Abstract
Innate interferons (IFN) are comprised of multiple Type I and III subtypes. The in vivo kinetics of subtype responses during human immunodeficiency virus (HIV) infection is not well defined. Using the acute simian immunodeficiency virus (SIV) infection model, we show that plasma IFNα levels peak at day 10 post-infection (pi) after which they rapidly declined. The mRNA expression of Type I and III IFN subtypes were significantly elevated in the lymph nodes (LN) at day 10 pi. Though the expression levels of all subtypes declined by day 14 – 31 pi, numerous subtypes remained elevated suggesting that ongoing viral replication in LN continues to drive induction of these subtypes. Interestingly, treatment with reverse transcriptase (RT) inhibitors at day 7 pi significantly suppressed plasma IFNα responses by day 10 pi that significantly correlated with cell-associated SIV DNA loads suggesting that RT byproducts such as viral DNA likely plays a role in driving IFN responses during acute SIV infection. Quantification of Type I and III subtype transcripts in sorted subsets of LN CD4+ and CD8+ T cells, CD14+/CD14− monocytes/macrophages, and total CD11c/CD123+ dendritic cells (DC) at day 10 pi showed that DC expressed ~3 – 4 log more subtype transcripts as compared to the other subsets. Taken together, our results provide new insights into the kinetics of innate interferon responses during early stages of infection, and provide evidence that DC’s are a major in vivo source of innate IFN during acute SIV infection.
Keywords: HIV, SIV, IFNα, IFNβ, Subtypes, Dendritic cells, Mucosa
Introduction
Human and simian immunodeficiency virus (HIV and SIV) infections are characterized by significant acute pathogenesis that is accompanied by massive viral replication, systemic loss of CD4 memory T cells, acute immune activation, and innate immune responses.
Innate interferons (IFN) are anti-viral cytokines that are released during viral infections in response to the stimulation of various pathogen recognition receptors (PRR) such as Toll-like receptors (TLR), RIG-I, MDA-5 etc. IFN exert their anti-viral action either by inducing the expression of interferon-stimulated genes (ISG) that directly suppresses viral replication or by modulating innate and adaptive immune responses[1; 2; 3; 4]. Numerous studies have shown that IFNα potently inhibits HIV by inhibiting reverse transcription and viral expression from integrated provirus, HIV replication in primary monocyte-derived macrophages, and virion release from infected cell lines[5; 6; 7; 8; 9; 10]. IFN-α treatment of HIV-1 infected cells suppressed viral replication[11; 12; 13; 14; 15; 16]. Others have shown that acute SIV infection is characterized by the release of IFN and upregulation of ISG[17; 18; 19; 20], whereas IFN-α treatment of HIV-1 infected cells suppressed viral replication during early stages of replication[11; 12; 13; 14; 15; 16]. Blocking of IFN signaling with antibodies to IFNR1 was shown to increase viremia whereas treatment with IFN-α2 increased the expression of ISG[21] in SIV infected macaques suggesting a protective role for IFN responses in controlling viral infection.
Innate IFN include two multi-gene families that code for numerous Type I and III IFN subtypes. Type I IFN gene family includes IFNβ, IFNω and multiple subtypes of IFN-α1, -α2, -α4, -α5, -α6, -α7, -α8, -α10, -α13, -α14, -α16, -α17 and -α21 subtypes in humans[22], and IFNα-01/13, 02, 06, 08, 14, 16, 23, 24, 25, 26, 27, 28, 29 in rhesus macaques[23]. Both humans and macaques express the type III IFN gene family that encodes for IFNλ-1, IFNλ-2, IFNλ-3 subtypes[22; 23]. Recent studies have identified IFN- λ4 as a new Type III subtype in humans[24] with antiviral activity[25].
All Type I IFN subtypes signal through a common cell-surface receptor that is composed of INFAR-1 and INFAR-2, whereas Type III IFN signals through a receptor complex consisting of the IL-28R and IL-10Rβ [26; 27; 28].
Interestingly, however, different subtypes have been shown to display different binding affinities for their receptors[29; 30; 31] that in turn was shown to influence their action and potency. Some subtypes such as IFNα-10 has been shown to bind the IFNAR1/2 receptor at 10 - 100 fold greater affinity that IFNα-01[32]. IFNα-10 was found to be highly effective against Semliki forest virus (SFV) and Vesicular stomatitis virus (VSV) whereas IFNα-02 was the least effective among the 9 different subtypes tested[33; 34]. On the other hand, IFNα-02 binding was found to induce chemotaxis genes and shown to be most effective against HIV-1[35] whereas IFNα-08 induced ISG’s that protected against HCV replication[36]. Other studies have reported that there were significant differences in the in vitro anti-viral and anti-proliferative effects between subtypes[33; 37; 38; 39; 40], with variable effect on T cells and dendritic cells (DC), and B cell proliferation[41; 42].
These studies raise the prospect that various subtypes may play a differential role in acute HIV infection. Harper et al[43] examined the expression of different IFNα subtypes in HIV-1 exposed plasmacytoid dendritic cells (pDC), and determined the potency of each IFNα subtype ex vivo using the Lamina propria aggregate culture model. They reported that IFN subtypes were highly expressed in pDC after exposure to HIV-1, and the relative potencies of subtypes was influenced by their binding affinities to the Type I IFNR with IFNα8 and IFNα14 being most potent at inhibiting HIV infectivity. Zaritsky et al[44] examined the expression of both total IFNα mRNA and the pattern of IFNα subtype mRNA expression in pigtailed macques (PTM) between day 7 and 21 pi and reported that expression and pattern of subtypes expressed differed between the brain, lung and spleen. There is limited information regarding the in vivo expression of various Type I and III subtypes in the lymph nodes, jejunal mucosa and PBMC and the source of the various Type I and III subtypes during the early acute stages of HIV infection.
We sought to address this question using the SIV infected rhesus macaque model and examined the kinetics of IFNα levels in the plasma during the 1st two weeks of infection and compared them to animals that were treated with reverser transcriptase (RT) inhibitors very early during the course of infection. To determine if acute SIV infection was characterized by differences in the expression of various subtypes, we examined the mRNA expression profile of both Type I and Type III IFN subtypes in peripheral blood mononuclear cells (PBMC), jejunal mucosa and lymph nodes (LN) during early stages of SIV infection using a quantitative RT-PCR assay. Our results showed that plasma IFNα levels were significantly elevated at day 10- post infection (pi) in untreated animals but was significantly suppressed in treated animals when ART was initiated at day 7 pi. Plasma IFNα levels significantly correlated with CD4 T cell associated SIV DNA loads suggesting a role of viral DNA in the induction of IFNα. Both Type I and III IFN subtypes were differentially expressed during acute SIV infection with most subtypes being significantly elevated in the LN as compared to the mucosa and peripheral blood. Analysis of various cellular sources in the LN at day 10 pi showed that dendritic cells were the primary in vivo source of all IFN subtypes as compared to other cellular subsets.
Materials and Methods
Animals, infection and samples
Archived cryopreserved cells from peripheral blood (PBMC; uninfected n = 10; day 10 pi n = 10; day 14 - 31 pi n = 8), lymph nodes (LN; uninfected n = 5; day 10 pi n = 8; day 14 - 31 pi n = 9) and jejunum (uninfected n = 5; day 10 PI n = 8; day 14 - 31 pi n = 5) that were collected from healthy and SIVmac251 infected rhesus macaques (Macaca mulatta) of Indian origin were used in this cross-sectional study. Additionally, archived cells and plasma from rhesus macaques that were treated with a combination of RT inhibitors PMPA (Tenofovir) and FTC (Emtricitabine) were used for analysis; PMPA and FTC were administered at 20-30 mg / Kg BW / day starting at day 7 pi. The animals were housed in accordance with the American Association for Accreditation of Laboratory Animal guidelines and were seronegative for SIV, simian retrovirus (SRV) and simian T-cell leukemia virus (STLV) type-1 prior to SIV challenge.
