Abstract
Protein acetyltransferases and deacetylases have been implicated in oncogenesis, apoptosis and cell cycle regulation. Most of the protein acetyltransferases described acetylate ε-amino groups of lysine residues within proteins. Mouse ARD1 (homologue of yeast Ard1p, where Ard1p stands for arrest defective 1 protein) is the only known protein acetyltransferase catalysing acetylation of proteins at both α- (N-terminus) and ε-amino groups. Yeast Ard1p interacts with Nat1p (N-acetyltransferase 1 protein) to form a functional NAT (N-acetyltransferase). We now describe the human homologue of Nat1p, NATH (NAT human), as the partner of the hARD1 (human ARD1) protein. Included in the characterization of the NATH and hARD1 proteins is the following: (i) endogenous NATH and hARD1 proteins are expressed in human epithelial, glioma and promyelocytic cell lines; (ii) NATH and hARD1 form a stable complex, as investigated by reciprocal immunoprecipitations followed by MS analysis; (iii) NATH–hARD1 complex expresses N-terminal acetylation activity; (iv) NATH and hARD1 interact with ribosomal subunits, indicating a co-translational acetyltransferase function; (v) NATH is localized in the cytoplasm, whereas hARD1 localizes both to the cytoplasm and nucleus; (vi) hARD1 partially co-localizes in nuclear spots with the transcription factor HIF-1α (hypoxia-inducible factor 1α), a known ε-amino substrate of ARD1; (vii) NATH and hARD1 are cleaved during apoptosis, resulting in a decreased NAT activity. This study identifies the human homologues of the yeast Ard1p and Nat1p proteins and presents new aspects of the NATH and hARD1 proteins relative to their yeast homologues.
Keywords: acetyltransferase, apoptosis, ARD1, hypoxia-inducible factor 1α (HIF-1α), MS, N-acetyltransferase human (NATH)
Abbreviations: ARD, arrest-defective (homologue of yeast Ard1p); hARD1, human ARD1; hARD1-t, truncated hARD1; mARD1, mouse ARD1; CS, cytosolic supernatant; HEK-293 cells, human embryonic kidney 293 cells; HIF-1α, hypoxia-inducible factor 1α; LC, liquid capillary; NAT, N-acetyltransferase; mNAT1, mouse NAT1; NATH, NAT human; NATH-t, truncated NATH; NLS, nuclear localization signal; P, polysomal pellet; RACK1, receptor of activated C kinase 1; TPR, tetratricopeptide repeat; US, post-ultracentrifugal supernatant; Z-VAD-FMK, benzyloxycarbonylvalylalanyl-DL-aspartylfluoromethane
INTRODUCTION
Protein acetylation and deacetylation are increasingly recognized as important post-translational modifications involved in the normal cell function and in cancer development [1–3]. Inhibitors of deacetylases have recently been recognized as potential cancer drug candidates [4]. One of the newly described mammalian protein acetyltransferases, mARD1 [mouse homologue of yeast Ard1p (arrest defective 1 protein)], expresses protein N-terminal α-acetylation activity in a complex with mNAT1 (mouse N-acetyltransferase 1) protein [5]. Importantly, it was demonstrated that mARD1 mediates ε-acetylation of a lysine residue of the transcription factor HIF-1α (hypoxia-inducible factor 1α) and thereby enhances HIF-1α ubiquitination and degradation [6]. ARD1-mediated HIF-1α acetylation results in a decrease in the expression of HIF-1α downstream genes regulating angiogenesis, apoptosis, cell proliferation and glucose metabolism [7]. Thus the mammalian ARD1 represents a novel type of enzyme with dual function capable of both α- and ε-protein acetylation.
In Saccharomyces cerevisiae, Nat1p forms a heterodimer with Ard1p to generate a functional protein NatA [8], Ard1p being the catalytic subunit. The human homologue of Nat1p, NATH (NAT human), is overexpressed in papillary thyroid carcinomas and found to be highly expressed in dividing tissues such as testis, embryonal brain and bone marrow [9]. It is therefore possible that NATH plays an important role in cell proliferation or apoptosis. NatA activity in yeast is linked to Go entry, cell growth and the ability to sporulate [10–13]. Proteins with an N-terminal serine, threonine, glycine or alanine residue are potential substrates of NatA [14]. Protein N-terminal α-acetylation is believed to be a co-translational process occurring when approx. 25–50 amino acid residues are extruding from the ribosome [15]. NatA was found to interact directly with nascent polypeptides as well as ribosomes through the binding of Nat1p to the large ribosomal subunit [16]. The molecular mechanisms of co-translational protein N-acetylation in humans have not been described. A human homologue of yeast Ard1p would be an obvious NAT candidate, probably complexed with NATH. The hARD1 (human Ard1p) homologue has not been studied, but a cDNA sequence, TE2, encoding the hARD1 is available in GenBank® (NM_003491) [17] and ARD1 mRNA expression has been demonstrated in human tissues [6]. To our knowledge, no evidence has been presented of complex formation between NATH and hARD1 and their activity in human cells. Thus the biological functions of NATH and hARD1 remain largely unexplored. The protein translational machinery is, during apoptosis, targeted by caspases, resulting in a substantial inhibition of protein synthesis [18–23]. In the present study, we show that NATH and hARD1 are cleaved in apoptotic cells and the protein NAT activity of the complex is significantly reduced. Thus NATH and hARD1 represent novel caspase targets associated with protein translation. The NATH and hARD1 proteins are described, including studies on protein interactions, NAT activity and localization potentially generating novel hypotheses of a multifunctional activity of NATH–hARD1.
EXPERIMENTAL
NATH and hARD1 cloning
Plasmid encoding V5-tagged hARD1 was constructed from cDNA made from total RNA isolated from human ARO cells. A plasmid encoding V5-tagged NATH was subcloned from a previously described plasmid [9]. PCR products were inserted into the TOPO TA vector pcDNA 3.1 V5His (Invitrogen). Deletion mutants of hARD1 were made using the respective full-length plasmid as template. cDNA was made as described previously [24].
Cell culture, transfection and apoptosis
All cell lines used were of human origin. HEK-293 cells (human embryonic kidney 293 cells; A.T.C.C. no. CRL-1573), HeLa cells (epithelial cervix adenocarcinoma; A.T.C.C. no. CCL-2), GaMg cells (glioblastoma; DSMZ no. ACC 242) and ARO cells (ARO, anaplastic thyroid carcinoma; Dr G. J. F. Juillard, University of California at Los Angeles, CA, U.S.A.) were cultured in 5% CO2 at 37 °C in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum and 3% L-glutamine. MCF-7 cells (epithelial mammary gland, breast adenocarcinoma; A.T.C.C. no. HTB-22) and NB4 cells (acute promyelocytic leukaemia; DSMZ no. ACC 207) were cultured as above, but with RPMI 1640. Transfections were performed using Fugene6 (Roche) according to the manufacturer's instructions. A stable HEK-293 cell line expressing the hARD1–V5 protein was generated using G418 (Gibco) selection at 1000 μg/ml. Apoptosis was induced by adding 200 nM or 10 μM daunorubicin (Sigma) to NB4 cells and HeLa cells respectively. Cells were incubated for 20–24 h before harvesting and analysed by SDS/PAGE and Western blotting. To inhibit specifically the caspase activities, 20 μM Z-VAD-FMK (benzyloxycarbonylvalylalanyl-DL-aspartylfluoromethane; Alexis Biochemicals, Lausen, Switzerland) was added 10 h before harvest. Arsenic trioxide (Sigma) was added to NB4 cell cultures at different concentrations for 24 h. Apoptosis was verified by Mayer–Grunwald staining and light microscopy (100×), to determine the percentage of cells with disrupted and pycnotic nuclei as described previously [25].
