Abstract
Phantom tooth pain (PTP) is one type of non-odontogenic neuropathic toothache, which rarely occurs after appropriate pulpectomy or tooth extraction. The cause of PTP is unknown. We investigated pain-related genetic factors that are associated with PTP. Four pain-associated genes, including G protein-coupled receptor 158 (GPR158) and phosphoribosyl transferase domain containing 1 (PRTFDC1), are adjacent to each other on the human genome. Some of these four genes or their genomic region may be related to PTP. We statistically analyzed associations between single-nucleotide polymorphisms (SNPs) in the genomic region and PTP in patients with PTP (PTP group), other orofacial pain (OFP group), and healthy control subjects. We then performed a database search of expression quantitative trait loci (eQTLs). For the seven SNPs that were significantly associated with PTP even after Bonferroni correction, we focused on the rs12411980 tag SNP (p = 9.42 × 10−4). Statistical analyses of the PTP group and healthy subject groups (group labels: NOC and TD) revealed that the rate of the GG genotype of the rs12411980 SNP was significantly higher in the PTP group than in the healthy subject groups (PTP group vs. NOC group: p = 2.92 × 10−4, PTP group vs. TD group: p = 5.46 × 10−4; percentage of GG: 30% in PTP group, 12% in NOC group, 11% in TD group). These results suggest that the GG genotype of the rs12411980 SNP is more susceptible to PTP. The rs2765697 SNP that is in strong linkage disequilibrium with the rs12411980 SNP is an eQTL that is associated with higher PRTFDC1 expression in the minor allele homozygotes in the healthy subject groups of the rs2765697 SNP. Thus, PRTFDC1 expression similarly increases in the minor allele homozygotes (GG genotype) in the healthy subject groups of the rs12411980 SNP, which would lead to greater susceptibility to PTP.
Keywords: Neuropathic pain, phantom tooth pain, single-nucleotide polymorphism, expression quantitative trait loci, gene expression
Introduction
As dental caries progress, treatment to remove the peripheral trigeminal nerve (i.e., the pulp nerve [pulpectomy]) is performed without residual neuropathic pain in most cases. Non-odontogenic neuropathic toothaches include phantom tooth pain (PTP), which rarely occurs after appropriate pulpectomy and tooth extraction procedures. PTP is still not widely recognized and often misdiagnosed as temporomandibular joint disorder, trigeminal neuralgia, sinusitis, denture incompatibility, neuralgia, or myofascial pain. 1 The cause of PTP is still unknown. PTP has essentially similar features to phantom pain after limb amputation, and these phenomena may be related.2,3 Genetic factors in phantom pain have been reported in animal studies. 1 In humans, the rs735055 single-nucleotide polymorphism (SNP) of the SLC17A9 gene, rs3732759 SNP of the P2RY12 gene, and rs216009 SNP of the CACNA1C gene were shown to be associated with PTP.4,5 However, the pathogenetic mechanism of PTP has not yet been elucidated.
There is a genomic region where pain-related genes are clustered on chromosome 10. Four pain-related genes (GPR158, PRTFDC1, ENKUR, and THNSL1) that are adjacent to each other exist in this region.
G protein-coupled receptor 158 (GPR158), one of the most abundant G protein-coupled receptors in the brain, 6 transduces signals by binding to regulator of G protein signaling (RGS) proteins. 7 It regulates signaling to the second messenger cyclic 3′,5′-adenosine monophosphate (cAMP) in nerves and regulates ion channels, kinases, and neurotrophic factors that are involved in neural excitation and synaptic transmission.8,9 GPR158 is associated with γ-aminobutyric acid (GABA) receptors, which are implicated in various neurological and psychiatric disorders, including epilepsy, anxiety disorders, schizophrenia, and depression. It is also implicated in the etiology of many affective disorders, including neurodegenerative disorders, memory loss, and stress-induced depression.7,10,11 In addition, chronic pain and depression are associated with lower cAMP concentrations and depressive-like behaviors under conditions of chronic stress. Glycine, an inhibitory neurotransmitter that is associated with neuropathic pain, is a ligand for GPR158 and inhibits activity of the GPR158-RGS complex, increasing neural excitability and cAMP levels in the prefrontal cortex. 12
A significant association was found between the rs6482463 SNP of the phosphoribosyl transferase domain containing 1 (PRTFDC1) gene and posttraumatic stress disorder (PTSD) in a genome-wide association study. 13 Many studies suggest that PTSD is associated with chronic pain.14–16 A range of traumatic experiences and PTSD is associated with higher pain detection and central sensitization. 17 Post-accident pain was shown to be associated with PTSD-related intrusive memories in Japanese patients. 18 These findings indicate that PRTFDC1 is associated with pain.
ENKUR protein, encoded by the Enkurin transient receptor potential canonical (TRPC) channel interacting protein (ENKUR) gene, interacts with TRPC channels. ENKUR functions as an adaptor protein and anchors signal transduction proteins to TRPC channels. 19 TRPC channels are known to be involved in pain transmission 20 ; thus, ENKUR may also be involved in pain via TRPC channels.
The L-threonine synthase gene threonine synthase like 1 (THNSL1) gene is a paralog of threonine synthase like 2 (THNSL2). THNSL2 expression causes a novel secreted osteoclastogenic factor of activated T cells (SOFAT) that acts to exacerbate inflammation and bone metabolic turnover under inflammatory conditions, such as rheumatoid arthritis, periodontitis, and estrogen deficiency. 21 SOFAT is associated with joint pain in rheumatoid arthritis, 22 and the THNSL1 gene, a paralog of the THNSL2 gene, may also be associated with pain.
