Abstract
Mammalian sulphatases (EC 3.1.6) are a family of enzymes that have a high degree of similarity in amino acid sequence, structure and catalytic mechanism. IDS (iduronate-2-sulphatase; EC 3.1.6.13) is a lysosomal exo-sulphatase that belongs to this protein family and is involved in the degradation of the glycosaminoglycans heparan sulphate and dermatan sulphate. An IDS deficiency causes the lysosomal storage disorder MPS II (mucopolysaccharidosis type II). To examine the structural alterations in heat-denatured and mutant IDS, a panel of four monoclonal antibodies was raised to the denatured protein and used as probes of protein conformation. The linear sequence epitope reactivity of a polyclonal antibody raised against the native protein and the monoclonal antibodies were defined and mapped to distinct regions on the IDS protein. The antigenicity of native IDS was higher in regions without glycosylation, but reactivity was not restricted to protein surface epitopes. One monoclonal epitope was relatively surface accessible and in close proximity to an N-linked glycosylation site, while three others required additional thermal energy to expose the epitopes. The monoclonal antibodies demonstrated the capacity to differentiate progressive structural changes in IDS and could be used to characterize the severity of MPS type II in patients based on variable denatured microstates.
Keywords: heat denaturation, iduronate-2-sulphatase (IDS), mucopolysaccharidosis type II (MPS II), mutant protein, protein conformation
Abbreviations: CHO cells, Chinese-hamster ovary cells; CNS, central nervous system; IDS, iduronate-2-sulphatase; LEC, liver endothelial cell; MPS II, mucopolysaccharidosis type II; PNGaseF, peptide N-glycosidase F
INTRODUCTION
Mammalian sulphatases (EC 3.1.6) are a family of enzymes that have a high degree of sequence identity (20–60%) [1], structural similarity [2–4] and common catalytic mechanism [4–8]. Currently, 11 different mammalian sulphatases have been identified, with eight having a lysosomal location and the other three being distributed to microsomes. These sulphatases catalyse the hydrolysis of sulphated ester bonds at the non-reducing end of sugars on glycosaminoglycans, glycolipids, glycoproteins and hydroxysteroids [9,10]. The human sulphatases display a high degree of substrate specificity and, when deficient, each has been associated with a specific disease state [11–13].
The common tertiary structure in sulphatases is involved in co-ordinating the conserved catalytic residues that form the catalytic site and specifically the Cα-formylglycine that is post-translationally generated during the processing of protein in the endoplasmic reticulum [2–8,10]. The crystal structures of both N-acetylgalactosamine-4-sulphatase (arylsulphatase B) [2] and arylsulphatase A [3] have been resolved. The monomeric proteins are composed of two domains: a large N-terminal domain that belongs to the α/β class of proteins and contains the active site and a smaller C-terminal domain. Arylsulphatase B has been shown to have a hydrophobic core that is typical of globular proteins and contains the recognition sites for the molecular chaperone BiP (immunoglobulin heavy-chain binding protein), which are used during the folding of the protein in the endoplasmic reticulum [14]. The sequence and structural similarity of human sulphatases has been used to justify the molecular modelling of other protein family members, including IDS (iduronate-2-sulphatase; EC 3.1.6.13) [15].
IDS is a lysosomal sulphatase that is involved in the catabolism of the glycosaminoglycan substrates heparan sulphate and dermatan sulphate. A full-length cDNA clone of the IDS gene was isolated and was predicted to encode an amino acid sequence of 550 residues [16]. Various molecular forms of IDS have been reported in different human tissues reflecting differential processing of the enzyme [17,18]. In human fibroblasts, a 76 kDa precursor is converted into a heavily glycosylated 90 kDa form before secretion [19].
N-linked oligosaccharides play a key role in the folding, function and stability of glycoproteins [20]. For example, in the endoplasmic reticulum, N-linked oligosaccharides interact with molecular chaperones to promote specific folding intermediates and this also forms a part of the quality control process that ensures correct protein folding. The IDS sequence contains eight potential N-linked glycosylation sites (NXS/T motif) at positions 31, 115, 144, 246, 280, 325, 513 and 537 (http://www.expasy.org/cgi-bin/sprot-search-de) [21]. Expression studies have shown that all these glycosylation sites can be utilized and that no single glycosylation site is essential for IDS stability, although the glycosylation site at position 280 is important for lysosomal targeting [21]. In circulation, IDS is highly sialylated [22] and this is presumed to prevent both antibody recognition and receptor recapture of the enzyme, maintaining a circulating pool of IDS.
A deficiency in IDS activity results in the accumulation of undegraded/partially degraded heparan and dermatan sulphate substrates in lysosomal organelles, causing the lysosomal storage disorder mucopolysaccharidosis type II (MPS II, Hunter syndrome; McKusick 309900) [11]. MPS II is an X-linked disorder with an autosomal recessive mode of inheritance. MPS II patients present within a spectrum of clinical phenotypes, ranging from severe to attenuated, based on the amount of residual IDS activity and storage product present in affected cells. We have previously reported the detection of residual mutant IDS protein in plasma samples from MPS II patients [23].
