Abstract
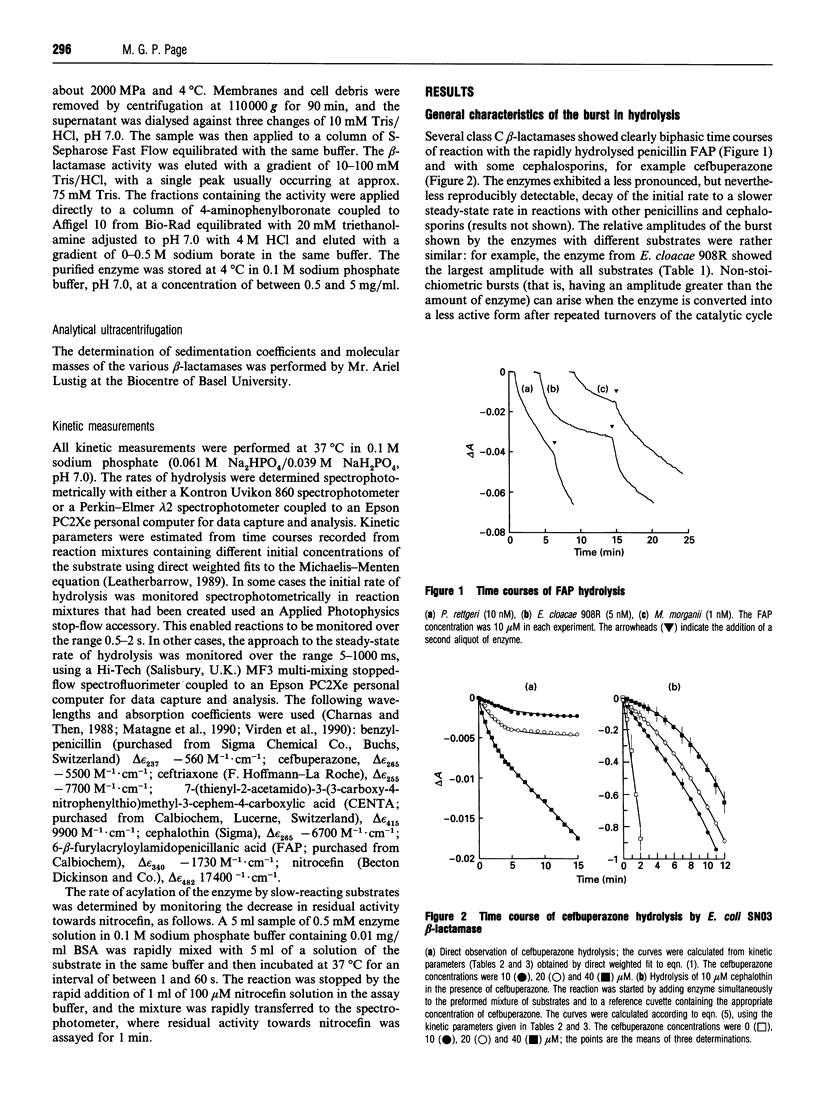
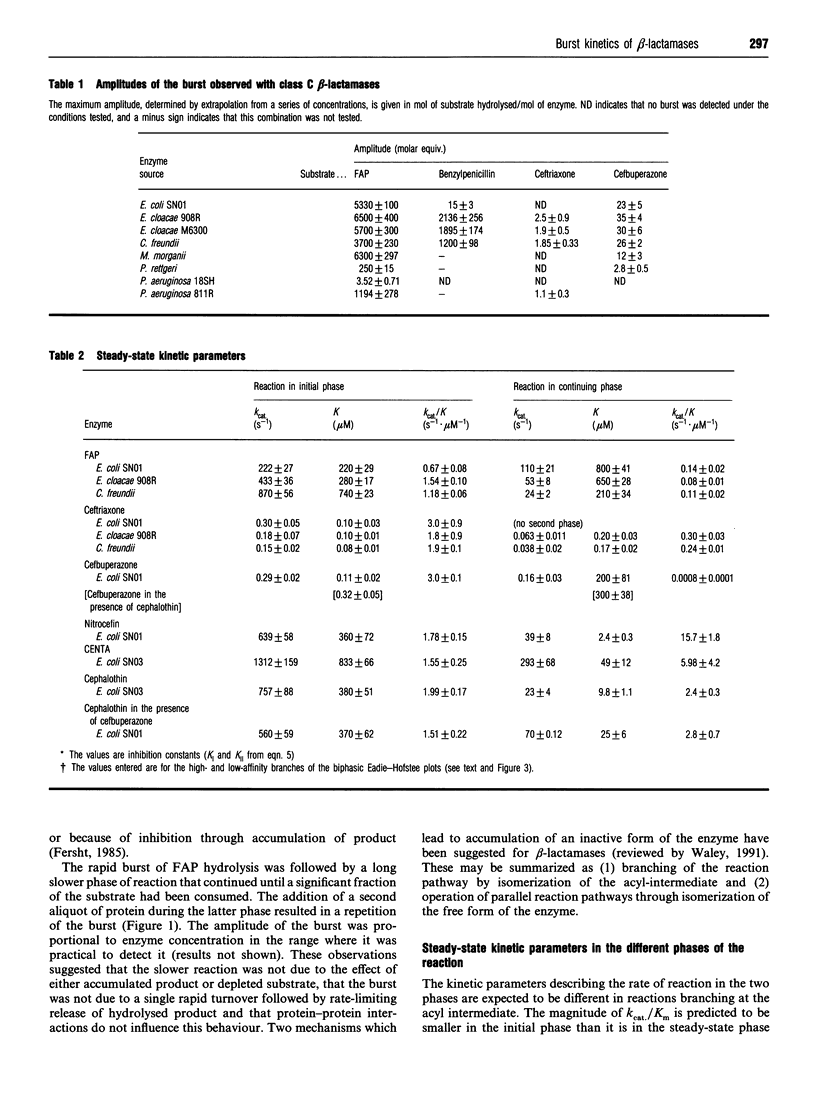
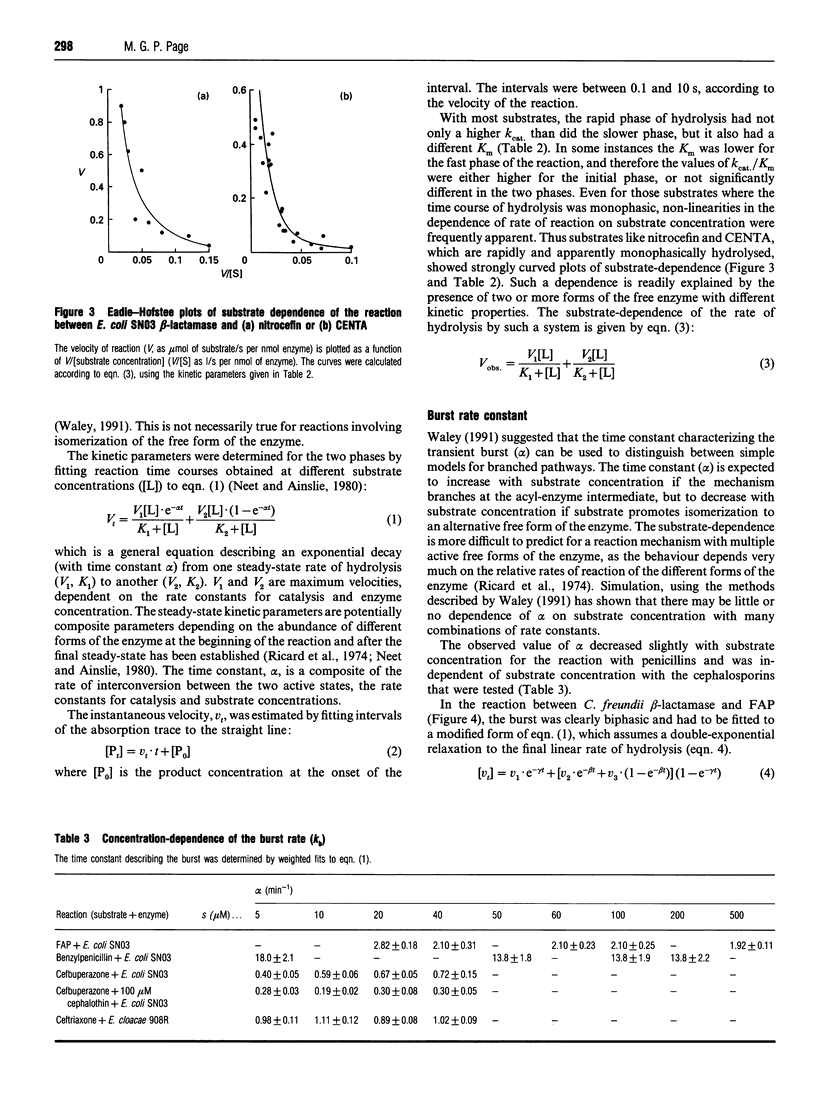
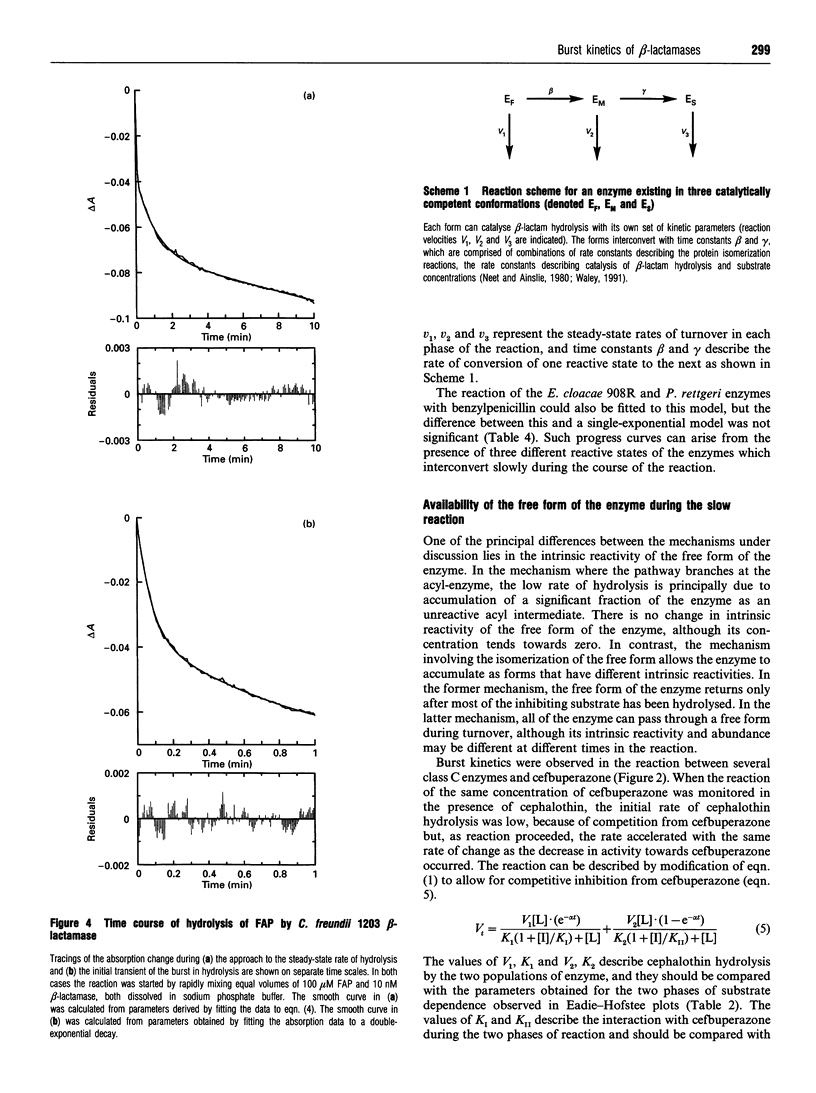
Class C beta-lactamases from Pseudomonas aeruginosa and several species of the Enterobacteriaceae have been observed to undergo a rapid burst in hydrolysis of beta-lactam antibiotics before relaxation to a steady-state rate of hydrolysis. The amplitude of the burst corresponds to the hydrolysis of between 1 and 10,000 mol of the substrate per mol of enzyme. The decay of the rate of hydrolysis in the burst phase comprises two exponential reactions, which indicates that there are three different reactive states of the enzymes. Examination of the kinetics of acylation by slowly reacting beta-lactams suggests that there are three forms of the free enzyme in slow equilibrium. Thus it would appear that the burst kinetics exhibited by class C enzymes can be attributed to redistribution of the enzyme between different conformations induced by the reaction with substrate.