The PRTFDC1, ENKUR, THNSL1, and GPR158 genes are located on chromosome 10, adjacent to each other in this order within a region of approximately 750 kilobase pairs (kbp). Although these genes could be associated with pain, no study has reported a significant association between SNPs of these genes and pain. The gene clustered region may be associated with pain as a whole. Therefore, we considered the possibility that these genes or this region may be genetically related to PTP. The association analysis of SNPs in this region in the PTP group and control groups revealed that the rs12411980 SNP is significantly associated with PTP. Thus, we focused on the rs12411980 SNP. The rs2765697 SNP, which is in strong linkage disequilibrium (LD) with the rs12411980 SNP, is an expression quantitative trait locus (eQTL) of PRTFDC1 according to the GTEx Portal database, indicating that both the rs12411980 and rs2765697 SNPs are associated with PRTFDC1 expression. The possibility that PRTFDC1 expression is associated with the susceptibility to PTP is discussed.
Materials and methods
Patients
The study was performed in accordance with provisions of the Declaration of Helsinki. All subjects provided written informed consent for the genetics studies. All of the subjects were Japanese.
The present study with PTP and OFP groups was approved by the Ethics Committees of Tokyo Dental College and Tokyo Metropolitan Institute of Medical Science (approval no. 1132 and 20-45 (1), respectively). Detailed demographic and clinical data of the subjects were provided in a previous report. 4 Briefly, the study enrolled 33 PTP patients (PTP group, 26-74 years old) and 118 patients without PTP with pain or dysesthesia in the orofacial region (OFP group, 23-89 years old) who visited Tokyo Dental College Suidobashi Hospital from May 2007 to November 2019. The patients were classified as having traumatic trigeminal neuropathy, trigeminal neuralgia, postherpetic neuralgia, neuralgia-inducing cavitational osteonecrosis, and nociplastic dental pain based on the International Classification of Orofacial Pain, 1st edition, 23 and International Statistical Classification of Diseases and Related Health Problems, 11th revision. 24
We enrolled 500 healthy adult volunteers who lived in the Kanto area of Japan and agreed to participate in the study (group label: NOC, American Society of Anesthesiologists Physical Status I, 20-72 years old, 253 males, 242 females, and five gender-unknown subjects). Detailed demographic data of the subjects and their statistics are provided in a previous report. 25 The study with NOC subjects was approved by the Ethics Committee of Tokyo Metropolitan Institute of Medical Science (approval no. 22-10). They were recruited between December 2004 and April 2005 as the control group. 11 samples were excluded because of a lack of genotyping data, and 489 samples were adopted as a control group (NOC) for the present study.
We also enrolled 361 healthy adult patients who underwent mandibular osteotomy at Tokyo Dental College Suidobashi Hospital as a control group (group label: TD) for the present study. Detailed demographic data of the subjects and their statistics are provided in previous reports.26,27 The study with TD subjects was approved by the Ethics Committees of Tokyo Dental College and Tokyo Metropolitan Institute of Medical Science (approval no. 811 and 21-21, respectively).
All of the Japanese patients and volunteers who enrolled in the study completed the study. The patients’ and volunteers’ demographic data are shown in Supplemental Table S1.
Genotyping and LD analysis
We examined SNPs in the region within a range of 20 kbp upstream of the PRTFDC1 gene and downstream of the GPR158 gene, including the PRTFDC1, ENKUR, THNSL1, and GPR158 genes with flanking regions. Genotype data from the whole-genome genotyping of 151 patients in the PTP or OFP group were used to analyze 146 SNPs within the region. Genomic DNA was extracted from whole blood samples using standard procedures. Extracted DNA was dissolved in TE buffer (10 mM Tris-HCl and 1 mM ethylenediaminetetraacetic acid, pH 8.0). Whole-genome genotyping and the data cleaning process were performed as described previously. 4 A total of 146 SNP markers survived the filtering process for this patient sample for the region that was investigated. The LD analysis was performed on 146 SNPs in the region that included the PRTFDC1, ENKUR, THNSL1, and GPR158 genes with flanking regions in the SNP array. The 26 SNPs with minor allele frequencies of zero were excluded from the LD analysis, and the remaining 120 SNPs were employed for further analysis (Supplemental Dataset S1). To estimate LD intensity between SNPs, the commonly used D′ and r2 values were pairwise calculated using the genotype dataset for each SNP. The LD block was defined among SNPs that showed “strong LD” based on the default algorithm of Gabriel et al. with an upper limit of 0.98 and a lower limit of 0.7 for the 95% confidence interval of D′ that indicated strong LD. 28 Tag SNPs in the LD blocks were determined using the Tagger software package that is incorporated in Haploview 4.2, which was detailed in a previous report. 28
Statistical analysis
For all genotype frequency data, deviations from the theoretical Hardy-Weinberg equilibrium distribution were examined. Fisher’s Exact Test and χ2 tests were performed to analyze associations with clinical data for PTP. The Hardy-Weinberg equilibrium distribution test was performed using Haploview 4.2 and Calculator of Hardy-Weinberg equilibrium. 29 Fisher’s exact test was performed using PLINK v1.07 software. 30 The χ2 tests were performed using SPSS 28 software (IBM Japan, Tokyo, Japan; Supplemental Dataset S2). For all statistical tests, the criterion for significance was p < .05. Bonferroni correction for multiple comparisons was performed for 120 SNPs in the trend, genotypic, dominant, and recessive models for each minor allele in the OFP group. For the allele frequency, the minor or major allele was defined in the OFP group in the present study if not otherwise specified.
Public database search
We extracted information on eQTLs of the SNPs using the GTEx Portal to examine effects of these SNPs on gene expression levels in humans.31,32 We extracted information on PRTFDC1 mRNA expression in human tissues and cells using The Human Protein Atlas. 33 We also extracted DNA sequence features plus additional chromatin accessibility (DNase I hypersensitive site [DHS]) and the DNase-Seq signal of the genes’ genomic regions using the ZENBU genome browser on December 18, 2023, to investigate transcriptional regulation around SNP regions in the genes. 34 The data source of the DHS was the NIH Roadmap Epigenomics Mapping Consortiums in 111 samples, showing promoter, enhancer, and dyadic regions. The data source of the DNase-Seq signal was the NIH Roadmap Epigenomics Mapping Consortiums in 127 samples, showing open chromatin regions only with a p-value signal ≥2. Data on promoters and enhancers were derived from DNase I-accessible regulatory regions that were defined by the NIH Roadmap Epigenomics Mapping Consortiums. Open chromatin regions correspond to gene expression control regions, such as promoters and enhancers.