Monoclonal antibodies have proven to be powerful tools for investigating protein conformations [24–27]. Here we have investigated the protein microstructure and glycosylation of IDS in relationship with antibody interaction. The specific linear sequence reactivities of a polyclonal antibody and monoclonal antibodies have been determined and mapped on to a sulphatase molecular model. The ability to expose specific epitopes by denaturation/mutation and deglycosylation has been defined. The antibody reactivities give insight into the antigenicity and structural organization of IDS and the structural alterations in mutant protein.
EXPERIMENTAL
Monoclonal and polyclonal antibodies
The monoclonal antibodies 2G3.2B9, 7B9.1B10, 1F7.2D11 and 2D3.1F9 were raised against reduced and denatured recombinant human IDS, in collaboration with CSL Limited (Melbourne, Victoria, Australia). The recombinant human IDS was purified as described previously [28]. The monoclonal antibodies were all IgG1 subclass, as defined by an isotyping kit (Roche Diagnostics, Sydney, NSW, Australia). The sheep polyclonal antibody was produced as described previously [23].
Determination of linear sequence epitopes on IDS
Peptide pins (Chiron Mimotopes, Clayton, Victoria, Australia) were used to determine the epitope reactivity of the mouse monoclonal antibodies and sheep polyclonal antibody, as described previously for other lysosomal proteins [29,30]. In defining antibody epitopes on IDS, we have assumed only a single epitope per 12-mer peptide and that the reactivities observed represent a single antibody interaction per peptide. The level of absorbance observed in the peptide ELISA therefore represents a qualitative measure of antibody affinity. ELISA results were expressed in absorbance units, where less than 0.699 absorbance units indicated regions of little or no epitope reactivity; 0.7–2.499 absorbance units represented low-affinity reactivity and absorbance units of 2.5 or greater represented high-affinity reactivity. All results were compared with positive and negative control pins to allow comparison with other epitope determinations.
Molecular modelling of the antibody epitopes
Using a sequence alignment of arylsulphatase B (Swiss-Prot database primary accession number P15848) and IDS (Swiss-Prot database primary accession number P22304) [31], and the molecular modelling program Swiss-PdbViewer [32], the linear sequence epitopes that had been defined for the monoclonal and polyclonal antibodies to IDS were mapped on to a structural model of arylsulphatase B [PDB (Protein Data Bank) code 1FSU]. The molecular modelling of IDS on arylsulphatase B was based on 18% sequence identity across the length of these two sulphatases, which restricted the protein alignment [9]. This has previously been used by another group to justify the molecular modelling of the two proteins [15]. The antibody epitopes were mapped to a region of sequence reactivity (reflecting the peptide pin sequence that contained the antibody epitope), accounting for any minor differences in structure/alignment.
PNGaseF (peptide N-glycosidase F) digest of IDS
Recombinant IDS (60 μg) was mixed with PNGaseF (3000 units) and incubated at 37 °C for 6 h, according to the manufacturer's instructions (New England Biolabs, Beverly, MA, U.S.A.). Digestion was confirmed by SDS/PAGE analysis.
Detection of native, heat-denatured and mutant IDS
An immunoassay was used to detect native, heat-denatured and mutant IDS, using a previously described method [23]. For heat denaturation, recombinant IDS (5 μg/ml), LEC (liver endothelial cell) IDS (0.125 μg/ml) and PNGaseF-digested IDS (0.125 μg/ml) were incubated at temperatures over the range 25–70 °C for 5 min, then centrifuged at 5000 g for 2 min to remove any protein aggregates before assay. A 100 μl aliquot of sheep polyclonal antibody at a concentration of 10 μg/ml was bound to ELISA plate wells and used to capture the IDS protein. The bound protein was detected using the monoclonal antibodies 2G3.2B9, 7B9.1B10, 1F7.2D11 and 2D3.1F9, and a peroxidase-labelled sheep anti-mouse immunoglobulin detection system (Silenus Laboratories, a subsidiary of Chemicon, Temecula, CA, U.S.A.).
Thermal denaturation of IDS activity
Aliquots (2 μg) of purified IDS were heat-treated for 5 min with temperatures ranging from 25 to 70 °C, and the enzyme activity was determined as described previously [18].
MPS II patients
Eight MPS II patient samples (leucocytes and plasma) were selected at random from submissions at the National Referral Laboratory (Women's and Children's Hospital, Adelaide, Australia) for diagnosis. IDS enzyme activity was determined as described previously [18] and expressed as pmol·min−1·(mg of total cell protein)−1 for leucocytes. The three MPS II patients with an attenuated clinical phenotype [no CNS (central nervous system) involvement and age of onset >3 years] had the mutations: c1094_1100delinsTT, 1246c→t and p.L259P. The MPS II patients with a severe clinical phenotype (CNS involvement and age of onset <3 years) had the mutations: p.S333L, p.C53X, p.E341K and p.P480R and in one patient we were unable to identify the pathogenic mutation.
