Abstract
The initial and the terminal three enzymes of the mammalian haem biosynthetic pathway are nuclear encoded, cytoplasmically synthesized and post-translationally translocated into the mitochondrion. The first enzyme, ALAS (5-aminolaevulinate synthase), occurs as an isoenzyme encoded on different chromosomes and is synthesized either as a housekeeping protein (ALAS-1) in all non-erythroid cell types, or only in differentiating erythroid precursor cells (ALAS-2). Both ALAS proteins possess mitochondrial targeting sequences that have putative haem-binding motifs. In the present study, evidence is presented demonstrating that two haem-binding motifs in the leader sequence, as well as one present in the N-terminus of the mature ALAS-1 function in vivo in the haem-regulated translocation of ALAS-1. Coproporphyrinogen oxidase, the antepenultimate pathway enzyme, possesses a leader sequence that is approx. 120 residues long. In contrast with an earlier report suggesting that only 30 residues were required for translocation of the coproporphyrinogen oxidase, we report that the complete leader is necessary for translocation and that this process is not haem-sensitive in vivo. PPO (protoporphyrinogen oxidase) lacks a typical mitochondrial targeting leader sequence and was found to be effectively targeted by just 17 N-terminal residues. Bacillus subtilis PPO, which is very similar to human PPO at its N-terminal end, is not targeted to the mitochondrion when expressed in mammalian cells, demonstrating that the translocation is highly specific with regard to both the length and spacing of charged residues in this targeting region. Ferrochelatase, the terminal enzyme, possesses a typical N-terminal leader sequence and no evidence of a role for the C-terminus was found in mitochondrial targeting.
Keywords: aminolaevulinate, ferrochelatase, haem, mitochondria, protoporphyrinogen, targeting
Abbreviations: ALA, aminolaevulinic acid; ALAS, 5-aminolaevulinate synthase; CPO, coproporphyrinogen oxidase; FC, ferrochelatase; GFP, green fluorescent protein; PPO, protoporphyrinogen oxidase; for brevity, the single-letter system for amino acids has been used; C8, for example means Cys8
INTRODUCTION
In animal cells, anabolic and catabolic pathways are frequently confined to a single cellular compartment unless the process involves import or export of products or substrates. In contrast, the tetrapyrrole biosynthetic pathway in animal cells has its first and its last three steps catalysed by mitochondrial enzymes, whereas the intermediate four steps are catalysed by cytosolic enzymes [1]. All pathway enzymes are nuclear-encoded and are synthesized in the cytosol with the mitochondrially located proteins being post-translationally translocated to their appropriate mitochondrial space. The initial pathway enzyme, ALAS (5-aminolaevulinate synthase), is located in the mitochondrial matrix where its substrates, glycine and succinyl CoA, are found [2,3]. CPO (coproporphyrinogen oxidase) and PPO (protoporphyrinogen oxidase), the antepenultimate and penultimate enzymes, are located in the space between the inner and outer mitochondrial membranes [4,5]. FC (ferrochelatase), the terminal enzyme, is associated with the matrix side of the inner mitochondrial membrane [6]. None of the mitochondrially associated haem synthetic enzymes contain any transmembrane segments.
The mitochondrially located enzymes present interesting and distinct examples of targeting mechanisms. ALAS, which is a homodimer, is translocated into the mitochondrion in a step that involves proteolytic removal of the leader sequence (region I) and subsequent dimerization and addition of its cofactor pyridoxal phosphate. Two isoenzymes of ALAS exist, one expressed only during erythroid differentiation (ALAS-2) and the second expressed in all other cell types (ALAS-1) [2]. The ALAS-2 enzyme has been found to associate with succinyl CoA synthetase in mitochondria [7]. In addition to the region I leader sequence, both ALAS-1 and -2 possess a segment of the protein coded by exons 2 and 3, which is not required for activity and whose function is currently unknown (region II), and the highly conserved catalytically active region III. Both the isoenzymes also contain a series of putative haem-binding motifs with the sequence R/L/N-C-P-L/V/I/F-L/M, where the cysteine and proline residues are invariant [8]. Two of these motifs are contained in the region I leader sequence, and the third is contained in region II. On the basis of in vitro translocation experiments with isolated mitochondria and erythroid ALAS protein, the two haem-binding motifs in the leader sequence have been proposed to bind haem and prevent translocation of apo-ALAS into the mitochondrion [8]. The third haem-binding motif in region II has not been previously studied for either of the isoenzymes.
