Abstract
Gene therapy holds promise for the treatment of a range of inherited diseases, such as cystic fibrosis. However, efficient delivery and expression of the therapeutic transgene at levels sufficient to result in phenotypic correction of cystic fibrosis pulmonary disease has proved elusive. There are many reasons for this lack of progress, both macroscopically in terms of airway defence mechanisms and at the molecular level with regard to effective cDNA delivery. This review of approaches to cystic fibrosis gene therapy covers these areas in detail and highlights recent progress in the field. For gene therapy to be effective in patients with cystic fibrosis, the cDNA encoding the cystic fibrosis transmembrane conductance regulator protein must be delivered effectively to the nucleus of the epithelial cells lining the bronchial tree within the lungs. Expression of the transgene must be maintained at adequate levels for the lifetime of the patient, either by repeat dosage of the vector or by targeting airway stem cells. Clinical trials of gene therapy for cystic fibrosis have demonstrated proof of principle, but gene expression has been limited to 30 days at best. Results suggest that viral vectors such as adenovirus and adeno-associated virus are unsuited to repeat dosing, as the immune response reduces the effectiveness of each subsequent dose. Nonviral approaches, such as cationic liposomes, appear more suited to repeat dosing, but have been less effective. Current work regarding non-viral gene delivery is now focused on understanding the mechanisms involved in cell entry, endosomal escape and nuclear import of the transgene. There is now increasing evidence to suggest that additional ligands that facilitate endosomal escape or contain a nuclear localization signal may enhance liposome-mediated gene delivery. Much progress in this area has been informed by advances in our understanding of the mechanisms by which viruses deliver their genomes to the nuclei of host cells.
Keywords: adenovirus, endosomal escape, liposome, molecular conjugate, nuclear import, vector
Abbreviations: AAV, adeno-associated virus; Ad2, etc., adenovirus 2, etc.; aGM1, asialoganglioside 1; CAR, Coxsackie B and adenovirus receptor; CF(TR), cystic fibrosis (transmembrane conductance regulator); CMV, cytomegalovirus; DC-Chol, 3b[N-(N′,N′-dimethylaminoethane)-carbamoyl] cholesterol; DMPE-PEG5000, dimyristoyl phosphatidylethanolamine-N-poly(ethylene glycol)-5000; DOPE, dioleoyl phosphatidylethanolamine; DOTAP, 1,2-bis(oleoyloxy)-3-(trimethylammonio)propane; DOTMA, N-[1-(2,3-dioleyloxy)propyl]-N,N,N,-trimethylammonium chloride; EMPDC, 1,2-dimyristoyl-P-O-ethylphosphatidylcholine; ENaC, epithelial Na+ channel; E1 etc., early region of human adenovirus 1, etc.; HAdV-2, human adenovirus serotype 2; ITR, inverted terminal repeat; LMD, liposome–Mu–DNA; NLS, nuclear localization signal; PEI, polyethyleneimine; SV40, simian virus 40
INTRODUCTION
Cystic fibrosis is the most common lethal hereditary disease among Caucasians, with a prevalence of approx. 1 in 2000 [1]. The condition is characterized by chronic lung inflammation that results in life expectancy being reduced to about 45 years at best [2,3]. The underlying cause for cystic fibrosis remained a mystery until important work by Quinton [4] demonstrated chloride impermeability across cystic fibrosis sweat duct epithelia. At the same time, three groups were independently using genetic linkage studies of families with two or more children affected with cystic fibrosis to determine the location of the cystic fibrosis gene. In November 1985, all three groups published simultaneously that the cystic fibrosis gene was located on the long arm of chromosome 7 [5–7]. Further examination of this region led to the identification of the gene itself and the prediction of the amino acid sequence of the encoded protein [8], which was termed the CFTR (cystic fibrosis transmembrane conductance regulator). An accompanying paper demonstrated the commonest mutation, a whole codon deletion, resulting in the loss of a phenylalanine residue at amino acid position 508 (ΔF508) in the protein [9]. The CFTR amino acid sequence contained 12 domains previously recognized as membrane-spanning sequences [10], and in vitro gene transfer experiments demonstrated restoration of chloride channel function in cystic fibrosis pancreatic cells [11]. Further work using purified CFTR protein in phospholipid vesicles confirmed that the protein functions as a cAMP-dependent chloride channel [12].
How mutations in this cAMP-dependent chloride channel lead to the phenotypic inflammatory changes that are characteristic of cystic fibrosis pulmonary disease remains controversial. However, there is now increasing evidence to support the view that there are two critical consequences of impaired CFTR function, namely a reduction in the volume of airway surface liquid [13], and increased bacterial adherence of organisms such as Pseudomonas aeruginosa to airway epithelial cells [14].
Airway surface liquid occupies the periciliary space on the apical surface of bronchial epithelial cells, and is thought to act as a ‘lubricant’, allowing the overlying mucus to be transported out of the lungs by beating cilia and/or cough [15]. Normal CFTR is partly responsible for maintaining this airway surface liquid volume by means of chloride secretion and down-regulation of the ENaC (epithelial Na+ channel) [16]. In vitro models of polarized human cystic fibrosis bronchial epithelial cells demonstrate an isotonic reduction in the volume of airway surface liquid, and consequently impaired movement of the overlying mucus [13]. The importance of CFTR's regulation of ENaC has been strengthened further by recent data demonstrating that overexpression of ENaC in a mouse model results in many of the key features of cystic fibrosis lung disease (e.g. reduction in airway surface liquid, reduced muco-ciliary clearance and increased mucus plugging of the lungs) without any direct impairment of CFTR function [17].
In addition to this mucus plugging, cystic fibrosis bronchial epithelial cells bind significantly more Ps. aeruginosa to their surface than matched cells rescued with wild-type CFTR [14]. This was found to be due to undersialylation of apical proteins and a consequent increase in aGM1 (asialoganglioside 1) on the apical membrane of cystic fibrosis cells [14]. The authors suggested that this undersialylation was due to a loss of CFTR from the intracellular glycosylation compartments impairing sialyltransferase activity, and that this adherence to aGM1 was a critical factor in the aetiology of chronic Ps. aeruginosa infection in cystic fibrosis patients.
GENE THERAPY IN CYSTIC FIBROSIS
Cystic fibrosis should be an ideal candidate for gene therapy, for four main reasons: (1) it is a single gene defect; (2) it is a recessive condition, with heterozygotes being phenotypically normal (suggesting gene dosage effects are not critical); (3) the main pathology is in the lung, which is accessible for treatment; and (4) it is a progressive disease with a virtually normal phenotype at birth, offering a therapeutic window.
It has been suggested that only 5–10% of normal CFTR function is required to reverse the chloride channel defect [18], although it is not clear whether this has to be achieved in the majority of the airway epithelial cells, or whether a minority of cells expressing much higher levels would suffice. In clinical trials to date, two main vector systems have been harnessed to deliver the CFTR cDNA with appropriate promoter into host cells (for reviews, see Flotte and Laube [19] and Driskell and Engelhardt [20]). First, viral vectors with the CFTR cDNA incorporated into the viral genome exploit the efficiency of viruses to enter host cells and achieve relatively high levels of gene expression. Secondly, cationic liposomes mixed with plasmid DNA encoding CFTR enhance the transport of the DNA into host cells. Although cationic liposomes seem to generate a lower immune response than current viral vector systems, the levels of CFTR expression using this delivery system have been relatively poor.
The ideal vector system would have the following characteristics: (1) an adequate carrying capacity; (2) to be undetectable by the immune system; (3) to be non-inflammatory; (4) to be safe to the patients with pre-existing lung inflammation; (5) to have an efficiency sufficient to correct the cystic fibrosis phenotype; and (6) to have long duration of expression and/or the ability to be safely re-administered.
The key components in this list are the carrying capacity, efficiency and low immunogenicity. The CFTR gene is 6.5 kb [10], although this can be reduced to 4.45 kb if a cDNA is used [21]. The physiological CFTR promoter including all known regulatory sequences is a further 3.9 kb, although the minimal CFTR promoter is only 250 bp [22]. To provide therapeutic benefit in vivo, the delivery of CFTR gene therapy must be extremely efficient to overcome the physical barriers such as thick tenacious mucus and pulmonary surfactant [23], as well as the logistical difficulties resulting from many viral receptors only being expressed on the basolateral membrane of bronchial epithelial cells [24]. Much of the morbidity and mortality seen in cystic fibrosis patients is related to pre-existing pulmonary inflammation [25]; consequently, it is important that any vector system does not cause further inflammation.
VIRAL VECTORS
Adenovirus vectors
Adenoviruses are able to transduce a wide variety of cell types, including non-dividing cells, and contain a double-stranded DNA genome of 30–40 kb [26]. These features, together with a genome that is relatively easy to genetically manipulate, resulted in recombinant adenoviruses being the first viral vectors used in clinical trials [27,28]. First- and second-generation adenovirus vectors are characterized by deletions within the genes encoding early transcripts [29].
The first part of the genome to be expressed, E1A (early region of human adenovirus 1A), encodes two proteins responsible for transcriptional activation of other viral promoters and facilitation of host cell entry into the S-phase of the cell cycle [26,30,31]. The E1B 19 kDa gene product prevents apoptosis induced by E1A [32], and the E1B 55 kDa protein forms a complex with both the E4 Orf6 gene product and the tumour suppressor gene p53, resulting in ubiquitination and degradation of p53, further preventing apoptosis [33]. The E2A region encodes the DNA-binding protein, and E2B encodes DNA polymerase and pre-terminal protein. Vectors deleted in the E2B region therefore show drastically reduced viral replication, and thus a reduction in immune response [29]. Products of the E3 region are involved in evasion of the host cell immune response, but are not necessary for virus production in cultured cells. For example, the E3 19 kDa protein binds MHC class I molecules within the lumen of the endoplasmic reticulum, preventing their trafficking to the cell surface [34]. The E4 region encodes proteins involved in a number of different processes [35]. E4 Orf3 and Orf6 gene products inhibit cellular mRNA export from the nucleus, and facilitate viral mRNA transport to the cytoplasm, and E4 Orf6 inhibits the p53 apoptotic pathway, as described above. E4-deleted adenovirus vectors appear to be particularly susceptible to down-regulation of transgene expression [36]. Late gene products are predominantly viral structural proteins [26].
When generating adenovirus vectors, most of the E1 region is deleted to prevent virus replication and consequent host cell lysis [26]. These replication-deficient adenoviruses require cells that express E1 proteins for successful propagation, such as the human embryonic kidney-derived 293 cell line [37,38]. Figure 1 shows schematically how an expression cassette, containing a promoter, transgene and polyadenylation signal, can be incorporated into replication-deficient adenovirus vectors.
Figure 1. Schematic diagram demonstrating methodology used to construct first generation E1 deletion adenovirus vectors.
E1-expressing cells, such as HEK-293 cells, are co-transfected with genomic adenovirus DNA and a shuttle plasmid containing the expression cassette with flanking sequences derived from sequences immediately upstream and downstream of the E1 region. The genomic adenovirus DNA is modified to confer a selection advantage for recombinant virus. Recombination between the plasmid and genomic DNA results in the generation of an adenovirus vector with an expression cassette replacing the E1 region. ITRs and the packaging signal are represented in purple. Reprinted from Advances in Virus Research, vol. 55, M. M. Hitt and F. L. Graham, “Adenovirus vectors for human gene therapy”, pp. 479–505, © 2000, with permission from Elsevier.
First-generation adenovirus vectors, adapted by deleting the E1 region and the E3 region, as the latter is non-essential for virus growth in cell culture, were the first viral vectors used in clinical trials for cystic fibrosis [27,28]. CFTR cDNA expression was under the control of the adenovirus major late promoter. The first human gene therapy trial for cystic fibrosis was commenced in New York in April 1993 [27]. Four patients had 2×106–2×109 plaque-forming units of the Ad2 (adenovirus 2) CFTR-encoding vector administered in liquid form, initially to the nasal epithelium, then one day later to a lower lobe bronchus via a bronchoscope. AdCFTR DNA and AdCFTR-directed CFTR mRNA were subsequently detected in some respiratory samples, and there was a modest increase in CFTR protein expression for approx. 10 days after administration. Unfortunately, dose-limiting systemic and pulmonary inflammation occurred, particularly at the higher dose, due to innate immunity (not epitope-specific), cell-mediated immunity against adenovirus protein products and humoral immune responses directed primarily against components of the viral capsid (for a review, see Vadolas et al. [23]). Soon afterwards, a group at the University of North Carolina reported their experiences giving an E1-deleted Ad5 vector encoding CFTR under the control of a CMV (cytomegalovirus) promoter nasally to 12 cystic fibrosis patients [39]. At doses from 2×107–2×1010, less than 1% of cells were transfected with no change in chloride conductance, but marked mucosal inflammation occurred, and the authors concluded that ‘local inflammatory responses limited the dose that could be administered to overcome the inefficiency of gene transfer’. A repeat dosing trial by the New York group, using an HAdV-2 (human adenovirus serotype 2) vector with E1a and E3 deleted, a partial deletion of E1b and a CMV promoter, was performed in 14 individuals with cystic fibrosis, using 3-monthly dosing cycles [40]. They demonstrated less local inflammation, but this may have been due to the much lower volume of fluid administered to the lungs (100 μl by bronchoscope spray, rather than 20 ml). However, vector-derived CFTR expression, although 5% of normal at higher vector doses, only persisted for approx. 4–30 days following the first dose. Expression was reduced following the second dose, and was undetectable following the third dose. As E3 gene products are known to suppress the cellular immune response against virus-infected cells [34], second- and third-generation adenovirus vectors were then developed, with retention of the E3 region. A vector with deletions in regions E1 and E4 was found to elicit a markedly reduced helper T cell response in murine and non-human primate lungs [41], and thus a similar vector expressing CFTR (H5.001CBCFTR) was tested in 11 human subjects with cystic fibrosis [42]. This dose-escalation trial from 2.1×109 to 2.1×1011 total viral particles delivered in a total volume of 7 ml via a bronchoscope demonstrated flu-like symptoms for all subjects, and dose-limiting pulmonary inflammation at the highest dose. Gene transfer was detected in only 1% of cells by in situ hybridization against vector-specific sequence 4 days after administration, and was undetectable at 43 days.