Full text
PDF
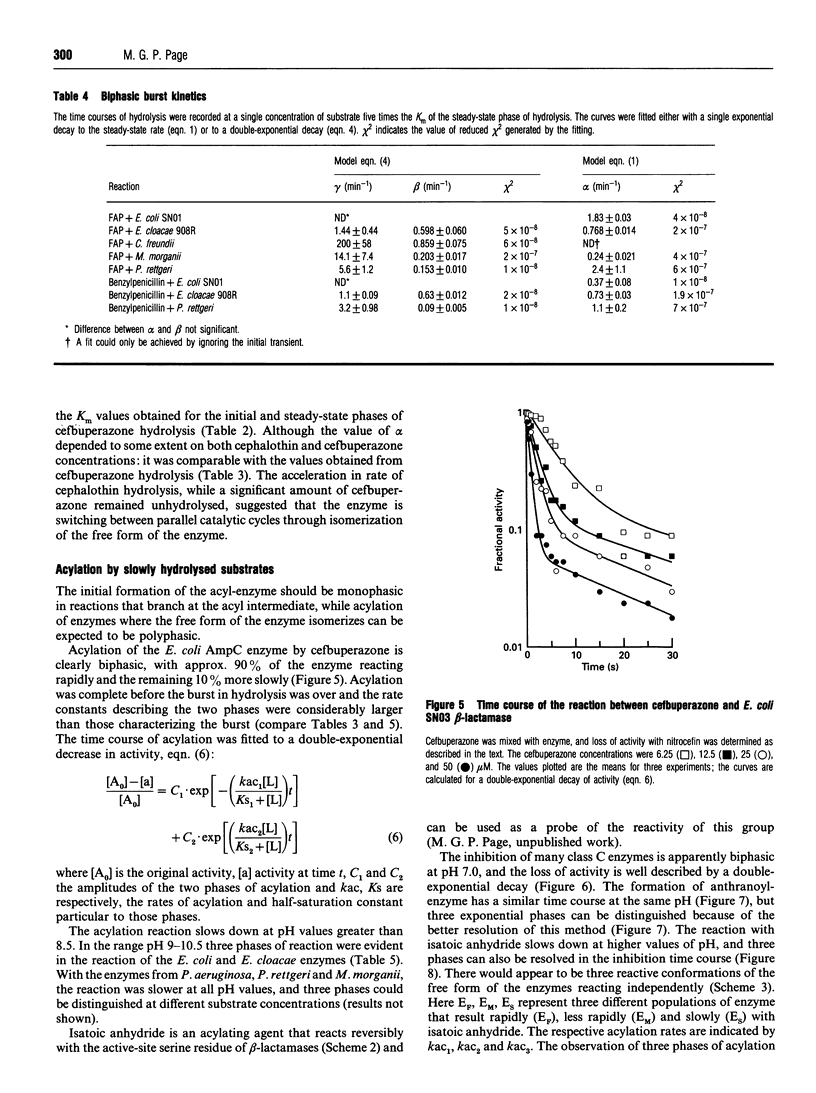
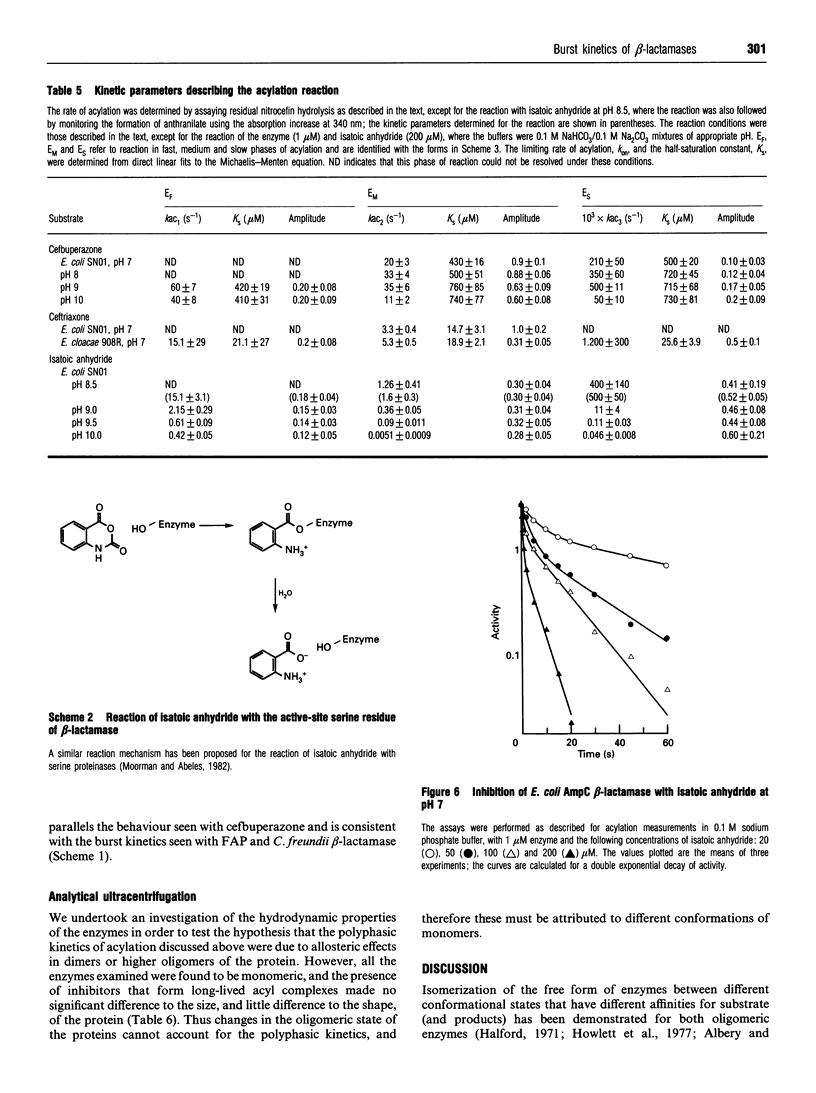
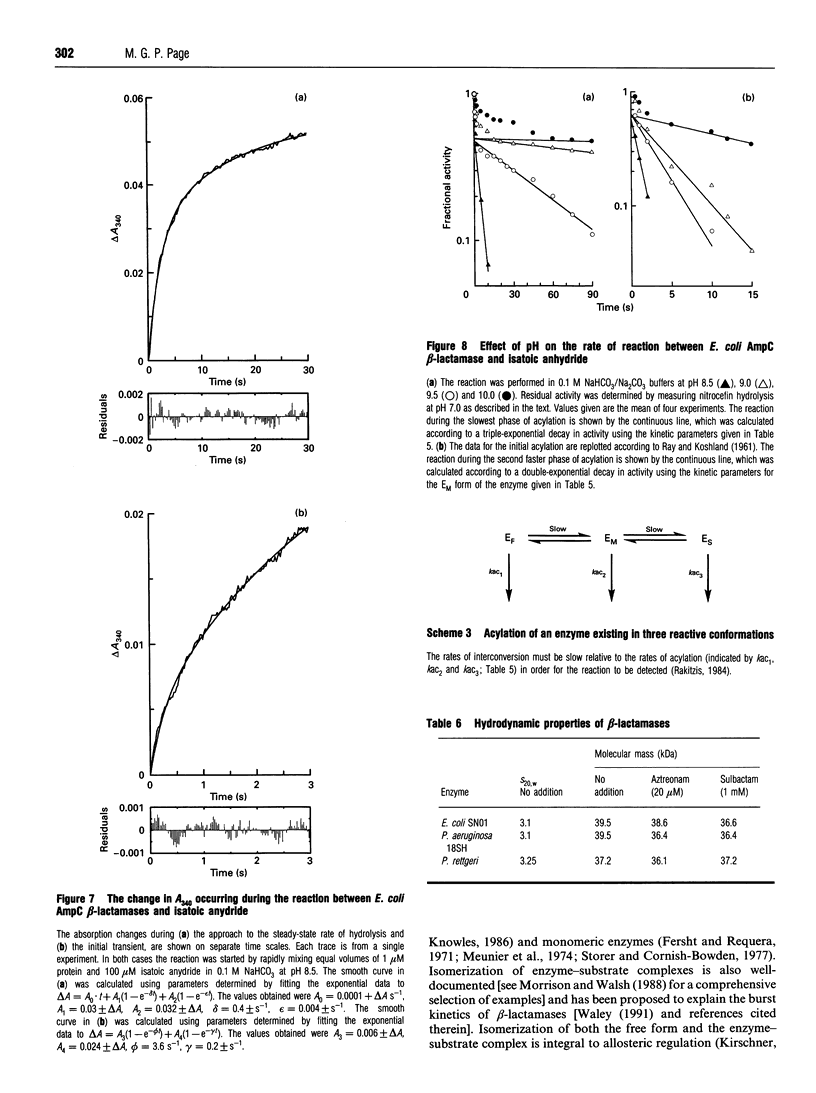
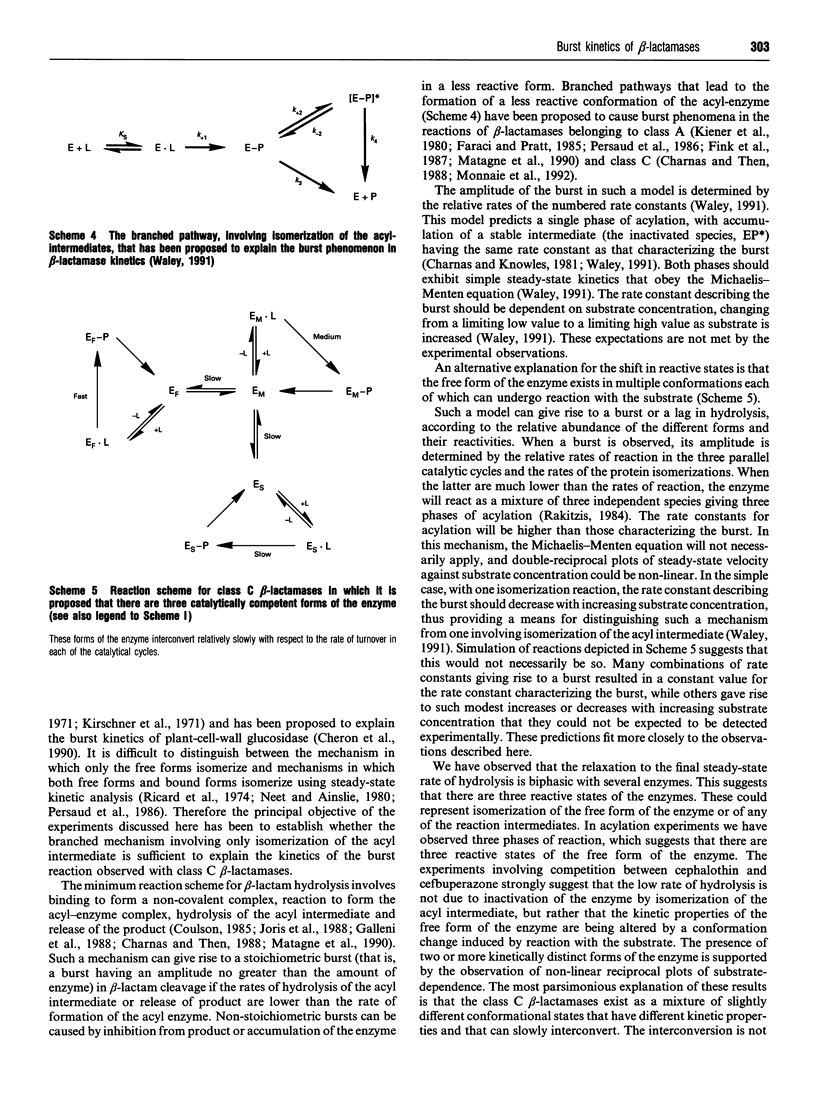

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albery W. J., Knowled J. R. Energetics and mechanism of proline racemase. Biochemistry. 1986 May 6;25(9):2572–2577. doi: 10.1021/bi00357a043. [DOI] [PubMed] [Google Scholar]
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Bush K. Characterization of beta-lactamases. Antimicrob Agents Chemother. 1989 Mar;33(3):259–263. doi: 10.1128/aac.33.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Classification of beta-lactamases: groups 1, 2a, 2b, and 2b'. Antimicrob Agents Chemother. 1989 Mar;33(3):264–270. doi: 10.1128/aac.33.3.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Classification of beta-lactamases: groups 2c, 2d, 2e, 3, and 4. Antimicrob Agents Chemother. 1989 Mar;33(3):271–276. doi: 10.1128/aac.33.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrey E. A., Virden R., Pain R. H. The reversible deactivation of beta-lactamase from Staphylococcus aureus by quinacillin and cephaloridine and its modification by antibodies. Biochim Biophys Acta. 1984 Mar 29;785(3):104–110. doi: 10.1016/0167-4838(84)90133-x. [DOI] [PubMed] [Google Scholar]
- Cartwright S. J., Tan A. K., Fink A. L. Trapping the acyl-enzyme intermediate in beta-lactamase I catalysis. Biochem J. 1989 Nov 1;263(3):905–912. doi: 10.1042/bj2630905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright S. J., Waley S. G. Purification of beta-lactamases by affinity chromatography on phenylboronic acid-agarose. Biochem J. 1984 Jul 15;221(2):505–512. doi: 10.1042/bj2210505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnas R. L., Knowles J. R. Inhibition of the RTEM beta-lactamase from Escherichia coli. Interaction of enzyme with derivatives of olivanic acid. Biochemistry. 1981 May 12;20(10):2732–2737. doi: 10.1021/bi00513a005. [DOI] [PubMed] [Google Scholar]
