Abstract
Neurotransmitter transporters of the SLC6 family play an important role in the removal of neurotransmitters in brain tissue and in amino acid transport in epithelial cells. Here we demonstrate that the mouse homologue of slc6a20 has all properties of the long-sought IMINO system. The mouse has two homologues corresponding to the single human SLC6A20 gene: these have been named XT3 and XT3s1. Expression of mouse XT3s1, but not XT3, in Xenopus laevis oocytes induced an electrogenic Na+-and-Cl−-dependent transporter for proline, hydroxyproline, betaine, N-methylaminoisobutyric acid and pipecolic acid. Expression of XT3s1 was found in brain, kidney, small intestine, thymus, spleen and lung, whereas XT3 prevailed in kidney and lung. Accordingly we suggest that the two homologues be termed ‘XT3s1 IMINOB’ and ‘XT3 IMINOK’ to indicate the tissue expression of the two genes.
Keywords: amino acid transport, iminoglycinuria, neurotransmitter transporter family, orphan transporter, XT3, XTRP3
Abbreviations: (Me)AIB, (N-methyl)aminoisobutyric acid; NCBI, National Center for Biotechnology Information; NMDG-Cl, N-methyl-D-glucamine chloride; RT, reverse transcription; XT3s1, X transporter protein 3 similar 1
INTRODUCTION
The transport of proline has been studied extensively in epithelial cells from kidney and intestine (for reviews, see [1–3]). The amino acid is completely resorbed in the proximal tubule of the kidney and also efficiently absorbed in the intestine. Several routes have been described for the epithelial transport of proline: first a specific Na+-dependent transporter for proline and hydroxyproline, called the IMINO system [4,5], secondly a transporter shared by proline and glycine and some other amino acids, such as GABA (γ-aminobutyric acid) and β-alanine, which has been termed the ‘imino acid carrier’ [6,7], and, thirdly, the Na+-dependent neutral-amino-acid-transport system, B0 [8]. Depending on the tissue and species, either the IMINO system or the imino acid carrier may carry the main load of proline transport in epithelia [9]. The molecular correlate of the imino acid carrier is the proton/amino acid transporter PAT1 [10]. System B0 is encoded by B0AT1 gene [11], but the molecular identity of the IMINO system has remained elusive until very recently [12].
The IMINO system has been characterized in some detail in vesicles from rabbit jejunum [4,5,9,13]. These studies indicate that proline is co-transported with Na+ and Cl− ions with a stoichiometry of 1:2:1. Na+ ions are thought to bind to the transporter before the substrate does. At physiological Na+ concentrations, the Km for proline is about 0.3 mM. Proline is the only proteinogenic amino acid accepted by the IMINO system. Additional substrates are L-pipecolate, hydroxyproline, proline methyl ester, betaine and MeAIB (N-methylaminoisobutyric acid). The transporter is stereoselective, preferring L-proline over D-proline [4].
The SLC6 family contains a number of orphan transporters. Recently we identified a new member of the SLC6 family (B0AT1, SLC6A19) that is closely related to the orphan transporters and transports neutral amino acids [11]. Following up on this observation, we assumed that other orphan transporters may be amino acid transporters as well. Here we show that mouse slc6a20 is indeed the long-sought IMINO system.
