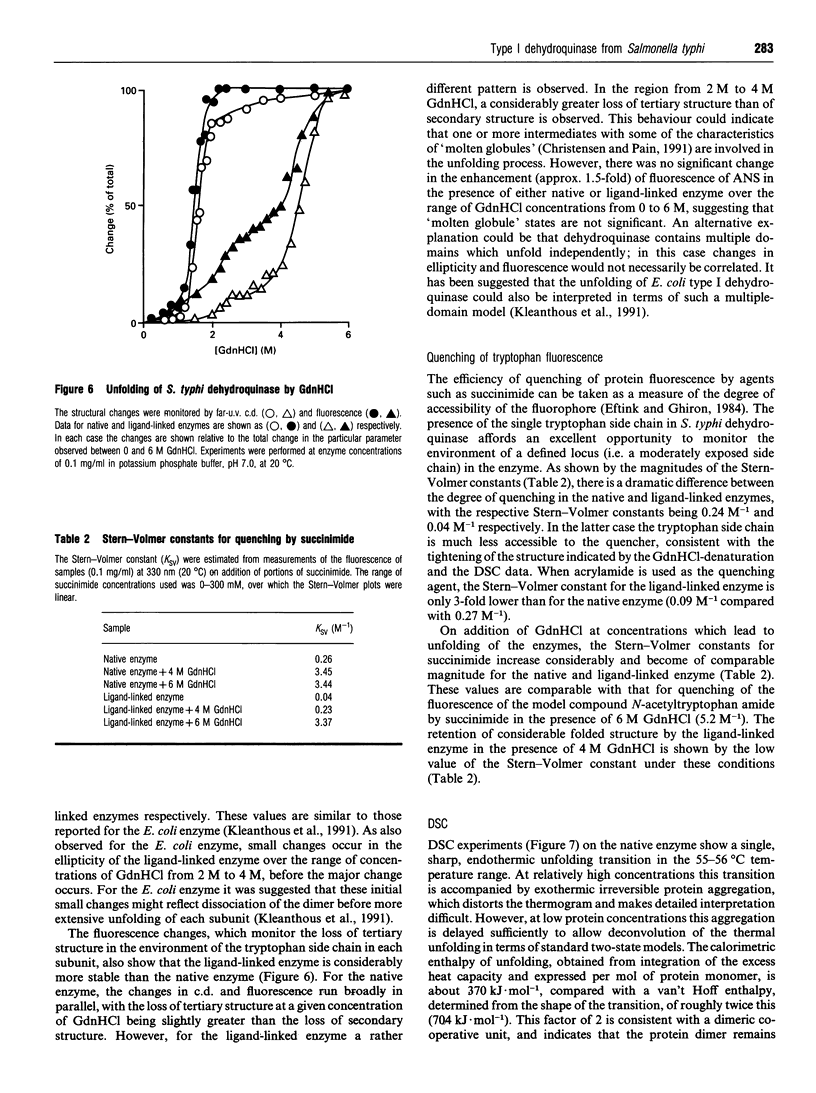
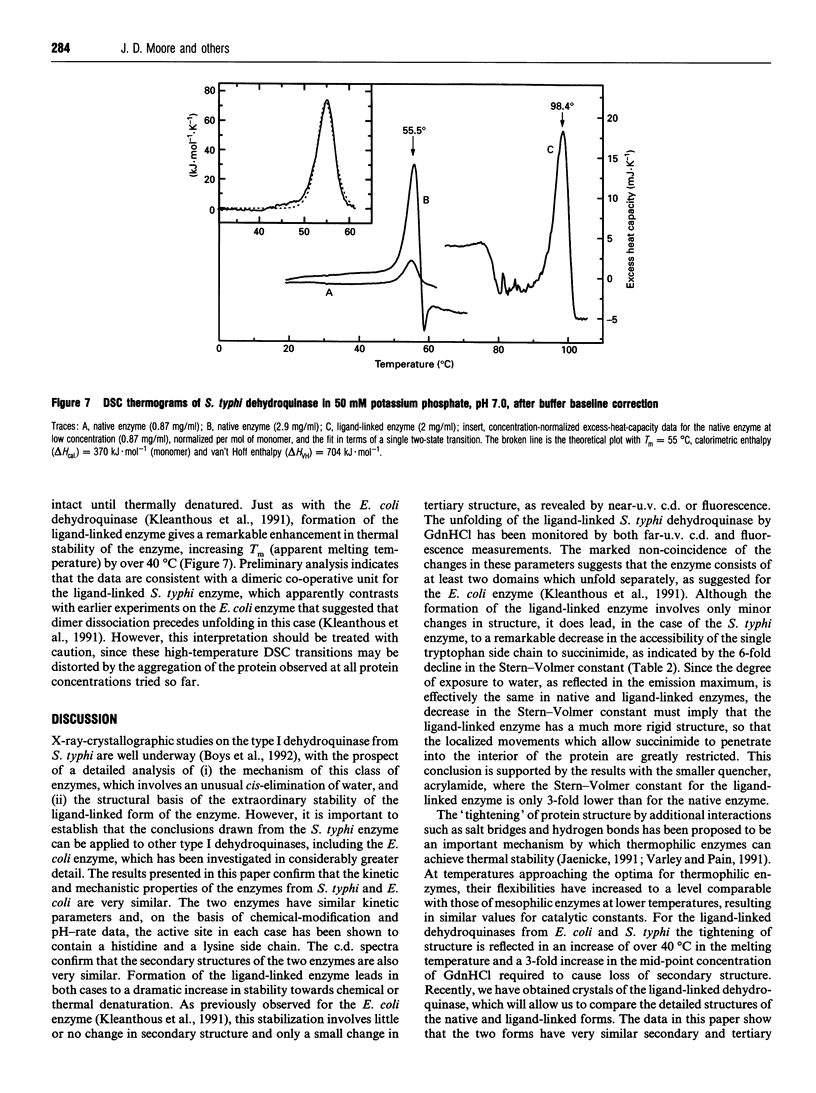
Abstract
The type I dehydroquinase from the human pathogen Salmonella typhi was overexpressed in an Escherichia coli host and purified to homogeneity. The S. typhi enzyme was characterized in terms of its kinetic parameters, important active-site residues, thermal stability and c.d. and fluorescence properties. In all important respects, the enzyme from S. typhi behaves in a very similar fashion to the well-characterized enzyme from E. coli, including the remarkable conformational stabilization observed on reduction of the substrate/product mixture by NaBH4. This gives confidence that the information from X-ray studies on the S. typhi enzyme [Boys, Fawcett, Sawyer, Moore, Charles, Hawkins, Deka, Kleanthous and Coggins (1992) J. Mol. Biol. 227, 352-355] can be applied to other type I dehydroquinases. Studies of the quenching of fluorescence of the S. typhi enzyme by succinimide show that NaBH4 reduction of the substrate/product imine complex involves a dramatic decrease in the flexibility of the enzyme, with only very minor changes in the overall secondary and tertiary structure.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger S. L. Diethyl pyrocarbonate: an examination of its properties in buffered solutions with a new assay technique. Anal Biochem. 1975 Aug;67(2):428–437. doi: 10.1016/0003-2697(75)90315-2. [DOI] [PubMed] [Google Scholar]
- Boys C. W., Bury S. M., Sawyer L., Moore J. D., Charles I. G., Hawkins A. R., Deka R., Kleanthous C., Coggins J. R. Crystallization of a type I 3-dehydroquinase from Salmonella typhi. J Mol Biol. 1992 Sep 5;227(1):352–355. doi: 10.1016/0022-2836(92)90704-n. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Charles I. G., Keyte J. W., Brammar W. J., Hawkins A. R. Nucleotide sequence encoding the biosynthetic dehydroquinase function of the penta-functional arom locus of Aspergillus nidulans. Nucleic Acids Res. 1985 Nov 25;13(22):8119–8128. doi: 10.1093/nar/13.22.8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles I. G., Keyte J. W., Brammar W. J., Smith M., Hawkins A. R. The isolation and nucleotide sequence of the complex AROM locus of Aspergillus nidulans. Nucleic Acids Res. 1986 Mar 11;14(5):2201–2213. doi: 10.1093/nar/14.5.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S., Duncan K., Graham L. D., Coggins J. R. Identification of the active-site lysine residues of two biosynthetic 3-dehydroquinases. Biochem J. 1991 Apr 1;275(Pt 1):1–6. doi: 10.1042/bj2750001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S., Lambert J. M., McColl L. A., Coggins J. R. Purification and characterization of 3-dehydroquinase from Escherichia coli. Biochem J. 1986 Nov 1;239(3):699–704. doi: 10.1042/bj2390699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen H., Pain R. H. Molten globule intermediates and protein folding. Eur Biophys J. 1991;19(5):221–229. doi: 10.1007/BF00183530. [DOI] [PubMed] [Google Scholar]
- Church F. C., Lundblad R. L., Noyes C. M. Modification of histidines in human prothrombin. Effect on the interaction of fibrinogen with thrombin from diethyl pyrocarbonate-modified prothrombin. J Biol Chem. 1985 Apr 25;260(8):4936–4940. [PubMed] [Google Scholar]
- Coggins J. R., Boocock M. R., Campbell M. S., Chaudhuri S., Lambert J. M., Lewendon A., Mousdale D. M., Smith D. D. Functional domains involved in aromatic amino acid biosynthesis. Biochem Soc Trans. 1985 Apr;13(2):299–303. doi: 10.1042/bst0130299. [DOI] [PubMed] [Google Scholar]