PBMC was isolated by density gradient centrifugation and cells from LN were isolated by mechanical disruption. Cells from the jejunal mucosa were isolated by enzymatic digestion and percoll gradient centrifugation as per procedures described previously[45; 46; 47; 48; 49; 50].
Plasma viral loads were determined by real-time PCR using reverse-transcribed viral RNA as the template, as previously described [51]. CD4 T cell associated viral loads were determined in sorted memory CD4 T cell subsets using a highly quantitative PCR assay for SIV-gag as described previously[52]. The level of IFNα in plasma was determined using a human IFNα pan ELISA kit as per manufacturers instructions (limit of detection: 4 pg/ ml).
Antibodies and flow cytometry
Cryopreserved cells were labeled with a panel of CD3-Cy-7APC, CD4-APC, CD8-Alexa700, CD95-FITC and CD28-Cy-5PE (BD Biosciences) and memory CD4 T cells were sorted using a Becton Dickinson Aria sorter and used for determining CD4 T cell-associated viral loads. Memory CD4 T cells was discriminated based on the expression of CD28 and CD95 as described previously[52; 53]. All the antibodies were titrated using rhesus macaque PBMC.
To determine the expression of IFN subtypes in cell subsets, live LN cells at day 10 pi were stained with Vivid live dead marker and anti-CD3-Cy-7APC, CD4-BV605, CD8-Alexa700, CD14-FITC, CD11c-APC and CD123-PE, and CD3+CD4+ T cells (CD4), CD3+CD8+ T cells (CD8), total CD3-CD11c-CD123-CD14+/CD14− monocytes/macrophages (CD14+/CD14−) and CD3-CD14-CD11c+CD123+ dendritic cells (DC) were sorted using BD Aria sorter. Dead cells were excluded and live cell subsets that were sufficient to yield a minimum of ~500 ng of RNA were sorted for each subset (>98% purity) and used for RNA extraction. The number of cells required to yield ~500 ng of RNA was determined in a cell titration experiment.
Absolute quantification of Macaca mulatta IFN subtype mRNA levels by qRT-PCR
RNA was extracted from whole cell populations (total PBMC, LN and jejunum cells), and sorted cell subsets (CD4, CD8, monocytes, macrophages, and DC) using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) and treated with RNase-free DNase (Qiagen). Total RNA was quantified using a Nanodrop spectrophotometer and 500 ng of RNA was reverse transcribed with the Verso cDNA Synthesis Kit (Thermo Scientific, Rockford, IL, USA) using a mixture of random hexamers and anchored oligo dT primers. The run conditions were as follows: 42°C for 30 min, 95°C for 2 min, 4°C for ∞ Samples were subsequently treated with RNase H (New England Biolabs, Ipswich, MA, USA) following the manufacturer’s instructions.
The copy numbers of type I and III IFN was determined by qRT-PCR as previously described[23; 54]. Briefly, Macaca mulatta type I and III interferon subtype (IFNα-01/13, 02, 06, 08, 14, 16, 23, 24, 25, 26, 27, 28, 29, IFNβ, IFNω, IFNλ-1 and IFNλ-3) specific primers and probes[23] were distributed into 384-well assay plates with the Solo automated multi-channel pipettor (Hudson Robotics, Inc., Springfield, NJ), dried, and stored in the dark at 4°C until use. TaqMan Fast Universal PCR Master Mix and the primer/probe sets for GAPDH and 18S were purchased from Applied Biosystems (Foster City, CA, USA). Primers for the IFN transcripts and Molecular Beacon probes were synthesized by the Facility for Biotechnology Resources at the Center for Biologics Evaluation and Research (Silver Spring, MD, USA). LNA probes were synthesized by Sigma-Aldrich (Saint Louis, MO, USA). All primer and probe stocks were purified by high-performance liquid chromatography. The master mix/water mixtures, sample cDNA, housekeeping gene primer/probe sets, and standards were added to each well using electronic multichannel pipettes (Thermo-Fisher Scientific, Waltham, MA, USA). No template controls (NTC) and four point standard curves of linearized plasmids containing the IFN sequences as inserts were included on each assay plate. The total volume of each PCR reaction per well was 7.5 μl (3.75 μl PCR Master Mix, 2.25 μl primer/probe sets and 1.5 μl cDNA template). Sealed plates were centrifuged at 1500 rpm, mixed with a MixMate two-dimensional plate vortexer at 2600 rpm (Eppendorf, Westbury, NY, USA), and centrifuged again. After centrifugation, the qRT-PCR assay plates were run on the ViiA 7 Real-Time PCR System (Life Technologies, Grand Island, NY, USA) using the following run conditions: 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 sec, 60°C for 1 min. Data was analyzed using the ViiA 7 RUO Software (Life Technologies) and exported into a Microsoft Excel Spreadsheet designed for in-house analysis. Each four point standard curve set was graphed and analyzed for linearity. Absolute copy numbers of each IFN transcript were calculated based on the four point standard curve and target gene transcripts were normalized to micrograms of RNA input per well.
Data analysis
Flow cytometric data was analyzed using FlowJo version 9.2 (Tree Star, Inc., Ashland, OR). Statistical analysis was performed using GraphPad Prism Version 5.0 software (GraphPad Prism Software, Inc. San Diego, CA). Mann Whitney U test was used to determine significance and spearman’s rank was used to determine correlations. A p < 0.05 was considered significant. Error bars represent standard error.
Results
Kinetics of plasma IFNα levels correlate with viral infection
Previous studies[19; 55] have shown that acute SIV infection was characterized by an increase in plasma IFNα levels. To confirm these findings, we examined the levels of IFNα in plasma collected prior to infection, and at day 7, 10 and 14 pi and correlated them with plasma viral loads. Our results showed that plasma viremia increased after infection and peaked at day 10 pi and continued to remain high at day 14 pi (Fig. 1a). As reported previously[19; 55; 56; 57], plasma IFNα levels were found to peak at day 10 pi that was followed by a significant decline to near baseline levels by day 14 pi (Fig. 1b). Interestingly, plasma viral loads (r = 0.4949; p = 0.0521) at day 10 pi did not significantly correlate with plasma IFNα levels at day 10 pi (Fig. 1c).
Figure 1. Reverse transcriptase inhibitors significantly suppressed plasma IFNα during acute SIV infection.