Isolation of polysomes
Polysomes were isolated using a modification of previously described methods [26,27]. Approximately 3×107 HEK-293 cells were cross-linked using dithiobis (succinimidyl propionate) (Pierce) according to the manufacturer's instructions. Cells were then lysed using 1.2 ml buffer P (10 mM triethanolamine, 130 mM KCl, 0.25% Triton X-100, 0.25 M sucrose, 5 mM magnesium acetate and 1 mM PMSF) and incubated on ice for 15 min. After 10 min centrifugation at 1500 g, the CS (cytosolic supernatant) was ultracentrifuged at 436000 g for 25 min in a TLA 102.2 rotor (Beckmann, Geneva, Switzerland) through a 0.4 ml 25% sucrose cushion giving a US (post-ultracentrifugal supernatant) and a P (polysomal pellet). The different fractions were analysed by SDS/PAGE and Western-blotting. Polysomes were also fractioned without cross-linking in free cytosolic polysomes, cytoskeleton-bound polysomes and membrane-bound polysomes as described in [26,27].
Immunofluorescence
Cells were grown on coverslips, washed in PBS and fixed for 20 min in 4% (v/v) formaldehyde. Fixed cells were washed three times in PBS, permeabilized in 0.1% Triton X-100 for 10 min and then incubated in blocking buffer (10% BSA) for 30 min. Cells were then incubated for 1 h with primary antibody diluted in 2% BSA. Dilutions: anti-NATH, 1:200; anti-hARD1, 1:100; anti-V5 (Invitrogen), 1:200; and anti-HIF-1α (clone OZ12; Sigma), 1:200. Rabbit polyclonal peptide-specific antibodies against NATH and hARD1 were produced by Biogenes GmBH (Berlin, Germany) and Sigma Genosys respectively. The immunogens correspond to amino acids 853–866 of NATH and amino acids 204–217 of hARD1. Antibody specificities were confirmed by peptide blocking on Western blots. Cells were washed at least three times for 5 min in washing buffer (0.01% Tween 20 in PBS) and then incubated with the fluorochrome-conjugated secondary antibody for 40 min (Alexa conjugated antibodies; Molecular Probes, Eugene, OR, U.S.A.). Cells were washed in washing buffer and finally with distilled water before mounting on glass slides in Vectashield H1200 (Vector Laboratories, Burlingame, CA, U.S.A.). The cells were studied using a Leica DMRXA microscope with a Leica DC500 camera and a Leica HCX PL APO ×40 oil immersion objective.
Western blotting
SDS/PAGE and Western blots were performed as described in [24]. Polyclonal rabbit antibodies against NATH and hARD1, described above, were used at 1:500 dilution, anti-V5 antibody at 1:1000, anti-RACK1 (receptor of activated C kinase 1; Affinity) at 1:1000 and anti-P (kindly provided by B. T. Gjertsen, Department of Internal Medicine, Haukeland University Hospital, Norway) at 1:500. For detection of RACK1, biotin-anti-mouse (Dako, Carpinteria, CA, U.S.A.) was used. Horseradish peroxidase-linked anti-mouse, anti-rabbit and streptavidin were from Amersham Biosciences (Little Chalfont, Bucks., U.K.).
Immunoprecipitation
Cells were harvested and lysed in 300 μl lysis buffer [50 mM Tris, pH 8, 50 mM NaCl, 0.5% Nonidet P40, 5 mM EDTA, 5 μM trichostatin A (Sigma), 1 mM Na3VO4 and 1 mM Pefabloc (Roche)]. Typically, 2×106 cells were used. The lysates were added to 40 μl of Protein A/G–agarose (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.) and incubated for 1 h at 4 °C. After centrifugation at 3000 g for 2 min, the supernatants were collected and incubated for another 16 h at 4 °C with specific antibody (3 μg). The samples were centrifuged as above and 40 μl Protein A/G–agarose was added to the supernatants. After incubation for 16 h, centrifugation and three periods of washing in the lysis buffer, the samples were subjected to SDS/PAGE and Western blotting.
MS
Thermofinnigan LCQ Deca XP Plus and Agilent 1100 Nanoflow HPLC were used for microcapillary LC/MS/MS analysis (where LC stands for liquid capillary) of tryptic peptides. Data analysis utilized SEQUEST [28], Medusa [29] and support vector machine learning [30]. For the preparation of samples for LC/MS/MS analysis, the immunprecipitation method was used with the following exceptions: approx. 5×107 cells were used per sample, 1 ml of lysis buffer was added to the cell pellet, 100 μl of Protein A/G–agarose was used and the antibody used was chemically cross-linked to the Protein A/G–agarose using dimethyl pimelimidate (Pierce) according to the manufacturer's instructions. The pellets of Protein A/G–agarose–antibody with the protein complexes were washed three times in PBS and twice in TEN buffer (0.02 M Tris/HCl, pH 7.5, 1 mM EDTA, 0.15 M NaCl). The agarose beads were resuspended in 100 μl TEN buffer and 140 μl of extraction solution (8 M urea, 20 mM methylamine, 1 M LiCl and 2 mM EDTA in 100 mM Tris/HCl, pH 8.5) was added. After incubation for 30 min at room temperature (20 °C), the samples were centrifuged at 12000 g for 2 min. The supernatants were added to 2 μl of 0.5 M dithiothreitol, incubated for 2 h at room temperature, and then added to 6 μl of 0.5 M iodoacetamide and incubated for another 2 h protected from light. Another 2 μl of 0.5 M dithiothreitol was added and after 1 h incubation, 90 μl of distilled water was added to the mixture. LysC endoprotease (Sigma) was added at a concentration of 1:50 (w/w) relative to the total protein content of the sample. After 15 h of incubation at 37 °C, 200 μl of 10 mM CaCl2 (pH 8.5) was added. Trypsin (Sigma) was added at a concentration of 1:10 (w/w) relative to the total protein content. After 10 h of incubation at 37 °C, 2 μl of acetic acid was added. A lysC endoprotease/trypsin digest of the hARD1–V5/NATH affinity extract was batch purified on a Vydac C18 Denali reversed-phase guard column, with elution of tryptic peptides with 80% acetonitrile plus 0.1% trifluoroacetic acid after washing with water plus 0.1% (v/v) trifluoroacetic acid. The peptide mixture was then freeze-dried, redissolved in water plus 0.1% formic acid, and loaded on to a self-packed 5 cm×75 μm internal diameter Picofrit (New Objective, Woburn, MA, U.S.A.) capillary C18 reversed-phase column with a 15 μm spray tip. The column was eluted with a 1%/min acetonitrile gradient from 5 to 85% acetonitrile, using an Agilent 1100 nanoflow HPLC at a flow rate of 300 nl/min. Mass spectra were collected as follows: a single full scan spectrum to detect peptide precursor ions was followed by collecting MS/MS spectra of the four most intense precursor ions at a relative collision energy of 35%. This cycle was repeated for the entire HPLC run. Dynamic exclusion was used to maximize the number of precursor ions examined. Data were analysed using a non-redundant human protein database constructed from the full NCBI non-redundant protein database.