- Da Silva A. J., Whittington H., Clements J., Roberts C., Hawkins A. R. Sequence analysis and transformation by the catabolic 3-dehydroquinase (QUTE) gene from Aspergillus nidulans. Biochem J. 1986 Dec 1;240(2):481–488. doi: 10.1042/bj2400481. [DOI] [PMC free article] [PubMed] [Google Scholar]
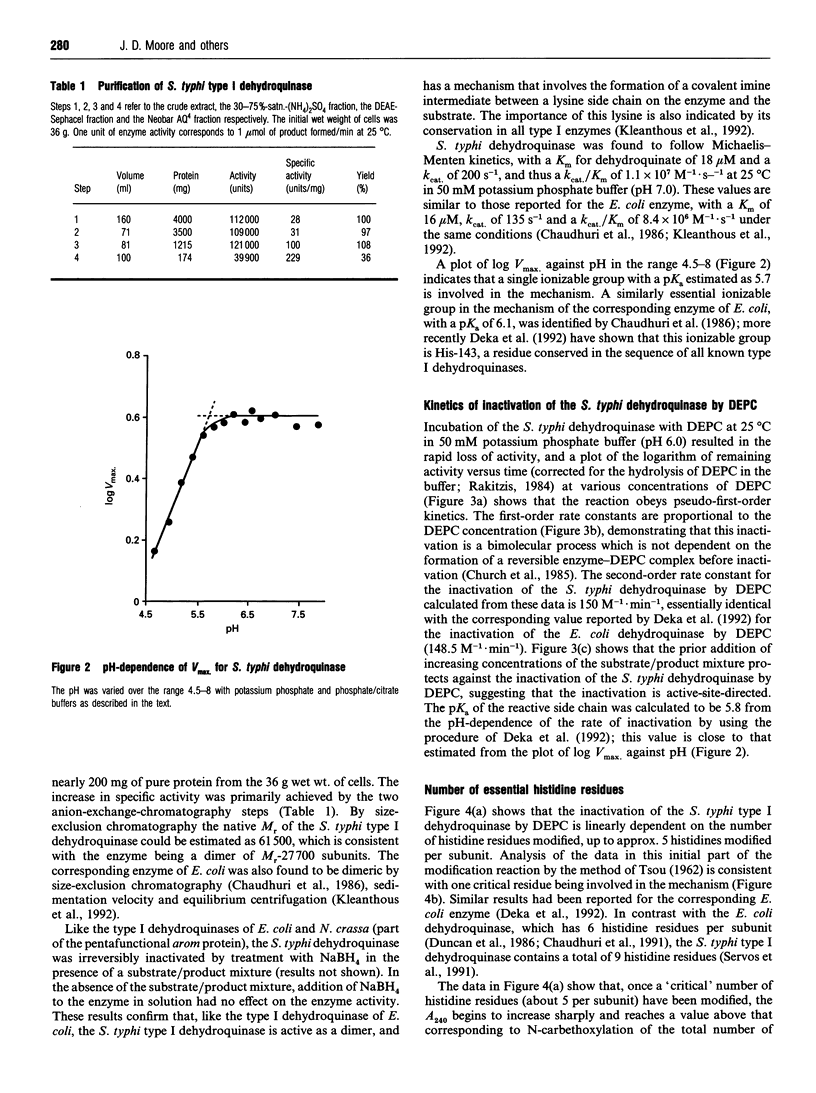
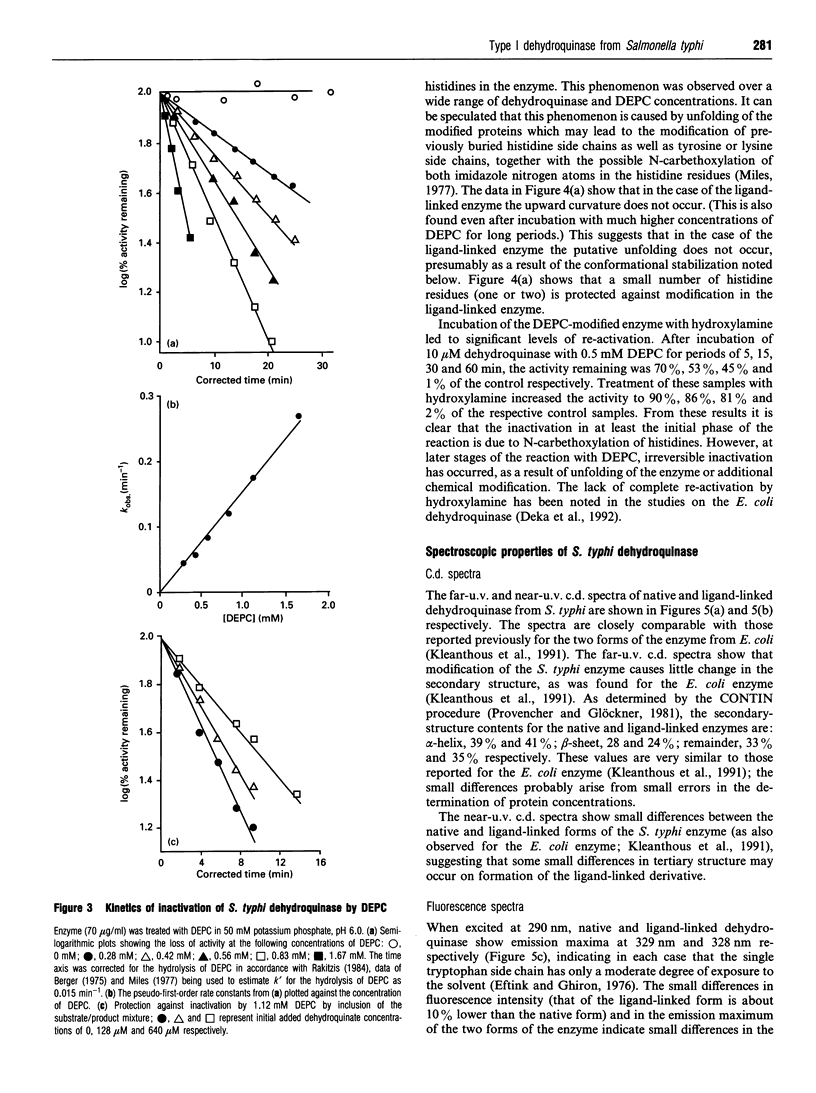
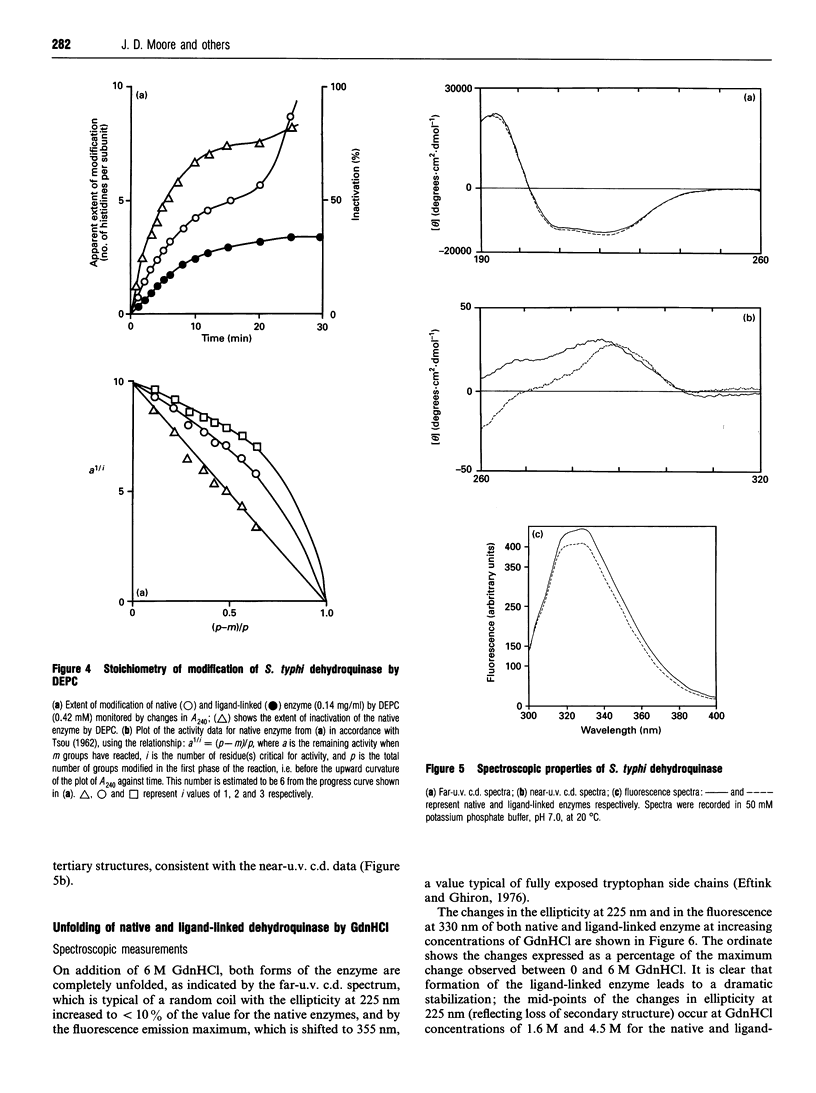
- Deka R. K., Kleanthous C., Coggins J. R. Identification of the essential histidine residue at the active site of Escherichia coli dehydroquinase. J Biol Chem. 1992 Nov 5;267(31):22237–22242. [PubMed] [Google Scholar]
- Duncan K., Chaudhuri S., Campbell M. S., Coggins J. R. The overexpression and complete amino acid sequence of Escherichia coli 3-dehydroquinase. Biochem J. 1986 Sep 1;238(2):475–483. doi: 10.1042/bj2380475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K., Edwards R. M., Coggins J. R. The pentafunctional arom enzyme of Saccharomyces cerevisiae is a mosaic of monofunctional domains. Biochem J. 1987 Sep 1;246(2):375–386. doi: 10.1042/bj2460375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftink M. R., Ghiron C. A. Exposure of tryptophanyl residues in proteins. Quantitative determination by fluorescence quenching studies. Biochemistry. 1976 Feb 10;15(3):672–680. doi: 10.1021/bi00648a035. [DOI] [PubMed] [Google Scholar]
- Euverink G. J., Hessels G. I., Vrijbloed J. W., Coggins J. R., Dijkhuizen L. Purification and characterization of a dual function 3-dehydroquinate dehydratase from Amycolatopsis methanolica. J Gen Microbiol. 1992 Nov;138(11):2449–2457. doi: 10.1099/00221287-138-11-2449. [DOI] [PubMed] [Google Scholar]