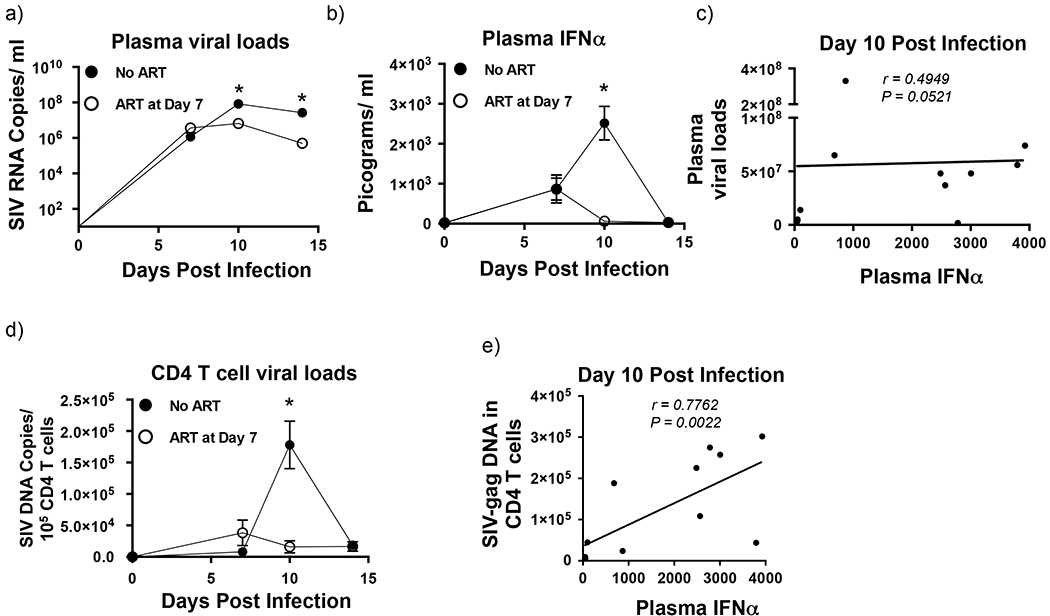
(a) Kinetics of plasma viral loads (Limit of detection is 30 copies /ml of plasma) and (b) plasma IFNα levels during the 1st 2 weeks of SIV infection in untreated (n = 8) and treated (n = 4) animals. Treated animals received reverse transcriptase inhibitors starting at day 7 post-infection. (c) Correlation between plasma viral loads and plasma IFNα levels at day 10 pi from untreated (n = 8) and treated (n = 4) animals. (d) Kinetics of SIV DNA loads in CD4 memory T cells in untreated and treated animals during the 1st 2 weeks of SIV infection. (e) Correlation between CD4 memory T cell-associated SIV DNA loads and plasma IFNα levels at day 10 pi from untreated (n = 8) and treated (n = 4) animals. Line of fit was determined using linear regression analysis and correlations were derived using Spearman’s rank test. Error bars represent standard error and * denotes p < 0.05.
Reverse transcriptase inhibitors administered at day 7 pi significantly suppresses plasma IFNα levels and CD4 T cell associated SIV-DNA loads at day 10 pi
Studies[58; 59] have shown that endosomal Toll-like-receptors (TLR) such as TLR-7, 8, and 9 play a key role in induction of IFNα. However, in vivo administration of TLR-7 and 9 antagonists had little or no effect on plasma IFNα levels[19] suggesting to a role for other mechanisms in this process. On the other hand, recent studies[60] have shown that a cytoplasmic DNA sensor, cGAS plays a role in induction of IFNα responses in macrophages raising the possibility that similar mechanisms may be driving early IFNα responses during SIV infection. To address this question we examined plasma IFNα levels in rhesus macaques that initiated anti-retroviral therapy with reverse transcriptase (RT) inhibitors at day 7 pi and compared them SIV infected untreated animals.
Our results show that initiation of therapy early during infection was associated with a significant decrease in peak plasma and cell-associated SIV DNA as compared to untreated animals (Fig. 1a and 1d). Suppression of infection was accompanied by a significant decline in plasma IFNα levels to near baseline levels (Fig. 1b). Though plasma viremia remained high in treated animals plasma IFNα levels remained at near baseline levels in treated animals at day 10 and 14 pi. These findings along with the lack of a significant correlation between plasma viral loads and plasma IFNα levels at day 10 pi suggest that byproducts of reverse transcription such as viral DNA maybe playing a key role in driving IFNα responses very early during SIV infection. In line with this argument, we found a significantly high positive correlation between cell-associated SIV DNA loads and plasma IFNα levels (r = 0.7762, p = 0.0022) at day 10 pi (Fig. 1e).
IFN subtypes are differentially expressed in PBMC, LN and Jejunal mucosa
SIV has been shown to extensively replicate in the LN and mucosa during acute stages of infection[52; 61; 62; 63; 64]. We hypothesized that these tissues were a likely source for the significant increase in IFN levels during the acute SIV infection. As IFN harbors numerous subtypes, we quantified the copy numbers of both Type I and III IFN subtype mRNA in total cells isolated from the PBMC, LN and jejunal mucosa at day 10, 14 – 31 pi and compared them to pre-infection levels (Fig. 2).
Figure 2. Most Type I and III IFN subtypes are significantly upregulated in the LN at day 10 post infection.
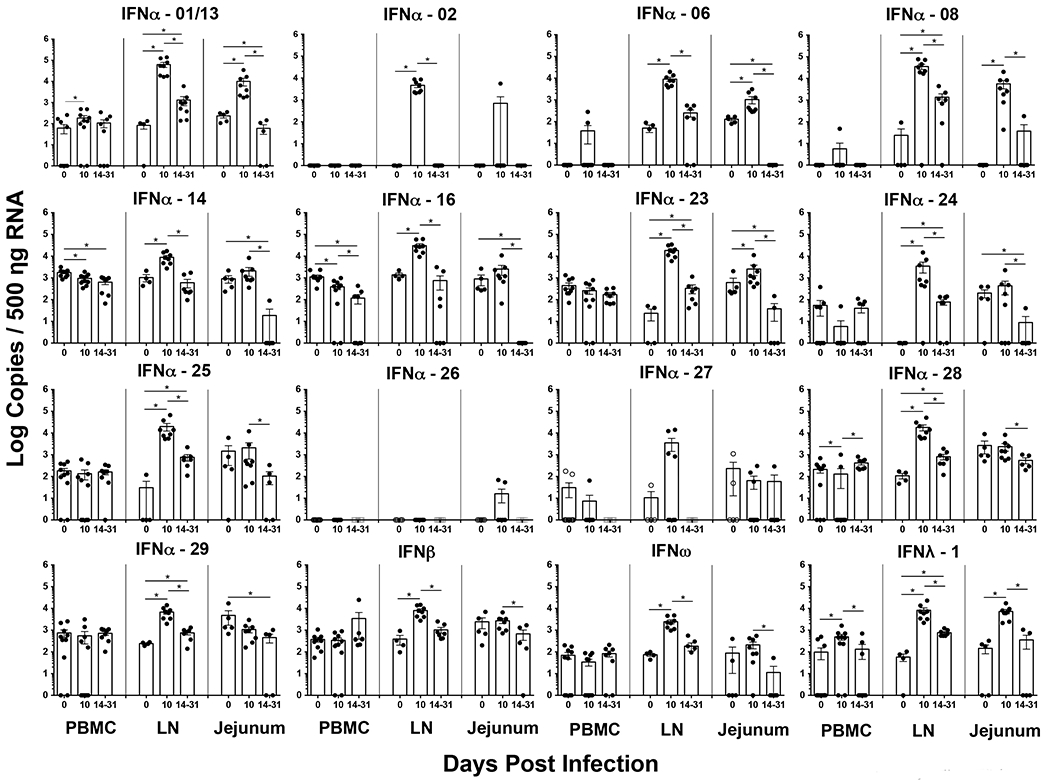
Absolute copies of Type I and III IFN subtypes was determined in total cells isolated from peripheral blood (PBMC), jejunum, and lymph nodes (LN) at day 10 and 14 – 31 post infection using a qRT-PCR assay and compared to pre-infection values. Absolute copy numbers were determined using rhesus macaque IFN subtype specific standards. Statistical analysis was performed using Mann-Whitney U test and a p < 0.05 (*) was considered significant.
Our results showed that the expression of IFN subtypes was highly restricted in PBMC with only IFNα-01/13 and IFNλ-1 being significantly upregulated at day 10 pi as compared to uninfected animals. Interestingly, IFNα-14 and 16 levels significantly declined by day 10 pi as compared to pre-infection levels. Like PBMC, cells from the jejunal mucosa at day 10 pi were found to selectively upregulate IFNα-01/13, 06, 08, 23, IFNω and IFNλ-1 subtypes as compared to preinfection values. Expression levels in both PBMC and jejunal mucosa declined significantly by day 14 – 31 pi with the loss being more pronounced in the jejunal mucosa that likely coincided with the massive loss of cells reported to occur at these sites during acute stages of infection.