Acetyltransferase assay
The N-terminal acetyltransferase assay was performed essentially as described in [5] using immunoprecipitated NATH and hARD1 complexes. Pellets of Protein A/G–agarose-bound NATH–hARD1 complex were added to 10 μl of corticotropin (ACTH; 0.5 mM; human corticotropin fragment 1–24; Calbiochem), 4 μl of [3H]acetyl-CoA (1 μCi and 107 GBq/mmol; Amersham Biosciences) and 136 μl of 0.2 M K2HPO4 (pH 8.1). The mixture was incubated at 37 °C and samples were collected after 2 h. After centrifugation, the supernatant was added to 150 μl of SP Sepharose (50% slurry in 0.5 M acetic acid; Sigma) and incubated on a rotor for 5 min. The mixture was centrifuged and the pellet was washed three times with 0.5 M acetic acid and finally with methanol. Radioactivity in the corticotropin-containing pellet was determined by scintillation counting. Immunoprecipitated samples from apoptosis experiments (Figure 8E) were analysed by SDS/PAGE and Western blotting after acetyltransferase assays. The intensities of hARD1 and hARD1-t (truncated hARD1) bands were measured using FUJIFILM Image Gauge v.3.45 and the activity data (c.p.m.) were adjusted according to the relative arbitrary units representing levels of hARD1/hARD1-t proteins. Experiments in Table 1 and Figure 8(E) were repeated at least three times and representative results are shown.
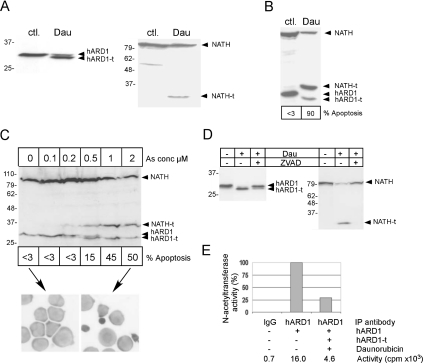
Figure 8. Cleavage of NATH and hARD1 in apoptosis.
(A) HeLa cells untreated (ctl.) or treated for 20 h with 10 μM daunorubicin (Dau). Shown in (A–D) are NATH, NATH-t, hARD1 and hARD1-t. (B) NB4 cells untreated (ctl.) or treated for 24 h with 200 nM Dau. % Apoptosis is the percentage of cells displaying apoptotic features as assessed by light microscopy. (C) NB4 cells untreated (0) or treated for 24 h with increasing concentrations of As2O3 as indicated. Bands representing hARD1-t and NATH-t are detected from 0.5 μM As2O3. Pictures display NB4 cells (100×) untreated, <3% apoptosis (left) or treated with 2 μM As0, approx. 50% apoptosis (right). (D) NB4 cells treated for 20 h with 200 nM Dau and for 10 h with 20 μM Z-VAD-FMK as indicated. (E) NATH–hARD1 complexes were immunoprecipitated from HeLa cells using limiting amounts of anti-hARD1 or rabbit IgG as a negative control. Apoptosis was induced by 10 μM daunorubicin for 20 h. The immunoprecipitates were incubated with corticotropin peptide amino acids 1–24 and [3H]acetyl-CoA, and the acetyl incorporation in corticotropin was determined by scintillation counting (c.p.m.). Percentage activity was adjusted according to the presence of hARD1/hARD1-t in the precipitates detected by Western blotting followed by densitometry. Five independent experiments were performed.
Table 1. NAT activity of the NATH–hARD1 complex.
NATH–hARD1 complexes were immunoprecipitated from HEK-293 cells using the indicated antibodies or rabbit IgG as a negative control. Immunoprecipitates were incubated with corticotropin peptide amino acids 1–24 and [3H]acetyl-CoA, and the acetyl incorporation in corticotropin was determined by scintillation counting.
| Immunoprecipitation antibody | NATH–hARD1 | Activity (×103 c.p.m.) |
|---|---|---|
| IgG | − | 1.1 |
| Anti-NATH | + | 38.2 |
| Anti-hARD1 | + | 88.3 |
RESULTS
Expression of NATH and hARD1 proteins
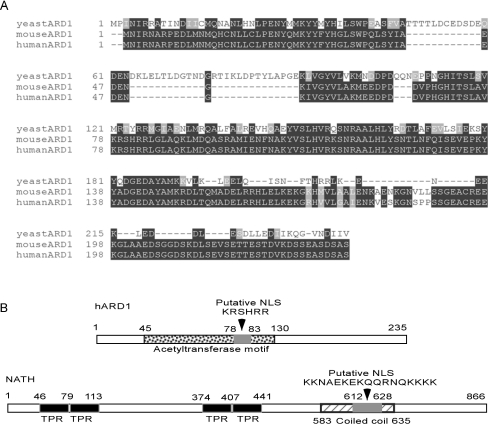
NATH was cloned and first identified as an mRNA overexpressed in papillary carcinomas of the thyroid [9], and the expression of hARD1 mRNA has recently been presented in [6]. To study hARD1, we cloned the full-length cDNA encoding the hARD1 from RNA purified from the thyroid anaplastic carcinoma cell line ARO. The hARD1 protein shares 27.8 and 96.2% sequence identity with its yeast and mouse homologues respectively (Figure 1A). The predicted acetyltransferase domain is located between amino acids 45 and 130, and a putative NLS (nuclear localization signal) is found at amino acids 78–83 (Figure 1B). Other functional domains were not identified by database searches, but an overall globular structure was suggested. To investigate the expression of endogenous NATH and hARD1 proteins in human cell lines originating from different tissues, Western-blot analyses of HEK-293 (kidney), HeLa (cervix), NB4 (promyelocytes) and ARO (thyroid) cell extracts were performed using rabbit polyclonal antibodies. Protein bands of the predicted sizes were detected as the major bands in all cases, approx. 30 kDa for hARD1 and 100 kDa for NATH (Figures 2A and 2B). The calculated molecular masses for hARD1 and NATH are 26.5 and 101.3 kDa respectively.
Figure 1. hARD1 and NATH.
(A) Alignment of yeast, mARD1 and hARD1. Identities are in black backgrounds and conservative substitutions are in grey backgrounds. (B) Schematic models (only showing sizes) of the 235 amino acids of hARD1 and the 866 amino acids of NATH. hARD1: acetyltransferase domain at amino acids 45–130 and the putative NLS at amino acids 78–83 are indicated. NATH: TPR motifs at amino acids 46–79, 80–113, 374–407, 408–441, and the putative NLS at amino acids 612–628 and a coiled-coil region at amino acids 583–635 are indicated.
Figure 2. Detection of endogenous NATH and hARD1.
(A) Different cell types were lysed and analysed by SDS/PAGE and Western blotting. The membrane was incubated with a rabbit hARD1-peptide-specific polyclonal antibody. *, A slowly migrating hARD1 variant, a hARD1 containing complex or probably an unspecific band. Lanes: 1, ARO; 2, HEK-293; 3, HeLa; and 4, NB4. (B) As in (A) using an NATH-specific polyclonal antibody. The experiment was repeated more than three times.
NATH interacts with hARD1
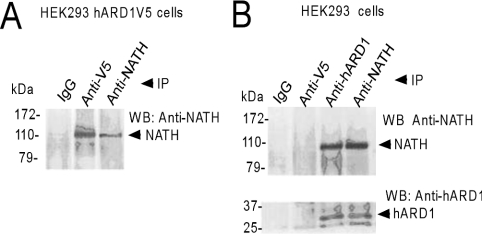
To study whether or not a stable NATH–hARD1 complex is formed in human cells, HEK-293 cells expressing V5-tagged hARD1 were lysed and subjected to immunoprecipitation and SDS/PAGE/Western-blotting analysis using either anti-V5 or anti-NATH antibodies. The presence of a strong NATH signal in the anti-V5 extract (hARD1–V5) was indicative of an NATH–ARD1 complex (Figure 3A). The interaction between endogenous NATH and hARD1 was confirmed by co-immunoprecipitation of untransfected HEK-293 cell lysates using either anti-hARD1 or anti-NATH antibodies (Figure 3B). NATH and hARD1 were simultaneously present in both these immunoprecipitates. The results from these reciprocal immunoprecipitation experiments with hARD1 and NATH strongly suggest that a stable complex of NATH–hARD1 exists in human cells.
