Abstract
Ydj1 is the major type I Hsp40 (heat-shock protein 40) family member in yeast. Ydj1 can pair with yeast Hsp70 Ssa1 to facilitate protein translocation and protein folding. Ydj1 itself can also function as a molecular chaperone to bind the non-native polypeptides and suppress protein aggregations in vitro. The crystal structure of Ydj1 complexed with its peptide substrate GWLYEIS reveals that a hydrophobic pocket located on Ydj1 domain I may play a major role in mediating the interactions between Ydj1 and the peptide substrate. To understand the mechanism by which Ydj1 interacts with non-native polypeptide, we have mutated the residues forming the hydrophobic pocket, based on the structural information. We have also constructed deletion mutations of the zinc-finger motifs within Ydj1. We have examined the functional consequences of these Ydj1 mutants by in vivo and in vitro assays. The results indicated that the hydrophobic pocket located on Ydj1 plays a critical role in its molecular chaperone activity by mediating interactions with the non-native polypeptides.
Keywords: crystal structure, heat-shock protein 40 (Hsp40), Hsp70, mutagenesis, substrate binding
Abbreviations: BS, borate-buffered saline; Hsp, heat-shock protein; ITC, isothermal titration calorimetry
INTRODUCTION
Molecular chaperone Hsp40 (heat-shock protein 40) plays critical roles in cell physiology by acting with molecular chaperone Hsp70 members to promote protein folding, assembly, translocation and degradation [1–3]. Hsp40 proteins can interact with the hydrophobic side chains of non-native polypeptides through the peptide-binding fragment and prevent the polypeptides from aggregating [4,5]. Hsp40s can then form transient complexes with Hsp70s and present the non-native polypeptides to Hsp70s for subsequent protein folding [6–8].
All Hsp40 proteins contain an N-terminal J-domain, which is approx. 70 amino acid residues long [9,10]. The J-domain can stimulate the ATPase activities of Hsp70 [1,11]. ATP hydrolysis will cause the conformational changes in the peptide-binding domain of Hsp70. These conformational changes will then stabilize the Hsp70 and peptide substrate complexes [1,12] and allow the subsequent protein folding to occur.
Both type I and type II Hsp40 have a peptide-binding fragment located at the C-terminus of the proteins. The N-terminal J-domains are connected to the peptide-binding fragments through a G/F rich linker in both type I and type II Hsp40s. Type I Hsp40 such as Escherichia coli DnaJ, yeast Ydj1 and human Hdj2 contain two zinc-finger-like motifs between the J-domain and the C-terminal peptide-binding fragment within their primary sequences, while type II Hsp40 proteins such as yeast Sis1 and human Hdj1 do not [13,14]. It has been reported that the ability to bind non-native polypeptides for the cytosolic Hsp40 is an essential function in vivo [19]. Type III Hsp40 only contains a J-domain and other specific domains and may not act as molecular chaperones [20].
The mechanism by which molecular chaperone Hsp40s interact with the non-native polypeptide substrates is still elusive. It was proposed that the zinc-finger-like motifs of Hsp40 type I DnaJ were involved in the peptide binding [13,15–18]. The NMR studies of DnaJ zinc-finger motifs indicated that the zinc-finger motifs have a V-shaped structure with two zinc atoms co-ordinated in the zinc-binding modules [21]. There is a well-conserved hydrophobic pocket in the middle part of zinc-finger motifs, which was suggested to be a potential peptide-binding site for DnaJ [21].
For type II yeast Hsp40 protein, which has no zinc-finger motif, the substrate binding site was found to be on its C-terminus [15,16]. The crystal structure of the C-terminus of Sis1, a yeast type II Hsp40 protein, revealed that the Sis1 functioned as a homodimer with a U-shaped molecular structure [22]. A hydrophobic depression about 5 Å deep (1 Å=10−10 m) was located on the molecular surface of domain I of Sis1 peptide-binding fragment monomer. It is proposed that yeast type II Hsp40 Sis1 dimer may interact with the non-native polypeptides through the two hydrophobic depressions [22]. Structure-based mutational analysis on the conserved residues constituting the hydrophobic patch supported this proposal [23]. Simultaneous binding of a non-native polypeptide at two sites on a Sis1 dimer might serve to hold the substrate in an extended conformation, which is preferred by Hsp70 [12,22,24].
Recently, we identified a peptide substrate GWLYEIS for type I Hsp40 Ydj1 using phage display library screening. The crystal structure of the complex of Ydj1 and the peptide substrate GWLYEIS has been determined [25]. The crystal structure of Ydj1 and the peptide substrate complex revealed the direct interaction between Ydj1 and the peptide substrate. The peptide substrate forms an extra β-strand with Ydj1 peptide-binding fragment domain I. The leucine residue in the middle of the peptide substrate plays important roles in mediating the specific binding of the peptide substrate to Hsp40 Ydj1. In the crystal structure, the side chain of the leucine residue of the peptide substrate is buried in a hydrophobic pocket on the molecular surface of Ydj1 [25]. The Sis1 peptide-binding site is also located at a similar position in the Sis1 peptide-binding fragment structure [22]. It is reasonable to believe that both type I and type II Hsp40 proteins may utilize these hydrophobic regions located on domain I to interact with the hydrophobic side chains of the non-native polypeptides. The complex crystal structure also revealed that two zinc-finger motifs are not directly involved in the peptide substrate binding. One zinc-finger motif (Zn II) formed by Cys159, Cys162, Cys185 and Cys188 was distal from the peptide-binding site, whereas the other zinc-finger (Zn I) formed by Cys143, Cys146, Cys201 and Cys204 connected directly to the β-strand that binds to the peptide substrate.