MATERIALS AND METHODS
cDNA cloning and plasmids
To isolate the cDNA for XT3 and XT3s1, RNA was purified from mouse kidney and brain (Nucleospin RNA-L kit; Macherey–Nagel, Düren, Germany). RT (reverse transcription) was carried out using Superscript II-RT according to the manufacturer's (Invitrogen, Mulgrave, VIC, Australia) instructions. The cDNA was purified (PCR purification kit; Qiagen, Clifton Hill, VIC, Australia) and used as a template for PCR. The coding sequence of both transporters was amplified (Pfu polymerase; Promega, Madison, WI, U.S.A.) during 30 PCR cycles using the following temperature profile: 94 °C, 30 s; 55 °C, 60 s; and 72 °C, 12 min. XT3s1 (X transporter protein 3 similar 1) was amplified from brain cDNA using the sense primer 5′-CCACCATGGAGAAGGCACGG and the antisense primer 5′-TCTTCTGCCAAGCTCCCTAA, corresponding to bases 39–54 and 1933–1952 of NCBI (National Center for Biotechnology Information) database accession no. NM_139142. XT3 was amplified from kidney cDNA using the sense primer 5′-CCACCATGGAATCACCTTCAG and the antisense primer 5′-TCTGTGGGACCTCCAGTTCT, corresponding to bases 118–133 and 2025–2044 of NCBI database accession no. NM_011731 [14]. Both sense primers included a Kozak sequence (underlined) in front of the start codon (in boldface). The amplified PCR products of XT3 (1932 bp) and XT3s1 (1918 bp) were purified by gel elution and cloned into pCR-BluntII-TOPO (Invitrogen). The sequence of XT3 and XT3s1 was determined by BigDye Terminator v3.1 cycle sequencing (the service being provided by the Biomolecular Resource Facility at the Australian National University). The sequences were found to be identical with those deposited in the NCBI database (XT3s1, NM_139142; XT3, NM_011731). For expression studies, mouse XT3 and XT3s1 were excised with EcoRI and inserted into the corresponding site of the oocyte expression vector pGEM-He-Juel [15].
Oocytes, injections and flux studies
Oocyte isolation and maintenance have been described in detail previously [16]. For expression, both plasmids were linearized with NotI and transcribed in vitro using the T7 mMessage mMachine Kit (Ambion, Austin, TX, U.S.A.). Oocytes were each injected with 20 ng of cRNA encoding mouse XT3s1 or XT3. Oocyte transport measurements were carried out 3–6 days after injection as has been recently described in detail [16]. ND96 (96 mM NaCl/2 mM KCl/1.8 mM CaCl2/1 mM MgCl2/5 mM Hepes, titrated with NaOH to pH 7.4) was used in all transport assays unless indicated otherwise. Uptake of [14C]proline increased linearly with time for more than 30 min. As a result, uptake was determined using an incubation period of 10–15 min. Radiochemicals were purchased from Amersham Pharmacia Biotech (Castle Hill, NSW, Australia).
Electrophysiological recordings
Amino-acid-induced currents were analysed by two-electrode voltage clamp recording. The recordings were performed with 1×LU and 10×MGU headstages connected to a Geneclamp 500B electronic amplifier (Axon Instruments, Union City, CA, U.S.A.). The output signal was amplified ten times and filtered at 50 Hz. Data were sampled at 3 Hz using a Digidata 1322A and pCLAMP software (Axon Instruments). Oocytes were chosen that had a membrane potential of more negative than −30 mV. Once a stable membrane potential was reached under current-clamp conditions, the amplifier was switched to voltage-clamp mode, holding the oocytes at −50 mV. Oocytes were superfused with ND96 containing different substrates as indicated. A complete change of the bath to a new solution was accomplished in about 10 s.
RT-PCR
Total RNA was isolated from male adult NRMI mouse tissues using the Nucleospin RNA-L Kit (Macherey–Nagel). RT was carried out using Superscript II-RT according to the manufacturer's (Invitrogen) instructions. A standard PCR protocol with 100 pmol of each primer and a 2 μl aliquot of the purified cDNA was used for amplification of the fragments during 30 cycles (95 °C, 30 s; 60 °C, 1 min; 72 °C, 2 min) in a Thermocycler using Platinum Taq polymerase (Invitrogen). The mXT3s1-specific fragment was amplified with sense primer [5′-TCCAGGAGAATGGAGGAGTG (corresponding to positions 518–537) and antisense primer [5′-TCTTCTGCCAAGCTCCCTAA (corresponding to positions 1933–1952)]. The XT3-specific fragment was amplified with sense primer [5′-ATCAACACCTGGGCTCTTTG (corresponding to positions 559–578) and antisense primer: 5′-AAACCTGAAATGGCCTCCTT (corresponding to positions 1534–1553)]. A 1 kb actin cDNA fragment was amplified during 30 cycles as a control using the sense primer 5′-GCTCACCATGGATGATGATATCGC-3′ and the antisense primer 5′-GGAGGAGCAATGATCTTGATCTTC-3′.
