultimately, the harvests resulted in the extirpation of the beluga population in southwest Greenland, which has still not returned to the area.
In our study, we find no change in diversity and a lack of temporal structuring in the Mackenzie Delta beluga whales based on paleo- and population genomic analysis of beluga individuals spanning seven centuries in age. Our integration of genetic sexing and stable isotope analysis reveal temporal shifts in the sex ratio of the harvested beluga whales, and in their δ13C values. Our findings suggest the impact of Inuvialuit subsistence harvests, which still form a social and economic focal point of the Inuvialuit community, has been negligible across the 700 y covered by our study.
Materials and Methods
Radiocarbon Dating.
The chronology of the archaeological contexts in this study is based primarily on 17 radiocarbon dates (SI Appendix, Table S1). One, BETA 201281, has been previously published (3); the other 16 are new to this study. All are on terrestrial mammal bone–caribou (Rangifer tarandus), moose (Alces alces), and Dall sheep (Ovis dalli)–which is widely considered the most accurate dating material in Arctic contexts (54). These new dates were processed at the W. M. Keck Carbon Cycle AMS facility at the University of California, Irvine. For bone samples, this lab uses pretreatment involving cleaning, decalcification, gelatinization, and ultrafiltration (55). Calibration of dates was performed in OxCal 4.4 (56), using the IntCal20 calibration curve (57).
Archaeological Sites and Samples.
Beluga whale petrous bones were collected from the excavations of two archeological sites: Cache Point and Kuukpak (Fig. 1 and SI Appendix, Table S1). Cache Point is the earliest large settlement on the Mackenzie River East Channel (3, 5, 58). Its early date is based on material culture traits relating to the “Thule Inuit” period, representing the original Inuit migrations from Alaska beginning ca. 1200 CE. These traits at Cache Point include open socket harpoon heads and distinctive Thule house forms with a single rear alcove and separate kitchen structure. The early date is also based on the site’s position as the southernmost of the major beluga whale hunting sites. Because the mouth of the Mackenzie River is gradually silting in, major occupations have been moving downstream (northward) over time, presumably to position settlements near to beluga whale aggregations.
Cache Point contains a minimum of 23 relatively small semisubterranean houses, as well as many other pit features. The present samples are drawn from two fully excavated houses, House 6 and House 8. Based on calibration of three radiocarbon dates from Cache Point House 6 that range from 620 ± 40 BP to 545 ± 20 BP, House 6 was occupied between ca. 1290 and 1430 CE. Two radiocarbon dates obtained for House 8 are at 530 ± 20 BP and 505 ± 20 BP; calibration suggests an occupation between ca. 1330 and 1440 CE. Note that four additional radiocarbon dates were previously reported for these houses (3), but are no longer considered reliable because they were not prepared with modern pretreatment techniques, including ultrafiltration, and are therefore more likely subject to contamination issues. Their inclusion would not significantly alter the dating of these houses.
The second site, Kuukpak, is located farther downstream and was occupied over a longer period (5, 8, 58). This very large site contains a minimum of 29 houses, most or all of which are of the large “cruciform” multifamily type (9). It extends for 750 m along the East Channel, and active erosion along much of this length reveals deep middens with large numbers of beluga whale bones visible. Based on oral histories, the site was occupied until the mid-19th century (59); its precise date of abandonment is difficult to reconstruct but may have been around 1860 to 1870 CE. The present samples are drawn from two fully excavated large houses with contrasting occupation histories. Area 3 House 5 (A3H5) contained only precontact archaeological remains from subsurface levels, with no European trade goods. Nine radiocarbon dates are tightly clustered from 375 ± 20 BP to 305 ± 20 BP; when calibrated these indicate an age range of ca. 1450 to 1650 CE (SI Appendix, Table S1).
The second excavated house from Kuukpak, Area 5 House 1 (A5H1), has a very different occupation history. Upper levels, including the house fill as well as the floor and sleeping benches, contained large numbers of European trade goods, consisting primarily of over 400 glass beads. Thus, the upper levels, which comprised most of the contexts excavated, date mainly to the 19th century. Due to limitations with radiocarbon dating of very recent samples, these recent levels were for the most part not radiocarbon dated. However, because the site history is complex and this house was likely constructed on top of earlier houses or middens, three radiocarbon dates were run on deeper levels or areas of the house suspected to contain mixed assemblages. Two of these dates relate to an earlier occupation, at 300 ± 25 BP and 345 ± 20 BP, confirming a precontact component roughly contemporaneous with A3H5 underlies the final A5H1 house construction (SI Appendix, Table S1). This includes one date on the floor level, indicating some mixing of earlier materials into later levels. The third date, at 140 ± 20 BP (UCIAMS 225638), which came from under the floor, calibrates to a very wide range of 1673 to 1944 CE, but likely relates to the historic period occupation.
In summary, we analyze the beluga whale specimens as belonging to three successive datasets in a temporal sequence, based on a combination of locational, typological, and radiocarbon information (Fig. 1). The early part of this period, from ca. 1300 to 1400 CE, is referred to by archaeologists as the “Thule period,” and after ca. 1400 CE as the Inuvialuit (or Mackenzie Inuit) period, due to changes in the form of houses and artifacts; however, all sites form an unbroken cultural continuum, which we refer to in aggregate as Inuvialuit (7). Specimens from the two Cache Point houses are combined to provide the earliest sample dating to the period ca. 1290 to 1440 CE, and represent Thule culture. The Kuukpak A3H5 sample dates to ca. 1450 to 1650 CE. The Kuukpak A5H1 sample dates mainly to the period ca. 1800 to 1870 CE, though due to some degree of mixing it may include some specimens from components as early as 1475 CE.