To reveal the mechanisms by which type I Hsp40 interacts with non-native polypeptides to facilitate protein folding, we performed a mutational analysis of the residues forming the hydrophobic pocket as well as the zinc-finger motifs of type I Hsp40 Ydj1. The results provided here suggest that the hydrophobic pocket located on Ydj1 plays critical roles in its molecular chaperone activity by mediating interactions with the non-native polypeptides.
EXPERIMENTAL
Subcloning and construction of site-directed mutagenesis
The DNA fragment encoding full-length ydj1 was amplified from Ydj1 cDNA by PCR using the 5′-primer ggaattccatatggttaaagaaactaagttttacgatattctagg and the 3′-primer ccgctcgagtcattgagatgcacattgaacaccttcgccaccttg. The PCR products were digested by using restriction endonucleases NdeI and XhoI (New England Biolabs, Beverly, MA, U.S.A.). The digested inserts were then ligated into the digested pET28b vector by T4 ligase.
The recombinant Ydj1 and its mutants were overexpressed in E. coli using the vector pET28b (Novagen, Madison, WI, U.S.A.). The vector pRS316 (from A.T.C.C., Manassas, VA, U.S.A.) was utilized to transform the Ydj1 mutant gene into yeast cells. The site mutations for pET28b-ydj1 and pRS316-ydj1 were made by use of Quik Change site-directed mutagenesis kit (Stratagene) and were verified through DNA sequencing. The pRS316-Y1 and pRS316-Y2 were constructed similarly to pET28b-Y1 and pET28b-Y2.
The expression plasmids for ydj1Δ(159–188) (Y1) and ydj1Δ(143–206) (Y2) were generated from the ligation of two DNA fragments (corresponding to peptides 1–142 and 207–409, 1–158 and 189–409 respectively) into plasmids pET28b, to generate pet28b-Y1 and pet28b-Y2. A short peptide sequence of GGSG was inserted in the position of the deleted zinc-finger motif in the Ydj1 sequence. For the pet28b-Y1 construction, DNA fragment 1 was PCR-amplified from the 5′-primer 5′-ggaattccatatggttaaagaaactaagttttacgatattctagg-3′ and the 3′-primer 5′-cgcggatccggtcacggtactggtgatatcattgatcctaaggatcg-3′ and digested with NdeI and BamHI. It encodes for wtYdj1 residues 1–158 and inserted residues GGS. DNA fragment 2 was PCR-amplified from the 5′-primer 5′-cgcggatccacccttcttgacggcgcctttcttaccaccacgaccttc-3′ and the 3′-primer 5′-ccgctcgagtcattgagatgcacattgaacaccttcgccaccttg-3′ and digested with BamHI and XhoI. It encodes for residues SGG and wtYdj1 residues 189–409. For the pET28b-Y2 construction, DNA fragment 1 was PCR-amplified from the 5′-primer 5′-ggaattccatatggttaaagaaactaagttttacgatattctagg-3′ and the 3′-primer 5′-cgcggatccggtaacggtaagaaagttgaaaacgaaaggaagatcc-3′ and digested with NdeI and BamHI. It encodes for wtYdj1 residues 1–205 and residues GGS. DNA fragment 2 was PCR-amplified from the 5′-primer 5′-cgcggatccacctaggatctgtttgttaagggctaacttagctgtcc-3′ and the 3′-primer 5′-ccgctcgagtcattgagatgcacattgaacaccttcgccaccttg-3′ and digested with BamHI and XhoI. It encodes for residues SGG and wtYdj1 residues 207–409. The plasmids were then transformed into E. coli strain BL21(DE3) for protein expression.
Protein expression and purification
The yeast Ssa1 was expressed by yeast strain MW141 grown in YPG media to D600 6.0 and purified using the method described in [25].
E. coli BL21(DE3) cells containing the pET28b-ydj1 or its mutant plasmids were grown at 37 °C in Luria–Bertani broth to D600 0.6 and then induced with 1 mM isopropyl β-D-thiogalactoside for 3 h. The cells were harvested 3 h later after induction. Purification was achieved by using a nickel column. The typical yield of soluble Ydj1 or mutants (∼95% pure from SDS/PAGE analysis) from 1 litre culture was approx. 15 mg. The N-terminal His-tag of Ydj1 was then digested by thrombin treatment. Thrombin (1 unit; Sigma) was utilized for 1 mg Ydj1 protein. The digestion was allowed for 12 h at room temperature and stopped by the addition of 0.2 mM PMSF. The protein was further purified on a gel filtration column Superdex 200 (Amersham Biosciences) mounted on an AKTA HPLC system (Amersham Biosciences) to remove thrombin and digested peptides.
