Abstract
Phenylalanine ammonia lyase (PAL) is a widely studied enzyme in plant biology due to its role in connecting primary metabolism to secondary phenylpropanoid metabolism, significantly influencing plant growth, development, and stress response. Although PAL genes have been extensively studied in various plant species but their exploration in cucumber has been limited. This study successfully identified 11 CsPAL genes in Cucumis sativus (cucumber). These CsPAL genes were categorized based on their conserved sequences revealing patterns through MEME analysis and multiple sequence alignment. Interestingly, cis-elements related to stress were found in the promoter regions of CsPAL genes, indicating their involvement in responding to abiotic stress. Furthermore, these gene’s promoters contained components associated with light, development and hormone responsiveness. This suggests that they may have roles in hormone developmental processes. MicroRNAs were identified as a key regulators for the CsPAL genes, playing a crucial role in modulating their expression. This discovery underscores the complex regulatory network involved in the plant’s response to various stress conditions. The influence of these microRNAs further highlights the complicated mechanisms that plants use to manage stress. Gene expression patterns were analyzed using RNA-seq data. The significant upregulation of CsPAL9 during HT3h (heat stress for 3 h) and the heightened upregulation of both CsPAL9 and CsPAL7 under HT6h (heat stress for 6 h) in the transcriptome study suggest a potential role for these genes in cucumber’s tolerance to heat stress. This comprehensive investigation aims to enhance our understanding of the PAL gene family’s versatility, offering valuable insights for advancements in cucumber genetics.
Keywords: Cucumis sativus, Cucumber, Phenylalanine ammonia-lyase (PAL), Transcriptomic analysis, Gene regulation, Abiotic stress, Heat stress
Introduction
The role of phenylalanine ammonia-lyase (PAL) is crucial for understanding its involvement in plant stress responses [1]. The enzyme plays a central role in key metabolic pathways that influence how plants adapt to and manage stress. In the biosynthetic pathways of numerous eukaryotes, a fundamental step is the removal of an amino group from L-phenylalanine, resulting in the formation of trans-cinnamic acid [2]. This reaction is catalyzed by the enzyme phenylalanine ammonia-lyase, which plays a crucial role in the process [3, 4]. PAL collaborates with tyrosine and the specialized compound dihydroxyphenylalidone to generate diverse secondary metabolites, including flavonoids and lignin [5, 6]. Among these metabolites, jasmonic acid (JA) and abscisic acid are crucial for regulating the plant’s genetic response to various stressors. These compounds help modulate the plant’s defense mechanisms and adaptive responses [7]. Phenylalanine ammonia-lyase (PAL) stands out due to its unique ability to be activated by multiple stress factors simultaneously, a feature that distinguishes it from many other enzymes involved in stress responses [8]. This multifunctional activation underscores PAL’s significant role in the plant’s comprehensive stress management system [9]. Consequently, PAL plays a pivotal role in the plant’s defense mechanisms, enabling it to effectively counter both biotic and abiotic stresses. By modulating the PAL enzyme activity, plants can better manage and respond to various environmental challenges. Enhancing the expression of the PAL gene can therefore be a strategic approach to boost the plant’s resilience, potentially leading to improved stress tolerance and overall plant health under diverse and challenging conditions [10, 11].
The phenylpropanoid pathway initiates with PAL, the enzyme responsible for removing an amino group from L-phenylalanine to produce trans-cinnamic acid This crucial reaction is integral to the synthesis of essential secondary metabolites influencing plant growth and development [9, 12]. Moreover, PAL plays a pivotal role in the plant’s stress response system, influenced by various factors. Changes in PAL expression occur in response to factors such as water scarcity from drought, fungal infections [13], injuries, excessive bleeding, and stressors specific to cut flowers or exposure to unfavorable temperatures.
The rapid response of the PAL gene underscores its critical importance for plants in managing diverse biotic and abiotic stresses [1]. The acceleration of PAL gene expression would represent a substantial breakthrough in enhancing plants’ resistance to a wide array of stress conditions. By increasing the activity of this crucial enzyme, plants could achieve improved tolerance to both biotic and abiotic stresses, including drought, salinity, and pathogen attacks. This advancement holds the promise of significantly boosting plant health and productivity, leading to more resilient crops capable of thriving in challenging environments [14]. As a pivotal enzyme in the synthesis of essential secondary metabolites, PAL holds a vital role in both plant growth and development. Moreover, it plays a significant part in a substantial portion of the plant’s stress response mechanisms [15].
PAL has different levels of activity depending on the developmental stages and the different cell and tissue types of a plant. These activity changes are more noticeable when the plant faces increased stress conditions in its life cycle [16]. For example, in Arabidopsis thaliana, there are four genes, called AtPAL1-4, that make active PAL isoforms [17]. These genes have different expression patterns in different plant parts. AtPAL1 is mainly found in vascular tissue, while seeds and are the main sites of expression for AtPAL2 and AtPAL4; how much they are expressed here depends a lot on the sensitivity criteria used [18–20].
Conversely, agricultural methods employing open fields may pose significant adverse consequences. Of the identified stressors, AtPAL1 and AtPAL2 demonstrated increased activity in response to low temperature and low nitrogen levels, respectively [21]. The expression of PAL genes exhibits variability among various plant species, specific plant components, and even distinct stress conditions, highlighting the dynamic nature of PAL gene regulation [22].
Cucumber (C. sativus) is a warm-season crop susceptible to both biotic and abiotic stress factors [23–26]. High temperatures, salinity, and pathogen attacks significantly impact cucumber yield and quality during summer cultivation or protected land production [27–29]. However, the specific PAL genes responsible for biotic and abiotic tolerance in cucumbers remain unidentified [30–33]. The recent update to the cucumber genome v3 offers an excellent opportunity for a comprehensive analysis of the PAL gene family [34]. In this investigation, we successfully identified 11 PAL genes within the Cucumis sativus v1.0 genome (https://phytozome-next.jgi.doe.gov/info/Csativus_v1_0), marking a significant step in understanding the genetic basis of stress responses in this species. Following this identification, we performed a different analysis of these genes, which included examining their sequence features to understand their structure and function, determining their chromosomal locations to map their distribution across the genome, and exploring their phylogenetic relationships to reveal evolutionary patterns and similarities with PAL genes in other plants. Additionally, we assessed the dynamic expression patterns of these genes under various biotic and abiotic stress conditions, providing insights into how they respond to environmental challenges.
This comprehensive analysis not only deepens our understanding of the functional roles of PAL genes in cucumber but also uncovers potential candidate genes that could be utilized in breeding programs. By targeting these candidate genes, we aim to develop new cucumber varieties with enhanced stress resistance, which could lead to improved resilience and productivity in challenging agricultural environments. This research opens avenues for advancing cucumber cultivation and could have broader implications for plant breeding strategies aimed at increasing crop durability and performance.
Methodology
Retrieve sequences from databases
The amino acid sequences of C. sativus were retrieved from the Phytozome v13 database (https://phytozome-next.jgi.doe.gov). The BLAST-P (Basic Local Alignment Search Tool for Protein Sequences) program was used to find the PAL genes of C. sativus, using the protein sequence with the PF00221 (Aromatic amino acid lyase) domain as a query at Phytozome v 13) [35]. Following this, the acquired amino acid sequences underwent verification through cross-checking against NCBI’s Conserved Domain Database (https://www.ncbi.nlm.nih.gov/genome/) [36].
Physicochemical properties, subcellular localization and cis elements
Two sources were used to collect data on 11 CsPAL proteins: Protparam (https://web.expasy.org/protparam/) and Phytozome. Phytozome gave information about the number and location of chromosomes as well as the direction (sense or antisense) of the gene, in that specific area. It also gave information on mRNA length (CDS) and peptide size. Protparam gave information such as theoretical pI, molecular weight, GRAVY (Grand Average of Hydropathy) and stability index, for these proteins. The WoLF PSORT database (https://wolfpsort.hgc.jp/ ) was used to find out the localization. The 1500-base pair upstream promoter regions were obtained from phytozomeV3 (https://phytozome-next.jgi.doe.gov). The web tool PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), was used to make a prediction based on 5 to 20 base pairs of upstream sequence from the first nucleotide in each case). These outputs were shown as a heat map, using TBtools for visualization [37].
Analysis of conserved motif domain and exon-intron arrangement
The MEME program, which can be found online at (http://meme.sdsc.edu/meme/website/intro .html ), was used to find motifs (motif = 10 default setting) in the amino acid sequences. Then, the amino acid sequences were added to the CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), which uses domains identified by NCBI as a database [35]. To analyze the exon and intron distribution, the GSDS web tool at (http://gsds.cbi.pku.edu.cn/) was used to process the genomic and CDS sequences of the CsPAL gene family [38].
Comparative phylogenetic analysis
The amino acid sequences of CsPAL proteins were aligned with those from C. sativus, O. sativa, A. thaliana and S. bicolor, to make a phylogenetic tree. The software MEGA 11 was used with the NJ (neighbor-joining) method, to build the tree and bootstrapping was done with 1000 replications. The tree was visually adjusted using the iTOL program (https://itol.embl.de/), which allows users to examine and annotate the phylogenetic relationships. [39].
miRNA analysis
The target sites of all CsPAL gene family were identified using the PmiREN website (https://www.pmiren.com/). There were three main groups of miRNAs (Csa-miRN967, Csa-miR166, and Csa-miR4414) that targeted the CsPAL genes. When considering their subgroups, there were 20 miRNAs in total that targeted the CsPAL genes. The CDS sequences of the genes and mature miRNA sequences were compared using the PsRNA online server tool (https://www.zhaolab.org/psRNATarget/) with default settings. Then, the Cytoscape program was used to show the interactions between target genes and the predicted miRNA [40].