Beluga Whale Faunal Material.
In total, we sampled 45 prehistoric and historic specimens from Cache Point and Kuukpak (Fig. 1 and SI Appendix, Table S2). Within each of the four house samples, beluga whale petrous bones were selected based on the most frequently occurring side, to ensure that all were from different individuals: Cache Point H6–right; Cache Point H8–right; Kuukpak A3H5–left; Kuukpak A5H1–left. None of these specimens were radiocarbon dated because of uncertainties regarding the regional marine reservoir correction. Ages were context dated based on the radiocarbon dates of associated terrestrial material, mentioned above (SI Appendix, Table S1).
To contextualize the zooarchaeological material with contemporary data from the delta, we included 10 tissue samples from beluga whales harvested around Hendrickson Island by the Inuvialuit inhabiting the region; samples were collected between 1997 and 2009 (Fig. 1 and SI Appendix, Table S2). We also included contemporary tissue samples from adjacent beluga whale populations; 10 samples from Anadyr, 7 samples from Cook Inlet, and 10 samples from Bristol Bay (Fig. 1 C and D and SI Appendix, Table S2). The contemporary tissue samples were only included in our DNA analysis, as we did not have bone material available for stable isotope analysis.
Community Consultations on Research and Sample Acquisition.
Archaeological samples were collected under Northwest Territories archaeology permits 98-664, 99-883, 2014-006, 2016-008, and 2017-006, and archaeological activities were performed in a collaborative manner with Inuvialuit organizations. In particular, the Kuukpak samples were collected as part of the Arctic Cultural Heritage at Risk (Arctic CHAR) project, which was performed as a partnership between the Inuvialuit Cultural Centre (ICC) and the University of Toronto. The ICC provided direction throughout the 5-y project, and indicated approval for further analyses of the large number of zooarchaeological samples recovered. Information flowed back to Inuvialuit communities through public reports, school visits, public talks, and meetings with the ICC and the Tuktoyaktuk Hunters and Trappers Committee (HTC).
Contemporary tissue samples were collected from beluga harvests annually as part of a collaborative regional sampling program between communities, the Fisheries Joint Management Committee, and Fisheries and Oceans Canada. Specific samples from harvested beluga whales from 1997 and 2004 were part of a regional genetics study led by Postma and supported by comanagement (Fisheries Joint Management Committee and Fisheries and Oceans Canada) to address community and scientific questions related to beluga kinship. These samples were shared with Lorenzen for global genomic research and were included in this study. Recognizing these samples were collected long ago, Loseto facilitated connecting Skovrind with the Tuktoyaktuk HTC to discuss the project. Attempts to hold virtual meetings were not successful due to problems with internet stability, so Skovrind sent a detailed letter describing the project and formally requesting permission to use these samples for the study. The Tuktoyaktuk HTC responded with support for the project and use of samples.
Stable Isotope Data Generation.
For the bone collagen stable carbon (δ13C) and nitrogen (δ15N) isotope analysis, powdered samples of the zooarchaeological petrous bone specimens were analyzed at the Water Quality Centre at Trent University, Canada (SI Appendix, Tables S2 and S3).
Approximately 100 to 150 µg of powdered bone from zooarchaeological samples were demineralized in 0.5 M HCl at room temperature under constant motion provided by an orbital shaker for 4 h. The samples were then rinsed to neutrality with Type I water and treated with 0.1 M NaOH for successive 20 min increments until there was no color change in the solution. The samples were rinsed to neutrality, and suspended in 0.01 M HCl at 75 °C for 36 h to solubilize the collagen. The solution containing the collagen was collected and transferred to preweighed glass vials, frozen, and freeze-dried. The collagen yields were calculated for each specimen.
Carbon and nitrogen isotopic and elemental compositions were determined using a Nu Horizon continuous-flow isotope-ratio mass spectrometer (CF-IRMS) coupled to a EuroVector 3300 elemental analyzer at Trent University. Sample measurements were calibrated relative to VPDB (δ13C) and AIR (δ15N) using USGS40 and USGS41, USGS63, or USGS66 (60, 61). The SD and number of calibration (quality control) standards used in all of the analytical sessions are listed in SI Appendix, Table S6.
Standard uncertainty for the δ13C and δ15N measurements of the samples was estimated following Szpak et al. (62), which largely follows the method presented in ref. 63. Systematic errors (u(bias)) were calculated to be ± 0.09 ‰ for δ13C and ± 0.23 for δ15N based on the known uncertainty in the check standards and the observed SD of those check standards from the known values (SI Appendix, Tables S7 and S8). Random errors (uR(w)) were calculated to be ± 0.18 ‰ for δ13C and ± 0.21 ‰ for δ15N based on the pooled SD of the check standards and sample replicates. Standard uncertainty, calculated as the root-sum-square of u(bias) and (uR(w)) was determined to be ± 0.20 for δ13C and ± 0.31 for δ15N.
Stable Isotope Analysis.