RESULTS
Sheep polyclonal antibody and mouse monoclonal antibody epitope mapping
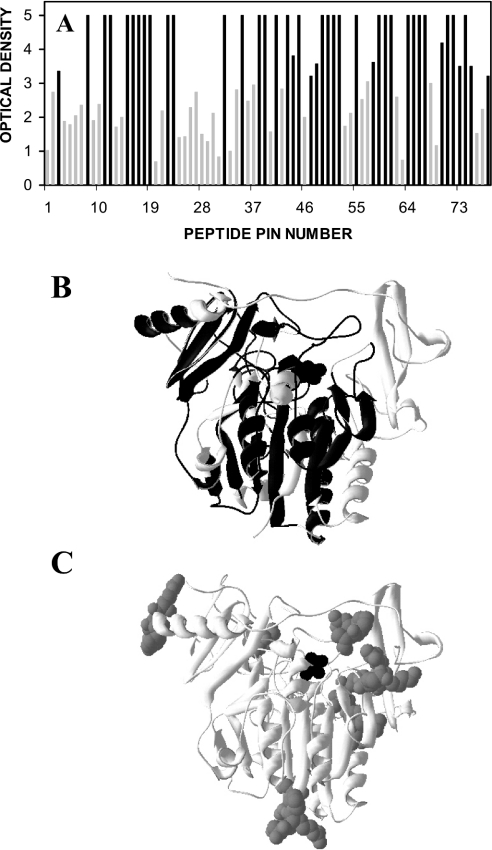
A sheep polyclonal antibody, which was raised against native IDS, produced high-affinity epitope reactivity to approx. 60% of the IDS protein (Figure 1A). The antibody reacted with linear sequence epitopes located both on the surface of the protein and within the internal core of the protein (Figure 1B), including a peptide sequence containing the active site residue (peptide #12; FAQQAVCAPSRVS). This reflected an approximately equivalent reactivity for the predicted β-sheet and α-helix structures within IDS. There was less antibody reactivity to the sites on IDS that contained N-linked glycosylation sites, but some peptides with N-linked glycosylation sites still had a high level of reactivity (Figures 1A and 1C).
Figure 1. Epitope reactivity of IDS polyclonal antibody.
(A) Epitope reactivity of a sheep polyclonal antibody raised against IDS, expressed as individual peptide sequences across the IDS protein. The level of ELISA reactivity to each peptide sequence was expressed in terms of A410 (‘optical density’). Black bars denote high-affinity epitope reactivity. (B) Epitopes detected by the sheep anti-IDS polyclonal antibody (black) mapped on to a structural representation of IDS. (C) The N-linked carbohydrate structures (grey spheres) depicted on the same structural representation of IDS (black spheres show the active site). N-linked glycosylation sites were located on the peptides of pins 4–5, 16–17, 20–21, 35–36, 40, 46–47, 73–74 and 77.
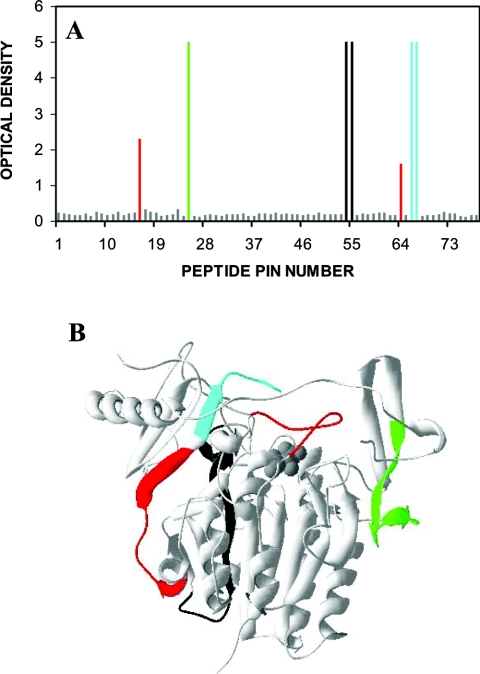
The monoclonal antibodies 2G3.2B9, 7B9.1B10, 1F7.2D11 and 2D3.1F9 were all generated to denatured IDS. Previous attempts to generate monoclonal antibodies to native IDS were all unsuccessful (E. Parkinson-Lawrence and D. A. Brooks, unpublished work) and this was presumed to be due to the high glycosylation/sialylation of IDS. Three of the monoclonal antibodies generated to denatured IDS, 2G3.2B9, 7B9.1B10 and 2D3.1F9, reacted with high affinity to linear-sequence epitopes on IDS (Figure 2A). The 2G3.2B9 epitope was apparently located near the surface of the IDS protein, but was in close proximity to and between two N-linked glycosylation structures (Figures 1C and 2B). The monoclonal antibody 7B9.1B10 was mapped to an internal location of IDS (Figure 2B). The 2D3.1F9 epitope was located between the two domains of IDS (Figure 2B). A fourth monoclonal antibody 1F7.2D11 reacted with a discontinuous sequence epitope (Figures 2A and 2B) that appeared to span the large and small domains of IDS. The individual linear sequence epitopes detected by the monoclonal antibodies 7B9.1B10, 1F7.2D11 and 2D3.1F9 (Figure 2A) were also reactive with the polyclonal antibody (Figure 1A). However, the polyclonal antibody demonstrated only a low level of reactivity to the epitope detected by the monoclonal antibody 2G3.2B9.
Figure 2. Epitope reactivity of IDS monoclonal antibodies.