- Charnas R. L., Then R. L. Mechanism of inhibition of chromosomal beta-lactamases by third-generation cephalosporins. Rev Infect Dis. 1988 Jul-Aug;10(4):752–760. doi: 10.1093/clinids/10.4.752. [DOI] [PubMed] [Google Scholar]
- Cheron G., Noat G., Ricard J. Hysteresis of plant cell-wall beta-glucosidase. Biochem J. 1990 Jul 15;269(2):389–392. doi: 10.1042/bj2690389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen H., Martin M. T., Waley S. G. Beta-lactamases as fully efficient enzymes. Determination of all the rate constants in the acyl-enzyme mechanism. Biochem J. 1990 Mar 15;266(3):853–861. [PMC free article] [PubMed] [Google Scholar]
- Citri N., Samuni A., Zyk N. Acquisition of substrate-specific parameters during the catalytic reaction of penicillinase. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1048–1052. doi: 10.1073/pnas.73.4.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson A. Beta-lactamases: molecular studies. Biotechnol Genet Eng Rev. 1985;3:219–253. doi: 10.1080/02648725.1985.10647814. [DOI] [PubMed] [Google Scholar]
- Faraci W. S., Pratt R. F. Mechanism of inhibition of the PC1 beta-lactamase of Staphylococcus aureus by cephalosporins: importance of the 3'-leaving group. Biochemistry. 1985 Feb 12;24(4):903–910. doi: 10.1021/bi00325a014. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Requena Y. Equilibrium and rate constants for the interconversion of two conformations of -chymotrypsin. The existence of a catalytically inactive conformation at neutral p H. J Mol Biol. 1971 Sep 14;60(2):279–290. doi: 10.1016/0022-2836(71)90294-4. [DOI] [PubMed] [Google Scholar]
- Fink A. L., Behner K. M., Tan A. K. Kinetic and structural characterization of reversibly inactivated beta-lactamase. Biochemistry. 1987 Jul 14;26(14):4248–4258. doi: 10.1021/bi00388a011. [DOI] [PubMed] [Google Scholar]
- Follath F., Costa E., Thommen A., Frei R., Burdeska A., Meyer J. Clinical consequences of development of resistance to third generation cephalosporins. Eur J Clin Microbiol. 1987 Aug;6(4):446–450. doi: 10.1007/BF02013108. [DOI] [PubMed] [Google Scholar]
