Abstract
The transcription of the human UCP3 (uncoupling protein-3) gene in skeletal muscle is tightly regulated by metabolic signals related to fatty acid availability. However, changes in thyroid status also modulate UCP3 gene expression, albeit by unknown mechanisms. We created transgenic mice bearing the entire human UCP3 gene to investigate the effect of thyroid hormones on human UCP3 gene expression. Treatment of human UCP3 transgenic mice with thyroid hormones induced the expression of the human gene in skeletal muscle. In addition, transient transfection experiments demonstrate that thyroid hormones activate the transcription of the human UCP3 gene promoter when MyoD and the TR (thyroid hormone receptor) were co-transfected. The action of thyroid hormones on UCP3 gene transcription is mediated by the binding of the TR to a proximal region in the UCP3 gene promoter that contains a direct repeat structure. An intact DNA sequence of this site is required for thyroid hormone responsiveness and TR binding. Chromatin immunoprecipitation assays revealed that the TR binds this element in vivo. The murine Ucp3 gene promoter was also dependent on MyoD and responsive to thyroid hormone in transient transfection assays. However, it was much less sensitive to thyroid hormone than the human UCP3 promoter. In summary, UCP3 gene transcription is activated by thyroid hormone treatment in vivo, and this activation is mediated by a TRE (thyroid hormone response element) in the proximal promoter region. Such regulation suggests a link between UCP3 gene expression and the effects of thyroid hormone on mitochondrial function in skeletal muscle.
Keywords: promoter regulation, thyroid hormone, transgenic mouse, uncoupling protein-3 (UCP3)
Abbreviations: C/EBPα, CCAAT/enhancer-binding protein; ChIP, chromatin immunoprecipitation; EMSA, electrophoretic mobility-shift assay; PPAR, peroxisome-proliferator-activated receptor; ROS, reactive oxygen species; RXR, retinoid X receptor; T3, thyroid hormone; TR, thyroid hormone receptor; TRE, thyroid hormone response element; UCP3, uncoupling protein-3; UCP3L, long form of UCP3; UCP3S, short form of UCP3
INTRODUCTION
Thyroid hormones regulate energy metabolism by increasing respiration and energy expenditure and by lowering metabolic efficiency. Although this has been known for years, the molecular mechanisms for these effects of thyroid hormones remain to be established. In rodents, thyroid hormones affect the energy metabolism by altering the proton leak across the inner mitochondrial membrane through unknown mechanisms [1,2]. Thyroid hormones exert most of their known biological effects through the stimulus of gene transcription, but the key target genes of thyroid hormones involved in energy metabolism and metabolic efficiency changes remain unidentified.
The discovery of mitochondrial UCPs (uncoupling proteins) 2 and 3, which are similar to the thermogenic brown-fat UCP1, suggested that these proteins are potential targets of thyroid hormone effects and mediators of the thyroid action promoting energy expenditure [3]. This research has been especially active after the finding that Ucp3 gene expression in muscle is extremely sensitive to the thyroid hormone status in rodents. Thus early after the discovery of UCP3, it was reported that the administration of thyroid hormones to rodents enhances the expression of Ucp3 in skeletal muscle, which is paralleled by a rise in resting metabolic rate [4,5]. Experiments using rodents at extreme thyroid status situations (profound hypo- or hyper-thyroid) support the major effect of thyroid status on Ucp3 gene expression [6,7].
Human UCP3 gene transcription is regulated by fatty acids and retinoic acid, as reported elsewhere [8,9], but the action of thyroid hormone has not been established. Although individual variations in UCP3 mRNA levels in skeletal muscle do not correlate with changes in circulating thyroid hormones in euthyroid healthy subjects [10], a doubling of T3 (thyroid hormone) levels induced in volunteers led to an up-regulation of UCP3 mRNA abundance in skeletal muscle [11]. Moreover, UCP3 has been recently reported as a thyroid-sensitive gene in a microarray study of cDNAs from human muscle [12]. However, the mechanisms by which thyroid status modulates UCP3 gene expression are unclear.
Results from cultured rodent myogenic cell lines indicate a positive effect of T3 on UCP3 mRNA levels, thus supporting a direct action of thyroid hormones in UCP3 gene expression [13]. In human myotubes, differentiated in culture, UCP3 mRNA expression is moderately increased after exposure to thyroid hormones [11]. The involvement of transcriptional and/or post-transcriptional mechanisms in the effects on steady-state UCP3 mRNA levels has not been elucidated, and the corresponding experimental approaches have been dampened by the extremely low levels of expression of the human UCP3 gene in cultured myotubes compared with skeletal muscle in vivo [8,11,14].
In the present study, we created a human UCP3 transgenic mouse and showed that thyroid hormones induce the tissue-specific expression of the human UCP3 gene. Moreover, T3 directly induced the expression of the human UCP3 gene through a multi-hormonal response element in the proximal promoter region of UCP3.
EXPERIMENTAL
Human UCP3 transgenic mice
Human UCP3 transgenic mice were generated using standard procedures described elsewhere [15]. A genomic P1 clone (plate 324 and well H6) containing human UCP3 [16] was mapped by restriction enzyme analysis and microinjected into pronuclei of fertilized FVB mouse oocytes. Transgenic offspring were identified using standard Southern blot analysis. Genomic DNA was isolated from the tail of mice and restriction analysis was performed using XbaI. Membranes were hybridized with a probe corresponding to human UCP3 exon 1 (200 bp, from −181 bp 5′ of the start codon to 18 bp 3′ of the start codon; GenBank accession number AF001787). Transgenic offspring were examined for the presence of a 2.9 kb band which was absent in the non-transgenic littermates. Two lines (TgA and TgB) from two different founders were analysed and used in the present study.
Analysis of human UCP3 mRNA transcripts by RNase protection assay
RNase assay was performed as previously described, using an in vitro-transcribed 32P-labelled RNA antisense probe corresponding to human UCP3L (long form of UCP3) [17], spanning exons 6 and 7 (+631 to +925 relative to ATG). Analysis was performed using 10 and 20 μg of RNA and protected bands for UCP3L and UCP3S (short form of UCP3) [16] were visualized by autoradiography.
Northern blot analysis of human UCP3 mRNA
Mice were housed with free access to standard rodent chow and water. For starvation experiments food was removed for 18 h. For thyroid hormone treatment, control and transgenic mice were treated either with saline or T3 (100 μg/kg; intraperitoneally) and studied 18 h later. In both cases, animals were killed and tissues were harvested. RNA was prepared with RNAzol (Cinna/Biotecx Laboratory, Houston, TX, U.S.A.), except for human skeletal muscle RNA which was obtained from Clontech and used for comparative purposes. RNA (20 μg) was loaded on to the gel for Northern blot analysis. Hybridization was performed by standard methods and ethidium bromide staining was used to confirm equal loading of RNA. Hybridization probe was generated by random priming from cDNA template of human UCP3 (described above). To quantify mRNA expression, Northern blots were analysed using PhosphorImager (Molecular Dynamics, Image Quant software).