Unlike PBMC and jejunal mucosa, however, the expression levels of all the Type I and III subtypes except for IFNα-26 were significantly upregulated at day 10 pi in the LN as compared to uninfected animals. The levels of IFN expression in LN were significantly higher in magnitude than both PBMC and jejunal mucosa suggesting that LN were a significant source of early IFN response. Though peak expression levels declined by day 14 – 31 as compared to day 10 pi, the expression levels of IFNα-01/13, 08, 23, 24, 25, 28, 29, IFNβ and IFNλ-1 remained significantly higher than preinfection levels suggesting that ongoing viral replication continues to drive innate IFN responses in organized lymphoid tissues after viral replication had peaked. IFNλ-3 was undetectable in all the three tissues we examined.
Dendritic cells are the primary producers of both Type I and III IFN during acute SIV infection
Our results showed that LN expressed ~ 3 – 4 logs more Type I and III IFN subtype mRNA transcripts at day 10 pi as compared other tissues. To determine the cellular source of the different subtypes, we sorted highly purified subsets of CD4 and CD8 T cells, total monocytes/macrophages (CD14+/CD14−) and total DC’s (CD11c+ and CD123+) from the LN at day 10 pi and quantified the expression of IFN subtypes in these subsets (Fig. 3). We were unable to obtain sufficient LN samples prior to infection for sorting cell subsets, hence restricted our analysis to samples collected at day 10 pi. Additionally, given the limited amount of samples we had and the low frequency of both CD11c+ myeloid DC and CD123+ plasmacytoid DC, we decided to sort total DC instead of individual subsets to extract sufficient RNA needed for qRT-PCR analysis of various IFN subtypes. A similar rationale was used to sort total monocyte/macrophage subsets. Our results showed that except for IFNα-26, at peak of infection DC subsets expressed significantly higher numbers of all IFN subtypes in vivo as compared to the other cell subsets. There was no significant difference between the other four subsets we examined.
Figure 3. Dendritic cells are the primary producers of all IFN subtypes at 10 days post infection.
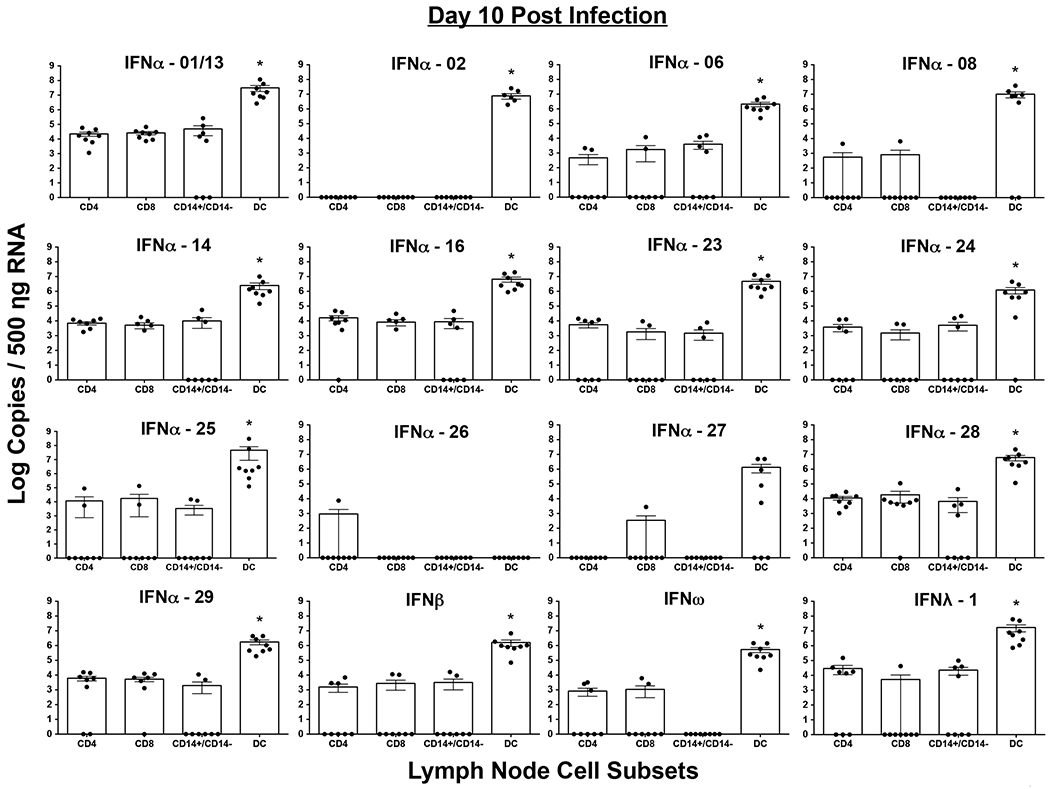
Highly purified populations of T cells (CD4+ and CD8+), monocytes/macrophages (CD14+/CD14−) and dendritic cells (DC) were sorted using a BD FACS Aria sorter and used for quantifying IFN subtype transcripts using a qRT-PCR assay. Absolute copy numbers were determined using rhesus macaque IFN subtype specific standards. Statistical analysis was performed using Mann-Whitney U test and a p < 0.05 (*) was considered significant.
Discussion
HIV-1 and SIV infections are characterized by a significant innate immune response early in infection[65]. Others have reported upregulated expression of interferon stimulating genes (ISG) that have been associated with control of infection[65]. The mechanisms that drive early IFN response have been a matter of intense investigation over a number of years though the exact cause for induction of Type I, and III IFN response in vivo remains unclear. Our results suggest that RT byproducts such as viral DNA may play a key role in this process; initiation of antiretroviral therapy with RT inhibitors at day 7 pi significantly suppressed plasma IFNα levels to that of baseline by day 10 pi as compared to ~3000 pg/ ml of IFNα at day 10 pi in SIV infected untreated animals. Interestingly, the plasma viral loads remained high (~6 logs) in treated animals at day 10 pi suggesting that endosomal compartmentalization of virus may not be the key driver of early IFN responses. Otherwise, we should have seen higher levels of plasma IFNα as viral RNA levels remained significantly high in the plasma from treated animals. In line with this argument, studies[19] have shown that treatment of SIV infected rhesus macaques with TLR7 and TLR9 antagonists had little or no effect on peak plasma IFNα levels during the first two weeks of SIV infection.
On the other hand, suppression of IFN responses by RT inhibitors suggests that RT byproducts such as the reverse transcribed viral DNA might be driving early IFN responses during acute SIV infection. Recent studies have demonstrated a key role of the cytoplasmic DNA sensor cGAS in macrophages infected with HIV and SIV[60]. Lahaye et al[66] showed that host sensing of HIV-2, a virus that is more similar to SIV, required viral cDNA synthesis rather than nuclear entry or genome integration, and this sensing in DC’s were mediated by cGAS. Martin-Gayo et al[67] showed that conventional DC from HIV infected elite controllers produce high levels of innate IFN that was associated with an accumulation of viral reverse transcripts and blocking of cGAS or reverse transcription inhibited these responses. Herzner et al[68] showed that single stranded HIV-1 DNA activates cGAS and HIV-1 reverse transcripts were the predominant viral DNA species in the cytoplasm of macrophages during early infection. The above studies along with our results suggest that reverse transcribed viral DNA and cytoplasmic sensors likely play an important role in the induction of innate IFN responses in vivo during acute SIV infection.