(A) Epitope reactivity of the monoclonal antibodies 2G3.2B9 (green), 7B9.1B10 (black), 1F7.2D11 (red) and 2D3.1F9 (turquoise) against IDS, expressed as the individual peptide sequences recognized within the IDS protein. The level of ELISA reactivity to each peptide sequence was expressed in terms of A410. (B) The epitopes detected by each of the monoclonal antibodies when mapped on to a structural representation of IDS (grey spheres depict the active site).
Reactivity of monoclonal antibodies to native and heat-denatured IDS
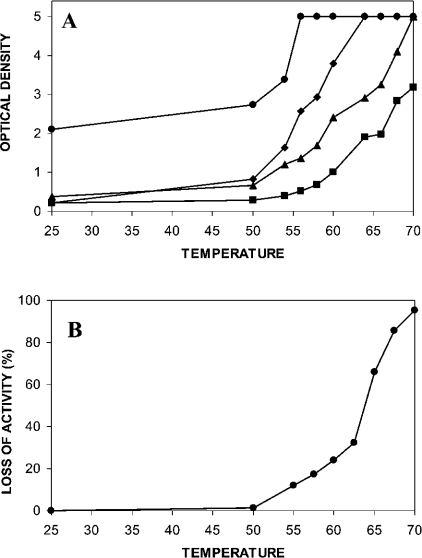
A thermal denaturation profile was defined for IDS by measuring the conformational alteration of the protein at different temperatures, using the exposure of specific monoclonal antibody reactive epitopes as a detection system (Figure 3A). The thermal profiles detected by the four monoclonal antibodies 2G3.2B9, 7B9.1B10, 1F7.2D11 and 2D3.1F9 were distinct, reflecting the thermal energy required to expose each epitope and indicating different locations within the IDS protein. The three monoclonal antibodies 7B9.1B10, 1F7.2D11 and 2D3.1F9 had similar base-line reactivity to IDS at 25 °C, but this was approx. 2-fold lower than the reactivity detected with the monoclonal antibody 2G3.2B9 at the same temperature. Only minimal change in the reactivity of each monoclonal antibody was observed for IDS temperature treatments up to 50 °C. At temperatures above 55 °C, the level of reactivity for each monoclonal antibody to IDS significantly increased to distinct plateaus. The thermal transition midpoints were at 53 °C for 2G3.2B9 (plateau at 55 °C), 58 °C for 7B9.1B10 (plateau at 63 °C), 63 °C for 1F7.2D11 (plateau at 70 °C) and 70 °C for 2D3.1F9 (plateau at 90 °C). These reactivities indicated that a significantly higher thermal energy was required to expose maximally each of the epitopes, with the order of 2G3.2B9<7B9.1B10<1F7.2D11<2D3.1F9. The thermal denaturation of IDS also showed a significant loss of enzyme activity for temperatures above 50 °C, with a thermal midpoint at 63 °C, which matched the midpoint determined for the monoclonal antibody 1F7.2D11 (Figure 3B). The reactivity of 1F7.2D11 must be considered to be different from the other three monoclonal antibodies as this represents the coming together of a split epitope as opposed to the exposure of an internal epitope. Each epitope represents different changes in the microstructure of IDS invoked by the progressive thermal denaturation of the protein.
Figure 3. Thermal denaturation of IDS.
(A) Thermal denaturation profiles for IDS detected by conformation changes in the protein at different temperatures, using the exposure of the specific monoclonal antibody reactive epitopes, 2G3.2B9 (●), 7B9.1B10 (◆), 1F7.2D11 (▲) and 2D3.1F9 (■), as a detection system. Results represent the level of ELISA reactivity at each temperature and were expressed in terms of A410. All samples were assayed in triplicate and the result presented as the mean (S.D. for all sample points was less than 0.2). (B) Percentage loss of IDS activity in response to heat denaturation.
Reactivity of monoclonal antibodies to CHO (Chinese-hamster ovary), LEC and PNGaseF-treated CHO IDS
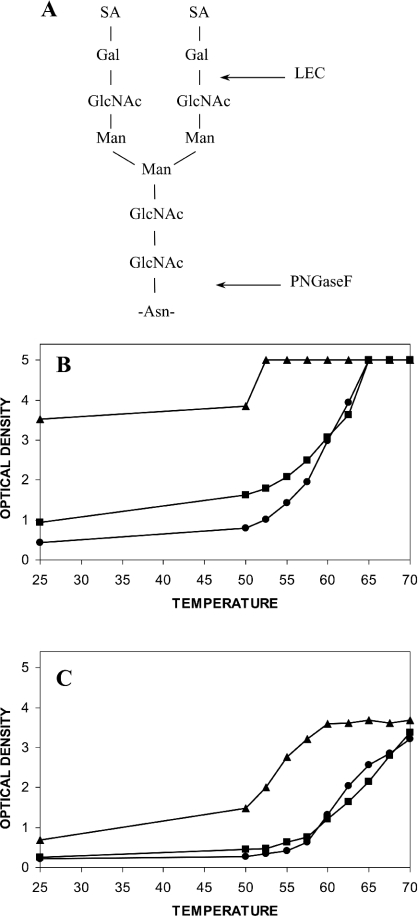
CHO, LEC and PNGaseF-treated CHO IDS were tested to determine the effect of N-linked carbohydrate structure (Figure 4A) on the epitope reactivity of the monoclonal antibodies 2G3.2B9, 7B9.1B10, 1F7.2D11 and 2D3.1F9. The thermal stability profile detected using the epitope 2G3.2B9 showed increased reactivity for LEC IDS compared with CHO IDS, and an even greater increase for PNGaseF-treated CHO IDS, in the temperature range of up to 60 °C (Figure 4B). The maximum reactivity for the 2G3.2B9 epitope was at 52.5 °C for PNGaseF-treated IDS, compared with 65 °C for CHO and LEC IDS. Treatment of CHO IDS with neuraminidase resulted in a similar increase in 2G3.2B9 epitope reactivity (results not shown) compared with that observed for LEC IDS (Figure 4B). There was little or no increase in epitope reactivity with all four monoclonal antibodies for LEC compared with CHO IDS, but a substantial increase in epitope reactivity across the temperature profile for PNGaseF-treated CHO IDS (Figures 4B and 4C). The monoclonal antibody 7B9.1B10 (Figure 4C) produced an almost identical pattern of reactivity to the other two monoclonal antibodies 1F7.2D11 and 2D3.1F9 (results not shown). The maximal epitope reactivity reached a peak at an absorbance of 3.5 and not 5 absorbance units for the monoclonal antibody 7B9.1B10, suggesting that the system was saturated.
