Abstract
Three-dimensionality (3D) was proven essential for developing reliable models for different anatomical compartments and many diseases. However, the neuronal compartment still poses a great challenge as we still do not understand precisely how the brain computes information and how the complex chain of neuronal events can generate conscious behavior. Therefore, a comprehensive model of neuronal tissue has not yet been found. The present work was conceived in this framework: we aimed to contribute to what must be a collective effort by filling in some information on possible 3D strategies to pursue. We compared directly different kinds of scaffolds (i.e., PDMS sponges, thermally crosslinked hydrogels, and glass microbeads) in their effect on neuronal network activity recorded using micro-electrode arrays. While the overall rate of spiking activity remained consistent, the type of scaffold had a notable impact on bursting dynamics. The frequency, density of bursts, and occurrence of random spikes were all affected. The examination of inter-burst intervals revealed distinct burst generation patterns unique to different scaffold types. Network burst propagation unveiled divergent trends among configurations. Notably, it showed the most differences, underlying that functional variations may arise from a different 3D spatial organization. This evidence suggests that not all 3D neuronal constructs can sustain the same level of richness of activity. Furthermore, we commented on the reproducibility, efficacy, and scalability of the methods, where the beads still offer superior performances. By comparing different 3D scaffolds, our results move toward understanding the best strategies to develop functional 3D neuronal units for reliable pre-clinical studies.
I. INTRODUCTION
Nowadays, it is widely acknowledged that in vitro brain models should embed morphological constraints, first and among everything, the one that enables the recreation of the intrinsic three-dimensional (3D) structure of the organ. In vivo, the 86 × 109 synaptically connected neurons are arranged in complex topologies that exist in the 3D space: the well-known modular,1 small-world,2 scale-free,3 and rich-club4 topologies lie in 3D. Morphologically, the spatial arrangement of neuronal assemblies in 2D and 3D has a significant impact on the shape of neurons, either flattened or round,5 on the growth of dendrites and axons, either forced in two directions or unbounded, and on the achieved cell densities, which are far from the in vivo values in the 2D case.6 The first evident consequence of a 3D vs a 2D layout is the richer connectivity that deeply modulates the electrophysiological7 as well as the biochemical properties of the neurons themselves.8,9
In vivo, 3D cell growth is possible thanks to the extracellular matrix (ECM), which is made up of proteins and polysaccharides acting as connective material to hold cells in a defined 3D space (Ref. 10 and references therein). ECM influences cellular and network development, as well as the protein expression, regulates cell sensitivity to external compounds (drugs and toxins), changes cell-to-cell interactions, drives cell differentiation, alters invasion patterns, and “participates in” the pathogenesis of various diseases.11–17 Considering the central role of the ECM, many studies have focused on developing both scaffold-free and scaffold-based techniques to mimic the 3D cell microenvironment. However, reproducing the mechanical and chemical properties of 3D neuronal networks, which are highly dependent on the intricate balance of biochemical components of the ECM, poses a significant challenge. Indeed, the pursuit of more and more realistic 3D brain-on-a-chip models boosted the synthesis of new materials (biomimetic polymeric compounds, hydrogels, etc.) and the development of new fabrication techniques (3D printing, bioprinting, etc.).
Synthetic and natural hydrogels have been used to develop 3D models by exploiting their hydrophilicity, biocompatibility, and tunable micro-porosity.18–20 Soft materials like collagen21–23 or mixtures of extracellular matrix protein like Matrigel24–26 have been successfully used as 3D scaffolds. The advancements in natural biomaterials were accompanied by the development of new fabricating techniques, such as manipulation techniques that allow for the creation of oriented and structured topologies26–28 or 3D bioprinting, which significantly improved the reproducibility of the scaffold. However, current techniques do not allow us to achieve the resolution necessary to design the fine structures of the neural tissues yet. Additionally, these materials may exhibit batch variability and incur degradation over time.15,29 Synthetic polymers hold the advantage in these aspects.11,30,31 In 2021, Koroleva and coworkers developed a 3D scaffold with a precise laser technique to support the long-term culturing of functional neuronal networks.11 These polymers, however, lack the ability to create the right biochemical environment for cells and neurites to grow, and they therefore need chemical modifications to allow for cell culturing. Other research studies have focused on the reproducibility of the porosity of the extracellular matrix by developing sponge-like structures using different materials such as poly- dimethyl-siloxane (PDMS),32 polyurethane-PLGA,33 and silk-collagen.34,35
Extracting reliable data for generating new information on 3D cultures is a challenging task due to the complexity of the system itself. The analysis of these types of constructs is mainly based on protein expression evaluation, RNA sequencing, and optical techniques to investigate both the morphological and the functional aspects of the networks.11,34,36 However, in the field of in vitro neuroscience studies on 2D networks, micro-electrode array (MEA) technology has proven to be invaluable for the recording of the electrical activity of the network. MEAs provide valuable insights into network dynamics and communication, development, disease progression, and drugs effect over time.37 Thus, interfacing the new 3D fabricated scaffolds with this technology would provide a precise characterization of spatiotemporal patterns of neuronal activity. Few works, however, focus on this aspect, as coupling 3D constructs to available 2D MEAs is not straightforward. The pioneering work that achieved this feat is dated 2014,38 where the authors exploited the self-assembly properties of glass microbeads to design a highly controllable and reproducible scaffold (in terms of obtained geometries and porosity) where hippocampal neurons grew in layers. The authors found substantial differences in the emerging spontaneous patterns of electrophysiological activity of 3D cultures, especially in the modulation of the bursting/network bursting activity. However, such experimental configuration suffered from the mechanical limit of the scaffold itself, which is intrinsically rigid. From that work, many attempts have been made to investigate the role of the scaffold in the emerging electrophysiological properties. In 2018, Bourke and coworkers created a 3D hippocampal neuronal network coupled to MEAs using a collagen gel as 3D support that permitted an unrestricted cell positioning and neurite outgrowth, defining a sort of brain tissue.39 This collagen structure affected the level of synchrony of the spiking activity and forced more extended bursting patterns than those found using microbeads-created 3D constructs. On the road of using soft materials as biocompatible scaffolds, in 2018, Tedesco et al. explored the use of chitosan, a natural polysaccharide promoting neuronal attachment and dendrites outgrowth, as a soft biological scaffold to develop cortical 3D assemblies coupled to MEAs.40 Once again, the achieved 3D assemblies showed significant differences in bursting dynamics compared to 2D sister cultures. In 2021, Yoon and coworkers presented a micromolding technique to build a micropattern of hydrogels to recreate engineered 3D networks over the active area of MEAs. Their focus was on controlling the design of neuronal connectivity, which was characterized both functionally and morphologically.41 In 2023, Yao et al. successfully applied a multi-bioink approach to create gray- and white-matter tracts reminiscent of cortical structures and carried out the basic physiological analysis to confirm spontaneous activity.42 In any case, a systematical characterization of network dynamics from extracellularly recorded data is still lacking.
In the present work, we explored in detail the effect of different scaffold implementations, with different mechanical and chemical properties, on the electrophysiological activity of 3D networks. We exploited soft and rigid materials with different porosity and stiffness properties to realize biocompatible scaffolds for dissociated neuronal networks coupled to MEAs. We spanned from the classic regular glass microbeads to a more amorphous PDMS sponge for synthetic materials, and we tested two different ECM-derived thermogels. The different techniques for obtaining 3D neuronal network scaffolds were evaluated in terms of performance, usability, efficacy, and scalability. While the spiking activity rate remained consistent, scaffold variations led to differences in burst frequency, density, and random spiking. Statistical analysis highlighted significant differences in firing and bursting parameters among configurations, with sponge configurations showing higher burst frequency but lower density, and beads displaying less frequent bursts with more spikes. Analysis of inter-burst intervals revealed distinct burst generation modalities across configurations. Additionally, investigation into network burst propagation showed unique trends among sponge, microbeads, and geltrex configurations. One-vs-one (OvO) approach for multi-class classification and support vector machine (SVM) algorithm allowed to achieve high accuracy in the classification, particularly for microbeads, reflecting differences at the functional network level that can be brought back to spatial organization variations in the 3D constructs.
II. RESULTS
In this section, the results of the fabrication and the electrophysiological recordings of 3D networks created using different 3D scaffolds are reported. For clarity of exposition and to simplify the reading, we may refer to the neuronal networks created using the PDMS sponge scaffolds as the sponge or sponge configuration, to the neuronal networks created using the ECM gel as ecm or ecm configuration, neuronal networks created using the Geltrex as geltrex/geltrex configuration, and to the neuronal networks created using the glass microbeads as beads or beads configuration.
A. Fabrication of the scaffolds and creation of 3D cultures
One of the goals of the present work is to compare different techniques to obtain 3D scaffolds for creating 3D neuronal networks, not only in terms of performance but also in terms of usability, efficacy, and scalability in view of standard use in routine and innovative pre-clinical studies. With this objective in mind, we evaluated the following parameters: duration of procedure, efficacy of sterilization, yield in terms of the number of specimens per experiment, availability, and user-friendliness for the experimenter (Table I).
TABLE I.
Summary of the usability, user-friendliness, and yield of the four presented techniques. The sterilization efficacy (%) is taken from Refs. 43–46.
| Availability | Sterilization | Protocol | Specimen loss | Cell loss | |
|---|---|---|---|---|---|
| sponge | Needs to be custom made | Autoclave (>99%) | 6 h | 30% | 37% |
| ecm | Commercially available | Already sterile | 5 h (required working on ice) | 30% | 0% |
| geltrex | Commercially available | Already sterile | 5 h (required working on ice) | 30% | 0% |
| beads | Commercially available | Ethanol (>70%) | 6 h + 12 h | <10% | 25% |
Glass microbeads come with the drawback of requiring an extensive cell seeding procedure (taking 6 h plus an additional 12 h for settling down and medium addition). In contrast, thermogels offer a simpler cell seeding process (5 h); however, they pose challenges in terms of yield, with a higher likelihood of specimen loss over time due to the gels detaching more easily from the surface compared to the set bead constructs (about 15%). Contrary to the previous, PDMS sponges have to be costume-made, but multiple scaffolds can be obtained from a single preparation. The cell seeding process for PDMS sponges follows a straightforward pipeline (6 h), with the application on the cell monolayer being the only step that requires extra attention to prevent sample loss. Additionally, PDMS sponges offer a more efficient sterilization method compared to glass microbeads.
