Abstract
The 12-lead electrocardiogram (ECG) is the most common front-line diagnosis tool for assessing cardiovascular health, yet traditional ECG analysis cannot detect many diseases. Machine learning (ML) techniques have emerged as a powerful set of techniques for producing automated and robust ECG analysis tools that can often predict diseases and conditions not detectable by traditional ECG analysis. Many contemporary ECG-ML studies have focused on utilizing the full 12-lead ECG; however, with the increased availability of single-lead ECG data from wearable devices, there is a clear motivation to explore the development of single-lead ECG-ML techniques. In this study we developed and applied a deep learning architecture for the detection of low left ventricular ejection fraction (LVEF), and compared the performance of this architecture when it was trained with individual leads of the 12-lead ECG to the performance when trained using the entire 12-lead ECG. We observed that single-lead-trained networks performed similarly to the full 12-lead-trained network. We also noted patterns of agreement and disagreement between network low LVEF predictions across the different lead-trained networks.
1. Introduction
The most common front-line tool for cardiovascular disease diagnosis is the electrocardiogram (ECG) because it is inexpensive, noninvasive, and ubiquitous.[1, 2] With the rise in availability of ECG recordings, development of machine learning tools for ECG analysis has shown promise in expanding the utility of the ECG to improve diagnostic power and address diseases that are not traditionally detectable via ECG, such as low left ventricular ejection fraction (LVEF).[3, 4]
Contemporary ML approaches have focused primarily on leveraging the full 12-lead ECG (which typically consists of between 8 to 10 unique measured electrograms).[3, 5] However, it is not clear if all leads are necessary to achieve acceptable accuracy, and numerous applications of these algorithms require training and deployment on much more limited data (i.e., a single-lead). Additionally, understanding the differences in the performance of ML techniques when applied to different individual leads is paramount to designing robust and accurate ML-ECG diagnosis techniques to better understand and optimize ML performance. Furthermore, ML techniques that leverage differences between leads require knowledge of how different leads perform in various ML tasks.
In this study, we developed and applied a deep learning architecture for the detection of reduced heart function (low left ventricular ejection fraction [LVEF]), and trained this network on each measured lead of the 12-lead ECGs individually. We then compared the performance of these individual lead networks to each other and also to a network trained using all the leads (the current standard). We found that several of the leads produced trained ML networks that accurately predict LVEF with comparable accuracy to using all the leads simultaneously. Additionally, we noted patterns of agreement and disagreement between ML diagnosis of low LVEF for networks trained on individual leads. This study seeks to pave the way for future research into the development of robust ML-ECG algorithms using limited ECG data, and to increase our understanding of how these algorithms work in the context of multivector ECG data.
2. Methods
Dataset:
Digital ECG recordings (8 measured channels, L1, L2, V1 through V6) from 24,868 patients were collected from the University of Utah health system from 2012 to 2021. Each ECG was associated with an LVEF measurement made within 4 weeks of the ECG recording. The ejection fraction was calculated by echocardiographic measurements. Each ECG recording consisted of 10 seconds of continuous simultaneous recording from each lead at 500 hz, resulting in an 8 × 5, 000 matrix for each ECG. ECG signals and associated LVEF measurements were split into a 90% training set (22,382 patients) and 10% (2,486 patients) testing set.
Machine Learning Architecture and Training:
Detection of low LVEF was framed as a binary classification task using a cutoff of below 40% as low LVEF, as seen in previous studies.[3] Our network architecture is based on a residual network that we have shown to be an effective structure for ML-ECG analysis [6]. In brief, the network consisted of temporal and spatial convolutional filters, batch normalization, dropout (probability = 0.5), a rectified linear unit (ReLU), fully connected layers, and a sigmoid output. Spatial and temporal convolutional layers were arranged into residual blocks, and their output features were concatenated before the fully connected layers. The architecture is depicted in Figure 1.
Figure 1.