Figure 4. Structure of a typical complex N-linked oligosaccharide and thermal denaturation profiles for CHO, LEC and PNGaseF-treated CHO IDS.
(A) The positions of the LEC and PNGaseF digest sites are marked. (B) The thermal denaturation profile for CHO (●), LEC (■) and PNGaseF-treated CHO (▲) IDS detected by the monoclonal antibody 2G3.2B9. (C) The thermal denaturation profile for CHO (●), LEC (■) and PNGaseF-treated CHO (▲) IDS detected by the monoclonal antibody 7B9.1B10. Results represent the level of ELISA reactivity at each temperature and were expressed in terms of A410. All samples were assayed in triplicate and the result presented as the mean (S.D. for all sample points was less than 0.2).
A difference was noted in the epitope reactivity for the monoclonal antibodies 2G3.2B9 and 7B9.1B10 against CHO IDS when comparing the data in Figures 3(A) and 4. There was a more than 2-fold difference in the level of IDS protein assayed in the two experiments (due to the dilutions required to control PNGaseF treatment), suggesting a protein-dependent effect on denaturation/epitope reactivity. This same effect was substantiated in subsequent experiments, where higher temperatures were required to achieve maximum (plateau) epitope reactivity with each of the monoclonal antibodies when assaying lower IDS protein concentrations (results not shown).
Epitope reactivity of normal and mutant IDS in human plasma samples
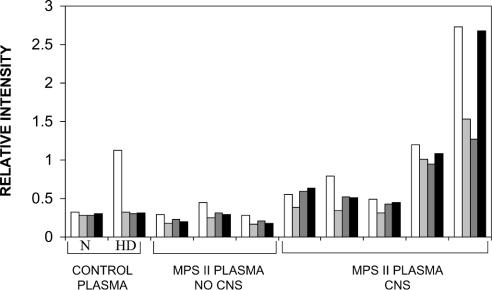
A previous study demonstrated increased levels of mutant IDS in the plasma samples from some MPS II patients [23]. Here, we have investigated the epitope reactivity of IDS in plasma samples from normal controls and MPS II patients with detectable immune protein (Figure 5). Treatment of normal control plasma at 60 °C resulted in an increase only in the 2G3.2B9 epitope, compared with control untreated plasma. In MPS II patients with an attenuated clinical phenotype (age of onset >3 years and no CNS involvement), there was a similar pattern of epitope reactivity to that detected for heat-denatured normal control plasma, with exposure of the 2G3.2B9 epitope. However, in MPS II patients with a severe clinical phenotype (age of onset <3 years and CNS involvement) there was both exposure of the 2G3.2B9 epitope as well as higher levels of reactivity for the internal epitopes 7B9.1B10, 1F7.2D11 and 2D3.1F9. This was consistent with greater structural changes in the mutant IDS protein for patients suffering from severe MPS II compared with those with attenuated MPS II.
Figure 5. Epitope reactivity of IDS in plasma samples from normal controls and MPS II patients.
ELISA reactivity for either 2G3.2B9 (white), 7B9.1B10 (light grey), 1F7.2D11 (dark grey) or 2D3.1F9 (black) against IDS in plasma from normal control and eight MPS II patient samples (Caucasian origin). The three MPS II patients with an attenuated clinical phenotype (no CNS involvement and age of onset >3 years) had ND–0.6 pmol·min−1·mg−1 IDS activity (where ND stands for none detected) in leucocytes and, from left to right, had the mutations: c1094_1100delinsTT, 1246c→t and p.L259P. The MPS II patients with a severe clinical phenotype (CNS involvement and age of onset <3 years) had ND–0.1 pmol·min−1·mg−1 IDS activity in leucocytes and from left to right, had the mutations: p.S333L, p.C53X, p.E341K and p.P480R, but in the last patient we were not able to identify the pathogenic mutation. This compared with n=10 normal controls who had a range of 10–85 pmol·min−1·mg−1 IDS activity. Plasma samples from normal controls (representative pattern shown) were either assayed direct (N) or after heat treatment at 60 °C (HD). Results were expressed as the relative intensity of ELISA reactivity determined at A410.