Results
Seven SNPs in the region spanning PRTFDC1, ENKUR, THNSL1, and GPR158 are significantly associated with PTP in the PTP and OFP groups
PRTFDC1, ENKUR, THNSL1, and GPR158 are reportedly associated with pain. Therefore, these genes and the region that spans these genes may be associated with PTP. We investigated the association between SNPs in the region that included the PRTFDC1, ENKUR, THNSL1, and GPR158 genes with the 20 kbp upstream and downstream flanking regions and PTP. Deviations from the theoretical Hardy-Weinberg equilibrium distribution were first examined. Supplemental Table S2 shows that many of the SNPs in the region have genotype frequencies that are close to zero (Supplemental Table S2); thus, we performed Fisher’s exact test for the 120 SNPs in the region for the PTP and OFP groups. Seven SNPs were significantly associated with PTP even after Bonferroni correction for multiple comparisons (p values after Bonferroni correction in the trend model: rs10828760, p = 2.11 × 10−2; rs12411696, p = 2.11 × 10−2; rs7921132, p = 2.11 × 10−2; rs7920241, p = 1.72 × 10−2; rs12411980, p = 9.42 × 10−4; rs11818773, p = 5.36 × 10−4; rs2765697, p = 1.20 × 10−3; Table 1, Supplemental Table S3). Of these seven significant SNPs, rs10828760, rs12411696, rs7921132, and rs7920241 were located near 10 kbp upstream of GPR158, and rs12411980, rs11818773, and rs2765697 were within the first intron region of GPR158.
Table 1.
SNPs in the region spanning from PRTFDC1 gene to GPR158 gene significantly associated with PTP after Bonferroni correction (PTP vs. OFP groups).
| SNP | Alleles (frequency in OFP group) | Genotypes AA/AB/BB (frequency (%)) | p (Trend model) | ||
|---|---|---|---|---|---|
| PTP | OFP | Original | after Bonferroni correction | ||
| rs10828760 | C < A | 9/19/5 (27/58/15) | 11/51/56 (10/43/47) | 1.76 × 10-4 | 2.11 × 10-2* |
| rs12411696 | A < G | 9/19/5 (27/58/15) | 11/51/56 (10/43/47) | 1.76 × 10-4 | 2.11 × 10-2* |
| rs7921132 | T < C | 9/19/5 (27/58/15) | 11/51/56 (10/43/47) | 1.76 × 10-4 | 2.11 × 10-2* |
| rs7920241# | A < G | 9/19/5 (27/58/15) | 11/50/57 (9/42/48) | 1.43 × 10-4 | 1.72 × 10-2* |
| rs12411980 | G < T | 10/18/5 (30/55/15) | 8/50/60 (7/42/51) | 7.85 × 10-6 | 9.42 × 10-4* |
| rs11818773 | T < C | 10/18/5 (30/55/15) | 7/51/60 (6/43/51) | 4.47 × 10-6 | 5.36 × 10-4* |
| rs2765697# | C < T | 10/18/5 (30/55/15) | 8/51/59 (7/43/50) | 9.98 × 10-6 | 1.20 × 10-3* |
PTP, phantom tooth pain; OFP, orofacial pain.
*p < 0.05.
#eQTL of PRTFDC1 gene.
AA, homozygote for the minor allele in each SNP in OFP group; AB, heterozygote for the major allele in each SNP in OFP group; BB, homozygote for the major allele in each SNP in OFP group.
LD analysis of SNPs in the region spanning PRTFDC1, ENKUR, THNSL1, and GPR158
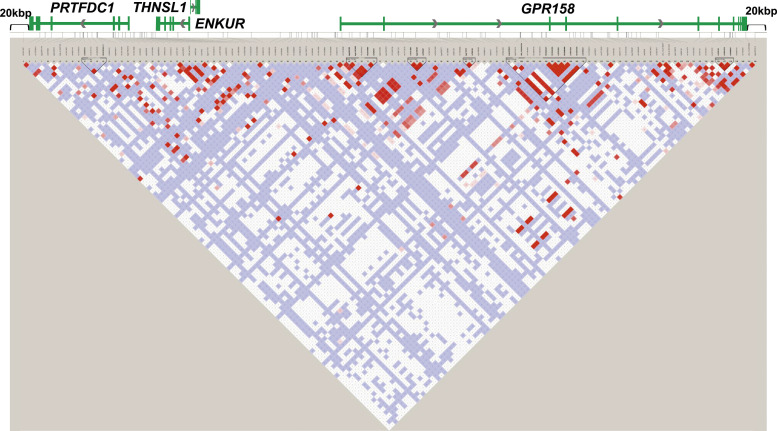
Next, an LD analysis was performed for 120 SNPs in the region (Figure 1, Supplemental Table S4).
Figure 1.
State of LD among SNPs in the region that spans from the PRTFDC1 gene to the GPR158 gene, including 20 kbp upstream and downstream (LD Plot-r2). D’ and r2 values are presented in Supplemental Table S4. White boxes represent D’ < 1, log of likelihood odds ratio (LOD) < 2. Pink and red boxes represent D’ < 1 and LOD ≥2. Blue boxes represent D’ = 1 and LOD <2. Bright red boxes represent D’ = 1 and LOD ≥2. The white horizontal line above the LD plot represents the genomic region that spans the PRTFDC1, ENKUR, THNSL1, and GPR158 genes, including 20 kbp upstream and downstream of the genes. The green boxes in the structure of the PRTFDC1, ENKUR, THNSL1, and GPR158 genes represent exons, and the green solid lines represent untranslated regions or introns. The gray arrows represent the direction of transcription.