Evolutionary analysis
The study investigated how PAL genes in cucumber have undergone changes through analyses of duplication and synteny. Protein sequences were aligned using the MUSCLE program, and the Tbtools 1.108 with default settings was employed to calculate the Ka/Ks substitution rates, revealing the evolutionary pace of each gene pair. The Ks value, incorporated into the formula T = Ks/2λ, where λ = 6.5 × 10− 9, was utilized to estimate the divergence time between these genes. Gene duplication events were identified using MCScanX v1.0 with default settings [35, 41]. Maps illustrating the synteny between paralogous genes in cucumber were created using Tbtools. This software was employed to visualize and analyze the gene relationships and genomic organization within the cucumber genome [40].
Analyzing gene structure
The gene structure display server (GSDS v2.0) (http://gsds.cbi.pku.edu.cn/) was used to analyze the gene structure and find out the intron-exon patterns. The PlantCare database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) was used to identify the cis-regulatory elements [42].
Gene Ontology (GO) term analysis
The research conducted a GO enrichment analysis to elucidate the functions of cucumber genes. Information regarding their activities and participation in biological processes was sourced from the Uniprot online database (https://www.uniprot.org/) [43]. Subsequently, the ShinyGo v0.741 web tool (http://bioinformatics.sdstate.edu/go/ ) was employed to perform the GO term enrichment analysis using the PAL gene sequences [40].
Protein-protein interaction
The study also confirmed the protein interactions among PAL genes by utilizing the String database v0.741 (https://string-db.org/ ). This online resource played a pivotal role in depicting the intricate network of interactions among proteins within the PAL genes of cucumber.
Expression analysis
RNA-seq data of CsPAL genes were obtained from the NCBI GEO database https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151055. (Accession: GSE151055) to analyze and investigate their expression profiles in response to heat stress. Three-week-old cucumber seedlings underwent heat treatment at 42℃, and leaves were collected at 0 (control), 3, and 6 h. This analysis aimed to identify variations between conditions and offer insights into potential targets for further investigation [44].
Results
Identifying PAL Genes C. sativus in and their Localization
The molecular features of the 11 CsPAL genes were extensively analyzed to uncover their specific characteristics. CsPAL10 had the lowest molecular weight of 17846.26 Da, while CsPAL8 had the highest of 78413.3 Da. The pI values varied from a low of 23.94 in CsPAL10 to a high of 38.61 in CsPAL8. All genes except CsPAL10 had negative Gravy scores, indicating that they were hydrophilic. CsPAL10 also had the shortest CDS length of 540, while CsPAL5 had the longest of 2154. Moreover, peptide length ranged from a low of 171 in CsPAL10 to a high of 717 in CsPAL5. We conducted a genomic-level investigation of the CsPAL genes, mapping their positions and orientations on chromosomes. Among the 11 CsPAL genes under scrutiny, CsPAL1, CsPAL2, CsPAL3, CsPAL4, CsPAL5, CsPAL6, CsPAL7, and CsPAL8 are situated on chromosome 1, CsPAL9 on chromosome 1, and CsPAL10 and CsPAL11 form a cluster on chromosome 4. Upon further examination of gene orientations, CsPAL1, CsPAL2, and CsPAL3 are oriented in the forward (F) direction, while CsPAL4, CsPAL5, CsPAL6, CsPAL7, CsPAL8, CsPAL9, CsPAL10, and CsPAL11 are oriented in the reverse (R) direction (Table 1)
Table 1.
Details on the PALgene family include information on 11 non-redundant genes identified in the cucumber genome. II: instability index, pl: isoelectric point, GRAVY: Grand Average of Hydropathy, pep: peptide, mw: Molecular Weight, Da: Dalton
| Rename | Phytozome | GenPept | CuGenDBv2 | PAC | no. | Scaffold | Start | End | Position | CDS | Pep | Mw (Da) | pI | II | GRAVY |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CSPAL1 | Cucsa.124,450 | XP_004143255.1 | CsaV3_6G039660.1 | 16,961,714 | 6 | scaffold01001 | 389,195 | 392,389 | F | 2145 | 714 | 78033.18 | 6.37 | 35.95 | -0.182 |
| CSPAL2 | Cucsa.124,460 | XP_031743814.1 | CsaV3_6G039670.1 | 16,961,715 | 6 | scaffold01001 | 398,860 | 401,204 | F | 2139 | 712 | 77793.65 | 5.8 | 35.29 | -0.194 |
| CSPAL3 | Cucsa.124,470 | XP_011657651.2 | CsaV3_6G039680.1 | 16,961,716 | 6 | scaffold01001 | 404,132 | 406,259 | F | 2127 | 708 | 77546.46 | 5.76 | 35.51 | -0.177 |
| CSPAL4 | Cucsa.124,480 | XP_004143258.1 | CsaV3_6G039690.1 | 16,961,717 | 6 | scaffold01001 | 408,087 | 410,360 | R | 2142 | 713 | 78054.03 | 6.15 | 36.57 | -0.191 |
| CSPAL5 | Cucsa.124,490 | XP_004143356.3 | CsaV3_6G039700.1 | 16,961,718 | 6 | scaffold01001 | 412,913 | 415,097 | R | 2154 | 717 | 78235.39 | 6.18 | 32.54 | -0.146 |
| CSPAL6 | Cucsa.124,500 | XP_031744164.1 | CsaV3_6G039710.1 | 16,961,719 | 6 | scaffold01001 | 417,206 | 419,324 | R | 2118 | 705 | 77236.4 | 6.12 | 37.04 | -0.152 |
| CSPAL7 | Cucsa.124,510 | XP_004143260.2 | CsaV3_6G039720.1 | 16,961,720 | 6 | scaffold01001 | 421,196 | 423,314 | R | 2118 | 705 | 77172.26 | 5.96 | 34.25 | -0.085 |
| CSPAL8 | Cucsa.137,590 | XP_004149916.1 | CsaV3_6G036550.1 | 16,963,222 | 6 | scaffold01053 | 349,339 | 352,789 | R | 2142 | 713 | 78413.3 | 6.26 | 38.61 | -0.229 |
| CSPAL9 | Cucsa.155,520 | XP_004144018.1 | CsaV3_1G040750.1 | 16,964,812 | 1 | scaffold01136 | 265,075 | 267,105 | R | 2030 | 675 | 74044.82 | 5.37 | 35.48 | -0.091 |
| CSPAL10 | Cucsa.350,120 | XP_004152155.3 | CsaV3_4G002330.1 | 16,979,871 | 4 | scaffold03490 | 26 | 540 | R | 514 | 171 | 17846.26 | 7.96 | 23.94 | 0.035 |
| CSPAL11 | Cucsa.385,970 | XP_031739733.1 | CsaV3_6G014060.1 | 16,982,658 | 4 | scaffold03904 | 1,153,049 | 1,157,200 | R | 2142 | 713 | 77640.7 | 6.23 | 28.22 | -0.177 |
In terms of subcellular localization, notable variations exist among the CsPAL genes. CsPAL7 exhibits the highest localization in chloroplasts, with a substantial presence of 4, contributing to a total of 16.5 (10%). Similarly, CsPAL2 shows significant localization in the Endoplasmic Reticulum, with 3, constituting 16 (10%) of its total. CsPAL3 demonstrates a diverse distribution, with 3 in chloroplasts and 2 in the cytoplasm, resulting in a total of 16 (10%). Conversely, CsPAL10, CsPAL4, and CsPAL9 display the minimum localization, each totaling 14 (8%). CsPAL10 is primarily localized in chloroplasts (7), while CsPAL4 shows multiple locations, including Cytoplasmic Nucleus, Peroxisomal, and Golgi apparatus, each with 1. CsPAL9 is predominantly found in the Endoplasmic Reticulum, accounting for its minimal localization (Fig. 1).
Fig. 1.