- Garbe T., Servos S., Hawkins A., Dimitriadis G., Young D., Dougan G., Charles I. The Mycobacterium tuberculosis shikimate pathway genes: evolutionary relationship between biosynthetic and catabolic 3-dehydroquinases. Mol Gen Genet. 1991 Sep;228(3):385–392. doi: 10.1007/BF00260631. [DOI] [PubMed] [Google Scholar]
- Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989 Nov 1;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R., Giles N. H., Kinghorn J. R. Genetical and biochemical aspects of quinate breakdown in the filamentous fungus Aspergillus nidulans. Biochem Genet. 1982 Apr;20(3-4):271–286. doi: 10.1007/BF00484424. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R., Smith M. Domain structure and interaction within the pentafunctional arom polypeptide. Eur J Biochem. 1991 Mar 28;196(3):717–724. doi: 10.1111/j.1432-1033.1991.tb15870.x. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R. The complex Arom locus of Aspergillus nidulans. Evidence for multiple gene fusions and convergent evolution. Curr Genet. 1987;11(6-7):491–498. doi: 10.1007/BF00384611. [DOI] [PubMed] [Google Scholar]
- Jaenicke R. Protein stability and molecular adaptation to extreme conditions. Eur J Biochem. 1991 Dec 18;202(3):715–728. doi: 10.1111/j.1432-1033.1991.tb16426.x. [DOI] [PubMed] [Google Scholar]
- Johnson C. M., Price N. C. Denaturation and renaturation of the monomeric phosphoglycerate mutase from Schizosaccharomyces pombe. Biochem J. 1987 Jul 15;245(2):525–530. doi: 10.1042/bj2450525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinghorn J. R., Hawkins A. R. Cloning and expression in Escherichia coli K-12 of the biosynthetic dehydroquinase function of the arom cluster gene from the eucaryote, Aspergillus nidulans. Mol Gen Genet. 1982;186(1):145–152. doi: 10.1007/BF00422927. [DOI] [PubMed] [Google Scholar]