DISCUSSION
Lysosomal sulphatases comprise a group of at least seven different proteins that have common structure but distinct function [9]. This common structure is also shared with non-lysosomal sulphatases and the phosphatase superfamily of enzymes [33]. Some of the structural similarity for sulphatases relates to a common catalytic mechanism, which depends on the specific location and processing of a rigidly conserved cysteine residue [2–6].
We presume that the high level of glycosylation on circulating IDS functions in part to mask its immune reactivity, and to promote the half-life of the protein in this compartment. Previous attempts to generate monoclonal antibodies to native IDS were unsuccessful, consistent with reduced antigenicity. One of the aims of the present study was to characterize the antigenicity of IDS using an epitope mapping strategy [29,30]. A polyclonal antibody reacted to epitopes located on the surface and internal core of IDS, reacting with both α-helix and β-sheet structures. However, there was a trend for less antibody reactivity to sites on IDS with N-linked glycosylation. The high level of epitope reactivity to peptides involving two of the eight N-linked glycosylation sites may indicate either an epitope in close proximity to the glycosylation site or antibody reactivity to the peptide on the internal face of the protein. Interestingly, we have been able to generate monoclonal antibodies to denatured IDS and these reacted with epitopes in the internal core of the protein. The masked nature of these epitopes in native IDS suggested that these monoclonal antibodies could be used for structural studies on IDS, with application for the investigation of mutant protein.
The monoclonal antibody 2G3.2B9 reacted with a linear-sequence epitope that was in close proximity to an N-linked glycosylation site on IDS. This antibody displayed more reactivity to native IDS than the three other monoclonal antibodies, suggesting the epitope was partially exposed and near the surface of the molecule. The epitope was readily exposed by heat denaturation, with a thermal transition midpoint at 53 °C. In contrast, the monoclonal antibodies 7B9.1B10, 1F7.2D11 and 2D3.1F9 only had residual reactivity to native IDS and had higher thermal transition midpoints at 58 °C for 7B9.1B10, 63 °C for 1F7.2D11 and 70 °C for 2D3.1F9. The thermal transition profile for the monoclonal antibody 1F7.2D11 closely matched the loss of IDS activity in response to heat denaturation and represented both the coming together of a discontinuous epitope and a major structural change in the protein. The dataset was consistent with the 2G3.2B9, 7B9.1B10 and 2D3.1F9 epitopes being progressively more buried within IDS, with each requiring more thermal energy for maximum exposure. The panel of monoclonal antibodies was therefore appropriate for the study of structural/conformational changes in IDS and was the opposite of a previously reported strategy that used conformation-sensitive monoclonal antibodies in another system as markers of protein denaturation (i.e. measuring the disruption of epitopes) [34]. The advantage of the method reported here was that the monoclonal antibodies were capable of giving a more detailed and progressive picture of structural change, reflecting the opening of the hydrophobic core of the protein and for 1F7.2D11, a change in the juxtaposition of two protein domains.
Having established the ability of the monoclonal antibodies to reflect the conformational alteration of native IDS in response to thermal denaturation, we investigated the role of glycosylation on the stability of the protein and its impact on epitope reactivity. The 2G3.2B9 epitope was located in close proximity to N-linked glycosylation sites and we postulated that it might be partly masked by these moieties. Neuraminidase treatment and LEC IDS partially increased the reactivity of this epitope relative to native CHO IDS, which was consistent with this hypothesis. In contrast, the 7B9.1B10, 1F7.2D11 and 2D3.1F9 epitope reactivities were relatively unchanged, consistent with being buried and distant from the surface N-linked glycosylation sites. Deglycosylation of IDS with PNGaseF produced a marked increase in 2G3.2B9 epitope reactivity, but there was still a response to thermal denaturation, suggesting that both glycosylation and protein conformation were involved in the masking of this epitope in native IDS. Thermal denaturation profiles for 7B9.1B10, 1F7.2D11 and 2D3.1F9 were altered in PNGaseF-treated IDS, suggesting that N-linked glycosylation contributed to the stability of IDS.
We have previously reported that neither age of disease onset, nor genotype, nor residual protein/activity in plasma samples was consistently informative for the prediction of clinical severity in MPS II patients [23]. In the present study, we have investigated the structure of mutant IDS in plasma samples of MPS II patients with immune-detectable protein. In three MPS II patients with an attenuated clinical phenotype (age of onset >3 years and no CNS involvement), we observed selective exposure of the 2G3.2B9 epitope. This was consistent with a moderate structural change in the mutant IDS, which resembled that observed for 60 °C heat-treated IDS. In five MPS II patients with a severe clinical phenotype (age of onset <3 years and CNS involvement), we observed the exposure of all four epitopes tested, which indicated greater structural change in the mutant IDS and exposure of the hydrophobic core of the protein. The ability of monoclonal antibodies to distinguish distinct steps in the denaturation of IDS and to reflect the degree of structural alteration in mutant protein, may be used to predict clinical severity in MPS II patients.