This persistent poor efficacy of adenovirus vectors to deliver the CFTR gene to terminally differentiated respiratory epithelium may be partly due to the CAR (Coxsackie B and adenovirus receptor) being predominantly located on the basolateral rather than the luminal surface of human respiratory cells [43]. Non-clinical studies have thus explored the potential of pseudotyping the adenovirus vectors to target other more suitable cell-surface receptors [44,45]. However, the tragic death of Jesse Gelsinger in September 1999, a volunteer in a dose-escalation clinical trial of an adenovirus vector containing the gene for ornithine transcarbamylase, as a result of an overwhelming immune response [46] has severely reduced further clinical research into this vector system. ‘Gutless’ adenovirus vectors, with no viral genes remaining, have been developed and show promise in animal models [47], but although their coding capacity is high and their susceptibility to cell-mediated immunity is reduced, the problem of an immune response to the viral capsid proteins remains.
Adeno-associated virus (AAV) vectors
AAV is a parvovirus that requires a helper virus (usually adenovirus) to replicate. Unlike adenovirus, AAV is not known to cause any illness in humans [20]. In the absence of a helper virus, wild-type AAV can produce a latent infection by stable integration of its genome into a site on human chromosome 19 [48], although, in contrast, recombinant AAV that lacks the replicase gene does not integrate at this specific locus [49]. Various cellular receptors have been identified for AAV, notably heparan sulphate proteoglycan for AAV-2 [50]. The first phase I clinical trial using AAV-2 containing the complete human CFTR coding sequence, with administration of the vector tgAAV-CF into the maxillary sinuses of 10 cystic fibrosis patients, showed no evidence of inflammation. However, although gene transfer was achieved for 1 in 10 respiratory epithelial cells for up to 10 weeks [51–53], no vector-derived RNA could be detected in any samples. A further dose escalation study of this vector with aerosolized delivery of a single dose of 1010–1013 vector particles into the lungs of 12 volunteers with cystic fibrosis also demonstrated safe gene transfer to approx. 1 in 10 epithelial cells at 30 days, but without detectable vector-derived mRNA [54]. Similar results were observed when 104–1012 tgAAV-CF particles were administered to 25 cystic fibrosis subjects via a bronchoscope [55]. This poor expression was attributed to lack of an efficient promoter due to AAV only having a packaging capacity of approx. 4.8 kb. A further concern was the rise in serum-neutralizing antibodies to AAV-2 observed in both studies of pulmonary administration [54,55], which raises concerns about possible reduced effectiveness in repeat dosing. This was assessed in the most recently reported clinical trial, with three doses of 1013 tgAAV-CFTR particles, or placebo, administered at 4-week intervals by aerosol to the lungs of 37 cystic fibrosis subjects [56]. Encouragingly, and for the first time in any trial of gene therapy in cystic fibrosis, clinical outcome measures were significantly improved in the treatment group when compared with placebo, with significant improvements in respiratory function and reduced levels of the pro-inflammatory cytokine interleukin-8. However, the effectiveness was reduced with each subsequent dose, associated with a rise in neutralizing antibodies.
Further work to improve the effectiveness of this promising vector is being undertaken. One strategy is to explore the use of other serotypes of AAV. Initial work demonstrated that serotypes AAV-5, AAV-1 and AAV-6 had increased pulmonary transduction efficiency when compared with AAV-2 [57], possibly due to the relative paucity of receptors for AAV-2 on the apical surface of lung epithelial cells [58]. More than 40 AAV serotypes have now been discovered, and are being tested for effectiveness in vivo [57], and a clinical trial using AAV-6 is planned [59]. The tgAAV-CFTR vector used so far in clinical trials relies on the ITR (inverted terminal repeat) promoter elements within the AAV-2 [60]. It has been shown that insertion of a further 83 bp transcriptional element increases transcription up to 5-fold in vitro [56], although further work with shortened CFTR cDNA sequences and alternative promoters is also under way [61]. Finally, agents that modulate proteasome function can re-route AAV-2 vector from the proteasome to the nucleus, increasing luciferase reporter expression up to 1000-fold [62,63].
Other viral vectors
There are other viral vector systems under development, the most promising being pseudotyped lentiviral vectors and parainfluenza virus type 3. Lentiviral vectors offer the possibility of providing sustained expression of the therapeutic gene, as they integrate their genome into the host DNA [57]. However, as with adenovirus and AAV-2, they are hampered by a lack of suitable receptors on the apical surface of bronchial epithelial cells. Recent work in vitro and in vivo suggests that lentiviral vectors pseudotyped with modified Ebola envelope glycoprotein demonstrate increased uptake and expression when delivered apically [64,65]. The potential problem of random integration into the host genome, which was the cause of T-cell leukaemia in two children treated for severe combined immunodeficiency with retrovirus-mediated gene therapy [66], will have to be solved prior to a clinical trial in cystic fibrosis patients [67].
Parainfluenza virus type 3 has the considerable advantage that it specifically targets ciliated airway epithelial cells from the apical surface, and a vector containing the CFTR gene efficiently corrects abnormalities seen in cystic fibrosis bronchial epithelial cells in tissue culture [68]. Current research with this vector is now directed at generating forms that can be safely delivered to the human airway in vivo.
NON-VIRAL GENE DELIVERY
Non-viral vectors have the potential to avoid some of the critical problems observed with viral vectors, such as the immune response, limited packaging capacity, and random integration (for a review, see Ratko et al. [69]). There are three main non-viral vector systems: cationic liposomes, DNA–polymer conjugates and naked DNA. To date, only cationic liposome-based systems have been tested in clinical trials in cystic fibrosis subjects.
Cationic liposomes
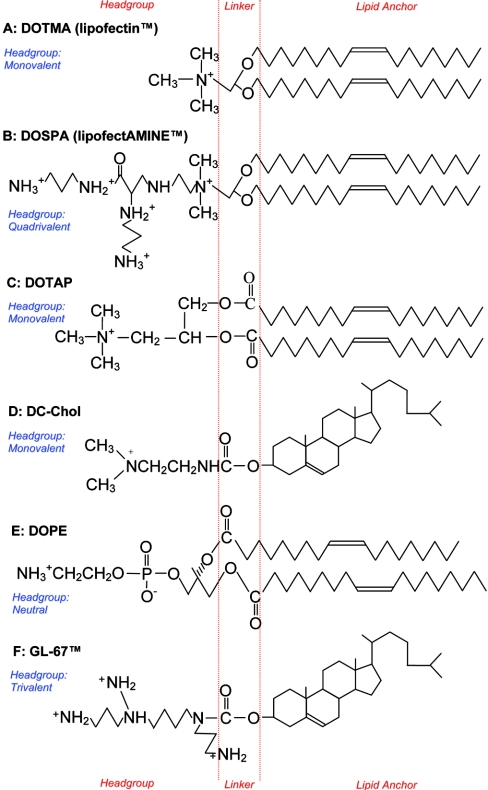
Cationic liposomes form large complexes in which their positively charged side chains interact with DNA and the hydrophobic lipid portion of the liposome enhances fusion with the host cell membrane (for a review, see Singhal and Huang [70]). Sufficient cationic liposome has to be mixed with the negatively charged DNA to ensure a net positive charge, facilitating this interaction with the cell membrane. Freeze–fracture electron microscopy suggests that the structure of cationic liposome–DNA complexes is intricate [71], with the liposome complexes fusing with each other to form a bilayered tube that coats the DNA. The formation of these bilayered tubes occurs slowly, over approx. 30 min following the addition of DNA to the liposomes. In contrast with viral vectors, there is no limitation to the length of DNA that can be incorporated. Most cationic liposomes used for transfection consist of a positively charged lipid mixed with a neutral helper lipid (co-lipid). The co-lipid facilitates the formation of stable lipid bilayers. The first commercially available cationic lipid, Lipofectin®, is a mixture of DOTMA {N-[1-(2,3-dioleyloxy)propyl]-N,N,N,-trimethylammonium chloride} and the colipid DOPE (dioleoyl phosphatidylethanolamine), and was described by Felgner et al. [72] (see Figure 2). DOTMA consists of a positively charged hydrophilic head-group, generated by a quaternary ammonium group, linked by an ether bond to two hydroxyalkyl chains which form the lipid anchor. Further development incorporated a spermine group into the hydrophilic head to increase the positive charge, resulting in DOSPA {2,3-dioleyloxy-N-[2(sperminecarboxamido)ethyl]-N,N-dimethyl-l-propanaminium trifluoroacetate} [73]. A 3:1 mix of this cationic lipid with the co-lipid DOPE is available commercially as Lipofectamine™. This cationic liposome has been extensively used in studies in vitro, but due to the risk of toxicity related to the spermine group it is unsuitable for use in vivo.
Figure 2. Chemical structures of lipids commonly used in the formation of cationic liposomes, highlighting hydrophilic head-groups, linker regions and hydrophobic lipid anchors.
Head-groups vary in degree of positive charge; the co-lipid DOPE has a neutral head-group. Lipid anchors consist of either two hydroxyalkyl chains or a modified cholesterol component (DC-Chol and GL-67™).
Cationic liposomes that have been used successfully in vivo include DOTAP [1,2-bis(oleoyloxy)-3-(trimethylammonio)propane], first synthesized by Leventis and Silvius [74], who suggested that the use of ester bonds to link the cationic head-group to the lipid anchor would reduce toxicity by facilitating degradation in eukaryotic cells. DOTAP is particularly effective when cholesterol is used as a co-lipid [75]. The liposome that has been most frequently used in cystic fibrosis clinical trials has been DC-Chol {3b[N-(N′,N′-dimethylaminoethane)-carbamoyl]cholesterol} mixed with DOPE. This was first produced in 1991 by Gao and Huang [76], who suggested that using cholesterol as a lipid anchor rather than hydroxyalkyl chains would lead to reduced cellular toxicity, because cholesterol stabilizes lipid bilayers. As well as demonstrating a 4-fold-improved tolerability over Lipofectin®, DC-chol/DOPE exhibited enhanced effectiveness and remarkable stability [77,78]. This is due to the carbamoyl linker that, although hydrolysed by esterases, is more stable than labile ester bonds [76]. Lipid GL-67™ has a similar structure to DC-Chol, with cholesterol as the lipid anchor and a carbamoyl linker, but with a triple amine head-group rather than DC-Chol's single amine head. When mixed with DOPE, lipid GL-67™ appeared 1000-fold more effective than naked DNA alone, and 100-fold more effective than DC-Chol/DOPE when delivered to murine airways, although it is not clear whether equivalent doses of DNA were delivered [79]. A systematic assessment of a number of related lipid structures generated important observations regarding the lipid structure required for optimized gene delivery to airway epithelial cells in vitro and in vivo. It was concluded that trivalent cationic head-groups, particularly in a ‘T’ formation as in GL-67™, were more effective than equivalent monovalent, bivalent or quadrivalent lipids. Similarly, carbamate linkers and cholesterol anchors appeared most effective when compared with alternative structures. Unfortunately, GL-67™ appeared to cause considerably greater pulmonary toxicity than DC-Chol.
The final cationic lipid of relevance to cystic fibrosis gene therapy is EDMPC (1,2-dimyristoyl-P-O-ethylphosphatidylcholine) mixed with cholesterol [80]. EDMPC has a lipid anchor consisting of two hydroxyalkyl chains, an ester linker region, and a monovalent head-group [81].
Clinical trials of cationic liposome-mediated gene therapy in cystic fibrosis
There have been eight clinical trials of liposome-mediated gene therapy in cystic fibrosis patients published to date [80,82–88] (results are summarized in Table 1).
Table 1. Summary of clinical trials of liposome-mediated gene therapy in cystic fibrosis subjects to date.
Abbreviation: P. D., potential difference.
| Trial/year/reference | Liposome | No. of patients | Target | Route | Efficacy | Side effects |
|---|---|---|---|---|---|---|
| Caplen et al., 1995 [82] | DC-Chol/DOPE | 15 | Nose | Aerosol | P.D. 20% correction towards normal | None |
| Zabner et al., 1997 [83] | GL-67™/DOPE | 12 | Nose | Direct installation | Partial correction nasal P.D. (equivalent to naked DNA) | None |
| Gill et al., 1997 [84] | DC-Chol/DOPE | 12 | Nose | Direct installation | Functional evidence in 6/8 treated patients | None |
| Porteous et al., 1997 [85] | DOTAP | 16 | Nose | Aerosol | P.D. 20% correction in two patients, no SPQ-positive | None |
| Alton et al., 1999 [86] | GL-67™/DOPE/DMPE-PEG5000 | 16 | Lungs | Nebulized | Lung P.D. 25% correction towards normal | Mild, flu-like symptoms |
| Hyde et al., 2000 [87] | DC-Chol/DOPE | 12 | Nose | Direct installation, repeat dosing | 6/10 patients positive after each dose | None |
| Noone et al., 2000 [80] | EDMPC-Chol | 11 | Nose | Aerosol | None | None |
| Ruiz et al., 2001 [88] | GL-67™/DOPE/DMPE-PEG5000 | 8 | Lungs | Nebulized | Vector-specific mRNA in 3/8 patients | Mild, flu-like symptoms |
The first trial used DC-Chol/DOPE liposomes and 10–300 μg of DNA delivered via aerosol to the nose [82]. The results have to be interpreted with caution, because three of the six control patients, receiving liposome alone, appeared to demonstrate vector DNA or CFTR mRNA with the assays used. However, the trial did demonstrate a 20% correction of nasal potential difference towards normal values in the treated group, without any significant toxicity. The delivery system was time-intensive, with the high-dose patients receiving an aerosol puff every 10 min for 7 h.