Figure 3. Immunoprecipitation of NATH–hARD1 complexes.
(A) HEK-293 cells stably expressing hARD1–V5 were harvested and the lysates were immunoprecipitated (IP) with the indicated antibodies (IgG denotes rabbit serum). The immunoprecipitates were analysed by SDS/PAGE and Western blotting. The membrane was incubated with anti-NATH. (B) A normal population of HEK-293 cells was immunoprecipitated and processed as in (A) with the addition of anti-hARD1. Experiments were repeated more than ten times.
To identify the hARD1 region that binds to NATH, we prepared four truncated forms of hARD1, N- and C-terminal, (Figure 4A) with V5-tags and used these for transfection of HEK-293 cells. The C-terminally truncated forms hARD1(1–174)–V5 and hARD1(1–222)–V5 immunoprecipitated endogenous NATH similar to the full-length hARD1–V5, whereas the N-terminally truncated hARD1(59–235)–V5 and hARD1(146–235)–V5 did not facilitate NATH pull-down (Figure 4B). These results show that the 61 C-terminal amino acid residues of hARD1 are dispensable for NATH binding, whereas the 58 N-terminal amino acid residues of hARD1 might be involved in the NATH interaction.
Figure 4. The C-terminal of hARD1 is not involved in NATH–hARD1 interaction.
(A) Some hARD1–V5 deletion proteins are outlined. The putative acetyltransferase motif (amino acids 45–130) is dotted, whereas the putative NLS (amino acids 78–83) is grey. (B) HEK-293 cells were transiently transfected with the indicated hARD1–V5 deletion plasmids. Cells were then harvested and the lysates were immunoprecipitated (IP) with anti-V5. The immunoprecipitates were analysed by SDS/PAGE and Western blotting. The membrane was incubated with anti-NATH and anti-V5. The experiment was repeated three times.
NAT activity of the NATH–hARD1 complex
We investigated whether or not endogenous NATH–hARD1 has protein NAT activity. Immunoprecipitated NATH–hARD1 complexes were prepared from HEK-293 cells and used in in vitro NAT assays (see the Experimental section and Table 1). The presence of NATH and hARD1 in the different protein preparations was verified by Western blots. A significant activity measured as 3H-acetyl incorporation was found. Thus, the human endogenous NATH–hARD1 complex has NAT activity, when using the N-terminal 24 amino acids of corticotropin as substrate (representative experiment in Table 1).
The NATH–hARD1 complex is associated with ribosomal subunits
To examine a potential interaction of NATH–hARD1 with the ribosome, as suggested by results in yeast [16], we performed a modified immunoprecipitation experiment on hARD1–V5-transfected HEK-293 cell extracts in which either anti-V5 or anti-NATH antibodies covalently cross-linked to the Protein A/G–agarose beads were used to prepare hARD1–NATH co-precipitates. After trypsin digestion of the extracted proteins, the tryptic peptides were separated and fragmented by microcapillary LC/MS/MS, and analysed using a series of software tools [28–30]. Both NATH- and hARD1-specific peptides were identified in the anti-V5/anti-NATH antibody-mediated pull-down, but were absent when the respective antibodies were omitted (Figures 5A and 5B). Importantly, several additional proteins were identified in the immunoprecipitates, indicating novel protein partners for NATH and hARD1. Of special interest were several ribosomal proteins (Table 2). In immunoprecipitates from HEK-293 cells expressing hARD1–V5, we identified peptides representing the ribosomal proteins P1, P2 (Figure 5C), S7, S15, L17, L23, L31 and L35. The eukaryotic translational elongation factor 1α1 and initiation factor 2B were also present. Each protein was identified by one or more specific peptides in at least half of the experiments (Table 2). The presence of ribosomal proteins in the NATH–hARD1 complex is a strong indication for a functional association with the ribosome. To verify the interaction between the NATH–hARD1 complex and the ribosome, we isolated polysomes from HEK-293 cells and analysed the polysomal fraction by SDS/PAGE and Western blotting. Both anti-NATH and anti-hARD1 antibodies confirmed the presence of NATH and hARD1 in the ribosomal pellet (Figure 6, lane P). However, visual comparison of the amount of NATH and hARD1 in corresponding amounts of extract before and after ultracentrifugation (Figure 6, lane CS versus US) indicated that the majority of NATH and hARD1 is non-polysomal. In contrast, the ribosomal protein P0 is significantly reduced from the CS fraction to the US fraction. In yeast, both Ard1p and Nat1p interact with Asc1p [31]. Recently, Asc1p was also found to interact with ribosomes, supporting the connection between Nat1p, Ard1p, Asc1p and ribosomes [32]. In our experiments, the human homologue of Asc1p, RACK1, was also found to interact with the ribosome (Figure 6). However, using co-immunoprecipitation, we were neither able to demonstrate the RACK1 interaction with NATH nor hARD1 (results not shown). This could result from technical difficulties or the Asc1p interaction with Nat1p and Ard1p has not been conserved between yeast and human. Subpolysomal fractionation demonstrated that NATH and hARD1 were present in the free cytosolic polysomes and the cytoskeleton-bound polysomes, whereas RACK1 was present in all polysomal fractions including the membrane-bound polysomes (results not shown).
Figure 5. MS identification of proteins.
HEK-293 cells stably expressing hARD1–V5 were harvested and lysates were immunoprecipitated and then subjected to serial lysC endoprotease/trypsin digestion and the peptides were identified by ion-trap MS. (A) MS/MS spectrum of the NATH-derived +2 peptide RLPLNFLSGEK, with the C-terminal y ions and N-terminal b ions labelled as assigned. All fragment ions had a +1 charge. The match between this spectrum and that predicted for the peptide had a cross-correlation coefficient (Xcorr) of 3.45. The y1 and b1 ions were below the low m/z detection limit in this experiment. The position of the fragmented parent ion is indicated by the double dagger on the x-axis for all three peptides. A second peptide, TQQTSPDKVDYEYSELLLYQNQVLR, sequenced separately twice, also indicated the presence of NATH in the affinity extract. (B) MS/MS spectrum of the hARD1 peptide AALHLYSNTLNFQISEVEPK, which was sequenced separately twice. This peptide had a charge of +2. The assigned y and b ion peaks (all +1) are indicated; the ions y1–y2 and b1–b3 were below the lower limit of detectable m/z in this experiment. The match between the predicted and observed spectrum was described by a very high Xcorr of 5.68. A second peptide assigned to hARD1 was MEEDPDDVPHGHITSLAVK. (C) MS/MS spectrum of the 60 S ribosomal protein P2-derived peptide YVASYLLAALGGNSSPSAK is shown, with the C-terminal y ions and N-terminal b ions labelled as assigned. All fragment ions had a +1 charge. The match between this spectrum and that predicted for the peptide had Xcorr of 4.29.
Table 2. Detection of NATH/hARD1 ribosome-associated proteins by LC/MS/MS.