We tested for dietary differences among beluga whales sampled in the three zooarchaeological time periods, and between sexes, by comparing δ13C using ANOVA and Tukey’s post hoc tests, and δ15N using Kruskal–Wallis tests. No contemporary samples were included in the δ13C and δ15N analyses, as no bone samples were available. Our data satisfied normality and homogeneity of variances for δ13C, but not for δ15N. We further tested for ecological differences between female and male beluga whales within each time period by comparing δ13C using Student’s t tests; the data satisfied normality and homogeneity of variances. All statistical analyses were performed in R v.4.0.5 (64).
We used the isotopic niche as a proxy for ecological niche (65, 66), and compared isotopic niches between female and male beluga whales within each time period using Bayesian multivariate ellipse-based metrics implemented in the packages SIBER and rjags (67, 68). We calculated standard ellipse areas corrected for sample size (SEAC), and Bayesian standard ellipses (SEAB) for each sex for each time period. We estimated SEAB using 105 posterior draws, a burn-in of 103 and a thinning of 10 and used SEAB to test for differences in niche width between female and male beluga whales within each time period (i.e., the proportion (p) of draws of the posterior distribution of the SEAB in which the area of one group was smaller than the other), when sample size allowed. We set the prediction ellipses to contain approximately 40% of the data. We evaluated isotopic niche similarity between two groups as the proportion (%) of the nonoverlapping area of the maximum likelihood fitted ellipses of the two.
DNA Data Generation.
Petrous bones were drilled to extract 30 to 70 µg bone powder for DNA extraction. To minimize contamination, drilling, DNA extractions, and library builds were conducted in the designated ancient DNA clean lab facilities at Globe Institute, University of Copenhagen. Postlibrary indexing was performed in a lab in a separate building, to prevent contamination with PCR products.
DNA was extracted from the bone powder using the extraction buffer described by Dabney et al. (69), with the inclusion of a 30-min predigestion step to increase the endogenous DNA yield (70). The extract was concentrated using 30 kDa Centrifugal Filter Units and further concentrated and cleaned using Qiagen Minelute tubes. Libraries were built on the DNA extract using the single-tube BEST protocol (71) and or the single-strand SCR protocol (72). To estimate the duplication rate and endogenous content, each library was indexed with a unique 6-base pair index and screened on an Illumina HiSeq 4000 using 80 bp SE technology, or a NovaSeq 6000 using 150 bp PE technology, at Novogene Europe. Libraries with more than 5% endogenous DNA and duplication rates below 20% were resequenced to reach a mean sequencing depth above 0.2×. Libraries with low amounts of endogenous DNA, that were not resequenced, were enriched for mitochondrial genomes using the hybridization capture myBaits Custom DNA-Seq kit from Daicel Arbor Biosciences (Ann Arbor, MI) (SI Appendix, Table S2). We applied the High Sensitivity conditions, which are optimized for ancient material, as described in the myBaits manual v/5.0. The enriched libraries were reamplified for 14 cycles and sequenced on the NovaSeq 6000 at Novogene Europe, using 150 bp PE technology. Throughout the DNA data generation, novel blanks were included and checked for contamination using Qubit 3.0 (Thermo Fisher Instruments, Waltham, USA) for DNA extracts and the Agilent Fragment Analyzer (Santa Clara, USA) for library builds and indexing. The raw sequencing data are available from the European Nucleotide Archive (ENA) under the project accession number PRJEB73809.
DNA Analysis.
Raw sequence reads were processed and mapped within the PALEOMIX pipeline 1.2.12 (73). Adapter sequences were removed from read ends using AdaptorRemoval v/2.2.0 (74) with default settings, except for minimum read length, which was set to 25 bp. Processed reads were mapped using BWA (75) applying the Backtrack algorithm, while disabling the seed function. A beluga whale genome assembly (76), which was improved to chromosome level as part of the DNA ZOO project (77) (accession: ASM228892v2_HiC), was used as nuclear reference, while a mitochondrial beluga whale assembly (78) was used as mitochondrial reference. Reads that mapped to multiple locations in the reference genome or had quality scores below 25 were excluded using SAMtools v/1.9 (79). Sequence duplicates were removed using the MarkDuplicates function in Picard v/2.18.26 (80) and indels were realigned using GATK v/3.8.1 (81). Final bam files were checked with the ValidateSamFile function in Picard v/2.18.26 and sequencing depth summaries were subsequently generated for all samples. All relevant DNA sequencing information for each sample is provided in Dataset S1.
To genetically determine the sex of each sampled individual we applied the SeXY pipeline (82), which uses the ratio of coverage at sites of the autosome and sex chromosomes. We used the human X and Y chromosomes (NCBI accessions CM000685.2 and CM000686.2) to identify sex-linked chromosomes in the beluga whale genome assembly. Individuals with a coverage ratio below 0.7 were identified as males and individuals with coverage ratios above 0.8 were identified as females (SI Appendix, Table S2).
We further filtered the data by excluding problematic genomic regions. Repetitive regions conserved in the cetartiodactyla group were identified in the beluga whale reference genome using RepeatMasker (83). Interspersed repeats were masked, while STRs, small RNAs, and low-complexity regions were retained. The X chromosome was identified using the SatsumaSynteny function in Satsuma v/3.10 (84), using the human X chromosome (accession: CM000685.2) as target. Masked repeat regions, putative X chromosome regions, and unplaced scaffolds were excluded from the bam files using the intersect function in Bedtools v2.29.0 (85).