B. Coupling with electrodes
A fundamental part of creating a workable model is its compatibility with available technologies for extracting data and therefore information. Although extensive research is currently being carried out to develop new recording chips, most of the available technologies for extracellular electrophysiological recordings are performed with planar electrodes. When considering 3D models, this means that the observable portion of the system is very limited. Moreover, compared to a 2D cell culture, there is a risk that 3D scaffolds hinder neuron-to-electrode coupling. Summing these factors, it becomes essential to consider the general “ability” of scaffolds to ensure a good interface between electronic and biological components. It is worth mentioning that although this aspect is crucial when dealing with planar microtransducers, also 3D ones need to maximize the coupling, especially when using ultra-thin tips.47,48
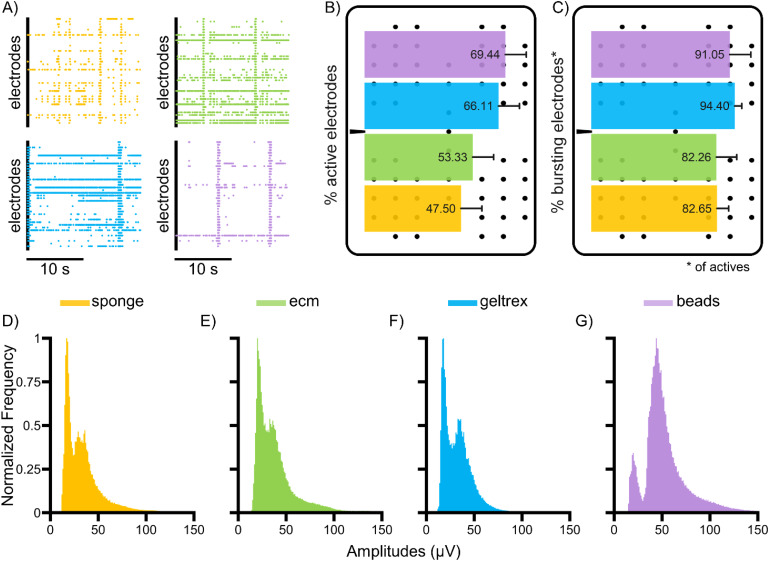
In general, all the considered scaffolds allow at least the recording of the typical features of cultures coupled to MEAs, as qualitatively demonstrated by the raster plot traces, where both spiking and bursting activity are clearly visible [Fig. 1(a)]. Three factors were taken into account to evaluate the effective coupling to electrodes: the percentage of (i) active and (ii) bursting electrodes (computed over the active ones), and (iii) the amplitudes of the recorded signals, which seem to indicate that beads detain the best performances in this perspective. In depth, these constructs generally guarantee a higher percentage of active electrodes (69.44 ± 16.49%), closely followed by geltrex (66.11 ± 9.98%), whereas, in the other two conditions, this percentage drops notably [Fig. 1(b)]. Despite these differences, the percentage of bursting electrodes (computed over the active ones) is maintained at high values (more than 80%) in all the configurations [Fig. 1(c)].
FIG. 1.
Coupling of the scaffolds to the electrodes. (a) 20-s spontaneous electrophysiological activity of the different configurations (color-coded). (b) Bar plot of the percentage of active electrodes for each configuration superimposed on a sketch of the layout of the electrodes of the MEAs. (c) Bar plot of the percentage of bursting electrodes over the active ones. (d)–(g) Normalized amplitude distributions of the recorded extracellular potential of the different scaffolds. Statistical values are reported in supplementary material Tables I and II.
Then, we proceeded to identify all the voltage amplitudes of the detected spikes whose normalized values are reported in the histograms of Figs. 1(d)–1(g). The range and the shape of the obtained curves for the amplitudes change considerably for the tested cases. The distributions were compared in terms of similarity (Jensen–Shannon divergence) and overlap (Bhattacharyya distance) (supplementary material Tables III and IV). Both parameters yield the following results: the beads retain the lowest similarity with the other curves, and their probability distribution overlaps the least with the others. Therefore, the beads are the most different and have the highest detected amplitude values, suggesting that they still allow better results in terms of coupling to the substrate.49 The other configurations have a similar shape of the distributions, shifted toward lower values of amplitude that are therefore more overlapping (as demonstrated by the Bhattacharyya distance, which is one order of magnitude down) and more similar among them (JSS over 0.92).
C. Spiking and Bursting activity: Reproducibility, richness of bursts, and separation of classes
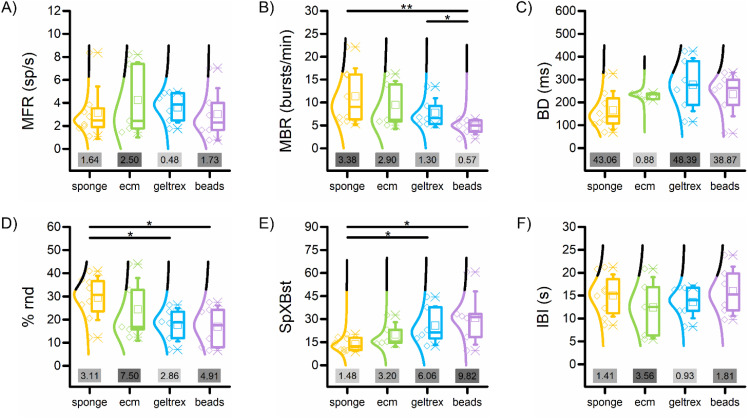
The impact of distinct topological organizations arising from varied internal structural arrangements of the scaffolds was investigated by assessing changes in electrophysiological parameters generated by the 3D models. Despite similar spiking activity, as evidenced by the mean firing rate (MFR) across different networks [Fig. 2(a)], the different scaffold types induced variations in the temporal distribution of these events, thus modulating the patterns of the spontaneous electrophysiological activity. Notably, the different spatial distribution of cells and mechanical properties of their environment brought differences to arise in the frequency [MBR, Fig. 2(b)] and in the “density” of the bursts [SpXBst, Fig. 2(e)], with a consequent effect also on the random spiking activity [% rnd, Fig. 2(d)]. To elaborate further, in the networks created using sponges (Fig. 2, yellow), bursts rise at a higher frequency, but they result to be less dense, leading to a higher random spiking activity. In contrast, the beads displayed a less frequent burst pattern with fewer spikes, while the gels had an intermediate behavior. Despite these differences, the bursts had comparable durations [BD, Fig. 2(c)] and inter-burst intervals [IBI, Fig. 2(f)].
FIG. 2.
Spiking and bursting parameters, with relative Fano factor (FF) values. (a) Mean firing rate (MFR), (b) mean bursting rate (MBR), (c) mean burst duration (BD), (d) percentage of random spiking, (e) spike per bursts (SpXBst), and (f) inter-burst interval (IBI) of the four configurations. The gray inset indicates the value of Fano factor of the relative configuration. The gray scale indicates the order value of FF. (* refers to 0.01 < p < 0.05, ** 0.001 < p < 0.01, Kruskal–Wallis non-parametric test). Statistical values are reported in supplementary material Tables V–X.
The fundamental firing and bursting parameters revealed significant differences among configurations. However, another critical feature to evaluate the different scaffolds is the reproducibility of the data generated within the same configuration, which was assessed by computing the relative Fano factor (i.e., the variance over the mean, FF). For the sponge configuration, the mean firing rate, spikes per burst, and inter-burst interval show only slight over-dispersion (FF < 2), while random spiking and mean bursting rate exhibit FF values exceeding 3. Excluding burst duration, the sponge configuration displays the lowest average FF and the smallest range among configurations. In contrast, the ecm configuration shows FF values within the range of 2 to 4, except for the MFR. Excluding BD, ecm presents the highest mean FF and a range comparable to geltrex. For the geltrex configuration, MFR, MBR, and IBI hover around 1, indicating variability consistent with a random process and showing minimal under/over-dispersion. However, the distribution of spikes is more dispersed, both in terms of spike per burst and random spiking. The beads configuration mirrors the characteristics of geltrex, with values diverging around one and higher values for random spiking and spike per burst. This disparity contributes to the largest range among configurations and the second-to-last mean. However, networks created with the beads show more reproducible values of MBR, with a FF of 0.57.
This suggests that not all 3D neuronal constructs can sustain the same level of richness of electrophysiological activity. In general, the distinct 3D scaffolds are intrinsically characterized by different topological structures, which (mostly) differ in the degree of spatial regularity of the construct itself. For example, the beads scaffold relies on the precise geometry of its components that result in a fixed packing fraction of its elements and consequently constitutes a regular environment where the networks can develop. On the contrary, PDMS sponges present an amorphous structure that arises from the irregular salt crystals that are leached out to form the pores. The regularity of the lattice should also reflect on the regularity of the morphological connections that are established within the 3D culture and consequently on the regularity of the activity patterns, particularly in burst generation. To further investigate the possible relation between the regularity of the scaffolds and the “richness” of the activity, we inspected the intrinsic temporal dynamics of the burst patterns, investigating both inside and outside bursting statistics.
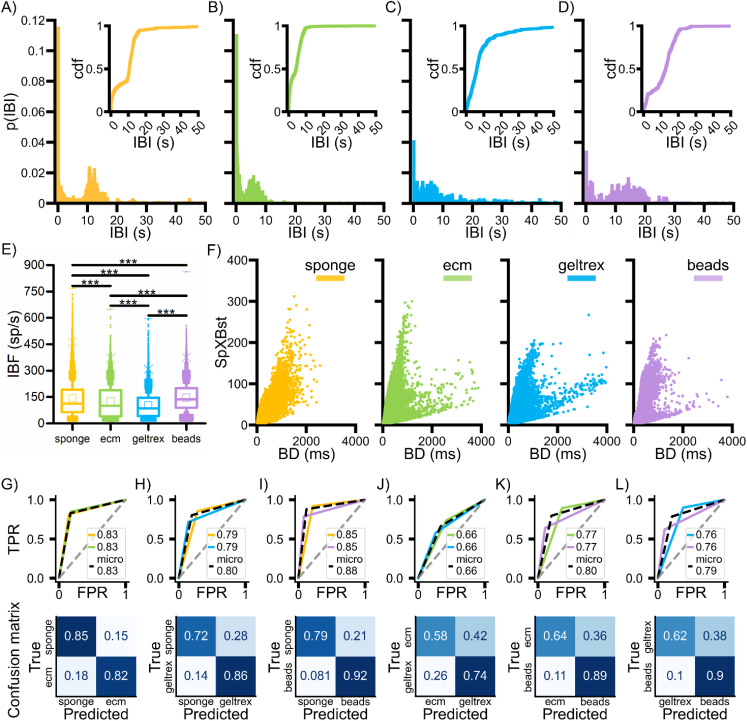
First of all, we evaluated the distribution of the inter-burst interval. Unlike the previous analysis, the values of this parameter were not averaged: we considered every value that was recorded from each bursting electrode separately to account for intra- as well as inter-specimen variability. The relative cumulative distributions of the IBI [Figs. 3(a)–3(d)] indicate a high similarity (computed from the Jensen–Shannon divergence) between the 3D networks generated by using the sponge and the microbeads, on the one hand, and the two thermogels, on the other (supplementary material Table XI). In general, all the distributions display a peak at short IBIs (<1 s) and a second one at larger IBIs (>5 s), which is less marked in the geltrex case [Fig. 3(c)].