That innate IFN response in plasma peaks very early during infection has been known for some time. Previous studies[19; 55] have shown that plasma IFNα levels peak between days 7 - 10 pi in SIV infected animals after which they rapidly decline to baseline levels. Our findings were in line with these earlier reports. What is, however, not clear is the kinetics of the various IFN subtype responses in vivo during acute stages of SIV infection, and if these responses differ between tissues. Our results showed that various subtypes were differentially expressed in mucosal and peripheral tissues with nearly all of the detectable subtypes being expressed at significantly high levels in the LN as compared to PBMC and jejunum; at day 10 pi only IFNα-01/13 and IFNλ-1 transcripts were significantly upregulated in PBMC, whereas IFNα-01/13, 06, 08, 23, IFNω and IFNλ-1 subtypes were significantly upregulated in the jejunal mucosa. IFNα-26 and IFNλ-3 were undetectable in all the tissues we examined. The expression levels in both PBMC and jejunal mucosa significantly declined by day 14 – 31 pi. Interestingly, there was a significant decline in IFNα-14 and 16 subtypes in PBMC by day 10 pi. Harper et al[43] using an ex vivo model of HIV infection showed that IFNα-06, 08 and 14 were the most potent of subtypes examined that showed significant HIV restriction in the mucosa. On the other hand, IFNα-01 and 02 had the weakest antiviral activity suggesting that decline of these highly restrictive subtypes may contribute to the increased pathogenesis and replication in the gut mucosa[43]. Likewise, Lavender et al[69] showed that IFNα-14 had significantly higher antiviral activity against HIV infection in humanized mouse models as compared to other subtypes.
In contrast to both PBMC and jejunal mucosa, the expression and magnitude of most subtypes were significantly upregulated in the LN at day 10 pi suggesting that LN was a significant source of early IFN response during acute SIV infection. Unlike the PBMC and jejunum, however, the expression levels of IFNα-01/13, 08, 23, 24, 25, 28, 29, IFNβ and IFNλ-1 transcripts remained significantly higher at day 14 – 31 pi as compared to pre-infection levels suggesting that ongoing replication continues to drive specific IFN subtype responses in the LN. The exact reason why only a subset of IFN subtypes were expressed is difficult to determine at this point. Previous studies[44] examined the relative expression of IFNα subtypes in SIV infected PTM during the 1st 3 weeks of infection and found tissue specific differences in the expression pattern of various IFNα subtypes; only subtypes 2, 6, and 13 were expressed in the brain of SIV infected PTM at day 7 pi, whereas only subtypes 6 and 13 were upregulated in the lung. In contrast to the brain and lung, all the 13 subtypes were expressed in the spleen with subtypes 2, 8, and 13 being the most abundantly expressed at day 7 pi whereas subtypes 4, 17 and 21 were least abundantly expressed.
It is unlikely that the lower levels of IFN subtypes expressed in the jejunum as compared to the LN was due to the lower frequency of IFN producing cells at these sites as fewer cells are likely to lower the magnitude of expression rather than the pattern of expression of IFN subtypes. It is, however, difficult to rule this out. The loss of DC subsets from peripheral blood may explain the decline in expression of subtypes in blood as numerous studies have documented that both DC and macrophages migrate to the gut mucosa during SIV infection[56; 70; 71; 72; 73; 74; 75]. The simultaneous decline in IFN expression in both mucosal and peripheral tissues, however, raises the possibility that DC are either lost or become refractive in both PBMC and the mucosa at the same time. Wonderlich et al[57] reported that macrophages and myeloid DC subsets loose stimulating function and was associated with decreased IFNα production during SIV infection.
Compared to PBMC and jejunum, LN is enriched for IFN producing cells such as DC that might explain the significantly higher levels of all IFN subtypes in the LN. In fact, we observed significantly higher levels of all detectable IFN subtypes in the DC’s from the LN as compared to other cell subsets; DC’s had ~100,000 fold more copies of IFN subtype transcripts than either T cells or monocyte/macrophage subsets. Our results for the first time provide in vivo evidence for previously reported in vitro studies[76; 77; 78; 79; 80] showing that DC produce significantly higher amounts of IFN that other cell types.
Previous studies have reported that LN DC’s were major producers of Type I IFNα during acute SIV infection[55; 81; 82] whereas others have shown that pDC’s were actively recruited from circulation into the LN during SIV infection[71; 83]. Bruel et al[55] showed that pDC were the major producers of IFNα in the gut and lymphoid tissues during acute SIV infection. Interestingly, there was no difference in the IFN transcript levels between CD4+ T cells and other non-DC subsets we examined even though CD4 T cells are highly infected at day 10 pi (Fig. 1d). There is little or no evidence in the literature showing that CD4 T cells were capable of making innate IFN responses during viral infection though they have been shown to upregulate the expression of ISG during acute stages of SIV infection. Like CD4 T cells, it was somewhat surprising that we did not see any significant upregulation of IFN transcripts monocytes/macrophage subsets even though previous studies have shown that macrophages carry SIV DNA[84]. The exact reason why macrophages did not express IFN in vivo very early in infection has to be examined further and additional studies are needed to clarify these questions in detail. It is difficult to determine from our studies if cellular subsets other than DC’s in the LN upregulated the expression of IFN subtypes at day 10 pi as we did not have sufficient cells from healthy LN to compare. However, there is little or no evidence of CD8 T cells producing IFNα during SIV infection and transcript levels in CD8 T cells did not differ from the other non-DC subsets suggesting that cell subsets other than DC’s likely did not upregulate the expression of innate IFN. Future studies will examine this question in greater detail.
In conclusion, our studies provide new insights into the kinetics of different Type I and III IFN subtypes during acute SIV infection and identifies DC’s as a major in vivo cellular source of IFN subtypes. The significant suppression of plasma IFNα with RT inhibitors suggest that viral DNA and cytoplasmic DNA sensors likely play a role in driving Type I and III IFN responses during acute SIV infection.
Highlights:
Plasma IFNα levels peak at day 10 post-SIV infection after which they rapidly decline to near base line levels.
Reverse transcriptase byproducts such as viral DNA play a major role in driving innate IFN responses during acute SIV infection.
Both Type I and III IFN subtypes are significantly elevated in the lymph nodes at day 10 post-SIV infection as compared to a restricted expression in PBMC and jejunal mucosa.
Dendritic cells are a major in vivo source of Type I and III IFN during acute SIV infection.
Acknowledgements
We thank Olusegun Onabajo, Sean Maynard and Sandra Bixler at the Uniformed Services University for assistance with processing the samples. Kateryna Lund at the Biomedical Instrumentation Center; Matt Collins at Bioqual Inc., Rockville, MD for expert assistance with the animals.
The described project was supported by funds (R0731976) from the Uniformed Services University of the Health Sciences to JJM. The opinions or assertions contained herein are the private ones of the authors and are not to be construed as official or reflecting the views of the Department of Defense, the Uniformed Services University of the Health Sciences or any other agency of the U.S. Government.