We used ANGSD v/0.935 (86) to identify variable sites in the nuclear dataset. Historic samples often exhibit DNA damage patterns and are prone to higher rates of sequencing errors, which can lead to false discovery of variable sites. To minimize the number of false variable sites, we excluded sequencing reads with quality scores and mapping quality below 30 (-minQ 30, -minMapQ 30), and sites where the minor allele frequency did not deviate from zero with a P-value <1e-6 based on a likelihood test (-SNP_pval 1e-6). We also removed triallelic sites (-skipTriallelic 1), sites with a minor allele frequency below 0.05 (-minMaf 0.05) as well as all transitions (-rmTrans 1). We excluded sites with total read depth below 40 (-setMinDepth 40) and above 80 (-setMaxDepth 80) and data in less than 30 individuals (minInd 30). In ANGSD, we used the GATK method to calculate genotype likelihood files (-GL 2). We also produced haploid files using a random read for each site (-doHaploCall 1) and a covariance matrix using the Identical By State function in ANGSD using a random base (-doIBS 1, -doCov 1).
To identify closely related individuals in the dataset, we used NgsRelate /v2.02 (87) on the genotype likelihood file to estimate the relatedness of all pairs of individuals. In pairs of related individuals (r > 0.3) the individual with the lowest coverage was excluded from the nuclear analyses (SI Appendix, Fig. S2).
To visualize the relationship between samples in the nuclear dataset, a principal component analysis (PCA) was performed using the Eigen function in R (64) with the covariance matrix produced by ANGSD as input.
To identify any changes in genetic diversity through time, we calculated the nucleotide diversity from the haploid call file for each of the four time periods (three past and one contemporary) as well as the three adjacent populations (Anadyr Gulf, Bristol Bay, Cook Inlet) in nonoverlapping, sliding 1-mb windows using the popgenWindows.py script available from https://github.com/simonhmartin/genomics_general.
To evaluate lineage continuity, the same script and window size was used to calculate the fixation index FST across the genome for each of the four time periods and three adjacent populations.
To estimate gene flow between the four Mackenzie Delta time periods—comprising three zooarchaeological and one contemporary—and the three adjacent contemporary populations, we estimated D statistics for all D[H1-Zooarchaeological, H2-Contemporary, H3-Adjacent, Outgroup] combinations of individuals. The D values and associated z scores are presented in SI Appendix, Figs. S5 and S6.
For mitochondrial genome analysis, fasta files were produced from the bam files mapped to the beluga whale mitochondrial reference genome (accession: KY888944) (88). Sites covered by more than four reads were included using the consensus base function in ANGSD (-doFasta 2). Sites with four or fewer reads were included as “N” indicating missing data. The mitochondrial sequences were aligned using MAFFT v/7.392 (89) using default settings.
Summary mitochondrial statistics including number of segregation sites (S), number of haplotypes (h), haplotype diversity (H), nucleotide diversity (π), and the fixation index (FST) were calculated using Arlequin v/3.5.2.2 (90) (SI Appendix, Table S5). To visualize the relationship between mitochondrial haplotypes, a median-spanning haplotype network was produced using Popart v/1.7 (91). The differences in haplotype diversity and nucleotide diversity among the three zooarchaeological and contemporary samples from Mackenzie Delta were tested using genetic_diversity_diffs v/1.0.6 (92).
Population Genetic Simulations.
We encoded a simplified demographic model of the beluga hunting history using the simulation R framework slendr (93), using its development version 0.9.1.9000. To investigate a range of plausible demographic scenarios, we conducted simulations across a grid of parameters which capture the most important aspects of the beluga history studied here.
First, for all simulation runs, we assumed the starting size of the population of 40,000 individuals, and recorded a snapshot of 50 diploid individuals every 50 y of simulation time between the year 1200 CE. and present-day in the output tree sequence using its msprime simulation engine (94). We simulated the uncertainty in the extent of hunting by removing 250, 500, or 1,000 individuals from the population in generation, and captured the range of plausible relationships between the census size and effective population size (Ne) of belugas, by scaling both the starting size and the number of individuals hunted each generation by a factor of 0.3, 0.5, or 0.7 (“Ne-to-census ratio”). For instance, in a scenario in which the starting population size was 40,000, the number of individuals hunted each generation was 500, and the Ne-to-census ratio was 0.3 lead to a simulation with the starting Ne of 40.000 × 0.3 = 12,000 and effective number of individuals hunted per generation of 500 × 0.3 = 150. A schematic representation of such a model is shown in SI Appendix, Fig. S4. For each simulation, we assumed a generation time of 32 y (41), mutation rate of 1.65 × 10−8 per bp per generation (95), and uniform recombination rate 1 × 10−8 per bp per generation. The total amount of simulated sequence was 50 megabases, chosen because it allowed for a reasonable computational time while minimizing stochastic noise given the number of individuals sampled. The total number of simulated scenarios across the grid of the three parameters was thus 1 × 3 × 3 = 9.