HEK-293 cells expressing hARD1–V5 were harvested and the lysates were immunoprecipitated, using anti-V5 or anti-NATH antibodies as indicated, and then subjected to a combined lysC endoprotease/trypsin digest. The peptides were identified by ion-trap tandem MS. Two and four independent parallel experiments were performed using anti-NATH and anti-V5 respectively. Peptides present in negative controls were omitted. SVM, support vector machine.
| Antibody | Identified protein | Reproducibility | Peptide | SVM probability |
|---|---|---|---|---|
| Anti-V5–hARD1 | El.F1α1 | 3/4 | VETGVLKPGMVVTFAPVNVTTEVK | 0.98 |
| VETGVLKPGM*VVTFAPVNVTTEVK | 0.87 | |||
| NM*ITGTSQADC†AVLIVAAGVGEFEAGISK | 0.59 | |||
| L23 | 2/4 | ISLGLPVGAVINC†ADNTGAK | 0.98 | |
| LNRLPAAGVGDMVMATVK | 0.34 | |||
| L17 | 2/4 | SAEFLLHMLK | 0.98 | |
| GLDVDSLVIEHIQVNK | 0.42 | |||
| EIF-2B | 2/4 | VLLGAHALLANGSVM*SR | 0.80 | |
| DEALEDEDNKKDDGISFSNQTGPAWAGSER | 0.44 | |||
| Anti-NATH | P1 | 1/2 | AAGVNVEPFWPGLFAK | 0.92 |
| ALANVNIGSLIC†NVGAGGPAP | 0.87 | |||
| AAGAAPAGGPAPSTAAAPAEEK | ||||
| P2 | 1/2 | YVASYLLAALGGNSSPSAK | 0.92 | |
| NIEDVIAQGIGK | 0.69 | |||
| S7 | 1/2 | TLTAVHDAILEDLVFPSEIVGK | 0.99 | |
| S15 | 1/2 | DMIILPEMVGSMVGVYNGK | 0.86 | |
| L35 | 1/2 | VLTVINQTQK | 0.79 | |
| L31 | 1/2 | LYTLVTYVPVTTFK | 0.64 | |
| El.F1α1 | 1/2 | KIGYNPNTVAFVPISGWNGDNMLEPSANMPWFK | 0.40 |
* Oxidized methionine.
† Carboxamidomethyl-cysteine.
Figure 6. Detection of NATH, hARD1 and RACK1 in polysomal fractions.
HEK-293 cells were treated with 20 μg/ml cycloheximide for 5 min before dithiobis(succinimidyl propionate) cross-linking and lysis. After an initial centrifugation, the CS was ultracentrifuged at 436000 g for 25 min through a 25% sucrose cushion, and the resultant supernatant (US) and P were analysed by SDS/PAGE and Western blotting. The membrane was incubated with the following antibodies: anti-NATH, anti-hARD1, anti-RACK1 and anti-P (ribosomal protein P0). Aliquots of the initial supernatant (S) and the US correspond to approx. 5% of the total material included in P. The experiment was repeated three times.
Subcellular localization of NATH and hARD1
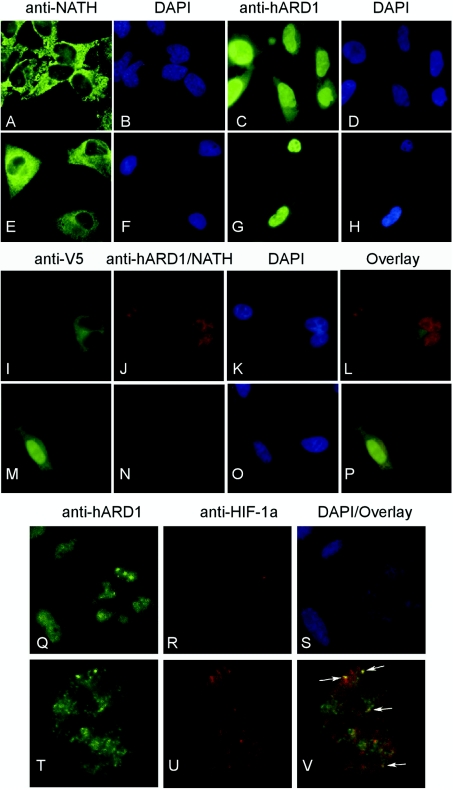
Detailed analysis of the NATH sequence indicates a possible bipartite NLS between amino acid residues 612–628 (KKNAEKEKQQRNQKKKK) in the predicted coiled-coil region (Figure 1B). Similarly, hARD1 contains a less conserved NLS between amino acids 78–83 (KRSHRR). Thus both proteins could be actively imported to the nucleus. Immunofluorescence analysis of hARD1 and NATH in HeLa and GaMg cell lines revealed that endogenous hARD1 was present both in the nuclei and the cytoplasm, whereas NATH predominantly localized in the cytoplasm (Figures 7A–7H). These results were verified in HEK-293, MCF-7 and NB4 cells (results not shown). In spite of the well-defined putative NLS in the NATH protein, only a very weak NATH protein staining could be detected in the nucleus. These findings were confirmed in independent experiments using confocal microscopy (results not shown). We also confirmed the localization patterns of NATH and hARD1 by expressing exogenous V5-tagged hARD1 and NATH proteins in HeLa cells (Figures 7I–7P). A fraction of exogenous NATH–V5 co-localized with endogenous hARD1 in the cytoplasm as assessed by double immunostaining (Figures 7I–7L). The same result was also found when studying exogenous hARD1–V5 and endogenous NATH (Figures 7M–7P). Neither leptomycin B nor actinomycin D significantly changed the localization patterns of hARD1 or NATH (results not shown). This might indicate that NATH is not shuttling between nucleus and cytoplasm and that hARD1 is not actively imported through importin β-dependent mechanisms. In addition to a general diffuse nuclear hARD1 staining, distinct spots were also observed. Previous results have suggested that mARD1 interacts with HIF-1α in the cytoplasm [6]. The nuclear localization of hARD1 suggested that the interaction may be nuclear. To analyse this hypothesis, we performed co-localization experiments using anti-hARD1 and anti-HIF-1α (Figures 7Q–7V). HeLa cells displayed strong nuclear hARD1 staining (Figure 7Q) in most cells, whereas HIF-1α is detectable only in a fraction of the cells (Figure 7R). A partial overlap of hARD1 and HIF-1α staining was found in approx. 50% of representative nuclei, where both proteins were present at detectable levels (Figures 7T–7V).
Figure 7. Localization of NATH and hARD1 by immunofluorescence.
HeLa cells (A–D) and GaMg cells (E–H) were fixed and labelled with the indicated primary antibodies and thereafter with Alexa-488-conjugated anti-rabbit antibodies (green). Blue DAPI (4′,6-diamidino-2-phenylindole) staining (B, D, F, H) displays the nuclei of the cells. HeLa cells were transiently transfected with plasmids expressing NATH–V5 (I–L) and hARD1–V5 (M–P). Anti-V5 antibodies and Alexa-488 conjugated anti-mouse antibodies (green) were used to visualize NATH–V5 (I) and hARD1–V5 (M). Anti-NATH/anti-hARD1 and Alexa-568 conjugated antibodies (red) were used to visualize the endogenous hARD1 (J) and NATH (N). Nuclear DAPI staining is seen in blue (K, O). Green and red staining were overlaid (L, P). (Q–V) HeLa cells were co-labelled with anti-HIF1-α and anti-hARD1. Alexa-488 conjugated antibodies (green) were used to visualize hARD1 (Q, T) and Alexa-568 antibodies (red) to visualize HIF-1α (R, U). (S) DAPI-staining (blue) of cells in images (Q/R). (V) hARD1/HIF-1α (green/red) overlay of one HeLa nucleus from images (T/U). At least three independent experiments were performed.
NATH and hARD1 are cleaved during apoptosis
The protein translational machinery is targeted by caspases in apoptotic cells, resulting in a substantial decrease in protein synthesis [18–23]. Therefore we wanted to investigate whether or not the co-translational NATH–hARD1 NAT activity is also modulated in apoptosis. HeLa cells were treated with 10 μM daunorubicin for 20 h, resulting in more than 90% apoptotic cells by the end of the treatment, as assessed by light microscopy. Both full-length NATH and hARD1 proteins were present in stressed cells as demonstrated by Western blots (Figure 8A), but in addition to the wild-type NATH and hARD1 forms, novel faster migrating forms of hARD1 (hARD1-t) and NATH (NATH-t, truncated NATH) were present in the daunorubicin-treated cells (Figure 8A). Similar results were obtained using 200 nM daunorubicin for 24 h in NB4 cells, verifying the presence of hARD1-t and NATH-t in apoptotic cells (Figure 8B). The weak hARD1-t band in apoptotic NB4 cells (Figure 8B) might indicate that hARD1-t represents an intermediate cleavage species that is further degraded. In addition, we treated NB4 cells for 24 h with increasing concentrations of arsenic trioxide known to either induce cell maturation or apoptosis in a concentration-dependent manner [33]. The hARD1-t/hARD1 and NATH-t/NATH ratios increased with increasing arsenic concentrations and apoptosis (Figure 8C).