A trial performed in 12 cystic fibrosis subjects used a 1:2 molar ratio of GL-67™ and DOPE mixed with 1.25 mg of CFTR plasmid DNA, with direct instillation on to the nasal epithelium via a catheter over 80 min [83]. They demonstrated no toxicity at lipid doses of up to 2 mg, and mild restoration of nasal potential difference towards normal values. Intriguingly, this trial used CFTR plasmid DNA alone as a control in the contralateral nostril, and found that plasmid DNA alone was as effective as plasmid DNA mixed with liposome, in marked contrast with previous data obtained in murine systems [79].
In the same year, Gill et al. [84] reported results of a trial in 12 cystic fibrosis subjects using a 6:4 molar ratio of DC-Chol/DOPE liposomes and 40–400 μg of CFTR plasmid DNA. Direct instillation over a total of 120 min into the nose resulted in transient correction of the chloride transport abnormality in two of the eight treated patients, as measured by nasal potential difference, lasting approx. 15 days. In addition, four further treated patients demonstrated CFTR function in an ex vivo assay. Low and high doses of plasmid DNA demonstrated comparable efficacy, and there was no evidence of significant local or systemic toxicity.
A study performed using DOTAP and 400 μg of CFTR DNA delivered via aerosol spray to the nose demonstrated vector-specific DNA in seven out of eight treated patients, with CFTR RNA detected in two [85]. Two patients demonstrated a 20% correction of nasal potential difference towards normal values, but ex vivo assays of chloride conductance were negative. Once again, there were no safety concerns.
The first trial of liposome-mediated gene delivery to the lungs of cystic fibrosis subjects was then performed [86]. This trial was particularly well designed in terms of outcome measures, introducing lung potential difference measurement using a bronchoscope and a Ps. aeruginosa bacterial adherence assay. Sixteen cystic fibrosis subjects received either GL-67™/DOPE/DMPEPEG5000 [dimyristoyl phosphatidylethanolamine-N-poly(ethylene glycol)-5000 (molar proportions 1:2:0.05)] mixed with 42.2 mg of CFTR plasmid DNA or the lipid alone, via a nebulizer. Seven of the eight patients in the treatment group developed influenzalike symptoms (high fever, headache and muscular ache) approx. six hours after nebulization. These transient symptoms were not seen with lipid alone, and as the plasmid DNA had been purified to remove endotoxins, the authors suggested that the high unmethylated cytosine and guanine content of bacterial DNA might have been responsible. Encouragingly, treated patients demonstrated chloride conductance significantly restored to 25% of normal values, decreased lung inflammatory markers and decreased Ps. aeruginosa adherence.
The only repeat dosing clinical trial of liposome-mediated gene therapy in cystic fibrosis was performed by Hyde et al. [87], using DC-Chol/DOPE and 400 μg of CFTR DNA. Doses were given by direct instillation to the nose, on three occasions, 4 weeks apart. Although there was no significant improvement in nasal potential difference, there was evidence of CFTR gene transfer in six out of ten treated patients after each dose. Encouragingly, there was no evidence of immune response, and no evidence of reduced efficacy with repeat dosing.
In contrast, Noone and colleagues [80] reported a total lack of efficacy following aerosol delivery of up to 4 mg of CFTR plasmid DNA to the nostrils of 11 cystic fibrosis subjects using EDMPC-Cholesterol. Despite earlier promising data regarding this vector system when delivered via aerosol to the lungs of non-human primates [89], in the clinical trial there was no change in nasal potential difference or any vector-specific CFTR mRNA detected. The group used a different methodology for nasal potential difference measurement, but their mRNA detection technique had a robust internal control, suggesting that in their hands EDMPC-Cholesterol is unlikely to be useful as a vector system in humans.
The most recently reported trial involved eight cystic fibrosis subjects receiving pulmonary delivery of GL-67™/DOPE/DMPE-PEG5000 (molar proportions 1:2:0.05) complexed to 7.9–21.1 mg of CFTR plasmid DNA [88]. Transient influenza-like symptoms were again described in four patients, which was not related to the dose received. An in vitro assay comparing liposome complexed with eukaryotic DNA with liposome complexed with the CFTR plasmid DNA suggested that the symptoms were not due to an immune response specifically to bacterial DNA, as suggested previously [86], but may rather be due to a more general response to this particular liposome–DNA complex. Vector-specific mRNA was positive in three patients, and negative in three, as assessed by reverse transcriptase-PCR. Two patients were not sampled due to the influenza-like syndrome.
Thus clinical trials of liposome-mediated gene therapy in cystic fibrosis patients have demonstrated evidence of vector-specific CFTR expression and some functional changes towards normality, but this has been variable and at low levels. Variation in end-point measurement techniques, particularly potential difference assessment and vector-specific mRNA detection, makes comparisons between studies difficult [80,90]. Although there has been no evidence of an immune response or any reduction in effectiveness in a repeat dosing clinical trial [87], current levels of gene transfer efficiency are probably too low to result in clinical benefit [90].
DNA–polymer conjugates
An alternative non-viral vector system uses cationic polymers such as polylysine, PEI (polyethyleneimine) and polyamidoamine. These polymers condense DNA, which is thought to offer protection from degradation and facilitate release from endosomes [91]. PEI, the organic macromolecule with the highest cationic charge density potential, has been extensively tested in animal models and shows promise, transfecting up to 5% of pulmonary cells after intravenous administration [92]. Lactosylated poly(L-lysine) enhanced reporter gene expression by 100-fold in primary human cystic fibrosis airway epithelial cells when compared with naked DNA, and confocal microscopy demonstrated improved trafficking of rhodamine-labelled plasmid DNA into the nucleus via the nuclear pore when complexed with this peptide [93]. Similarly, polyarginine (Arg16) increased gene expression by 7 times more than Lipofectin® in HEK-293T cells in 10% serum in vitro [94].
Other groups have studied receptor-mediated endocytosis as a mechanism for gene delivery, for instance using the integrin receptor-binding ligand RGD linked to the short DNA-binding peptide polylysine [95]. This peptide has shown transfection efficacy comparable with the commercial liposome Lipofectamine™, but as it targets a broad range of integrins this system is relatively non-specific. Furthermore, integrins are predominantly expressed on the basolateral, rather than apical, surface of bronchial epithelial cells [96]. The serpin enzyme complex receptor (sec-R) is expressed on the apical surface of airway epithelial cells, and administration of CFTR plasmid DNA complexed to polylysine linked to the sec-R ligand to the lungs of cystic fibrosis mice resulted in correction of chloride channel activity [97].
Similarly, the integrin α9β1 is mainly restricted to airway epithelial cells, but attempts to use the specific ligand PLAEIDGIEL linked to oligolysine proved 200-fold less effective than Lipofectamine™ [98].
In addition to facilitating cell entry, peptides can also assist in nuclear targeting of plasmid DNA; for example, polylysine can be linked to NLSs (nuclear localization sequences) [99]. Polylysine alone doubled the transfection efficacy of naked DNA, but linked to the NLS from SV40 (simian virus 40) T-antigen there was a further 50% enhancement, and evidence of interaction with nuclear import factors importin α and importin β. Similarly, a polylysine peptide linked to two motifs from adenovirus fibre protein, an NLS and an internalization signal for receptor-mediated endocytosis, demonstrated a 2-fold enhancement of gene delivery when compared with a DOTAP control [100]. A series of deletion mutants demonstrated that all three domains of this transfecting peptide contribute to this enhancement, although it is possible that charge/mass ratio was as important as the particular motifs. Unfortunately, the peptide was only effective in the presence of up to 5% serum, limiting the potential in vivo use to topical or direct instillation.
More recently, it has been demonstrated that the HIV-1 tat protein (GRKKRRQRRRPPQ) increased nuclear delivery of rhodamine-labelled plasmid DNA 30-fold [101]. This appeared to be partly mediated by an interaction with cell-surface proteoglycans. Work with similar synthetic arginine-rich peptides suggested that optimum translocation across the cell membrane occurred with peptides containing eight arginine residues [102]. Spacing the arginine residues with histidine and glycine residues [e.g. CG(RHGH)5RGC compared with CR7C] resulted in a further 50-fold improvement of reporter gene expression in COS-7 cells [103].
To date, there has been one clinical trial of gene therapy using DNA–polymer conjugates in cystic fibrosis subjects. Nasal application of a system using poly(L-lysine) modified on an N-terminal cysteine with PEG complexed with plasmid DNA encoding CFTR demonstrated a modest improvement in nasal potential difference [104,105].
Naked DNA
As many of the serious side effects seen in clinical trials of gene therapy have been related to the vector [27,46], the delivery of naked DNA by injection, gene gun or electroporation has potential safety benefits. Manufacture and storage considerations also favour this system [69]. Clinical trials of intramuscular or endo-arterial injection of plasmid DNA encoding vascular endothelial growth factor for coronary artery disease [106] or peripheral arterial disease [107], and ex vivo electroporation of factor VIII plasmid DNA for haemophilia A [108], have been performed with some positive short-term results. However, despite one report of efficacy in the nose [83], these methods are unlikely to be suited to gene delivery to the lung.
METHODS TO ENHANCE LIPOSOME-MEDIATED GENE THERAPY
Of the non-viral vector systems studied so far, cationic liposomes have shown the most promise to date (for reviews, see Audouy et al. [109] and Smyth-Templeton [110,111]). However, further improvements are required to achieve the efficacy necessary to offer the prospect of a cure for cystic fibrosis. Ongoing work towards this goal is based on a detailed understanding of the barriers facing plasmid DNA delivered by this means.
Overview of liposome–cell interaction
There are two main pathways by which cationic liposomes deliver plasmid DNA to the intracellular compartment (Figure 3). It has been suggested that endocytosis, with the liposome–DNA complex taken up into the early endosome (Figure 3A), is the predominant pathway for DOTAP [74]. Mixing of plasmid DNA with DOTAP inhibited the fusion of the liposome with artificial lipid membranes, yet the same mixture was effective in transfecting cultured mammalian cells. Furthermore, inhibitors of endocytosis prevented uptake of DC-Chol and DOTAP liposomes, suggesting that both DC-Chol and DOTAP enter mainly by this route [112]. In contrast, fusion of the cationic liposome with the cell membrane (Figure 3B) seemed to be the main cell-entry mechanism in the case of Lipofectin® [72], since fluorescently labelled DOPE was widely dispersed within the cell membranes early following transfection, rather than the punctate intracellular localization seen soon after endocytosis [112].
Figure 3. Cell-entry mechanisms for cationic liposome–plasmid DNA complexes.
The two major pathways are by endocytosis (A) and direct fusion with the cell membrane (B). The endocytic pathway results in the formation of endosomes, which can lead to eventual destruction of plasmid DNA before release into the cytoplasm. Reprinted from Bioscience Reports, vol. 22(2), © 2002, “Cationic liposome-mediated gene delivery in vivo” by N. Smyth Templeton, Fig. 3, with kind permission of both Springer Sciences and Business Media, and the author, Nancy Smyth Templeton.
Different liposome formulations and preparation techniques are thought to alter which of these two pathways predominate. It is suggested that fusion is the most advantageous cell-entry mechanism, since this results in the majority of the plasmid DNA avoiding entry to the endosome and consequent potential destruction by the lysosomal system [110]. Extrusion of cationic liposomes through 0.1 μm filters, resulting in conversion of large multi-lamellar vesicles into small bilamellar invaginated structures, has been shown to double the effectiveness of DOTAP–cholesterol [75]. The authors suggested that the main reason for this enhancement was that the overall surface area of the liposome suspension was increased by extrusion, resulting in more efficient coating of the plasmid DNA. They have further suggested that extruded DOTAP–cholesterol delivers plasmid DNA to the intracellular compartment predominantly by the fusion pathway [110], although the supportive data has not been published.
Various adjuvants have been mixed with the liposome–DNA complex with the aim of facilitating cell entry. A promising approach is to combine the benefits of liposomes with cationic or receptor-ligand proteins. Gao and Huang [113] showed that polylysine enhanced the efficacy of Lipofectin®, Lipofectamine™ and Dc-Chol/DOPE liposome-mediated gene therapy by 10–30 times in vitro, and suggested that, since polylysine condensed the liposomes by approx. 20-fold, endocytosis might be enhanced. Liposome–DNA complexes of approx. 200 nm diameter are thought to be the largest that can be endocytosed efficiently via the clathrin-coated pathway [78]. RGD-polylysine enhanced the transfection efficacy of Lipofectin® by 4–70-fold in vitro, depending on cell type [114], and a series of peptide mutants suggested that the RGD motif, rather than the polylysine domain, was chiefly responsible for this effect. Similarly, the integrin α9β1 ligand PLAEIDGIEL linked to oligolysine, although ineffective as a gene-delivery agent on its own, enhanced Lipofectamine™-mediated transfection 10-fold in vitro [98], and Lipofectin®-mediated transfection 4-fold in vivo [115]. However, this appears to be mainly by an α9β1-independent mechanism, as it is also occurs in cells not expressing α9β1.
ENDOSOMAL ESCAPE
For cationic liposomes that enter the cell via the endocytic pathway, a mechanism of endosomal escape is vital to prevent eventual destruction of the plasmid DNA in the lysosome. For DC-Chol/DOPE liposomes, the neutral co-lipid DOPE appears to be critically important in this regard. Experiments using the similar co-lipid DOPC and chloroquine to prevent endosomal maturation demonstrated that DOPE appeared to destabilize the endosome membrane, triggering release into the cytosol [116]. The cationic polymer PEI is believed to efficiently buffer endosomes, leading to chloride influx, osmotic swelling and endosome rupture [91], and plasmid DNA delivery to the nucleus is thus enhanced. A co-polymer of histidine and lysine appears to be 3–50-fold more effective than poly(L-lysine) in enhancing DOTAP-mediated gene delivery, possibly due to the histidine residues buffering the endosome [117]. Amphipathic peptides can also be designed to facilitate endosomal escape, such as a peptide (GLFEALLELLESLWELLLEA) with hydrophobic and hydrophilic domains at each extremity of an α-helical structure stable at pH 7.4 [118]. Under low-pH conditions, as found in early endosomes, there was a loss of the α-helix, resulting in membrane destabilization, endosome rupture and a 10-fold increase in reporter gene expression.