From each simulated tree sequence, we computed nucleotide diversity at each recorded time snapshot (i.e., across the 50 sampled diploid individuals) using the slendr function ts_diversity(). For ease of comparison with the result in Fig. 3A, the expected nucleotide diversities across time in SI Appendix, Fig. S3 are presented using individuals sampled at approximately the same time points to those featured in empirical data. Because we were interested in the trajectory of expected nucleotide diversity over time rather than its absolute values, nucleotide diversity at each time point shown in SI Appendix, Fig. S3 has been normalized by the mean diversity in the first time point. For each combination of parameters in each individual scenario, we fitted a linear model using the R function lm, modeling the nucleotide diversity across all recorded individuals as a function of time. SI Appendix, Table S4 shows the P-value, the R2 (variance explained), and the slope of each linear model. All the simulation, analysis, and visualization code can be found at https://github.com/bodkan/beluga-simulations/.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
This study was funded by the Villum Fonden YIP+ grant no 37352, the Independent Research Fund Denmark Sapere Aude grant no 9064-00025B, and the Carlsberg Foundation Semper Ardens Accelerate grant no CF23-1061 to E.D.L. Archaeological excavations were performed in partnership with the Inuvialuit Cultural Centre, and were funded by grants to T.M.F. from the Social Sciences and Humanities Research Council of Canada (Insight Grant 435-2012-0641), the Polar Continental Shelf Program (Grants 61914 and 62816), and the Aurora Research Institute. We thank and acknowledge the board of the Tuktoyaktuk Hunters and Trappers Committee for supporting the study, as well as the Fisheries Joint Management Committee and Fisheries and Oceans Canada for the Hendrickson Island beluga sampling program, and the monitors for their dedication in collecting samples along with the harvesters who kindly shared their catches for research.
Author contributions
P.S., T.M.F., and E.D.L. designed research; M. Skovrind, M.L., S.H.F., D.M.G., D.I.L., I.G.M., M.M.M., M.P., L.P., V.V.R., M. Scott, M.V.W., T.M.F., and P.S. performed research; M. Skovrind and M.L. analyzed data; S.H.F., D.M.G., D.I.L., I.G.M., L.P., and V.V.R. provided samples; and M. Skovrind, L.L., M.P., P.S., T.M.F., and E.D.L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Mikkel Skovrind, Email: mikkel.skovrind@sund.ku.dk.
T. Max Friesen, Email: max.friesen@utoronto.ca.
Eline D. Lorenzen, Email: elinelorenzen@sund.ku.dk.
Data, Materials, and Software Availability
Raw sequencing data in fastq format; simulation, analysis, and visualization code data have been deposited in European Nucleotide Archive; GitHub (PRJEB73809; https://github.com/bodkan/beluga-simulations) (96, 97).
Supporting Information
References
- 1.Mason O. K., Friesen T. M., Out of the Cold: Archaeology on the Arctic Rim of North America (University Press of Colorado, 2018). [Google Scholar]
- 2.Friesen T. M., Arnold C. D., “Prehistoric Beluga Whale Hunting at Gupuk, Mackenzie Delta, Northwest Territories, Canada” in Native Whaling in the Western Arctic and Subarctic: Hunting the Largest Animals, McCartney A., Ed. (Canadian Circumpolar Institute, Edmonton, 1995), pp. 109–126.
- 3.Friesen T. M., “The cache point site: An early Thule occupation in the Mackenzie delta” in On the Track of the Thule Culture from Bering Strait to East Greenland, Grønnow B., Ed. (National Museum of Denmark, 2009), pp. 63–74. [Google Scholar]
- 4.Friesen T. M., O’Rourke M. J. E., Biogeographic barriers and coastal erosion: Understanding the lack of interaction between the Eastern and Western Regions of the North American Arctic. World Archaeol. 51, 484–501 (2019). [Google Scholar]
- 5.Arnold C. D., “Archaeological investigations on Richards Island” in Bridges Across Time: The NOGAP Archaeology Project, Pilon J. L., Ed. (Canadian Archaeological Association Occasional Paper, 1994), pp. 85–94. [Google Scholar]
- 6.McGhee R., Beluga Hunters: An Archaeological Reconstruction of the History and Culture of the Mackenzie Delta Kittegaryumiut (Institute of Social and Economic Research, Memorial University of Newfoundland, 1974). [Google Scholar]
- 7.Arnold C. D., “Development of Mackenzie Inuit culture” in The Oxford Handbook of the Prehistoric Arctic, Friesen T. M., Mason O. K., Eds. (Oxford University Press, 2016), pp. 585–606. [Google Scholar]
- 8.Friesen T. M., Arnold C. D., Zooarchaeology of a focal resource: Dietary importance of beluga whales to the Precontact Mackenzie Inuit. Arctic 48, 22–30 (1995). [Google Scholar]
- 9.Friesen T. M., Méreuze R., An Igluryuaq unearthed: A pre-contact Inuvialuit cruciform house from Arctic Canada. J. Field Archaeol. 45, 464–478 (2020). [Google Scholar]
- 10.Lowe R., Siglit Inuvialuit Uqautchiita Nutaat Kipuktirutait Aglipkaqtat. Siglit Inuvialuit Eskimo Dictionary (ditions Nota Bene, ed. 2, 2001). [Google Scholar]