Construction of transfection plasmids
Fragments from −4511 to +47 and from −2903 to +47 of the human UCP3 promoter were amplified by PCR and were cloned into pGL3 basic to generate −4511hUCP3-Luc (where Luc is luciferase) and −2903hUCP3-Luc respectively. From −2903hUCP3-Luc, a SacI restriction and re-ligation generate the −1588hUCP3-Luc construct, and the rest of the constructs were generated according to a previous study [8]. A fragment of the mouse Ucp3 gene was amplified using 200 ng of mouse genomic DNA. The complementary 3′ primer corresponded to bases from +60 to +35 downstream to the transcription initiation site, according to GenBank (accession number AB011070). The complementary 5′ primer corresponded to −1946 to −1924. The 3′ and 5′ complementary primers generated included 6 bp non-complementary extensions capable of generating KpnI and HindIII restriction sites. Reaction was performed using the Expand Long Term PCR System (Roche) in a 50 μl final volume containing 15 pmol of each primer, 350 μM of each dNTP, 1.75 mM MgCl2 and 2.5 units of Taq DNA polymerase. Ten cycles were performed at 94 °C for 10 s, 60 °C for 30 s and 68 °C for 2 min, and twenty cycles were performed at 94 °C for 10 s, 60 °C for 30 s and 68 °C for 2 min 30 s. The resulting DNA product of approx. 2 kb was digested with KpnI and HindIII, purified and ligated into pGL3-basic (Promega), which contains the cDNA for firefly (Photinus pyralis) luciferase as a reporter gene. The whole fragment was sequenced by the dideoxy method. The insertion of the fragment from −1946 to +60 of the mouse Ucp3 gene into pGL3-basic generated −1946mUCP3-Luc. Point mutation constructs were generated using a QuikChange® site-directed mutagenesis kit (Stratagene). In summary, two complementary oligonucleotides containing the desired mutations, flanked by unmodified nucleotide sequence, were synthesized. Of each oligonucleotide, 50 ng of double-stranded −1588hUCP3-Luc or −1946mUCP3-Luc was incubated with 2.5 units of Pfu Turbo DNA polymerase. PCR reaction was performed for 17 cycles at 95 °C for 30 s, 55 °C for 1 min and 68 °C for 14 min. DpnI restriction enzyme (10 units) was then added to each PCR reaction and incubated at 37 °C for at least 1 h to digest parental DNA. DpnI-treated DNA (1 μl) was used for the transformation. To generate point mutation of the complete TRE1 (thyroid hormone response element 1) (both half sites) of the −165hUCP3-Luc and −1588hUCP3-Luc to create −165mutTRE1hUCP3-Luc and −1588mutTRE1hUCP3-Luc, the oligonucleotide used was: GTCAACCAACTTCTCTAGGATAtcGTTTCAaaTCAGCCTGTGTG. To generate point mutation of the mouse TRE in −1946mUCP3-Luc, the oligonucleotide used was GCTTCTCAGAATTCCGTTTGATATCAGCTGGTGCACAGGGCC to create −1946mmutTREUCP3-Luc. Point-mutated constructs were checked by direct DNA sequencing.
Cell culture and transient transfection assays
Rat myoblastic L6 cells were obtained from the A.T.C.C. and grown in DMEM (Dulbecco's modified Eagle's medium) containing 10% FBS (fetal bovine serum). Transfection experiments were carried out in L6 cells at 50% confluence using FuGene6 Transfection Reagent (Roche) in accordance with the manufacturer's instructions. For L6 transfection, each point was assayed (unless otherwise indicated) in triplicate in a 6-well plate and contained 1.5 μg of luciferase reporter vector, 0.3 μg of the mammalian expression vectors pCMV-MyoD [18], pRSV-human TRβ1 (thyroid hormone receptor β1) ([19], pRSV-chicken TRα [20] and 3 ng of pRL-CMV (Promega), an expression vector for the sea pansy (Renilla reniformis) luciferase used as an internal transfection control. When indicated 0.3 μg of pCMV-rat TRα and pCMV-rat TRβ1 expression vectors [21] were included. Cells were incubated for 48 h after transfection and, when indicated, treated for 24 h before harvest with or without T3 treatment was performed at 50 nM unless otherwise indicated.
Firefly luciferase and Renilla luciferase activities were measured in a Turner Designs Luminometer (model TD20/20) using the Dual Luciferase Reporter assay system kit (Promega). Homogenates from cells were prepared with 500 μl of PLB (passive lysis buffer; Promega). Luciferase activity elicited by UCP3 promoter constructs was normalized for variation in transfection efficiency using Renilla luciferase as an internal standard.
EMSAs (electrophoretic mobility-shift assays)
Nuclear protein extracts from L6 cells were isolated as reported elsewhere [22], and protein concentration was determined by the micro method of Bio-Rad (Richmond, CA, U.S.A.) using BSA as standard. For gel retardation assays, the double-stranded human UCP3-TRE1 oligonucleotide corresponding to the −79 to −50 sequence of the human UCP3 gene or the mutated ones, UCP3-TRE1m1 or UCP3-TRE1m2 (see Figure 5), or mouse UCP3-TRE corresponding to −59 to −30 of mouse Ucp3 gene were end-labelled using [α-32P]dCTP and Klenow enzyme. The labelled DNA probe (25000 c.p.m.) was incubated for 30 min at 25 °C with 5 μg of nuclear protein extract or 200 ng of recombinant chicken TRα (Santa Cruz; #sc-4087) plus or minus in vitro-translated RXR (retinoid X receptor) (TNT Coupled Reticulocyte Lysate System; Promega). Reactions were carried out in a final volume of 25 μl containing 20 mM Hepes (pH 7.6), 0.1 mM EDTA, 1 mM dithiothreitol, 50 mM NaCl, 10% glycerol and 2.5 μg of poly(dI-dC). Samples were analysed by electrophoresis at 4 °C for 60–80 min in non-denaturing 5% polyacrylamide gels in 0.5×TBE. In the competition experiments, 20-, 50- and 100-fold molar excess of unlabelled double-stranded oligonucleotides was included in each respective binding reaction. When indicated 2 μl of sera against both with rat TRα and TRβ (Santa Cruz; #sc-772x) or C/EBPα (CCAAT/enhancer-binding protein α) (Santa Cruz; #sc-9314) was added to the incubation media for 15 min.
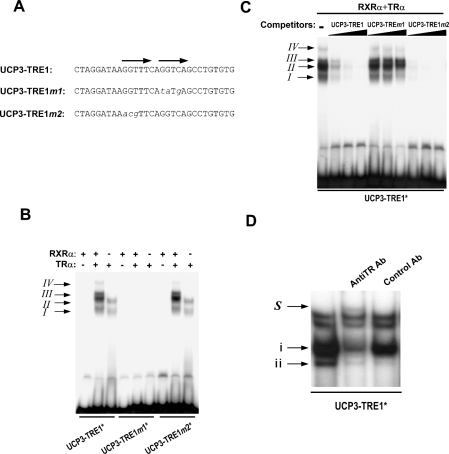
Figure 5. TR binds to the TRE1 in the proximal region of the human UCP3 gene promoter.