We analysed further whether or not NATH-t and hARD1-t resulted from caspase-cleavage by Z-VAD-FMK-mediated inhibition of caspase activity. NB4 cells were treated with daunorubicin for 20 h with or without Z-VAD-FMK for the last 10 h (Figure 8D). The addition of caspase inhibitor to stressed cells substantially decreased the amount of NATH-t and hARD1-t, strongly indicating that NATH and hARD1 are cleaved by caspases in cells undergoing apoptosis. Functional consequences of the cleavages of NATH and hARD1 were studied by preparing immunoprecipitates of hARD1 from apoptotic and normal HeLa cells and assaying for NAT activity. The amount of anti-hARD1 used in these immunoprecipitations was limited to ensure that equal amounts of hARD1 were present in the assays. Furthermore, the intensity of hARD1 and hARD1-t bands was determined by Western blotting and the acetyltransferase activity was adjusted accordingly (see the Experimental section). Compared with control extracts, the N-acetylase activity in extracts from apoptotic cells was reduced by approx. 40–80% (Figure 8E). Western blotting using anti-hARD1 in the apoptotic extracts identified both NATH and NATH-t (results not shown), demonstrating that the NATH-t can still interact with hARD1. The possibility therefore exists that different complexes between NATH–hARD1, NATH–hARD1-t, NATH-t–hARD1 and NATH-t–hARD1-t may be formed during apoptosis. The anti-NATH antibody recognizes the oligopeptide between amino acids 853 and 866, thus the NATH-t (apparently ∼37 kDa) represents the C-terminal part of NATH including the putative coiled-coil domain (between amino acids 583 and 635) (Figure 1B). The NATH–hARD1 interaction therefore takes place between hARD1 and the C-terminal end of NATH.
DISCUSSION
The NATH and hARD1 proteins are the human homologues of the components of the most common yeast NAT, NatA. Various aspects of protein N-acetylation and biological function of the NatA complex have been studied in yeast, but much less is described in higher eukaryotes [34]. NATH was first identified as overexpressed in papillary carcinomas of the thyroid gland [9], indicating that increased levels of the protein may provide a growth or survival advantage to the cancer cells. Protection against apoptotic cell death would be one such possibility [35]. Alternatively, a high protein synthesis rate in aggressive cancer cells would require an increase in NAT activity to ensure proper processing of newly synthesized proteins. To study further the biological function of NATH, we are the first to clone hARD1 and we show in this paper that endogenous NATH and hARD1 proteins are simultaneously expressed in several different human cell lines and they specifically interact to form a very stable complex that can easily be co-immunoprecipitated. This complex has protein N-terminal acetyltransferase activity. Thus these human proteins share at least some of the properties of their yeast homologues in terms of interaction and activity.
Our observation that ribosomal proteins interact with the NATH–hARD1 complex points to a potential functional link between the ribosome and the NAT. Specific NATH and/or hARD1 interactions with ribosomal proteins were repeatedly identified, possibly indicating that a specific interaction with one or more of these proteins occurs. For instance, the acidic P1 and P2 proteins are present at the surface of the ribosome [36], making them possible interaction partners, and they directly interact with each other [37]. Also the S15 protein is located at the ribosomal surface [38]. The anti-hARD1–V5 and anti-NATH antibodies mainly facilitated co-immunoprecipitation of proteins specific for either hARD1 or NATH (Table 2). This might reflect direct NATH or hARD1 interactions with different ribosomal proteins or different stabilities between the various complexes. There could also be a competition between antibody and ribosomal protein–NATH–hARD1 complex binding, resulting in the exclusion of specific protein binding. Since ribosomal proteins are immunoprecipitated with both NATH and hARD1, it is possible that both NATH and hARD1 directly interact with the ribosome. Alternatively, and perhaps quite probably, the ribosomal binding of one of the components occurs simultaneously, similar to the binding to the NAT partner. Either way, we therefore believe that the NATH and hARD1 proteins are bound to the polyribosome as one unit. The possibility that hARD1–V5 and NATH simply co-immunoprecipitated ribosomal proteins as they were being translated is excluded by the fact that the antibodies used are targeted against the C-terminal regions of the proteins and thus bind only to fully synthesized proteins. Protein N-acetylation is a co-translational process [39,40] and the observed NATH–hARD1-ribosome interaction may facilitate protein acetylation. Our results linking the NATH–ARD1 complex to the ribosome is in good agreement with the recent finding that the yeast NAT1–ARD1 complex is anchored to the ribosome [16]. The substantial amount of NATH and hARD1 found in the non-polyribosomal fraction indicates that both proteins are dynamically associated with the ribosomes and that NATH and hARD1 may have individual biological functions other than the ribosome-associated NAT activity. For hARD1, this is quite probable, considering its nuclear localization.
Additional functions of NATH and hARD1 may have evolved from yeast to human. The reported ARD1-mediated lysine ε-acetylation of HIF-1α is probably such an example [6]. The acetylation of HIF-1α was believed to occur in the cytoplasm because overexpressed tagged mARD1 was found in this compartment by biochemical fractionation [6]. As our results demonstrated that a substantial fraction of hARD1 is present in the nucleus, it may well be that the HIF-1α acetylation by ARD1 occurs in the nucleus and could be independent of NATH. Our co-localization studies using anti-hARD1 and anti-HIF1α in a normal population of HeLa cells demonstrated that hARD1 and HIF1α partially co-localize in nuclear spots. These results support the previously described interaction between hARD1 and HIF-1α [6], but indicate that the interaction occurs at distinct nuclear localizations and not in the cytoplasm as believed. Furthermore, a connection between hARD1 and HIF-1α in the nucleus, where no NATH is present could explain why the addition of mNAT1 did not affect the hARD1-mediated acetylation of HIF-1α [6].
NATH, also termed Tubedown 100, has been reported to act as a nuclear transcriptional cofactor, regulating expression from the osteocalcin promoter [41]. The presence of NATH in the nucleus is not confirmed by our data, but this localization may be tightly regulated and probably cell-type specific.
The caspase-mediated cleavage of NATH and hARD1 is probably another example of how the apoptotic cell shuts down vital biological processes. Several of the known caspase substrates are involved in protein translation [18–23]. The cleavage of hARD1, resulting in an approx. 1 kDa loss, most probably occurs in the C-terminal of the protein due to the presence of putative aspartate cleavage sites in this region. Two possible alternative cleavage sites are at Asp-223 (ESTD) and Asp-226 (DVKD). An observation of two-dimensional-gel patterns of hARD1 in apoptotic cells supported the presence of a far C-terminal cleavage site, in that hARD1-t displays a basic shift relative to hARD1 (results not shown). This indicates a loss of acidic residues found in the C-terminus. In comparison, the N-terminus of hARD1 is neutral. In addition, the only aspartate residue present in the N-terminus of hARD1, Asp-10 (RPED), has a very unfavourable surrounding sequence, with arginine at position P4 and proline at position P3 [42]. According to the mapping of the NATH interaction domain within hARD1, demonstrating that hARD1(1–222)–V5 and hARD1(1–174) bind NATH (Figure 4B), the cleavage of hARD1 probably does not affect NATH-binding. Since the anti-NATH antibody is directed against 14 amino acids at the C-terminal end, the NATH cleavage product detected under apoptosis must represent an approx. 35 kDa C-terminal NATH fragment. At present, we do not have tools to analyse the existence of a stable N-terminal fragment of NATH in apoptotic cells.