- 11.Waugh D., et al. , Inuvialuit traditional ecological knowledge of beluga whale (Delphinapterus leucas) under changing climatic conditions in Tuktoyaktuk, NT. Arct. Sci., 1–17 (2018). [Google Scholar]
- 12.Nuligak I., Metayer M., Ed. (Peter Martin Associates, Toronto, 1966). [Google Scholar]
- 13.Krech S., A Victorian Earl in the Arctic: The Travels and Collections of the Fifth Earl of Lonsdale 1888–89 (University of Washington Press, 1989). [Google Scholar]
- 14.Whittaker C. E., Arctic Eskimo, a Record of Fifty Years’ Experience & Observation among the Eskimo (Seeley, Service & Company, Limited, London, 1937). [Google Scholar]
- 15.Friesen T. M., Kitigaaryuit: A portrait of the Mackenzie Inuit in the 1890s, based on the journals of Isaac O. Stringer. Arctic Anthropol. 41, 222–237 (2004). [Google Scholar]
- 16.Usher P., The Canadian Western Arctic: A century of change. Anthropologica 13, 169–183 (1971). [Google Scholar]
- 17.Harwood L. A., et al. , “Research, monitoring and hunter knowledge in support of the assessment of the Eastern Beaufort Sea beluga stock” (Res. Doc. 2020/075, Canadian Science Advisory Secretariat, 2020). [Google Scholar]
- 18.DFO, “Eastern Beaufort Sea beluga (Delphinapterus leucas) population abundance estimate in 2019” (Rep. 2023/047, DFO Canadian Science Advisory Secretariat Science Advisory Report, 2023). [Google Scholar]
- 19.Hobbs R. C., Reeves R. R., Prewitt J. S., Global review of the conservation status of monodontid stocks. Marine Fish. 81, 1–53 (2019). [Google Scholar]
- 20.Skovrind M., et al. , Circumpolar phylogeography and demographic history of beluga whales reflect past climatic fluctuations. Mol. Ecol. 30, 2543–2559 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Harwood L. A., Innes S., Norton P., Kingsley M. C. S., Distribution and abundance of beluga whales in the Mackenzie estuary, southeast Beaufort Sea, and west Amundsen Gulf during late July 1992. Can. J. Fish. Aquat. Sci. 53, 2262–2273 (1996). [Google Scholar]
- 22.Harwood L. A., Iacozza J., Auld J. C., Norton P., Loseto L., Belugas in the Mackenzie River estuary, NT, Canada: Habitat use and hot spots in the Tarium Niryutait marine protected area. Ocean Coast. Manag. 100, 128–138 (2014). [Google Scholar]
- 23.Richard P. R., Martin A. R., Orr J. R., Summer and autumn movements of belugas of the Eastern Beaufort Sea stock. Arctic 54, 223–236 (2001). [Google Scholar]
- 24.Hauser D. D. W., et al. , Habitat selection by two beluga whale populations in the Chukchi and Beaufort seas. PLoS One 12, e0172755 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storrie L., et al. , Year-round dive characteristics of male beluga whales from the Eastern Beaufort Sea population indicate seasonal shifts in foraging strategies. Front. Mar. Sci. 8, 715412 (2022). [Google Scholar]
- 26.Citta J. J., et al. , Satellite telemetry reveals population specific winter ranges of beluga whales in the Bering Sea. Mar. Mamm. Sci. 33, 236–250 (2017). [Google Scholar]
- 27.DeNiro M. J., Postmortem preservation and alteration of in vivo bone collagen isotope ratios in relation to palaeodietary reconstruction. Nature 317, 806–809 (1985). [Google Scholar]
- 28.Ambrose S. H., Preparation and characterization of bone and tooth collagen for isotopic analysis. J. Archaeol. Sci. 17, 431–451 (1990). [Google Scholar]
- 29.Colbeck G. J., et al. , Groups of related belugas (Delphinapterus leucas) travel together during their seasonal migrations in and around Hudson Bay. Proc. Biol. Sci. 280, 20122552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint] (2013). 10.48550/arXiv.1303.3997 (Accessed 25 July 2024). [DOI]
- 31.Bowers P. M., "The archaeology of Deering, Alaska: Final report on the Deering village safe water archaeological program" (Northern Land Use Research Inc., Fairbanks, 2009). [Google Scholar]
- 32.Heide-Jørgensen M. P., Teilmann J., Growth, reproduction. age structure and feeding habits of white whales (Delphinapterus leucas) in West Greenland Waters. Meddelelser Grønland Biosci. 39, 195–212 (1994). [Google Scholar]
- 33.O’Corry-Crowe G., et al. , Group structure and kinship in beluga whale societies. Sci. Rep. 10, 11462 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harwood L. A., Norton P., Day B., Hall P. A., The harvest of beluga whales in Canada’s Western Arctic: Hunter-based monitoring of the size and composition of the catch. Arctic 55, 10–20 (2002). [Google Scholar]
- 35.Szpak P., et al. , Sexual differences in the foraging ecology of 19th century beluga whales (Delphinapterus leucas) from the Canadian High Arctic. Mar. Mamm. Sci. 36, 451–471 (2020). [Google Scholar]
- 36.Louis M., et al. , Population-specific sex and size variation in long-term foraging ecology of belugas and narwhals. R. Soc. Open Sci. 8, 202226 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choy E. S., Rosenberg B., Roth J. D., Loseto L. L., Inter-annual variation in environmental factors affect the prey and body condition of beluga whales in the eastern Beaufort Sea. Mar. Ecol. Prog. Ser. 579, 213–225 (2017). [Google Scholar]
- 38.Louis M., Routledge J., Heide-Jørgensen M. P., Szpak P., Lorenzen E. D., Sex and size matter: Foraging ecology of offshore harbour porpoises in waters around Greenland. Mar. Biol. 169, 140 (2022). [Google Scholar]