The authors declare no financial conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Dusheiko G, Side effects of alpha interferon in chronic hepatitis C. Hepatology 26 (1997) 112S–121S. [DOI] [PubMed] [Google Scholar]
- [2].Koyama Y, Tanaka Y, Oda S, Yamashita U, and Eto S, Antiviral and antiproliferative activities of recombinant human interferon alpha 2, beta and gamma on HTLV-I and ATL cells in vitro. J UOEH 12 (1990) 149–61. [DOI] [PubMed] [Google Scholar]
- [3].Witter F, Barouki F, Griffin D, Nadler P, Woods A, Wood D, and Lietman P, Biologic response (antiviral) to recombinant human interferon alpha 2a as a function of dose and route of administration in healthy volunteers. Clin Pharmacol Ther 42 (1987) 567–75. [DOI] [PubMed] [Google Scholar]
- [4].Zhu H, Butera M, Nelson DR, and Liu C, Novel type I interferon IL-28A suppresses hepatitis C viral RNA replication. Virol J 2 (2005) 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lapenta C, Santini SM, Proietti E, Rizza P, Logozzi M, Spada M, Parlato S, Fais S, Pitha PM, and Belardelli F, Type I interferon is a powerful inhibitor of in vivo HIV-1 infection and preserves human CD4(+) T cells from virus-induced depletion in SCID mice transplanted with human cells. Virology 263 (1999) 78–88. [DOI] [PubMed] [Google Scholar]
- [6].Leissner P, Calenda V, Marigliano M, Sanhadji K, Touraine JL, Pavirani A, and Mehtali M, [In vitro and in vivo inhibition of HIV1 replication by retroviral transfer of interferon alpha, beta, or gamma genes: application to gene therapy of AIDS]. Ann Biol Clin (Paris) 56 (1998) 167–73. [PubMed] [Google Scholar]
- [7].Poli G, Biswas P, and Fauci AS, Interferons in the pathogenesis and treatment of human immunodeficiency virus infection. Antiviral Res 24 (1994) 221–33. [DOI] [PubMed] [Google Scholar]
- [8].Popik W, and Pitha PM, Exploitation of cellular signaling by HIV-1: unwelcome guests with master keys that signal their entry. Virology 276 (2000) 1–6. [DOI] [PubMed] [Google Scholar]
- [9].Sanhadji K, Leissner P, Firouzi R, Pelloquin F, Kehrli L, Marigliano M, Calenda V, Ottmann M, Tardy JC, Mehtali M, and Touraine JL, Experimental gene therapy: the transfer of Tat-inducible interferon genes protects human cells against HIV-1 challenge in vitro and in vivo in severe combined immunodeficient mice. AIDS 11 (1997) 977–86. [DOI] [PubMed] [Google Scholar]
- [10].Weiden M, Tanaka N, Qiao Y, Zhao BY, Honda Y, Nakata K, Canova A, Levy DE, Rom WN, and Pine R, Differentiation of monocytes to macrophages switches the Mycobacterium tuberculosis effect on HIV-1 replication from stimulation to inhibition: modulation of interferon response and CCAAT/enhancer binding protein beta expression. J Immunol 165 (2000) 2028–39. [DOI] [PubMed] [Google Scholar]
- [11].Baca-Regen L, Heinzinger N, Stevenson M, and Gendelman HE, Alpha interferon-induced antiretroviral activities: restriction of viral nucleic acid synthesis and progeny virion production in human immunodeficiency virus type 1-infected monocytes. J Virol 68 (1994) 7559–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Coccia EM, Krust B, and Hovanessian AG, Specific inhibition of viral protein synthesis in HIV-infected cells in response to interferon treatment. J Biol Chem 269 (1994) 23087–94. [PubMed] [Google Scholar]
- [13].Fernie BF, Poli G, and Fauci AS, Alpha interferon suppresses virion but not soluble human immunodeficiency virus antigen production in chronically infected T-lymphocytic cells. J Virol 65 (1991) 3968–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kinzl P, Otani T, Benz R, and Minowada J, Interferon-alpha and -gamma differentially reduce rapid immature T-cell death by contact with HIV-1 carrier cell clones in vitro. Microbiol Immunol 41 (1997) 709–16. [DOI] [PubMed] [Google Scholar]
- [15].Meylan PR, Guatelli JC, Munis JR, Richman DD, and Kornbluth RS, Mechanisms for the inhibition of HIV replication by interferons-alpha, -beta, and - gamma in primary human macrophages. Virology 193 (1993) 138–48. [DOI] [PubMed] [Google Scholar]
- [16].Shirazi Y, and Pitha PM, Alpha interferon inhibits early stages of the human immunodeficiency virus type 1 replication cycle. J Virol 66 (1992) 1321–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Abel K, Alegria-Hartman MJ, Rothaeusler K, Marthas M, and Miller CJ, The relationship between simian immunodeficiency virus RNA levels and the mRNA levels of alpha/beta interferons (IFN-alpha/beta) and IFN-alpha/beta-inducible Mx in lymphoid tissues of rhesus macaques during acute and chronic infection. J Virol 76 (2002) 8433–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bosinger SE, Li Q, Gordon SN, Klatt NR, Duan L, Xu L, Francella N, Sidahmed A, Smith AJ, Cramer EM, Zeng M, Masopust D, Carlis JV, Ran L, Vanderford TH, Paiardini M, Isett RB, Baldwin DA, Else JG, Staprans SI, Silvestri G, Haase AT, and Kelvin DJ, Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J Clin Invest 119 (2009) 3556–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kader M, Smith AP, Guiducci C, Wonderlich ER, Normolle D, Watkins SC, Barrat FJ, and Barratt-Boyes SM, Blocking TLR7- and TLR9-mediated IFN-alpha production by plasmacytoid dendritic cells does not diminish immune activation in early SIV infection. PLoS Pathog 9 (2013) e1003530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schaefer TM, Fuller CL, Basu S, Fallert BA, Poveda SL, Sanghavi SK, Choi YK, Kirschner DE, Feingold E, and Reinhart TA, Increased expression of interferon-inducible genes in macaque lung tissues during simian immunodeficiency virus infection. Microbes Infect 8 (2006) 1839–50. [DOI] [PubMed] [Google Scholar]
- [21].Sandler NG, Bosinger SE, Estes JD, Zhu RT, Tharp GK, Boritz E, Levin D, Wijeyesinghe S, Makamdop KN, del Prete GQ, Hill BJ, Timmer JK, Reiss E, Yarden G, Darko S, Contijoch E, Todd JP, Silvestri G, Nason M, Norgren RB Jr., Keele BF, Rao S, Langer JA, Lifson JD, Schreiber G, and Douek DC, Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature 511 (2014) 601–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Szubin R, Chang WL, Greasby T, Beckett L, and Baumgarth N, Rigid interferon-alpha subtype responses of human plasmacytoid dendritic cells. J Interferon Cytokine Res 28 (2008) 749–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schramm LM, Kirschman KD, Heuer M, Chen AA, Verthelyi D, Puig M, and Rabin RL, High-throughput quantitative real-time polymerase chain reaction array for absolute and relative quantification of rhesus macaque types I, II, and III interferon and their subtypes. J Interferon Cytokine Res 32 (2012) 407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I, Chen S, Brand N, Tarway M, Liu L, Sheikh F, Astemborski J, Bonkovsky HL, Edlin BR, Howell CD, Morgan TR, Thomas DL, Rehermann B, Donnelly RP, and O’Brien TR, A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet 45 (2013) 164–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, and Donnelly RP, IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol 4 (2003) 69–77. [DOI] [PubMed] [Google Scholar]
- [26].Chen J, Baig E, and Fish EN, Diversity and relatedness among the type I interferons. J Interferon Cytokine Res 24 (2004) 687–98. [DOI] [PubMed] [Google Scholar]
- [27].Kumaran J, Wei L, Kotra LP, and Fish EN, A structural basis for interferon-alpha-receptor interactions. FASEB J 21 (2007) 3288–96. [DOI] [PubMed] [Google Scholar]
- [28].Pestka S, Langer JA, Zoon KC, and Samuel CE, Interferons and their actions. Annu Rev Biochem 56 (1987) 727–77. [DOI] [PubMed] [Google Scholar]