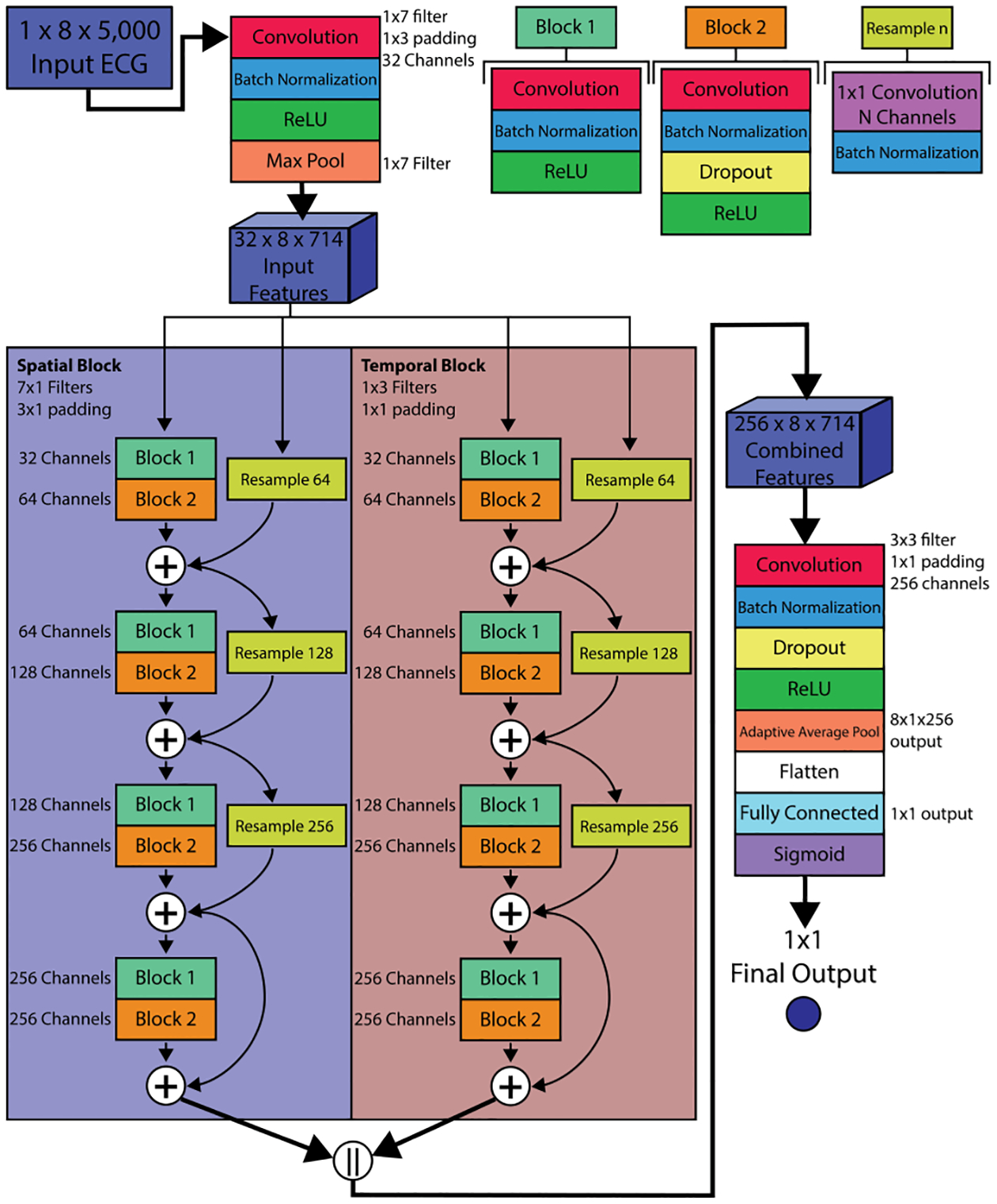
ECG low LVEF detection architecture. This network consists of an input stage, temporal and spatial residual blocks, and an output stage. Each residual block consists of four layers of residual blocks, similar to the common resnet structure. In cases where the number of input channels is less than the output channels (layers 1 through 3), the input is re-sampled using a 1×1 convolutional layer. The spatial residual block uses 7×1 convolutional filters whereas the temporal uses 1×3 filters. The features from the two residual blocks are concatenated before the output stage. When single-leads are used, the spatial blocks instead use 1×1 convolutional filters.
A separate instance of the architecture was trained for each individual lead, as well as using all 8 unique leads as input, resulting in 9 total training scenarios (Lead I, II, V1, V2, V3, V4, V5, V6, and All Leads). For each training scenario, the binary cross entropy loss was evaluated between the network prediction and target LVEF label using the ADAM optimizer to tune weights. At each of the 50 training iterations, area under the receiver operator curve (AUC) was computed for the test dataset and used as a selection criterion for preventing overfitting of the parameters. This training process was replicated 5 times for each lead scenario using the same training and test datasets to minimize the effect of weight and bias initialization on network performance. The result was 45 trained networks (5 per lead group).