Nuclear import
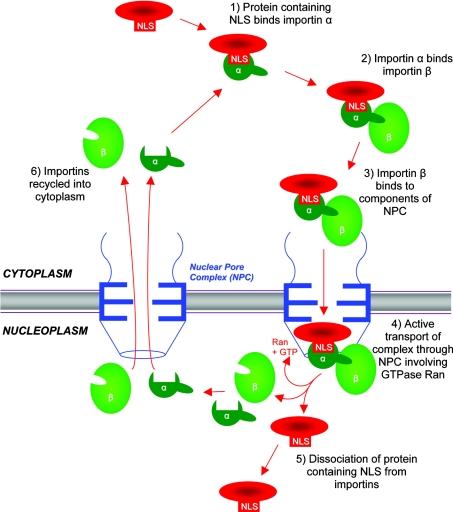
Plasmid DNA must be delivered to the nucleus for transcription to occur. Entry to the nucleus in non-dividing cells is via the nuclear pore complex [119]. Small molecules up to 10 nm, or 50–60 kDa in size, can pass through by passive diffusion, but larger molecules require a peptide NLS for active transport through the pore [120]. Such NLSs, typically comprising clusters of four or more basic amino acids flanked by α-helix breakers proline or glycine, principally interact with importin α, which subsequently interacts with importin β, resulting in energy-dependent uptake of the complex through the nuclear pore (Figure 4) [121]. When supercoiled, plasmid DNA is approx. 25 nm in diameter, suggesting such active transport is likely to be required [120]. Indeed, Texas Red-labelled linear double-stranded DNA of up to 310 bp diffused passively across the nuclear pore complex, but larger fragments required active transport facilitated by binding to peptide containing the SV40 large T antigen NLS (PKKKRKVED) [122]. The transport of isolated DNA from the cytoplasm to the nucleus is extremely inefficient: less than 0.1% of naked cDNA copies were delivered to the nucleus when micro-injected into the cytoplasm of COS-7 cells [123]. Furthermore, co-injection of cationic lipids into the cytoplasm did not enhance this process [123,124], and injection of plasmid DNA, alone or in combination with cationic lipids, directly into the nucleus suggested that cationic lipids inhibit expression of the transgene. This would suggest that once plasmid DNA is delivered to the cytoplasm, cationic lipids have no role in ongoing transport into the nucleus and must dissociate from the DNA prior to nuclear entry. This critical stage of plasmid DNA transport from the cytoplasm to the nucleus is generally recognized as a major limiting step in the efficiency of gene transfer using non-viral vectors [90,119,123,124]. As cationic lipids are unlikely to contribute to this step, various alternative adjuvant molecules have been proposed as facilitators of transport into the nucleus.
Figure 4. Schematic representation of nuclear import process involving importin α and importin β.
Reprinted from Advanced Drug Delivery Reviews, vol. 34, C. W. Pouton, “Nuclear import of polypeptides, polynucleotides and supramolecular complexes”, pp. 51–64, © 1998, with permission from Elsevier.
PEI and polylysine enhance the delivery of plasmid DNA from the cytoplasm to the nucleus approx. 10-fold [123]. PEI appears to facilitate nuclear transport due to compaction, rather than by overall ionic charge of the complex [124], although the exact mechanism remains unclear. Protection of plasmid DNA from degradation by cytoplasmic nucleases is suggested as one important factor [113,119]. A more specific system for facilitating nuclear translocation of liposome-delivered plasmid DNA involves the use of NLS-containing peptides. Fritz et al. [125] used the NLS of the SV40 large T antigen (PKKKRKVEDK) fused to a 94-amino-acid sequence from the C-terminus of human histone H1. When mixed with plasmid DNA and Lipofectin®, expression of the transgene was increased by between 3 and 20 times when compared with DNA and Lipofectin® alone. Surprisingly, however, their micro-injection data demonstrate that transport from the cytoplasm to the nucleus was not enhanced, and that the overall increase in effectiveness was due to improved internalization into cells. The same group subsequently assessed the benefits of covalently linking the SV40 large T antigen NLS to plasmid DNA, but demonstrated that, to increase nuclear accumulation of the plasmid, 100 NLS peptides per 1 kb of DNA were required [126]. This level of covalent modification of the plasmid completely abolished reporter gene expression. In contrast, Zanta et al. [127], using the cationic lipid Transfectam, demonstrated that a single SV40 large T antigen-derived NLS fused to a 3.3 kb DNA fragment encoding luciferase under the control of a CMV promoter, with hairpin cap termini, led to a 10–1000-fold enhancement of luciferase expression when compared with an identical DNA fragment without an NLS. This did seem to be related to the functional interaction of the NLS with nuclear import proteins, because a single mutation of Lys3 to threonine abolished the effect. Importantly, although both groups had used the same NLS in HeLa cells, the latter group had the NLS linked C-terminally to the DNA, rather than N-terminally. The oligonucleotide, although containing a similar quantity of DNA as a plasmid, had an estimated diameter of 3 nm, which may have eased transport once the single NLS had been recognized and imported. Zanta et al. [127] suggested, when comparing their studies with others, that multiple NLSs associated with the full length of a plasmid may hamper import, since it may be recognized by importins at more than one nuclear pore. The SV40 large T antigen NLS linked to a short polylysine tail enhanced DC-Chol/DOPE-mediated transfection 8-fold in a human cystic fibrosis epithelial cell line in vitro, although the exact mechanism was not determined [128]. Schwartz et al. [129] demonstrated that the SV40 large T antigen NLS fused via its C-terminus to sequences of 9–27 amino acids in length from human histone H1 did not offer any additional benefit to liposome-mediated gene delivery when compared with the same peptides without the NLS. Despite this, the histone-derived peptides, with or without the NLS, did enhance liposome-mediated gene delivery up to 50-fold compared with liposome and plasmid DNA alone. This may be due to either non-specific DNA condensation and nuclease protection properties of the peptides together with dissociation from the DNA prior to arrival at the nuclear pore, or alternatively the peptides may remain associated with the DNA but with the SV40 NLS masked. Alternatively, the histone-derived sequence (KTPKKAKKP) could itself also be acting as an NLS.
Transgene expression
The original study describing GL-67™/DOPE delivery of plasmid DNA to murine airways [79] demonstrated impressive peak reporter gene expression at day 2 following instillation, but this had diminished to 20% by day 7, and only 5% by day 21. This was not due to an immune response, because the same pattern was seen in mice lacking humoral or cell-mediated immunity. This raised the concern that the CMV promoter used might be undergoing down-regulation. The same fall in reporter gene expression was seen with the CMV promoter in human cystic fibrosis bronchial epithelial cells in vitro [130]. This group then demonstrated that pulmonary expression in mice was sustained for 3 months when an identical plasmid was used containing CMV promoter enhancer sequences linked to a human ubiquitin promoter [130]. Similarly, the human elongation factor 1α promoter and ubiquitin C promoters resulted in persistence of luciferase expression far in excess of that seen with CMV, rous sarcoma virus and SV40 promoters following topical instillation into the murine lung [131]. The ongoing research into promoters suitable to drive sustained and qualitatively physiological levels of therapeutic transgene is of great importance to the potential success of gene therapy.
Adenovirus core proteins
In order for adenovirus to replicate, it must deliver its DNA genome to the nucleus (for a review, see Shenk [31]). Understanding the role that adenovirus core proteins play in this delivery process may facilitate the rational design of improved gene therapy adjuvant proteins.
Enclosed by 12 structural proteins, the linear double-stranded DNA adenovirus genome of approx. 36 kb is non-covalently bound to the viral core proteins Mu, V and VII [132]. Protein VII and Mu are tightly associated with the viral DNA [132–134], whereas protein V, found only in mastadenoviruses [135], may form a link between the viral DNA–core protein complex and the viral capsid [134]. These core proteins all contain highly basic arginine and/or lysine-rich domains [136,137]. Proteins V, preVII and preMu are encoded via late transcription unit L2 [138].
HAdV-2 protein V is 369 amino acids in length, and contains at least three highly basic sequences, rich in arginine and lysine, that can independently target the nucleolus [136]. Previous studies demonstrated that protein V is associated with the nucleolus during the late phase of infection and, in the nucleus, V is excluded from the viral DNA-binding, protein-rich centres where single-stranded DNA accumulates [134]. Protein V may disrupt nucleolar function by affecting the subcellular localization of the nucleolar antigens, nucleolin and B23 [136]. Why adenovirus might disrupt nucleolar function is unknown, but recent data suggest that B23, for example, may have importance in initiating adenovirus DNA replication [139].
HAdV-2 protein VII contains 174 amino acids, and is formed from its precursor by adenovirus-encoded protease-mediated cleavage of a 24-amino-acid N-terminal segment [137]. The mature protein has four highly basic domains containing arginine- and lysine-rich sequences, separated by predicted α-helices. Protein VII has homologues in every adenovirus studied so far, isolated either from mammals or birds or from lower vertebrates [135,140–142], and has significant functional and sequence similarity with histone H3 [143].
Mu protein (pX) is also highly conserved across all four recognized adenovirus genera [135]. In HAdV-2, Mu is made as a 79-amino-acid precursor, undergoing both N- and C-terminal cleavage to form Mu [144]. Of Mu's 19 amino acids, nine are arginine, possibly explaining the observation that Mu protein precipitates DNA in vitro [144]. The interaction of proteins V, VII and Mu with adenovirus DNA appears to be charge-based between these basic regions and the phosphate backbone of the DNA [145], resulting in the DNA being considerably condensed as a result of superfolding [146,147]. Protein VII associates more efficiently with double-stranded DNA than single-stranded DNA [147]. Binding of both proteins VII and Mu to DNA is not DNA-sequence-specific [148].
Adenovirus entry into host cells
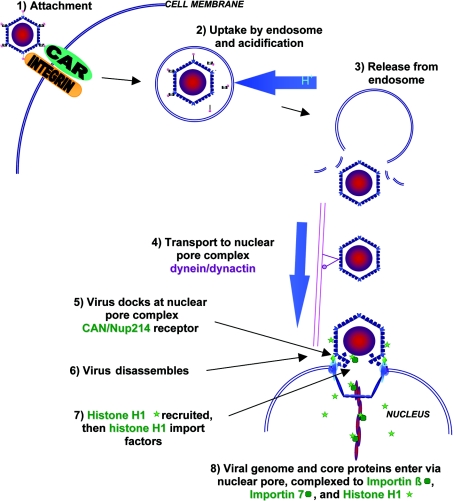
The mechanisms by which Ad2 and Ad5 enter host cells and transport their genome to the nucleus have become increasingly well understood over the last 10 years, particularly since the discovery of the CAR protein in 1997 [149]. An overview is shown in Figure 5 (for reviews, see Shenk [31] and Meier and Greber [150]). CAR functions as the cellular receptor for most human adenoviruses, other than subgroup B [151]. Adenovirus binds to CAR via the C-terminal knob of the fibre protein [152], but for internalization to occur a second interaction between the RGD motif on the penton base of the virus particle and cell-surface αv integrins is necessary [153]. This triggers clathrin-mediated endocytosis of the adenovirus particle [154], and uptake into the acidic early endosome. The virus particle then escapes from the endosome, with approx. 90% of the endocytosed virus being released into the cytosol within 5 min [155]. This critical step is poorly understood, but is thought to depend on viral protease-mediated modification of capsid proteins, as viruses deficient in protease fail to escape from the endosome [156]. Penton base and αv integrins are also thought to activate endosomal release [155]. Once in the cytoplasm, virus particles are transported to the nucleus by the dynein/dynactin motor complex along microtubules [157], and reach the nuclear pore complex approx. 40 min after cell entry [155]. Attachment of the virus particle to the nuclear pore complex receptor CAN/Nup214, and subsequent recruitment of nuclear histone H1, are then both required for virus disassembly and nuclear import of viral DNA [158]. The import of histone H1 by the nuclear import factors importin β and importin 7 has been suggested as the mechanism by which the viral genome is then co-transported into the nucleus via the nuclear pore complex [150,158]. Adenovirus DNA enters the nucleus together with core protein VII and the terminal protein [159], but whether these proteins perform any active role in nuclear entry has not yet been determined. Protein VII inhibits adenovirus DNA synthesis [160] and transcription in vitro [161], and thus is then likely to dissociate from DNA following entry of the complex into the nucleus to allow these processes to occur. It is not known whether Mu protein remains part of this complex during nuclear entry, as antibodies raised against Mu protein cross react with protein VII, possibly due to an homologous arginine-rich sequence present in both proteins [162].
Figure 5. Adenovirus 2 entry into host cells during infection.
Following binding to CAR and αv-integrin, clathrin-mediated endocytosis occurs. The virus particle is taken up into the acidic endosome, but escapes to the cytosol. Transport to the nuclear pore complex along the microtubules is dynein/dynactin-dependent. The virus then docks to CAN/Nup214 on the nuclear pore complex, disassembles and recruits histone H1. The adenovirus genome and core proteins are then imported with the H1 import factors importin β and importin 7. Reproduced from the Journal of Gene Medicine, “Adenovirus endocytosis”, O. Meier and U. F. Greber, pp. 451–462, 2003, with permission. © John Wiley and Sons Limited.
Adenovirus core proteins and gene therapy
Whether proteins VII and Mu have any role beyond that of DNA binding proteins remains unclear, and their potential as gene therapy adjuvants remains largely unexplored. Recent work has demonstrated that protein VII and Mu contain nuclear and/or nucleolar targeting sequences [163,164], and protein VII co-localizes with host cell chromosomes when expressed in cells undergoing mitosis [163]. In 1987, Wienhues et al. [165] reported that mature protein VII enhanced the uptake and expression of naked DNA 4-fold when used alone as a transfection agent for human HeLa and hamster BHK21 cells, suggesting that it may be actively involved in the transport of DNA to the nucleus. Only one other group has published data regarding the ability of adenovirus core proteins to enhance liposome-mediated gene delivery [166–169].