- 39.Choy E. S., et al. , Variation in the diet of beluga whales in response to changes in prey availability: Insights on changes in the Beaufort Sea ecosystem. Mar. Ecol. Prog. Ser. 647, 195–210 (2020). [Google Scholar]
- 40.Loseto L. L., Richard P., Stern G. A., Orr J., Ferguson S. H., Segregation of Beaufort Sea beluga whales during the open-water season. Can. J. Zool. 84, 1743–1751 (2006). [Google Scholar]
- 41.Garde E., et al. , Life history parameters of narwhals (Monodon monoceros) from Greenland. J. Mammal. 96, 866–879 (2015). [Google Scholar]
- 42.Ferguson S. H., et al. , Reproductive parameters for female beluga whales (Delphinapterus leucas) of Baffin Bay and Hudson Bay, Canada. Arctic 73, 405–420 (2020). [Google Scholar]
- 43.Rocha R. C. Jr., Clapham P. J., Ivashchenko Y., Emptying the oceans: A summary of industrial whaling catches in the 20th century. Mar. Fish. Rev. 76, 37–48 (2015). [Google Scholar]
- 44.Alter S. E., Newsome S. D., Palumbi S. R., Pre-whaling genetic diversity and population ecology in eastern Pacific gray whales: Insights from ancient DNA and stable isotopes. PLoS One 7, e35039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Béland S. L., Frasier B. A., Darling J. D., Frasier T. R., Using pre- and postexploitation samples to assess the impact of commercial whaling on the genetic characteristics of eastern North Pacific gray and humpback whales and to compare methods used to infer historic demography. Mar. Mamm. Sci. 36, 398–420 (2020). [Google Scholar]
- 46.Frasier B. A., et al. , Genetic examination of historical North Atlantic right whale (Eubalaena glacialis) bone specimens from the eastern North Atlantic: Insights into species history, transoceanic population structure, and genetic diversity. Mar. Mamm. Sci. 38, 1050–1069 (2022). [Google Scholar]
- 47.Mills K. K., et al. , Ancient DNA indicates a century of overhunting did not reduce genetic diversity in Pacific Walruses (Odobenus rosmarus divergens). Sci. Rep. 14, 8257 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolf M., de Jong M., Halldórsson S. D., Árnason Ú., Janke A., Genomic impact of whaling in North Atlantic Fin Whales. Mol. Biol. Evol. 39, msac094 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nigenda-Morales S. F., et al. , The genomic footprint of whaling and isolation in fin whale populations. Nat. Commun. 14, 5465 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Greef E., et al. , Unraveling the genetic legacy of commercial whaling in bowhead whales and narwhals. bioRxiv [Preprint] (2024). 10.1101/2024.01.04.574243 (Accessed 25 July 2024). [DOI]
- 51.Westbury M. V., et al. , Four centuries of commercial whaling eroded 11,000 years of population stability in bowhead whales. bioRxiv [Preprint] (2024). 10.1101/2024.04.10.588858 (Accessed 25 July 2024). [DOI]
- 52.Heide-Jørgensen M. P., Rosing-Asvid A., Catch statistics for belugas in West Greenland 1862 to 1999. NAMMCOSP 4, 127–142 (2002). [Google Scholar]
- 53.Heide-Jørgensen M.-P., Laidre K., Greenland’s Winter Whales: The Beluga, the Narwhal and the Bowhead Whale (Ilinniusiorfik Undervisningsmiddelforlag, 2006). [Google Scholar]
- 54.Friesen T. M., Radiocarbon evidence for fourteenth-century dorset occupation in the Eastern North American Arctic. Am. Antiq. 85, 222–240 (2020). [Google Scholar]
- 55.dos Santos G. M., KCCAMS prep-laboratory personnel, UCI AMS facility chemical pretreatment for bone: Ultrafiltration method. Electronic Document 1–9 (2011). https://sites.ps.uci.edu/kccams/education/protocols/. [Google Scholar]
- 56.Ramsey C. B., Bayesian analysis of radiocarbon dates. Radiocarbon 51, 337–360 (2009). [Google Scholar]
- 57.Reimer P. J., Composition and consequences of the IntCal20 radiocarbon calibration curve. Quat. Res. 96, 22–27 (2020). [Google Scholar]
- 58.Arnold C. D., Vanishing villages of the past: Rescue archaeology in the Mackenzie Delta. North. Rev. 1, 40–58 (1988). [Google Scholar]
- 59.Stefansson V., The Stefánsson-Anderson Arctic expedition of the American Museum: Preliminary ethnological report. Anthropol. Pap. Am. Mus. Nat. Hist. 14, 1–395 (1919). [Google Scholar]
- 60.Qi H., Coplen T. B., Geilmann H., Brand W. A., Böhlke J. K., Two new organic reference materials for delta13C and delta15N measurements and a new value for the delta13C of NBS 22 oil. Rapid Commun. Mass Spectrom. 17, 2483–2487 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Schimmelmann A., et al. , Organic reference materials for hydrogen, carbon, and nitrogen stable isotope-ratio measurements: Caffeines, n-alkanes, fatty acid methyl esters, glycines, L-valines, polyethylenes, and oils. Anal. Chem. 88, 4294–4302 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Szpak P., Metcalfe J. Z., Macdonald R. A., Best practices for calibrating and reporting stable isotope measurements in archaeology. J. Archaeol. Sci.: Rep. 13, 609–616 (2017). [Google Scholar]
- 63.Magnusson B., Näykki T., Hovind H., Krysell M., Handbook for Calculation of Measurement Uncertainty in Environmental (Nordtest, 2017).