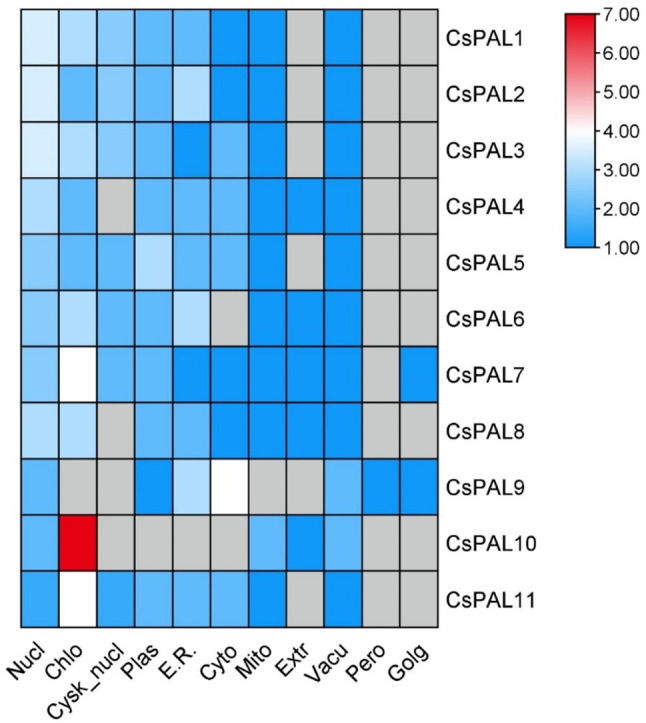
Heat map displays 11 CsPAL gene distribution in plant cell components; red indicates higher functional significance, revealing specific gene presence in nucleus, cytoplasm, chloroplast, Golgi apparatus, mitochondria, plasmid, and peroxisomes
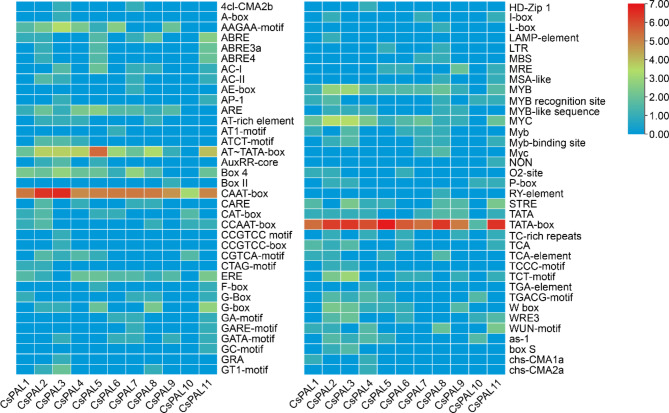
Conserved Cis elements
In examining the promoter regions of the 11 CsPAL genes, a diverse array of regulatory motifs controlling their transcription was identified. The TATA-box motif emerged as a prevalent feature across all genes, constituting 39% of the discovered motifs. Additionally, the CAAT-box, another recurrent motif, represented 26% of the motifs. Various motifs such as the MYB recognition site, MYC, and W box displayed distinct frequencies, suggesting potential roles in gene regulation. Notably, the STRE motif, associated with stress response, was present in 1% of the motifs. This comprehensive analysis underscored crucial motifs associated with stress response and plant defense mechanisms within the CsPAL gene family. With 451 occurrences, the CAAT-box emerged as a dominant cis-acting element, underscoring its significance in orchestrating CsPAL expression during stress [45]. Furthermore, the identification of elements such as the W box, known for its role in responding to pathogen attacks, and the STRE motif emphasized the participation of these genes in stress-responsive pathways. The significant occurrences of Myb, MYB-like sequence, and WRE3 motifs, linked to transcriptional responses associated with drought and low oxygen, offered additional valuable insights (Fig. 2)
Fig. 2.
Cis regulatory analysis of the CsPAL gene and the relative importance of its functions
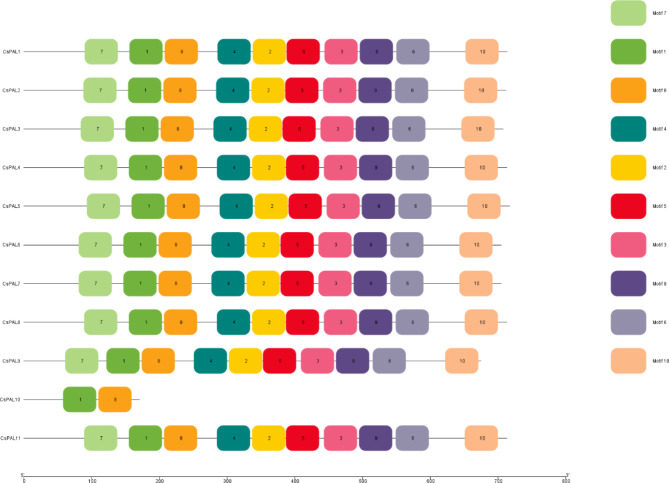
Conserved motif analysis and domain prediction
In the analysis of conserved patterns among the 11 CsPAL genes, it became apparent that Motifs 1 and 8 were consistently present throughout the entire gene set. Notably, Motifs 2, 3, 4, 5, 6, 7, 9, and 10 showed conservation in all genes with the exception of CsPAL10. It is worth mentioning that Demain PLN02457 exhibited the presence of Motifs 2, 3, 4, 5, 6, 7, 9, and 10, while the Lyase_I_like superfamily featured Motifs 1 and 8 Fig. 3.
Fig. 3.
The ten motifs’ distribution across the ten CsPAL protein family members. The CsPAL proteins reveals consistent presence of specific motifs across the protein set, highlighting distinct patterns in domain and superfamily structures
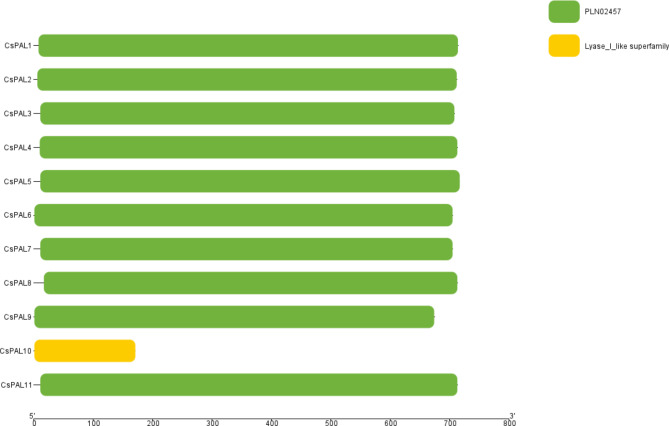
Our analysis of conserved domains among the 11 CsPAL genes revealed a striking uniformity, as a single domain identified as the Lyase_I_like superfamily (Accession: cl00013) was consistently present in all genes. Notably, CsPAL1, CsPAL2, CsPAL3, CsPAL4, CsPAL5, CsPAL6, CsPAL7, CsPAL8, CsPAL9, and CsPAL11 exhibited an additional conserved domain, PLN02457 (Accession: PLN02457), which is part of the Lyase_I_like superfamily. CsPAL10, on the other hand, belonged to the Lyase_I_like superfamily. This underscores a high level of conservation of the Lyase_I_like superfamily domain across all 11 CsPAL genes Fig. 4.
Fig. 4.
The conserved domain analysis revealed the consistent presence of the Lyase_I_like superfamily across all 11 poteins genes, with an additional PLN02457 domain found in most genes
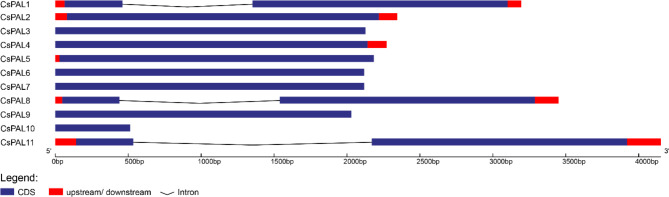
Exon intron analysis
The examination of the exon-intron structures of the CsPAL gene family revealed distinct patterns. CsPAL2, CsPAL3, CsPAL4, CsPAL5, CsPAL6, CsPAL7, CsPAL9, and CsPAL10 exhibited a unique profile, characterized by a solitary exon and the absence of introns. In contrast, CsPAL1, CsPAL8, and CsPAL11 shared a conserved structure, marked by two exons and one intron. These findings underscore notable genomic variations within the CsPAL gene family, highlighting diversity in their exon-intron organization Fig. 5.
Fig. 5.
The CsPAL gene family exhibits distinct exon-intron structures, with some genes having a single exon and no introns, while others have two exons and one intron. Exons are represented in blue and introns in black line
Analysis of the CsPAL gene family’s phylogeny
In the phylogenetic analysis aimed at identifying conserved patterns, PAL genes from four plant species O. sativa, C. sativus, A. thaliana, and S. bicolor were systematically grouped into three distinct clades denoted as I-III. The analysis included a total of 35 PAL genes, comprising 10 from O. sativa, 11 from C. sativus, 5 from A. thaliana, and 9 from S. bicolor. To enhance clarity and facilitate a comprehensive understanding of the phylogenetic relationships, each clade was assigned a specific color scheme: green for Clade I, red for Clade II, and blue for Clade III Fig. 6.
Fig. 6.