(A) Sequence of the three oligonucleotides corresponding to −79 to −50 of the promoter region, the wild type (UCP3-TRE1) and the ones with two point mutations (UCP3-TRE1m1 and UCP3-TRE1m2). (B) EMSA analysis of the three labelled oligonucleotides depicted above. When indicated, 5 μl of the RXRα product from a transcription–translation in vitro reaction and/or 200 ng of recombinant TR was added to the assay. Four retarded complexes, denoted I, II, III and IV, were formed. (C) Competition analysis. EMSA was performed using UCP3-TRE1 as labelled probe and in the presence of RXRα plus TR as described above. Increasing concentrations of non-labelled UCP3-TRE1, UCP3-TRE1m1 and UCP3-TRE1m2 (25, 50 and 100 ng respectively) were added to the incubation mixture. (D) EMSA using nuclear extract. UCP3-TRE1 was incubated with 5 μg of L6 nuclear extracts and when indicated an antibody against TRα or C/EBPα was added. i and ii are two of the bands modified by the addition of TR antibody and S represents the new band that appears owing to the addition of the TR antibody.
ChIP (chromatin immunoprecipitation) assay
L6 cells were transfected with −1588hUCP3-Luc or −1588mutTRE1hUCP3-Luc in the presence of MyoD and TR expression vectors (pCMV-MyoD and pRSV human TRβ1). Protein–DNA cross-linking was achieved by adding formaldehyde (final concentration 1%) to the culture medium for 10 min at 37 °C. The cells were washed twice with cold PBS containing 1 mM PMSF, 1 μg/ml aprotinin and 1 μg/ml pepstatin A (protease inhibitors). Cells were then scraped into PBS, centrifuged and re-suspended in 0.2 ml of cell lysis buffer [5 mM Pipes (KOH), pH 8.0, 85 mM KCl, 0.5% (v/v) Nonidet P40 plus the protease inhibitors]. Samples were incubated on ice for 10 min and centrifuged for 10 min at 4 °C. Pellets were re-suspended in 0.2 ml of SDS buffer (1% SDS, 10 mM EDTA, 50 mM Tris/HCl, pH 8.1, plus the protease inhibitors) and incubated on ice for 10 min. The samples were sonicated, centrifuged for 10 min at 4 °C and supernatants were diluted 10-fold with immunoprecipitation dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris/HCl, pH 8.1, 167 mM NaCl, plus the protease inhibitors). To reduce non-specific binding, samples were incubated at 4 °C for 1 h, with 80 μl of Protein G–Sepharose slurry in the form of 50% suspension in TE, which was pre-treated with salmon sperm DNA.
Pre-cleared chromatin solutions were incubated overnight at 4 °C with 20 μg of anti-Trα1 antibody (Santa Cruz; #sc-772) or an equal amount of an unrelated immunoglobulin (Santa Cruz; #sc-9314). The immunocomplex was then collected by binding to 60 μl of Potein G–Sepharose slurry as described above. After incubation for 1 h at 4 °C, the Sepharose beads were collected by centrifugation and washed with 1 ml of low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris/HCl, pH 8.1, 150 mM NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris/HCl, pH 8.1, 500 mM NaCl), LiCl buffer (0.25 M LiCl, 1% Nonidet P40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris/HCl, pH 8.1) and twice with TE. The complexes were eluted by two successive 15-min incubations with 250 μl of elution buffer (1% SDS, 0.1 M NaHCO3) at room temperature (25 °C). The pooled eluates were heated to 65 °C for 4 h in presence of 20 μl of NaCl (5 M) to reverse the formaldehyde cross-links and then treated with Proteinase K for 1 h at 45 °C. After phenol/chloroform extraction, DNA was used for PCR analysis.
Statistical analysis
Where appropriate, statistical analysis was performed by Mann–Whitney non-parametric U test. Significance differences are indicated in the text.
RESULTS
Human UCP3 gene expression in transgenic mice: effects of starvation
To study the regulation of human UCP3 in vivo, transgenic mice bearing 80 kb of human genomic P1 clone containing the UCP3 gene sequence were created. The P1 clone includes the entire human UCP3 sequence, as well as 50 kb upstream sequence and 30 kb downstream sequence relative to the start codon. We obtained several transgenic lines and all of them showed similar levels of human UCP3 mRNA expression in skeletal muscle. No apparent phenotype (body mass, growth rate, etc.) was observed in any of them. This is consistent with the phenotype of transgenic mice bearing the human UCP3 gene, obtained using the same experimental strategy as reported by Horvath et al. [23], which showed a mild reduction in body mass.
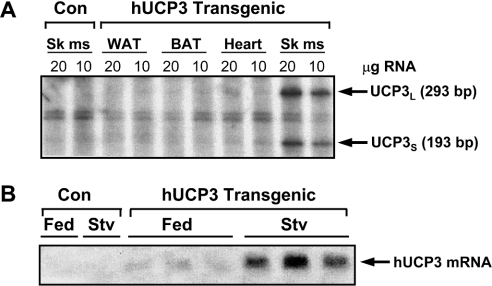
Further experiments were performed using two transgenic lines, A and B. The expression of human UCP3 mRNA was predominant in skeletal muscle in transgenic mice and no detectable levels of human UCP3 mRNA were found in white adipose tissue, brown adipose tissue or heart (Figure 1A). The levels of human UCP3 mRNA in transgenic mice were similar to those observed in human skeletal muscle samples (82±15%). Moreover, both the long and the short form of human UCP3 mRNA transcripts were expressed in human UCP3 transgenic mice, as revealed by the RNase protection assay (Figure 1A). This finding is in agreement with the observation that human skeletal muscle contains both forms of UCP3 transcripts [16].
Figure 1. Expression of the human UCP3 gene in transgenic mice.
(A) Tissue distribution of human UCP3 (hUCP3) expression in transgenic mice. Total RNA, 10 or 20 μg, was analysed by RNase protection assay in white fat (WAT), brown fat (BAT), heart and skeletal muscle (Sk ms). UCP3S and UCP3L forms of human UCP3 mRNA [16] were detected as described in the Experimental section. (B) Induction of human UCP3 mRNA expression by starvation in transgenic mice. Northern blotting was performed with 20 μg of total RNA from skeletal muscle and hybridized with a specific human DNA probe that hybridizes exclusively with human, but not mouse, UCP3 mRNA. Control mice were used to avoid cross-hybridization of the human probe with mouse UCP3 mRNA. Control and human UCP3 transgenic mice were either fed ad libitum (Fed) or starved for 24 h (Stv).