Acknowledgments
This work was supported by an NH&MRC Program Grant and an NH&MRC Fellowship Grant in Australia. Special thanks to L. Melville and P. Clements for the provision of purified LEC IDS, V. Muller for mutation analysis and C. Boulter for assistance with tissue culture. CSL Limited is acknowledged for the production of the monoclonal antibodies.
References
- 1.Boltes I., Czapinska H., Kahnert A., von Bulow R., Dierks T., Schmidt B., von Figura K., Kertesz M. A., Uson I. 1.3 Å structure of arylsulfatase from Pseudomonas aeruginosa establishes the catalytic mechanism of sulfate ester cleavage in the sulfatase family. Structure. 2001;9:483–491. doi: 10.1016/s0969-2126(01)00609-8. [DOI] [PubMed] [Google Scholar]
- 2.Bond C. S., Clements P. R., Ashby S. J., Collyer C. A., Harrop S. J., Hopwood J. J., Guss J. M. Structure of a human lysosomal sulfatase. Structure. 1997;5:277–289. doi: 10.1016/s0969-2126(97)00185-8. [DOI] [PubMed] [Google Scholar]
- 3.Lukatela G., Krauss N., Theis K., Selmer T., Gieselmann V., von Figura K., Saenger W. Crystal structure of human arylsulfatase A: the aldehyde function and the metal ion at the active site suggest a novel mechanism for sulfate ester hydrolysis. Biochemistry. 1998;37:3654–3664. doi: 10.1021/bi9714924. [DOI] [PubMed] [Google Scholar]
- 4.von Bulow R., Schmidt B., Dierks T., von Figura K., Uson I. Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis. J. Mol. Biol. 2001;305:269–277. doi: 10.1006/jmbi.2000.4297. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt B., Selmer T., Ingendoh A., von Figura K. A novel amino acid modification in sulfatases that is defective in multiple sulfatase deficiency. Cell (Cambridge, Mass.) 1995;82:271–278. doi: 10.1016/0092-8674(95)90314-3. [DOI] [PubMed] [Google Scholar]
- 6.Selmer T., Hallmann A., Schmidt B., Sumper M., von Figura K. The evolutionary conservation of a novel protein modification, the conversion of cysteine to serinesemialdehyde in arylsulfatase from Volvox carteri. Eur. J. Biochem. 1996;238:341–345. doi: 10.1111/j.1432-1033.1996.0341z.x. [DOI] [PubMed] [Google Scholar]
- 7.Miech C., Dierks T., Selmer T., von Figura K., Schmidt B. Arylsulfatase from Klebsiella pneumoniae carries a formylglycine generated from a serine. J. Biol. Chem. 1998;273:4835–4837. doi: 10.1074/jbc.273.9.4835. [DOI] [PubMed] [Google Scholar]
- 8.Dierks T., Miech C., Hummerjohann J., Schmidt B., Kertesz M. A., von Figura K. Posttranslational formation of formylglycine in prokaryotic sulfatases by modification of either cysteine or serine. J. Biol. Chem. 1998;273:25560–25564. doi: 10.1074/jbc.273.40.25560. [DOI] [PubMed] [Google Scholar]
- 9.Parenti G., Meroni G., Ballabio A. The sulfatase gene family. Curr. Opin. Genet. Dev. 1997;7:386–391. doi: 10.1016/s0959-437x(97)80153-0. [DOI] [PubMed] [Google Scholar]
- 10.von Figura K., Schmidt B., Selmer T., Dierks T. A novel protein modification generating an aldehyde group in sulfatases: its role in catalysis and disease. Bioessays. 1998;20:505–510. doi: 10.1002/(SICI)1521-1878(199806)20:6<505::AID-BIES9>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 11.Neufeld E. F., Muenzer J. The mucopolysaccharidoses. In: Scriver C. R., Beaudet A. L., Sly W. S., Valle D., editors. The Metabolic and Molecular Bases of Inherited Diseases. 8th edn. New York: McGraw-Hill; 2001. pp. 3421–3452. [Google Scholar]
- 12.Jobsis A. C., De Groot W. P., Tigges A. J., De Bruijn H. W., Rijken Y., Meijer A. E., Marinkovic-Ilsen A. X-linked ichthyosis and X-linked placental sulfatase deficiency: a disease entity. Histochemical observations. Am. J. Pathol. 1980;99:279–289. [PMC free article] [PubMed] [Google Scholar]
- 13.Crawfurd M. A. Review: genetics of steroid sulphatase deficiency and X-linked ichthyosis. J. Inherit. Metab. Dis. 1982;5:153–163. doi: 10.1007/BF01800171. [DOI] [PubMed] [Google Scholar]
- 14.Bradford T. M., Gething M. J., Davey R., Hopwood J. J., Brooks D. A. Processing of normal lysosomal and mutant N-acetylgalactosamine 4-sulphatase: BiP (immunoglobulin heavy-chain binding protein) may interact with critical protein contact sites. Biochem. J. 1999;341:193–201. [PMC free article] [PubMed] [Google Scholar]
- 15.Kim C. H., Hwang H. Z., Song S. M., Paik K. H., Kwon E. K., Moon K. B., Yoon J. H., Han C. K., Jin D. K. Mutational spectrum of the iduronate 2 sulfatase gene in 25 unrelated Korean Hunter syndrome patients: identification of 13 novel mutations. Hum. Mutat. 2003;21:449–450. doi: 10.1002/humu.9128. [DOI] [PubMed] [Google Scholar]