ITC (isothermal titration calorimetry) assay
Measurement of the binding affinities between Ydj1 and peptide substrates was performed by using an isothermal titration calorimeter (MicroCal, Northampton, MA, U.S.A.) at room temperature. Ydj1 (or the mutant) and the peptides were dialysed against the same buffer (10 mM Mes, pH 6.0, 100 mM NaCl, 1 mM 2-mercaptoethanol). Ydj1 (or the mutant) was placed in the calorimetric cell and the synthetic peptide was injected into the cell by a 250 μl injection syringe. The released heat was calculated by integrating the calorimetric output curves. Pure buffers were injected into the Ydj1 protein as control experiments. The heat releases from the control experiments were subtracted from the experimental results before the results were utilized for Kd fitting. The Kd values and the binding ratios were calculated using the software supplied with the calorimeter.
Luciferase refolding assay
Luciferase (15 mg/ml; Promega, Madison, WI, U.S.A.) was diluted 42 times with the denaturant solution (30 mM Hepes, pH 7.5, 50 mM KCl, 10 mM MgCl2, 7 M urea) and incubated at room temperature for 40 min. Then the denatured luciferase was diluted 125 times with 125 μl of refolding solution (30 mM Hepes, pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM ATP, 1.6 μM Ydj1 or various mutants, 0.8 μM Hsp70 Ssa1). Luciferase activity was determined by luciferase assay kit (Promega).
Rhodanese/luciferase aggregation assay
Rates of rhodanese/luciferase aggregation were determined by light scattering as described in [15]. Bovine rhodanese or insect recombinant luciferase (50 μM; Sigma) was denatured for 1 h at 25 °C in 6 M guanidinium/HCl buffered with 20 mM Tris (pH 7.4). Denatured rhodanese was diluted 100-fold into reaction buffer composed of 20 mM Tris (pH 7.4) and 150 mM NaCl. At the time of reaction, Ydj1 or its mutants (5 μM) were added into buffers before denatured rhodanese/luciferase. Rates of rhodanese/luciferase aggregation were determined by monitoring increases in light scattering over time with a spectrophotometer set at 320 nm at 25 °C.
Measurement of polypeptide binding to Hsp40 by ELISA
The ELISA was used to measure complex formation between Hsp40 proteins and denatured luciferase [23]. Hsp40s were diluted to 50 nM in 50 mM borate, 100 mM NaCl (pH 8.4) (BS; borate-buffered saline). Then, 100 μl aliquots of Ydj1 or mutant solutions were added to the wells of 96-well microtitre plates. The proteins were incubated in the wells for 1 h at room temperature. Wells were then washed to remove unbound Hsp40 using BS. Wells were then blocked with 200 μl of 1% BSA in BS for a 1 h. Guanidinium/HCl denatured luciferase (0.4 μg) in BS with 0.5% BSA was then added to each well. After 1 h incubation at 25 °C, the unbound luciferase was then taken out and the wells were washed three times with BS. Luciferase bound in the wells was then detected by an ELISA kit (Cortex Biochem, San Leandro, CA, U.S.A.). In the kit, goat α-rabbit serum coupled with alkaline phosphatase (AP) was used to detect the α-luciferase that was retained in the wells. Colour density was measured by use of a microplate reader.
Function of Ydj1 mutants in vivo
The wild-type Ydj1 is essential to support yeast growth at 37 °C. To test the function of mutant Ydj1 in vivo, we use a Δydj1 yeast strain JJ160 (ydj1::HIS3) and transform it with Ydj1 or its mutants supplied on the low copy Ura3 plasmid pRS316. The 3.6-kb fragment of ydj1 was cloned into the centromeric plasmid pRS316 to create pRS316-ydj1. The mutagenized 3.6 kb fragment encompasses 500 bp upstream of the ATG through 1.4 kb downstream of the stop codon [27]. The growth defects were examined for the yeast strains transformed with Ydj1 mutants. To select Ydj1 or its mutants existing in the URA plasmids, transformants were grown on drop-out media without uracil. Strains were grown at 30 or 37 °C for 4–5 days, and the plates were then scanned.
Western-blot analysis of Ydj1 expression
The expression levels of Ydj11 mutants were detected by Western blot using the yeast extracts with polyclonal Ydj1 antibody from rabbits. Yeast strains were grown in selective media without uracil to D600 3.0. Yeast cells were broken by glass beads in the buffers with protease inhibitor cocktails. The cell lysate was mixed with the same volume of SDS/PAGE sample buffer and then centrifuged for 5 min. The supernatant was loaded on to SDS/13% polyacrylamide gel, electrophoresed, and then transferred on to PVDF membranes for blotting.