NATH-t is present when analysing the anti-hARD1-immunoprecipitates by Western blotting (results not shown). This indicates that hARD1 binds within this C-terminal portion of NATH. Thus, the formation of a stable complex appears to be dependent on the C-terminal region of NATH, but not the C-terminal region of hARD1. These results diverge from the previously published results for yeast, where the C-terminal portion of ARD1 seems to be necessary for the interaction with NAT1 [8]. However, hARD1 is most similar by sequence comparison with its yeast homologue in the N-terminal and acetyltransferase domains, whereas most of the C-terminal region of hARD1 does not have a counterpart in yeast ARD1 (Figure 1A). The very N-terminal of hARD1 is not necessarily directly involved in the NATH interaction. It is possible that the deletion of the 58 N-terminal amino acids of hARD1 affects the folding of the protein so that the interaction with NATH is disrupted on this account. hARD1 may have different biological activities depending on subcellular localization or post-translational modifications. By inspection of the hARD1 amino acids sequence, it is apparent that it contains several potential phosphorylation sites in the C-terminus, thus hARD1 may be regulated by multiple phosphorylation/dephosphorylation events. The putative phosphorylation sites in hARD1 are not present in the yeast ARD1, indicating a higher level of functional control in the hARD1 compared with yeast. The loss of an approx. 63 kDa N-terminal fragment of NATH and a minor (∼1 kDa) C-terminal hARD1 fragment in stressed cells does not appear to affect significantly the complex formation. The loss of these fragments may, however, affect the acetyltransferase activity. The possibility also exists that the acetyltransferase specificity will be altered by these truncations. TPR (tetratricopeptide repeat) motifs are known to be involved in protein–protein interactions [43]. The cleavage of NATH separates the TPR motifs from the C-terminus, and if the TPR motifs are responsible for ribosome binding, whereas the C-terminal binds hARD1, then this might separate the ribosomes and hARD1 and thereby disrupt the co-translational peptide N-acetylation by hARD1. This effect would not be observed in the in vitro assay used in the present study due to the availability of the substrates. However, our activity studies using enzyme complexes from apoptotic cells clearly demonstrate a significant decrease in enzyme activity. The truncations present may also affect possible individual, independent NATH and hARD1 functions. It cannot be ruled out that the caspase cleavage of NATH and hARD1 creates truncated proteins with new functions or with a change in, to date, unknown functions.
In summary, the NATH and hARD1 proteins represent the first described NATH complex. The ribosome-associated N-terminal acetyltransferase activity of the NATH–hARD1 complex may be evolutionarily conserved from yeast to humans, although a substantial fraction of the proteins seems not to be ribosome-associated. Both hARD1 and NATH are regulated by caspases in stressed cells, and stable proteolytic forms of these proteins are created. The hARD1 protein might have different functions depending on its localization and its partners. A nuclear activity of ARD1 has not been described for any organism, and identifying additional substrates of this acetyltransferase will give further insight into the biological importance of ARD1.
Acknowledgments
We thank K. Jacobsen for excellent technical assistance and F. Pendino and M. Ziegler for assistance with the manuscript. This work was supported by The Norwegian Cancer Society (grants to T.A., J.E.V. and J.R.L.), the Locus of Experimental Cancer Research (University of Bergen) and the Meltzer Foundation. Work at the University of Oregon was supported by a grant from the M.J. Murdock Charitable Trust.
References
- 1.Cohen H. Y., Lavu S., Bitterman K. J., Hekking B., Imahiyerobo T. A., Miller C., Frye R., Ploegh H., Kessler B. M., Sinclair D. A. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol. Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 2.Fu M., Wang C., Wang J., Zafonte B. T., Lisanti M. P., Pestell R. G. Acetylation in hormone signaling and the cell cycle. Cytokine Growth Factor Rev. 2002;13:259–276. doi: 10.1016/s1359-6101(02)00003-5. [DOI] [PubMed] [Google Scholar]
- 3.Mahlknecht U., Hoelzer D. Histone acetylation modifiers in the pathogenesis of malignant disease. Mol. Med. 2000;6:623–644. [PMC free article] [PubMed] [Google Scholar]
- 4.Di Gennaro E., Bruzzese F., Caraglia M., Abruzzese A., Budillon A. Acetylation of proteins as novel target for antitumor therapy: review article. Amino Acids. 2004;26:435–441. doi: 10.1007/s00726-004-0087-3. [DOI] [PubMed] [Google Scholar]
- 5.Sugiura N., Adams S. M., Corriveau R. A. An evolutionarily conserved N-terminal acetyltransferase complex associated with neuronal development. J. Biol. Chem. 2003;278:40113–40120. doi: 10.1074/jbc.M301218200. [DOI] [PubMed] [Google Scholar]
- 6.Jeong J. W., Bae M. K., Ahn M. Y., Kim S. H., Sohn T. K., Bae M. H., Yoo M. A., Song E. J., Lee K. J., Kim K. W. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell (Cambridge, Mass.) 2002;111:709–720. doi: 10.1016/s0092-8674(02)01085-1. [DOI] [PubMed] [Google Scholar]
- 7.Semenza G. L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 8.Park E. C., Szostak J. W. Ard1 and Nat1 proteins form a complex that has N-terminal acetyltransferase activity. EMBO J. 1992;11:2087–2093. doi: 10.1002/j.1460-2075.1992.tb05267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fluge O., Bruland O., Akslen L. A., Varhaug J. E., Lillehaug J. R. NATH, a novel gene overexpressed in papillary thyroid carcinomas. Oncogene. 2002;21:5056–5068. doi: 10.1038/sj.onc.1205687. [DOI] [PubMed] [Google Scholar]
- 10.Lee F. J. S., Lin L. W., Smith J. A. N-alpha acetylation is required for normal growth and mating of Saccharomyces cerevisiae. J. Bacteriol. 1989;171:5795–5802. doi: 10.1128/jb.171.11.5795-5802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullen J. R., Kayne P. S., Moerschell R. P., Tsunasawa S., Gribskov M., Colavitoshepanski M., Grunstein M., Sherman F., Sternglanz R. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 1989;8:2067–2075. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whiteway M., Szostak J. W. The Ard1 gene of yeast functions in the switch between the mitotic cell-cycle and alternative developmental pathways. Cell (Cambridge, Mass.) 1985;43:483–492. doi: 10.1016/0092-8674(85)90178-3. [DOI] [PubMed] [Google Scholar]
- 13.Whiteway M., Freedman R., Vanarsdell S., Szostak J. W., Thorner J. The yeast Ard1 gene-product is required for repression of cryptic mating-type information at the Hml locus. Mol. Cell. Biol. 1987;7:3713–3722. doi: 10.1128/mcb.7.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polevoda B., Norbeck J., Takakura H., Blomberg A., Sherman F. Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. EMBO J. 1999;18:6155–6168. doi: 10.1093/emboj/18.21.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Driessen H. P., de Jong W. W., Tesser G. I., Bloemendal H. The mechanism of N-terminal acetylation of proteins. CRC Crit. Rev. Biochem. 1985;18:281–325. doi: 10.3109/10409238509086784. [DOI] [PubMed] [Google Scholar]