FIG. 3.
Bursting richness. (a)–(d) Inter-burst interval (IBI) probability density distributions and (inset) relative cumulative distribution functions for the four configurations. (e) Intra-burst frequency (IBF) of each recorded burst event grouped by configuration (***p < 0.001, Kruskal–Wallis non-parametric test). (f) Scatter plot of the spikes per burst over burst duration for the four configurations. (g)–(l) ROC curves (above) and confusion matrices (below) obtained with the OvO SVM classification of the data represented in (f). The legend in the ROC curves indicates the AUC values for the considered color-coded configurations and for the micro-average, which aggregates performance metrics across classes (dashed black). The dashed gray like indicates the random choice. The color-coded confusion matrices are normalized in the [0–1] interval over the true values (per row). The configurations are represented in yellow for sponge, green for ecm, light blue for geltrex, purple for beads.
Such bimodal distributions suggest the possibility of two mechanisms in burst generation. In the case of beads [Fig. 3(d)] and sponges [Fig. 3(a)], where the second peak is at higher values of IBI (>10 s), the two activation modalities are well separated, with high frequency bursts separated by long silent periods. A broader distribution of IBIs suggests a higher aperiodicity and temporal complexity of the events, as is the case of the geltrex [Fig. 3(c)], followed by the sponges and the beads. The ecm scaffolds [Fig. 3(b)] seem to sustain bursts at more regular time intervals. Finally, there is one more notable feature, that is, the high peak at very low IBIs in the sponge and ecm cases, indicating that many bursts are temporally very close, a behavior less evident in the other two configurations. The variability of the bursting dynamics was further investigated by evaluating the intra-burst spike frequency [IBF, Fig. 3(e)], computed over all the generated bursts for each 3D scaffold. The 3D networks developed with the beads resulted in the configuration with highest mean intra burst frequency, suggesting a general higher excitability of the neurons during these events. Generally, all the configurations turned out to be significantly different from this point of view, i.e., when associating the information of the duration of the burst with the number of spikes that were generated within the event. By plotting these two quantities in a scatterplot representation [Fig. 3(f)], we observed different trends. At comparable burst duration, the different configurations do not present always the same number of spikes, resulting in two different types of bursts, namely, higher and lower frequency ones. At the same time, the thermogels and partly also the beads seem to present bursts with longer duration and increasing spikes per burst, with spikes at different frequencies, suggesting yet again a different variability in the generated patterns of bursting activity. These observations led us to consider the possibility of using this information to discriminate between the different scaffolds. The analysis to verify this hypothesis was conducted using the OvO SVM approach to preprocessed data. The classification highlighted that the sponge configuration is the one that is set apart from the others with the highest accuracy [>80%, Figs. 3(g)–3(i), supplementary material Table XII], the highest values achieved when compared to the beads [Fig. 3(i)]. This result is confirmed by the ROC curves, with an AUC around 0.90. The next best performance is detained by the beads that guarantee accuracy values just below 80% with the two thermogels [Figs. 3(k) and 3(l), supplementary material Table XII]. The classification fails consistently when the ecm and geltrex bursting data are considered, with accuracy of about 66% [supplementary material Table XII] and low AUC values [Fig. 3(J)], suggesting that in terms of internal structure of the bursts, the two gels cannot be recognized and separated efficiently.
D. Population analysis and network involvement
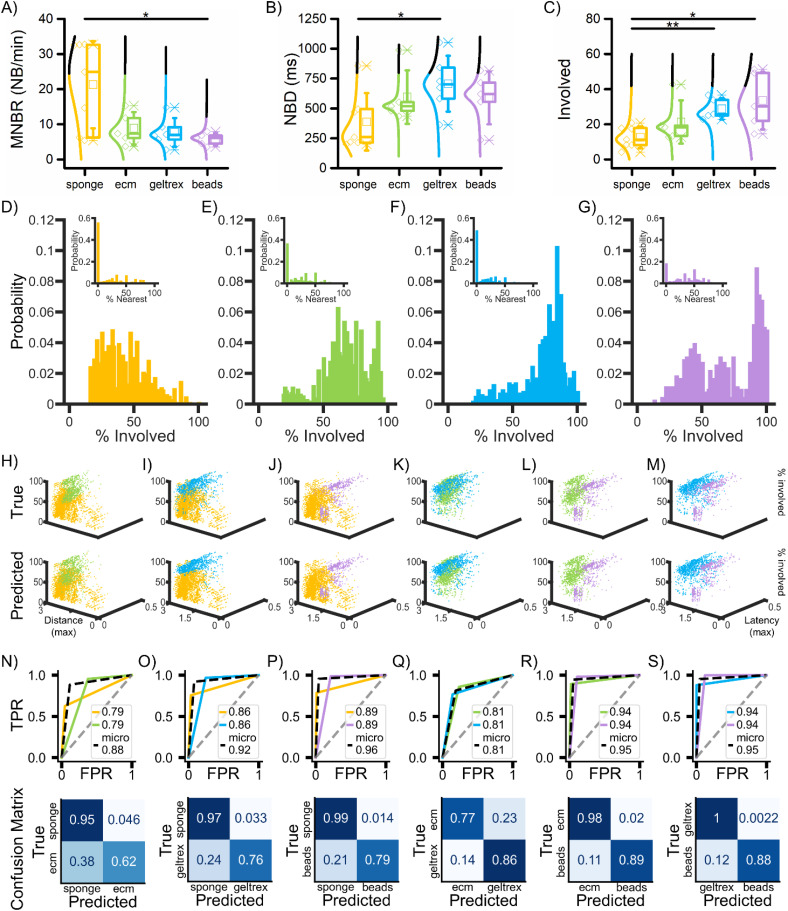
The structure of a neuronal network is closely related to the functional outcomes it generates. Therefore, a different spatial regularity in the scaffolds would also result in different patterns of network events, particularly affecting their propagation, which we evaluated by analyzing the features of the network bursts. These population events in the different configurations rise at rates [Fig. 4(a)] and have durations [Fig. 4(b)] that follow a similar trend to the single channel bursts [Figs. 2(b) and 2(c)]. The MNBR and the NBD show a statistical difference in the case of the sponge configuration when compared to beads and geltrex configurations, respectively (supplementary material Tables XIII and XIV). The number of involved electrodes [Fig. 4(c)] in each network burst is lower in the sponges, a difference that becomes significant in the case of geltrex and beads networks (supplementary material Table XVI). When evaluating the network involvement in population events and, consequently, the network burst propagation, it is essential to consider a feature of the overall experimental setup: the electrodes are only a 2D readout layer of a 3D construct. Never as in this case, a lower activity and fewer involved electrodes could imply a different organization of the network, resulting in a lower signal on the observable portion of the model. In this framework, we decided to evaluate the distribution of the percentage of involved electrodes [Figs. 4(d)–4(g)], as well as the position of the first involved electrodes with respect to the originating point of the network event [Figs. 4(d)–4(g), insets]. In fact, their dispersion and shape could give information on the regularity of the network pathways in the networks. Qualitatively, the distributions of involved electrodes follow trends that vary depending on the configurations. In the sponges [Fig. 4(d)], the distribution is right-skewed with a relatively low peak, indicating that each NB generally involves fewer electrodes, and their number varies a lot for each population event. Additionally, the first activated electrodes are often not the nearest neighbors to the originating point, as demonstrated by the very high peak at 0% in the distribution of the percentage of nearest electrodes (inset). The other three configurations have generally left-skewed distributions but with great differences (supplementary material Tables XVI and XVII). As in the case of sponges, the ecm configuration (JSS = 0.72) does not present a high mode, indicating a high variability in the number of involved electrodes from NB to NB, but the mean percentage is higher. A similar trend is observed in the case of geltrex, which has the highest overlap to the ecm case (JSS = 0.78 and BC = 0.41). However, a high mode (the highest among configurations) at high percentages is present, suggesting that most of the time, about 85% of the electrodes are involved. The beads have a yet different behavior. They show a similar peak to the one of geltrex (JSS = 0.78). However, in this case, the distribution presents a bimodal trend. The second peak shows similarities to the sponge configuration (JSS = 0.88). This indicates that most of the time, about 90% of the electrodes are involved, suggesting the highest involvement among the configurations. However, a second set of NBs, confined to only a part of the network, is present. Combining this information with the fact that the beads configuration has the highest number of nearest electrodes involved in the initial part of the population events, the observations suggest the highest regularity. Concerning the initiation of the events (supplementary material Tables XVIII and XIX), the other configurations show a similar behavior among them (JSS = 0.92, 0.95, and 0.93), where the NB spreads further away from the origin, with only a few nearest neighbors involved in the first part of the NB.
FIG. 4.
Population activity and propagation. (a) Mean network bursting rate (MNBR), (b) mean burst duration (NBD), and (c) mean number of involved electrodes per network burst. (d)–(g) Probability distributions of the percentage of involved electrodes and (inset) of the percentage of nearest involved electrodes. (h)–(m) First row: Scatter plots of the percentage of involved electrodes (z-axis), maximum latency (y-axis), and maximum distance (x-axis) reached by each NB of the different color-coded configurations. Last row: Scatterplot of the predicted classes using a OvO SVM algorithm. (n)–(s) ROC curves (above) and confusion matrices (below) obtained in the classification process. The legend in the ROC curves indicates the AUC values for the considered color-coded configurations and for the micro-average, which aggregates performance metrics across classes (dashed black). The dashed gray like indicates the random choice. The color-coded confusion matrices are normalized in the [0–1] interval over the true values (per row). The configurations are represented in yellow for sponge, green for ecm, light blue for geltrex, and purple for beads. Statistical values are reported in supplementary material Tables XIII–XV.