In situ hybridization
Tissue specimens of brain, kidney, liver, pancreas, small intestine, spleen, heart and skeletal muscle from C57BL/6, SWR/J and DBA1/J mice were fixed in 4% (w/v) paraformaldehyde/0.1 M sodium phosphate buffer, pH 7.2, for 4 h and embedded in paraffin. Tissue sections, 5 μm in thickness, were dewaxed and hybridized as previously described [11,17]. Hybridization probes were generated by in vitro transcription of mXT3 cDNA cloned into pCR-blunt II-TOPO using SP6 polymerase (antisense) and T7 polymerase (sense). Following washing steps, the slide preparations were dipped in NTB2 emulsion (Kodak, Rochester, NY, U.S.A.) and exposed at 4 °C for 6 weeks. After development, the slides were stained with haematoxylin/eosin and photographed with a Sony DSC digital camera.
Calculations, statistics and computer analysis
Each datapoint or bar in flux experiments represents the activity (mean±S.D.) for eight XT3s1- or XT3-expressing oocytes minus the activity (mean±S.D.) for eight non-injected oocytes. For electrophysiological recordings, all experiments were performed with at least seven different oocytes, each from at least two different oocyte batches. The number of independent experiments (e) is indicated in the Figure legends. Kinetic constants were derived by non-linear curve-fitting using Origin7.0 software (OriginLab Corporation, Northampton, MA, U.S.A.). To determine K0.5 (half-saturation constant), Imax (maximum current) and h (Hill coefficient), the modified Michaelis–Menten equation:
 |
was used, where I is actual current.
RESULTS
Tissue distribution and cellular distribution of XT3 and XT3s1
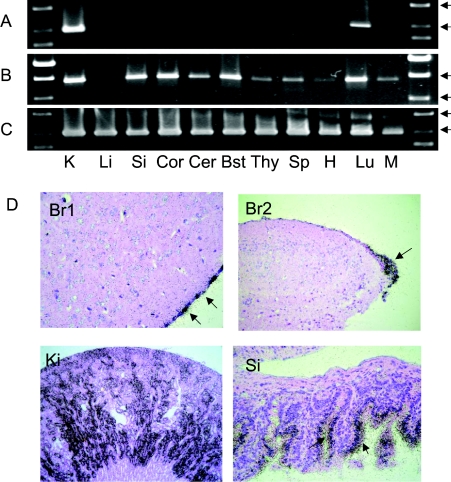
In the human genome, SLC6A20 is a unique gene located on chromosome 3p21.3. Instead of a single gene, two highly homologous genes are found in mouse. They are referred to as XT3 [14] and XT3s1 (‘similar to XT3’) [18] and lie next to each other on chromosome 9F4. To reveal the possible function of the two isoforms, we determined their tissue distribution by RT-PCR (Figures 1A–1C). XT3 was expressed in kidney and lung only (Figure 1A), whereas XT3s1 was found in brain, kidney, small intestine, thymus, spleen and lung (Figure 1B). In the brain, XT3s1 was found in cerebellum, cortex and brain stem, but not in liver, muscle and heart.
Figure 1. Expression of XT3 and XT3s1 in mouse tissues.
Total RNA was isolated from different mouse tissues and reverse-transcribed into cDNA. Specific fragments were amplified for XT3 (A), XT3s1 (B) and for actin (C). Tissues are labelled as follows: K, kidney; Li, liver; Si, small intestine; Cor, brain cortex; Cer, brain cerebellum; Bst, brain stem; Thy, thymus; Sp, spleen; H, heart; Lu, lung; M, muscle. Markers were loaded on each side of the gel. The arrows in all three panels point to standards of 1000 and 1500 bp in length. (D) In situ hybridization of XT3/XT3s1 in mouse tissues. The antisense probe was hybridized to mRNA in sections from brain (Br), small intestine (Si), kidney (Ki). In the brain the highest expression was found in the pia mater (arrow, panel Br1) and the plexus choroideus (arrow, panel Br2). In the kidney, XT3/XT3s1 mRNA was detected throughout the cortex. In the small intestine mRNA localized in villus enterocytes, with the highest expression in basal cells (arrows).