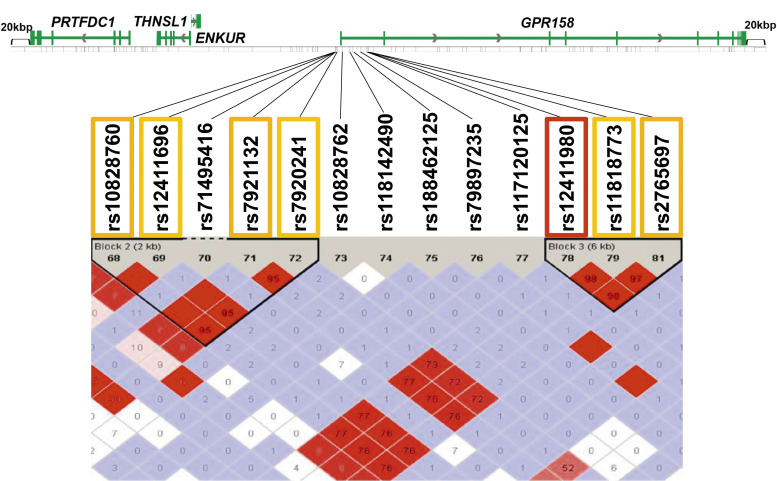
Four SNPs (rs10828760, rs12411696, rs7921132, and rs7920241) in Block 2 were in strong LD with each other with r2 > 0.95, and three SNPs (rs12411980, rs11818773, and rs2765697) in Block 3 were in strong LD with each other with r2 > 0.97 (Figure 2, Supplemental Table S4). The rs10828760, rs12411696, rs7921132, and rs7920241 SNPs in Block 2 were in LD with the rs12411980, rs11818773, and rs2765697 SNPs in Block 3 with r2 > 0.76. The SNPs in Block 3 showed lower p values than those in Block 2. Among the SNPs in Block 3, the rs12411980 tag SNP was selected for further analysis because Tagger software calculated the SNP as a representative of SNPs in this block.
Figure 2.
Enlarged view of Figure 1. In Block 2, four SNPs (rs10828760, rs12411696, rs7921132, and rs7920241) were in LD at r2 > 0.95. In Block 3, three SNPs (rs12411980, rs11818773, and rs2765697) were in LD at r2 > 0.97. rs10828760, rs12411696, rs7921132, and rs7920241 in Block 2 were in LD with rs12411980, rs11818773, and rs2765697 in Block 3 at r2 > 0.76. The orange and red squares represent the SNPs that were significant after Bonferroni correction for multiple comparisons. The red square represents the SNP on which we focused in the present study. LD, linkage disequilibrium; SNP, single-nucleotide polymorphism; LOD, likelihood odds ratio.
rs12411980 SNP is significantly associated with PTP in the PTP group and healthy controls
To confirm whether the rs12411980 SNP is significantly associated with PTP even in the PTP group and healthy control groups (NOC and TD) instead of the OFP group, we performed χ2 tests for the PTP, NOC, and TD groups (Table 2). Significant differences were found in the rs12411980 SNP in the PTP group compared with healthy controls (PTP group vs. NOC group: TT/TG/GG, p = 2.92 × 10−4; TT + TG/GG, p = 2.32 × 10−3; TT/TG + GG, p = 3.98 × 10−4; PTP group vs. TD group: TT/TG/GG, p = 5.46 × 10−4; TT + TG/GG, p = 1.50 × 10−3; TT/TG + GG, p = 1.86 × 10−3).
Table 2.
Association analysis of rs12411980 SNP.
| Genotype groups | p | ||||
|---|---|---|---|---|---|
| PTP vs. OFP groups | PTP vs. NOC groups | PTP vs. TD groups | OFP vs. NOC groups | OFP vs. TD groups | |
| TT/TG/GG | 3.62 × 10−5* | 2.92 × 10−4* | 5.46 × 10−4* | 2.72 × 10−1 | 2.08 × 10−1 |
| TT + TG/GG | 2.28 × 10−4* | 2.32 × 10−3* | 1.50 × 10−3* | 1.12 × 10−1 | 1.77 × 10−1 |
| TT/TG + GG | 2.51 × 10−4* | 3.98 × 10−4* | 1.86 × 10−3* | 4.33 × 10−1 | 1.34 × 10−1 |
PTP, phantom tooth pain; OFP, orofacial pain; NOC and TD, healthy subjects.
*p < 0.05.
To examine whether there is a significant difference in the rs12411980 SNP between the OFP group and healthy control groups (NOC and TD), OFP group vs. NOC or TD group comparisons were statistically analyzed using χ2 tests (Table 2). No significant difference in the rs12411980 SNP was found between the OFP group and NOC or TD group (OFP group vs. NOC group: TT/TG/GG, p = 2.72 × 10−1; TT + TG/GG, p = 1.12 × 10−1; TT/TG + GG, p = 4.33 × 10−1; OFP group vs. TD group: TT/TG/GG, p = 2.08 × 10−1; TT + TG/GG, p = 1.77 × 10−1; TT/TG + GG, p = 1.34 × 10−1). These results suggest that the OFP group is similar to healthy subjects for the rs12411980 SNP.
To identify which genotype of the rs12411980 SNP is more susceptible to PTP, genotype distributions of the rs12411980 SNP were examined in each group (Table 3). There were significantly more individuals with the GG genotype in the PTP group than in the OFP group (percentage of GG: 30% in PTP group, 7% in OFP group). There were also significantly more individuals with the GG genotype in the PTP group than in the healthy control groups (percentage of GG: 30% in PTP group, 12% in NOC group, 11% in TD group). Thus, a larger rate of the GG genotype existed in the PTP group than in the OFP and healthy control groups. These results suggest that individuals with the GG genotype of the rs12411980 SNP are more susceptible to PTP.
Table 3.