After considering the initial phase of the event and the recruitment, we further investigated the propagation of the NB to the whole network. We considered the percentage of involved electrodes, the maximum distance reached, and the maximum delay [depicted in a scatterplot in Figs. 4(h)–4(m), first row] to evaluate how the NBs spread. Generally, the sponge scaffold (yellow), which had more variability in the number of involved electrodes, generated two clusters based on the maximum reached distance. They have common low maximum delay, which could be a result of a high velocity in recruiting and/or shorter NB. The ecm (green) involved a varying number of electrodes, high maximum distance, with varying maximum delay. In the geltrex configuration (light blue), which involved most of the electrodes in the population events, the maximum distance was higher, with different maximum delay. Finally, in the beads (purple), two clusters formed depending on the number of involved electrodes. The maximum reached distance was lower than in geltrex, and the maximum registered delay varied. We classified the data using the OvO SVM approach, which resulted in a good separation of the classes, as shown in Figs. 4(h)–4(m). All accuracy values (supplementary material Table XX) were around or above 90%, except for thermogels, where it settled at 81%. Generally, the classification reached higher performances when the beads were involved, as confirmed by the ROC curves, with an AUC around 0.95 [Figs. 4(p), 4(r), and 4(s)], suggesting that the beads show a distinctive behavior in terms of NB propagation. The sponges also exhibited good classification performances, with an AUC of 0.88 with ecm [Fig. 4(n)] and 0.92 with geltrex [Fig. 4(o)], respectively. As mentioned, the classification on NB propagation exhibited lower performance in the case of the two gels [Fig. 4(q)], with an AUC of about 0.81. Nonetheless, in all cases, we were able to separate the classes (with an accuracy of at least 81%) using NB propagation data. This suggests that the differences among the configurations are most evident at the network level, which is indeed the most influenced by the different spatial organization of the 3D construct.
III. DISCUSSION AND CONCLUSION
In the present work, we assessed some methods for fabricating 3D neuronal networks based on their performance, ease of use, effectiveness, and scalability. This evaluation aimed to gauge the suitability of each technique for creating robust and versatile neuronal models, which are fundamental characteristics when 3D in vitro models are used in innovative pre-clinical studies.
Glass microbeads require a longer cell seeding process, whereas thermogels offer simplicity but pose challenges in yield. PDMS sponges, while requiring customization, offer multiple scaffolds per preparation and efficient sterilization. Despite scaffold variations, all allowed the recording of the typical neuronal features (i.e., spikes, bursts, and network bursts), with beads showing superior electrode coupling. Beads exhibited distinct spike voltage distributions, indicating better coupling with the substrate than other configurations. This higher performance could be due to the higher weights of the glass microbeads, which press neurons to the electrode increasing the sealing conditions and thus the amplitude of the recording signal.49 In the other types of scaffolds, the induced pressure is lower, decreasing the coupling of the scaffold to the substrate; this is partially due to the geometry of the sponge and the adhesion properties of PDMS to glass, on the one hand, and the adhesion properties of thermogels.
We investigated how different scaffold types influence electrophysiological parameters in 3D neuronal models. While spiking activity remained similar across configurations [Fig. 2(a)], scaffold variations induced differences in burst frequency, density, and random spiking [Figs. 2(b), 2(d), and 2(e)]. Sponge configurations exhibited higher burst frequency but lower density, leading to increased random spiking. Conversely, beads showed less frequent bursts with more spikes, while gels displayed intermediate behavior. Statistical analysis revealed significant differences in firing and bursting parameters among configurations, with reproducibility assessed through the Fano factor. The results suggest that scaffold regularity impacts the richness and regularity of electrophysiological activity. Additionally, analysis of inter-burst intervals revealed distinct burst generation modalities across configurations, with sponge and beads configurations exhibiting well-separated bursts and geltrex displaying greater temporal complexity (Fig. 3).
We explored how scaffold regularity affects network burst propagation (Fig. 4). Significant differences were observed in mean network bursting rate and network burst duration among sponge, beads, and geltrex configurations. Electrode involvement distributions revealed unique trends: sponges showed lower involvement with dispersed activation, geltrex exhibited high involvement, and beads demonstrated bimodal distributions with intense network engagement. OvO SVM classification achieved high accuracy (>90%), except for thermogels (81%), with beads showing distinctive NB propagation behavior. ROC curves confirmed robust classification, reflecting differences at the network level due to spatial organization variations in 3D constructs (Fig. 4).
The different scaffolds considered in our study exhibit distinct architectural structures and material compositions that influence key factors such as porosity, stiffness, and gas exchange, all of which are critical for cell viability and network function. Understanding the interplay between these biophysical and biochemical properties is essential for interpreting the different behaviors observed in our experiments. Nowadays, it is well known that the formation of connections within a network is influenced by the structure of the lattice in which it is embedded.50–52 This aspect was elucidated both in vitro and in silico, as current experimental techniques do not allow a complete study of the interplay between structural and functional connectivity.53–55 A more regular lattice is more likely to produce regular connections, whereas a more amorphous structure would reduce the probability of similar connections forming.24,31,56,57 Our results indicate that different scaffolds create networks that exhibit different burst frequency, density, and propagation. From these electrophysiological findings, we speculate that the spatial distribution of cells within the scaffolds significantly influences the activity of the networks. Indeed, this aspect is reflected in activity patterns, and particularly in the generation of bursts and network events. In the recorded electrophysiological data, this hypothesis can be supported at the first instance by the “regularity” of the frequency of the bursts, on the one hand, and by the regularity of the propagation of population events, on the other. Beads scaffolds, with their precise shape and geometrical arrangement, form networks where the bursting rate is very regular. Indeed, scaffolds with regular geometry can provide uniform and reproducible microenvironments, where the morphological connections can grow in a more consistent way across experiments. Their population events involve most of the network, with activity spreading evenly from the initiating point to farther regions. By contrast, sponge networks are expected to have more variability in their bursting due to the irregular pore lattice on which connections develop.18 The network burst spread is more saltatory, with varying numbers of electrodes involved, highlighting once again the possible link between the functional and structural aspects of the network. Larger pores in scaffolds, such as the ones that may be present in our PDMS sponges, can facilitate the formation of rich club networks, which are crucial for efficient communication within the network. The bursting variability and propagation parameters in the two thermogels are in between the other two conditions, which could be consistent as they do not present physical constraints to the connections that are free to grow unbounded. Moreover, in the thermogels, cells tend to sediment during the gelation process, potentially leading to uneven cell distribution and impacting the number of cells interfacing with the electrodes. This uneven distribution can result in varied electrophysiological readouts, complicating the interpretation of data. Their soft nature may produce limited mechanical support, affecting long-term stability and network integrity, but it could also pose an advantage for cell culture maturation. It is known that the stiffness of the scaffold can influence cell behavior through mechano-transduction pathways. Softer scaffolds like the thermogels promote neuronal differentiation and neurite outgrowth, while stiffer scaffolds like PDMS induce different adhesion and spreading behaviors. The gels provide a compliant environment that closely mimics the natural extracellular matrix, promoting cell proliferation, differentiation, and neurite extension. The presence of proteins and growth factors in these thermogels significantly influences cell behaviors, enhancing cell adhesion, survival, and differentiation. Highly porous structures facilitate better nutrient flow and waste removal, supporting higher cell density and network robustness, allowing for efficient gas exchange and may support the long-term survival of neuronal cultures. Due to their high porosity, ECM hydrogels facilitate efficient nutrient and gas exchange, crucial for cell survival and metabolic activity, especially in dense neuronal cultures. In contrast, bead-based scaffolds can have limited porosity, affecting nutrient and gas exchange and potentially leading to hypoxic conditions in dense cultures. This aspect is particularly relevant when considering the rise of long-term neuronal cultures derived from human induced pluripotent stem cells. As previously mentioned, the use of three-dimensional structures is becoming increasingly important in in vitro models that are used for innovative pre-clinical studies. The development of scaffold-free structures, like neurospheroids and brain organoids,58–60 and the engineering of the 3D connectivity by means of ad hoc scaffolds (in this work we proved the relevance in terms of the emerging patterns of electrophysiological activity), together with the use of human induced pluripotent stem cells (hiPSC), are nowadays considered a valid experimental model59,61 to study the physio-pathogenesis of brain diseases or for testing ad hoc drugs.62–64 Indeed, it has been demonstrated that the development and tailoring of specific scaffold properties promote specific gene expression and cell differentiation,52,65 which are essential for using this kind of model for precision medicine applications. In this perspective, next-generation brain-on-a-chip will use personalized hiPSC to build 3D neural systems to find patient-specific treatment strategies to create a personalized “in vitro brain” for the screening of novel drugs and performing pre-clinical testing of novel compounds.66 Given the substantial differences in scaffold properties, it is challenging to pinpoint which specific factors are driving the observed behaviors. Overall, the biophysical and biochemical properties of scaffolds play crucial roles in determining cell behavior and network function, with each type of scaffold offering unique advantages and limitations. Understanding these interactions can help optimize scaffold designs for specific biological applications, ultimately advancing the development of 3D tissue models in neuroscience research. Future work could involve systematic variations of individual scaffold properties to isolate their specific effects on cellular behavior. This approach will help to further elucidate the mechanisms driving the observed differences and guide the optimization of scaffold design for specific biological applications.
IV. METHODS
A. 3D cell cultures with different scaffolds
In the present work, we used dissociated cortical tissues obtained from Sprague Dawley rat embryos on gestational day 18 (E18). The protocols for 3D were the ones presented in Ref. 67 and detailed in the following. To record and thus analyze the electrophysiological activity of the 3D neuronal networks, cells were plated on 4Q Micro-Electrode Arrays (MEAs, Multi Channel Systems, MCS, Reutlingen, Germany) made up of 60 electrodes in a 4-compartment grid.