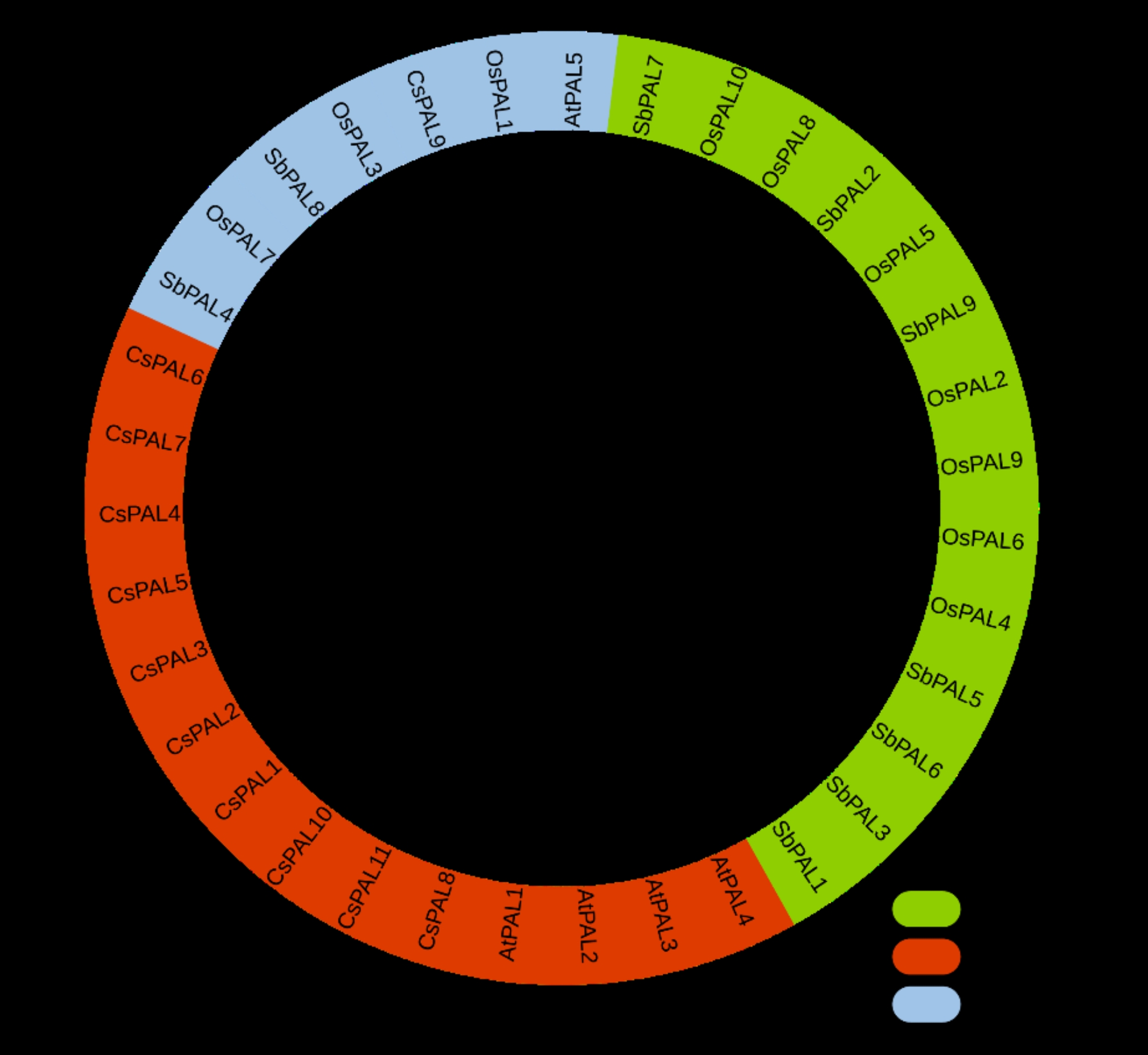
The phylogenetic analysis of 35 PAL genes from four plant species O. sativa, C. sativus, A. thaliana, and S. bicolor were systematically grouped into three distinct clades: Clade I (green), Clade II (red), and Clade III (blue)
MicroRNA (miRNA) analysis
The miRNA analysis of CsPAL genes revealed a range of interactions with distinct miRNAs, each characterized by varying lengths. The shortest miRNA, Csa-miR166, covered positions 1 to 21, while the longest, Csa-miRN967, extended from position 1 to 22. All targeted miRNAs play a role in cleavage inhibition. Specifically, Csa-miRN967 targeted the genes CsPAL3 and CsPAL10, Csa-miR166 targeted CsPAL2 and CsPAL4, and Csa-miRN967 targeted CsPAL5.
PAL gene duplication and synteny analysis
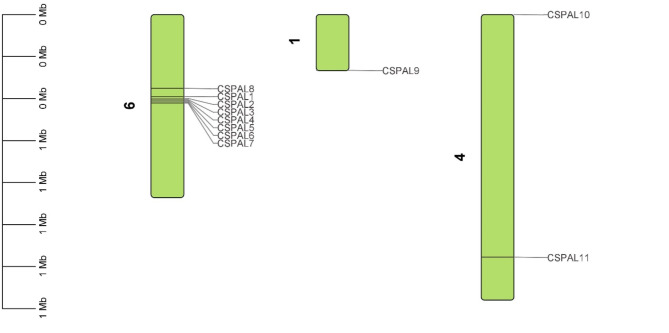
The chromosomal arrangement of PAL genes reveals their distribution across multiple chromosomes. PAL genes were detected on chromosomes 1, 4, and 6. Significantly, the predominant location for PAL genes was identified on chromosome 6, encompassing CsPAL1, CsPAL2, CsPAL3, CsPAL4, CsPAL5, CsPAL6, CsPAL7, and CsPAL8. Furthermore, chromosome 1 retained CsPAL9, while CsPAL10 and CsPAL11 were located on chromosome 4 (Fig. 7)
Fig. 7.
Chromosomal distribution of PAL genes shows their presence on chromosomes 1, 4, and 6, with chromosome 6 hosting the majority
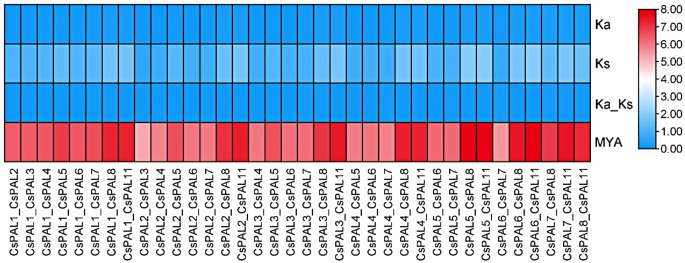
The Ka/Ks analysis revealed that the CsPAL6_CsPAL7 pair exhibits a higher Ka/Ks ratio of 0.10617, indicating a relatively higher rate of non-synonymous substitutions compared to synonymous substitutions. In contrast, the CsPAL6_CsPAL11 and CsPAL8_CsPAL11 pairs show lower Ka/Ks ratios of 0.04391 and 0.08776, respectively, suggesting a more conserved evolutionary pattern with fewer non-synonymous changes.
These findings are further supported by divergence time estimates, expressed in millions of years ago (MYA). The CsPAL2_CsPAL3 pair has lower MYA values, indicating a more recent divergence from a common ancestor. Conversely, the CsPAL6_CsPAL11 pair has higher MYA values, reflecting a more ancient divergence (Fig. 8).
Fig. 8.
The figure shows higher non-synonymous substitution rates in CsPAL6_CsPAL7 and greater conservation in CsPAL6_CsPAL11 and CsPAL8_CsPAL11. Divergence times confirm recent divergence for CsPAL2_CsPAL3 and ancient divergence for CsPAL6_CsPAL11.
The syntenic analysis shows that there were tandem duplications among CsPAL1, CsPAL2, CsPAL3, CsPAL4, CsPAL5, CsPAL6, CsPAL7, and CsPAL8 genes, and they were all located on chromosome 9. Additionally, CsPAL11 and CsPAL10 were found on chromosome 4, while CsPAL9 was on chromosome 1, suggesting segmental duplications for these genes (Fig. 9)
Fig. 9.
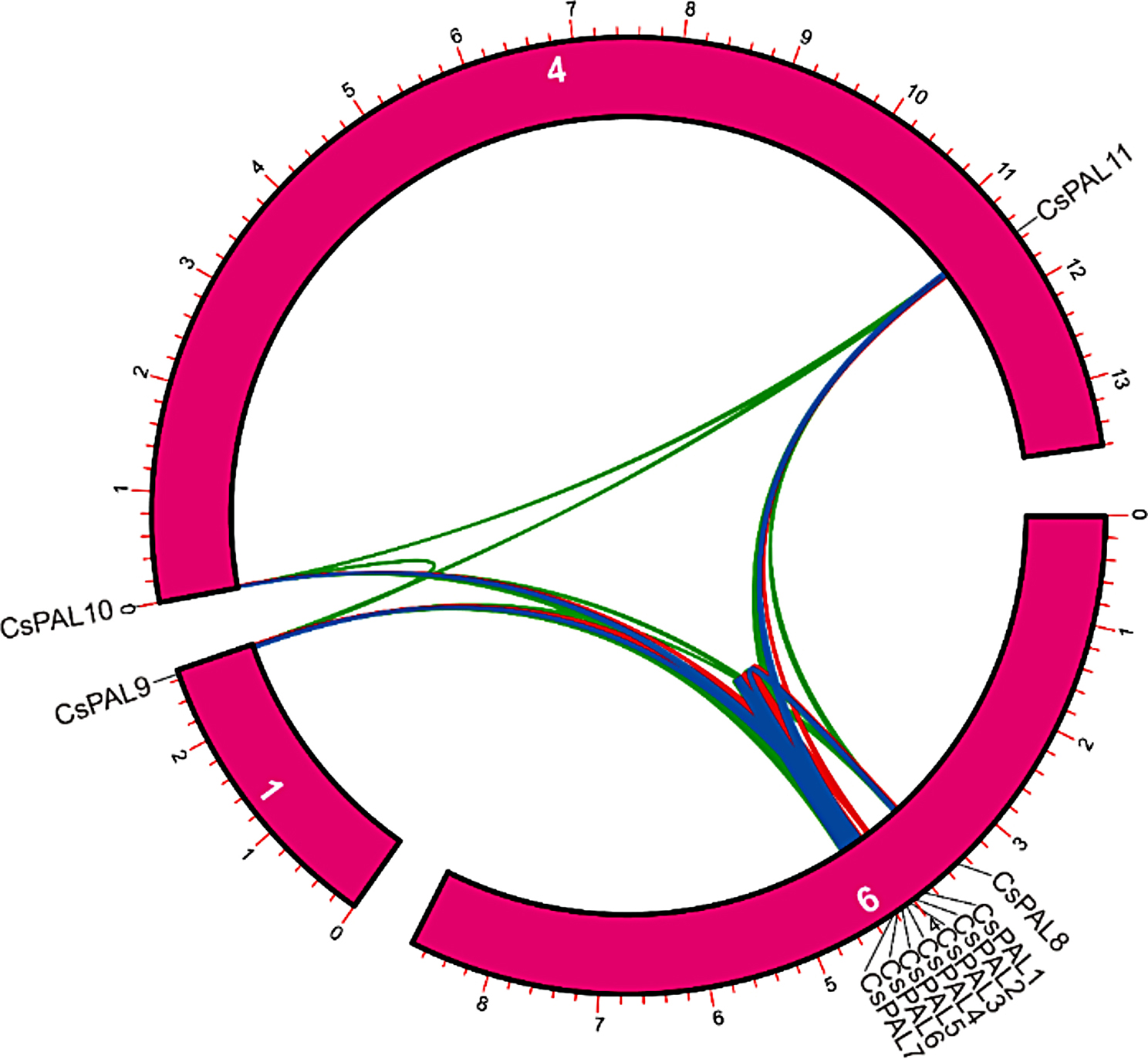
Syntenic analysis of CsPAL genes shows tandem duplications on chromosome 9 and segmental duplications on chromosomes 1 and 4
Go annotation and Orthologue Identification
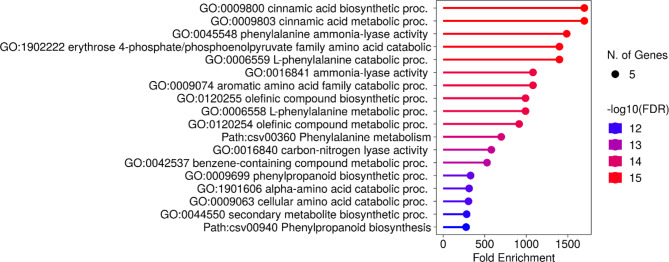
The GO enrichment analysis performed in this study provided insights into the roles of CsPAL genes. Specifically, these genes were found to be involved in the biosynthesis of cinnamic acid (GO:0009800), cinnamic acid metabolic process (GO:0009803), phenylalanine ammonia-lyase activity (GO:0045548), erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process (GO:1902222), L-phenylalanine catabolic process (GO:0006559), ammonia-lyase activity (GO:0016841), aromatic amino acid family catabolic process (GO:0009074), olefinic compound biosynthetic process (GO:0120255), L-phenylalanine metabolic process (GO:0006556), olefinic compound metabolic process (GO:0120254), phenylalanine metabolism (Path: csv00360), carbon-nitrogen lyase activity (GO:0016840), benzene-containing compound metabolic process (GO:0042537), phenylpropanoid biosynthetic process (GO:0009699), alpha-amino acid catabolic process (GO:1901606), cellular amino acid catabolic process (GO:0009063), secondary metabolite biosynthetic process (GO:0044550), and phenylpropanoid biosynthesis (Path: csv00940) (Fig. 10).