Mouse Ucp3 gene expression is up-regulated in skeletal muscle during starvation [4]. Thus we tested whether the expression of the human UCP3 gene was also regulated during starvation in transgenic mice. For this purpose, food was removed from transgenic mice for 18 h and RNA was isolated from skeletal muscle. Northern blot analysis was performed using a probe specific for human UCP3 mRNA, generated as described in the Experimental section, to avoid cross-hybridization of the human transcripts with mouse UCP3 mRNA (Figure 1B). Northern blot analysis detected the expression of the human UCP3 mRNA transcripts as a single band, given the small difference in size between UCP3L and UCP3S mRNAs (100 bp). In both lines, A and B, human UCP3 mRNA levels from starved transgenic mice were a 5.8- and 6.5-fold higher respectively than human UCP3 mRNA from fed transgenic littermates (Figure 1B). Using a specific probe to detect exclusively the mouse UCP3 transcripts, a similar induction was observed for mouse UCP3 mRNA due to fasting (results not shown). Therefore, the human UCP3 transgene is not only expressed in a tissue-specific manner, but is also regulated by changes in the physiological status.
T3 treatment induces human UCP3 gene expression in transgenic mice
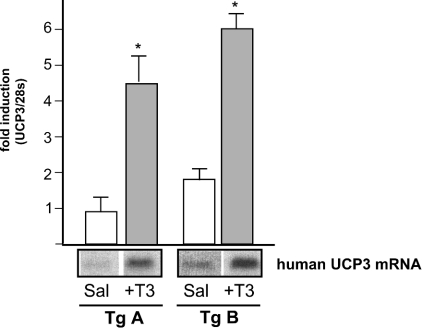
We then analysed the effect of thyroid hormone treatment in vivo on the human gene in the transgenic mice. A dose of 100 μg of T3/kg or saline as a control was injected intraperitoneally to transgenic mice. Mice were killed 18 h later, hindlimb skeletal muscle was extracted and RNA was analysed by Northern blotting using the human-specific probe as described above. Human UCP3 mRNA in skeletal muscle was significantly induced in both transgenic lines (A by 4.5-fold and B by 3.3-fold respectively) (Figure 2). As performed for starvation experiments, a specific probe was used to detect the mouse UCP3 mRNA. In our conditions, mouse UCP3 mRNA was not significantly induced after the treatment with T3 (1.3±0.25-fold induction by T3 treatment versus saline). Thus, although starvation induced both the human and mouse UCP3 mRNA, the single dose of T3 used selectively induced the expression of the human gene.
Figure 2. Thyroid hormones induce human UCP3 gene expression in transgenic mice.
Two different lines of transgenic mice (Tg A and Tg B) were injected intraperitoneally with either saline or T3 (100 μg/kg) and studied after 18 h. Northern blotting was performed as described in Figure 1(B). The results are expressed as the means±S.E.M. for 6 different mice for each transgenic line. Differences were considered significant when *P<0.05.
Thyroid hormone activates the human UCP3 promoter through a proximal element in the promoter
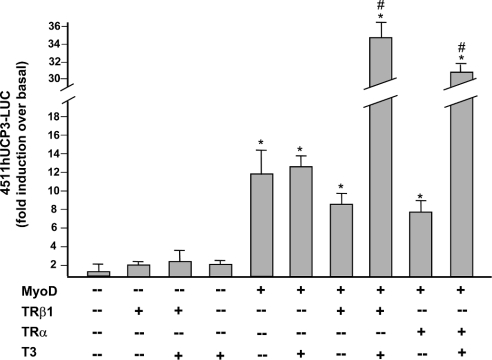
To determine the molecular basis for T3 responsiveness of the human UCP3 gene, we performed transient transfection experiments using various constructs containing the human UCP3 promoter fused to the luciferase reporter gene (Figure 3). Because the human UCP3 promoter requires MyoD for basal and fatty acid stimulated activity [9], the effects of T3 were determined in the absence or presence of co-transfected MyoD. As expected, in the absence of MyoD, a construct containing a 4.5 kb fragment (from −4511 to +47) of the 5′ non-coding region of the human UCP3 promoter was poorly expressed and not sensitive to T3, either in the absence or presence of TR. In the presence of MyoD, T3 treatment induced the expression of the construct several fold and this effect was completely dependent upon co-transfection with the expression vector for either TRβ1 or TRα.
Figure 3. Effects of T3 on human UCP3 promoter activity in L6 cells.
Construct (1.5 μg) containing 4511 bp of human UCP3 promoter region (4511hUCP3-Luc) was co-transfected where indicated with 0.3 μg of MyoD, TRβ1 or TRα expression vectors. Treatment with T3 at dose of 50 nM was performed for 24 h. The results are expressed as the fold induction of luciferase activity compared with transfection of 4511hUCP3-Luc alone, and are the means±S.E.M. of at least 3 independent experiments performed in triplicate. Statistically significant differences due to the co-transfection with MyoD expression vector are indicated by * and those due to the addition of T3 by # (P<0.05).
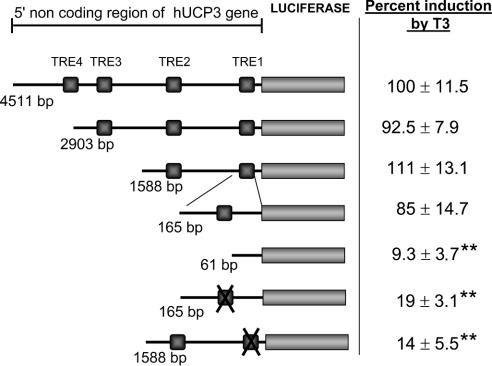
Data from computer analysis of the 5′ non-coding region of the human UCP3 gene revealed four potential TREs in the promoter [24,25] (Figure 4). Thereafter, we assayed serial deletions of the human UCP3 5′ region from −4.5 kb to a −165 bp construct, which were activated by T3 with a similar fold induction with respect to the 4.5 kb hUCP3-Luc. When the fragment between −165 to −61 was deleted, responsiveness to T3 was lost (Figure 4). These results indicate that the element responsible for T3 response is located within the proximal region. This region (from −165 to +47) of the human UCP3 promoter contains TRE1, a DNA element responsive to retinoic acid, as well as to PPAR (peroxisome-proliferator-activated receptor) agonists, with an imperfect structure of direct repeat with 1 bp spacing [8,9]. Point-mutant analysis of this TRE1 element was undertaken to establish whether it is also responsible for mediating the effect of T3. The wild-type construct −165hUCP3Luc responded to T3, whereas the mutant −165hUCP3mutTRE1hUCP3-Luc was not significantly induced by the hormone (Figure 4). The same result was obtained when the point mutation was introduced into the −1588hUCP3 plasmid, thus indicating that the TRE1 element located at −71 to −59 is essential for the human UCP3 gene to respond to thyroid hormone.
Figure 4. Deletion and point mutation analysis of the T3 responsiveness of the 5′ non-coding region of the human UCP3 gene.
Potential TREs, on the basis of computer-assisted analysis [24], are shown. TRE1 is located in between −71/−59, TRE2 in −1506/−1492, TRE3 in −2143/−2136 and TRE4 in −3358/−3343. Crossed boxes represent a point mutation performed in the TRE1 (see the Experimental section). The results are expressed as the percentage induction of the constructs by T3 compared with the induction observed in −4511hUCP3-Luc (100%). Transfections were performed in the presence of co-transfected MyoD and TRβ expression vectors. The results are expressed as the means±S.E.M. of at least 3 independent experiments performed in triplicate. Statistical significance with respect to −4511hUCP3-Luc is shown as ** (P<0.01).