In order to localize the transcriptional activity of XT3 and XT3s1 in mouse organs, we performed radioactive in situ hybridization assays on paraffin-embedded tissue sections. It is worth noting that it is not possible to discriminate between XT3 and XT3s1 in these experiments, because their DNA sequences are 92% identical (including uninterrupted stretches of identical sequence of up to 100 bp). Hybridization with the 35S-labelled antisense XT3 probe revealed significant expression of XT3/XT3s1 in brain (Figure 1D, panels Br1 and Br2), kidney (panel Ki), small intestine (panel Si), and spleen (result not shown); other organs, such as skeletal muscle, heart, liver or pancreas gave negative results (not shown). No autoradiographic signals were detected when using the 35S-labelled sense control probe (results not shown). In kidney, XT3/XT3s1 transcripts were confined to the renal cortex. The signal was broadly distributed throughout the cortex, suggesting that XT3/XT3s1 was localized in all three segments of the proximal tubule. The medulla and the glomeruli were consistently hybridization-negative. In the small intestine, XT3/XT3s1 mRNA localized to villus enterocytes, with the highest expression in basal cells. Other cell types in the small intestine gave a negative response. In the brain, XT3/XT3s1 transcripts were consistently detected in epithelial cells of the pia mater covering the brain (panel Br1) and in the plexus choroideus (panel Br2). In addition, single hybridization-positive cells of unidentified origin were observed in brain tissue.
Cloning and functional properties of mouse XT3 and XT3s1
As a result of the expression data, full-length cDNAs of XT3 and XT3s1 were isolated from kidney and brain respectively. Expression of mouse XT3s1 in Xenopus laevis oocytes resulted in a more than 30-fold increase of proline-uptake activity compared with control oocytes (Figure 2A). Other amino acids, such as glutamine, glutamate, histidine, glycine and arginine, were not actively transported. A very small uptake activity was observed for phenylalanine, leucine and alanine. XT3-expressing oocytes, by contrast, did not show increased uptake of any of the tested amino acids (Figure 2A). To determine the substrate specificity of XT3s1 more precisely, we used competition experiments and electrophysiological recordings. Uptake of 50 μM [14C]proline was strongly inhibited by a 100-fold excess of proline itself, by MeAIB and betaine. A slight inhibition was caused by AIB (aminoisobutyric acid), but not by other amino acids (Figure 2B). Superfusion of XT3s1-expressing oocytes with substrates resulted in robust inward currents (Figure 2C). Amino acids with secondary, tertiary or quarternary amine groups were the preferred substrates of the transporter (Figure 2D). Neutral amino acids, such as phenylalanine, leucine and alanine, and the amino acid analogue AIB induced smaller, but significant, currents. Analysis of the substrate K0.5 values (Table 1) further revealed that the transporter was stereoselective and that piperidine-2-carboxylic acid (or pipecolic acid, a homologue of proline with a six-atom ring) had the highest affinity for the transporter. Movement of the carboxy group out of the α-position (nipecotic acid or piperidine-3-carboxylic acid) drastically decreased the affinity for the transporter.
Figure 2. Substrate specificity of XT3s1.