The first of these studies (Murray et al. [166]) demonstrated that Mu protein enhanced transfection efficiency up to 11-fold when used as an adjunct to DC-Chol/DOPE liposome-mediated gene therapy on differentiated hybrid rat×mouse ND7 neuronal cells in vitro. This enhancement was attributed to Mu's ability to bind DNA tightly as a result of its strong positive charge. The DNA binding protein Vp1 from polyomavirus bound DNA less tightly in gel-retardation assays, and proved disappointing as an adjunct to liposome-mediated gene delivery, despite containing a classical monopartite NLS (APKRKSGVSK).
This liposome–Mu–DNA (LMD) transfection complex was characterized further by Tagawa et al. [167]. Buffer conditions and ratios of DC-Chol/DOPE liposomes, peptide and plasmid DNA proved critical to achieve maximum enhancement of DNA expression. These particles appeared stable and amenable to long-term storage, in contrast with similar complexes formed using poly(L-lysine) or protamine instead of Mu. In vitro data using ND7, COS7 and Panc-1 cells suggested that LMD complexes were associated with 10–35-times-greater reporter gene expression under serum-free or 10% serum conditions; however, these data are difficult to interpret because the liposome–DNA complex controls contained different amounts of liposome than within the LMD complexes. Confocal data using cyanine dye-labelled plasmid DNA within the LMD complexes on cells arrested in S-phase by the DNA polymerase inhibitor aphidicolin demonstrated DNA was effectively transported to the cytoplasm within 15 min. However, the DNA then accumulated at the nuclear pore complex, suggesting that the addition of Mu to the liposome–DNA complex may aid endocytosis and endosomal escape, but not nuclear import. Finally, this study [167] presented in vivo data demonstrating a 6-fold-greater chloramphenicol acetyltransferase activity in mouse lungs with LMD compared with the current in vivo ‘gold standard’ GL-67/DOPE/DMPE-PEG5000 following topical administration.
The intracellular trafficking of plasmid DNA delivered by LMD complexes was investigated further by Keller et al. [168]. They compared LMD to a similar complex containing an SV40-derived NLS peptide with arginine-flanking residues (RRRPKKKRKVSRRR). Mu peptide was labelled at its N-terminus by tetramethylrhodamine, NLS peptide was N-terminally labelled with green fluorescent 5-carboxyfluorescein (FAM), fluorescently labelled lipid FAM-Lp-24 was incorporated at a 3% molar ratio into the CD-Chol/DOPE liposomes, and plasmid DNA was labelled with cyanine dye. Although the potential effects of the labelling process on the dynamics of complex formation have to be considered, the data suggest that, when complexes are transfected into cells, both Mu and the NLS peptide are predominantly located within the nucleus after 15 min, but both liposome and DNA are predominantly in the cytoplasm. By 45 min following transfection, the labelled DNA is observed in the nucleus, but as found by Tagawa et al. [167] above, aphidicolin treatment appeared to result in the plasmid DNA being arrested at the nuclear pore complex. Taken as a whole, this study suggests that the major barrier to effectiveness of LMD complex-mediated transfection is the nuclear pore, and although Mu has nuclear-targeting properties, it does not appear to efficiently augment the transport of plasmid DNA through the nuclear pore. This may be due to Mu disassociating from the plasmid DNA too readily in the cytoplasm of the cell, although this may be partly, or even wholly, due to the labelling methodology employed.
The fourth paper from this group [169] contrasts slightly with the previous three. LMD complexes were compared with similar complexes containing a 23-amino-acid nuclear/nucleolar localization signal from adenovirus core protein V (RPRRRATTRRRTTTGTRRRRRRR). Importantly, the positive control liposome–DNA had exactly the same ratio of DC-Chol/DOPE to plasmid DNA as in the LMD and protein V-containing complexes. The protein V sequence bound to plasmid DNA less strongly than Mu protein, on the basis of ethidium bromide exclusion assays, but formed complexes with liposome and DNA in a similar manner to LMD. Assays of relative transfection efficacy were performed in vitro using HeLa cells and a β-galactosidase reporter plasmid. Under serum-free conditions, LMD showed no benefit over liposome–DNA complex, in contrast with the previous studies [166–168], although the complex containing the sequence from protein V did appear to demonstrate a 50% improvement over liposome–DNA. The authors concluded that Mu and the sequence from protein V had great potential as gene delivery adjuvant proteins, but require further characterization, and possibly modification, to enhance nuclear targeting of the plasmid DNA.
Conclusions
Clinical trials of gene therapy for cystic fibrosis lung disease have demonstrated proof of principle, but so far levels of gene expression have been considered too low to offer therapeutic benefit [24,90]. The augmentation of gene delivery to the nucleus is dependent on the efficiency of a number of key processes, such as cell entry, endosomal escape, trafficking to the nuclear pore and nuclear import. Various approaches have been used to address individual components of this system, but to date examination of the effect of gene delivery adjuvants on successive steps has been limited. The airway epithelium in vivo is mainly composed of non-dividing cells with intact nuclear membranes [90]. Thus, although ligands such as RGD, to enhance cell entry, may offer modest improvements in gene expression, it is likely that adjuvants that enhance nuclear entry through the nuclear pore will have considerably greater impact when used in liposome-mediated gene delivery. For this reason, data from in vitro studies using subconfluent cells, a proportion of which will be in mitosis, are unlikely to predict which candidate adjuvants are likely to be most effective in vivo. The methods that viruses use to deliver genomic material to the nucleus of host cells offer useful insights into potential routes that might be exploited by further vector development.
References
- 1.Bobadilla J. L., Macek M., Fine J. P., Farrell P. M. Cystic fibrosis: a worldwide analysis of CFTR mutations — correlation with incidence data and application to screening. Hum. Mutat. 2002;19:575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]
- 2.FitzSimmons S. C. The changing epidemiology of cystic fibrosis. J. Pediatr. 1993;122:1–9. doi: 10.1016/s0022-3476(05)83478-x. [DOI] [PubMed] [Google Scholar]
- 3.Frederiksen B., Lanng S., Koch C., Hoiby N. Improved survival in the Danish centre treated cystic fibrosis patients: results of aggressive treatment. Pediatr. Pulmonol. 1996;21:153–158. doi: 10.1002/(SICI)1099-0496(199603)21:3<153::AID-PPUL1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 4.Quinton P. M. Chloride impermeability in cystic fibrosis. Nature (London) 1983;301:421–422. doi: 10.1038/301421a0. [DOI] [PubMed] [Google Scholar]
- 5.Knowlton R. G., Cohen-Haguenauer O., Van Cong N., Frezal J., Brown V. A., Barker D., Braman J. C., Schumm J. W., Tsui L., Buchwald M., Donis-Keller H. A polymorphic DNA marker linked to cystic fibrosis is located on chromosome 7. Nature (London) 1985;318:380–382. doi: 10.1038/318380a0. [DOI] [PubMed] [Google Scholar]
- 6.White R., Woodward S., Leppert M., O'Connell P., Hoff M., Herbst J., Lalouel J., Dean M., Van de Woude G. A closely linked genetic marker for cystic fibrosis. Nature (London) 1985;318:382–384. doi: 10.1038/318382a0. [DOI] [PubMed] [Google Scholar]
- 7.Wainwright B. J., Scambler P. J., Schmodtke J., Watson E. A., Law H., Farrall M., Cooke H. J., Eiberg H., Williamson R. Localization of cystic fibrosis locus to human chromosome 7cen-q22. Nature (London) 1985;318:384–385. doi: 10.1038/318384a0. [DOI] [PubMed] [Google Scholar]
- 8.Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J., et al. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 9.Kerem B., Rommens J. M., Buchanan J. A., Markiewicz D., Cox T. K., Chakravarti A., Buckwald M., Tsui L. Identification of the cystic fibrosis gene: Genetic analysis. Science. 1989;245:1073–1079. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 10.Riordan J. R. CFTR function. In: Dodge J. A., Brock D. J. H., Widdicombe J. H., editors. Cystic Fibrosis: Current Topics. London: John Wiley and Sons Ltd; 1993. pp. 157–173. [Google Scholar]
- 11.Drumm M. L., Pope H. A., Cliff W. H., Rommens J. M., Marvin S. A., Tsui L., Collins F. S., Frizzell R. A., Wilson J. M. Correction of the cystic fibrosis defect in vitro by retrovirus-mediated gene transfer. Cell. 1990;62:1227–1233. doi: 10.1016/0092-8674(90)90398-x. [DOI] [PubMed] [Google Scholar]
- 12.Bear C. E., Li C. H., Kartner N., Bridges R. J., Jensen T. J., Ramjeesingh M. Purification and functional reconstitution of the cystic fibrosis transmembrane conductance regulator (CFTR) Cell. 1992;68:809–818. doi: 10.1016/0092-8674(92)90155-6. [DOI] [PubMed] [Google Scholar]
- 13.Matsui H., Grubb B. R., Tarran R., Randell S. H., Gatzy J. T., Davis W., Boucher R. C. Evidence for periciliary liquid layer depletion, not abnormal ion composition, in the pathogenesis of cystic fibrosis airways disease. Cell. 1998;95:1005–1015. doi: 10.1016/s0092-8674(00)81724-9. [DOI] [PubMed] [Google Scholar]
- 14.Imundo L., Barasch J., Prince A., Al-Awqati Q. Cystic fibrosis epithelial cells have a receptor for pathogenic bacteria on their apical surface. Proc. Natl. Acad. Sci. U.S.A. 1995;92:3019–3023. doi: 10.1073/pnas.92.7.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zahm J. M., Pierrot D., Vaquez-Girod S., Duvivier C., King M., Puchelle E. The role of mucus sol phase in clearance by simulated cough. Biorheology. 1989;26:747–752. doi: 10.3233/bir-1989-26407. [DOI] [PubMed] [Google Scholar]
- 16.Mall M., Hipper A., Greger R., Kunzelmann K. Wild type but not ΔF508 CFTR inhibits Na+ conductance when coexpressed in Xenopus oocytes. FEBS Lett. 1996;381:47–52. doi: 10.1016/0014-5793(96)00079-8. [DOI] [PubMed] [Google Scholar]
- 17.Mall M. Overexpression of ENaC in mouse airways: a novel animal model for chronic bronchitis and CF lung disease. Pediatr. Pulmonol. Suppl. 2003;25:121. [Google Scholar]
- 18.Johnson L. G., Boyles S. E., Wilson J., Boucher R. C. Normalisation of raised sodium absorption and raised calcium-mediated chloride secretion by adenovirus-mediated expression of cystic fibrosis transmembrane conductance regulator in primary human cystic fibrosis airway epithelial cells. J. Clin. Invest. 1995;95:1377–1382. doi: 10.1172/JCI117789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flotte T. R., Laube B. L. Gene therapy in cystic fibrosis. Chest. 2001;120:124S–131S. doi: 10.1378/chest.120.3_suppl.124s. [DOI] [PubMed] [Google Scholar]
- 20.Driskell R. A., Engelhardt J. F. Current status of gene therapy for inherited lung diseases. Annu. Rev. Physiol. 2003;65:585–612. doi: 10.1146/annurev.physiol.65.092101.142426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L., Wang D., Fischer H., Fan P., Widdicombe J. H., Wai Kan Y., Dong J. Efficient expression of CFTR function with adeno-associated virus vectors that carry shortened CFTR genes. Proc. Natl. Acad. Sci. U.S.A. 1998;95:10158–10163. doi: 10.1073/pnas.95.17.10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romey M., Guittard C., Carles S., Demaille J., Claustres M., Ramsey M. First putative sequence alterations in the minimal CFTR promoter region. J. Med. Genet. 1999;36:263–264. [PMC free article] [PubMed] [Google Scholar]
- 23.Vadolas J., Williamson R., Ioannou P. A. Gene therapy for inherited lung disorders: An insight into pulmonary defence. Pulmon. Pharm. Ther. 2002;15:61–72. doi: 10.1006/pupt.2001.0316. [DOI] [PubMed] [Google Scholar]
- 24.Stern M., Geddes D. M., Alton E. W. F. W. Taking stock of gene therapy for cystic fibrosis. Respir. Res. 2000;1:78–81. doi: 10.1186/rr16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennan A. L., Geddes D. M. Cystic fibrosis. Curr. Opin. Infect. Dis. 2002;15:175–182. doi: 10.1097/00001432-200204000-00013. [DOI] [PubMed] [Google Scholar]