CPO, the antepenultimate enzyme, is present in the inter-membrane space of mitochondria in higher animal cells [4]. In comparison with ALAS-1 and -2 and FC, which both have leaders approx. 60 residues long, this protein possesses an unusually long (>120 amino acids) N-terminal leader sequence that is proteolytically removed [9]. The mature protein possesses no known cofactor and is a homodimer [10]. The penultimate enzyme, PPO, is associated with the outer surface of the inner mitochondrial membrane [5]. Association with the membrane has been proposed to involve a protein-bound fatty acid [11], although recently it has been suggested that PPO forms a transmembrane complex with FC [12]. This homodimeric protein does not contain a typical N-terminal targeting sequence and is not proteolytically processed, but does have a dinucleotide-binding motif at the N-terminal end that is essential for binding of the enzyme's cofactor, FAD [13]. The terminal pathway enzyme, FC, is found in its mature homodimeric form bound to the matrix side of the inner mitochondrial membrane [14]. The cytoplasmically synthesized apoprotein possesses a targeting sequence that is required for translocation into the mitochondrion [14]. This step is energy-dependent, and involves proteolysis to remove the leader sequence, and assembly of the [2Fe-2S] cluster [15].
We present below data that characterize features of intracellular translocation for all of the mitochondrially localized haem synthetic pathway enzymes in intact cells. These data demonstrate that all three putative haem-binding motifs in ALAS can function in situ as haem-responsive regulators of intracellular translocation. Also shown is that the complete putative leader sequence of CPO is necessary for protein translocation and that the truncated version previously reported as sufficient [16] does not support protein targeting. The N-terminal 17 residues of PPO are sufficient to target a reporter protein to the mitochondrion, and protein lacking this region is not targeted to the mitochondrion. Effective targeting of FC requires only the N-terminal leader sequence. Of all the proteins examined only the translocation of ALAS is sensitive to inhibition by haem.
MATERIALS AND METHODS
Cloning of mouse CPO exon 1
Initial attempts to PCR exon 1 of CPO from human libraries were unsuccessful. Mouse Hepa1c1c7 cells were harvested and genomic DNA was prepared using Trizole reagent (Invitrogen, Carlsbad, CA, U.S.A.). This genomic DNA (100 ng) was used as a template for PCR with the following primers: sense, 5′-GTTGGGTCTGCAGGATCGGAT-3′ and antisense 5′-TTGGTGTTCATGTCCTCCGGC-3′; the antisense primer is contained in the coding region for the processed form of mouse CPO. The resulting product, when sequenced, identified 12 base miscalls and a frameshift in the original mouse CPO exon 1 sequence submitted to GenBank® (accession no. AB011243). The corrected sequence has been submitted to GenBank® (accession no. AY382578). This CPO PCR product was used as a template for the targeting constructs.
Cloning of human ALAS-1
The full-length ALAS-1 was obtained by PCR from a human liver library (Stratagene), which was mass-excised into pBluescript. The sense primer used was the universal primer pUCR (to maximize the amount of 5′-untranslated region obtained). The antisense primer contained 7 bp of the 3′-end of the ALAS-1 gene and 18 bp of the 3′-untranslated region; specifically, the antisense primer was 5′-AAATAATTGAGGTCATGCTCAGGCC-3′, where the underlined portion corresponds to the 3′-end of the gene, including the termination codon. This PCR product was cloned into pGEM-T vector (Promega) for amplification and sequencing. The resulting sequence contains 44 additional base pairs of the 5′-untranslated sequence composed with the sequence contained in GenBank® (accession no. X56531).
Targeting constructs
Wherever possible, existing restriction sites were used for subcloning into the GFP (green fluorescent protein) expression vectors (see below). Most often, however, PCR primers containing restriction sites were used to amplify the regions of interest for subsequent cloning into the GFP vectors.
For ALAS-1, eight reporter constructs were made. The first construct (pd2ALASAS2) contained regions I and II (residues 1–203 as annotated in Swiss-Prot entry P13196) fused to a destabilized version of GFP (pd2-EGFPN1; ClonTech). The pd2ALASAS2 construct, which contains all three of the putative haem-binding motifs, was used as a template for Quik Change site-directed mutagenesis (Stratagene) to generate a series of mutants which contained two of the three haem-binding motifs (C8S, C33S and C108S), one of the three haem-binding motifs (C8S/C33S, C8S/C108S and C33S/C108S) and none of the haem-binding motifs (C8S/C33S/C108S).
Three CPO targeting constructs were produced using the PCR product described above. The first contained residues 1–132 of mouse CPO (as annotated in Swiss-Prot entry P36552) fused to the N-terminus of the GFP in the expression vector pd2EGFPN1. The second contained residues 90–132 of mouse CPO fused to pd2EGFPN1 and the third contained residues 68–132 of mouse CPO fused to pd2EGFPN1.
For FC, two constructs were made using the plasmid pHFwt (a gift from D. Brenner, University of North Carolina at Chapel Hill, Chapel Hill, NC, U.S.A.) as a template. The first contained residues 7–68 of human FC (as annotated in Swiss-Prot entry P22830) fused to the N-terminus of GFP in the expression vector pEGFPN1 (ClonTech). The second contained residues 390–423 of FC fused to the C-terminal end of the GFP in the expression vector pEGFPC1 (ClonTech).