The substrate was dry oven sterilized at 120 °C for 3 h. Next, the surface underwent a coating process using Poly-L-Ornithine at a concentration of 100 μg/ml, which took place overnight at 37 °C, followed by rinsing with sterile water. Cortical tissues were subjected to digestion using an enzymatic solution containing 0.125% Trypsin and 0.03% DNAse (Sigma-Aldrich), which was diluted in Hanks' solution. The enzymatic digestion was halted after 20 min at 37 °C with a medium supplemented with 10% FBS, and the tissues were mechanically dissociated using a fine-tipped Pasteur pipette. Subsequently, single cells were suspended in culture medium consisting of Neurobasal Medium (Sigma-Aldrich), 1% GlutaMAX (Sigma-Aldrich), 2% B-27 supplement (Sigma-Aldrich), and 1% penicillin–streptomycin solution (Sigma-Aldrich). A monolayer of cells was created on the substrate to serve as an interface between the 3D cell culture and the recording electrodes. Following standard procedures,68 cortical cells were directly plated onto the active area of the MEA at a final cell density of 1800 cells/mm2. Cells were incubated for at least 3 h to ensure monolayer adhesion before introducing the 3D scaffold (Fig. S1). After the 3D culture was established (specifics for various scaffolding methods are outlined below), the cultures were maintained in an incubator with a controlled environment set at 37 °C, 5% CO2, and 95% humidity for 18 days in vitro. Half the medium was replaced first at day in vitro (DIV) 5 and subsequently twice a week. This replacement medium was BrainPhys™ (Stemcell Technologies) neuronal medium, supplemented with 2% NeuroCult SM1 Neuronal Supplement (Stemcell Technologies), 1% penicillin–streptomycin solution (Sigma-Aldrich), and 1% GlutaMAX (Sigma-Aldrich).
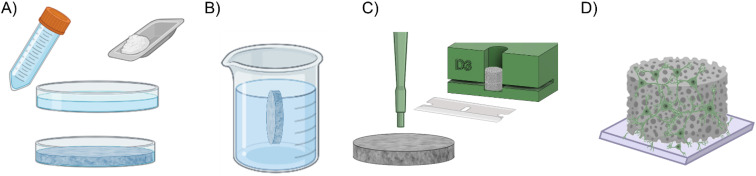
1. PDMS sponge scaffold
The PDMS sponges were fabricated by adapting the procedure described in Ref. 34. It is essential to create these sponges at least a week before preparing the cells. Initially, granules of NaCl with a size smaller than 600 μm were obtained through sieving. A PDMS solution (with a curing agent at a 1:10 weight ratio in prepolymer) was poured into a Petri dish, and the selected salt was added and thoroughly mixed to form a uniform layer. Subsequently, the Petri dish was placed in a vacuum pump to remove any trapped air bubbles and then cured in a dry oven at 80 °C for at least 30 min [Fig. 5(a)]. After removing the polymerized mixture from the dish, any excess material on the top or bottom layers (if present) was trimmed. The resulting compound was placed in a beaker filled with distilled water on a stirrer for at least 72 h to leach out the embedded salt, with the water being changed 2–3 times daily [Fig. 5(b)]. Following, scaffolds were cut out using a biopsy punch (with a diameter of 3 mm) and adjusted to the desired height using a custom 3D-printed mold [Fig. 5(c)]. These 3D scaffolds were then stored in de-ionized water and sterilized in an autoclave (using a wet cycle at 121 °C for 20 min). Subsequently, the scaffolds underwent plasma oxygen treatment (at 100 W for 60 s) and were coated with Poly-L-Ornithine at a concentration of 100 μg/ml (overnight at 37 °C). On the day of cell preparation, the 3D sponges were rinsed with sterile water and then with culture medium. About 700 000 cells were then seeded on each scaffold, accounting for an expected 37% cell loss. The obtained structures were placed in the incubator for at least 5 h to ensure robust cellular adhesion. The scaffolds were then transferred onto the cell monolayers [Fig. 5(d)], incubated for an additional hour, and flooded with culture medium.
FIG. 5.
Fabrication of the PDMS sponge scaffold and plating protocol for the formation of the 3D cultures. (a) Sieved NaCl was added in a PDMS solution (Sylgard 184, curing agent 1:10 w/w in prepolymer) to create a uniform mixture. Air bubbles were removed with a vacuum pump, and then, the compound was cured in dry oven at 80 °C for at least 30 min. (b) The salt was leached out to expose/create the cavities. (c) The scaffolds were cut out with a biopsy punch (Ø = 3 mm) and trimmed to height with an ad hoc 3D-printed mold. (d) On the day before cell preparation, the obtained scaffolds were autoclaved for sterilization and the surface of the pores was hydrophilized by plasma oxygen treatment and coated with Poly-L-Ornithine overnight. After rinsing with sterile water and culture medium, the scaffolds were ready for cell seeding.
2. Thermogels scaffolds
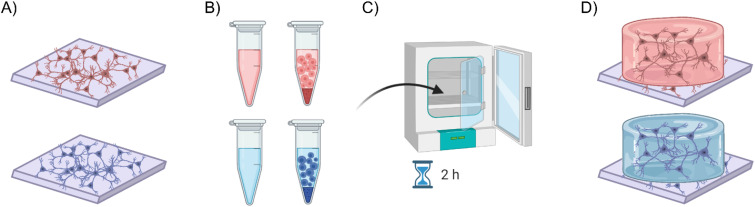
We tested two different thermogels, ECM gel (Sigma-Aldrich) and Geltrex (Thermo Fisher Scientific), as 3D scaffolds for neuronal networks. The experimental procedure for dealing with such gel materials was identical for both, except for the final cell concentration, which was determined by the specific recommendations provided for each product. The ECM gel is derived from mice Engelbreth-Holm-Swarm sarcoma. It contains a mixture of laminin, collagen type IV, heparin sulfate proteoglycan, entactin, and other minor components, resulting in a final protein concentration of 8–12 mg/ml (which was then diluted 1:2 as per datasheet in cell and culture medium solution). On the other hand, Geltrex™ LDEV-Free Reduced Growth Factor Basement Membrane Matrix is a similar soluble basement membrane sourced from murine EHS tumors, free from viruses such as lactose dehydrogenase elevating virus (LDEV), but with a concentration of 12–18 mg/ml (which was then diluted 2:3 as per datasheet in cell and culture medium solution). After the cell monolayers were established [Fig. 6(a)], the medium was removed to prevent excessive dilution of the gel. While working on ice to prevent early gelation, single cells (60 000) were evenly mixed into the gels according to the specified dilution factors [Fig. 6(b)]. Subsequently, the gel-cell solution was plated onto the substrates and allowed to polymerize adequately by incubating for at least 2 h [Fig. 6(c)] before adding culture medium to the formed 3D structure [Fig. 6(d)].
FIG. 6.
Creation process of the 3D cultures with the two thermogels, ECM gel (top) and Geltrex (bottom). (a) After the consolidation of the monolayer and the removal of excessive medium, (b) working on ice cells were mixed with the liquid thermogels and (c) stored in the incubator to allow for polymerization for about 2 h. (d) The obtained scaffolds were then inspected to ensure the creation of a 3D structure before adding culture medium.
3. Glass microbeads scaffold
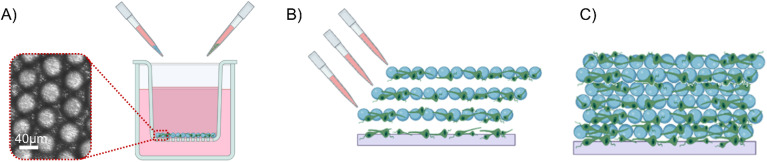
On the day before preparation, glass microbeads with a diameter of 40 μm (certified mean diameter of 42.3 ± 1.1 μm, Thermo Fisher) were sterilized in 70% ethanol for 3 h (the ethanol and containing Eppendorf were changed every hour). After being rinsed thrice with sterile water, they were placed in the adhesion factor solution (Poly-L-Ornithine at 100 μg/ml) and incubated at 37 °C overnight. On the day of cell dissection, the microbeads were once again rinsed and then transferred to a Transwell® (with an area of 33 mm2), where they formed a single and uniform layer with a hexagonal geometric structure, which facilitated the determination of the number of microbeads required to cover the porous membrane (approximately 20 000 beads) and the total surface area available for cell seeding (approximately 40 mm2). Each Transwell® was submerged in culture medium to help in the seeding procedure. Disassociated cells were then introduced into the Transwell®, taking into account two important factors: (i) approximately 25% of the cells would be lost when transferred to the MEA and (ii) a portion of the cells may not come into contact with the microbeads and would not adhere. The Transwell® was subsequently incubated for approximately 6 h to ensure robust cell adhesion to the glass surface, enabling their transfer [Fig. 7(a)]. The 3D structure was then created by stacking multiple layers of seeded microbeads over the monolayer [Fig. 7(b)]. In total, four to six layers of seeded microbeads were placed in the culture area (70 000 cell). A 500 μl drop of culture medium was added to the culture overnight to ensure cell survival. The following day, the overall structure was sufficiently stable to allow for the addition of the remaining medium for long-term culturing [Fig. 7(c)]. This protocol was adapted from Ref. 69.
FIG. 7.
Schematics of the 3D protocol with glass microbeads. (a) The microbeads and cells in the Transwell®, where they self-assemble in a monolayer with a precise structure (zoom), which allows estimating the plating area. (b) After 6 h, cells and microbeads were collected and stacked on the monolayer to progressively form layers, and a drop of culture medium was added to ensure survival overnight. (c) The day after, the scaffold was stable enough to add the remaining culture medium.
B. Electrophysiological data collection and analysis
We extracellularly recorded the spontaneous activity of 3D cultures in their mature developmental stage (DIV 18). Electrophysiological recordings were performed using the MEA2100 system (Multi Channel Systems, Reutlingen, Germany) with a sampling frequency of 10 kHz. The raw data were subsequently processed to extract spiking activity using the adaptive algorithm described in Ref. 70. In summary, this algorithm used individualized thresholds for each channel, set at eight times the standard deviation of the noise, both biological and thermal, along with the peak lifetime period (2 ms) and the refractory period (1 ms). This process was employed to extract the spike trains. Subsequently, bursts in the neural activity were identified using the string method outlined in Ref. 71. The criteria for the identification of a burst included a minimum of five spikes within a burst and a maximum inter-spike interval of 100 ms. The analysis of spiking and bursting activity was characterized using several parameters, which included: (i) mean firing rate (MFR), representing the average number of spikes per second (spikes per second); (ii) mean bursting rate (MBR), corresponding to the rate of bursts (bursts per minute); (iii) spike per bursts (SpXBst), indicating the average number of spikes within each burst; (iv) percentage of random spiking (%rnd), which indicates the proportion of spikes that do not belong to a burst; (v) burst duration (BD), i.e., the duration of the burst event; (vi) inter-burst interval (IBI), that is the temporal distance between two consecutive burst events; (vii) inter-spike interval (ISI), or the distance between two consecutive spikes; and (viii) intra-burst frequency (IBF), i.e., the spike frequency within a burst. Specific thresholds were established to determine whether a channel was active from a spiking and/or bursting point of view. A channel was considered active if its MFR exceeded 0.1 spikes/s and was categorized as bursting if its MBR exceeded 0.4 bursts/min. The population events defined as network bursts (NBs) were identified using the self-adaptive algorithm devised in Ref. 72 by setting two parameters: (i) the maximum inter-burst event interval for burst events within a network burst (100 ms), and (ii) the minimum percentage of recording electrodes involved in the network burst (in the present study, the total number of active channels was set at 20%). From the network burst trains, we computed the mean network bursting rate (MNBR), which represents the average number of network bursts per minute, and the network burst duration (NBD), indicating their temporal span. Further considered metrics were the percentage of active electrodes during each NB, the percentage of nearest neighbors (in a radius of 600 μm, corresponding to about three electrodes in each direction) that activate in the initial chunk of the NB (1/8 of the NBD); the maximum latency necessary to activate each involved electrode; the maximum distance reached by each NB.