Genotype distributions of rs12411980 SNP.
| Patiants and control subjects | Genotype | |||
|---|---|---|---|---|
| TT | TG | GG | ||
| PTP group | n a | 5 | 18 | 10 |
| Rate (%) | 15 | 55 | 30 | |
| OFP group | n | 60 | 50 | 8 |
| Rate (%) | 51 | 42 | 7 | |
| NOC group | n | 230 | 203 | 59 |
| Rate (%) | 46.7 | 41.3 | 12 | |
| TD group | n | 155 | 166 | 40 |
| Rate (%) | 43 | 46 | 11 | |
PTP, phantom tooth pain; OFP, orofacial pain; NOC and TD, healthy subject.
aThe number of participants.
LD SNPs of rs12411980 SNP are eQTLs of PRTFDC1
To investigate the association of the seven significant SNPs with gene expression, we performed a database search using the GTEx Portal. The rs7920241 SNP in Block 2 upstream of GPR158 and the rs2765697 SNP in Block 3 within the GPR158 first intron region are eQTLs that are associated with the human gene expression of PRTFDC1 (rs7920241 SNP, p = 1.13 × 10−4; rs2765697 SNP, p = 1.61 × 10−4).31,32 On the SNPs, PRTFDC1 expression is upregulated in minor allele homozygotes (Supplemental Figure S1 and S2). The rs7920241 and rs2765697 SNPs are in LD with the rs12411980 SNP (between rs7920241 and rs12411980 SNPs, r2 = 0.77; between rs2765697 and rs12411980 SNPs, r2 = 0.98, Supplemental Table S4), suggesting that the rs12411980 SNP is also associated with PRTFDC1 expression. The minor allele homozygote (GG) of the rs12411980 SNP is also expected to show higher PRTFDC1 expression.
Discussion
The present results suggest that the GG genotype (the minor allele homozygote in the OFP, NOC, and TD group) of the rs12411980 SNP is significantly associated with PTP. The database and LD analyses showed that the rs12411980 SNP would also be associated with PRTFDC1 expression. PRTFDC1 expression is upregulated in minor allele homozygotes of the LD SNPs, and minor allele homozygotes of the rs12411980 SNP are expected to show higher PRTFDC1 expression. The high PRTFDC1 expression in minor allele homozygotes of the rs12411980 SNP would thus lead to PTP.
PRTFDC1 protein, similar to human hypoxanthine-guanine phosphoribosyltransferase (HPRT), converts hypoxanthine to inosine monophosphate, although its converting activity is over 300 times lower than HPRT. 35 Hypoxanthine was shown to accumulate in HPRT-deficient mice. 36 Although the converting activity of PRTFDC1 protein is much lower than HPRT, PRTFDC1 would have a competitive inhibitory function against hypoxanthine conversion through the efficient binding affinity of hypoxanthine as a substrate for PRTFDC1. Thus, high PRTFDC1 protein expression may competitively inhibit hypoxanthine conversion and cause hypoxanthine accumulation. Hypoxanthine inhibits the benzodiazepine (BDZ)-binding site of the GABAA receptor. 37 In Lesch-Nyhan syndrome, which is characterized by a deficiency of HPRT activity, above-normal concentrations of purines, including hypoxanthine, in the brain may be sufficient to significantly affect the ability of BDZ receptors to modulate GABA-mediated brain mechanisms. 38 Thus, GABAA receptor signaling could be modulated by hypoxanthine through the BDZ binding site. Hypoxanthine also displaces nuclear flunitrazepam (which reversibly binds to the BDZ receptor with high affinity) binding with greater potency than membrane flunitrazepam binding in brain tissues, and a major portion of nuclear flunitrazepam binding sites are located on chromatin. 39 Together with the fact that BDZs increase the transcription of human immunodeficiency virus 1 and alter chromatin at the long terminal repeat (LTR) 40 and that LTR retrotransposons exist in the human genome, 41 hypoxanthine binding on nuclear binding sites on chromatin could affect the transcription of particular genes. Altogether, high PRTFDC1 expression in the GG genotype (the minor allele homozygote in the OFP, NOC, and TD groups) of the rs12411980 SNP may cause PTP through higher hypoxanthine levels through an indeterminate pathway, including the BDZ-binding site. However, further pharmacological and pharmacotherapeutic studies are needed to investigate this possibility.
Data extraction using the ZENBU genome browser revealed that the rs12411980 and rs11818773 SNPs were located in enhancer regions in brain tissue and neurons (hippocampus middle inferior temporal lobe, substantia nigra, fetal brain, and neurosphere cultured cells derived from cortex or ganglionic eminence; Supplemental Table S5). 34 The rs2765697 SNP was located around the enhancer region on the chromosome of cells in the anterior caudate. The rs10828760, rs12411696, rs7921132, and rs7920241 SNPs were located around the enhancer region on the chromosome of cells in the male fetal brain (Supplemental Table S5). 34 The DNase-Seq results showed that the signal was detected in regions that included the rs10828760, rs12411980, and rs11818773 SNPs on the chromosome of cells in human brain tissues and neurons (angular gyrus, anterior caudate, cingulate gyrus, dorsolateral prefrontal cortex, germinal matrix, middle hippocampus, inferior temporal lobe, substantia nigra, fetal brain, and neurosphere-cultured cells derived from the cortex or ganglionic eminence; Supplemental Table S6). 34 The DNase-Seq signals indicated that the regions are open chromatin and considered to contribute to transcription. The regions on the chromosome that included the SNPs on which we focused with the signal in brain tissues and neurons may contribute to transcription. No DNase-Seq signal was detected in the regions on the chromosome in astrocytes (Supplemental Table S6), suggesting that these regions do not contribute to transcription in astrocytes. Thus, the regions on the chromosome that included the SNPs on which we focused in the present study would contribute to transcriptional activity as an enhancer in human brain tissues and neurons and not in astrocytes.
Higher P2RY12 expression in microglia may lead to PTP with the aid of SLC17A9. 4 Furthermore, neurons and microglia are mainly responsible for the production and release of hypoxanthine and other purines after stresses in the brain. 42 Consistent with the fact that the region that included the SNPs on which we focused does not contribute to transcription in astrocytes (Supplemental Table S6) 34 but contributes to PRTFDC1 expression and that PRTFDC1 is expressed in excitatory neurons, inhibitory neurons, and microglial cells, 33 neurons and microglia other than astrocytes may play important roles in PTP with regard to PRTFDC1 transcription.