RESULTS
Identification of a hydrophobic pocket important for peptide binding in the Ydj1–peptide complex structure
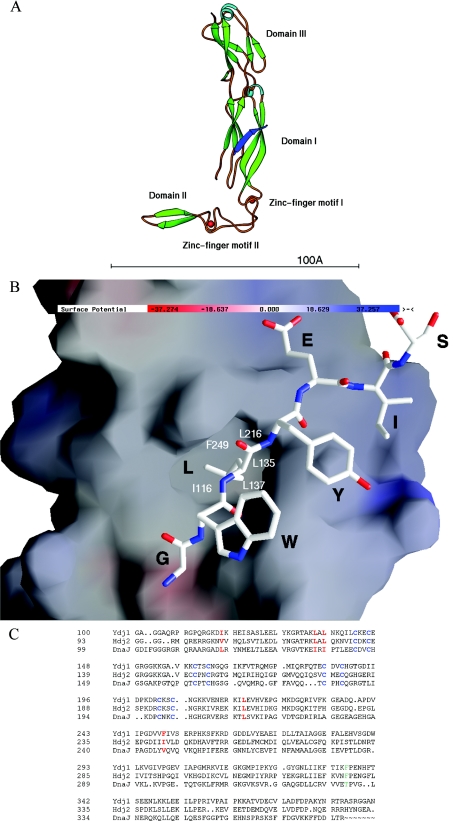
The crystal structure of Hsp40 Ydj1 complexed with the peptide substrate GWLYEIS indicated that the bound peptide substrate GWLYEIS forms an anti-parallel β-strand with the β-sheet from domain I of theYdj1 structure [25] (Figure 1A). The leucine residue in the middle of the peptide substrate GWLYEIS makes most of the contacts with Ydj1 among the seven amino acid residues. The side chain of this leucine is fully buried in a hydrophobic pocket formed on the surface of domain I of Ydj1 (Figure 1B). The hydrophobic pocket is about 5 Å×7 Å×7 Å (width×length×depth; Figure 1B) in dimension. This pocket is constructed by a number of hydrophobic residues that include Ile116 from B1, Leu135 and Leu137 from B2, Leu216 from B5 and Phe249 from B7. The hydrophobicity of these residues is fairly conserved among the family members of type I Hsp40s, indicating that this hydrophobic pocket may be a common feature for this molecular chaperone family (Figure 1C). It is reasonable to postulate that this hydrophobic pocket located in domain I of Ydj1 may play important roles in mediating the interactions between type I Hsp40 and the non-native polypeptides.
Figure 1. The structure of Hsp40 Ydj1 complexed with the peptide substrate and the sequence alignment of the C-terminal regions from Hsp40 family members.
(A) The ribbon drawing of the Ydj1 peptide-binding fragment complexed with the peptide substrate GWLYEIS. For Ydj1 structure, the α-helices are shown in light blue and the β-strands are shown in green. The bound peptide GWLYEIS is shown in blue. The two zinc-finger motifs are labelled. The two zinc atoms are shown as red spheres. The bar at the bottom of the Figure indicates 100 Å. (B) GRASP presentations of the Ydj1 and the peptide substrate complex structure. The orientation of the complex molecule in (B) is similar to that in (A). Blue and red denote positively and negatively charged regions respectively. The bound peptide GWLYEIS is shown as a rod model. The residues of the peptide substrate GWLYEIS are labelled in black. In the rod model, carbon atoms are shown in white, oxygen atoms are shown in red and the nitrogen atoms are shown in blue. The residues involved in forming the hydrophobic pocket are labelled in white. (C) Sequence alignment of the C-terminal regions from Hsp40 family members. Program Pileup from the GCG package was utilized to align the C-terminal peptide-binding fragment of type I Hsp40 Ydj1 from Saccharomyces cerevisiae with similar regions of type I Hsp40 proteins from Homo sapiens Hdj-2 and E. coli DnaJ. The zinc-finger motifs (CXXC) within type I Hsp40s are labelled in blue. The residues that are involved in forming the hydrophobic pocket on Ydj1 domain I are marked in red.
Ydj1 mutants in hydrophobic pocket exhibit severe defects in molecular chaperone activities
To test whether the hydrophobic pocket located on the surface of Ydj1 domain I plays an important role in type I Hsp40 chaperone function, we have mutated the hydrophobic residues Ile116, Leu135, Leu137, Leu216 and Phe249, which are involved in forming the pocket, to alanine residues to reduce the hydrophobic nature of the pocket. The structure-based Ydj1 mutants were expressed as the recombinant form in E. coli and purified to homogeneity.
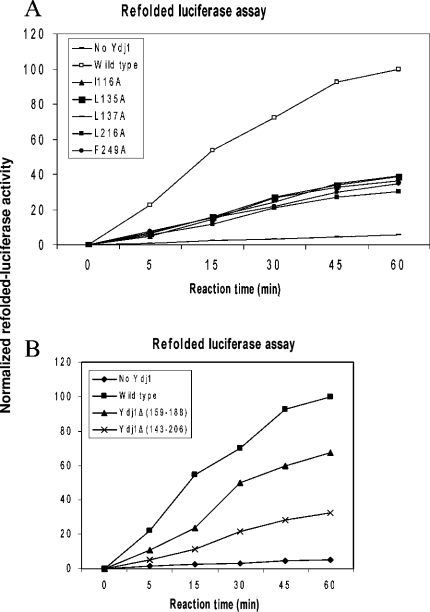
The molecular chaperone activities of these Ydj1 mutants to assist Hsp70 in refolding denatured proteins are measured by the luciferase refolding assay (Figure 2A). Wild-type Ydj1 can co-operate with the yeast Hsp70 Ssa1 to refold denatured forms of the model protein luciferase. When the abilities of the missense mutants to assist Ssa1 to refold chemically denatured luciferase were examined, all of the mutants exhibited severe defects in their molecular chaperone activities. These Ydj1 mutants retain only approx. 25–35% of their wild-type molecular chaperone ability to facilitate Hsp70 Ssa1 to refold denatured luciferase (Figure 2A). Therefore the hydrophobic residues involved in forming the hydrophobic pocket of Ydj1 are critical for type I Hsp40 molecular chaperone activity.