TR binds to the multi-hormonal response element TRE1 in human UCP3 promoter
To test whether TR binds to the TRE1 element, EMSA was performed. An oligonucleotide corresponding to nucleotides from −79 to −57 named UCP3-TRE1 was labelled together with those containing mutations in one or the other half site of the direct repeat structure of the TRE1 (UCP3-TRE1m1 and UCP3-TRE1m2) (see Figure 5A for sequence). The presence of RXR from a transcription–translation reaction alone did not result in the formation of any protein–DNA retarded band, indicating that the direct repeat structure of the TRE1 element is not compatible with binding RXR monomers or homodimers (Figure 5B). When recombinant TR was present in the reaction mixture, two bands (I and II) were formed, in agreement with the known binding of TR to TREs as a monomer and homodimer [20]. The addition of the RXR in the reaction mixture containing the recombinant TR resulted in the formation of another two retarded complexes (bands III and IV). Therefore, bands III and IV contain protein–DNA complexes formed by the presence of RXR plus TR, which are likely to include the RXR–TR heterodimer, which is thought to mediate most of the effects of T3 on gene transcription [26]. When UCP3-TRE1m1 labelled probe was incubated, with TR plus or minus RXR, none of the bands was formed, indicating that this half site is crucial to allow the binding of TR or TR–RXR complexes. The results obtained using UCP3-TRE1m2 were similar to those obtained with the wild-type oligonucleotide, suggesting that, although the site 2 was mutated, binding of the TR alone or TR–RXR still occurred. In addition, we performed competition experiments using labelled UCP3-TRE1 always in presence of RXR and TR (Figure 5C). When the non-labelled UCP3-TRE1 oligonucleotide was used for competition, the amount of the DNA–protein complex decreased as increasing amounts of non-labelled oligonucleotide were added. The bands disappeared completely at 100-fold excess of unlabelled oligonucleotide, pointing to the specificity of the binding of the TR and TR–RXR complexes to the TRE1 element. When the competition assay was performed using UCP3-TRE1m1, even at 100-fold excess, all retarded DNA–protein complexes were still formed. Thus there was no competition between the oligonucleotides by the nuclear proteins bound at the specific site when the AGGTCA half site was mutated. As expected from the previous results, competition with UCP3-TRE1m2 showed the same results as the wild-type.
To further support these results, we checked whether the TR present in L6 nuclear extracts binds to this site. Briefly, the UCP3-TRE1 labelled oligonucleotide was incubated with 5 μg of protein extracts from L6. Upon incubation with nuclear extracts, one major band and three other retarded bands were formed. When a specific antibody against TR was added to the incubation media, the intensity of the bands labelled i and ii decreased, and a new band of lower mobility, marked as S, was formed (Figure 5D). Of note, only the reduction in the i band was specific, since band ii also disappeared when anti-C/EBPα antibody was added as a negative control. Therefore, band i is formed by a DNA–protein complex containing TR, thus indicating that TR binds the hUCP3-TRE1 site.
In vivo binding of TR to the TRE1 element by ChIP assay
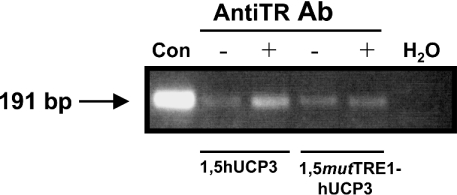
We next examined the interaction of TR with the human UCP3 promoter in vivo. We performed ChIP assays on transfected L6 cells with two human UCP3 reporter constructs, the wild-type (−1588hUCP3-Luc) and the one containing the mutation in the TRE1 site (−1588mutTRE1hUCP3-Luc) always in the presence of MyoD and TRβ1. Immunoprecipitation of protein–DNA complexes with the TR antibody caused a specific enrichment of the 191 bp PCR product corresponding to the −166 to +26 region of the human UCP3 gene in the wild-type construct when compared with the control with IgG antibody (Figure 6). Correspondingly, when the TR antibody was added to the protein–DNA complexes from cells transfected with the mutated TRE1 construct, the specific band was not enriched. These results confirm that TR binds in vivo to TRE1 in the human UCP3 promoter.
Figure 6. ChIP analysis of TR binding to the human UCP3 promoter in L6 cells.
ChIP was performed on −1588hUCP3-Luc construct and on −1588hUCP3mutTRE1-Luc, a construct containing a point mutation in TRE1 (see Figure 2). The experiment was performed in presence of co-transfected MyoD (pCMV-MyoD) and TRβ1 (pRSV-human TRβ1) expression vectors. Immunoprecipitation and PCR were performed as described in the Experimental section. The arrow indicates the 191 bp PCR product from human UCP3 gene (hUCP3). H2O, negative (no DNA); Con, positive (plasmid amplification) PCR controls. The results are representative of two independent experiments.
Mouse Ucp3 promoter is also activated by thyroid hormone, but it is less sensitive than the human promoter
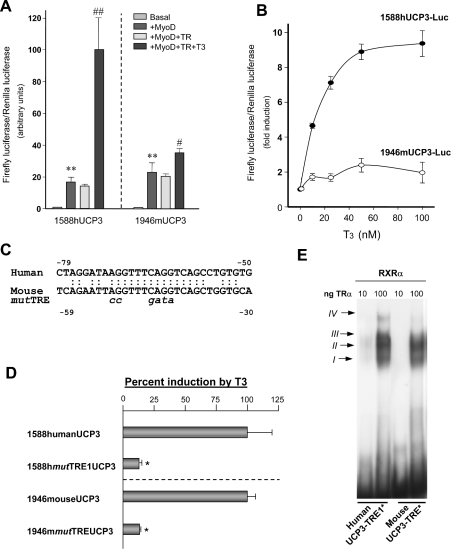
In order to explore the basis of the higher effects of T3 observed in the human UCP3 transgene with respect to the endogenous mouse Ucp3 gene, we cloned a 2 kb fragment of the 5′ non-coding region of the mouse Ucp3 gene upstream of the transcription initiation site. When transiently transfected into L6 cells, the mouse Ucp3 promoter showed low basal expression levels, but it was strongly induced by co-transfected MyoD to a similar extent to that observed in the human UCP3 promoter. When TR was co-transfected, T3 induced the mouse Ucp3 promoter activity 2-fold. This induction is significantly lower than the induction observed in the human UCP3 promoter (Figure 7A). This result was also confirmed in dose–response curves where induction of the mouse Ucp3 promoter by T3 was lower than in the human UCP3 promoter at all T3 concentrations tested (Figure 7B). This lower sensitivity did not depend on the subtype of TR co-transfected, as it was equally observed when co-transfecting chicken TRα (3.8±0.2-fold induction by T3 in mouse versus 14.6±1.80-fold induction in human), rat TRα (2.5±0.1-fold induction by T3 in mouse versus 13.0±1.0-fold in human) and rat TRβ1 (2.1±0.1-fold induction by T3 in mouse versus 5.7±0.1-fold in human) expression vectors. Computer-assisted analysis (MatInspector program) of the 5′ non-coding region of the mouse Ucp3 gene did not reveal any consensus site for a TRE, in agreement with previous observations [27]. However, comparing the human UCP3 and mouse Ucp3 promoter sequences, a site was detected in the mouse proximal Ucp3 promoter region with a similar sequence to the TRE1 previously characterized in the human gene (see Figure 7C). Point mutation of this region abolished the responsiveness of the mouse Ucp3 promoter to T3 (Figure 7D) (as also happened with the human UCP3 promoter, Figure 4), thus indicating that this site behaves as a TRE in the mouse Ucp3 promoter. Band-shift analysis of this site in comparison with the human TRE1 indicated that the binding observed in the mouse TRE was weaker, especially the bands III and IV corresponding to TR–RXR heterodimers which were almost undetectable (Figure 7E).