Data were analyzed using a hierarchical approach, grouping, and averaging data at varying levels of integration (i.e., all observations, averaged over the electrodes, averaged over the MEA). This process involves successive steps to generate descriptive metrics, providing insights into the dataset's characteristics at different scales, from finer nuances that highlight internal variability, to overarching trends that describe a particular condition.
For the comparison of probability distributions, namely, A and B defined in the same domain , we used two different metrics: the Bhattacharyya distance,73,74
| (1) |
which indicates how well two distributions overlap, and the similarity (JSS) computed from the Jensen–Shannon divergence75,76 (JSD),
| (2) |
where is a mixture distribution defined as
| (3) |
Finally, we quantified the amplitudes of the detected signals as a (indirect) measure of the degree of coupling between the 3D network and the active area of the MEAs.53
All algorithms used for these analyses were developed in MATLAB (The MathWorks, Natick, US).
C. Classification
A classification algorithm was used to separate the different configurations based on different sets of parameters at different scale levels.
In the evaluation of the richness of the bursting activity, all observed bursts were studied in terms of spikes per burst and burst duration. We applied a support vector machine (SVM) algorithm, a supervised machine learning algorithm employed for classification tasks, on these features. Data were preprocessed in order to eliminate the most common bursts, i.e., those bursts that have lower burst duration (<1000) and lower spikes per burst (<100), and were standardized by centering and scaling each predictor variable by the corresponding weighted mean and standard deviation. Similarly, the same SVM algorithm was applied to the evaluation of the propagation of the network bursts throughout the network (we considered the propagation parameters of each single population event for each configuration). We implemented a one-vs-one (OvO) approach for multi-class classification, which consists in fitting one classifier per configuration pair. The algorithm was trained to use 30% of the data for testing and uses a radial basis function kernel for computation.
The performance of the classification algorithm was evaluated with different metrics: (i) the accuracy, as the percentage of true positives over all the outcomes; (ii) receiver operating characteristic (ROC) curves, which is the trade-off between the true positive rate (sensitivity) and the false positive rate (1— specificity) across various classification thresholds and visually represent a model's discriminatory power; (iii) the relative area under the curve (AUC), which provides a quantitative measure of this discrimination; and (iv) confusion matrices (normalized over the true conditions), which offers a detailed breakdown of the model's classification performance across different outcome configurations. The metrics were evaluated using Python.
D. Immunocytochemistry
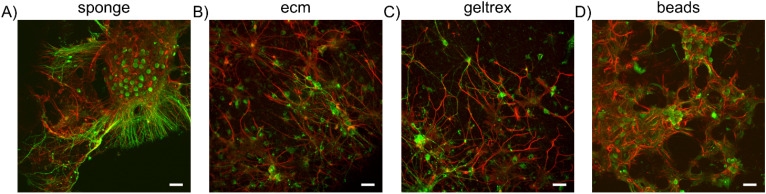
Cells were fixed in 4% paraformaldehyde (PFA) at 4 °C overnight, followed by permeabilization with a 0.2% Triton-X100 solution in phosphate buffer solution (PBS) at room temperature for 10 min. To minimize nonspecific antibody binding, cells were then treated with a blocking buffer solution (BBS) composed of PBS and 5% fetal bovine serum (FBS) (Sigma-Aldrich) for 40 min. Subsequently, the cultures were incubated with primary antibodies diluted in BBS in a humidified atmosphere at 4 °C overnight. The primary antibodies used were Anti-MAP2 (1:500, polyclonal, rabbit, Synaptic System) and Anti-GFAP (1:500, monoclonal, mouse, Synaptic System). The samples were then exposed to the secondary antibody's solution (1:700 Alexa Fluor 488, goat anti rabbit; 1:1000 Alexa Fluor 546, goat anti mouse) for 1 h at room temperature. The samples were rinsed twice in PBS after each step of the protocol except after the BBS incubation. Imaging was performed using a Leica TCS SP5 AOBS Tandem DM6000 upright microscope equipped with a Leica HCX IRAPO L 25 × 0.95 N.A., water immersion objective (Leica Microsystems S.r.l. Italy). Z-stack sequences were collected at different heights of the samples as depicted in supplementary material movies S1–S4. The Z-project of the same images is depicted in Fig. 8 (Multimedia view).
FIG. 8.
E. Dataset
We recorded the spontaneous activity of n = 25 3D cortical cultures in their mature stage of development (18 days in vitro, DIVs). The n = 25 MEAs came from ten preparations, each exploited to plate the different types of constructs. In particular, the dataset consists of n = 8 neuronal networks created using the PDMS sponge scaffolds, n = 5 neuronal networks created using the ECM gel, n = 6 neuronal networks created using the Geltrex, and n = 6 neuronal networks created using the glass microbeads.
F. Statistical significance
Due to the non-normal distribution of the data evaluated by means of Kolmogorov–Smirnov normality test, statistical analysis was conducted using a non-parametric Kruskal–Wallis test through the software Origin (Origin Lab in Northampton, MA) with statistical significance set at p < 0.05.
SUPPLEMENTARY MATERIAL
See the supplementary material for (i) supplementary Fig. S1, a schematic of cell-to-electrode coupling, and (ii) for the overall results of the performed statistical analysis, divided for each section of the results and the z-stack movies relative to Fig. 8.
ACKNOWLEDGMENTS
This work was supported by #NEXTGENERATIONEU (NGEU) and funded by the Ministry of University and Research (MUR), National Recovery and Resilience Plan (NRRP), Project MNESYS (PE0000006)—A Multiscale integrated approach to the study of the nervous system in health and disease (DN. 1553 11.10.2022).
AUTHOR DECLARATIONS
Conflict of Interest
Martina Brofiga was employed by ScreenNeuroPharm S.r.l. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics Approval
Ethics approval for experiments reported in the submitted manuscript on animal or human subjects was granted. The experimental protocol aimed at reducing the number of sacrificed animals and minimizing their suffering. It was approved by the European Animal Care Legislation (2010/63/EU), by the Italian Ministry of Health in accordance with the D.L. 116/1992 and by the guidelines of the University of Genova (Prot. 75F11.N.6JI, 08/08/18).
Author Contributions
Francesca Callegari: Data curation (lead); Formal analysis (lead); Methodology (equal); Software (lead); Visualization (equal); Writing – original draft (lead). Martina Brofiga: Conceptualization (equal); Data curation (supporting); Methodology (equal); Writing – original draft (supporting). Mariateresa Tedesco: Methodology (supporting). Paolo Massobrio: Conceptualization (equal); Project administration (lead); Resources (equal); Supervision (equal); Writing – original draft (equal).
DATA AVAILABILITY
The data that support the findings of this study are openly available in Zenodo at https://doi.org/10.5281/zenodo.13259912, Ref. 77.