- Galleni M., Amicosante G., Frère J. M. A survey of the kinetic parameters of class C beta-lactamases. Cephalosporins and other beta-lactam compounds. Biochem J. 1988 Oct 1;255(1):123–129. doi: 10.1042/bj2550123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S. E. Escherichia coli alkaline phosphatase. An analysis of transient kinetics. Biochem J. 1971 Nov;125(1):319–327. doi: 10.1042/bj1250319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett G. J., Blackburn M. N., Compton J. G., Schachman H. K. Allosteric regulation of aspartate transcarbamoylase. Analysis of the structural and functional behavior in terms of a two-state model. Biochemistry. 1977 Nov 15;16(23):5091–5100. doi: 10.1021/bi00642a023. [DOI] [PubMed] [Google Scholar]
- Jaurin B., Grundström T., Edlund T., Normark S. The E. coli beta-lactamase attenuator mediates growth rate-dependent regulation. Nature. 1981 Mar 19;290(5803):221–225. doi: 10.1038/290221a0. [DOI] [PubMed] [Google Scholar]
- Kiener P. A., Knott-Hunziker V., Petursson S., Waley S. G. Mechanism of substrate-induced inactivation of beta-lactamase I. Eur J Biochem. 1980 Aug;109(2):575–580. doi: 10.1111/j.1432-1033.1980.tb04830.x. [DOI] [PubMed] [Google Scholar]
- Kirschner K. Co-operative binding of nicotinamide-adenine dinucleotide to yeast glyceraldehyde-3-phosphate dehydrogenase. II. Stopped-flow studies at pH 8-5 and 40 degrees C. J Mol Biol. 1971 May 28;58(1):51–68. doi: 10.1016/0022-2836(71)90231-2. [DOI] [PubMed] [Google Scholar]
- Kirschner K., Gallego E., Schuster I., Goodall D. Co-operative binding of nicotinamide-adenine dinucleotide to yeast glyceraldehyde-3-phosphate dehydrogenase. I. Equilibrium and temperature-jump studies at pH 8-5 and 40 degrees C. J Mol Biol. 1971 May 28;58(1):29–50. doi: 10.1016/0022-2836(71)90230-0. [DOI] [PubMed] [Google Scholar]