Figure 7. Effects of thyroid hormones on the mouse Ucp3 gene promoter.
(A) Transient transfection assays included 1.5 μg of the constructs containing 1588 bp of the human UCP3 (1588hUCP3-Luc) and 1946 bp of the mouse Ucp3 (1946mUCP3-Luc) promoter regions. When indicated 0.3 μg of MyoD and TRβ1 (TR) expression vectors were co-transfected. T3 treatment was performed at a dose of 50 nM for 24 h. The results are expressed as fold induction of luciferase activity with respect to basal promoter values, and are the means±S.E.M. of at least 3 independent experiments performed in triplicate. Statistically significant differences due to the co-transfection with the MyoD expression vector were shown as *P<0.01 and those due to the addition of T3 as #P<0.05 and ##P<0.01. (B) Human UCP3 (1588hUCP3-Luc) (1.5 μg) and mouse Ucp3 (1946mUCP3-Luc) promoter regions were co-transfected with 0.3 μg of MyoD and TR expression vectors and treated with T3 at the indicated concentrations for 24 h. The results are expressed as the fold induction of luciferase activity achieved by the addition of T3, and are the means±S.E.M. of at least 3 independent experiments performed in triplicate. (C) Comparison of the human TRE1 and mouse TRE sites in the proximal promoter regions. (D) Functional analysis of the TRE present in the mouse Ucp3 promoter in comparison with the human TRE1 was performed by point mutation of the site, as shown by italics in (C), and assayed in transient transfections. The results are expressed as the percentage induction by T3 of each construct when co-transfected with MyoD and TR. Maximal induction (100%) was assigned to the non-mutated human UCP3-Luc and mouse UCP3-Luc constructs. Statistically significant differences due to mutation in the TRE site was shown as *P<0.01. (E) EMSA analysis of the human UCP3 TRE1 and the TRE region in the mouse Ucp3 gene. Assay conditions and indications of the four retarded complexes I, II, III and IV were as described in Figure 5.
DISCUSSION
We have determined the effect of thyroid hormones on the human UCP3 gene. The study of the regulation of the UCP3 gene in humans is particularly complex owing to the extremely low levels of expression in human myotubes in culture. Therefore, we created transgenic mice bearing a human P1 clone that contains the entire UCP3 gene. The expressed transgene in mice retained the tissue-specific regulation of the UCP3 gene in humans, i.e. highly preferential expression in skeletal muscle. It is noteworthy that the human UCP3 transgene expression was almost absent in heart and brown fat, in which the rodent Ucp3 gene is substantially expressed [28]. This finding, together with the lack of UCP3 mRNA detection in human brown fat and extremely low levels in human heart [28,29], indicates a more strict tissue specificity of the UCP3 gene expression in skeletal muscle in humans with respect to rodents. Moreover, human UCP3 transgene expression is up-regulated by starvation, similarly to the murine Ucp3 gene, which may be due to the fatty acid-mediated stimulation of human UCP3 gene transcription, as reported previously [9]. The results obtained after T3 administration in transgenic mice demonstrate that the human UCP3 gene is up-regulated by thyroid hormones in vivo, in agreement with previous findings in muscle biopsies from human volunteers [11]. Most of the reported experiments in which UCP3 mRNA levels are induced by a single T3 injection were carried out in hypothyroid rats [7]. When we applied T3 to euthyroid mice, only the human UCP3 transgene was induced, but not the endogenous murine UCP3 mRNA. This suggested that the human UCP3 gene is more sensitive to thyroid-dependent stimulation than the mouse gene. The construct of the mouse Ucp3 gene promoter transiently transfected to the same cells in identical conditions to those used for the human UCP3 promoter, was responsive to T3 stimulation, but to a lesser extent than the human UCP3 promoter construct. Both the human UCP3 and mouse Ucp3 gene promoters required a similar TRE in the proximal promoter region, but the mouse TRE appeared to be weaker than the human TRE. By contrast, other features of regulation, such as dependency on MyoD, were equally shared by both promoters.
Thyroid hormones can induce lipolysis and fatty acid oxidation [30], and hyperthyroidism increases the levels of non-esterified fatty acids. Most of the physio-pathological situations reported to date in which UCP3 expression is modified in skeletal muscle are associated to parallel changes in fatty acid availability to skeletal muscle. The effects of fatty acids on UCP3 gene transcription are mediated by PPARα and PPARδ which interact with the promoter region of the human UCP3 gene [9]. Therefore, it could not be excluded that the effects of T3 on UCP3 gene expression in vivo may be due to the action of fatty acids. Moreover, thyroid hormones induce master transcription factors for mitochondrial gene expression, such as nuclear respiratory factors, which can affect UCP3 gene expression [31]. However, our present results establish that the action of thyroid hormones is direct and mediated by TR binding to the proximal region of the UCP3 gene promoter. In the human promoter, the TRE element required for responsiveness is coincident with the reported site responsible for responsiveness to PPAR and retinoids, thus indicating that it behaves as a multihormonal responsive element. Moreover, the action of T3 on the human UCP3 promoter requires MyoD, as does the PPAR- and RAR (retinoic acid receptor)-dependent activation [8,9]. This confirms that MyoD acts as permissive factor for basal and hormone-dependent transcriptional activity of the human UCP3 promoter.
The finding that responsiveness to thyroid hormone shares a common DNA region with the retinoic acid and PPAR-dependent pathways suggests a cross-talk between these regulatory pathways in the human UCP3 promoter, as reported for other genes [32,33]. Despite a recent report showing that a single injection of T3 to fasted rats can further induce UCP3 mRNA expression [34], the interaction between the thyroid hormone and fatty acid-dependent regulation of the UCP3 gene in vivo has been poorly explored. Research is currently under way to determine the mutual influence of PPAR and thyroid-dependent regulation of UCP3 gene transcription and the involvement of the common multi-hormonal responsive site in their cross-talk.