Fig. 10.
GO enrichment analysis of CsPAL genes, highlighting their functional roles with red indicating high gene function values and blue representing low gene function values
Protein-protein Interaction
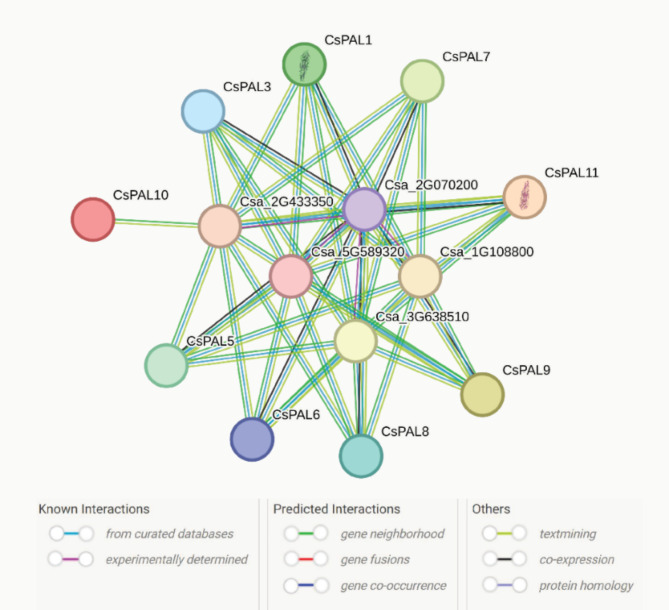
During a protein-protein interaction investigation, 14 nodes and 45 edges were observed in the network. The calculated average node degree was 6.43, and the average local clustering coefficient was 0.395. Notably, the expected number of edges was 5, and the p-value for protein-protein interaction enrichment was remarkably high at < 1.0e-16, indicating a significant enrichment of interactions. To meet the minimum required interaction score, a low confidence threshold of 0.700 was applied. Specifically, CsPAL was found to interact with other proteins, while proteins showed no associations within themselves (Fig. 11)
Fig. 11.
Protein-protein interaction analysis of CsPAL shows 14 nodes and 45 edges, with significant enrichment (p < 1.0e-16) and an average node degree of 6.43. CsPAL interacts with other proteins but not with itself
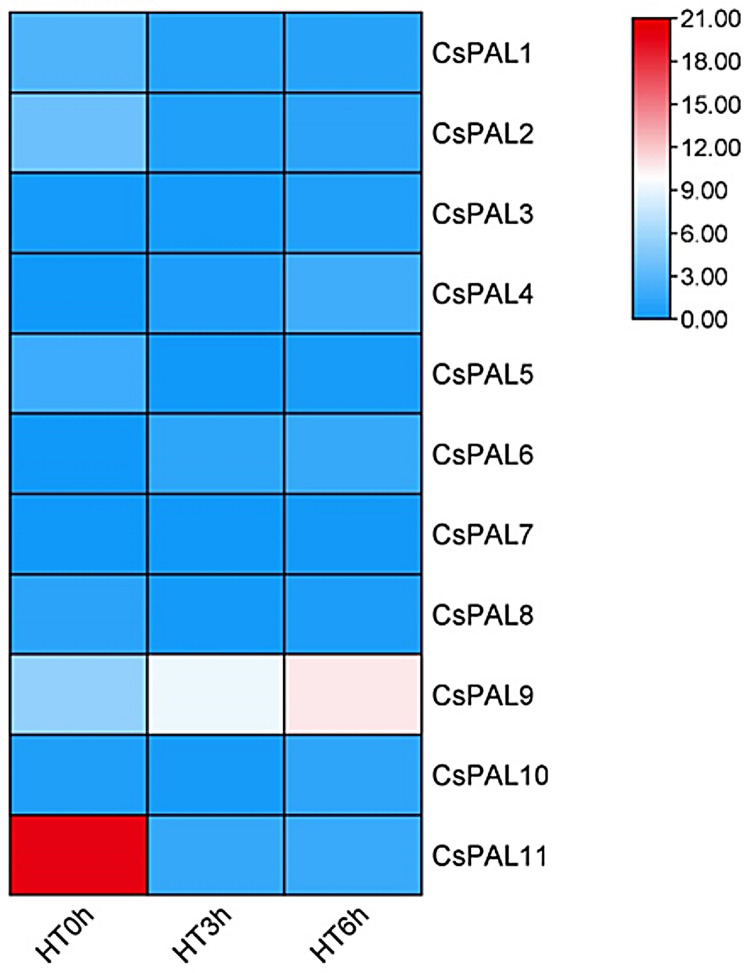
Transcriptomic analysis
In transcriptomic analysis aiming to the role of CsPAL genes under heat stress, significant regulation was observed for CsPAL1, CsPAL2, CsPAL5, CsPAL8, CsPAL9, and CsPAL11 across different temperature conditions (HT0h, HT3h, and HT6h). Specifically, CsPAL9 exhibited significant upregulation during HT3h, while both CsPAL9 and CsPAL7 showed high upregulation under HT6h, with CsPAL7 being upregulated 6-fold and CsPAL9 2-fold (Fig. 12).
Fig. 12.
Transcriptomic analysis of CsPAL genes under heat stress, showing significant regulation and notable upregulation of CsPAL9 at HT3h (heat stress for 3 h), and CsPAL7 and CsPAL9 at HT6h (heat stress for 6 h)
Discussion
The PAL (Phenylalanine Ammonia Lyase) gene family in cucumber (C. sativus) plays a pivotal role in regulating the phenylpropanoid pathway, influencing the biosynthesis of secondary metabolites, including lignin and phenolic compounds [46]. This family contributes to plant defense against both biotic and abiotic stresses, with PAL serving as a key enzyme in the production of antimicrobial compounds, such as phytoalexins [47]. Additionally, PAL is involved in the synthesis of flavonoids and acts as a mediator in the response to abiotic stresses, providing antioxidative properties. Understanding the PAL gene family in cucumbers is crucial for enhancing genetic diversity, developing disease-resistant varieties, and improving the nutritional quality of cucumber crops [48].
A genome-wide investigation was carried out to find and understand the PAL genes in cucumber. The physicochemical properties of eleven CsPAL genes in the cucumber genome were investigated to observe their distinctions within a clade of proteins [49]. All the CsPAL proteins that were identified showed hydrophilic properties, which means they have a tendency to interact with water and their electrical charges depend on pl levels. This can be observed from their GRAVY values [50]. Upon examination using the instability index, it was observed that all proteins of CsPAL proteins were stable. These consistent localization patterns underscore the unique roles and potential functional specialization of CsPAL genes within distinct cellular compartments, contributing to the overall complexity of the gene family’s involvement in cellular processes. The maximum localization of CsPAL7 in chloroplasts suggests its primary involvement and potentially critical role in chloroplast-specific processes. The analysis of conserved motifs among the 11 CsPAL genes in cucumber reveals the consistent presence of Motifs 1 and 8 across the entire gene set, suggesting essential functional elements in these regions. The presence of these conserved motifs is crucial in understanding gene expression regulation in cucumber. Conserved motifs often represent regulatory elements involved in transcriptional control, affecting gene expression levels and patterns [51]. The identification of shared motifs provides insights into potential regulatory networks and common regulatory mechanisms among CsPAL genes, shedding light on their roles in cucumber development, stress responses, or other biological processes [52]. The analysis of conserved domains among the 11 CsPAL genes revealed a remarkable consistency, with all genes sharing a common Lyase_I_like superfamily domain (Accession: cl00013). The presence of these conserved domains is significant in understanding gene expression and functional specificity. Conserved domains often play crucial roles in determining the function of genes [53]. In this case, the Lyase_I_like superfamily and PLN02457 domains may contribute to common biochemical activities or structural elements essential for the phenylalanine ammonia-lyase (PAL) enzyme, which is encoded by CsPAL genes. The identification of conserved domains provides insights into the evolutionary relationships and potential functional roles of CsPAL genes in plant biology [54].