A total of five human PPO targeting constructs were made. The first contained residues 1–178 of human PPO (as annotated in Swiss-Prot entry P50336) fused to the N-terminus of the GFP in pEGFPN1. Three additional constructs containing residues 1–59, 1–34 and 1–17 of human PPO fused to the N-terminal end of GFP were also made. The fifth construct contained residues 420–477 of human PPO fused to the C-terminal end of pEGFPC1.
Four Bacillus subtilis PPO targeting constructs were made. The first contained residues 1–20 of the wild-type enzyme (as annotated in Swiss-Prot entry P32397) fused to the N-terminal end of the GFP in pEGFPN1. This construct was used as a template for site-directed mutagenesis to create the ΔK5-H7, D3R and D3R/ΔK5-H7 mutants.
Cell culture and transfection
Hepa1c1c7 or Hepa 1–6 cells were grown to confluency in BITSC [17] +1% defined bovine serum, trypsinized, rinsed once with defined PBS, and electrophorated at 1650 V with 50 μg of the GFP fusion constructs. Cells were plated into sterile tissue culture dishes containing sterile glass coverslips and incubated at 37 °C in 5% CO2 for 24 h before examination or media additions. Where appropriate, a sterile solution of 100 μM final concentration haemin in DMSO and/or 400 μM final concentration ALA (aminolaevulinic acid) in water was added and the cells were incubated for an additional 4 h before examination.
Microscopy
For the confocal images, cells were treated with the mitochondrial specific stain Mitotracker Red (Molecular Probes) immediately before examination. The cells were then examined with a BioRad MRC 600 scanning confocal microscope equipped with an Ar/Kr laser. Images were collected simultaneously using an excitation/emission wavelength of 488 nm/522 nm for GFP and of 568 nm/585 nm for Mitotracker Red. The emission data for each fluor was recorded individually and overlaid to produce a colour photograph.
Cells were also visualized using a Nikon Eclipse E600 fluorescence microscope equipped with an FITC filter for the visualization of GFP localization. To study the effects of ALA and haem on the translocation of the ALAS-destabilized GFP constructs, cells were grown, transfected and incubated as described above. For each construct, a total of 50 GFP-positive cells were scored for the presence or absence of GFP targeting to the mitochondria.
RESULTS
The results of the targeting experiments, described in detail below, are summarized in Table 1.
Table 1. Targeting of reporter constructs to mitochondria.
| Enzyme | Residues in fusion construct* | Targeted | Not targeted |
|---|---|---|---|
| ALAS | 1–203: destabilized GFP | × | |
| CPO | 1–132: destabilized GFP | × | |
| 68–132: destabilized GFP | × | ||
| 90–132: destabilized GFP | × | ||
| PPO | 1–178: GFP | × | |
| 1–59: GFP | × | ||
| 1–34: GFP | × | ||
| 1–17: GFP | × | ||
| GFP: 420–477 | × | ||
| FC | 7–68: GFP | × | |
| GFP: 390–423 | × |
* Residues are numbered according to the Swiss-Prot entry corresponding to each enzyme (see text for details). The GFP reporter placement is shown for each construct by its position in column 2 (e.g. all of the CPO constructs are N-terminal fusions to destabilized GFP).
ALAS-1 targeting
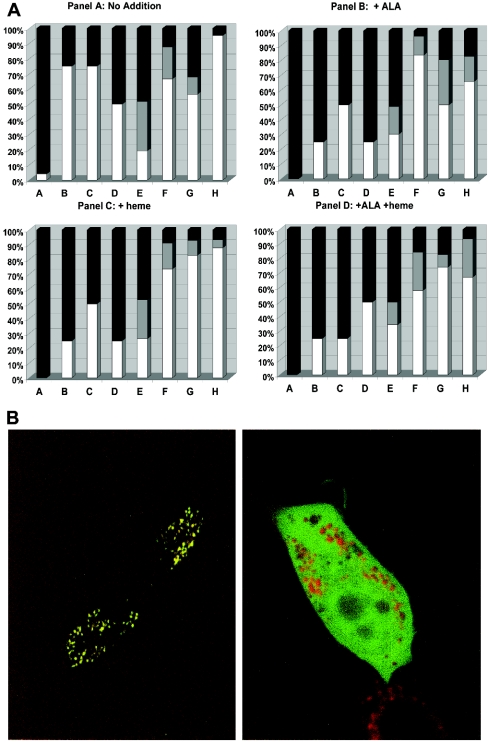
Both ALAS-1 and -2 are described as being composed of three regions: region I is the mitochondrial targeting sequence, region II is the N-terminal portion of the mature, proteolytically processed protein and region III is the highly conserved catalytic region of the enzyme [2]. Within region I are two previously identified putative haem-binding motifs [8], involving C8 and C33. Within region II, there is another putative motif involving C108. All three of these motifs were investigated as potential haem regulatory motifs by examining the targeting of GFP reporter constructs in cultured cells (Figure 1). For these constructs a destabilized GFP protein with a half-life of 2 h was employed to minimize cellular accumulation of background GFP fluorescence. Additions to culture media were made 4 h before observations.