- [29].Lamken P, Lata S, Gavutis M, and Piehler J, Ligand-induced assembling of the type I interferon receptor on supported lipid bilayers. J Mol Biol 341 (2004) 303–18. [DOI] [PubMed] [Google Scholar]
- [30].Piehler J, Roisman LC, and Schreiber G, New structural and functional aspects of the type I interferon-receptor interaction revealed by comprehensive mutational analysis of the binding interface. J Biol Chem 275 (2000) 40425–33. [DOI] [PubMed] [Google Scholar]
- [31].Roisman LC, Jaitin DA, Baker DP, and Schreiber G, Mutational analysis of the IFNAR1 binding site on IFNalpha2 reveals the architecture of a weak ligand-receptor binding-site. J Mol Biol 353 (2005) 271–81. [DOI] [PubMed] [Google Scholar]
- [32].Yamaoka T, Kojima S, Ichi S, Kashiwazaki Y, Koide T, and Sokawa Y, Biologic and binding activities of IFN-alpha subtypes in ACHN human renal cell carcinoma cells and Daudi Burkitt’s lymphoma cells. J Interferon Cytokine Res 19 (1999) 1343–9. [DOI] [PubMed] [Google Scholar]
- [33].Foster GR, Rodrigues O, Ghouze F, Schulte-Frohlinde E, Testa D, Liao MJ, Stark GR, Leadbeater L, and Thomas HC, Different relative activities of human cell-derived interferon-alpha subtypes: IFN-alpha 8 has very high antiviral potency. J Interferon Cytokine Res 16 (1996) 1027–33. [DOI] [PubMed] [Google Scholar]
- [34].Palmer P, Tovey MG, Raschilas F, Brassart L, Meritet JF, Porcher R, and Lebon P, Type I interferon subtypes produced by human peripheral mononuclear cells from one normal donor stimulated by viral and non-viral inducing factors. Eur Cytokine Netw 18 (2007) 108–14. [DOI] [PubMed] [Google Scholar]
- [35].Sperber SJ, Gocke DJ, Haberzettl C, Kuk R, Schwartz B, and Pestka S, Anti-HIV-1 activity of recombinant and hybrid species of interferon-alpha. J Interferon Res 12 (1992) 363–8. [DOI] [PubMed] [Google Scholar]
- [36].Koyama T, Sakamoto N, Tanabe Y, Nakagawa M, Itsui Y, Takeda Y, Kakinuma S, Sekine Y, Maekawa S, Yanai Y, Kurimoto M, and Watanabe M, Divergent activities of interferon-alpha subtypes against intracellular hepatitis C virus replication. Hepatol Res 34 (2006) 41–9. [DOI] [PubMed] [Google Scholar]
- [37].Pestka S, Krause CD, and Walter MR, Interferons, interferon-like cytokines, and their receptors. Immunol Rev 202 (2004) 8–32. [DOI] [PubMed] [Google Scholar]
- [38].Scagnolari C, Trombetti S, Selvaggi C, Carbone T, Monteleone K, Spano L, Di Marco P, Pierangeli A, Maggi F, Riva E, and Antonelli G, In vitro sensitivity of human metapneumovirus to type I interferons. Viral Immunol 24 (2011) 159–64. [DOI] [PubMed] [Google Scholar]
- [39].Weck PK, Apperson S, May L, and Stebbing N, Comparison of the antiviral activities of various cloned human interferon-alpha subtypes in mammalian cell cultures. J Gen Virol 57 (1981) 233–7. [DOI] [PubMed] [Google Scholar]
- [40].Yanai Y, Sanou O, Kayano T, Ariyasu H, Yamamoto K, Yamauchi H, Ikegami H, and Kurimoto M, Analysis of the antiviral activities of natural IFN-alpha preparations and their subtype compositions. J Interferon Cytokine Res 21 (2001) 835–41. [DOI] [PubMed] [Google Scholar]
- [41].Hibbert L, and Foster GR, Human type I interferons differ greatly in their effects on the proliferation of primary B cells. J Interferon Cytokine Res 19 (1999) 309–18. [DOI] [PubMed] [Google Scholar]
- [42].Hilkens CM, Schlaak JF, and Kerr IM, Differential responses to IFN-alpha subtypes in human T cells and dendritic cells. J Immunol 171 (2003) 5255–63. [DOI] [PubMed] [Google Scholar]
- [43].Harper MS, Guo K, Gibbert K, Lee EJ, Dillon SM, Barrett BS, McCarter MD, Hasenkrug KJ, Dittmer U, Wilson CC, and Santiago ML, Interferon-alpha Subtypes in an Ex Vivo Model of Acute HIV-1 Infection: Expression, Potency and Effector Mechanisms. PLoS Pathog 11 (2015) e1005254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zaritsky LA, Dery A, Leong WY, Gama L, and Clements JE, Tissue-specific interferon alpha subtype response to SIV infection in brain, spleen, and lung. J Interferon Cytokine Res 33 (2013) 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bixler SL, Sandler NG, Douek DC, and Mattapallil JJ, Suppressed Th17 Levels Correlate with Elevated PIAS3, SHP2, and SOCS3 Expression in CD4 T cells During Acute Simian Immunodeficiency Virus Infection. J Virol (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Eberly MD, Kader M, Hassan W, Rogers KA, Zhou J, Mueller YM, Mattapallil MJ, Piatak M Jr., Lifson JD, Katsikis PD, Roederer M, Villinger F, and Mattapallil JJ, Increased IL-15 production is associated with higher susceptibility of memory CD4 T cells to simian immunodeficiency virus during acute infection. J Immunol 182 (2009) 1439–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].George J, Cofano EB, Lybarger E, Louder M, Lafont BA, Mascola JR, Robert-Guroff M, and Mattapallil JJ, Early short-term antiretroviral therapy is associated with a reduced prevalence of CD8(+)FoxP3(+) T cells in simian immunodeficiency virus-infected controller rhesus macaques. AIDS Res Hum Retroviruses 27 (2011) 763–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kader M, Hassan WM, Eberly M, Piatak M, Lifson JD, Roederer M, and Mattapallil JJ, Antiretroviral therapy prior to acute viral replication preserves CD4 T cells in the periphery but not in rectal mucosa during acute simian immunodeficiency virus infection. J Virol 82 (2008) 11467–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kader M, Wang X, Piatak M, Lifson J, Roederer M, Veazey R, and Mattapallil JJ, Alpha4(+)beta7(hi)CD4(+) memory T cells harbor most Th-17 cells and are preferentially infected during acute SIV infection. Mucosal Immunol 2 (2009) 439–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Onabajo OO, George J, Lewis MG, and Mattapallil JJ, Rhesus Macaque Lymph Node PD-1(hi)CD4(+) T Cells Express High Levels of CXCR5 and IL-21 and Display a CCR7(lo)ICOS(+)Bcl6(+) T-Follicular Helper (Tfh) Cell Phenotype. PLoS One 8 (2013) e59758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cline AN, Bess JW, Piatak M Jr., and Lifson JD, Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J Med Primatol 34 (2005) 303–12. [DOI] [PubMed] [Google Scholar]
- [52].Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, and Roederer M, Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 434 (2005) 1093–7. [DOI] [PubMed] [Google Scholar]
- [53].Pitcher CJ, Hagen SI, Walker JM, Lum R, Mitchell BL, Maino VC, Axthelm MK, and Picker LJ, Development and homeostasis of T cell memory in rhesus macaque. J Immunol 168 (2002) 29–43. [DOI] [PubMed] [Google Scholar]
- [54].Renn LA, Theisen TC, Navarro MB, Mane VP, Schramm LM, Kirschman KD, Fabozzi G, Hillyer P, Puig M, Verthelyi D, and Rabin RL, High-throughput quantitative real-time RT-PCR assay for determining expression profiles of types I and III interferon subtypes. J Vis Exp (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bruel T, Dupuy S, Demoulins T, Rogez-Kreuz C, Dutrieux J, Corneau A, Cosma A, Cheynier R, Dereuddre-Bosquet N, Le Grand R, and Vaslin B, Plasmacytoid dendritic cell dynamics tune interferon-alfa production in SIV-infected cynomolgus macaques. PLoS Pathog 10 (2014) e1003915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wonderlich ER, Kader M, Wijewardana V, and Barratt-Boyes SM, Dissecting the role of dendritic cells in simian immunodeficiency virus infection and AIDS. Immunol Res 50 (2011) 228–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wonderlich ER, Wu WC, Normolle DP, and Barratt-Boyes SM, Macrophages and Myeloid Dendritic Cells Lose T Cell-Stimulating Function in Simian Immunodeficiency Virus Infection Associated with Diminished IL-12 and IFN-alpha Production. J Immunol 195 (2015) 3284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Beignon AS, McKenna K, Skoberne M, Manches O, DaSilva I, Kavanagh DG, Larsson M, Gorelick RJ, Lifson JD, and Bhardwaj N, Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor-viral RNA interactions. J Clin Invest 115 (2005) 3265–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Mandl JN, Barry AP, Vanderford TH, Kozyr N, Chavan R, Klucking S, Barrat FJ, Coffman RL, Staprans SI, and Feinberg MB, Divergent TLR7 and TLR9 signaling and type I interferon production distinguish pathogenic and nonpathogenic AIDS virus infections. Nat Med 14 (2008) 1077–87. [DOI] [PubMed] [Google Scholar]