Cis elements in the promoter regions of genes play a crucial role in regulating gene expression and determining functional specificity [55]. In the case of the 11 CsPAL genes, the diverse array of identified regulatory motifs provides key insights into the intricate control mechanisms governing their transcription. The prevalence of the TATA-box and CAAT-box motifs, constituting 39% and 26% of the discovered motifs, respectively, highlights their significance in initiating transcriptional processes. The specific enrichment of motifs such as MYB recognition sites, MYC, W box, and STRE suggests their potential roles in finely tuning gene regulation, particularly in response to stress [56]. Overall, the presence and frequency of these cis-acting elements provide valuable clues about the molecular mechanisms underlying the regulation of CsPAL genes, shedding light on their roles in stress adaptation and plant defense mechanisms [57]. The exon-intron structures of genes play a pivotal role in determining gene expression and functional of genes [58]. In the context of the CsPAL gene family, the observed patterns reveal distinct genomic variations. The presence of a single exon in CsPAL2, CsPAL3, CsPAL4, CsPAL5, CsPAL6, CsPAL7, CsPAL9, and CsPAL10 suggests a simplified gene structure without introns. This streamlined organization can facilitate rapid and efficient transcription and translation processes, potentially contributing to a swift response to environmental stress [59]. On the other hand, the conserved two-exon and one-intron structure shared by CsPAL1, CsPAL8, and CsPAL11 may introduce additional regulatory complexity, allowing for fine-tuned control of gene expression. The observed diversity in exon-intron organization within the CsPAL gene family emphasizes the importance of these structural elements in shaping the functional repertoire of genes, influencing their expression levels, and contributing to the adaptability and specificity of their biological functions [60].
In a phylogenetic analysis focused on identifying conserved patterns, PAL genes from four plant species, namely O. sativa, C. sativus, A. thaliana, and S. bicolor, were systematically classified into three distinct clades labeled as I-III. The all member of same clade have similarity in function. The systematic grouping of PAL genes into these clades suggests evolutionary relationships and potential functional similarities or differences among the PAL genes from the analyzed plant species [61]. The color scheme serves as an effective organizational tool, allowing for an intuitive grasp of the phylogenetic landscape and contributing to a more accessible interpretation of the study’s findings. This chromosomal distribution of PAL genes across chromosomes 1, 4, and 6 suggests a non-random arrangement, possibly indicating a degree of genomic organization or functional specialization. Understanding the chromosomal arrangement of PAL genes contributes valuable insights into the genomic architecture and organization of the plant species under investigation, shedding light on potential gene clusters, regulatory elements, or functional relationships among the PAL genes [62]. This information holds significant importance for investigators seeking to unravel the complexities underlying PAL gene evolution and their functional contributions to the physiological processes of these plants [62].
The Ka/Ks analysis provides insights into the selective pressures acting on pairs of CsPAL genes. Specifically, CsPAL6_CsPAL7 exhibits a higher Ka/Ks value (0.10617), indicating potential positive selection or relaxed purifying selection. In contrast, CsPAL6_CsPAL11 and CsPAL8_CsPAL11 display lower Ka/Ks values (0.04391 and 0.08776, respectively), suggestive of purifying selection. The divergence time estimation, measured in million years ago (MYA), aligns with these findings [63]. CsPAL2_CsPAL3 shows lower MYA values, suggesting a more recent divergence, while CsPAL6_CsPAL11 has higher MYA values, indicating an ancient divergence from a common ancestor. The syntenic analysis reveals tandem duplications among CsPAL1 to CsPAL8 genes, all located on chromosome 9, suggesting a shared evolutionary origin for this gene cluster. Furthermore, CsPAL11 and CsPAL10 on chromosome 4, and CsPAL9 on chromosome 1, indicate segmental duplications, supporting the notion of diversification through genomic rearrangements. These findings provide valuable insights into the evolutionary history of CsPAL genes, indicating both ancient and more recent divergence events, as well as the contribution of tandem and segmental duplications to their genomic organization [34]. The varied selective pressures and duplication events highlight the dynamic nature of PAL gene evolution in the studied plant species [64].
The miRNA-mediated regulation discovered in the analysis adds a complex layer to the control of CsPAL gene expression. This regulatory mechanism suggests that alterations in miRNA abundance or activity directly influence the levels of PAL gene expression, thereby impacting the synthesis of phenylpropanoid compounds [65]. This additional layer of regulation contributes complexity to the broader gene expression landscape, playing a crucial role in fine-tuning plant responses to diverse environmental stress and developmental stages [66]. In essence, the miRNA analysis, approached from an expression perspective, illuminates the intricate post-transcriptional regulatory network governing CsPAL genes within the context of phenylpropanoid metabolism [67].
The Go enrichment analysis conducted in this study has provided valuable insights into the functional roles of CsPAL genes. These findings underscore the diverse functions of CsPAL genes, particularly their contributions to the synthesis of acids and metabolic processes associated with amino acids. Protein-protein interaction analysis revealed that CsPAL interacts with other proteins, emphasizing its role in forming complexes or participating in molecular pathways. Interestingly, the absence of associations among the interacting proteins themselves suggests that CsPAL may act as a central hub or connector in the network, potentially orchestrating various cellular processes [68].
The significant upregulation of CsPAL9 during HT3h and the heightened upregulation of both CsPAL9 and CsPAL7 under HT6h in the transcriptome study suggest a potential role for these genes in cucumber’s tolerance to heat stress. The 2-fold upregulation of CsPAL9 and the substantial 6-fold upregulation of CsPAL7 indicate an active response to the heat conditions [69]. This upregulation might imply an involvement in heat stress tolerance mechanisms, potentially contributing to the plant’s ability to withstand and adapt to elevated temperatures. Further investigations into the specific functions of CsPAL9 and CsPAL7 could provide insights into the molecular pathways associated with cucumber’s heat tolerance. The comprehensive genome-wide identification and analysis conducted in this study establish a foundation for upcoming examinations, aiding in gene cloning, marker assistant breeding and further investigations.
Conclusion
This study identifies 11 PAL genes in cucumber, highlighting their conserved sequences, motifs, and regulatory elements. It emphasizes their roles in stress responses, hormone signaling, and development, with microRNAs playing a key regulatory role. RNA-seq data reveals specific expression patterns, notably the significant upregulation of CsPAL9 during HT3h and both CsPAL9 and CsPAL7 during HT6h, suggesting their involvement in heat stress tolerance. These insights open avenues for improving agronomic traits and developing heat-tolerant cucumber varieties by targeting CsPAL9 and CsPAL7 expression.
Acknowledgements
We would like to thank the Key Laboratory of kiwifruit resources development and utilization of Guizhou Universities (Qian Jiaoji [2022] 054), projects of Liupanshui Normal University (Biological Science, LPSSYYlzy2003, LPSSY2023XKTD09, Lpssyzxxm202304, LPSSYKYJJ201601) and the Science and Technology project of Liupanshui City (Grant #52020-2020-0906).
Author contributions
MA, YW, and SH wrote the original draft and performed data analysis. MZH and AS did in-silico experiments, and data analysis, and edited the manuscript, MS, and AB edited the manuscript and performed data analysis, AA, and JD edited the manuscript, and acquired the funding. IAS, MAM, and Q. Ali co-supervised the work, conceptualized, reviewed, and edited the manuscript, and provided the infrastructure.
Funding
This work was supported by the Key Laboratory of kiwifruit resources development and utilization of Guizhou Universities (Qian Jiaoji [2022] 054), projects of Liupanshui Normal University (Biological Science, LPSSYYlzy2003, LPSSY2023XKTD09, Lpssyzxxm202304, LPSSYKYJJ201601) and the Science and Technology project of Liupanshui City (Grant #52020-2020-0906).
Data availability
The data associated with this study are available in the manuscript.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Data transparency
The authors ensured that all data, materials, software applications, and custom codes supported the claims made in this article and fully complied with field standards. The authors have also considered the possibility of individual journal policies regarding research data sharing, considering the norms and expectations of our discipline. Therefore, the data is available in the supplementary materials or deposited in online databases.
Conflict of interest
The authors have no potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yuexia Wang and Shiming Han contributed equally to this work.
Contributor Information
Shiming Han, Email: hanshiliang888@163.com.
Muhammad Zeshan Haider, Email: zeech280@gmail.com.
Adnan Sami, Email: adnansami4949@gmail.com.
Qurban Ali, Email: saim1692@gmail.com.
Irfan Ali Sabir, Email: irfanalisabir@sjtu.edu.cn.
Muhammad Aamir Manzoor, Email: aamirmanzoor1@hotmail.com.