Figure 2. Molecular chaperone activities of Ydj1 mutants to assist Hsp70 Ssa1 in folding chemically denatured luciferase.
The horizontal axis indicates the reaction time in minutes and the vertical axis shows the normalized refolded luciferase activities. Wild-type Ydj1 was used as the positive control and the refolded luciferase activity with wild-type Ydj1 and Hsp70 Ssa1 after 60 min reaction time is defined as 100% in this Figure. The refolded luciferase activity with no Ydj1 in the reaction is used as the negative control. (A) The molecular chaperone activities of the Ydj1 missense mutants. The residues involved in forming the hydrophobic pocket located on Ydj1 domain are mutated to alanine residues and their chaperone activities to assist Hsp70 Ssa1 to refold chemically denatured luciferase are measured. (B) The molecular chaperone activities of the Ydj1 zinc-finger motif deletion mutants to assist Hsp70 Ssa1 in folding the chemically denatured luciferase.
Ydj1 missense mutants show severe defects in peptide GWLYEIS binding
The binding affinities of these missense mutants to the Ydj1 peptide substrate GWLYEIS were measured by ITC studies. None of these Ydj1 mutants showed detectable binding affinity to the peptide substrate GWLYEIS (Table 1). This result strongly suggested that the hydrophobicity of the Ydj1 peptide-binding pocket plays a critical role in mediating Hsp40 and peptide substrate interactions. It is quite probable that the substantial loss of the molecular chaperone activities of these Ydj1 mutants resulted from their much reduced ability to bind to the peptide substrates. Our results strongly support that the hydrophobic pocket located on Ydj1 domain I contributes significantly to the molecular chaperone activity by interacting with the hydrophobic side chains of the polypeptide substrates.
Table 1. The binding affinities between the wild-type Ydj1 and mutants and the Ydj1 peptide substrate GWLYEIS measured by ITC studies.
The ITC studies also generated the molar binding ratios between the peptides and the Ydj1 monomer by fitting the experimental results to the standard curve. ND, not detectable by ITC.
| Ydj1 and mutants | Dissociation constant (μM) | Binding molar ratio |
|---|---|---|
| Wild-type | 12.5 | 1.06 |
| I116A | ND | ND |
| L135A | ND | ND |
| L137A | ND | ND |
| L216A | ND | ND |
| F249A | ND | ND |
| Ydj1Δ(159–188) | 24.0 | 0.91 |
| Ydj1Δ(143–206) | 58.4 | 1.26 |
Ydj1 missense mutants show severe defects in Ydj1 Hsp70-independent chaperone activity
Hsp40 Ydj1 contains Hsp70-independent chaperone function to suppress non-native protein aggregations, which relies on the direct interactions between Ydj1 and the unfolded polypeptides. To test whether the hydrophobic pocket in Ydj1 domain I is responsible for interactions with non-native polypeptides, the aforementioned Ydj1 mutants were examined for their ability to suppress the aggregation of denatured luciferase and rhodanese (Figures 3A and 3B). The results indicate that the ability of Ydj1 missense mutants to prevent rhodanese and luciferase aggregation is compromised compared with the wild-type Ydj1. These results suggest that the hydrophobic pocket is a region capable of binding unfolded protein substrates.
Figure 3. The Hsp70-independent molecular chaperone activities of Ydj1 mutants in suppressing aggregations of the chemically denatured substrates (luciferase and rhodanese).
The horizontal axis indicates the reaction time in min and the vertical axis shows the normalized light scattering values (D320). Wild-type (WT) Ydj1 was used as the positive control. The residues involved in forming the hydrophobic pocket located on Ydj1 domain are mutated to alanine residues and their independent chaperone activities to prevent chemically denatured luciferase (A) and rhodanese (B) aggregation are measured. (C) Binding of Ydj1 to unfolded luciferase measured by ELISA.
Ydj1 mutants exhibit defects in polypeptide binding
To further confirm that the Ydj1 mutants exhibited defects in non-native polypeptide binding, we also utilized an ELISA assay to analyse their ability to form stable complexes with unfolded luciferase directly (Figure 3C). Compared with wild-type Ydj1, all the missense mutants in the hydrophobic pocket have significantly lower binding capacity (∼50%) to unfolded luciferase. The results provided direct evidence to support the notion that residues forming the hydrophobic pocket are required for non-native polypeptide binding. These results strongly indicate that these Ydj1 missense mutants are defective in molecular chaperone activity because they have a much reduced ability to associate with denatured luciferase.