The analysis of the regulation of the UCP3 gene in humans does not provide direct information on the function of the UCP3 protein. However, the high sensitivity of human UCP3 transcription to thyroid hormones is consistent with a potential role of UCP3 in the response of skeletal muscle mitochondrial functions to the action of thyroid hormones. Although thyroid hormones increase body temperature in rodents and humans by unknown mechanisms, current information on the function of UCP3 is poorly supportive of a role for this protein in thermogenesis. It is possible that thyroid stimulation of UCP3 gene expression was related to the regulation of production of ROS (reactive oxygen species), a major current hypothesis for UCP3 protein function. Activation of mitochondrial respiration by thyroid hormones results in increased ROS production, at least in liver cells [35], and it has been also proposed that a high production of ROS is on the basis of muscular injury caused by hyperthyrodism [36]. In this context, induction of UCP3 gene expression by thyroid hormones could be viewed as a protective mechanism for excessive ROS production when cell respiration is enhanced by a physiological increase in these hormones.
In summary, we conclude that thyroid hormones directly activate the human and mouse UCP3 genes through interaction of TRs with the proximal promoter regions. This suggests that UCP3 upregulation is a relevant component of the regulation of mitochondrial function in human skeletal muscle in response to thyroid hormones.
Acknowledgments
This work was supported by grant SAF2002-03648 from Ministerio de Ciencia y Tecnología, Fundació la Marató de TV3 and grant C03/08 from Instituto Carlos III, Ministerio de Sanidad y Consumo, Spain. G.S. is under the ‘Ramon y Cajal’ Investigator program. We thank Dr R. Evans, Dr H.H. Samuels, Dr H.C. Towle and Dr A.B. Lassar for kindly supplying expression vectors.
References
- 1.Brand M. D., Chien L. F., Ainscow E. K., Rolfe D. F., Porter R. K. The causes and functions of mitochondrial proton leak. Biochim. Biophys. Acta. 1994;1187:132–139. doi: 10.1016/0005-2728(94)90099-x. [DOI] [PubMed] [Google Scholar]
- 2.Harper M. E., Brand M. D. Hyperthyroidism stimulates mitochondrial proton leak and ATP turnover in rat hepatocytes but does not change the overall kinetics of substrate oxidation reactions. Can. J. Physiol. Pharmacol. 1994;72:899–908. doi: 10.1139/y94-127. [DOI] [PubMed] [Google Scholar]
- 3.Lanni A., Moreno M., Lombardi A., Goglia F. Thyroid hormone and uncoupling proteins. FEBS Lett. 2003;543:5–10. doi: 10.1016/s0014-5793(03)00320-x. [DOI] [PubMed] [Google Scholar]
- 4.Gong D. W., He Y., Karas M., Reitman M. Uncoupling protein-3 is a mediator of thermogenesis regulated by thyroid hormone, β3-adrenergic agonists, and leptin. J. Biol. Chem. 1997;272:24129–24132. doi: 10.1074/jbc.272.39.24129. [DOI] [PubMed] [Google Scholar]
- 5.Larkin S., Mull E., Miao W., Pittner R., Albrandt K., Moore C., Young A., Denaro M., Beaumont K. Regulation of the third member of the uncoupling protein family, UCP3, by cold and thyroid hormone. Biochem. Biophys. Res. Commun. 1997;240:222–227. doi: 10.1006/bbrc.1997.7636. [DOI] [PubMed] [Google Scholar]
- 6.Lanni A., Beneduce L., Lombardi A., Moreno M., Boss O., Muzzin P., Giacobino J. P., Goglia F. Expression of uncoupling protein-3 and mitochondrial activity in the transition from hypothyroid to hyperthyroid state in rat skeletal muscle. FEBS Lett. 1999;444:250–254. doi: 10.1016/s0014-5793(99)00061-7. [DOI] [PubMed] [Google Scholar]
- 7.De Lange P., Lanni A., Beneduce L., Moreno M., Lombardi A., Silvestri E., Goglia F. Uncoupling protein-3 is a molecular determinant for the regulation of resting metabolic rate by thyroid hormone. Endocrinology. 2001;142:3414–3420. doi: 10.1210/endo.142.8.8303. [DOI] [PubMed] [Google Scholar]
- 8.Solanes G., Pedraza N., Iglesias R., Giralt M., Villarroya F. The human uncoupling protein-3 gene promoter requires MyoD and is induced by retinoic acid in muscle cells. FASEB J. 2000;14:2141–2143. doi: 10.1096/fj.00-0363fje. [DOI] [PubMed] [Google Scholar]
- 9.Solanes G., Pedraza N., Iglesias R., Giralt M., Villarroya F. Functional relationship between MyoD and peroxisome proliferator-activated receptor-dependent regulatory pathways in the control of the human uncoupling protein-3 gene transcription. Mol. Endocrinol. 2003;17:1944–1958. doi: 10.1210/me.2002-0395. [DOI] [PubMed] [Google Scholar]
- 10.Boivin M., Camirand A., Carli F., Hoffer L. J., Silva J. E. Uncoupling protein-2 and -3 messenger ribonucleic acids in adipose tissue and skeletal muscle of healthy males: variability, factors affecting expression, and relation to measures of metabolic rate. J. Clin. Endocrinol. Metab. 2000;85:1975–1983. doi: 10.1210/jcem.85.5.6585. [DOI] [PubMed] [Google Scholar]
- 11.Barbe P., Larrouy D., Boulanger C., Chevillotte E., Viguerie N., Thalamas C., Oliva T. M., Roques M., Vidal H., Langin D. Triiodothyronine-mediated up-regulation of UCP2 and UCP3 mRNA expression in human skeletal muscle without coordinated induction of mitochondrial respiratory chain genes. FASEB J. 2001;15:13–15. doi: 10.1096/fj.00-0502fje. [DOI] [PubMed] [Google Scholar]
- 12.Clement K., Viguerie N., Diehn M., Alizadeh A., Barbe P., Thalamas C., Storey J. D., Brown P. O., Barsh G. S., Langin D. In vivo regulation of human skeletal muscle gene expression by thyroid hormone. Genome Res. 2002;12:281–291. doi: 10.1101/gr.207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagase I., Yoshida S., Canas X., Irie Y., Kimura K., Yoshida T., Saito M. Up-regulation of uncoupling protein 3 by thyroid hormone, peroxisome proliferator-activated receptor ligands and 9-cis retinoic acid in L6 myotubes. FEBS Lett. 1999;461:319–322. doi: 10.1016/s0014-5793(99)01477-5. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Martinez C., Sibille B., Solanes G., Darimont C., Mace K., Villarroya F., Gomez-Foix A. M. Overexpression of UCP3 in cultured human muscle lowers mitochondrial membrane potential, raises ATP/ADP ratio, and favors fatty acid vs. glucose oxidation. FASEB J. 2001;15:2033–2035. doi: 10.1096/fj.00-0828fje. [DOI] [PubMed] [Google Scholar]