References
- 1.Cass CL, Peraldi A, Dowd PF, Mottiar Y, Santoro N, Karlen SD, Bukhman YV, Foster CE, Thrower N, Bruno LC. Effects of PHENYLALANINE AMMONIA LYASE (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. J Exp Bot. 2015;66(14):4317–35. 10.1093/jxb/erv269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gho Y-S, Kim S-j, Jung K-H. Phenylalanine ammonia-lyase family is closely associated with response to phosphate deficiency in rice. Genes Genomics. 2020;42:67–76. 10.1007/s13258-019-00879-7 [DOI] [PubMed] [Google Scholar]
- 3.Jones DH. Phenylalanine ammonia-lyase: regulation of its induction, and its role in plant development. Phytochemistry. 1984;23(7):1349–59. 10.1016/S0031-9422(00)80465-3 [DOI] [Google Scholar]
- 4.Weisshaar B, Jenkins GI. Phenylpropanoid biosynthesis and its regulation. Curr Opin Plant Biol. 1998;1(3):251–7. 10.1016/S1369-5266(98)80113-1 [DOI] [PubMed] [Google Scholar]
- 5.Whetten R, Sederoff R. Lignin biosynthesis. Plant Cell. 1995;7(7):1001. 10.2307/3870053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo J, Wang M-H. Characterization of the phenylalanine ammonia-lyase gene (SlPAL5) from tomato (Solanum lycopersicum L). Mol Biol Rep. 2009;36:1579–85. 10.1007/s11033-008-9354-9 [DOI] [PubMed] [Google Scholar]
- 7.Huber AE, Bauerle TL. Long-distance plant signaling pathways in response to multiple stressors: the gap in knowledge. J Exp Bot. 2016;67(7):2063–79. 10.1093/jxb/erw099 [DOI] [PubMed] [Google Scholar]
- 8.Duque AS, de Almeida AM, da Silva AB, da Silva JM, Farinha AP, Santos D, Fevereiro P, de Sousa Araújo S. Abiotic stress responses in plants: unraveling the complexity of genes and networks to survive. Abiotic stress-plant Responses Appl Agric 2013:49–101.
- 9.Dixon RA, Achnine L, Kota P, Liu CJ, Reddy MS, Wang L. The phenylpropanoid pathway and plant defence—a genomics perspective. Mol Plant Pathol. 2002;3(5):371–90. 10.1046/j.1364-3703.2002.00131.x [DOI] [PubMed] [Google Scholar]
- 10.Sami A, Haider MZ, Shafiq M. Microbial nanoenzymes: Features and applications. In: Fungal Secondary Metabolites Elsevier; 2024: 353–367.
- 11.Xu F, Deng G, Cheng S, Zhang W, Huang X, Li L, Cheng H, Rong X, Li J. Molecular cloning, characterization and expression of the phenylalanine ammonia-lyase gene from Juglans regia. Molecules. 2012;17(7):7810–23. 10.3390/molecules17077810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rui-Fang M, Qian-Zi L, Ying X, Zhang L, Qing L, Jun Y, Wan-Sheng C. The phenylalanine ammonia-lyase gene family in Isatis Indigotica Fort.: molecular cloning, characterization, and expression analysis. Chin J Nat Med. 2016;14(11):801–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Li F, Tian L, Huang M, Deng R, Li X, Chen W, Wu P, Li M, Jiang H. The phenylalanine ammonia lyase gene LjPAL1 is involved in plant defense responses to pathogens and plays diverse roles in Lotus japonicus-rhizobium symbioses. Mol Plant Microbe Interact. 2017;30(9):739–53. 10.1094/MPMI-04-17-0080-R [DOI] [PubMed] [Google Scholar]
- 14.Dehghan S, Sadeghi M, Pöppel A, Fischer R, Lakes-Harlan R, Kavousi HR, Vilcinskas A, Rahnamaeian M. Differential inductions of phenylalanine ammonia-lyase and chalcone synthase during wounding, salicylic acid treatment, and salinity stress in safflower, Carthamus tinctorius. Biosci Rep. 2014;34(3):e00114. 10.1042/BSR20140026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartwal A, Mall R, Lohani P, Guru S, Arora S. Role of secondary metabolites and brassinosteroids in plant defense against environmental stresses. J Plant Growth Regul. 2013;32:216–32. 10.1007/s00344-012-9272-x [DOI] [Google Scholar]
- 16.Liang X, Dron M, Cramer C, Dixon RA, Lamb CJ. Differential regulation of phenylalanine ammonia-lyase genes during plant development and by environmental cues. J Biol Chem. 1989;264(24):14486–92. 10.1016/S0021-9258(18)71704-3 [DOI] [PubMed] [Google Scholar]
- 17.Dong C-J, Shang Q-M. Genome-wide characterization of phenylalanine ammonia-lyase gene family in watermelon (Citrullus lanatus). Planta. 2013;238:35–49. 10.1007/s00425-013-1869-1 [DOI] [PubMed] [Google Scholar]
- 18.Yu X-Z, Fan W-J, Lin Y-J, Zhang F-F, Gupta DK. Differential expression of the PAL gene family in rice seedlings exposed to chromium by microarray analysis. Ecotoxicology. 2018;27:325–35. 10.1007/s10646-018-1897-5 [DOI] [PubMed] [Google Scholar]
- 19.Javed MM, Sami A, Haider MZ, Abbas A, Ali MH, Naeem S, Amjad M, Ahmad A, Bostani R. THE CONTRIBUTION OF TRANSGENIC RICE TO ENHANCE GRAIN YIELD. Bull Biol Allied Sci Res. 2024;2024(1):65. 10.54112/bbasr.v2024i1.65 [DOI] [Google Scholar]
- 20.Abbas A, Arshad A, Rehman AU, Bukhari MS, Zaman S, REVOLUTIONIZING PLANT BREEDING PROGRAMS WITH ADVANCEMENTS IN MOLECULAR MARKER-ASSISTED SELECTION. Bull Biol Allied Sci Res. 2024;2024(1):57. 10.54112/bbasr.v2024i1.57 [DOI] [Google Scholar]
- 21.Hahlbrock K, Scheel D. Physiology and molecular biology of phenylpropanoid metabolism. Annu Rev Plant Biol. 1989;40(1):347–69. 10.1146/annurev.pp.40.060189.002023 [DOI] [Google Scholar]
- 22.Ngumbi E, Dady E, Calla B. Flooding and herbivory: the effect of concurrent stress factors on plant volatile emissions and gene expression in two heirloom tomato varieties. BMC Plant Biol. 2022;22(1):1–18. 10.1186/s12870-022-03911-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Wang Z, Tang R, Wang L, Chen C, Ren Z. Genome-wide identification and expression analysis of Hsf and Hsp gene families in cucumber (Cucumis sativus L). Plant Growth Regul. 2021;95(2):223–39. 10.1007/s10725-021-00739-z [DOI] [Google Scholar]
- 24.Abbas A, Rashad A, Rehman AU, Bukhari MS, EXPLORING THE RESPONSE MECHANISMS OF RICE TO SALINITY STRESS. Bull Biol Allied Sci Res. 2024;2024(1):58. 10.54112/bbasr.v2024i1.58 [DOI] [Google Scholar]
- 25.ALI S. Response of rice under salt stress. Biol Agricultural Sci Res J. 2022;2022(1):6–6. 10.54112/basrj.v2022i1.6 [DOI] [Google Scholar]
- 26.BASHIR H, ZAFAR S, REHMAN R, KHALID M, AMJAD I:, BREEDING POTENTIAL OF SESAME FOR WATERLOGGING STRESS IN ASIA. Biol Agricultural Sci Res J. 2023;2023(1):10–10. 10.54112/basrj.v2023i1.10 [DOI] [Google Scholar]
- 27.Manzoor N, Masoodi UH, Mufti S, Anayat R. Effect of changing climatic scenario on vegetable production and mitigation strategies: a review. SKUAST J Res. 2023;25(2):159–72. 10.5958/2349-297X.2023.00024.7 [DOI] [Google Scholar]
- 28.Fatima S, CHEEMA K, Shafiq M, Manzoor M, Ali Q, Haider M, Shahid M. The genome-wide bioinformatics analysis of 1-aminocyclopropane-1-carboxylate synthase (acs), 1-aminocyclopropane-1-carboxylate oxidase (aco) and ethylene overproducer 1 (eto1) gene family of fragaria vesca (woodland strawberry). Bull Biol Allied Sci Res. 2023;2023(1):38–38. 10.54112/bbasr.v2023i1.38 [DOI] [Google Scholar]
- 29.Junaid MD, Gokce AF, GLOBAL AGRICULTURAL LOSSES AND THEIR CAUSES. Bull Biol Allied Sci Res. 2024;2024(1):66. 10.54112/bbasr.v2024i1.66 [DOI] [Google Scholar]
- 30.Xiao X, Li J, Lyu J, Hu L, Wu Y, Tang Z, Yu J, Calderón-Urrea A. Grafting-enhanced tolerance of cucumber to toxic stress is associated with regulation of phenolic and other aromatic acids metabolism. PeerJ. 2022;10:e13521. 10.7717/peerj.13521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasheed M, Malik A. Mechanism of drought stress tolerance in wheat. Bull Biol Allied Sci Res. 2022;2022(1):23–23. 10.54112/bbasr.v2022i1.23 [DOI] [Google Scholar]
- 32.REHMAN K, KHALID M, NAWAZ M, PREVALENCE OF POTATO LEAF ROLL VIRUS DISEASE IMPACTS AND SEVERAL MANAGEMENT STRATEGIES TO HALT THE DAMAGE. Bull Biol Allied Sci Res. 2020;2020(1):21–21. 10.54112/bbasr.v2020i1.21 [DOI] [Google Scholar]
- 33.Haider M, Sami A, Mazhar H, Akram J, NISA B, Umar M, Meeran M. Exploring morphological traits variation in Gomphrena globosa: a multivariate analysis. Biol Agricultural Sci Res J. 2023;2023(1):21–21. 10.54112/basrj.v2023i1.21 [DOI] [Google Scholar]
- 34.DONG C-j, Ning C, ZHANG, Z-g. SHANG Q-m: phenylalanine ammonia-lyase gene families in cucurbit species: structure, evolution, and expression. J Integr Agric. 2016;15(6):1239–55. 10.1016/S2095-3119(16)61329-1 [DOI] [Google Scholar]