- 16.Wilson P. J., Morris C. P., Anson D. S., Occhiodoro T., Bielicki J., Clements P. R., Hopwood J. J. Hunter syndrome: isolation of an iduronate-2-sulfatase cDNA clone and analysis of patient DNA. Proc. Natl. Acad. Sci. U.S.A. 1990;87:8531–8535. doi: 10.1073/pnas.87.21.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniele A., Di Natale P. Hunter syndrome: presence of material cross-reacting with antibodies against iduronate sulphatase. Hum. Genet. 1987;75:234–238. doi: 10.1007/BF00281065. [DOI] [PubMed] [Google Scholar]
- 18.Bielicki J., Freeman C., Clements P. R., Hopwood J. J. Human liver iduronate-2-sulphatase: purification, characterization and catalytic properties. Biochem. J. 1990;271:75–86. doi: 10.1042/bj2710075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Froissart R., Millat G., Mathieu M., Bozon D., Maire I. Processing of iduronate 2-sulphatase in human fibroblasts. Biochem. J. 1995;309:425–430. doi: 10.1042/bj3090425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond C., Braakman I., Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. U.S.A. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millat G., Froissart R., Maire I., Bozon D. Characterization of iduronate sulphatase mutants affecting N-glycosylation sites and the cysteine-84 residue. Biochem. J. 1997;326:243–247. doi: 10.1042/bj3260243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Archer I. M., Harper P. S., Wusteman F. S. Multiple forms of iduronate 2-sulphate sulphatase in human tissues and body fluids. Biochim. Biophys. Acta. 1982;708:134–140. doi: 10.1016/0167-4838(82)90213-8. [DOI] [PubMed] [Google Scholar]
- 23.Parkinson E. J., Muller V., Hopwood J. J., Brooks D. A. Iduronate-2-sulphatase protein detection in plasma from mucopolysaccharidosis type II patients. Mol. Genet. Metab. 2004;81:58–64. doi: 10.1016/j.ymgme.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Kato A., Shimizu T., Saga S. Conformational changes in mutant lysozymes detected with monoclonal antibody. FEBS Lett. 1995;371:17–20. doi: 10.1016/0014-5793(95)00846-2. [DOI] [PubMed] [Google Scholar]
- 25.Korth C., Streit P., Oesch B. Monoclonal antibodies specific for the native, disease-associated isoform of the prion protein. Methods Enzymol. 1999;309:106–122. doi: 10.1016/s0076-6879(99)09010-2. [DOI] [PubMed] [Google Scholar]
- 26.Borromeo V., Gaggioli D., Berrini A., Secchi C. Monoclonal antibodies as a probe for the unfolding of porcine growth hormone. J. Immunol. Methods. 2003;272:107–115. doi: 10.1016/s0022-1759(02)00433-7. [DOI] [PubMed] [Google Scholar]
- 27.Brooks D. A., McCourt P. A., Gibson G. J., Ashton L. J., Shutter M., Hopwood J. J. Analysis of N-acetylgalactosamine-4-sulfatase protein and kinetics in mucopolysaccharidosis type VI patients. Am. J. Hum. Genet. 1991;48:710–719. [PMC free article] [PubMed] [Google Scholar]
- 28.Bielicki J., Hopwood J. J., Wilson P. J., Anson D. S. Recombinant human iduronate-2-sulphatase: correction of mucopolysaccharidosis-type II fibroblasts and characterization of the purified enzyme. Biochem. J. 1993;289:241–246. doi: 10.1042/bj2890241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner C. T., Hopwood J. J., Bond C. S., Brooks D. A. Immune response to enzyme replacement therapy: 4-sulfatase epitope reactivity of plasma antibodies from MPS VI cats. Mol. Genet. Metab. 1999;67:194–205. doi: 10.1006/mgme.1999.2859. [DOI] [PubMed] [Google Scholar]
- 30.Kakavanos R., Turner C. T., Hopwood J. J., Kakkis E. D., Brooks D. A. Immune tolerance after long-term enzyme-replacement therapy among patients who have mucopolysaccharidosis I. Lancet. 2003;361:1608–1613. doi: 10.1016/S0140-6736(03)13311-9. [DOI] [PubMed] [Google Scholar]
- 31.Litjens T. Adelaide: University of Adelaide; 1994. The molecular genetics of mucopolysaccharidosis type VI; pp. 1–281. Ph.D. thesis. [Google Scholar]
- 32.Guex N., Peitsch M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 33.Galperin M. Y., Jedrzejas M. J. Conserved core structure and active site residues in alkaline phosphatase superfamily enzymes. Proteins. 2001;45:318–324. doi: 10.1002/prot.1152. [DOI] [PubMed] [Google Scholar]
- 34.Pfund W. P., Bourdage J. S., Farley K. A. Structural analysis of bovine somatotropin using monoclonal antibodies and the conformation-sensitive immunoassay. J. Biol. Chem. 1996;271:14055–14061. doi: 10.1074/jbc.271.24.14055. [DOI] [PubMed] [Google Scholar]