Analysis metrics:
AUC, F1 score, sensitivity, and specificity were computed for each network using the training data and averaged across the 5 replicates for each of the 9 training scenarios (leads 1 through 8, and the scenario that used all 8 simultaneously). Sensitivity and specificity were computed at a network output threshold corresponding to the maximum F1 score. Unthresholded network outputs were then compared between each trained network according to correlation defined as
| (1) |
where Pn was the 1 × 2, 486 mean subtracted vector of network predictions for the nth network for the 2,486 test samples. Here, T denotes the vector transpose, and ||−||2 denotes the Euclidean 2 norm. Each network was provided as input the same lead(s) used to train it.
3. Results
Detection of low LVEF using all 8 recorded leads (the current standard) resulted in an average AUC of 0.93 ± 0.00, a mean F1 score of 0.60 ± 0.02, a mean sensitivity of 0.62 ± 0.04, and a mean specificity of 0.96 ± 0.01. Training using each individual lead produced low LVEF detection networks with AUCs within 0.07 of the networks trained using all 8 recorded ECG leads. Lead V6 produced the highest performing networks with a mean AUC of 0.91 ± 0.00, mean F1 score of 0.57 ± 0.02, mean sensitivity of 0.58 ± 0.05, and mean specificity of 0.96 ± 0.01. Lead I was not far behind with a slightly lower F1 score, sensitivity, and specificity. The numerical results are summarized in Table 1.
Table 1.
Low LVEF classification metrics for each network training scenario. For each lead used, 5 separate networks were trained to detect low LVEF. Metrics are reported as mean plus or minus one standard deviation.
| Lead Used | AUC | F1 Score | sensitivity | specificity |
|---|---|---|---|---|
| I | 0.91 ± 0.00 | 0.54 ± 0.00 | 0.56 ± 0.09 | 0.95 ± 0.02 |
| II | 0.89 ± 0.01 | 0.50 ± 0.01 | 0.56 ± 0.02 | 0.94 ± 0.01 |
| V1 | 0.88 ± 0.00 | 0.48 ± 0.01 | 0.50 ± 0.04 | 0.95 ± 0.01 |
| V2 | 0.86 ± 0.01 | 0.48 ± 0.01 | 0.55 ± 0.08 | 0.93 ± 0.03 |
| V3 | 0.87 ± 0.00 | 0.49 ± 0.01 | 0.57 ± 0.08 | 0.93 ± 0.02 |
| V4 | 0.88 ± 0.00 | 0.51 ± 0.01 | 0.57 ± 0.04 | 0.94 ± 0.01 |
| V5 | 0.90 ± 0.00 | 0.53 ± 0.01 | 0.58 ± 0.07 | 0.95 ± 0.02 |
| V6 | 0.91 ± 0.00 | 0.57 ± 0.02 | 0.58 ± 0.05 | 0.96 ± 0.01 |
| all | 0.93 ± 0.00 | 0.60 ± 0.02 | 0.62 ± 0.04 | 0.96 ± 0.01 |
Agreement between network outputs (unthresholded values) varied between networks trained with different leads. Figure 2 shows a heat map of the average correlation between network outputs for each network training scenario. A high value in this heat map indicates that, on average, networks trained with the two indicated lead sets produced outputs that agreed or were similar. A low value indicated that these networks produced outputs that were dissimilar. Excluding comparisons between networks trained on the same leads, we observed relatively high agreement (average correlation above 0.7) between leads V1 and V2, between leads V2 and V3, and between leads V5 and V6. Networks trained using leads I, V2, V3, V5, and V6 all showed a high average correlation of outputs (above 0.7) with networks trained using all leads.
Figure 2.