Figure 1. Fusion translocation to mitochondria.
(A) Bar graph of ALAS-GFP fusion constructs showing targeting to mitochondria. Cells in which the GFP fluorescence was located in the mitochondria are shown in white; cells in which fluorescence was located in both the mitochondria and the cytoplasm are shown in grey; cells in which fluorescence was located in the cytoplasm are shown in black. Panel A shows the results for cultures incubated with no ALA or haemin addition. Panel B shows the results for cultures incubated with 400 μM ALA. Panel C shows the results for cultures incubated with 100 μM haemin. Panel D shows the results for cultures incubated with both 400 μM ALA and 100 μM haemin. All culture additions were made 4 h before examination. For each construct, at least 50 GFP-positive cells were scored for the presence or absence of GFP targeting to the mitochondria. (B) Representative pictures showing (i) GFP fluorescence targeted to the mitochondria as determined by co-localization with the mitochondria-specific dye Mitotracker Red and (ii) GFP fluorescence diffused throughout the cell in contrast with the mitochondrial localization of the Mitotracker Red fluorescence. These images were obtained for targeted and non-targeted ALAS, but are characteristic of all other targeted and non-targeted proteins examined in this study.
It was found that wild-type regions I and II of ALAS-1 were targeted very poorly to mitochondria in cells maintained in normal medium containing 1% defined bovine serum (Figure 1). These results indicate that under normal cell culture conditions, the intracellular haem content is sufficient to allow only minimal amounts of ALAS-1 to be translocated to the mitochondria. Addition of exogenous haem, ALA, or haem plus ALA to the culture medium had no additional impact on the targeting frequency of the wild-type protein. Targeting of the construct in which all three motifs were disrupted (e.g. C8S/C33S/C108S) occurred at a high frequency under all culture conditions (Figure 1), indicating that the leader sequence is effective in targeting and that it is the presence of the motifs which results in the inhibition of mitochondrial targeting.
Reporter constructs in which any single motif, including the one in region II, was mutated (e.g. C8S, C33S or C108S) had significantly reduced sensitivity to in vivo intracellular haem (Figure 1). All three single mutants do respond to increased intracellular haem levels that occur when cultures are supplemented with haem or ALA, but none recovers the wild-type level of targeting inhibition. These results are consistent with a model in which all three putative motifs are functional as haem sensors and imply that all three are essential for maximal sensitivity and response to low haem concentrations. Reporter constructs in which two motifs have been altered also show decreased sensitivity to haem and the two double mutants in which C108 is mutated (C8S/C108S and C33S/C108S) possess little haem responsiveness making it clear that even though this motif is located outside of the leader sequence, it still plays a synergistic role with the leader region-located motifs.
CPO targeting
CPO is targeted to the space between the inner and outer mitochondrial membranes. Three targeting reporter constructs were prepared. In one the proposed 31 amino acid residue leader sequence proposed by Kohno et al. [16] was fused to GFP. TargetP analysis predicts that the mitochondrial targeting sequence consisted of the 67 N-terminal residues [18]. For the second construct, this putative leader sequence was removed thus leaving a 53 residue extension. The third construct contained the complete 120 residue N-terminal segment including the TargetP-predicted leader sequence. Of these three constructs, only the full-length sequence targeted the GFP reporter into the mitochondrion. Since the leader sequence was quite large and because there have been several reports on the potential regulation of haem biosynthesis at the site of CPO [19,20], we examined the effects of culture haem content on translocation of the 120 residue targeting sequence. The addition of haem or ALA had no effect on the targeting of the CPO–GFP reporter.
PPO targeting
Since PPO lacks any readily identifiable mitochondrial targeting sequence, several putative targeting constructs were examined. Constructs containing the N-terminal 178, 59, 34 and 17 amino acid residues were fused to GFP and their targeting examined. It was found that all these fusions, including the short, 17-residue-long fragment, targeted GFP to mitochondria. Monoamine oxidase, which is highly similar to PPO, has previously been proposed to possess a targeting motif at its C-terminus [21]. To determine if PPO has a similar motif, the C-terminal 57 residues of PPO were fused to GFP. This construct did not target the reporter GFP to mitochondria.