Oocytes were injected with XT3 or XT3s1 cRNA or remained uninjected (n.i.) in the controls. (A) Uptake of [14C]glutamine, [14C]glutamate, [14C]leucine, [14C]histidine, [14C]alanine, [14C]phenylalanine, [14C]glycine, [14C]proline and [14C]arginine (100 μM each) was determined 4 days after injection in oocytes expressing XT3 (grey bars), XT3s1 (black bars) and in non-injected oocytes (white bars). (B) Uptake of 50 μM [14C]proline was challenged by 5 mM unlabelled amino acids and their analogues. Each bar represents the transport activity (mean±S.D.) for ten oocytes (e=3). Abbreviations: BCH, 2-aminobicyclo[2.2.1]-heptane-2-carboxylic acid; Bet, betaine. (C) Original tracings of substrate-induced currents (0.5 mM) in XT3s1-expressing oocytes after 4 days incubation. Superfusion intervals are indicated by bars. Abbreviation: HO-Pro, hydroxyproline. (D) Summary of substrate-induced inward currents. Substrate-induced currents (0.5 mM) were compiled from experiments of the type shown in (C) and normalized to proline-induced currents. The experiment was performed with seven oocytes (e=3). Noninjected oocytes showed inward currents of 2–3 nA in response to the same panel of substrates. Abbreviations: Nip, nipecotic acid; Pip, pipecolic acid.
Table 1. Substrate affinity of XT3s1.
Oocytes were injected with XT3s1 cRNA. After incubation for 4 days, oocytes were superfused with different substrates ranging in concentration from 0 to 10 mM. K0.5-values were determined for each oocyte and subsequently averaged for seven oocytes (e=3).
| Substrate | K0.5 (mM) |
|---|---|
| L-Proline | 0.13±0.03 |
| D-Proline | 0.46±0.02 |
| Hydroxyproline | 0.19±0.03 |
| Betaine | 0.20±0.02 |
| D/L-Pipecolate | 0.09±0.01 |
| Nipecotic acid | 2.80±0.28 |
| L-Phenylalanine | 3.0±0.7 |
Uptake of [14C]proline was Na+- and Cl−-dependent; replacement of NaCl by NMDG-Cl (N-methyl-D-glucamine chloride) or LiCl completely abolished the transport activity, as did replacement of chloride with gluconate (Figure 3A). These data demonstrate that XT3s1 is an electrogenic Na+-and-Cl−-dependent transporter. An activation analysis of proline transport as a function of the Na+ concentration showed a hyperbolic dependence. Half-maximal transport velocity was reached at an Na+ concentration of 22±2 mM (Figure 3B), and an h value of 1.1±0.1 was determined. The chloride-binding site had an apparent K0.5 of 2.3±1 mM (Figure 3C). Similar results were obtained by using flux experiments (results not shown). To investigate further the stoichiometry of the transporter, we determined uptake of 0.5 mM [14C]proline under voltage-clamp conditions. In eight oocytes we determined a charge/proline ratio of 0.9±0.1.
Figure 3. Ion activation of proline transport by XT3s1.
Oocytes were injected with XT3s1 cRNA or remained uninjected (n.i.) in the controls. (A) [14C]Proline (100 μM) uptake was determined 4 days after injection in buffer containing NaCl or in buffer where NaCl was replaced by LiCl, NMDG-Cl or sodium gluconate. Each bar represents the transport activity (mean±S.D.) for ten oocytes (e=3). (B) Proline-induced currents (5 mM) plotted as a function of the extracellular Na+ concentration. (C) Proline-induced currents plotted as a function of the extracellular Cl− concentration. Curves display the transport activity (mean±S.D.) for seven oocytes (e=3).
DISCUSSION
General properties
The IMINO system is one of the major systems for the resorption of proline in the intestine and kidney and has been extensively characterized in the brush-border membranes of the jejunum [4,5,9,13]. It is an Na+-and-Cl−-dependent transporter. Amino acids with secondary or tertiary amine groups are the preferred substrates of the transporter, including proline, hydroxyproline, MeAIB and betaine. Mouse XT3s1, when expressed in X. laevis oocytes, shows all the properties of this transport system. The cloning of mouse XT3 from kidney has been reported previously [14]. Similarly, rat rB21a, which is homologous with mouse XT3s1, has been isolated from brain [19]. These studies neither recognized the presence of two highly homologous genes in these two species nor identified their function. In humans only one XT3 isoform, namely SLC6A20, is found, and its gene is located on chromosome 3p21.3. To simplify the confusing nomenclature, we suggest the use of the terms ‘IMINOK’ (‘highly expressed in kidney’) for mouse XT3 and its equivalent in rat (unfortunately named XT3s1) and IMINOB (‘highly expressed in brain’) to refer to XT3s1 and its rat homologue rB21a. The high similarity between IMINOK and IMINOB suggests that a gene duplication event may have occurred in mice and rat after separation from humans in evolution. Interestingly, we found active proline transport in oocytes expressing IMINOB, but not in oocytes expressing IMINOK. This suggests that the extended N-terminus of IMINOK may be required to interact with as yet unknown proteins.