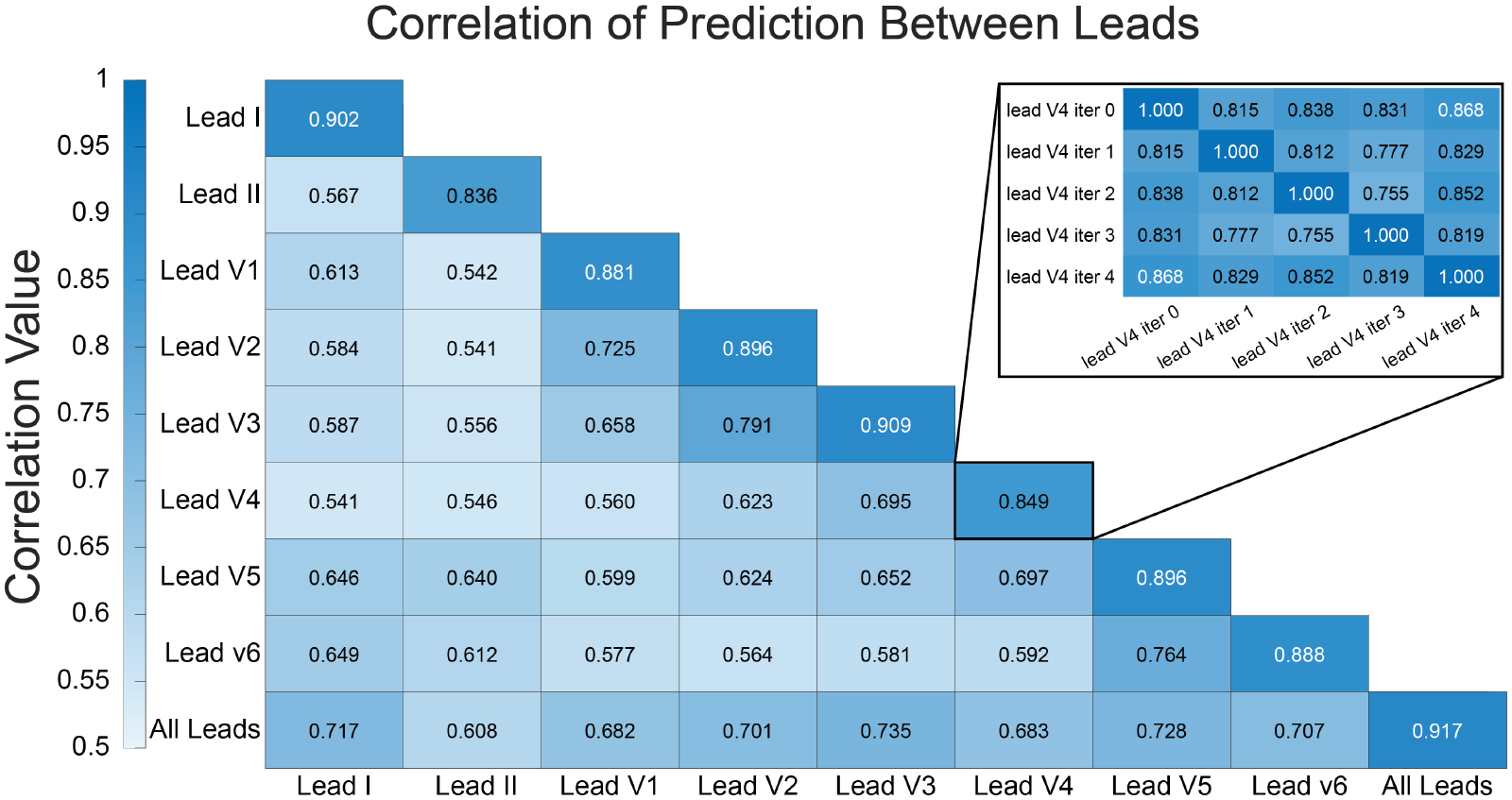
Average correlation of network outputs between each lead scenario. Each correlation in the larger heat map is an average of the correlations between the 5 instances of each lead-specific network. The identify correlations (along the diagonal for the same lead to same lead comparisons) were excluded from the average. The inset shows the individual per iteration comparisons for lead V4 compared to lead V4.
4. Discussion and Conclusions
In this study we report the detection of low LVEF using a custom residual-based ML architecture, and compare the performance of that network when trained using limited leads to using a full ECG lead set. We also compare the resulting network predictions across training scenarios. When trained using all 8 unique ECG leads of a 12-lead ECG, our novel residual-based ML architecture depicted in Figure 1 achieves network performance that matches contemporary published implementations according to AUC (average AUC of 0.93 over 5 instances).[3]
Low LVEF detection performance did not drop substantially when using only a single-lead compared to using all available leads for training and inference, with the largest drop in performance observed when using lead V2 (average AUC 0.86, average F1 0.48, average sensitivity 0.55, average specificity 0.93). In some cases such as when using lead I, the limited lead networks performed comparably to the full lead-set networks. With the rising popularity of wearable ECG devices, these results suggest that research should continue emphasize designing wearable ECGs that target recording these high performing leads – many of the most popular devices already record an ECG equivalent to lead I. The results here indicate that recordings from such wearable devices would be sufficient to accurately detect low LVEF.