Because of the sequence similarity between human and B. subtilis PPO near their N-terminal ends [13,22], we prepared B. subtilis PPO–GFP reporter constructs that were analogous to the human N-terminal 17-mer targeting sequence. Because of a three residue insert, the analogous B. subtilis construct was 20 residues long (Figure 2). This construct did not target GFP to mitochondria. Removal of the three additional residues found in the bacterial PPO did not result in targeting. Similarly the alteration of aspartate 3 to arginine (found in human) did not result in targeting. Only the removal of the three residues combined with the alteration of residue three to arginine resulted in a construct that was targeted to the mitochondrion.
Figure 2. Human/Bacillus PPO chimaeras.
The N-terminal sequences of human and Bacillus PPO, as well as three chimaeras, are shown. Positively charged amino acid residues are in boldface, negatively charged residues are unmarked, neutral polar residues are double underlined and hydrophobic residues are single underlined. *, Sequences that correctly target the fusion to the mitochondria.
FC targeting
Previously, in yeast, we have shown that removal of the mitochondrial targeting segment results in FC being bound to all cellular membranes and not targeted specifically to the mitochondrion [23]. However, it was not clear if targeting might also involve some portion other than the N-terminus, in particular the C-terminal end that is characteristic of those FCs located within the eukaryotic cellular organelles.
Two GFP constructs were prepared, one with the N-terminal 54 residue putative leader peptide and the second with the C-terminal 34 amino acid residues. Of these constructs only the signal peptide with 54 residues was targeted.
DISCUSSION
The transcriptional regulation of ALAS is well documented and post-translational regulation at the level of haem-mediated control of intracellular translocation has been proposed [24] and supported by in vitro experiments with ALAS-2 [8]. Three putative haem-binding motifs are present in ALAS, two of which are located in the targeting sequence and a third near the N-terminus of the mature protein. Previously, it was demonstrated that in vitro uptake by isolated mitochondria of a fusion protein possessing the ALAS-2 leader sequence (region I) was inhibited by 25 μM haemin only when one of the two putative motifs was intact [8].
We have demonstrated above that intracellular translocation of ALAS-1, as monitored with a short half-life GFP fusion, is haem-sensitive and that this sensitivity is dependent on the presence of at least one of the haem-binding motifs. Interestingly, not only do the two region I sites impart haem-responsive translocation, but the single haem-binding motif in region II of the mature protein is sufficient to block translocation into the mitochondrion. This haem-binding motif is located over 50 amino acid residues from the proteolytic processing site in ALAS-1 and 20 residues distant in ALAS-2. Because of this distance from the targeting sequence, it would seem to be an unusual site for the regulation of translocation. However, given that little haem is present in the mitochondrial matrix compartment, it would seem that any regulatory role for this particular haem-binding motif would be for translocation and not for enzyme inhibition or inhibition of cofactor binding by the apoprotein. Although a specific role for this internal motif is not obvious, it may be that the binding of haem at this site prevents the protein from folding properly, which makes it more susceptible to proteolytic processing in the cytoplasm.
CPO, which catalyses the oxidative decarboxylation of two propionate side-chains to vinyl groups, is located in the space between the inner and outer mitochondrial membranes [4]. The current literature contains conflicting reports related to the size of the CPO mitochondrial targeting sequence. The initial report for cloning mouse CPO proposed a 31 long residue leader that was reported to target CPO to mitochondria in COS cells. Delfau-Larue et al. [9] reported the full-length cDNA for human CPO showing that it contained a leader sequence comprising 120 residues. Examination of the putative leader sequence comprising 120 residues by the program TargetP, however, predicted that the mitochondrial targeting sequence was comprised of only the N-terminal 67 residue long region. When the truncated CPO leader constructs in the present study were produced and examined, two significant observations were made. First, it was found that the 31 amino acid leader sequence identified by Kohno et al. [16] was not sufficient to target any detectable amount of GFP reporter to the mitochondrion. Neither did a construct containing an additional 20 residues, but lacking the complete TargetP predicted sequence, target CPO to mitochondria. Only the 120 residues leader sequence effectively targeted the GFP reporter to mitochondria. Secondly, we found that the leader sequence of mouse CPO published by the Kohno et al. [16] contained nucleotide sequence errors coding for the putative leader sequence that resulted in 12 different amino acid assignments from what was found in the current study as well as a frameshift in the translated coding region that would result in an early termination of protein synthesis.