- 64.R Core Team, R: A Language and Environment for Statistical Computing (R Core Team, R: A Language and Environment for Statistical Computing, Vienna, 2021). https://www.R-project.org. Accessed 25 July 2024.
- 65.Bearhop S., Adams C. E., Waldron S., Fuller R. A., Macleod H., Determining trophic niche width: A novel approach using stable isotope analysis. J. Anim. Ecol. 73, 1007–1012 (2004). [Google Scholar]
- 66.Newsome S. D., Martinez del Rio C., Bearhop S., Phillips D. L., A niche for isotopic ecology. Front. Ecol. Environ. 5, 429–436 (2007). [Google Scholar]
- 67.Jackson A. L., Inger R., Parnell A. C., Bearhop S., Comparing isotopic niche widths among and within communities: SIBER–Stable isotope Bayesian Ellipses in R. J. Anim. Ecol. 80, 595–602 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Plummer M., rjags: Bayesian graphical models using MCMC (R package version 4, 2016). https://mcmc-jags.sourceforge.io. Accessed 25 July 2024.
- 69.Dabney J., et al. , Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U.S.A. 110, 15758–15763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Damgaard P. B., et al. , Improving access to endogenous DNA in ancient bones and teeth. Sci. Rep. 5, 11184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carøe C., et al. , Single-tube library preparation for degraded DNA. Methods Ecol. Evol. 9, 410–419 (2018). [Google Scholar]
- 72.Kapp J. D., Green R. E., Shapiro B., A fast and efficient single-stranded genomic library preparation method optimized for ancient DNA. J. Hered. 112, 241–249 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schubert M., et al. , Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat. Protoc. 9, 1056–1082 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Schubert M., Lindgreen S., Orlando L., AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 9, 88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones S. J. M., et al. , The genome of the beluga whale (Delphinapterus leucas). Genes 8, 378 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dudchenko O., et al. , De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Skovrind M., Samaniego Castruita J. A., Heide-Jørgensen M. P., Dalén L., Lorenzen E., Mitochondrial genome divergence between beluga whales in Baffin Bay and the Sea of Okhotsk. Mitochondr. DNA B Resour. 2, 257–258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li H., et al. , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Broad Institute, Picard Toolkit. GitHub Repository (2016). http://broadinstitute.github.io/picard/. Accessed 25 July 2024. [Google Scholar]
- 81.McKenna A., et al. , The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cabrera A. A., et al. , How low can you go? Introducing SeXY: Sex identification from low-quantity sequencing data despite lacking assembled sex chromosomes. Ecol. Evol. 12, e9185 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smit A. F. A., Hubley R., Green P., Repeat-Masker Open-4.0 (2013–2015). http://www.repeatmasker.org. Accessed 25 July 2024.
- 84.Grabherr M. G., et al. , Genome-wide synteny through highly sensitive sequence alignment: Satsuma. Bioinformatics 26, 1145–1151 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Korneliussen T. S., Albrechtsen A., Nielsen R., ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hanghøj K., Moltke I., Andersen P. A., Manica A., Korneliussen T. S., Fast and accurate relatedness estimation from high-throughput sequencing data in the presence of inbreeding. Gigascience 8, giz034 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Skovrind M., Samaniego Castruita J. A., Heide-Jørgensen M. P., Dalén L., Lorenzen E., Mitochondrial genome divergence between beluga whales in Baffin Bay and the Sea of Okhotsk. mtDNA Part B 2, 257–258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Excoffier L., Lischer H. E. L., Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010). [DOI] [PubMed] [Google Scholar]
- 91.Leigh J. W., Bryant D., popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116 (2015). [Google Scholar]
- 92.Alexander A., et al. , What influences the worldwide genetic structure of sperm whales (Physeter macrocephalus)? Mol. Ecol. 25, 2754–2772 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Petr M., Haller B. C., Ralph P. L., Racimo F., slendr: A framework for spatio-temporal population genomic simulations on geographic landscapes. Peer Commun. J. 3, e121 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baumdicker F., et al. , Efficient ancestry and mutation simulation with msprime 1.0. Genetics 220, iyab229 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Westbury M. V., Petersen B., Garde E., Heide-Jørgensen M. P., Lorenzen E. D., Narwhal genome reveals long-term low genetic diversity despite current large abundance size. iScience 15, 592–599 (2019), 10.1016/j.isci.2019.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Skovrind M., Lorenzen E. D., Project accession number PRJEB73809, Beluga whale raw sequencing data for Skovrind et al 2024. European Nucleotide Archive. https://www.ebi.ac.uk/ena/browser/view/PRJEB73809. Deposited 1 August 2024.
- 97.Petr M., Beluga simulations. GitHub. https://github.com/bodkan/beluga-simulations. Deposited 1 August 2024.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
Raw sequencing data in fastq format; simulation, analysis, and visualization code data have been deposited in European Nucleotide Archive; GitHub (PRJEB73809; https://github.com/bodkan/beluga-simulations) (96, 97).