- Matagne A., Misselyn-Bauduin A. M., Joris B., Erpicum T., Granier B., Frère J. M. The diversity of the catalytic properties of class A beta-lactamases. Biochem J. 1990 Jan 1;265(1):131–146. doi: 10.1042/bj2650131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier J. C., Buc J., Navarro A., Ricard J. Regulatory behavior of monomeric enzymes. 2. A wheat-germ hexokinase as a mnemonical enzyme. Eur J Biochem. 1974 Nov 1;49(1):209–223. doi: 10.1111/j.1432-1033.1974.tb03826.x. [DOI] [PubMed] [Google Scholar]
- Monnaie D., Virden R., Frère J. M. A rapid-kinetic study of the class C beta-lactamase of Enterobacter cloacae 908R. FEBS Lett. 1992 Jul 20;306(2-3):108–112. doi: 10.1016/0014-5793(92)80979-q. [DOI] [PubMed] [Google Scholar]
- Morrison J. F., Walsh C. T. The behavior and significance of slow-binding enzyme inhibitors. Adv Enzymol Relat Areas Mol Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- Neet K. E., Ainslie G. R., Jr Hysteretic enzymes. Methods Enzymol. 1980;64:192–226. doi: 10.1016/s0076-6879(80)64010-5. [DOI] [PubMed] [Google Scholar]
- Normark S., Burman L. G. Resistance of Escherichia coli to penicillins: fine-structure mapping and dominance of chromosomal beta-lactamase mutations. J Bacteriol. 1977 Oct;132(1):1–7. doi: 10.1128/jb.132.1.1-7.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oefner C., D'Arcy A., Daly J. J., Gubernator K., Charnas R. L., Heinze I., Hubschwerlen C., Winkler F. K. Refined crystal structure of beta-lactamase from Citrobacter freundii indicates a mechanism for beta-lactam hydrolysis. Nature. 1990 Jan 18;343(6255):284–288. doi: 10.1038/343284a0. [DOI] [PubMed] [Google Scholar]
- Persaud K. C., Pain R. H., Virden R. Reversible deactivation of beta-lactamase by quinacillin. Extent of the conformational change in the isolated transitory complex. Biochem J. 1986 Aug 1;237(3):723–730. doi: 10.1042/bj2370723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAY W. J., Jr, KOSHLAND D. E., Jr A method for characterizing the type and numbers of groups involved in enzyme action. J Biol Chem. 1961 Jul;236:1973–1979. [PubMed] [Google Scholar]