In this study we also sought to compare the predictions for low LVEF classification both within networks trained on the same lead and between networks trained with different leads. Perhaps surprisingly, we found that for some leads such as V4, the agreement between repeated instances of the same network architecture trained on this lead produced predictions in the test set that agreed only with an average correlation of 0.849. These precordial leads are known to be subject to high variability in their placement on the chest, which may be an explanation for their poorer performance. Because these correlation metrics are based on unthresholded network outputs, assessment of network performance differences from this correlation is indirect. The actual performance of these networks with respect to false positive, false negative, true positive, and true negative rates may turn out to be minimally different once the outputs are thresholded, as evidenced by the low standard deviation on the other performance metrics (see Table 1). However, it is still note-worthy that the raw outputs show such variability between networks trained in the same manner.
Perhaps as expected, leads that are spatially co-located (V1 to V2, V2 to V3, etc) showed better agreement than those spaced more distantly. Identification of similarly performing leads can be an important first step in assessing an approach to apply contrastive and self-supervised learning techniques to ECG data by leveraging relative differences in the information content of each lead. Such techniques can then be used to leverage large unlabeled datasets for pretraining to improve ML performance on tasks with smaller dataset sizes. This lead comparison analysis may also provide useful insight into which leads are redundant, and allow for the design of a streamlined lead set for use in wearable ECG systems and future ML-ECG studies, where reduction in network size and computational cost is a key factor in determining feasibility for clinical translation. Future studies may also explore leveraging the disparate information of each lead to both optimize the design of ECG-Ml tools and possibly explore how they leverage information in the leads to produce clinical decisions or diagnoses.
Future studies may expand on this research by examining what features of these different individual leads contribute to the differences in detection of low LVEF. Low LVEF is not a feature detectable by traditional ECG analysis, yet ML techniques are able to predict low LVEF. Understanding which leads contain relevant information for that prediction is a first step proposed in this study, which should be followed up with an investigation on what information is present in those leads (i.e., ‘explainability’). Furthermore, this study was limited to investigation of low LVEF detection, and future applications of these same techniques to different clinical pathologies would provide further insight into selection of ideal ML approaches and leads for ML-ECG analysis.
Acknowledgments
Support for this research came from the Center for Integrative Biomedical Computing (www.sci.utah.edu/cibc), NIH/NIGMS grants P41 GM103545 and R24 GM136986, NIH/NIBIB grant U24EB029012, NIH/NHLBI grants T32HL007576 (to JAB), K23HL143156 (to BAS), F30HL149327 (to BZ) and the Nora Eccles Harrison Foundation for Cardiovascular Research.
References
- [1].Bergquist JA, Rupp L, Zenger B, Brundage J, Busatto A, MacLeod R. Body surface potential mapping: Contemporary applications and future perspectives. Hearts 2021;2:514–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rafie N, Kashou AH, Noseworthy PA. Ecg interpretation: Clinical relevance, challenges, and advances. Hearts 2021; 2(4):505–513. ISSN 2673–3846. [Google Scholar]
- [3].Jentzer JC, Kashou AH, Attia ZI, Lopez-Jimenez F, Kapa S, Friedman PA, Noseworthy PA. Left ventricular systolic dysfunction identification using artificial intelligence-augmented electrocardiogram in cardiac intensive care unit patients. International jJournal of Cardiology 3 2021; 326:114–123. ISSN 1874–1754. [DOI] [PubMed] [Google Scholar]
- [4].Trayanova NA, Popescu DM, Shade JK. Machine Learning in Arrhythmia and Electrophysiology. Circulation Research 2021;128(4):544–566. ISSN 15244571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mahayni AA, Attia ZI, Medina-Inojosa JR, Elsisy MF, Noseworthy PA, Lopez-Jimenez F, Kapa S, Asirvatham SJ, Friedman PA, Crestenallo JA, Alkhouli M. Electrocardiography-based artificial intelligence algorithm aids in prediction of long-term mortality after cardiac surgery. Mayo Clinic Proceedings 12 2021;96:3062–3070. ISSN 1942–5546. [DOI] [PubMed] [Google Scholar]
- [6].Bergquist JA, Zenger B, Brundage J, MacLeod RS, Ranjan R, Tasdizen T, Steinberg B. Performance of off-the-shelf machine learning arhitectures and biases in their performance in detection of low left ventircular ejection fraction. medRxiv 2023;. [DOI] [PMC free article] [PubMed] [Google Scholar]