- 35.Haider MZ, Sami A, Shafiq M, Anwar W, Ali S, Ali Q, Muhammad S, Manzoor I, Shahid MA, Ali D. Genome-wide identification and in-silico expression analysis of carotenoid cleavage oxygenases gene family in Oryza sativa (rice) in response to abiotic stress. Front Plant Sci 2023, 14. [DOI] [PMC free article] [PubMed]
- 36.Zhang X, Liu H, Guo Q, Zheng C, Li C, Xiang X, Zhao D, Liu J, Luo J, Zhao D. Genome-wide identification, phylogenetic relationships, and expression analysis of the carotenoid cleavage oxygenase gene family in pepper. Genet Mol Res. 2016;15(4):15048695. 10.4238/gmr.15048695 [DOI] [PubMed] [Google Scholar]
- 37.Bülow L. R Hehl 2016 Bioinformatic identification of conserved cis-sequences in coregulated genes. Plant Synth Promoters: Methods Protocols 233 245. [DOI] [PubMed] [Google Scholar]
- 38.Barre A, Culerrier R, Granier C, Selman L, Peumans WJ, Van Damme EJ, Bienvenu F, Bienvenu J, Rougé P. Mapping of IgE-binding epitopes on the major latex allergen Hev b 2 and the cross-reacting 1, 3β-glucanase fruit allergens as a molecular basis for the latex-fruit syndrome. Mol Immunol. 2009;46(8–9):1595–604. 10.1016/j.molimm.2008.12.007 [DOI] [PubMed] [Google Scholar]
- 39.Rehman OU, Uzair M, Chao H, Fiaz S, Khan MR, Chen M. Role of the type-B authentic response regulator gene family in fragrant rice under alkaline salt stress. Physiol Plant. 2022;174(3):e13696. 10.1111/ppl.13696 [DOI] [PubMed] [Google Scholar]
- 40.Mazhar HS-U-D, Shafiq M, Ali H, Ashfaq M, Anwar A, Tabassum J, Ali Q, Jilani G, Awais M, Sahu R. Genome-wide identification, and In-Silico expression analysis of YABBY Gene Family in response to biotic and abiotic stresses in Potato (Solanum tuberosum). Genes. 2023;14(4):824. 10.3390/genes14040824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo R, Pan W, Liu W, Tian Y, Zeng Y, Li Y, Li Z, Cui L. The barley DIR gene family: an expanded gene family that is involved in stress responses. Front Genet 2022, 13. [DOI] [PMC free article] [PubMed]
- 42.Abdulla MF, Mostafa K, AYDIN A, Kavas M. GATA transcription factor in common bean: a comprehensive genome-wide functional characterization, identification, and abiotic stress response evaluation. bioRxiv. 2023;2023:2004–8. 536124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langenbacher AD, Shimizu H, Hsu W, Zhao Y, Borges A, Koehler C, Chen J-N. Mitochondrial calcium uniporter deficiency in zebrafish causes cardiomyopathy with arrhythmia. Front Physiol. 2020;11:617492. 10.3389/fphys.2020.617492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C, Chen X, Han J, Lu W, Ren Z. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 2020;20(1):443. 10.1186/s12870-020-02625-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sami A, Haider MZ, Shafiq M, Sadiq S, Ahmad F. Genome-Wide Identification and In-silico Expression Analysis of CCO Gene Family in Sunflower (Helianthus annnus). 2023. [DOI] [PMC free article] [PubMed]
- 46.Wang B, Wu C, Wang G, He J, Zhu S. Transcriptomic analysis reveals a role of phenylpropanoid pathway in the enhancement of chilling tolerance by pre-storage cold acclimation in cucumber fruit. Sci Hort. 2021;288:110282. 10.1016/j.scienta.2021.110282 [DOI] [Google Scholar]
- 47.Tuladhar P, Sasidharan S, Saudagar P. Role of phenols and polyphenols in plant defense response to biotic and abiotic stresses. Biocontrol agents and secondary metabolites. Elsevier; 2021. pp. 419–41.
- 48.Tiku AR. Antimicrobial compounds (phytoanticipins and phytoalexins) and their role in plant defense. Co-Evolution Secondary Metabolites 2020:845–68.
- 49.Walker KR. Regulation of candidate genes in black point formation in barley. 2012.
- 50.Ju Z, 762 PB 190 RELATIONSHIP OF PHENYLALANINE AMMONIA LYASE AND ANTHOCYANIN SYNTHESIS IN APPLE, HortScience. 1994, 29(5):b542–542.
- 51.Boeva V. Analysis of genomic sequence motifs for deciphering transcription factor binding and transcriptional regulation in eukaryotic cells. Front Genet. 2016;7:24. 10.3389/fgene.2016.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13(9):613–26. 10.1038/nrg3207 [DOI] [PubMed] [Google Scholar]
- 53.Gutbrod MJ, Martienssen RA. Conserved chromosomal functions of RNA interference. Nat Rev Genet. 2020;21(5):311–31. 10.1038/s41576-019-0203-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen X, Wang P, Gu M, Hou B, Zhang C, Zheng Y, Sun Y, Jin S, Ye N. Identification of PAL genes related to anthocyanin synthesis in tea plants and its correlation with anthocyanin content. Hortic Plant J. 2022;8(3):381–94. 10.1016/j.hpj.2021.12.005 [DOI] [Google Scholar]
- 55.Hernandez-Garcia CM, Finer JJ. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014;217:109–19. 10.1016/j.plantsci.2013.12.007 [DOI] [PubMed] [Google Scholar]
- 56.Abdullah SNA, Azzeme AM, Yousefi K. Fine-tuning cold stress response through regulated cellular abundance and mechanistic actions of transcription factors. Front Plant Sci. 2022;13:850216. 10.3389/fpls.2022.850216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L, Ahammed GJ, Li X, Yan P, Zhang L, Han W-Y. Plant hormones as mediators of stress response in tea plants. Stress Physiol Tea Face Clim Change 2018:269–88.
- 58.Jacob AG, Smith CW. Intron retention as a component of regulated gene expression programs. Hum Genet. 2017;136(9):1043–57. 10.1007/s00439-017-1791-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Razi K, Muneer S. Drought stress-induced physiological mechanisms, signaling pathways and molecular response of chloroplasts in common vegetable crops. Crit Rev Biotechnol. 2021;41(5):669–91. 10.1080/07388551.2021.1874280 [DOI] [PubMed] [Google Scholar]
- 60.Gloss BS, Dinger ME. The specificity of long noncoding RNA expression. Biochim et Biophys Acta (BBA)-Gene Regul Mech. 2016;1859(1):16–22. 10.1016/j.bbagrm.2015.08.005 [DOI] [PubMed] [Google Scholar]
- 61.Li G, Wang H, Cheng X, Su X, Zhao Y, Jiang T, Jin Q, Lin Y, Cai Y. Comparative genomic analysis of the PAL genes in five Rosaceae species and functional identification of Chinese white pear. PeerJ. 2019;7:e8064. 10.7717/peerj.8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, Li J, Li Z, Liu B, Zhang L, Guo D, Huang S, Qian W, Guo L. Genomic insights into longan evolution from a chromosome-level genome assembly and population genomics of longan accessions. Hortic Res. 2022;9:uhac021. 10.1093/hr/uhac021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chernikova D, Motamedi S, Csürös M, Koonin EV, Rogozin IB. A late origin of the extant eukaryotic diversity: divergence time estimates using rare genomic changes. Biol Direct. 2011;6:1–18. 10.1186/1745-6150-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carretero-Paulet L, Fares MA. Evolutionary dynamics and functional specialization of plant paralogs formed by whole and small-scale genome duplications. Mol Biol Evol. 2012;29(11):3541–51. 10.1093/molbev/mss162 [DOI] [PubMed] [Google Scholar]
- 65.Owusu Adjei M, Zhou X, Mao M, Rafique F, Ma J. MicroRNAs roles in plants secondary metabolism. Plant Signal Behav. 2021;16(7):1915590. 10.1080/15592324.2021.1915590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reddy AS, Marquez Y, Kalyna M, Barta A. Complexity of the alternative splicing landscape in plants. Plant Cell. 2013;25(10):3657–83. 10.1105/tpc.113.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yadav V, Wang Z, Wei C, Amo A, Ahmed B, Yang X, Zhang X. Phenylpropanoid pathway engineering: an emerging approach towards plant defense. Pathogens. 2020;9(4):312. 10.3390/pathogens9040312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Di Martino C, Palumbo G, Di Iorio E, Di Santo P, Fuggi A. Nitrogen assimilation in mycorrhized durum wheat plants. In: Abstracts of Plant Biology Europe EPSO/FESPB 2016 Congress: 2016. CS.
- 69.Chen C, Chen X, Han J, Lu W, Ren Z. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 2020;20:1–19. 10.1186/s12870-020-02625-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data associated with this study are available in the manuscript.