- Kishore G. M., Shah D. M. Amino acid biosynthesis inhibitors as herbicides. Annu Rev Biochem. 1988;57:627–663. doi: 10.1146/annurev.bi.57.070188.003211. [DOI] [PubMed] [Google Scholar]
- Kleanthous C., Campbell D. G., Coggins J. R. Active site labeling of the shikimate pathway enzyme, dehydroquinase. Evidence for a common substrate binding site within dehydroquinase and dehydroquinate synthase. J Biol Chem. 1990 Jul 5;265(19):10929–10934. [PubMed] [Google Scholar]
- Kleanthous C., Coggins J. R. Reversible alkylation of an active site methionine residue in dehydroquinase. J Biol Chem. 1990 Jul 5;265(19):10935–10939. [PubMed] [Google Scholar]
- Kleanthous C., Deka R., Davis K., Kelly S. M., Cooper A., Harding S. E., Price N. C., Hawkins A. R., Coggins J. R. A comparison of the enzymological and biophysical properties of two distinct classes of dehydroquinase enzymes. Biochem J. 1992 Mar 15;282(Pt 3):687–695. doi: 10.1042/bj2820687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleanthous C., Reilly M., Cooper A., Kelly S., Price N. C., Coggins J. R. Stabilization of the shikimate pathway enzyme dehydroquinase by covalently bound ligand. J Biol Chem. 1991 Jun 15;266(17):10893–10898. [PubMed] [Google Scholar]
- Koshiba T. Purification of two forms of the associated 3-dehydroquinate hydro-lyase and shikimate:NADP+ oxidoreductase in Phaseolus mungo seedlings. Biochim Biophys Acta. 1978 Jan 12;522(1):10–18. doi: 10.1016/0005-2744(78)90317-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lumsden J., Coggins J. R. The subunit structure of the arom multienzyme complex of Neurospora crassa. A possible pentafunctional polypeptide chain. Biochem J. 1977 Mar 1;161(3):599–607. doi: 10.1042/bj1610599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles E. W. Modification of histidyl residues in proteins by diethylpyrocarbonate. Methods Enzymol. 