The rs7920241 and rs2765697 SNPs are eQTLs for PRTFDC1 and not for GPR158 or other genes.31,32 The rs7920241 and rs2765697 SNPs are located in upstream and intron regions of the GPR158 gene, respectively (Figure 2), implying that these SNPs may also affect the expression of GPR158 or other genes. However, because eQTLs of the GPR158 gene reside in different regions, 43 the regions that included the rs7920241 and rs2765697 SNPs mainly contribute to the transcription of PRTFDC1 rather than GPR158, although the genomic position of PRTFDC1 is approximately 250 kbp from the rs2765697 SNP. A topological operon was recently proposed, and transcription factor clusters were shown to be shared during the co-activation of distant genes. 44 Likely attributable to the chromatin level control of gene expression, a change in the gene expression of one gene likely affects the expression evolution of neighboring genes. 45 Thus, the rs7920241 and rs2765697 SNPs appear to contribute primarily to PRTFDC1 expression, which may also affect the overall expression of pain-related genes in the region and may influence the susceptibility to PTP. However, because the eQTL is based on traditional bulk studies that typically assess average expression levels across millions of cells from whole tissues, possible regulatory relationships that are specific to certain cell types, if any, could not be detected.
In the present study, minor allele homozygotes (GG) of the rs12411980 SNP had a greater susceptibility to PTP. The subjects in the present study were Japanese, and the subjects’ allele frequencies of the rs12411980 SNP were 62% for the T allele and 38% for the G allele (PTP + OFP + NOC + TD groups), and healthy subjects’ allele frequencies of the rs12411980 SNP were 67% for the T allele and 33% for the G allele (NOC + TD groups). In East Asians, the rs12411980 SNP showed a 70% T-allele frequency and 30% G-allele frequency according to the 1000 Genomes Project in the dbSNP database, 46 suggesting that allele frequencies of the subjects in the present study were similar to East Asians generally. Allele frequencies in the OFP group were similar to East Asians (OFP group: 72% for T allele, 28% for G allele). PTP showed a 42% T-allele frequency and 58% G-allele frequency, suggesting a higher G-allele frequency compared with East Asians, the OFP group, and subjects in other geographical regions (Americans: 84% T allele, 16% G allele; Africans: 86% T allele, 14% G allele; Europeans: 89% T allele, 11% G allele; South Asians: 78% T allele, 22% G allele). In the present study, the GG genotype of the rs12411980 SNP had a higher incidence of PTP, suggesting that the G allele of the rs12411980 SNP is associated with the susceptibility to PTP. Thus, East Asians, including Japanese, may have a higher risk of PTP compared with other regions because of their higher G-allele frequency.
The present results showed that the GG genotype (the minor allele homozygote in the OFP, NOC, and TD groups) of the rs12411980 SNP, which is associated with PRTFDC1 expression, was significantly associated with PTP, suggesting that PRTFDC1 expression would contribute to PTP. These results elucidate mechanisms of PTP and may contribute to individualized medicine to predict the vulnerability to PTP.
Supplemental Material
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Acknowledgements
We thank Mr. Michael Arends for his assistance with editing the manuscript. We are grateful to the volunteers for their participation in the study and anesthesiologists and surgeons for collecting the clinical data. This work was supported by grants from the Japan Society for the Promotion of Science (JSPS) KAKENHI (no. 23K21457 [KI], 23K06839 [SO], 22K10202 [KF], 21H03028 [KI], JP22H04922 [AdAMS] [KI], 20K07774 [SO], 20K09259 [DN], 17K08970 [DN], and 17H04324 [KI]) and Japan Agency for Medical Research and Development (AMED; no. JP19ek0610011 [KI]).
Author contributions: JA, SO, DN, KF, and KI conceived the study and designed the experiments. JA, SO, KN, YE, and DN performed the statistical analyses. JA wrote the manuscript. MS, KF, and YO collected clinical samples and data. JA and JH performed the genotyping procedures. SO, MS, DN, KF, and KI supervised the experiments and finalized the manuscript. All authors contributed to writing the manuscript, and all authors read and approved the final manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (no. 23K21457 [KI], 23K06839 [SO], 22K10202 [KF], 21H03028 [KI], JP22H04922 [AdAMS] [KI], 20K07774 [SO], 20K09259 [DN], 17K08970 [DN], and 17H04324 [KI]) and Japan Agency for Medical Research and Development (AMED; no. JP19ek0610011 [KI]).
Supplemental Material: Supplemental material for this article is available online.