One notable discrepancy between the functional properties of the intestinal IMINO system and the properties of IMINOB is the Na+ stoichiometry. For the intestinal IMINO system a 2 Na+/1 Cl−/proline co-transport has been proposed [5,9]. Activation analysis in oocytes, by contrast, indicates a 1 Na+/1 Cl−/proline co-transport, which would be electroneutral. Uptake experiments under voltage-clamp conditions, however, suggest translocation of 1 charge per proline molecule. A plausible explanation for this apparent discrepancy is that chloride transport will occur by the way of an exchange process because of the very high affinity of its binding site. As a result, chloride transport will not affect the electrogenicity of the transporter.
Role of IMINOB in the brain
Over recent years a number of proline transporters have been identified in brain tissue. They includes SLC6A7, a vesicular proline transporter [20], PAT1, which is found in the plasma membrane and in the lysosomes of neurons [21] and, as reported here, IMINOB. The location of IMINOB in the pia mater and plexus choroideus suggests that it is involved in the transport of proline into and out of the brain. Given that proline is a precursor for glutamate, proline transporters may have a role in providing precursors for glutamate biosynthesis to certain neuronal subpopulations [22]. Owing to the lack of anaplerotic enzymes for the tricarboxylic acid cycle, neurons have to rely on precursors for glutamate biosynthesis [23]. Interestingly, it has been reported that mutations of proline dehydrogenase affect sensorimotor gating in mice [24]. The highly specific location of different proline transporters in brain supports role of proline in neuromodulation.
Implications for inherited aminoacidurias
Proline and glycine have served as model substrates for amino acid resorption in kidney and intestine. The resorption of these two amino acids is driven by the Na+ electrochemical gradient [25] and the protonmotive force [7]. In relation to the inherited disease aminoaciduria iminoglycinuria [26], resorption of proline and glycine appears to be particularly important. Iminoglycinuria is an autosomal-recessive disorder characterized by increased excretion of proline and glycine in the urine [27]. Functional and molecular analysis suggests that at least three transporters contribute to proline and glycine uptake in kidney and intestine, namely the proline-specific IMINO transporter described here, the proton/amino acid transporter PAT1 [10] and the neutral amino acid transporter B0AT1 [11]. The contribution of each transporter to the resorption of these amino acids and their involvement in the phenotypical variability of iminoglycinuria will require the clarification of the genotype of iminoglycinuric patients.
While this manuscript was being revised, we became aware of a publication by Takanaga et al. [12] describing the functional properties of rat slc6a20. The results of both our studies are similar and agree in the assignment of SLC6A20 as the IMINO transporter.
Acknowledgments
This study was supported by grants from the Australian Research Council (ARC Discovery Project Grant DP 0559104) and the National Health and Medical Research Council (NHMRC; Project Grant 224229) to S.B.