- Rakitzis E. T. Kinetics of protein modification reactions. Biochem J. 1984 Jan 15;217(2):341–351. doi: 10.1042/bj2170341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard J., Meunier J. C., Buc J. Regulatory behavior of monomeric enzymes. 1. The mnemonical enzyme concept. Eur J Biochem. 1974 Nov 1;49(1):195–208. doi: 10.1111/j.1432-1033.1974.tb03825.x. [DOI] [PubMed] [Google Scholar]
- Storer A. C., Cornish-Bowden A. Kinetic evidence for a 'mnemonical' mechanism for rat liver glucokinase. Biochem J. 1977 Jul 1;165(1):61–69. doi: 10.1042/bj1650061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Then R. L., Angehrn P. Multiply resistant mutants of Enterobacter cloacae selected by beta-lactam antibiotics. Antimicrob Agents Chemother. 1986 Nov;30(5):684–688. doi: 10.1128/aac.30.5.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Then R. L., Charnas R. L., Kocher H. P., Manneberg M., Röthlisberger U., Stocker J. Biochemical characterization of type A and type B beta-lactamase from Enterobacter cloacae. Rev Infect Dis. 1988 Jul-Aug;10(4):714–720. doi: 10.1093/clinids/10.4.714. [DOI] [PubMed] [Google Scholar]
- Tsukamoto K., Nishida N., Tsuruoka M., Sawai T. Function of the conserved triad residues in the class C beta-lactamase from Citrobacter freundii GN346. FEBS Lett. 1990 Oct 1;271(1-2):243–246. doi: 10.1016/0014-5793(90)80416-g. [DOI] [PubMed] [Google Scholar]
- Tsukamoto K., Tachibana K., Yamazaki N., Ishii Y., Ujiie K., Nishida N., Sawai T. Role of lysine-67 in the active site of class C beta-lactamase from Citrobacter freundii GN346. Eur J Biochem. 1990 Feb 22;188(1):15–22. doi: 10.1111/j.1432-1033.1990.tb15365.x. [DOI] [PubMed] [Google Scholar]
- Virden R., Tan A. K., Fink A. L. Cryoenzymology of staphylococcal beta-lactamase: trapping a serine-70-linked acyl-enzyme. Biochemistry. 1990 Jan 9;29(1):145–153. doi: 10.1021/bi00453a018. [DOI] [PubMed] [Google Scholar]
- Waley S. G. The kinetics of substrate-induced inactivation. Biochem J. 1991 Oct 1;279(Pt 1):87–94. doi: 10.1042/bj2790087. [DOI] [PMC free article] [PubMed] [Google Scholar]