The conclusion from data presented above is that only the full-length CPO leader reporter construct was effective in mitochondrial targeting. What purpose this extended motif serves is not clear since translocation of the GFP reporter was not sensitive to cellular haem synthesis even when the culture medium was spiked with ALA to increase haem synthesis or with haem itself. Recently, Susa et al. [25] reported that full-length human CPO produced in a cDNA-based in vitro transcription–translation coupled system was imported into isolated rat liver mitochondria. This import was inhibited by micromolar levels of haem with 99% inhibition of translocation occurring at 20 μM haem. This level of haem is similar to that reported to inhibit ALAS-2 preprotein import into isolated mitochondria in a similar system [8] and is well above what would be expected in vivo. Whereas these data may seem to be in conflict with those reported above, which found no inhibition of CPO translocation in the presence of exogenous haem, it should be noted that the import of ALAS-1/GFP reporter construct into mitochondria is completely blocked under conditions of haem elevation produced by ALA spiked medium and the complete 120 residue CPO leader/GFP reporter construct under identical culture conditions is effectively targeted. These data indicate that haem-regulated CPO translocation in situ must be of minimal, if any, significance. It should be noted that this does not rule out a role for CPO in overall pathway regulation as has been suggested by other studies [19,20], but only indicates that haem regulation of CPO translocation through interaction with the leader sequence is not of physiological consequence.
Animal PPO does not contain a typical mitochondrial targeting sequence even though the cytoplasmically synthesized protein is translocated to the cytoplasmic site of the inner mitochondrial membrane [13]. Whereas some computer algorithms predict the N-terminal dinucleotide-binding motif as a transmembrane segment, it is clear from analogy with a number of other FAD-containing proteins that this motif does not participate in membrane attachment [26]. Indeed, there is no evidence that PPO has a transmembrane motif and it has been proposed that a protein-bound lipid mediates membrane attachment [11], although the recently published structure does not show the presence of a lipid in the enzyme from tobacco [12].
We have examined above the requirements for mitochondrial targeting of human PPO through the use of GFP reporter constructs. The data clearly demonstrate that targeting of a reporter protein can be mediated by a minimal sequence of only 17 amino acid residues at the N-terminus. That such a small region was sufficient for targeting was somewhat surprising given that the entire 120 residue leader of CPO was necessary for targeting of CPO. It also raised the question of whether a water soluble bacterial PPO that has considerable sequence homology to the human PPO would be targeted to the mitochondrion of a mammalian cell. This was examined with a fusion of the wild-type B. subtilis PPO N-terminus to GFP. This fusion was not targeted to the mitochondria.
In previous studies, the functionality of other signal sequences was shown to be dependent on the water chemistry of their amino acid residues, including acid–base chemistry [27]. The human PPO signal sequence consists of a 17 amino acid sequence of alternating stretches of hydrophobic and hydrophilic residues. In the event that this signal sequence is an α-helix, as many other sequences capable of inducing mitochondrial translocation are [27], the targeting sequence will be an amphiphilic hydrophobic–hydrophilic α-helix with a positively charged region at its N-terminus. Unlike the human PPO N-terminus, the wild-type B. subtilis PPO N-terminus would not have the capacity to be an amphiphilic helix, a possible explanation for the fact that it does not target proteins to the mitochondria. However, one of the Bacillus PPO/GFP fusions produced (ΔK5-H7) might form an amphiphilic helix, but did not target GFP to the mitochondria, suggesting that a hydrophobic–hydrophilic amphiphilic helix alone is not sufficient to function as a targeting sequence. This ΔK5-H7 mutant PPO is nearly identical with the human targeting sequence in relation to the general water chemistry of its bases, with the significant exception of the third residue. In the human protein, this is a basic arginine, but in B. subtilis PPO it is an acidic aspartate. It seems probable, therefore, that this residue plays a role in targeting. To further investigate this possibility, two additional mutants, D3R and D3R/ΔK5-H7, were constructed from the wild-type Bacillus PPO/GFP fusion and studied. Of the three mutants examined only the mutant with the same length and charge distribution as human PPO (D3R/ΔK5-H7) was targeted to the mitochondrion.
Fraunberg et al. [28] proposed that the 28 N-terminal residues contain an ‘independently functioning signal for mitochondrial targeting’. Interestingly, they reported that constructs lacking the N-terminal 24 residues still targeted to the mitochondrion. Furthermore, it was shown that the PPO mutants I12T, L8Q and L15Q did not target to the mitochondrion. These observations are consistent with the data presented above for the targeting of modified B. subtilis PPO. Recently, Morgan et al. [29] provided evidence that an internal set of residues located between amino acids 150 and 250 are involved in mitochondrial targeting. These authors used the approach of truncating inward from the N-terminus, which is complementary to our approach of truncating from the C-terminus to identify the smallest necessary fragment that will target a reporter to the mitochondria. It is clear from our data along with those of Morgan et al. [29] and Fraunberg et al. [28] that elements at both the N-terminus as well as internal elements are involved in the targeting of PPO to the correct mitochondrial compartment. Now that a structure is available for PPO, it is clear that the internal ‘targeting’ sequence is in fact the membrane association region comprising residues 150–213 [12]. Thus we propose that the N-terminal 17 amino acid segment is responsible for the initial targeting of PPO to the proper mitochondrial compartment and the region of residues 150–213 assures proper membrane association. This is similar to what has been reported previously for yeast FC, where membrane association with the mitochondria occurs in the absence of a functional N-terminal leader segment. In that system specificity for correct insertion of the enzyme into the proper membrane occurs only when the leader sequence is present [30].