References
- 1.Munck L. K., Munck B. G. Amino acid transport in the small intestine. Physiol. Res. 1994;43:335–345. [PubMed] [Google Scholar]
- 2.Silbernagl S., Foulkes E. C., Deetjen P. Renal transport of amino acids. Rev. Physiol. Biochem. Pharmacol. 1975;74:105–167. doi: 10.1007/3-540-07483-x_20. [DOI] [PubMed] [Google Scholar]
- 3.Stevens B. R., Kaunitz J. D., Wright E. M. Intestinal transport of amino acids and sugars: advances using membrane vesicles. Annu. Rev. Physiol. 1984;46:417–433. doi: 10.1146/annurev.ph.46.030184.002221. [DOI] [PubMed] [Google Scholar]
- 4.Stevens B. R., Wright E. M. Substrate specificity of the intestinal brush-border proline/sodium (IMINO) transporter. J. Membr. Biol. 1985;87:27–34. doi: 10.1007/BF01870696. [DOI] [PubMed] [Google Scholar]
- 5.Stevens B. R., Wright E. M. Kinetics of the intestinal brush border proline (Imino) carrier. J. Biol. Chem. 1987;262:6546–6551. [PubMed] [Google Scholar]
- 6.Munck B. G., Munck L. K., Rasmussen S. N., Polache A. Specificity of the imino acid carrier in rat small intestine. Am. J. Physiol. 1994;266:R1154–R1161. doi: 10.1152/ajpregu.1994.266.4.R1154. [DOI] [PubMed] [Google Scholar]
- 7.Roigaard-Petersen H., Jacobsen C., Iqbal Sheikh M. H+–L-proline cotransport by vesicles from pars convoluta of rabbit proximal tubule. Am. J. Physiol. 1987;253:F15–F20. doi: 10.1152/ajprenal.1987.253.1.F15. [DOI] [PubMed] [Google Scholar]
- 8.Ganapathy V., Roesel R. A., Howard J. C., Leibach F. H. Interaction of proline, 5-oxoproline, and pipecolic acid for renal transport in the rabbit. J. Biol. Chem. 1983;258:2266–2272. [PubMed] [Google Scholar]
- 9.Munck L. K., Munck B. G. Chloride-dependent intestinal transport of imino and β-amino acids in the guinea pig and rat. Am. J. Physiol. 1994;266:R997–R1007. doi: 10.1152/ajpregu.1994.266.3.R997. [DOI] [PubMed] [Google Scholar]
- 10.Anderson C. M., Grenade D. S., Boll M., Foltz M., Wake K. A., Kennedy D. J., Munck L. K., Miyauchi S., Taylor P. M., Campbell F. C., et al. H+/amino acid transporter 1 (PAT1) is the imino acid carrier: an intestinal nutrient/drug transporter in human and rat. Gastroenterology. 2004;127:1410–1422. doi: 10.1053/j.gastro.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 11.Bröer A., Klingel K., Kowalczuk S., Rasko J. E., Cavanaugh J., Bröer S. Molecular cloning of mouse amino acid transport system B0, a neutral amino acid transporter related to Hartnup disorder. J. Biol. Chem. 2004;279:24467–24476. doi: 10.1074/jbc.M400904200. [DOI] [PubMed] [Google Scholar]
- 12.Takanaga H., Mackenzie B., Suzuki Y., Hediger M. A. Identification of a mammalian proline transporter (SIT1,slc6a20) with characteristics of classical system IMINO. J. Biol. Chem. 2005 doi: 10.1074/jbc.M413027200. http://www.jbc.org/cgi/reprint/M413027200v1. [DOI] [PubMed]
- 13.Stevens B. R., Wright E. M. Kinetic model of the brush-border proline/sodium (IMINO) cotransporter. Ann. N.Y. Acad. Sci. 1985;456:115–117. doi: 10.1111/j.1749-6632.1985.tb14853.x. [DOI] [PubMed] [Google Scholar]
- 14.Nash S. R., Giros B., Kingsmore S. F., Kim K. M., el-Mestikawy S., Dong Q., Fumagalli F., Seldin M. F., Caron M. G. Cloning, gene structure and genomic localization of an orphan transporter from mouse kidney with six alternatively-spliced isoforms. Receptors Channels. 1998;6:113–128. [PubMed] [Google Scholar]