1977;47:431–442. doi: 10.1016/0076-6879(77)47043-5. [DOI] [PubMed] [Google Scholar]
- Moore J. D., Hawkins A. R. Overproduction of, and interaction within, bifunctional domains from the amino- and carboxy-termini of the pentafunctional AROM protein of Aspergillus nidulans. Mol Gen Genet. 1993 Jul;240(1):92–102. doi: 10.1007/BF00276888. [DOI] [PubMed] [Google Scholar]
- Moore J. D., Lamb H. K., Garbe T., Servos S., Dougan G., Charles I. G., Hawkins A. R. Inducible overproduction of the Aspergillus nidulans pentafunctional AROM protein and the type-I and -II 3-dehydroquinases from Salmonella typhi and Mycobacterium tuberculosis. Biochem J. 1992 Oct 1;287(Pt 1):173–181. doi: 10.1042/bj2870173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki Y. The preparation of guanidine hydrochloride. Methods Enzymol. 1972;26:43–50. doi: 10.1016/s0076-6879(72)26005-0. [DOI] [PubMed] [Google Scholar]
- O'Callaghan D., Maskell D., Liew F. Y., Easmon C. S., Dougan G. Characterization of aromatic- and purine-dependent Salmonella typhimurium: attention, persistence, and ability to induce protective immunity in BALB/c mice. Infect Immun. 1988 Feb;56(2):419–423. doi: 10.1128/iai.56.2.419-423.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V. B., Giles N. H. Purification of the arom multienzyme aggregate from Euglena gracilis. Biochim Biophys Acta. 1979 Mar 16;567(1):24–34. doi: 10.1016/0005-2744(79)90168-2. [DOI] [PubMed] [Google Scholar]