- 16.Gautschi M., Just S., Mun A., Ross S., Rucknagel P., Dubaquie Y., Ehrenhofer-Murray A., Rospert S. The yeast N(alpha)-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol. Cell. Biol. 2003;23:7403–7414. doi: 10.1128/MCB.23.20.7403-7414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tribioli C., Mancini M., Plassart E., Bione S., Rivella S., Sala C., Torri G., Toniolo D. Isolation of new genes in distal Xq28: transcriptional map and identification of a human homologue of the ARD1 N-acetyl transferase of Saccharomyces cerevisiae. Hum. Mol. Genet. 1994;3:1061–1067. doi: 10.1093/hmg/3.7.1061. [DOI] [PubMed] [Google Scholar]
- 18.Bushell M., Wood W., Carpenter G., Pain V. M., Morley S. J., Clemens M. J. Disruption of the interaction of mammalian protein synthesis eukaryotic initiation factor 4B with the poly(A)-binding protein by caspase- and viral protease-mediated cleavages. J. Biol. Chem. 2001;276:23922–23928. doi: 10.1074/jbc.M100384200. [DOI] [PubMed] [Google Scholar]
- 19.Fischer U., Janicke R. U., Schulze-Osthoff K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 2003;10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marissen W. E., Lloyd R. E. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol. Cell. Biol. 1998;18:7565–7574. doi: 10.1128/mcb.18.12.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marissen W. E., Guo Y., Thomas A. A., Matts R. L., Lloyd R. E. Identification of caspase 3-mediated cleavage and functional alteration of eukaryotic initiation factor 2 alpha in apoptosis. J. Biol. Chem. 2000;275:9314–9323. doi: 10.1074/jbc.275.13.9314. [DOI] [PubMed] [Google Scholar]
- 22.Morley S. J., McKendrick L., Bushell M. Cleavage of translation initiation factor 4G (eIF4G) during anti-Fas IgM-induced apoptosis does not require signalling through the p38 mitogen-activated protein (MAP) kinase. FEBS Lett. 1998;438:41–48. doi: 10.1016/s0014-5793(98)01269-1. [DOI] [PubMed] [Google Scholar]
- 23.Satoh S., Hijikata M., Handa H., Shimotohno K. Caspase-mediated cleavage of eukaryotic translation initiation factor subunit 2 alpha. Biochem. J. 1999;342:65–70. [PMC free article] [PubMed] [Google Scholar]
- 24.Fluge O., Haugen D. R., Akslen L. A., Marstad A., Santoro M., Fusco A., Varhaug J. E., Lillehaug J. R. Expression and alternative splicing of c-ret RNA in papillary thyroid carcinomas. Oncogene. 2001;20:885–892. doi: 10.1038/sj.onc.1204161. [DOI] [PubMed] [Google Scholar]
- 25.Belmokhtar C. A., Hillion J., Dudognon C., Fiorentino S., Flexor M., Lanotte M., Segal-Bendirdjian E. Apoptosome-independent pathway for apoptosis. Biochemical analysis of APAF-1 defects and biological outcomes. J. Biol. Chem. 2003;278:29571–29580. doi: 10.1074/jbc.M302924200. [DOI] [PubMed] [Google Scholar]
- 26.Vedeler A., Pryme I. F., Hesketh J. E. The characterization of free, cytoskeletal and membrane-bound polysomes in Krebs II ascites and 3T3 cells. Mol. Cell. Biochem. 1991;100:183–193. doi: 10.1007/BF00234167. [DOI] [PubMed] [Google Scholar]
- 27.Vedeler A., Hollas H. Annexin II is associated with mRNAs which may constitute a distinct subpopulation. Biochem. J. 2000;348:565–572. [PMC free article] [PubMed] [Google Scholar]
- 28.Eng J. K., Mccormack A. L., Yates J. R. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 29.Gururaja T., Li W. Q., Bernstein J., Payan D. G., Anderson D. C. Use of MEDUSA-based data analysis and capillary HPLC-ion-trap mass spectrometry to examine complex immunoaffinity extracts of RbAp48. J. Proteome Res. 2002;1:253–261. doi: 10.1021/pr0255147. [DOI] [PubMed] [Google Scholar]
- 30.Anderson D. C., Li W. Q., Payan D. G., Noble W. S. A new algorithm for the evaluation of shotgun peptide sequencing in proteomics: support vector machine classification of peptide MS/MS spectra and SEQUEST scores. J. Proteome Res. 2003;2:137–146. doi: 10.1021/pr0255654. [DOI] [PubMed] [Google Scholar]
- 31.Gavin A. C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature (London) 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 32.Baum S., Bittins M., Frey S., Seedorf M. Asc1p, a WD40-domain containing adaptor protein, is required for the interaction of the RNA-binding protein Scp160p with polysomes. Biochem. J. 2004;380:823–830. doi: 10.1042/BJ20031962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen G. Q., Zhu J., Shi X. G., Ni J. H., Zhong H. J., Si G. Y., Jin X. L., Tang W., Li X. S., Xong S. M., et al. In vitro studies on cellular and molecular mechanisms of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia: As2O3 induces NB4 cell apoptosis with downregulation of Bcl-2 expression and modulation of PML-RAR alpha/PML proteins. Blood. 1996;88:1052–1061. [PubMed] [Google Scholar]
- 34.Polevoda B., Sherman F. Composition and function of the eukaryotic N-terminal acetyltransferase subunits. Biochem. Biophys. Res. Commun. 2003;308:1–11. doi: 10.1016/s0006-291x(03)01316-0. [DOI] [PubMed] [Google Scholar]
- 35.Mitsiades N., Poulaki V., Tseleni-Balafouta S., Koutras D. A., Stamenkovic I. Thyroid carcinoma cells are resistant to FAS-mediated apoptosis but sensitive tumor necrosis factor-related apoptosis-inducing ligand. Cancer Res. 2000;60:4122–4129. [PubMed] [Google Scholar]
- 36.Agafonov D. E., Kolb V. A., Spirin A. S. Proteins on ribosome surface: measurements of protein exposure by hot tritium bombardment technique. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12892–12897. doi: 10.1073/pnas.94.24.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tchorzewski M., Boldyreff B., Issinger O., Grankowski N. Analysis of the protein-protein interactions between the human acidic ribosomal P-proteins: evaluation by the two hybrid system. Int. J. Biochem. Cell Biol. 2000;32:737–746. doi: 10.1016/s1357-2725(00)00017-0. [DOI] [PubMed] [Google Scholar]
- 38.Marion M. J., Marion C. Ribosomal proteins S2, S6, S10, S14, S15 and S25 are localized on the surface of mammalian 40 S subunits and stabilize their conformation. A study with immobilized trypsin. FEBS Lett. 1988;232:281–285. doi: 10.1016/0014-5793(88)80753-1. [DOI] [PubMed] [Google Scholar]
- 39.Pestana A., Pitot H. C. Acetylation of nascent polypeptide chains on rat liver polyribosomes in vivo and in vitro. Biochemistry. 1975;14:1404–1412. doi: 10.1021/bi00678a010. [DOI] [PubMed] [Google Scholar]
- 40.Yamada R., Bradshaw R. A. Rat liver polysome N alpha-acetyltransferase: isolation and characterization. Biochemistry. 1991;30:1010–1016. doi: 10.1021/bi00218a018. [DOI] [PubMed] [Google Scholar]
- 41.Willis D. M., Loewy A. P., Charlton-Kachigian N., Shao J. S., Ornitz D. M., Towler D. A. Regulation of osteocalcin gene expression by a novel Ku antigen transcription factor complex. J. Biol. Chem. 2002;277:37280–37291. doi: 10.1074/jbc.M206482200. [DOI] [PubMed] [Google Scholar]
- 42.Thornberry N. A., Rano T. A., Peterson E. P., Rasper D. M., Timkey T., Garcia-Calvo M., Houtzager V. M., Nordstrom P. A., Roy S., Vaillancourt J. P., et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 43.Blatch G. L., Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]