FC, the terminal pathway enzyme, appears to be typical of many mitochondrially targeted proteins. It possesses an average-sized N-terminal leader sequence that does not impart haem-sensitive translocation as is found in ALAS. Previously, we have reported for yeast FC that removal of the leader sequence resulted in distribution among all cell membrane fractions, but without proper intramitochondrial targeting ([23] and above). This is consistent with the current data that show the C-end of the protein does not participate in targeting as well as with previous data by Gora et al. [30], which demonstrated that while the leader sequence of yeast FC would allow the targeting of B. subtilis FC to the yeast mitochondrial matrix space, it did not convert this soluble bacterial enzyme into a membrane-associated protein.
Overall the data presented above demonstrate that all mitochondrially located haem biosynthetic pathway enzymes possess sequence features necessary for proper intracellular targeting. Clearly, ALAS-1 also possesses haem-responsive elements in both its leader sequence and near the N-terminus of the mature protein, which inhibit translocation into the mitochondrion. The translocation of no other protein examined was sensitive in situ to the same levels of haem as was ALAS-1. This demonstrates that at a physiologically relevant concentration of haem only ALAS-1 translocation is important in the regulation of tetrapyrrole synthesis in non-erythroid cells by haem.
Acknowledgments
We thank Dr J. Shields and Dr M. Farmer of the University of Georgia's Center for Advanced Ultrastructural Research for assistance with the confocal microscopy. This work was supported by grant DK 32303 from the National Institutes of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
References
- 1.Dailey H. A. Enzymes of heme biosynthesis. J. Biol. Inorg. Chem. 1997;2:411–417. [Google Scholar]
- 2.Sadlon T. J., Dell'Oso T., Surinya K. H., May B. K. Regulation of erythroid 5-aminolevulinate synthase expression during erythropoiesis. Int. J. Biochem. Cell Biol. 1999;31:1153–1167. doi: 10.1016/s1357-2725(99)00073-4. [DOI] [PubMed] [Google Scholar]
- 3.Ades I. Z., Harpe K. G. Biogenesis of mitochondrial proteins: identification of the mature and precursor forms of the subunit of δ-aminolevulinic acid synthase from embryonic chick liver. J. Biol. Chem. 1981;250:9329–9333. [PubMed] [Google Scholar]
- 4.Elder G. H., Evans J. O. Evidence that coproporphyrinogen oxidase activity of rat liver is situated in the intermembrane space of mitochondria. Biochem. J. 1978;172:345–347. doi: 10.1042/bj1720345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira G. C., Andrew T., Karr S. W., Dailey H. A. Organization of the terminal two enzymes of the heme biosynthetic pathway. Orientation of protoporphyrinogen oxidase and evidence for a membrane complex. J. Biol. Chem. 1988;263:3835–3839. [PubMed] [Google Scholar]
- 6.Harbin B. M., Dailey H. A. Orientation of ferrochelatase in bovine liver mitochondrial. Biochemistry. 1985;24:366–370. doi: 10.1021/bi00323a019. [DOI] [PubMed] [Google Scholar]
- 7.Furuyama K., Harigae H., Sassa S. Structure of the human succinyl CoA synthetase bet A-subunit gene and increased gene expression during erythroid differentiation. Blood. 2001;98:29. [Google Scholar]
- 8.Lathrop J. T., Timko M. P. Regulation by heme of mitochondrial protein transport through a conserved amino acid motif. Science. 1993;259:522–525. doi: 10.1126/science.8424176. [DOI] [PubMed] [Google Scholar]
- 9.Delfau-Larue M. H., Martasek P., Grandchamp B. Coproporphyrinogen oxidase: gene organization and description of a mutation leading to exon 6 skipping. Hum. Mol. Genet. 1994;3:1325–1330. doi: 10.1093/hmg/3.8.1325. [DOI] [PubMed] [Google Scholar]
- 10.Medlock A., Dailey H. A. Human coproporphyrinogen oxidase is not a metalloenzyme. J. Biol. Chem. 1996;271:32507–32510. doi: 10.1074/jbc.271.51.32507. [DOI] [PubMed] [Google Scholar]