- Provencher S. W., Glöckner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981 Jan 6;20(1):33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- Rakitzis E. T. Kinetics of protein modification reactions. Biochem J. 1984 Jan 15;217(2):341–351. doi: 10.1042/bj2170341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servos S., Chatfield S., Hone D., Levine M., Dimitriadis G., Pickard D., Dougan G., Fairweather N., Charles I. Molecular cloning and characterization of the aroD gene encoding 3-dehydroquinase from Salmonella typhi. J Gen Microbiol. 1991 Jan;137(1):147–152. doi: 10.1099/00221287-137-1-147. [DOI] [PubMed] [Google Scholar]
- Smith D. D., Coggins J. R. Isolation of a bifunctional domain from the pentafunctional arom enzyme complex of Neurospora crassa. Biochem J. 1983 Aug 1;213(2):405–415. doi: 10.1042/bj2130405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSOU C. L. Relation between modification of functional groups of proteins and their biological activity. I.A graphical method for the determination of the number and type of essential groups. Sci Sin. 1962 Nov;11:1535–1558. [PubMed] [Google Scholar]
- Varley P. G., Pain R. H. Relation between stability, dynamics and enzyme activity in 3-phosphoglycerate kinases from yeast and Thermus thermophilus. J Mol Biol. 1991 Jul 20;220(2):531–538. doi: 10.1016/0022-2836(91)90028-5. [DOI] [PubMed] [Google Scholar]
- Ward L. D. Measurement of ligand binding to proteins by fluorescence spectroscopy. Methods Enzymol. 1985;117:400–414. doi: 10.1016/s0076-6879(85)17024-2. [DOI] [PubMed] [Google Scholar]
- White P. J., Young J., Hunter I. S., Nimmo H. G., Coggins J. R. The purification and characterization of 3-dehydroquinase from Streptomyces coelicolor. Biochem J. 1990 Feb 1;265(3):735–738. doi: 10.1042/bj2650735. [DOI] [PMC free article] [PubMed] [Google Scholar]