- 26.Hitt M. M., Graham F. L. Adenovirus vectors for human gene therapy. Adv. Virus Res. 2000;55:479–505. doi: 10.1016/s0065-3527(00)55014-3. [DOI] [PubMed] [Google Scholar]
- 27.Crystal R. G., McElvaney N. G., Rosenfeld M. A., Chu C. S., Mastrangeli A., Hay J. G., Brody S. L., Jaffe H. A., Eissa N. T., Danel C. Administration of an adenovirus containing the human CFTR cDNA to the respiratory tract of individuals with cystic fibrosis. Nat. Genet. 1994;8:42–51. doi: 10.1038/ng0994-42. [DOI] [PubMed] [Google Scholar]
- 28.Crystal R. G. Gene therapy for cystic fibrosis: lessons learned and hurdles to success; Selected proceedings of the Ninth Annual American Cystic Fibrosis Conference, Bethesda, MD, U.S.A., Cystic Fibrosis Foundation; 1995. pp. 11–15. [Google Scholar]
- 29.Imperiale M. J., Kochanek S. Adenovirus vectors: biology, design and production. Curr. Top. Microbiol. Immunol. 2003;273:335–357. doi: 10.1007/978-3-662-05599-1_10. [DOI] [PubMed] [Google Scholar]
- 30.Pfeifer A., Verma I. M. Virus vectors and their applications. In: Fields B. N., Knipe D. M., Howley P. M., editors. Fields – Virology, vol. 2. Philadelphia: Lippincott-Raven; 2001. pp. 469–491. [Google Scholar]
- 31.Shenk T. Adenoviridae: the viruses and their replication. In: Fields B. N., Knipe D. M., Howley P. M., editors. Fields – Virology, vol. 2. Philadelphia: Lippincott-Raven; 2001. pp. 2265–2299. [Google Scholar]
- 32.Chinnadurai G. Control of apoptosis by human adenovirus genes. Semin. Virol. 1998;8:399–408. [Google Scholar]
- 33.Querido E., Blanchette P., Yan Q., Kamura T., Morrison M., Boivin D., Kaelin W. G., Conaway R. C., Conaway J. W., Branton P. E. Degradation of p53 by adenovirus E4orf6 and E1B 55 K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 2001;15:3104–3117. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wold W. S. M., Doronin K., Toth K., Kuppuswamy M., Lichtenstein D. L., Tollefson A. E. Immune responses to adenoviruses: viral evasion mechanisms and their implications for the clinic. Curr. Opin. Immunol. 1999;11:380–386. doi: 10.1016/S0952-7915(99)80064-8. [DOI] [PubMed] [Google Scholar]
- 35.Leppard K. N. E4 gene function in adenovirus, adenovirus vector and adeno-associated virus infections. J. Gen. Virol. 1997;78:2131–2138. doi: 10.1099/0022-1317-78-9-2131. [DOI] [PubMed] [Google Scholar]
- 36.Armentano D., Zabner J., Sacks C., Sookdeo C. C., Smith M. P., St George J. A., Wadsworth S. C., Smith A. E., Gregory R. J. Effects of the E4 region on the persistence of transgene expression from adenovirus vectors. J. Virol. 1997;71:2408–2416. doi: 10.1128/jvi.71.3.2408-2416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graham F. L., Smiley J., Russell W. C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 38.Kootstra N. A., Verma I. M. Gene therapy with viral vectors. Annu. Rev. Pharmacol. Toxicol. 2002;43:413–439. doi: 10.1146/annurev.pharmtox.43.100901.140257. [DOI] [PubMed] [Google Scholar]
- 39.Knowles M. R., Hohneker K. W., Zhou Z., Olsen J. C., Noah T. L., Hu P., Leigh M. W., Engelhardt J. F., Edwards L. J., Jones K. R., et al. A controlled study of adenoviral-vector mediated gene transfer in the nasal epithelium of patients with cystic fibrosis. N. Engl. J. Med. 1995;333:823–831. doi: 10.1056/NEJM199509283331302. [DOI] [PubMed] [Google Scholar]
- 40.Harvey B. G., Leopold P. L., Hackett N. R., Grasso T. M., Williams P. M., Tucker A. L., Kaner R. J., Ferris B., Gonda I., Sweeney T. D., et al. Airway epithelial CFTR mRNA expression in cystic fibrosis patients after repetitive administration of a recombinant adenovirus. J. Clin. Invest. 1999;104:1245–1255. doi: 10.1172/JCI7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chirmule N., Hughes J. V., Gao G. P., Raper S. E., Wilson J. M. Role of E4 in eliciting CD4 T-cell and B-cell responses to adenovirus vectors delivered to human and nonhuman primate lungs. J. Virol. 1998;72:6138–6145. doi: 10.1128/jvi.72.7.6138-6145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuckerman J. B., Robinson C. B., McCoy K. S., Shell R., Sferra T. J., Chirmule N., Magosin S. A., Propert K. J., Brown-Parr E. C., Hughes J. V., et al. A phase 1 study of adenovirus-mediated transfer of the human cystic fibrosis transmembrane conductance regulator gene to a lung segment of individuals with cystic fibrosis. Hum. Gene Ther. 1999;10:2973–2985. doi: 10.1089/10430349950016384. [DOI] [PubMed] [Google Scholar]
- 43.Pickles R. J., Fahrner J. A., Petrella J. M., Boucher R. C., Bergelson J. M. Retargeting the coxsackievirus and adenovirus receptor to the apical surface of polarised epithelial cells reveals the glycocalyx as a barrier to adenovirus mediated gene transfer. J. Virol. 2000;74:6050–6057. doi: 10.1128/jvi.74.13.6050-6057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wickham T. J., Roelvink P. W., Brough D. E., Kovesdi I. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat. Biotechnol. 1996;14:1570–1573. doi: 10.1038/nbt1196-1570. [DOI] [PubMed] [Google Scholar]
- 45.Dmitriev I., Krasnykh V., Miller C. R., Wang M., Kashentseva E., Mikheeva G., Belousova N., Curiel D. T. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J. Virol. 1998;72:9706–9713. doi: 10.1128/jvi.72.12.9706-9713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raper S. E., Yudkoff M., Chirmule N., Guang-Ping G., Nunes F., Haskal Z. J., Furth E. E., Propert K. J., Robinson M. B., Magosin S., et al. A pilot study of in vivo liver-directed gene transfer with an adenoviral vector in partial ornithine transcarbamylase deficiency. Hum. Gene Ther. 2002;13:163–175. doi: 10.1089/10430340152712719. [DOI] [PubMed] [Google Scholar]
- 47.Schiedner G., Morral N., Parks R. J., Wu Y., Koopmans S. C., Langston C., Graham F. L., Beaudet A. L., Kochanek S. Genomic DNA transfer with a high capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nat. Genet. 1998;18:180–183. doi: 10.1038/ng0298-180. [DOI] [PubMed] [Google Scholar]
- 48.Samulski R. J., Zhu X., Xiao X., Brook J. D., Housman D. E., Epstein N., Hunter L. A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kearns W. G., Afione S. A., Fulmer S. B., Pang M. C., Erikson D., Egan M., Landrum M. J., Flotte T. R., Cutting G. R. Recombinant adeno-associated virus (AAV-CFTR) vectors do not integrate in a site-specific fashion in an immortalized epithelial cell line. Gene Ther. 1996;3:748–755. [PubMed] [Google Scholar]
- 50.Summerford C., Samulski R. J. Membrane associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner J. A., Moran M. L., Messner A. H., Daifuku R., Conrad C. K., Reynolds T., Guggino W. B., Moss R. B., Carter B. J., Wine J. J., et al. A phase I/II study of tgAAV-CF for the treatment of chronic sinusitis in patients with cystic fibrosis. Hum. Gene Ther. 1998;9:889–909. doi: 10.1089/hum.1998.9.6-889. [DOI] [PubMed] [Google Scholar]
- 52.Wagner J. A., Reynolds T., Moran M. L., Moss R. B., Wine J. J., Flotte T. R., Gardner P. Efficient and persistent gene transfer of AAV-CFTR in maxillary sinus. Lancet. 1998b;351:1702–1703. doi: 10.1016/S0140-6736(05)77740-0. [DOI] [PubMed] [Google Scholar]
- 53.Wagner J. A., Messner A. H., Moran M. L., Daifuku R., Kouyama K., Desch J. K., Manley S., Norbash A. M., Conrad C. K., Friborg S., et al. Safety and biological efficacy of an adeno-associated virus vector-cystic fibrosis transmembrane regulator (AAV-CFTR) in the cystic fibrosis maxillary sinus. Laryngoscope. 1999;109:266–274. doi: 10.1097/00005537-199902000-00017. [DOI] [PubMed] [Google Scholar]
- 54.Aitken M. L., Moss R. B., Waltz D. A., Dovey M. E., Tonelli M. R., McNamara S. C., Gibson R. L., Ramsey B. W., Carter B. J., Reynolds T. C. A phase I study of aerosolized administration of tgAAVCF to cystic fibrosis subjects with mild lung disease. Hum. Gene Ther. 2001;12:1907–1916. doi: 10.1089/104303401753153956. [DOI] [PubMed] [Google Scholar]
- 55.Flotte T. R., Zeitlin P. L., Reynolds T. C., Heald A. E., Pedersen P., Beck S., Conrad C. K., Brass-Ernst L., Humphries M., Sullivan K., et al. Phase I trial of intranasal and endobronchial administration of a recombinant adeno-associated virus serotype 2 (rAAV2)-CFTR vector in adult cystic fibrosis patients: a two-part clinical study. Hum. Gene Ther. 2003;14:1079–1088. doi: 10.1089/104303403322124792. [DOI] [PubMed] [Google Scholar]
- 56.Carter B. J., Munson K., Burstein H., Peluso R., Gerard C., Guggino W., Flotte T., Moss R. AAV-CFTR gene therapy for cystic fibrosis: retrospect and prospect. Pediatr. Pulmonol. Suppl. 2003;25:159–160. [Google Scholar]
- 57.Wilson J. M. Novel vectors for gene therapy of cystic fibrosis. Pediatr. Pulmonol. Suppl. 2003;25:157–158. [Google Scholar]
- 58.Flotte T. R. CF gene therapy with AAV. Pediatr. Pulmonol. Suppl. 2003;25:156–157. [Google Scholar]
- 59.Miller A. D., Halbert C. L., Aitken M. L., Liggitt D. H., Ramsey B. W. AAV reporter vector trial. Pediatr. Pulmonol. Suppl. 2003;25:158. [Google Scholar]
- 60.Flotte T. R., Afione S. A., Solow R., Drumm M. L., Markakis D., Guggino W. B., Zeitlin P. L., Carter B. J. Expression of the cystic fibrosis transmembrane regulator from a novel adeno-associated virus promoter. J. Biol. Chem. 1993;268:3781–3790. [PubMed] [Google Scholar]
- 61.Wang D., Fischer H., Zhang L., Fan P., Ding R. X., Dong J. Efficient CFTR expression from AAV vectors packaged with promoters – the second generation. Gene Ther. 1999;6:667–675. doi: 10.1038/sj.gt.3300856. [DOI] [PubMed] [Google Scholar]
- 62.Duan D., Yue Y., Yan Z., Yang J., Engelhardt J. F. Endosomal processing limits gene transfer to polarized epithelia by adeno-associated virus. J. Clin. Invest. 2000;105:1573–1587. doi: 10.1172/JCI8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang N., Karp P., Gerard C. J., Munson K., Yan Z., Godwin S., Zabner J., Peluso R., Carter B., Engelhardt J. F. Intracellular barriers to rAAV-mediated CFTR correction in the CF airway can be overcome using proteosome modulating agents. Pediatr. Pulmonol. Suppl. 2003;25:160–162. [Google Scholar]
- 64.Kobinger G. P., Schumer G. P., Medina M. F., Weiner D. J., Wilson J. M. Stable and efficient gene transfer in airway of non-human primates with HIV vector pseudotyped with deletion mutant of the Ebola envelope glycoproteins. Pediatr. Pulmonol. Suppl. 2003;25:256. [Google Scholar]
- 65.Sinn P., Williams G., Davidson B., McCray P. Targeting apical entry in airway epithelia using pseudotyped FIV-based lentiviral vectors. Pediatr. Pulmonol. Suppl. 2003;25:257. [Google Scholar]
- 66.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M. P., Wulffraat N., Leboulch P., Lim A., Osbourne C. S., Pawliuk R., Morillon E., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 67.Kohn D. B., Sadelain M., Glorioso J. C. Occurrence of leukaemia following gene therapy of X-linked SCID. Nat. Rev. Cancer. 2003;3:477–488. doi: 10.1038/nrc1122. [DOI] [PubMed] [Google Scholar]
- 68.Zhang L., Button B., Skiadopoulos M. H., Dang Y., Bukreyev A., Gabriel S. E., Boucher R. C., Collins P. L., Pickles R. J. Restoration of periciliary liquid volume to normal levels in human CF airway epithelium after targeted expression of CFTR in ciliated cells with a parainfluenza virus type 3-CFTR gene transfer vector. Pediatr. Pulmonol. Suppl. 2003;25:256–257. [Google Scholar]
- 69.Ratko T. A., Cummings J. P., Blebea J., Matuszewski K. A. Clinical gene therapy for non-malignant disease. Am. J. Med. 2003;115:560–569. doi: 10.1016/s0002-9343(03)00447-9. [DOI] [PubMed] [Google Scholar]
- 70.Singhal A., Huang L. Gene transfer in mammalian cells using liposomes as carriers. In: Wolff J. A., editor. Gene Therapeutics. Boston: Birkhauser; 1994. pp. 118–142. [Google Scholar]
- 71.Sternberg B., Sorgi F. L., Huang L. New structures in complex formation between DNA and cationic liposomes visualized by freeze-fracture electron microscopy. FEBS Lett. 1994;356:361–366. doi: 10.1016/0014-5793(94)01315-2. [DOI] [PubMed] [Google Scholar]
- 72.Felgner P. L., Gadek T. R., Holm M., Roman R., Chan H. W., Wenz M., Northrop J. P., Ringold G. M., Danielson M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. U.S.A. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hawley-Nelson P., Ciccarone V., Gebeyehu G., Jessee J., Felgner P. L. LipofectAMINE™ reagent: a new, higher efficiency polycationic liposome transfection reagent. Focus. 1993;15:73–79. [Google Scholar]