The functions of the Ydj1 zinc-finger motifs
To investigate the possible roles that the Ydj1 zinc-finger motifs may play in type I Hsp40 molecular chaperone activities, we have constructed two deletion mutants Ydj1Δ(159–188) (Y1) and Ydj1Δ(143–206) (Y2). In Y1, the zinc-finger motif (Zn II) that is distal to the peptide-binding site was deleted whereas both zinc-finger motifs (Zn I and Zn II) were removed in Y2, based on the information provided by the crystal structure of Ydj1 ([25], Figure 1A). A short flexible loop with the sequence of GGSG was inserted in the position of the deleted zinc-finger motifs in the Ydj1 structure to maintain the correct protein folding. The two Ydj1 deletion mutants were expressed as the recombinant form in E. coli and purified to homogeneity. The molecular chaperone activities of the Ydj1 deletion mutants to assist Hsp70 Ssa1 to refold denatured luciferase were measured by the luciferase refolding assay (Figure 2B). The binding affinities of the mutants to the Ydj1 peptide substrate GWLYEIS were measured by ITC studies (Table 1). The deletion mutant Y1 (Ydj1ΔZn II) showed considerable binding affinity to the peptide substrate GWLYEIS while Y2 [Ydj1Δ(Zn I and Zn II)] had a significantly reduced binding affinity. The deletion mutant Y1, which does not contain the distal zinc-finger motif from the peptide-binding site, possessed approx. 60% of the wild-type molecular chaperone activity to assist Hsp70 to refold polypeptides. The deletion mutant Y2 with both zinc-finger motifs removed possessed only approx. 25% of the wild-type molecular chaperone activity. Apparently the zinc-finger motifs in type I Hsp40 play significant roles in its molecular chaperone activity to assist Hsp70 to refold denatured proteins.
To monitor the direct binding between Ydj1 zinc-finger motifs with non-native polypeptides, we examined the abilities of deletion mutants to suppress the aggregation of denatured luciferase and rhodanese (Figures 3A and 3B). We have also performed ELISAs to test the interactions of the deletion mutants with unfolded luciferase (Figure 3C). The results showed that both deletion mutants Y1 and Y2 were defective in suppressing luciferase aggregation but they have capabilities similar the wild-type to suppress rhodanese aggregation. The ELISA results indicated that Y1 had almost the same binding affinity as the wild-type with unfolded luciferase, whereas Y2 possessed only approx. 50% of the wild-type binding affinity as the to unfolded luciferase.
Combining all the results above, we propose that the distal zinc-finger motif Zn II from the peptide-binding site may not be involved in non-native polypeptide binding, while the other zinc-finger motif Zn I may play a role in non-native polypeptide binding. The zinc-finger motifs seem to have specificity for polypeptide substrates when Hsp40 Ydj1 functions to suppress protein aggregations.
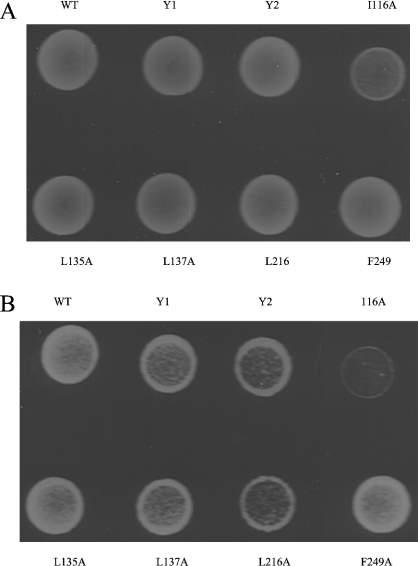
Some Ydj1 mutations compromised yeast growth at 37 °C in vivo
To test the importance of the residues constituting the hydrophobic pocket in vivo, we have transformed the missense mutants into the yeast stain Δydj1 (Figure 4). Almost no growth defects can be observed in the ydj1 mutants at 30 °C (Figure 4A). This is probably because Ydj1 functions to sustain the cell viability at 37 °C. At 37 °C, yeast strain Δydj1 transformed with ydj1 I116A (Ile116→Ala) is not viable (Figure 4B). Growth defects were seen in the yeast strain Δydj1 transformed with mutants L137A and L216A. These results indicate that the hydrophobic pocket located on Ydj1 domain I may contribute significantly to maintain the normal cell growth in vivo possibly through interacting with the non-native polypeptides. The yeast strain Δydj1 transformed with mutants L135A and F249A grew normally at 37 °C. Residues Leu135 and Phe249 were shown to be essential for chaperone activity in vitro but not essential for cell growth in vivo (Figure 4B). It is possible that the mutations of these two residues impair Ydj1 binding affinity to the natural polypeptide substrates to a lesser extent than to luciferase and rhodanese.
Figure 4. Growth phenotypes of ydj1 mutants.
The wild-type or mutant ydj1 genes were transformed into the yeast strain Δydj1 by the plasmid shuffle method and selected on media without Uracil. Strains were grown at (A) 30 °C or (B) 37 °C for 4 days.