- 15.Bröer S., Rahman B., Pellegri G., Pellerin L., Martin J. L., Verleysdonk S., Hamprecht B., Magistretti P. J. Comparison of lactate transport in astroglial cells and monocarboxylate transporter 1 (MCT 1) expressing Xenopus laevis oocytes. Expression of two different monocarboxylate transporters in astroglial cells and neurons. J. Biol. Chem. 1997;272:30096–30102. doi: 10.1074/jbc.272.48.30096. [DOI] [PubMed] [Google Scholar]
- 16.Bröer S. Xenopus laevis Oocytes. Methods Mol. Biol. 2003;227:245–258. doi: 10.1385/1-59259-387-9:245. [DOI] [PubMed] [Google Scholar]
- 17.Klingel K., Hohenadl C., Canu A., Albrecht M., Seemann M., Mall G., Kandolf R. Ongoing enterovirus-induced myocarditis is associated with persistent heart muscle infection: quantitative analysis of virus replication, tissue damage, and inflammation. Proc. Natl. Acad. Sci. U.S.A. 1992;89:314–318. doi: 10.1073/pnas.89.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiss H., Darai E., Kiss C., Kost-Alimova M., Klein G., Dumanski J. P., Imreh S. Comparative human/murine sequence analysis of the common eliminated region 1 from human 3p21.3. Mamm. Genome. 2002;13:646–655. doi: 10.1007/s00335-002-3037-y. [DOI] [PubMed] [Google Scholar]
- 19.Smith K. E., Fried S. G., Durkin M. M., Gustafson E. L., Borden L. A., Branchek T. A., Weinshank R. L. Molecular cloning of an orphan transporter. A new member of the neurotransmitter transporter family. FEBS Lett. 1995;357:86–92. doi: 10.1016/0014-5793(94)01328-x. [DOI] [PubMed] [Google Scholar]
- 20.Fremeau R. T., Jr, Caron M. G., Blakely R. D. Molecular cloning and expression of a high affinity L-proline transporter expressed in putative glutamatergic pathways of rat brain. Neuron. 1992;8:915–926. doi: 10.1016/0896-6273(92)90206-s. [DOI] [PubMed] [Google Scholar]
- 21.Wreden C. C., Johnson J., Tran C., Seal R. P., Copenhagen D. R., Reimer R. J., Edwards R. H. The H+-coupled electrogenic lysosomal amino acid transporter LYAAT1 localizes to the axon and plasma membrane of hippocampal neurons. J. Neurosci. 2003;23:1265–1275. doi: 10.1523/JNEUROSCI.23-04-01265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson S. G., Wong P. T., Leong S. F., McGeer E. G. Regional distribution in rat brain of 1-pyrroline-5-carboxylate dehydrogenase and its localization to specific glial cells. J. Neurochem. 1985;45:1791–1796. doi: 10.1111/j.1471-4159.1985.tb10535.x. [DOI] [PubMed] [Google Scholar]
- 23.Hertz L., Dringen R., Schousboe A., Robinson S. R. Astrocytes: glutamate producers for neurons. J. Neurosci. Res. 1999;57:417–428. [PubMed] [Google Scholar]
- 24.Gogos J. A., Santha M., Takacs Z., Beck K. D., Luine V., Lucas L. R., Nadler J. V., Karayiorgou M. The gene encoding proline dehydrogenase modulates sensorimotor gating in mice. Nat. Genet. 1999;21:434–439. doi: 10.1038/7777. [DOI] [PubMed] [Google Scholar]
- 25.Sacktor B. Membranes and Transport, vol. 2. In: Martinosi A., editor. New York: Plenum Press; 1982. pp. 197–206. [Google Scholar]
- 26.Scriver C. R., Schafer I. A., Efron M. L. New renal tubular amino-acid transport system and a new hereditary disorder of amino-acid metabolism. Nature (London) 1961;192:672–673. doi: 10.1038/192672a0. [DOI] [PubMed] [Google Scholar]
- 27.Scriver C. R., Efron M. L., Schafer I. A. Renal tubular transport of proline, hydroxyproline, and glycine in health and in familial hyperprolinemia. J. Clin. Invest. 1964;43:374–385. doi: 10.1172/JCI104922. [DOI] [PMC free article] [PubMed] [Google Scholar]