- 74.Leventis R., Silvius J. R. Interactions of mammalian cells with lipid dispersions containing novel metabolizable cationic amphiphiles. Biochim. Biophys. Acta. 1990;1023:124–132. doi: 10.1016/0005-2736(90)90017-i. [DOI] [PubMed] [Google Scholar]
- 75.Smyth-Templeton N., Lasic D. D., Frederik P. M., Strey H. H., Roberts D. D., Pavlakis G. N. Improved DNA: liposome complexes for increased systemic delivery and gene expression. Nat. Biotech. 1997;15:647–652. doi: 10.1038/nbt0797-647. [DOI] [PubMed] [Google Scholar]
- 76.Gao X., Huang L. A novel cationic liposome reagent for efficient transfection of mammalian cells. Biochem. Biophys. Res. Commun. 1991;179:280–285. doi: 10.1016/0006-291x(91)91366-k. [DOI] [PubMed] [Google Scholar]
- 77.Farhood H., Bottega R., Epand R. M., Huang L. Effect of cationic cholesterol derivatives on gene transfer and protein kinase C activity. Biochim. Biophys. Acta. 1992;1111:239–246. doi: 10.1016/0005-2736(92)90316-e. [DOI] [PubMed] [Google Scholar]
- 78.Gao X., Huang L. Cationic liposome-mediated gene transfer. Gene Ther. 1995;2:710–722. [PubMed] [Google Scholar]
- 79.Lee E. R., Marshall J., Siegel C. S., Jiang C., Yew N. S., Nichols M. R., Nietupski J. B., Ziegler R. J., Lane M. B., Wang K. X., et al. Detailed analysis of structures and formulations of cationic lipids for efficient gene transfer to lung. Hum. Gene Ther. 1996;7:1701–1717. doi: 10.1089/hum.1996.7.14-1701. [DOI] [PubMed] [Google Scholar]
- 80.Noone P. G., Hohneker K. W., Zhou Z., Johnson L. G., Foy C., Gipson C., Jones K., Noah T. L., Leigh M. W., Schwartzbach C., et al. Safety and biological efficacy of a lipid-CFTR complex for gene transfer in the nasal epithelium of adult patients with cystic fibrosis. Mol. Ther. 2000;1:105–114. doi: 10.1006/mthe.1999.0009. [DOI] [PubMed] [Google Scholar]
- 81.MacDonald R. C., Ashley G. W., Shida M. M., Rakhmanova V. A., Tarahovsky Y. S., Pantazatos D. P., Kennedy M. T., Pozharski E. V., Baker K. A., Jones R. D., et al. Physical and biological properties of cationic trimesters of phosphatidylcholine. Biophys. J. 1999;77:2612–2629. doi: 10.1016/S0006-3495(99)77095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Caplen N. J., Alton E. W., Middleton P. G., Dorin J. R., Stevenson B. J., Gao X., Durham S. R., Jeffery P. K., Hodson M. E., Coutelle C., et al. Liposome-mediated CFTR gene transfer to the nasal epithelium of patients with cystic fibrosis. Nat. Med. 1995;1:39–46. doi: 10.1038/nm0195-39. [DOI] [PubMed] [Google Scholar]
- 83.Zabner J., Cheng S. H., Meeker D., Launspach J., Balfour R., Perricone M. A., Morris J. E., Marshall J., Fasbender A., Smith A. E., Welsh M. J. Comparison of DNA-lipid complexes and DNA alone for gene transfer to cystic fibrosis airway epithelia in vivo. J. Clin. Invest. 1997;100:1529–1537. doi: 10.1172/JCI119676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gill D. R., Southern K. W., Mofford K. A., Seddon T., Huang L., Sorgi F., Thomson A., MacVinish L. J., Ratcliff R., Bilton D., et al. A placebo-controlled study of liposome-mediated gene transfer to the nasal epithelium of patients with cystic fibrosis. Gene Ther. 1997;4:199–209. doi: 10.1038/sj.gt.3300391. [DOI] [PubMed] [Google Scholar]
- 85.Porteous D. J., Dorin J. R., McLachlan G., Davidson-Smith H., Davidson H., Stevenson B. J., Carothers A. D., Wallace W. A., Moralee S., Hoenes C., et al. Evidence for safety and efficacy of DOTAP cationic liposome mediated CFTR gene transfer to the nasal epithelium of patients with cystic fibrosis. Gene Ther. 1997;4:210–218. doi: 10.1038/sj.gt.3300390. [DOI] [PubMed] [Google Scholar]
- 86.Alton E. W. F. W., Stern M., Farley R., Jaffe A., Chadwick S. L., Phillips J., Davies J., Smith S. N., Browning J., Davies M. G., et al. Cationic lipid-mediated CFTR gene transfer to the lungs and nose of patients with cystic fibrosis: a double blind placebo controlled trial. Lancet. 1999;353:947–954. doi: 10.1016/s0140-6736(98)06532-5. [DOI] [PubMed] [Google Scholar]
- 87.Hyde S. C., Southern K. W., Gileadi U., Fitzjohn E. M., Mofford K. A., Waddell B. E., Gooi H. C., Goddard C. A., Hannavy K., Smyth S. E., et al. Repeat administration of DNA/liposomes to the nasal epithelium of patients with cystic fibrosis. Gene Ther. 2000;7:1156–1165. doi: 10.1038/sj.gt.3301212. [DOI] [PubMed] [Google Scholar]
- 88.Ruiz F. E., Clancy J. P., Perricone M. A., Bebok Z., Hong J. S., Cheng S. H., Meeker D. P., Young K. R., Schoumacher R. A., Weatherly M. R., et al. A clinical inflammatory syndrome attributable to aerosolized lipid-DNA administration in cystic fibrosis. Hum Gene Ther. 2001;12:751–761. doi: 10.1089/104303401750148667. [DOI] [PubMed] [Google Scholar]
- 89.MacDonald R. J., Liggitt H. D., Roche L., Nguyen H. T., Pearlman R., Raabe O. G., Bussey L. B., Gorman C. M. Aerosol delivery of lipid:DNA complexes to the lungs of Rhesus monkeys. Pharm. Res. 1998;15:671–679. doi: 10.1023/a:1011902532163. [DOI] [PubMed] [Google Scholar]
- 90.Ferrari S., Geddes D. M., Alton E. W. F. W. Barriers to and new approaches for gene therapy and gene delivery in cystic fibrosis. Adv. Drug Delivery Rev. 2002;54:1373–1393. doi: 10.1016/S0169-409X(02)00145-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Boussif O., Lezoualc'h F., Zanta M. A., Mergny M., Scherman D., Demeneix B., Behr J. P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zou S. M., Erbacher P., Remy J. S., Behr J. P. Systemic linear polyethylenimine (L-PEI)-mediated gene delivery in the mouse. J. Gene Med. 2000;2:128–134. doi: 10.1002/(SICI)1521-2254(200003/04)2:2<128::AID-JGM95>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 93.Klink D. T., Chao S., Glick M. C., Scanlin T. F. Nuclear translocation of lactosylated poly-L-lysine/cDNA complex in cystic fibrosis airway epithelial cells. Mol. Ther. 2001;3:831–841. doi: 10.1006/mthe.2001.0332. [DOI] [PubMed] [Google Scholar]
- 94.Kim H. H., Lee W. S., Yang J. M., Shin S. Basic peptide system for efficient delivery of foreign genes. Biochim. Biophys. Acta. 2003;1640:129–136. doi: 10.1016/s0167-4889(03)00028-4. [DOI] [PubMed] [Google Scholar]
- 95.Hart S. Use of adhesion molecules for gene delivery. Exp. Nephrol. 1999;7:193–199. doi: 10.1159/000020600. [DOI] [PubMed] [Google Scholar]
- 96.Harbottle R. P., Cooper R. G., Hart S. L., Ladhoff A., McKay T., Knight A. M., Wagner E., Miller A. D., Coutelle C. An RGD-oligolysine peptide: a prototype construct for integrin-mediated gene delivery. Hum. Gene Ther. 1998;9:987–988. doi: 10.1089/hum.1998.9.7-1037. [DOI] [PubMed] [Google Scholar]
- 97.Ziady A. G., Kelley T., Davis P. B. Molecular conjugate mediated CFTR gene transfer in CF mice affects secondary CF defects. Pediatr. Pulmonol. Suppl. 2001;22:254. [Google Scholar]
- 98.Schneider H., Harbottle R. P., Yokosaki Y., Jost P., Coutelle C. Targeted gene delivery into α9β1-integrin-displaying cells by a synthetic peptide. FEBS Lett. 1999;458:329–332. doi: 10.1016/s0014-5793(99)01181-3. [DOI] [PubMed] [Google Scholar]
- 99.Chan C-K., Senden T., Jans D. A. Supramolecular structure and nuclear targeting efficiency determine the enhancement of transfection by modified polylysines. Gene Ther. 2000;7:1690–1697. doi: 10.1038/sj.gt.3301275. [DOI] [PubMed] [Google Scholar]
- 100.Zhang F., Andreassen P., Fender P., Geissler E., Hernandez J. F., Chroboczek J. A transfecting peptide derived from adenovirus fiber protein. Gene Ther. 1999;6:171–181. doi: 10.1038/sj.gt.3300801. [DOI] [PubMed] [Google Scholar]
- 101.Sandgren S., Cheng F., Belting M. Nuclear targeting of macromolecular polyanions by an HIV-Tat derived peptide. J. Biol. Chem. 2002;277:38877–38883. doi: 10.1074/jbc.M205395200. [DOI] [PubMed] [Google Scholar]
- 102.Futaki S., Suzuki T., Ohashi W., Yagami T., Tanaka S., Ueda K., Sugiura Y. Arginine-rich peptides. J. Biol. Chem. 2001;276:5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- 103.Siprashvili Z., Scholl F. A., Oliver S. F., Adams A., Contag C. H., Wender P. A., Khavari P. A. Gene transfer via reversible plasmid condensation with cysteine flanked, internally spaced arginine-rich peptides. Hum. Gene Ther. 2003;14:1225–1233. doi: 10.1089/104303403767740768. [DOI] [PubMed] [Google Scholar]
- 104.Ziady A. G., Gedeon C. R., Muhammad O., Stillwell V., Oette S. M., Fink T. L., Quan W., Kowalczyk T. H., Hyatt S. L., Payne J., et al. Minimal toxicity of compacted DNA nanoparticles in the murine lung. Mol. Ther. 2003;8:948–956. doi: 10.1016/j.ymthe.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 105.Davis P. B., Konstan M. W. Developing CF therapies: from the laboratory to the patient. Pediatr. Pulmonol. Suppl. 2004;27:18. [Google Scholar]
- 106.Losardo D. W., Vale P. R., Hendel R. C., Milliken C. E., Fortuin F. D., Cummings N., Schatz R. A., Asahara T., Isner J. M., Kuntz R. E. Phase 1/2 placebo-controlled, double-blind, dose-escalating trial of myocardial vascular endothelial growth factor 2 gene transfer by catheter delivery in patients with chronic myocardial ischaemia. Circulation. 2002;105:2012–2018. doi: 10.1161/01.cir.0000015982.70785.b7. [DOI] [PubMed] [Google Scholar]
- 107.Simovic D., Isner J. M., Ropper A. H., Pieczek A., Weinberg D. H. Improvement in chronic ischaemic neuropathy after intramuscular phVEGF165 gene transfer in patients with critical limb ischaemia. Arch. Neurol. 2001;58:761–768. doi: 10.1001/archneur.58.5.761. [DOI] [PubMed] [Google Scholar]
- 108.Roth D. A., Tawa N. E., Jr, O'Brien J. M., Treco D. A., Selden R. F. the Factor VIII Transkaryotic Therapy Study Group. Nonviral transfer of the gene encoding coagulation factor VIII in patients with severe haemophilia A. N. Engl. J. Med. 2001;344:1735–1742. doi: 10.1056/NEJM200106073442301. [DOI] [PubMed] [Google Scholar]
- 109.Audouy S. A. L., de Leij L. F. M. H., Hoekstra D., Molema G. In vivo characteristics of cationic liposomes as delivery vectors for gene therapy. Pharm. Res. 2002;19:1599–1605. doi: 10.1023/a:1020989709019. [DOI] [PubMed] [Google Scholar]
- 110.Smyth Templeton N. Cationic liposome-mediated gene delivery in vivo. Biosci. Report. 2002;22:283–295. doi: 10.1023/a:1020142823595. [DOI] [PubMed] [Google Scholar]
- 111.Smyth Templeton N. Liposomal delivery of nucleic acids in vivo. DNA Cell Biol. 2002;21:857–867. doi: 10.1089/104454902762053828. [DOI] [PubMed] [Google Scholar]
- 112.Wrobel I., Collins D. Fusion of cationic liposomes with mammalian cells occurs after endocytosis. Biochim. Biophys. Acta. 1995;1235:296–304. doi: 10.1016/0005-2736(95)80017-a. [DOI] [PubMed] [Google Scholar]
- 113.Gao X., Huang L. Potentiation of cationic liposome-mediated gene delivery by polycations. Biochemistry. 1996;35:1027–1036. doi: 10.1021/bi952436a. [DOI] [PubMed] [Google Scholar]
- 114.Hart S. L., Arancibia-Carcamo C. V., Wolfert M. A., Mailhos C., O'Reilly N. J., Ali R. R., Coutelle C., George A. J., Harbottle R. P., Knight A. M., et al. Lipid-mediated enhancement of transfection by a non-viral integrin targeted vector. Hum. Gene Ther. 1998;9:575–585. doi: 10.1089/hum.1998.9.4-575. [DOI] [PubMed] [Google Scholar]
- 115.Jenkins R. G., Meng Q. H., Hodges R. J., Lee L. K., Bottoms S. E. W., Laurent G. J., Willis D., Ayazi Shamlou P., McAnulty R. J., Hart S. L. Formation of LID vector complexes in water allows physicochemical properties and enhances pulmonary gene expression in vivo. Gene Ther. 2003;10:1026–1034. doi: 10.1038/sj.gt.3301963. [DOI] [PubMed] [Google Scholar]
- 116.Farhood H., Serbina N., Huang L. The role of dioleoyl phosphatidylethanolamine in cationic liposome mediated gene transfer. Biochim. Biophys. Acta. 1995;1235:289–295. doi: 10.1016/0005-2736(95)80016-9. [DOI] [PubMed] [Google Scholar]
- 117.Chen Q.-R., Zhang L., Stass S. A., Mixson A. J. Co-polymer of histidine and lysine markedly enhances transfection efficiency of liposomes. Gene Ther. 2000;7:1698–1705. doi: 10.1038/sj.gt.3301294. [DOI] [PubMed] [Google Scholar]
- 118.Duguid J. G., Li C., Shi M., Logan M. J., Alila H., Rolland A., Tomlinson E., Sparrow J. T., Smith L. C. A physiochemical approach for predicting the effectiveness of peptide-based gene delivery systems for use in plasmid-based gene therapy. Biophys. J. 1998;74:2802–2814. doi: 10.1016/S0006-3495(98)77987-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brown M. D., Schätzlein A. G., Uchegbu I. F. Gene delivery with synthetic (non viral) carriers. Int. J. Pharm. 2001;229:1–21. doi: 10.1016/s0378-5173(01)00861-4. [DOI] [PubMed] [Google Scholar]