References
- 1. Sporns O. and Betzel R. F., “ Modular brain networks,” Annu. Rev. Psychol. 67(1), 613–640 (2016). 10.1146/annurev-psych-122414-033634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sporns O. and Zwi J. D., “ The small world of the cerebral cortex,” Neuroinformatics 2(2), 145–162 (2004). 10.1385/NI:2:2:145 [DOI] [PubMed] [Google Scholar]
- 3. Gal E., Amsalem O., Schindel A., London M., Schürmann F., Markram H., and Segev I., “ The role of hub neurons in modulating cortical dynamics,” Front. Neural Circuits 15, 718270 (2021). 10.3389/fncir.2021.718270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van den Heuvel M. P. and Sporns O., “ Rich-club organization of the human connectome,” J. Neurosci. 31(44), 15775–15786 (2011). 10.1523/JNEUROSCI.3539-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cullen D. K., Wolf J. A., Vernekar V. N., Vukasinovic J., and la Placa M. C., “ Neural tissue engineering and biohybridized microsystems for neurobiological investigation in vitro (Part 1),” Crit. Rev. Biomed. Eng. 39(3), 201–240 (2011). 10.1615/CritRevBiomedEng.v39.i3.30 [DOI] [PubMed] [Google Scholar]
- 6. Schüz A. and Palm G., “ Density of neurons and synapses in the cerebral cortex of the mouse,” J. Comput. Neurol. 286(4), 442–455 (1989). 10.1002/cne.902860404 [DOI] [PubMed] [Google Scholar]
- 7. Severino F. P. U., Ban J., Song Q., Tang M., Bianconi G., Cheng G., and Torre V., “ The role of dimensionality in neuronal network dynamics,” Sci. Rep. 6(1), 29640 (2016). 10.1038/srep29640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baker B. M. and Chen C. S., “ Deconstructing the third dimension-how 3D culture microenvironments alter cellular cues,” J. Cell Sci. 125(13), 3015–3024 (2012). 10.1242/jcs.079509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hopkins A. M., DeSimone E., Chwalek K., and Kaplan D. L., “ 3D in vitro modeling of the central nervous system,” Prog. Neurobiol. 125, 1–25 (2015). 10.1016/j.pneurobio.2014.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yue B., “ Biology of the extracellular matrix: An overview,” J. Glaucoma 23(8), S20–S23 (2014). 10.1097/IJG.0000000000000108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koroleva A., Deiwick A., El-Tamer A., Koch L., Shi Y., Estévez-Priego E., Ludl A. A., Soriano J., Guseva D., Ponimaskin E., and Chichkov B., “ In vitro development of human iPSC-derived functional neuronal networks on laser-fabricated 3D scaffolds,” ACS Appl. Mater. Interfaces 13(7), 7839–7853 (2021). 10.1021/acsami.0c16616 [DOI] [PubMed] [Google Scholar]
- 12. Cenini G., Hebisch M., Iefremova V., Flitsch L. J., Breitkreuz Y., Tanzi R. E., Kim D. Y., Peitz M., and Brüstle O., “ Dissecting Alzheimer's disease pathogenesis in human 2D and 3D models,” Mol. Cell. Neurosci. 110, 103568 (2021). 10.1016/j.mcn.2020.103568 [DOI] [PubMed] [Google Scholar]
- 13. Wittrahm R., Takalo M., Kuulasmaa T., Mäkinen P. M., Mäkinen P., Končarević S., Fartzdinov V., Selzer S., Kokkola T., Antikainen L., Martiskainen H., Kemppainen S., Marttinen M., Jeskanen H., Rostalski H., Rahunen E., Kivipelto M., Ngandu T., Natunen T., Lambert J. C., Tanzi R. E., Kim D. Y., Rauramaa T., Herukka S. K., Soininen H., Laakso M., Pike I., Leinonen V., Haapasalo A., and Hiltunen M., “ Protective Alzheimer's disease-associated APP A673T variant predominantly decreases sAPPβ levels in cerebrospinal fluid and 2D/3D cell culture models,” Neurobiol. Dis. 182, 106140 (2023). 10.1016/j.nbd.2023.106140 [DOI] [PubMed] [Google Scholar]
- 14. Kang Y. J., Diep Y. N., Tran M., Tran V. T. A., Ambrin G., Ngo H., and Cho H., “ Three-dimensional human neural culture on a chip recapitulating neuroinflammation and neurodegeneration,” Nat. Protoc. 18(9), 2838–2867 (2023). 10.1038/s41596-023-00861-4 [DOI] [PubMed] [Google Scholar]
- 15. Fabbri R., Cacopardo L., Ahluwalia A., and Magliaro C., “ Advanced 3D models of human brain tissue using neural cell lines: State-of-the-art and future prospects,” Cells 12(8), 1181 (2023). 10.3390/cells12081181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park D. H., He M. T., Cho E. J., Morten K., and Go J. S., “ Development of a novel microfluidic perfusion 3D cell culture system for improved neuronal cell differentiation,” Biomed. Microdevices 25(3), 22 (2023). 10.1007/s10544-023-00660-4 [DOI] [PubMed] [Google Scholar]
- 17. Edmondson R., Broglie J. J., Adcock A. F., and Yang L., “ Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors,” Assay Drug Dev. Technol. 12(4), 207–218 (2014). 10.1089/adt.2014.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fan D., Staufer U., and Accardo A., “ Engineered 3D polymer and hydrogel microenvironments for cell culture applications,” Bioengineering 6(4), 113 (2019). 10.3390/bioengineering6040113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cadamuro F., Nicotra F., and Russo L., “ 3D printed tissue models: From hydrogels to biomedical applications,” J. Controlled Release 354, 726–745 (2023). 10.1016/j.jconrel.2023.01.048 [DOI] [PubMed] [Google Scholar]
- 20. Nikolova M. P. and Chavali M. S., “ Recent advances in biomaterials for 3D scaffolds: A review,” Bioact. Mater. 4, 271–292 (2019). 10.1016/j.bioactmat.2019.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koutsopoulos S. and Zhang S., “ Long-term three-dimensional neural tissue cultures in functionalized self-assembling peptide hydrogels, Matrigel and Collagen I,” Acta Biomater. 9(2), 5162–5169 (2013). 10.1016/j.actbio.2012.09.010 [DOI] [PubMed] [Google Scholar]
- 22. Honegger T., Thielen M. I., Feizi S., Sanjana N. E., and Voldman J., “ Microfluidic neurite guidance to study structure-function relationships in topologically-complex population-based neural networks,” Sci. Rep. 6(1), 28384 (2016). 10.1038/srep28384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang W. H., Ding S. L., Zhao X. Y., Li K., Guo H. T., Zhang M. Z., and Gu Q., “ Collagen for neural tissue engineering: Materials, strategies, and challenges,” Mater. Today Bio 20, 100639 (2023). 10.1016/j.mtbio.2023.100639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lam D., Enright H. A., Peters S. K. G., Moya M. L., Soscia D. A., Cadena J., Alvarado J. A., Kulp K. S., Wheeler E. K., and Fischer N. O., “ Optimizing cell encapsulation condition in ECM-Collagen I hydrogels to support 3D neuronal cultures,” J. Neurosci. Methods 329, 108460 (2020). 10.1016/j.jneumeth.2019.108460 [DOI] [PubMed] [Google Scholar]
- 25. Sun G., Liu W., Fan Z., Zhang D., Han Y., Xu L., Qi J., Zhang S., Gao B. T., Bai X., Li J., Chai R., and Wang H., “ The three-dimensional culture system with matrigel and neurotrophic factors preserves the structure and function of spiral ganglion neuron in vitro,” Neural Plast. 2016, 2070–2084. 10.1155/2016/4280407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bang S., Na S., Jang J. M., Kim J., and Jeon N. L., “ Engineering‐aligned 3D neural circuit in microfluidic device,” Adv. Healthcare Mater. 5(1), 159–166 (2016). 10.1002/adhm.201500397 [DOI] [PubMed] [Google Scholar]
- 27. Antman-Passig M., Levy S., Gartenberg C., Schori H., and Shefi O., “ Mechanically oriented 3D collagen hydrogel for directing neurite growth,” Tissue Eng., Part A 23(9–10), 403–414 (2017). 10.1089/ten.tea.2016.0185 [DOI] [PubMed] [Google Scholar]
- 28. Liu C., Nguyen R. Y., Pizzurro G. A., Zhang X., Gong X., Martinez A. R., and Mak M., “ Self-assembly of mesoscale collagen architectures and applications in 3D cell migration,” Acta Biomater. 155, 167–181 (2023). 10.1016/j.actbio.2022.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lovett M. L., Nieland T. J. F., Dingle Y. T. L., and Kaplan D. L., “ Innovations in 3D tissue models of human brain physiology and diseases,” Adv. Funct. Mater. 30(44), 1909146 (2020). 10.1002/adfm.201909146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li S., Severino F. P. U., Ban J., Wang L., Pinato G., Torre V., and Chen Y., “ Improved neuron culture using scaffolds made of three-dimensional PDMS micro-lattices,” Biomed. Mater. 13(3), 034105 (2018). 10.1088/1748-605X/aaa777 [DOI] [PubMed] [Google Scholar]
- 31. Timashev P. S., Vedunova M. V., Guseva D., Ponimaskin E., Deiwick A., Mishchenko T. A., Mitroshina E. V., Koroleva A. V., Pimashkin A. S., Mukhina I. V., Panchenko V. Y., Chichkov B. N., and Bagratashvili V. N., “ 3D in vitro platform produced by two-photon polymerization for the analysis of neural network formation and function,” Biomed. Phys. Eng. Express 2(3), 035001 (2016). 10.1088/2057-1976/2/3/035001 [DOI] [Google Scholar]
- 32. Bosi S., Rauti R., Laishram J., Turco A., Lonardoni D., Nieus T., Prato M., Scaini D., and Ballerini L., “ From 2D to 3D: Novel nanostructured scaffolds to investigate signalling in reconstructed neuronal networks,” Sci. Rep. 5(1), 9562 (2015). 10.1038/srep09562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Du B., Yin H., Chen Y., Lin W., Wang Y., Zhao D., Wang G., He X., Li J., Li Z., Luo F., Tan H., and Fu Q., “ A waterborne polyurethane 3D scaffold containing PLGA with a controllable degradation rate and an anti-inflammatory effect for potential applications in neural tissue repair,” J. Mater. Chem. B 8(20), 4434–4446 (2020). 10.1039/D0TB00656D [DOI] [PubMed] [Google Scholar]
- 34. Dingle Y. T. L., Liaudanskaya V., Finnegan L. T., Berlind K. C., Mizzoni C., Georgakoudi I., Nieland T. J. F., and Kaplan D. L., “ Functional characterization of three-dimensional cortical cultures for in vitro modeling of brain networks,” iScience 23(8), 101434 (2020). 10.1016/j.isci.2020.101434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Subia B., Rao R. R., and Kundu S. C., “ Silk 3D matrices incorporating human neural progenitor cells for neural tissue engineering applications,” Polym. J. 47(12), 819–825 (2015). 10.1038/pj.2015.69 [DOI] [Google Scholar]
- 36. Kwak S. S., Washicosky K. J., Brand E., von Maydell D., Aronson J., Kim S., Capen D. E., Cetinbas M., Sadreyev R., Ning S., Bylykbashi E., Xia W., Wagner S. L., Choi S. H., Tanzi R. E., and Kim D. Y., “ Amyloid-β42/40 ratio drives tau pathology in 3D human neural cell culture models of Alzheimer's disease,” Nat. Commun. 11(1), 1377 (2020). 10.1038/s41467-020-15120-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Taketani M. and Baudry M., Advances in Network Electrophysiology ( Springer US, Boston, MA, 2006). [Google Scholar]