- 11.Arnould S., Takahashi M., Camadro J.-M. Acylation stabilizes a protease-resistant conformation of protoporphyrinogen oxidase, the molecular target of diphenyl ether-type herbicides. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14825–14830. doi: 10.1073/pnas.96.26.14825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch M., Breithaupt C., Kiefersauer R., Greigang J., Huber R., Messerschmidt A. Crystal structure of protoporphyrinogen IX oxidase: a key enzyme in haem and chlorophyll biosynthesis. EMBO J. 2004;23:1–9. doi: 10.1038/sj.emboj.7600189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dailey T. A., Dailey H. A. Human protoporphyrinogen oxidase. Expression, purification, and characterization of the cloned enzyme. Protein Sci. 1996;5:98–105. doi: 10.1002/pro.5560050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karr S. R., Dailey H. A. The synthesis of murine ferrochelatase in vitro and in vivo. Biochem. J. 1988;254:799–803. doi: 10.1042/bj2540799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dailey H. A., Dailey T. A., Wu C.-K., Medlock A. E., Wang K.-F., Rose J. P., Wang B.-C. Ferrochelatase at the millennium: structures, mechanisms, and [2Fe-2S] clusters. Cell. Mol. Life Sci. 2000;57:1909–1926. doi: 10.1007/PL00000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohno H., Furukawa T., Yoshinaga T., Tokunaga R., Taketani S. Coproporphyrinogen oxidase: purification, molecular cloning, and induction of mRNA during erythroid differentiation. J. Biol. Chem. 1993;268:21359–21363. [PubMed] [Google Scholar]
- 17.Eldridge M. G., Dailey H. A. Serum free, defined medium for the growth and differentiation of murine erythroleukemia cells. Biotechniques. 1992;12:848–852. [PubMed] [Google Scholar]
- 18.Emanuelsson O., Nielsen H., Brunak S., von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 19.Conder L. H., Woodard S. I., Dailey H. A. Multiple mechanisms for regulation of haem synthesis during erythroid cell differentiation. Possible role for coproporphyrinogen oxidase. Biochem. J. 1991;275:321–326. doi: 10.1042/bj2750321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi S., Taketani S., Akasaka J. E., Kobayashi A., Hayashi N., Yamamoto M., Nagai T. Differential regulation of coproporphyrinogen oxidase gene between erythroid and nonerythroid cells. Blood. 1998;92:3436–3444. [PubMed] [Google Scholar]
- 21.Binda C., Newton-Vinson P., Hubalek F., Edmondson D. E., Mattevi A. Structure of human monoamine oxidase B, a drug target for the treatment of neurological disorders. Nat. Struct. Biol. 2002;9:22–26. doi: 10.1038/nsb732. [DOI] [PubMed] [Google Scholar]
- 22.Dailey T. A., Meissner P., Dailey H. A. Expression of a cloned protoporphyrinogen oxidase. J. Biol. Chem. 1994;269:813–815. [PubMed] [Google Scholar]
- 23.Prasad A. R. K., Dailey H. A. Effect of cellular location on the function of ferrochelatase. J. Biol. Chem. 1995;270:18198–18200. doi: 10.1074/jbc.270.31.18198. [DOI] [PubMed] [Google Scholar]
- 24.Yamauchi K., Hayashi N., Kikuchi G. Translocation of 5-aminolevulinate synthase from the cytosol to the mitochondria and its regulation by hemin in rat liver. J. Biol. Chem. 1980;255:1746–1751. [PubMed] [Google Scholar]
- 25.Susa S., Daimon M., Ono H., Li S., Yoshida T., Kato T. Heme inhibits the mitochondrial import of coproporphyrinogen oxidase. Blood. 2002;100:4678–4679. doi: 10.1182/blood-2002-08-2621. [DOI] [PubMed] [Google Scholar]
- 26.Dailey T. A., Dailey H. A. Identification of an FAD superfamily containing protoporphyrinogen oxidase, monoamine oxidases, and phytoene desaturase. Expression and characterization of phytoene desaturase of Myxococcus xanthus. J. Biol. Chem. 1998;273:13658–13662. doi: 10.1074/jbc.273.22.13658. [DOI] [PubMed] [Google Scholar]
- 27.Lithgow T. Targeting of proteins to mitochondria. FEBS Lett. 2000;476:22–26. doi: 10.1016/s0014-5793(00)01663-x. [DOI] [PubMed] [Google Scholar]
- 28.Fraunberg M. V., Nyronen T., Kauppinen R. Mitochondrial targeting of normal and mutant protoporphyrinogen oxidase. J. Biol. Chem. 2003;278:13376–13381. doi: 10.1074/jbc.M300151200. [DOI] [PubMed] [Google Scholar]
- 29.Morgan R. R., Errington R., Elder G. H. Identification of sequences required for the import of human protoporphyrinogen oxidase to mitochondria. Biochem. J. 2004;377:281–287. doi: 10.1042/BJ20030978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gora M., Rytka J., Labbe-Bois R. Activity and cellular location in Saccharomyces cerevisiae of chimeric mouse/yeast and Bacillus subtilis/yeast ferrochelatases. Arch. Biochem. Biophys. 1999;361:231–240. doi: 10.1006/abbi.1998.0990. [DOI] [PubMed] [Google Scholar]