- 120.Tachibana R., Harashima H., Shinohara Y., Kiwada H. Quantitative studies on the nuclear import of plasmid DNA and gene expression employing nonviral vectors. Adv. Drug Delivery Rev. 2001;52:219–226. doi: 10.1016/s0169-409x(01)00211-3. [DOI] [PubMed] [Google Scholar]
- 121.Pouton C. W. Nuclear import of polypeptides, polynucleotides and supramolecular complexes. Adv. Drug. Delivery Rev. 1998;34:51–64. doi: 10.1016/s0169-409x(98)00050-7. [DOI] [PubMed] [Google Scholar]
- 122.Ludtke J. J., Zhang G., Sebestyén M. G., Wolff J. A. A nuclear localization signal can enhance both the nuclear transport and expression of 1 kb DNA. J. Cell Sci. 1999;112:2033–2041. doi: 10.1242/jcs.112.12.2033. [DOI] [PubMed] [Google Scholar]
- 123.Pollard H., Remy J. S., Loussouarn G., Demolombe S., Behr J. P., Escande D. Polyethylenimine but not cationic lipids promotes transgene delivery to the nucleus in mammalian cells. J. Biol. Chem. 1998;273:7507–7511. doi: 10.1074/jbc.273.13.7507. [DOI] [PubMed] [Google Scholar]
- 124.Zabner J., Fasbender A. J., Moninger T., Poellinger K. A., Welsh M. J. Cellular and molecular barriers to gene transfer by a cationic lipid. J. Biol. Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- 125.Fritz J. D., Herweijer H., Zhang G., Wolff J. A. Gene transfer into mammalian cells using histone-condensed plasmid DNA. Hum. Gene Ther. 1996;7:1395–1404. doi: 10.1089/hum.1996.7.12-1395. [DOI] [PubMed] [Google Scholar]
- 126.Sebestyén M. G., Ludtke J. J., Bassik M. C., Zhang G., Budker V., Lukhtanov E. A., Hagstrom J. E., Wolff J. A. DNA vector chemistry: the covalent attachment of signal peptides to plasmid DNA. Nat. Biotech. 1998;16:80–85. doi: 10.1038/nbt0198-80. [DOI] [PubMed] [Google Scholar]
- 127.Zanta M. A., Belguise-Valladier P., Behr J. P. Gene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc. Natl. Acad. Sci. U.S.A. 1999;96:91–96. doi: 10.1073/pnas.96.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Conary J. T., Erdos G., McGuire M., Faulks R., Gagné L., Price P., Christman B., Brigham K., Ebner F., Schreier H. Cationic liposome plasmid DNA complexes: in vitro cell entry and transgene expression augmented by synthetic signal peptides. Eur. J. Pharm. Biopharm. 1996;42:277–285. [Google Scholar]
- 129.Schwartz B., Ivanov M.-A., Pitard B., Escriou V., Rangara R., Byk G., Wils P., Crouzet J., Scherman D. Synthetic DNA-compacting peptides derived from human sequence enhance cationic lipid-mediated gene transfer in vitro and in vivo. Gene Ther. 1999;6:282–292. doi: 10.1038/sj.gt.3300795. [DOI] [PubMed] [Google Scholar]
- 130.Yew N. S., Przybylska M., Ziegler R. J., Liu D., Cheng S. H. High and sustained expression in vivo from plasmid vectors containing a hybrid ubiquitin promoter. Mol. Ther. 2001;4:75–82. doi: 10.1006/mthe.2001.0415. [DOI] [PubMed] [Google Scholar]
- 131.Gill D. R., Smyth S. E., Goddard C. A., Pringle I. A., Higgins C. F., Colledge W. H., Hyde S. C. Increased persistence of lung gene expression using plasmids containing the ubiquitin C or elongation factor 1α promoter. Gene Ther. 2001;8:1539–1546. doi: 10.1038/sj.gt.3301561. [DOI] [PubMed] [Google Scholar]
- 132.Vayda M. E., Rogers A. E., Flint S. J. The structure of nucleoprotein cores released from adenovirus. Nucleic Acids Res. 1983;11:441–451. doi: 10.1093/nar/11.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chatterjee P. K., Vayda M. E., Flint S. J. Interactions among the three adenovirus core proteins. J. Gen. Virol. 1985;55:379–386. doi: 10.1128/jvi.55.2.379-386.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Matthews D. A., Russell W. C. Adenovirus core protein V is delivered by the invading virus to the nucleus of the infected cell and later in infection is associated with the nucleoli. J. Gen. Virol. 1998;79:1671–1675. doi: 10.1099/0022-1317-79-7-1671. [DOI] [PubMed] [Google Scholar]
- 135.Davison A. J., Benkő M., Harrach B. Genetic content and evolution of adenoviruses. J. Gen. Virol. 2003;84:2895–2908. doi: 10.1099/vir.0.19497-0. [DOI] [PubMed] [Google Scholar]
- 136.Matthews D. A. Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J. Virol. 2001;75:1031–1038. doi: 10.1128/JVI.75.2.1031-1038.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sung M. T., Cao T. M., Coleman R. T., Budelier K. A. Gene and protein sequences of adenovirus core protein VII, a hybrid basic chromosomal protein. Proc. Nat. Acad. Sci. U.S.A. 1983;80:2902–2906. doi: 10.1073/pnas.80.10.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Alestrom P., Akusjärvi G., Lager M., Yeh-kai L., Pettersson U. Genes encoding the core proteins of adenovirus type 2. J. Biol. Chem. 1984;259:13980–13985. [PubMed] [Google Scholar]
- 139.Okuwaki M., Iwamatsu A., Tsujimoto M., Nagata K. Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J. Mol. Biol. 2001;311:41–55. doi: 10.1006/jmbi.2001.4812. [DOI] [PubMed] [Google Scholar]
- 140.Zhang C. L., Nagaraja K. V., Sivanandan V., Newman J. A. Identification and characterization of viral polypeptides from type-II avian adenoviruses. Am. J. Vet. Res. 1991;52:1137–1141. [PubMed] [Google Scholar]
- 141.Tarassishin L., Szawlowski P., McLay J., Kidd A. H., Russell W. C. Adenovirus core protein VII displays a linear epitope conserved in a range of human adenoviruses. J. Gen. Virol. 1999;80:47–50. doi: 10.1099/0022-1317-80-1-47. [DOI] [PubMed] [Google Scholar]
- 142.Farkas S. L., Benkő M., Élő P., Ursu K., Dán Á., Ahne W., Harrach B. Genomic and phylogenetic analyses of an adenovirus isolated from a corn snake (Elaphe guttata) imply a common origin with members of the proposed new genus Atadenovirus. J. Gen. Virol. 2002;83:2403–2410. doi: 10.1099/0022-1317-83-10-2403. [DOI] [PubMed] [Google Scholar]
- 143.Cai F., Weber J. M. Primary structure of the canine adenovirus PVII protein: functional implications. Virology. 1993;193:986–988. [PubMed] [Google Scholar]
- 144.Anderson C. W., Young M. E., Flint S. J. Characterisation of the adenovirus 2 virion protein, mu. Virology. 1989;172:506–512. doi: 10.1016/0042-6822(89)90193-1. [DOI] [PubMed] [Google Scholar]
- 145.Vayda M. E., Flint S. J. Isolation and characterisation of adenovirus core nucleoprotein subunits. J. Virol. 1987;61:3335–3339. doi: 10.1128/jvi.61.10.3335-3339.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Black B. C., Center M. S. DNA-binding properties of the major core proteins of adenovirus 2. Nucleic Acids Res. 1979;6:2339–2353. doi: 10.1093/nar/6.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sato K., Hosokawa K. Analysis of the interaction between the DNA and major core protein in adenovirus chromatin by circular dichroism and ultraviolet light induced cross-linking. J. Biol. Chem. 1984;95:1031–1039. doi: 10.1093/oxfordjournals.jbchem.a134690. [DOI] [PubMed] [Google Scholar]
- 148.Russell W. C., Precious B. Nucleic acid binding properties of adenovirus structural polypeptides. J. Gen. Virol. 1982;63:69–79. doi: 10.1099/0022-1317-63-1-69. [DOI] [PubMed] [Google Scholar]
- 149.Bergelson J. M., Cunningham J. A., Droguett G., Kurt-Jones E. A., Krithivas A., Hong J. S., Horwitz M. S., Crowell R. L., Finberg R. W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 150.Meier O., Greber U. F. Adenovirus endocytosis. J. Gene Med. 2003;5:451–462. doi: 10.1002/jgm.409. [DOI] [PubMed] [Google Scholar]
- 151.Roelvink P. W., Lizonova A., Lee J. G., Li Y., Bergelson J. M., Finberg R. W., Brough D. E., Kovesdi I., Wickham T. J. The coxsackievirus-adenovirus receptor protein can function as cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 1998;72:7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Devaux C., Caillet-Boudin M. L., Jacrot B., Boulanger P. Crystallization, enzymatic cleavage, and the polarity of adenovirus type 2 fibre. Virology. 1987;161:121–128. doi: 10.1016/0042-6822(87)90177-2. [DOI] [PubMed] [Google Scholar]
- 153.Wickham T. J., Mathias P., Cheresh D. A., Nemerow G. R. Integrins αv β5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 154.Meier O., Boucke K., Hammer S. V., Keller S., Stidwill R. P., Hemmi S., Greber U. F. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J. Cell Biol. 2002;158:1119–1131. doi: 10.1083/jcb.200112067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Greber U. F., Willets M., Webster P., Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75:477–486. doi: 10.1016/0092-8674(93)90382-z. [DOI] [PubMed] [Google Scholar]
- 156.Greber U. F., Webster P., Weber J., Helenius A. The role of the adenovirus protease on virus entry into cells. EMBO J. 1996;15:1766–1777. [PMC free article] [PubMed] [Google Scholar]
- 157.Suomalainen M., Nakano M. Y., Boucke K., Keller S., Stidwill R. P., Greber U. F. Microtubule-dependent minus and plus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Trotman L. C., Mosberger N., Fornerod M., Stidwill R. P., Greber U. F. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 2001;3:1092–1100. doi: 10.1038/ncb1201-1092. [DOI] [PubMed] [Google Scholar]
- 159.Greber U. F., Suomalainen M., Stidwell R. P., Boucke K., Ebersold M. W., Helenius A. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 1997;16:5998–6007. doi: 10.1093/emboj/16.19.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Korn R., Horwitz M. S. Adenovirus DNA synthesis in vitro is inhibited by the virus-coded major core protein. Virology. 1986;150:342–351. doi: 10.1016/0042-6822(86)90299-0. [DOI] [PubMed] [Google Scholar]
- 161.Nakanishi Y., Maeda K., Ohtsuki M., Hosokawa K., Natori S. In vitro transcription of a chromatin like complex of major core protein VII and DNA of adenovirus serotype 2. Biochem. Biophys. Res. Commun. 1986;136:86–93. doi: 10.1016/0006-291x(86)90880-6. [DOI] [PubMed] [Google Scholar]
- 162.Lunt R., Vayda M. E., Young M., Flint S. J. Isolation and characterisation of monoclonal antibodies against the adenovirus core proteins. Virology. 1988;164:275–279. doi: 10.1016/0042-6822(88)90645-9. [DOI] [PubMed] [Google Scholar]
- 163.Lee T. W. R., Blair G. E., Matthews D. A. Adenovirus core protein VII contains distinct sequences that mediate targeting to the nucleus and nucleolus, and colocalization with human chromosomes. J. Gen. Virol. 2003;84:3423–3428. doi: 10.1099/vir.0.19546-0. [DOI] [PubMed] [Google Scholar]
- 164.Lee T. W. R., Lawrence F. J., Dauksaite V., Akusjärvi G., Blair G. E., Matthews D. A. Precursor of adenovirus core polypeptide Mu targets the nucleolus and modulates the expression of E2 proteins. J. Gen. Virol. 2004;85:185–196. doi: 10.1099/vir.0.19352-0. [DOI] [PubMed] [Google Scholar]
- 165.Wienhues U., Hosokawa K., Hoveler A., Siegmann B., Doerfler W. A novel method for transfection and expression of reconstituted DNA–protein complexes in eukaryotic cells. DNA. 1987;6:81–89. doi: 10.1089/dna.1987.6.81. [DOI] [PubMed] [Google Scholar]
- 166.Murray K. D., Etheridge C. J., Shah S. I., Matthews D. A., Russell W. C., Gurling H. M., Miller A. D. Enhanced cationic liposome-mediated transfection using the DNA-binding peptide Mu from the adenovirus core. Gene Ther. 2001;8:453–460. doi: 10.1038/sj.gt.3301401. [DOI] [PubMed] [Google Scholar]
- 167.Tagawa T., Manvell M., Brown N., Keller M., Perouzel E., Murray K. D., Harbottle R. P., Tecle M., Booy F., Brahimi-Horn M. C., Coutelle C., et al. Characterisation of LMD virus-like nanoparticles self-assembled from cationic liposomes, adenovirus core peptide μ (mu) and plasmid DNA. Gene Ther. 2002;9:564–576. doi: 10.1038/sj.gt.3301686. [DOI] [PubMed] [Google Scholar]
- 168.Keller M., Harbottle R. P., Perouzel E., Colin M., Shah I., Rahim A., Vaysse L., Bergau A., Moritz S., Brahimi-Horn C., et al. Nuclear localization sequence templated nonviral gene delivery vectors: investigation of intracellular trafficking events of LMD and LD vector systems. Chembiochem. 2003;4:286–298. doi: 10.1002/cbic.200390049. [DOI] [PubMed] [Google Scholar]
- 169.Preuss M., Tecle M., Shah I., Matthews D. A., Miller A. D. Comparison between the interactions of adenovirus-derived peptides with plasmid DNA and their role in gene delivery mediated by liposome-peptide-DNA virus-like nanoparticles. Org. Biomol. Chem. 2003;1:2430–2438. doi: 10.1039/b302361c. [DOI] [PubMed] [Google Scholar]