- 38. Frega M., Tedesco M., Massobrio P., Pesce M., and Martinoia S., “ Network dynamics of 3D engineered neuronal cultures: A new experimental model for in-vitro electrophysiology,” Sci. Rep. 4(1), 5489 (2014). 10.1038/srep05489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bourke J. L., Quigley A. F., Duchi S., O'Connell C. D., Crook J. M., Wallace G. G., Cook M. J., and Kapsa R. M. I., “ Three-dimensional neural cultures produce networks that mimic native brain activity,” J. Tissue Eng. Regener. Med. 12(2), 490–493 (2018). 10.1002/term.2508 [DOI] [PubMed] [Google Scholar]
- 40. Tedesco M. T., Di Lisa D., Massobrio P., Colistra N., Pesce M., Catelani T., Dellacasa E., Raiteri R., Martinoia S., and Pastorino L., “ Soft chitosan microbeads scaffold for 3D functional neuronal networks,” Biomaterials 156, 159–171 (2018). 10.1016/j.biomaterials.2017.11.043 [DOI] [PubMed] [Google Scholar]
- 41. Yoon D., Son J., Park J. K., and Nam Y., “ Development of the micro-patterned 3D neuronal-hydrogel model using soft-lithography for study a 3D neural network on a microelectrode array,” in 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) ( IEEE, 2021). [DOI] [PubMed] [Google Scholar]
- 42. Yao Y., Coleman H. A., Meagher L., Forsythe J. S., and Parkington H. C., “ 3D functional neuronal networks in free-standing bioprinted hydrogel constructs,” Adv. Healthcare Mater. 12(28), 2300801 (2023). 10.1002/adhm.202300801 [DOI] [PubMed] [Google Scholar]
- 43. Venkatasubramanian R., Jayanthi, Das U., and Bhatnagar S., “ Comparison of the effectiveness of sterilizing endodontic files by 4 different methods: An in vitro study,” J. Indian Soc. Pedod. Prev. Dent. 28(1), 2–5 (2010). 10.4103/0970-4388.60478 [DOI] [PubMed] [Google Scholar]
- 44. Holy C. E., Cheng C., Davies J. E., and Shoichet M. S., “ Optimizing the sterilization of PLGA scaffolds for use in tissue engineering,” Biomaterials 22(1), 25–31 (2000). 10.1016/S0142-9612(00)00136-8 [DOI] [PubMed] [Google Scholar]
- 45. Ingram L. O. N. and Buttke T. M., “ Effects of alcohols on micro-organisms,” Adv. Microb. Physiol. 25, 253–300 (1985). 10.1016/S0065-2911(08)60294-5 [DOI] [PubMed] [Google Scholar]
- 46. Sahu P. K., Tilgam J., Mishra S., Hamid S., Gupta A., Jayalakshmi K., Verma S. K., and Kharwar R. N., “ Surface sterilization for isolation of endophytes: Ensuring what (not) to grow,” J. Basic Microbiol. 62(6), 647–668 (2022). 10.1002/jobm.202100462 [DOI] [PubMed] [Google Scholar]
- 47. Choi J. S., Lee H. J., Rajaraman S., and Kim D. H., “ Recent advances in three-dimensional microelectrode array technologies for in vitro and in vivo cardiac and neuronal interfaces,” Biosens. Bioelectron. 171, 112687 (2021). 10.1016/j.bios.2020.112687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cho Y. H., Park Y. G., Kim S., and Park J. U., “ 3D electrodes for bioelectronics,” Adv. Mater. 33(47), 2005805 (2021). 10.1002/adma.202005805 [DOI] [PubMed] [Google Scholar]
- 49. Massobrio P., Massobrio G., and Martinoia S., “ Multi-program approach for simulating recorded extracellular signals generated by neurons coupled to microelectrode arrays,” Neurocomputing 70(13–15), 2467–2476 (2007). 10.1016/j.neucom.2006.09.008 [DOI] [Google Scholar]
- 50. Li W., Tang Q. Y., Jadhav A. D., Narang A., Qian W. X., Shi P., and Pang S. W., “ Large-scale topographical screen for investigation of physical neural-guidance cues,” Sci. Rep. 5(1), 8644 (2015). 10.1038/srep08644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhuang P., Sun A. X., An J., Chua C. K., and Chew S. Y., “ 3D neural tissue models: From spheroids to bioprinting,” Biomaterials 154, 113–133 (2018). 10.1016/j.biomaterials.2017.10.002 [DOI] [PubMed] [Google Scholar]
- 52. Nguyen R. Y., Cabral A. T., Rossello-Martinez A., Zulli A., Gong X., Zhang Q., Yan J., and Mak M., “ Tunable mesoscopic collagen island architectures modulate stem cell behavior,” Adv. Mater. 35(16), 2207882 (2023). 10.1002/adma.202207882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tibau E., Ludl A. A., Rudiger S., Orlandi J. G., and Soriano J., “ Neuronal spatial arrangement shapes effective connectivity traits of in vitro cortical networks,” IEEE Trans. Network Sci. Eng. 7(1), 435–448 (2020). 10.1109/TNSE.2018.2862919 [DOI] [Google Scholar]
- 54. Ludl A. A. and Soriano J., “ Impact of physical obstacles on the structural and effective connectivity of in silico neuronal circuits,” Front. Comput. Neurosci. 14, 557191 (2020). 10.3389/fncom.2020.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Callegari F., Brofiga M., and Massobrio P., “ Modeling the three-dimensional connectivity of in vitro cortical ensembles coupled to micro-electrode arrays,” PLoS Comput. Biol. 19(2), e1010825 (2023). 10.1371/journal.pcbi.1010825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jakobsson A., Ottosson M., Zalis M. C., O'Carroll D., Johansson U. E., and Johansson F., “ Three-dimensional functional human neuronal networks in uncompressed low-density electrospun fiber scaffolds,” Nanomed.: Nanotechnol., Biol. Med. 13(4), 1563–1573 (2017). 10.1016/j.nano.2016.12.023 [DOI] [PubMed] [Google Scholar]
- 57. Crowe J. A., El-Tamer A., Nagel D., Koroleva A. V., Madrid-Wolff J., Olarte O. E., Sokolovsky S., Estevez-Priego E., Ludl A., Soriano J., Loza-Alvarez P., Chichkov B. N., Hill E. J., Parri H. R., and Rafailov E. U., “ Development of two-photon polymerised scaffolds for optical interrogation and neurite guidance of human iPSC-derived cortical neuronal networks,” Lab Chip 20(10), 1792–1806 (2020). 10.1039/C9LC01209E [DOI] [PubMed] [Google Scholar]
- 58. Kanton S. and Pasça S. P., “ Human assembloids,” Development 149(23), dev201120 (2022). 10.1242/dev.201120 [DOI] [PubMed] [Google Scholar]
- 59. Castiglione H., Vigneron P. A., Baquerre C., Yates F., Rontard J., and Honegger T., “ Human brain organoids-on-chip: Advances, challenges, and perspectives for preclinical applications,” Pharmaceutics 14(11), 2301 (2022). 10.3390/pharmaceutics14112301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Park J. H., Kim J., Walter J., and Kim C. Y., “ Use of neural 3D organoid with MEA in neurotoxicity testing: Comparison to traditional in vitro cell culture and in vivo methods,” Mol. Cell. Toxicol. 18(1), 17–21 (2022). 10.1007/s13273-021-00184-z [DOI] [Google Scholar]
- 61. Teli P., Kale V., and Vaidya A., “ Beyond animal models: Revolutionizing neurodegenerative disease modeling using 3D in vitro organoids, microfluidic chips, and bioprinting,” Cell Tissue Res. 394(1), 75–91 (2023). 10.1007/s00441-023-03821-2 [DOI] [PubMed] [Google Scholar]
- 62. Silva M. C. and Haggarty S. J., “ Human pluripotent stem cell–derived models and drug screening in CNS precision medicine,” Ann. N. Y. Acad. Sci. 1471(1), 18–56 (2020). 10.1111/nyas.14012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pasteuning-Vuhman S., de Jongh R., Timmers A., and Pasterkamp R. J., “ Towards advanced iPSC-based drug development for neurodegenerative disease,” Trends Mol. Med. 27(3), 263–279 (2021). 10.1016/j.molmed.2020.09.013 [DOI] [PubMed] [Google Scholar]
- 64. Cerutti L. and Brofiga M., “ Unraveling brain diseases: The promise of brain-on-a-chip models,” J. Neurosci. Methods 405, 110105 (2024). 10.1016/j.jneumeth.2024.110105 [DOI] [PubMed] [Google Scholar]
- 65. Moxon S. R., Corbett N. J., Fisher K., Potjewyd G., Domingos M., and Hooper N. M., “ Blended alginate/collagen hydrogels promote neurogenesis and neuronal maturation,” Mater. Sci. Eng. C 104, 109904 (2019). 10.1016/j.msec.2019.109904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brofiga M. and Massobrio P., “ Brain-on-a-chip: Dream or reality?,” Front. Neurosci. 16, 837623 (2022). 10.3389/fnins.2022.837623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Callegari F., Brofiga M., Tedesco M., and Massobrio P., in 45th Annual International Conference of the IEEE Engineering in Medicine and Biology Society ( IEEE, 2023). [DOI] [PubMed] [Google Scholar]
- 68. Brofiga M., Pisano M., Tedesco M., Raiteri R., and Massobrio P., “ Three-dimensionality shapes the dynamics of cortical interconnected to hippocampal networks,” J. Neural Eng. 17(5), 056044 (2020). 10.1088/1741-2552/abc023 [DOI] [PubMed] [Google Scholar]
- 69. Tedesco M., Frega M., Martinoia S., Pesce M., and Massobrio P., “ Interfacing 3D engineered neuronal cultures to micro-electrode arrays: An innovative in vitro experimental model,” J. Vis. Exp. 2015(104), e53080. 10.3791/53080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Maccione A., Gandolfo M., Massobrio P., Novellino A., Martinoia S., and Chiappalone M., “ A novel algorithm for precise identification of spikes in extracellularly recorded neuronal signals,” J. Neurosci. Methods 177(1), 241–249 (2009). 10.1016/j.jneumeth.2008.09.026 [DOI] [PubMed] [Google Scholar]
- 71. Chiappalone M., Novellino A., Vajda I., Vato A., Martinoia S., and van Pelt J., “ Burst detection algorithms for the analysis of spatio-temporal patterns in cortical networks of neurons,” Neurocomputing 65–66, 653–662 (2005). 10.1016/j.neucom.2004.10.094 [DOI] [Google Scholar]
- 72. Pasquale V., Martinoia S., and Chiappalone M., “ A self-adapting approach for the detection of bursts and network bursts in neuronal cultures,” J. Comput. Neurosci. 29(1–2), 213–229 (2010). 10.1007/s10827-009-0175-1 [DOI] [PubMed] [Google Scholar]
- 73. Kailath T., “ The divergence and Bhattacharyya distance measures in signal selection,” IEEE Trans. Commun. Technol. 15(1), 52–60 (1967). 10.1109/TCOM.1967.1089532 [DOI] [Google Scholar]
- 74. Nielsen F. and Boltz S., “ The Burbea-Rao and Bhattacharyya centroids,” IEEE Trans. Inf. Theory 57(8), 5455–5466 (2011). 10.1109/TIT.2011.2159046 [DOI] [Google Scholar]
- 75. Briët J. and Harremoës P., “ Properties of classical and quantum Jensen-Shannon divergence,” Phys. Rev. A 79(5), 052311 (2009). 10.1103/PhysRevA.79.052311 [DOI] [Google Scholar]
- 76. Nielsen F., “ On a generalization of the Jensen-Shannon divergence and the Jensen-Shannon centroid,” Entropy 22(2), 221 (2020). 10.3390/e22020221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martina Brofiga F. (2024). “ScreenNeuroPharm/3DNeuronalScaffolds: 3DNeuronalScaffolds_v2,” Zenodo. 10.5281/zenodo.13259912 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Martina Brofiga F. (2024). “ScreenNeuroPharm/3DNeuronalScaffolds: 3DNeuronalScaffolds_v2,” Zenodo. 10.5281/zenodo.13259912 [DOI]
Data Availability Statement
The data that support the findings of this study are openly available in Zenodo at https://doi.org/10.5281/zenodo.13259912, Ref. 77.