We have also examined the significance of the zinc-finger motifs in vivo by transforming the zinc-finger motif deletion mutants Y1 and Y2 into the yeast strain Δydj1. Severe growth defects were found for both of the zinc-finger motif deletion mutants at 37 °C (Figure 4B), which indicates that the zinc-finger motifs play important roles for type I Hsp40 molecular chaperone activity to support the cell growth.
It is not likely that the loss of function of these structure-based mutants results from misfolding of the proteins. All the recombinant proteins eluted from a gel-filtration column at the expected size (results not shown). They all possessed capabilities similar to the wild-type to stimulate the ATPase activities of yeast Hsp70 Ssa1 (results not shown).
Expressions of these structure-based Ydj1 mutants are under the control of the ydj1 promoter, therefore the expression levels of these mutants should be similar to the wild-type. To check these mutant expression levels in vivo, we examined the protein expressions of the transformed mutants in the yeast strain Δydj1 cell lysates by Western blotting using a polyclonal antibody against the wild-type Ydj1 (Figure 5). Western blotting clearly showed that all the missense mutants and the deletion mutants of Ydj1 were expressed in the yeast at similar levels as the wild-type Ydj1. This suggested that the growth defects of the Δydj1 yeast strain transformed by the missense and deletion mutants resulted from loss of function, not from the low expression levels of the mutants.
Figure 5. Western blotting of the yeast lysates reveals the expression levels of the yeast strains transformed with ydj1 mutants.
The ydj1 wild-type (WT) and the mutants are labelled in the Figure.
DISCUSSION
The crystal structure of Hsp40 Ydj1 complexed with the peptide substrate revealed that the Ydj1 peptide substrate forms an extra β-strand within domain I of Ydj1 [25]. The crystal structure of the complex identifies a hydrophobic pocket located on domain I of Ydj1 that plays important roles in mediating peptide substrate interactions. This pocket is mainly constituted by five hydrophobic residues Ile116, Leu135, Leu137, Leu216 and Phe249. Mutations of these hydrophobic residues abolished the peptide substrate binding ability for Hsp40 Ydj1. These mutants also exhibited severe defects in their molecular chaperone activities due to their compromised polypeptide binding capabilities. The structural and mutagenesis results demonstrated that the hydrophobic pocket located on Ydj1 domain I plays significant roles in type I Hsp40 molecular chaperone activity by mediating non-native polypeptide interactions.
This hydrophobic pocket may bind hydrophobic side chains from the non-native polypeptide. The ITC experiments for Ydj1 and synthetic peptides indicated that Ydj1 prefers a hydrophobic residue in the middle of the peptide substrate with the minimum length of six residues [25]. Therefore the pocket may define the peptide substrate specificity for type I Hsp40. Mutations in the pocket may reduce the hydrophobicity of this area, which may result in the changes in the type I Hsp40 specificity for binding the non-native polypeptides.
To test the possible functions of the Ydj1 zinc-finger motifs, we have constructed two mutants by deleting the distal zinc-finger motif Zn II from the binding site (Y1) or both of the zinc-finger motifs (Y2). Functional studies showed that both deletion mutants exhibit defects in their molecular chaperone activities to assist Hsp70 to refold the denatured luciferase. The deletion of the distal zinc-finger motif Zn II from the peptide-binding site had little effect on polypeptide substrate binding while deletion of the two zinc-finger motifs compromised Ydj1 polypeptide binding ability. The results suggest that the distal zinc-finger motif Zn II may not be involved in peptide substrate binding while the other zinc-finger motif Zn I may play a role in mediating substrate binding. It has been suggested that the Zn I of E. coli type I Hsp40 DnaJ is involved in peptide binding and the Zn II of DnaJ may play roles for locking-in substrate proteins on to Hsp70 DnaK [26]. This is consistent with our observations. Interestingly, the deletions of zinc-finger motifs of Ydj1 did not affect capability of Ydj1 in suppressing Rhodanese aggregation, whereas they compromised the capability of Ydj1 in suppressing Luciferase aggregation. We hypothesize that the zinc-finger motifs seem to have specificities for polypeptide substrates when Hsp40 Ydj1 functions to suppress protein aggregations.
It has been reported that the C-terminal substrate-binding regions for both type I and II Hsp40s play essential roles for yeast viability in vivo [19]. In this study, we further demonstrate that the hydrophobic pocket located in the peptide-binding fragment of type I Hsp40 may sustain the yeast normal growth at 37 °C by mediating the interactions between Hsp40 and the non-native polypeptides in vivo. A hydrophobic depression located in the peptide-binding fragment of type II Hsp40 Sis1 was discovered to be the putative peptide-binding site [22,25]. The structure-based mutagenesis of this hydrophobic depression for Sis1 caused growth defect [23]. Therefore both type I and II Hsp40 may utilize similar hydrophobic regions to interact with non-native polypeptides. The peptide-binding activities for both type I and II Hsp40s may play important roles to achieve their molecular chaperone activities and to sustain the normal cell growth in vivo.