- 15.Ito M., Grujic D., Abel E. D., Vidal-Puig A., Susulic V. S., Lawitts J., Harper M. E., Himms-Hagen J., Strosberg A. D., Lowell B. B. Mice expressing human but not murine β3-adrenergic receptors under the control of human gene regulatory elements. Diabetes. 1998;47:1464–147. doi: 10.2337/diabetes.47.9.1464. [DOI] [PubMed] [Google Scholar]
- 16.Solanes G., Vidal-Puig A., Grujic D., Flier J. S., Lowell B. B. The human uncoupling protein-3 gene. Genomic structure, chromosomal localization, and genetic basis for short and long form transcripts. J. Biol. Chem. 1997;272:25433–25436. doi: 10.1074/jbc.272.41.25433. [DOI] [PubMed] [Google Scholar]
- 17.Chung W. K., Luke A., Cooper R. S., Rotini C., Vidal-Puig A., Rosenbaum M., Gordon D., Leal S. M., Caprio S., Goldsmith R., et al. The long isoform uncoupling protein-3 (UCP3L) in human energy homeostasis. Int. J. Obes. Relat. Metab. Disord. 1999;23(Suppl. 6):S49–S50. doi: 10.1038/sj.ijo.0800945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crescenzi M., Fleming T. P., Lassar A. B., Weintraub H., Aaronson S. A. MyoD induces growth arrest independent of differentiation in normal and transformed cells. Proc. Natl. Acad. Sci. U.S.A. 1990;87:8442–8446. doi: 10.1073/pnas.87.21.8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass C. K., Lipkin S. M., Devary O. V., Rosenfeld M. G. Positive and negative regulation of gene transcription by a retinoic acid-thyroid hormone receptor heterodimer. Cell. 1989;59:697–708. doi: 10.1016/0092-8674(89)90016-0. [DOI] [PubMed] [Google Scholar]
- 20.Forman B. M., Casanova J., Raaka B. M., Ghysdael J., Samuels H. H. Half-site spacing and orientation determines whether thyroid hormone and retinoic acid receptors and related factors bind to DNA response elements as monomers, homodimers, or heterodimers. Mol. Endocrinol. 1992;6:429–442. doi: 10.1210/mend.6.3.1316541. [DOI] [PubMed] [Google Scholar]
- 21.Murray M. B., Zilz N. D., McCreary N. L., MacDonald M. J., Towle H. C. Isolation and characterization of rat cDNA clones for two distinct thyroid hormone receptors. J. Biol. Chem. 1988;263:12770–12777. [PubMed] [Google Scholar]
- 22.Swick A. G., Blake M. C., Kahn J. W., Azizkhan J. C. Functional analysis of GC element binding and transcription in the hamster dihydrofolate reductase gene promoter. Nucleic Acids Res. 1989;17:9291–9304. doi: 10.1093/nar/17.22.9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horvath T. L., Diano S., Miyamoto S., Barry S., Gatti S., Alberati D., Livak F., Lombardi A., Moreno M., Goglia F., et al. Uncoupling proteins-2 and 3 influence obesity and inflammation in transgenic mice. Int. J. Obes. Relat. Metab. Disord. 2003;27:433–442. doi: 10.1038/sj.ijo.0802257. [DOI] [PubMed] [Google Scholar]
- 24.Tu N., Chen H., Winnikes U., Reinert I., Pirke K. M., Lentes K. U. Functional characterization of the 5′-flanking and the promoter region of the human UCP3 (hUCP3) gene. Life Sci. 2000;67:2267–2279. doi: 10.1016/s0024-3205(00)00802-x. [DOI] [PubMed] [Google Scholar]
- 25.Acin A., Rodriguez M., Rique H., Canet E., Boutin J. A., Galizzi J. P. Cloning and characterization of the 5′ flanking region of the human uncoupling protein 3 (UCP3) gene. Biochem. Biophys. Res. Commun. 1999;258:278–283. doi: 10.1006/bbrc.1999.0530. [DOI] [PubMed] [Google Scholar]
- 26.Yen P. M. Physiological and molecular basis of thyroid hormone action. Physiol. Rev. 2001;81:1097–1142. doi: 10.1152/physrev.2001.81.3.1097. [DOI] [PubMed] [Google Scholar]
- 27.Yoshitomi H., Yamazaki K., Tanaka I. Cloning of mouse uncoupling protein 3 cDNA and 5′-flanking region, and its genetic map. Gene. 1998;215:77–84. doi: 10.1016/s0378-1119(98)00279-0. [DOI] [PubMed] [Google Scholar]
- 28.Vidal-Puig A., Solanes G., Grujic D., Flier J. S., Lowell B. B. UCP3: an uncoupling protein homologue expressed preferentially and abundantly in skeletal muscle and brown adipose tissue. Biochem. Biophys. Res. Commun. 1997;235:79–82. doi: 10.1006/bbrc.1997.6740. [DOI] [PubMed] [Google Scholar]
- 29.Boss O., Samec S., Paoloni-Giacobino A., Rossier C., Dulloo A., Seydoux J., Muzzin P., Giacobino J. P. Uncoupling protein-3: a new member of the mitochondrial carrier family with tissue-specific expression. FEBS Lett. 1997;408:39–42. doi: 10.1016/s0014-5793(97)00384-0. [DOI] [PubMed] [Google Scholar]
- 30.Freake H. C., Oppenheimer J. H. Thermogenesis and thyroid function. Annu. Rev. Nutr. 1995;15:263–291. doi: 10.1146/annurev.nu.15.070195.001403. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez-Pena A., Escriva H., Handler A. C., Vallejo C. G. Thyroid hormone increases transcription of GA-binding protein/nuclear respiratory factor-2 α-subunit in rat liver. FEBS Lett. 2002;514:309–314. doi: 10.1016/s0014-5793(02)02389-x. [DOI] [PubMed] [Google Scholar]
- 32.Chu R., Madison L. D., Lin Y., Kopp P., Rao M. S., Jameson J. L., Reddy J. K. Thyroid hormone (T3) inhibits ciprofibrate-induced transcription of genes encoding beta-oxidation enzymes: cross talk between peroxisome proliferator and T3 signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 1995;92:11593–11597. doi: 10.1073/pnas.92.25.11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunter J., Kassam A., Winrow C. J., Rachubinski R. A., Capone J. P. Crosstalk between the thyroid hormone and peroxisome proliferator-activated receptors in regulating peroxisome proliferator-responsive genes. Mol. Cell. Endocrinol. 1996;116:213–221. doi: 10.1016/0303-7207(95)03717-9. [DOI] [PubMed] [Google Scholar]
- 34.Moreno M., Lombardi A., De Lange P., Silvestri E., Ragni M., Lanni A., Goglia F. Fasting, lipid metabolism, and triiodothyronine in rat gastrocnemius muscle: interrelated roles of uncoupling protein 3, mitochondrial thioesterase, and coenzyme Q. FASEB J. 2003;17:1112–1114. doi: 10.1096/fj.02-0839fje. [DOI] [PubMed] [Google Scholar]
- 35.Venditti P., De Rosa R., Di Meo S. Effect of thyroid state on H2O2 production by rat liver mitochondria. Mol. Cell. Endocrinol. 2003;205:185–192. doi: 10.1016/s0303-7207(02)00332-5. [DOI] [PubMed] [Google Scholar]
- 36.Asayama K., Kato K. Oxidative muscular injury and its relevance to hyperthyroidism. Free Radical Biol. Med. 1990;8:293–303. doi: 10.1016/0891-5849(90)90077-v. [DOI] [PubMed] [Google Scholar]