ORCID iDs
Seii Ohka https://orcid.org/0000-0003-2394-6440
Kazutaka Ikeda https://orcid.org/0000-0001-8342-0278
References
- 1.Marbach JJ, Raphael KG. Phantom tooth pain: a new look at an old dilemma. Pain Med 2000; 1: 68–77. DOI: 10.1046/j.1526-4637.2000.00012.x. [DOI] [PubMed] [Google Scholar]
- 2.Marbach JJ. Is phantom tooth pain a deafferentation (neuropathic) syndrome? Part I: evidence derived from pathophysiology and treatment. Oral Surg Oral Med Oral Pathol 1993; 75: 95–105. DOI: 10.1016/0030-4220(93)90413-x. [DOI] [PubMed] [Google Scholar]
- 3.Marbach JJ, Hulbrock J, Hohn C, Segal AG. Incidence of phantom tooth pain: an atypical facial neuralgia. Oral Surg Oral Med Oral Pathol 1982; 53: 190–193. DOI: 10.1016/0030-4220(82)90285-7. [DOI] [PubMed] [Google Scholar]
- 4.Soeda M, Ohka S, Nishizawa D, Hasegawa J, Nakayama K, Ebata Y, Fukuda KI, Ikeda K. Single-nucleotide polymorphisms of the SLC17A9 and P2RY12 genes are significantly associated with phantom tooth pain. Mol Pain 2022; 18: 17448069221089592. DOI: 10.1177/17448069221089592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morii M, Ohka S, Nishizawa D, Hasegawa J, Nakayama K, Ebata Y, Soeda M, Fukuda KI, Yoshida K, Koshika K, Ichinohe T, Ikeda K. The rs216009 single-nucleotide polymorphism of the CACNA1C gene is associated with phantom tooth pain. Mol Pain 2023; 19: 17448069231193383. DOI: 10.1177/17448069231193383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li N, Zhang Y, Sidlauskas K, Ellis M, Evans I, Frankel P, Lau J, El-Hassan T, Guglielmi L, Broni J, Richard-Loendt A, Brandner S. Inhibition of GPR158 by microRNA-449a suppresses neural lineage of glioma stem/progenitor cells and correlates with higher glioma grades. Oncogene 2018; 37: 4313–4333. DOI: 10.1038/s41388-018-0277-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orlandi C, Sutton LP, Muntean BS, Song C, Martemyanov KA. Homeostatic cAMP regulation by the RGS7 complex controls depression-related behaviors. Neuropsychopharmacology 2019; 44: 642–653. DOI: 10.1038/s41386-018-0238-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutton LP, Orlandi C, Song C, Oh WC, Muntean BS, Xie K, Filippini A, Xie X, Satterfield R, Yaeger JDW, Renner KJ, Young SM, Jr., Xu B, Kwon H, Martemyanov KA. Orphan receptor GPR158 controls stress-induced depression. Elife 2018; 7: e33273. DOI: 10.7554/eLife.33273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song C, Orlandi C, Sutton LP, Martemyanov KA. The signaling proteins GPR158 and RGS7 modulate excitability of L2/3 pyramidal neurons and control A-type potassium channel in the prelimbic cortex. J Biol Chem 2019; 294: 13145–13157. DOI: 10.1074/jbc.RA119.007533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ando E, Higashi S, Mizokami A, Watanabe S, Hirata M, Takeuchi H. Osteocalcin promotes proliferation, differentiation, and survival of PC12 cells. Biochem Biophys Res Commun 2021; 557: 174–179. DOI: 10.1016/j.bbrc.2021.03.146. [DOI] [PubMed] [Google Scholar]
- 11.Bjarnadottir TK, Fredriksson R, Schioth HB. The gene repertoire and the common evolutionary history of glutamate, pheromone (V2R), taste(1) and other related G protein-coupled receptors. Gene 2005; 362: 70–84. DOI: 10.1016/j.gene.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 12.Laboute T, Zucca S, Holcomb M, Patil DN, Garza C, Wheatley BA, Roy RN, Forli S, Martemyanov KA. Orphan receptor GPR158 serves as a metabotropic glycine receptor: mGlyR. Science 2023; 379: 1352–1358. DOI: 10.1126/science.add7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nievergelt CM, Maihofer AX, Mustapic M, Yurgil KA, Schork NJ, Miller MW, Logue MW, Geyer MA, Risbrough VB, O'Connor DT, Baker DG. Genomic predictors of combat stress vulnerability and resilience in U.S. Marines: a genome-wide association study across multiple ancestries implicates PRTFDC1 as a potential PTSD gene. Psychoneuroendocrinology 2015; 51: 459–471. DOI: 10.1016/j.psyneuen.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 14.Asmundson GJ, Coons MJ, Taylor S, Katz J. PTSD and the experience of pain: research and clinical implications of shared vulnerability and mutual maintenance models. Can J Psychiatry 2002; 47: 930–937. DOI: 10.1177/070674370204701004. [DOI] [PubMed] [Google Scholar]
- 15.Fishbain DA, Pulikal A, Lewis JE, Gao J. Chronic pain types differ in their reported prevalence of post-traumatic stress disorder (PTSD) and there is consistent evidence that chronic pain is associated with PTSD: an evidence-based structured systematic review. Pain Med 2017; 18: 711–735. DOI: 10.1093/pm/pnw065. [DOI] [PubMed] [Google Scholar]
- 16.Siqveland J, Hussain A, Lindstrom JC, Ruud T, Hauff E. Prevalence of posttraumatic stress disorder in persons with chronic pain: a meta-analysis. Front Psychiatry 2017; 8: 164. DOI: 10.3389/fpsyt.2017.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nanavaty N, Thompson CG, Meagher MW, McCord C, Mathur VA. Traumatic life experience and pain sensitization: meta-analysis of laboratory findings. Clin J Pain 2023; 39: 15–28. DOI: 10.1097/AJP.0000000000001082. [DOI] [PubMed] [Google Scholar]
- 18.Uchiumi C, Kato H, Ishida M, Nakataki M, Ohmori T. Peritraumatic reactions, PTSD symptoms, and pain : a study of train disasters in Japan. J Med Invest 2021; 68: 85–89. DOI: 10.2152/jmi.68.85. [DOI] [PubMed] [Google Scholar]
- 19.Sutton KA, Jungnickel MK, Wang Y, Cullen K, Lambert S, Florman HM. Enkurin is a novel calmodulin and TRPC channel binding protein in sperm. Dev Biol 2004; 274: 426–435. DOI: 10.1016/j.ydbio.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 20.Sun ZC, Ma SB, Chu WG, Jia D, Luo C. Canonical transient receptor potential (TRPC) channels in nociception and pathological pain. Neural Plast 2020; 2020: 3764193. DOI: 10.1155/2020/3764193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rifas L, Weitzmann MN. A novel T cell cytokine, secreted osteoclastogenic factor of activated T cells, induces osteoclast formation in a RANKL-independent manner. Arthritis Rheum 2009; 60: 3324–3335. DOI: 10.1002/art.24877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Napimoga MH, Dantas Formiga WD, Abdalla HB, Trindade-da-Silva CA, Venturin CM, Martinez EF, Rossaneis AC, Verri WA, Jr., Clemente-Napimoga JT. Secreted osteoclastogenic factor of activated T cells (SOFAT) is associated with rheumatoid arthritis and joint pain: initial evidences of a new pathway. Front Immunol 2020; 11: 1442. DOI: 10.3389/fimmu.2020.01442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International Classification of Orofacial Pain. Cephalalgia 2020; 40(2): 129–221. DOI: 10.1177/0333102419893823. [DOI] [PubMed] [Google Scholar]
- 24.World Health Organization portal . ICD-11 2022 release. Geneva: WHO, https://icd.who.int/browse11/l-m/en. [Google Scholar]
- 25.Nishizawa D, Fukuda K, Kasai S, Ogai Y, Hasegawa J, Sato N, Yamada H, Tanioka F, Sugimura H, Hayashida M, Ikeda K. Association between KCNJ6 (GIRK2) gene polymorphism rs2835859 and post-operative analgesia, pain sensitivity, and nicotine dependence. J Pharmacol Sci 2014; 126: 253–263. DOI: 10.1254/jphs.14189fp. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi D, Nishizawa D, Takasaki Y, Kasai S, Kakizawa T, Ikeda K, Fukuda K. Genome-wide association study of sensory disturbances in the inferior alveolar nerve after bilateral sagittal split ramus osteotomy. Mol Pain 2013; 9: 34. DOI: 10.1186/1744-8069-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aoki Y, Yoshida K, Nishizawa D, Kasai S, Ichinohe T, Ikeda K, Fukuda K. Factors that affect intravenous patient-controlled analgesia for postoperative pain following orthognathic surgery for mandibular prognathism. PLoS One 2014; 9: e98548. DOI: 10.1371/journal.pone.0098548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet 2005; 37: 1217–1223. DOI: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 29.WpCalc . Equilibrium hardy-weinberg, https://wpcalc.com/en/equilibrium-hardy-weinberg/ (accessed 6 December 2023).