Acknowledgments
We thank Professor Elizabeth Craig, University of Wisconsin-Madison, Madison, WI, U.S.A., for providing yeast strain MW141 for Ssa1 overexpression and yeast strains JJ160 for studies in vivo. We also thank Dr Douglas Cyr, University of North Carolina School of Medicine, Chapel Hill, NC, U.S.A., for the gift of cDNA of Ydj1. The work is supported by grants from NIH R01 DDK56203 and R01 GM65959 and NASA to B.D.S.
References
- 1.Bukau B., Horwich A. L. The Hsp70 and Hsp60 chaperone machines. Cell (Cambridge, Mass.) 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 2.Hartl F. U. Molecular chaperones in cellular protein folding. Nature (London) 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 3.Hartl F. U., Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 4.Langer T., Lu C., Echols H., Flanagan J., Hayer M. K., Hartl F. U. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature (London) 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 5.Schmid D., Baici A., Gehring H., Christen P. Kinetics of molecular chaperone action. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- 6.Laufen T., Mayer M. P., Beisel C., Klostermeier D., Mogk A., Reinstein J., Bukau B. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. U.S.A. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misselwitz B., Staeck O., Rapoport T. A. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol. Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- 8.Qian X., Hou W., Zhengang L., Sha B. Direct interactions between molecular chaperones heat-shock protein (Hsp) 70 and Hsp40: yeast Hsp70 Ssa1 binds the extreme C-terminal region of yeast Hsp40 Sis1. Biochem. J. 2002;361:27–34. doi: 10.1042/0264-6021:3610027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zylicz M., Yamamoto T., McKittrick N., Sell S., Georgopoulos C. Purification and properties of the dnaJ replication protein of Escherichia coli. J. Biol. Chem. 1985;260:7591–7598. [PubMed] [Google Scholar]
- 10.Georgopoulos C. P., Lundquist-Heil A., Yochem J., Feiss M. Identification of the E. coli dnaJ gene product. Mol. Gen. Genet. 1980;178:583–588. doi: 10.1007/BF00337864. [DOI] [PubMed] [Google Scholar]
- 11.Wall D., Zylicz M., Georgopoulos C. The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J. Biol. Chem. 1994;269:5446–5451. [PubMed] [Google Scholar]
- 12.Zhu X., Zhao X., Burkholder W. F., Gragerov A., Ogata C. M., Gottesman M. E., Hendrickson W. A. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banecki B., Liberek K., Wall D., Wawrzynow A., Georgopoulos C., Bertoli E., Tanfani F., Zylicz M. Structure-function analysis of the zinc finger region of the DnaJ molecular chaperone. J. Biol. Chem. 1996;271:14840–14848. doi: 10.1074/jbc.271.25.14840. [DOI] [PubMed] [Google Scholar]
- 14.Caplan A. J., Douglas M. G. Characterization of YDJ1: a yeast homologue of the bacterial dnaJ protein. J. Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Z., Cyr D. M. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J. Biol. Chem. 1998;273:5970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- 16.Lu Z., Cyr D. M. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J. Biol. Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- 17.Szabo A., Korszun R., Hartl F. U., Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- 18.Goffin L., Georgopoulos C. Genetic and biochemical characterization of mutations affecting the carboxy-terminal domain of the Escherichia coli molecular chaperone DnaJ. Mol. Microbiol. 1998;30:329–340. doi: 10.1046/j.1365-2958.1998.01067.x. [DOI] [PubMed] [Google Scholar]
- 19.Johnson J. L., Craig E. A. An essential role for the substrate-binding region of Hsp40s in Saccharomyces cerevisiae. J. Cell Biol. 2001;152:851–856. doi: 10.1083/jcb.152.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheetham M. E., Caplan A. J. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Yamout M., Legge G. B., Zhang O., Wright P. E., Dyson H. J. Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ. J. Mol. Biol. 2000;300:805–818. doi: 10.1006/jmbi.2000.3923. [DOI] [PubMed] [Google Scholar]
- 22.Sha B., Lee S., Cyr D. M. The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Structure Fold. Des. 2000;8:799–807. doi: 10.1016/s0969-2126(00)00170-2. [DOI] [PubMed] [Google Scholar]
- 23.Lee S., Fan C. Y., Younger J. M., Ren H., Cyr D. M. Identification of essential residues in the type II Hsp40 Sis1 that function in polypeptide binding. J. Biol. Chem. 2002;277:21675–21682. doi: 10.1074/jbc.M111075200. [DOI] [PubMed] [Google Scholar]
- 24.Flynn G. C., Pohl J., Flocco M. T., Rothman J. E. Peptide-binding specificity of the molecular chaperone BiP. Nature (London) 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 25.Li J., Qian X., Sha B. D. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure. 2003;11:1475–1483. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 26.Linke K., Wolfram T., Bussemer J., Jakob U. The roles of the two zinc binding sites in DnaJ. J. Biol. Chem. 2003;278:44457–44466. doi: 10.1074/jbc.M307491200. [DOI] [PubMed] [Google Scholar]
- 27.Johnson J. L., Craig E. A. A role for the Hsp40 Ydj1 in repression of basal steroid receptor activity in yeast. Mol. Cell. Biol. 2000;20:3027–3036. doi: 10.1128/mcb.20.9.3027-3036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]