- [60].Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, and Chen ZJ, Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341 (2013) 903–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, and Douek DC, CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med 200 (2004) 749–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Li Q, Duan L, Estes JD, Ma ZM, Rourke T, Wang Y, Reilly C, Carlis J, Miller CJ, and Haase AT, Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 434 (2005) 1148–52. [DOI] [PubMed] [Google Scholar]
- [63].Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, Hogan C, Boden D, Racz P, and Markowitz M, Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med 200 (2004) 761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, and Lackner AA, Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280 (1998) 427–31. [DOI] [PubMed] [Google Scholar]
- [65].Doyle T, Goujon C, and Malim MH, HIV-1 and interferons: who’s interfering with whom? Nat Rev Microbiol 13 (2015) 403–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Lahaye X, Satoh T, Gentili M, Cerboni S, Conrad C, Hurbain I, El Marjou A, Lacabaratz C, Lelievre JD, and Manel N, The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity 39 (2013) 1132–42. [DOI] [PubMed] [Google Scholar]
- [67].Martin-Gayo E, Buzon MJ, Ouyang Z, Hickman T, Cronin J, Pimenova D, Walker BD, Lichterfeld M, and Yu XG, Potent Cell-Intrinsic Immune Responses in Dendritic Cells Facilitate HIV-1-Specific T Cell Immunity in HIV-1 Elite Controllers. PLoS Pathog 11 (2015) e1004930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Herzner AM, Hagmann CA, Goldeck M, Wolter S, Kubler K, Wittmann S, Gramberg T, Andreeva L, Hopfner KP, Mertens C, Zillinger T, Jin T, Xiao TS, Bartok E, Coch C, Ackermann D, Hornung V, Ludwig J, Barchet W, Hartmann G, and Schlee M, Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat Immunol 16 (2015) 1025–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lavender KJ, Gibbert K, Peterson KE, Van Dis E, Francois S, Woods T, Messer RJ, Gawanbacht A, Muller JA, Munch J, Phillips K, Race B, Harper MS, Guo K, Lee EJ, Trilling M, Hengel H, Piehler J, Verheyen J, Wilson CC, Santiago ML, Hasenkrug KJ, and Dittmer U, Interferon Alpha Subtype-Specific Suppression of HIV-1 Infection In Vivo. J Virol 90 (2016) 6001–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Brown KN, Trichel A, and Barratt-Boyes SM, Parallel loss of myeloid and plasmacytoid dendritic cells from blood and lymphoid tissue in simian AIDS. J Immunol 178 (2007) 6958–67. [DOI] [PubMed] [Google Scholar]
- [71].Brown KN, Wijewardana V, Liu X, and Barratt-Boyes SM, Rapid influx and death of plasmacytoid dendritic cells in lymph nodes mediate depletion in acute simian immunodeficiency virus infection. PLoS Pathog 5 (2009) e1000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kwa S, Kannanganat S, Nigam P, Siddiqui M, Shetty RD, Armstrong W, Ansari A, Bosinger SE, Silvestri G, and Amara RR, Plasmacytoid dendritic cells are recruited to the colorectum and contribute to immune activation during pathogenic SIV infection in rhesus macaques. Blood 118 (2011) 2763–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Malleret B, Maneglier B, Karlsson I, Lebon P, Nascimbeni M, Perie L, Brochard P, Delache B, Calvo J, Andrieu T, Spreux-Varoquaux O, Hosmalin A, Le Grand R, and Vaslin B, Primary infection with simian immunodeficiency virus: plasmacytoid dendritic cell homing to lymph nodes, type I interferon, and immune suppression. Blood 112 (2008) 4598–608. [DOI] [PubMed] [Google Scholar]
- [74].Reeves RK, Evans TI, Gillis J, Wong FE, Kang G, Li Q, and Johnson RP, SIV infection induces accumulation of plasmacytoid dendritic cells in the gut mucosa. J Infect Dis 206 (2012) 1462–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Swan ZD, Wonderlich ER, and Barratt-Boyes SM, Macrophage accumulation in gut mucosa differentiates AIDS from chronic SIV infection in rhesus macaques. Eur J Immunol 46 (2016) 446–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Cella M, Jarrossay D, Facchetti F, Alebardi O, Nakajima H, Lanzavecchia A, and Colonna M, Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat Med 5 (1999) 919–23. [DOI] [PubMed] [Google Scholar]
- [77].Lehmann C, Harper JM, Taubert D, Hartmann P, Fatkenheuer G, Jung N, van Lunzen J, Stellbrink HJ, Gallo RC, and Romerio F, Increased interferon alpha expression in circulating plasmacytoid dendritic cells of HIV-1-infected patients. J Acquir Immune Defic Syndr 48 (2008) 522–30. [DOI] [PubMed] [Google Scholar]
- [78].Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, Ho S, Antonenko S, and Liu YJ, The nature of the principal type 1 interferon-producing cells in human blood. Science 284 (1999) 1835–7. [DOI] [PubMed] [Google Scholar]
- [79].Swiecki M, and Colonna M, Unraveling the functions of plasmacytoid dendritic cells during viral infections, autoimmunity, and tolerance. Immunol Rev 234 (2010) 142–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Theofilopoulos AN, Baccala R, Beutler B, and Kono DH, Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol 23 (2005) 307–36. [DOI] [PubMed] [Google Scholar]
- [81].Campillo-Gimenez L, Laforge M, Fay M, Brussel A, Cumont MC, Monceaux V, Diop O, Levy Y, Hurtrel B, Zaunders J, Corbeil J, Elbim C, and Estaquier J, Nonpathogenesis of simian immunodeficiency virus infection is associated with reduced inflammation and recruitment of plasmacytoid dendritic cells to lymph nodes, not to lack of an interferon type I response, during the acute phase. J Virol 84 (2010) 1838–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Harris LD, Tabb B, Sodora DL, Paiardini M, Klatt NR, Douek DC, Silvestri G, Muller-Trutwin M, Vasile-Pandrea I, Apetrei C, Hirsch V, Lifson J, Brenchley JM, and Estes JD, Downregulation of robust acute type I interferon responses distinguishes nonpathogenic simian immunodeficiency virus (SIV) infection of natural hosts from pathogenic SIV infection of rhesus macaques. J Virol 84 (2010) 7886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Jacquelin B, Petitjean G, Kunkel D, Liovat AS, Jochems SP, Rogers KA, Ploquin MJ, Madec Y, Barre-Sinoussi F, Dereuddre-Bosquet N, Lebon P, Le Grand R, Villinger F, and Muller-Trutwin M, Innate immune responses and rapid control of inflammation in African green monkeys treated or not with interferon-alpha during primary SIVagm infection. PLoS Pathog 10 (2014) e1004241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Moore AC, Bixler SL, Lewis MG, Verthelyi D, and Mattapallil JJ, Mucosal and peripheral Lin- HLA-DR+ CD11c/123− CD13+ CD14− mononuclear cells are preferentially infected during acute simian immunodeficiency virus infection. J Virol 86 (2012) 1069–78. [DOI] [PMC free article] [PubMed] [Google Scholar]