- 30.PLINK v1.07, https://www.bwh.harvard.edu/plink/.
- 31.GTEx portal . rs7920241 of PRTFDC1 gene, Multi-tissue eQTL plot, variant page, https://gtexportal.org/home/snp/rs7920241 (accessed 14 October 2023).
- 32.GTEx portal . rs2765697 of PRTFDC1 gene, Multi-tissue eQTL plot, variant page, https://gtexportal.org/home/snp/rs2765697.
- 33.The human protein Atlas, PRTFDC1 mRNA, https://www.proteinatlas.org/ENSG00000099256-PRTFDC1 (accessed October 3, 2023).
- 34.ZENBU 3.0, GPR158 gene, hg19::chr10:25421602..25581679+, https://fantom.gsc.riken.jp/zenbu/gLyphs/index.html#config=rrGagFwdGxmsqx5Mg_OFEC;loc=hg19::chr10:25421602..25581679+ (accessed December 18, 2023).
- 35.Welin M, Egeblad L, Johansson A, Stenmark P, Wang L, Flodin S, Nyman T, Tresaugues L, Kotenyova T, Johansson I, Eriksson S, Eklund H, Nordlund P. Structural and functional studies of the human phosphoribosyltransferase domain containing protein 1. FEBS J 2010; 277: 4920–4930. DOI: 10.1111/j.1742-4658.2010.07897.x. [DOI] [PubMed] [Google Scholar]
- 36.Keebaugh AC, Mitchell HA, Gaval-Cruz M, Freeman KG, Edwards GL, Weinshenker D, Thomas JW. PRTFDC1 is a genetic modifier of HPRT-deficiency in the mouse. PLoS One 2011; 6: e22381. DOI: 10.1371/journal.pone.0022381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daval JL, Barberis C, Vert P. In vitro and in vivo displacement of [3H]-diazepam binding by purine derivatives in developing rat brain. Dev Pharmacol Ther 1984; 7: 169–176. DOI: 10.1159/000457160. [DOI] [PubMed] [Google Scholar]
- 38.Kish SJ, Fox IH, Kapur BM, Lloyd K, Hornykiewicz O. Brain benzodiazepine receptor binding and purine concentration in Lesch-Nyhan syndrome. Brain Res 1985; 336: 117–123. DOI: 10.1016/0006-8993(85)90422-6. [DOI] [PubMed] [Google Scholar]
- 39.Dalezios Y, Matsokis N. Nuclear benzodiazepine binding: possible interaction with thyroid hormone receptors. Neurochem Res 1993; 18: 305–311. DOI: 10.1007/BF00969087. [DOI] [PubMed] [Google Scholar]
- 40.Elbezanti W, Lin A, Schirling A, Jackson A, Marshall M, Duyne RV, Maldarelli F, Sardo L, Klase Z. Benzodiazepines drive alteration of chromatin at the integrated HIV-1 LTR. Viruses 2020; 12: 191. DOI: 10.3390/v12020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet 2009; 10: 691–703. DOI: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson EK, Kotermanski SE, Menshikova EV, Dubey RK, Jackson TC, Kochanek PM. Adenosine production by brain cells. J Neurochem 2017; 141: 676–693. DOI: 10.1111/jnc.14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.GTEx portal, GTEx Locus Browser (Gene-centric) . GPR158 gene, https://www.gtexportal.org/home/locusBrowserPage/ENSG00000151025.9.
- 44.Kawasaki K, Fukaya T. Functional coordination between transcription factor clustering and gene activity. Mol Cell 2023; 83: 1605–1622. DOI: 10.1016/j.molcel.2023.04.018. [DOI] [PubMed] [Google Scholar]
- 45.Ghanbarian AT, Hurst LD. Neighboring genes show correlated evolution in gene expression. Mol Biol Evol 2015; 32: 1748–1766. DOI: 10.1093/molbev/msv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.NCBI dbSNP . rs12411980 of GPR158 gene, frequency, https://www.ncbi.nlm.nih.gov/snp/rs12411980#frequency_tab.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental Material for rs12411980 single-nucleotide polymorphism related to PRTFDC1 expression is significantly associated with phantom tooth pain by Jun Araida, Seii Ohka, Moe Soeda, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Yasukazu Ogai, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain