Abstract
The common octopus, Octopus vulgaris, is the first invertebrate species that was shown to possess two oxytocin/vasopressin (OT/VP) superfamily peptides, octopressin (OP) and cephalotocin (CT). Previously, we cloned a GPCR (G-protein-coupled receptor) specific to CT [CTR1 (CT receptor 1)]. In the present study, we have identified an additional CTR, CTR2, and a novel OP receptor, OPR. Both CTR2 and OPR include domains and motifs typical of GPCRs, and the intron– exon structures are in accord with those of OT/VP receptor genes. CTR2 and OPR expressed in Xenopus oocytes induced calcium-mediated inward chloride current in a CT- and OP-specific manner respectively. Several regions and residues, which are requisite for binding of the vertebrate OT/VP receptor family with their ligands, are highly conserved in CTRs, but not in OPR. These different sequences between CTRs and OPR, as well as the amino acid residues of OP and CT at positions 2–5, were presumed to play crucial roles in the binding selectivity to their receptors, whereas the difference in the polarity of OT/VP family peptide residues at position 8 confers OT and VP with the binding specificity in vertebrates. CTR2 mRNA was present in various peripheral tissues, and OPR mRNA was detected in both the nervous system and peripheral tissues. Our findings suggest that the CT and OP genes, similar to the OT/VP family, evolved through duplication, but the ligand–receptor selectivity were established through different evolutionary lineages from those of their vertebrate counterparts.
Keywords: cephalotocin, cloning, octopressin, G-protein-coupled receptor, genome structure
Abbreviations: CT, cephalotocin; CTR, cephalotocin receptor; GPCR, G-protein-coupled receptor; LSCPR, Lys-conopressin receptor; ORF, open reading frame; OP, octopressin; OPR, octopressin receptor; OT, oxytocin; OxyR, oxytocin receptor; RACE, rapid amplification of cDNA ends; RT, reverse transcriptase; TM, transmembrane; UTR, untranslated region; VP, vasopressin; V1aR, V1a receptor; V1bR, V1b receptor; V2R, V2 receptor
INTRODUCTION
Oxytocin (OT) and vasopressin (VP) are structurally related neurohypophysial peptide hormones in mammals. The major endocrine functions of OT are the contraction of the uterus in reproduction and milk ejection, and those of VP are the regulation of osmotic balance and contraction of smooth muscle cells in arteries [1,2]. The difference in the polarity of OT/VP family peptide residues at position 8 is believed to enable OT and VP to interact selectively with their respective receptors (Table 1) [3]. All vertebrate species, except for cyclostomes [4,5], contain at least one OT family peptide and one VP family peptide. It has been proposed that vertebrate OT and VP genes have evolved via duplication from a common ancestral gene. In contrast, only a single OT/VP superfamily peptide was isolated from proterostomes [6]. Consequently, the OT/VP family gene might have been present in the Archaemetazoa, a stem group from which both vertebrates and invertebrates diverged about 600 million years ago [6].
Table 1. Structures of OT/VP superfamily.
| Hormone | Sequence | Organism |
|---|---|---|
| Vertebrate OT family | ||
| OT | Cys-Tyr-Ile-Gln-Asn-Cys-Pro-Leu-Gly-NH2 | Mammal |
| Mesotocin | Cys-Tyr-Ile-Gln-Asn-Cys-Pro-Ile-Gly-NH2 | Marsupial, non-mammalian tetrapod, lungfish |
| Isotocin | Cys-Tyr-Ile-Ser-Asn-Cys-Pro-Ile-Gly-NH2 | Osteichthye (bony fish) |
| Vertebrate VP family | ||
| Arg-VP | Cys-Tyr-Phe-Gln-Asn-Cys-Pro-Arg-Gly-NH2 | Mammal |
| Lys-VP | Cys-Tyr-Phe-Gln-Asn-Cys-Pro-Lys-Gly-NH2 | Mammal |
| Vasotocin | Cys-Tyr-Ile-Gln-Asn-Cys-Pro-Arg-Gly-NH2 | Non-mammalian vertebrate, cyclostome (jawless fish) |
| Invertebrate OT/VP superfamily | ||
| Lys-conopressin | Cys-Phe-Ile-Arg-Asn-Cys-Pro-Lys-Gly-NH2 | Leech, geography cone (sea-snail), pond snail, sea hare, imperial cone (sea-snail) |
| Arg-conopressin | Cys-Ile-Ile-Arg-Asn-Cys-Pro-Arg-Gly-NH2 | Striped cone (sea-snail) |
| Diuretic hormone | Cys-Leu-Ile-Thr-Asn-Cys-Pro-Arg-Gly-NH2 | Locust |
| Annetocin | Cys-Phe-Val-Arg-Asn-Cys-Pro-Thr-Gly-NH2 | Earthworm |
| CT | Cys-Tyr-Phe-Arg-Asn-Cys-Pro-Ile-Gly-NH2 | Octopus |
| OP | Cys-Phe-Trp-Thr-Ser-Cys-Pro-Ile-Gly-NH2 | Octopus |
OT/VP family peptide receptors are typical members of the rhodopsin-type (class I) GPCRs (G-protein-coupled receptors). To date, four vertebrate OT/VP receptors have been identified. The OT receptor (OxyR) is selective for OT, and the V1a, V1b and V2 receptors (V1aR, V1bR, and V2R) are all selective for VP. OxyR, V1aR and V1bR, coupled to Gq/11, activate the inositol 1,4,5-trisphosphate–calcium signal transduction cascade [7–9], whereas V2R is coupled to adenylate cyclase via G-proteins followed by production of cAMP [10]. Although many GPCR genes do not contain an intron, the OxyR, V1aR, V1bR, and V2R genes [7–10] contain an intron at the same location between the sixth and seventh transmembrane (TM) domains.
Octopuses are one of the most advanced invertebrates in the aspects of intelligence, sensory systems and physical ability [11], suggesting that octopuses have some unique neuropeptidergic regulation comparable with those of vertebrates. In fact, we have characterized two members of the OT/VP superfamily peptide, namely cephalotocin (CT) [2] and octopressin (OP) [12], in Octopus vulgaris. The high conservation of sequence and gene organization between OP and CT suggests that they occurred via gene duplication [13]. Moreover, we have cloned CTR1, a CT receptor (CTR) from octopus brain, which is specific to CT, apart from an isoleucine residue conserved in OP and CT at position 8 (Table 1) [14], allowing us to presume that octopus possess more than two receptors specific for OP and/or CT.
In the present study, we report the sequence, activity and localization of a CT receptor subtype (CTR2) and an OP receptor (OPR) and genome structure of those genes. Our results suggest unprecedented structural evolution of invertebrate OT/VP superfamily peptides and ligand–receptor selectivity.
EXPERIMENTAL
Animals
Adult octopuses (approx. 2 kg), Octopus vulgaris, were purchased from a local fish shop, and kept in artificial seawater at 18 °C.
Total RNA and mRNA preparation
Total RNA was extracted from various tissues using Sepasol-RNA I Super (Nacalai Tesque, Kyoto, Japan) and mRNA was prepared using an Oligotex™-dT30 mRNA Purification Kit (Takara Bio, Kyoto, Japan) according to the manufacturer's instructions.
Oligonucleotide primers
Oligonucleotide primers were purchased from Qiagen (Tokyo, Japan) and Proligo (Kyoto, Japan). Degenerate primers, OP/CT-R TM6N, OP/CT-R TM7N and OP/CT-R TM7a-2, were designed based on sequences in the sixth and seventh TM domains respectively, both of which are conserved among the receptors of the OT/VP superfamily in vertebrates, CTR1 and the Lys-conopressin receptors (LSCPR1 and LSCPR2). All primers used are listed in the supplementary data (http://www.BiochemJ.org/bj/387/bj3870085add.htm).
Cloning of the partial-length cDNA
All PCR amplifications were carried out in a reaction mixture containing Ex Taq™ polymerase (Takara Bio) and 200 μM dNTPs in a PerkinElmer GeneAmp PCR System 9700 thermal cycler (Applied Biosystems Japan, Tokyo, Japan). Total RNA was isolated from various tissues of octopus and reverse-transcribed using Oligo (dT)12–18 primer and Superscript II supplied in the SuperScript Choice System (Invitrogen, Groningen, The Netherlands). The first PCR was performed using OP/CT-R TM6N and OP/CT-R TM7a-2 under the following conditions: 5 min at 94 °C, 40 cycles of 30 s at 94 °C, 30 s at 50 °C, 1 min at 72 °C (7 min for the last cycle). The second PCR was performed using OP/CT-R TM6N and OP/CT-R TM7aN under the following conditions: 94 °C for 5 min, 30 cycles of 30 s at 94 °C, 30 s at 50 °C, 1 min at 72 °C (7 min for the last cycle). PCR products separated by 1.5% agarose gel electrophoresis were purified using a Qiaquick™ Gel Extraction kit (Qiagen), and subcloned into the pDrive cloning vector using the Qiagen PCR cloning kit according to the manufacturer's instructions. Subcloned inserts were sequenced on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems Japan) using a Big-Dye sequencing kit version 1.1 (Applied Biosystems Japan) and analysed using GENETYX-MAC software (Software Development, Tokyo, Japan). Universal M13 primers or gene-specific primers were used to sequence both strands.
3′-RACE (rapid amplification of cDNA ends) and 5′-RACE
The transcriptional start site was determined by oligo-capping RACE methods by use of the GeneRacer kit (Invitrogene). First strand cDNA was synthesized from mRNA with the GeneRacer Oligo dT Primer supplied in the GeneRacer kit (Invitrogene) according to the manufacturer's instructions. The first PCR was performed using the GeneRacer 3′ primer and CTR2 3′-1F for CTR2 cDNA (or OPR 3′-1F for OPR cDNA) (for primer sequences, see http://www.BiochemJ.org/bj/387/bj3870085add.htm) under the following conditions: 5 min at 94 °C, 35 cycles of 30 s at 94 °C, 30 s at 52 °C, 2 min at 72 °C (7 min for the last cycle). The second PCR was performed using the GeneRacer 3′ nested primer and CTR2 3′-2F (or OPR 3′-2F) under the following conditions: 94 °C for 5 min, 35 cycles of 30 s at 94 °C, 30 s at 52 °C, 2 min at 72 °C (7 min for the last cycle). The second PCR products were subcloned and sequenced as described above. The 5′-ends of the cDNAs were determined as follows: the first template was amplified using the GeneRacer 5′ primer and CTR2 5′-1R (or OPR 5′-1R). Each of the first PCR products was re-amplified using the GeneRacer 5′ nested primer and CTR2 5′-2R (or OPR 5′-2R). Both first and second PCR reactions were performed for 5 min at 94 °C, 35 cycles for 30 s at 94 °C, 30 s at 52 °C, and 2 min at 72 °C (7 min for the last cycle). The second PCR products were subcloned and sequenced as described above.
RT (reverse transcriptase)-PCR and Southern blot analysis
Each of total RNAs (1 μg) extracted from various tissues was reverse transcribed using SuperScript Choice System (Invitrogene). The PCR was performed using CTR2 Fw1 and CTR2 Rv1 for CTR2 (or OPR 3′-1F and OPR Rv1 for OPR) (for primer sequences, see http://www.BiochemJ.org/bj/387/bj3870085add.htm) under the following conditions: 5 min at 94 °C, 35 cycles of 30 s at 94 °C, 30 s at 52 °C, 1 min at 72 °C (7 min for the last cycle). The PCR products were separated by 1.5% agarose gel electrophoresis, and then transferred on to Hybond-N+ membrane (Amersham Bioscience, Piscataway, NJ, U.S.A.) and cross-linked by UV irradiation. Digoxigenin-labelled oligonucleotide probes, digoxigenin–CTR2 Fw2 for CTR2 (or OPR 3′-2F for OPR), were hybridized with the CTR2 cDNA or the OPR cDNA respectively at 50 °C. The methods for detection have been described previously [14]. The expression of octopus β-actin (AB053937) was also tested as an internal control.
Expression of receptor in Xenopus oocytes
The ORF (open reading frame) region of the orphan receptor cDNAs were amplified and inserted into a pSP64 poly(A) vector (Promega, Madison, WI, U.S.A.). The plasmid was linearized with EcoRI, and cRNA was prepared using SP6 RNA polymerase (Ambion, Austin, TX, U.S.A.). The methods for assay were the same as those described previously [14]. OP and CT were synthesized with a solid-phase peptide synthesizer using standard Fmoc™ chemistry (Model 433A, Applied Biosystems Japan). Peptides of the OT/VP superfamily were purchased from Bachem (Bubendorf, Switzerland).
Determination of the intron–exon structure and promoter region
High-molecular-mass DNA from the Octopus vena cava was isolated using a Qiagen Genomic-tip system according to the manufacturer's instructions. A genomic DNA library was constructed using the GenomeWalker Universal kit (BD Biosciences Clonthech, Tokyo, Japan). Octopus genomic DNA digested with DraI, EcoRV, PvuII and StuI was subjected to PCR amplification in the 5′ and 3′ directions using adaptor primers and gene-specific primers according to the manufacturer's instructions. The amplified products were subcloned and sequenced using several gene-specific primers.
RESULTS
Cloning and structure of the putative GPCRs
The sixth and seventh TM domains are highly conserved among known receptors of the OT/VP superfamily in vertebrates, as well as invertebrates. To identify receptors for OP and CT in octopus, three degenerate primers were designed based on the conserved regions, and RT-PCRs were performed between these primers. BLAST searches of the two PCR products showed the highest homology of the resultant fragments with OT/VP superfamily receptors. A full-length cDNA sequence (2130 bp) encoding the CTR2 was determined by the 5′-/3′-RACE methods from the octopus branchial vessel. CTR2 was shown to comprise an ORF of 426 amino acids and 395 bp of 5′-UTR (untranslated region) and 544 bp of 3′-UTR. The other full-length cDNA sequence (1833 bp) encoding the OPR was also determined using the same method from the salivary gland, revealing that OPR has an ORF of 419 amino acids flanked by 220 bp of 5′-UTR and 351 bp of 3′-UTR.
As shown in Figure 1, both CTR2 and OPR contain the seven hydrophobic TM regions and several potential sites for N-linked glycosylation and phosphorylation that are the most typical characteristic of GPCRs. Consensus sequences for N-linked glycosylation sites (Asn-Xaa-Ser/Thr; two sites in CTR2, three sites in OPR) were found in the extracellular N-terminal domain; and consensus sequences for phosphorylation by protein kinase A (Lys/Arg-Xaa-(Xaa)-Ser/Thr; twelve sites in CTR2, thirteen sites in OPR), by protein kinase C (Ser/Thr-Xaa-Lys/Arg; eight sites in CTR2, four sites in OPR), and by protein kinase CK2 (Ser/Thr-Xaa-Xaa-Asp/Glu; two sites in CTR2, one site in OPR). These are located exclusively in the third intracellular loop and the C-terminus, suggesting that phosphorylation is involved in the modulation of G-protein coupling and receptor function [15]. An aspartate in TM2 (Asp98 in CTR2, Asp80 in OPR) and the consensus tripeptide (Glu/Asp-Arg-Tyr and Asp-Arg-Tyr in CTR2 and OPR) at the interface of TM3, both of which are believed to play a pivotal role in the receptor activation in the class I GPCR family, were also conserved. Two cysteine residues responsible for a disulphide bridge in most GPCRs were present (at positions 119 and 198 in CTR2, at positions 107 and 182 in OPR) in the first and second extracellular loops in both the putative octopus GPCRs. These results indicated that CTR2 and OPR belong to the class I GPCR family.
Figure 1. Sequence alignment of CTR2, OPR and receptors of the OT/VP superfamily.
The amino acid sequence of CTR2 and OPR are aligned with those of the Octopus CTR1 [14], Lymnaea LSCPR1 and LSCPR2 [16,17], and the rat receptors (OxyR, V1aR and V2R [7,8,10]). Amino acid residues that are identical in all receptors are indicated by an asterisk. N-linked glycosylation sites are underlined. Potential phosphorylated serine or threonine residues are marked by open circles. Amino acid residues in shaded boxes are believed to play a pivotal role in the GPCR activation, and those in boxes are highly conserved in the vertebrate receptors of the OT/VP superfamily, but not in other GPCRs, and have been suggested to be important in receptor–ligand interactions [19]. Residues in the agonist-binding pocket, as described in the text, are shown by arrows. Horizontal lines above the sequnce indicate the seven putative TM domains. The nucleotide sequences have been deposited into the DDBJ, EMBL and GeneBank® databases under the accession numbers: CTR2, AB112347; OPR, AB116233.
Comparative amino acid sequence between the octopus receptors and OT/VP receptor family
The total amino acid sequence of the CTR2 is 33.1–48.1% homologous to the sequences of other OT/VP receptor family members, and OPR is 24.9–40.5% homologous (Table 2). The sequence of similarity of CTR2 and OPR to the OT receptor family is not so different from their similarity to VP receptors. When combined with these findings, the sequence analysis revealed that CTR2 and OPR are octopus counterparts for the OT/VP receptors. Several residues in the TM and extracellular domains are known to play a crucial role in the binding of the OT/VP receptors to their ligands. Five glutamine residues in TM domains 2, 3, 4 and 6 (at positions 92, 96, 119, 171 and 294 in the rat OxyR) and a lysine residue localized in TM3 (Lys128 in the rat V1R) are known to play a crucial role in the binding of the OT/VP receptors to their ligands [18]. Lys128 in the V1aR is proposed to interact with the glutamine residue at position 4 in VP. These residues are highly conserved in CTR1 and CTR2, but not in OPR. In OPR, three glutamine and lysine residues are substituted by histidine, threonine, leucine and glutamine residues. The sequences Phe-Gln-Val-Leu-Pro-Gln-Leu at the C-terminal end of TM2, Gly-Pro-Asp in the first extracellular loop, Asp-Cys-Trp-Ala and Pro-Trp-Gly in the second extracellular loop in the OT/VP receptors (Figure 1), which are presumed to have a crucial role in ligand binding [19], are substituted by Phe-Asn-Ile-Leu-Pro-Gln-Leu, Ala-Gly-Asp, Asp-Cys-Trp-Val and Ala-Trp-Val, and Phe-His-Ile-Leu-Pro-Thr-Ile, Gly-Asp-Ile, Leu-Cys-Arg-Pro and Glu-Leu-Ala sequences in the corresponding regions of CTR2 and OPR respectively.
Table 2. Total amino acid sequence homology of CTR2 and OPR to other OT/VP superfamily receptors.
VTR, vasotocin receptor; MTR, mesotocin receptor; ITR, isotocin receptor.
| (a) | |
|---|---|
| Receptor | Homology compared with CTR2 (%) |
| CTR1 | 40.2 |
| OPR | 33.1 |
| LSCPR1 | 41.7 |
| LSCPR2 | 48.1 |
| Rat OxyR | 34.8 |
| Rat V1aR | 37.8 |
| Rat V1bR | 37.3 |
| Rat V2R | 35.5 |
| Chicken VTR | 34.3 |
| Toad MTR | 38.4 |
| White sucker ITR | 38.3 |
| White sucker VTR | 40.9 |
| (b) | |
| Receptor | Homology compared with OPR (%) |
| CTR1 | 38.1 |
| CTR 2 | 33.1 |
| LSCPR1 | 40.5 |
| LSCPR2 | 31.5 |
| Rat OxyR | 30.6 |
| Rat V1aR | 31.5 |
| Rat V1bR | 30.1 |
| Rat V2R | 24.9 |
| Chicken VTR | 28.8 |
| Toad MTR | 29.9 |
| White sucker ITR | 31.5 |
| White sucker VTR | 28.4 |
Intron–exon structure of the CTR1, CTR2 and OPR genes
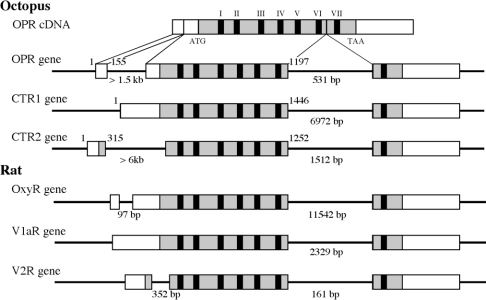
Although many GPCR genes do not contain any introns, the OxyR, V1aR, V1bR and V2R genes [7–10] contain an intron at the same location between TM6 and TM7. Thus we determined the intron–exon structure of CTR1, CTR2 and OPR. The CTR1 gene consists of two exons and one intron with 6972 bp between TM6 and TM7 (Figure 2), which occurred at position 1446–1467 in the CTR1 cDNA. Both OPR and CTR2 genes comprise three exons and two introns. The introns longer than 1.5 kb and 531 bp occurred at positions 155–156 and 1197–1978 in the OPR cDNA. The introns longer than 6 kb and 1512 bp occurred at positions 315–316 and 1252–1253 in the CTR2 cDNA. CTR2 and OPR genes were also found to harbour an intron between TM6 and TM7. This comparative analysis of intron–exon structure suggests that the mammalian OT/VP receptor gene and the octopus CTR1, CTR2 and OPR genes were derived from a common ancestral gene. In addition, the sequence around the intron–exon junctions of these receptor genes was in accordance with the GT–AG consensus splicing site.
Figure 2. Structure organization of the Octopus CTR 1, CTR2 and OPR genes compared with the rat OT, V1a and V2 receptor genes.
Exons are indicated by the open boxes, ORFs are in the shaded boxes, and TMs are black boxes. The gene sequences have been deposited into the DDBJ, EMBL and GeneBank® databases under the accession numbers: OPR gene, AB158496 and AB158497; CTR1 gene, AB158493; CTR2 gene, AB158494 and AB158495.
Functional expression of the cloned receptors in Xenopus oocytes
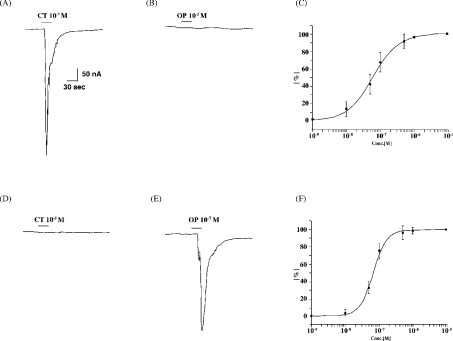
To evaluate binding affinity and selectivity of CTR2 and OPR to CT and OP, each of these receptors was expressed in Xenopus oocytes, since this system has been employed for functional analysis of OT/VP receptors other than V2 [7–10]. The voltage-clamped oocytes expressing CTR2 or OPR displayed typical inward membrane currents upon application of CT at 100 nM (Figure 3A) or of OP at 100 nM (Figure 3E). In contrast, OP failed to trigger current even at levels higher than 10 μM on CTR2 (Figure 3B). Similarly, OPR showed no response to CT (Figure 3D). The EC50 for CT on CTR2 and OP on OPR were estimated to be 67.2±8.9 nM and 68.2±9.3 nM respectively from the dose–response curves (Figures 3C and 3F). We also tested vertebrate and other invertebrate OT/VP superfamily peptides, such as OT, mesotocin, isotocin, VP, vasotocin, annetocin, Lys-conopressin or Arg-conopressin, but they had no effect, even at levels higher than 10 μM (results not shown).
Figure 3. Functional expression of CTR2 and OPR in Xenopus oocytes.
The ligands were applied during the period indicated by the black bar. (A, B) Traces of membrane current induced by CT at 100 nM and OP at 10 μM in an oocyte expressing CTR2. (D, E) Traces of membrane currents induced by CT at 10 μM and OP at 100 nM in an oocyte expressing OPR. (C, F) Dose–response curve over the concentration range 1 nM to 10 μM CT (C) or OP (F) in CTR2 and OPR. Maximum membrane currents elicited by the ligands are plotted. The results are expressed as the means±S.E.M. (n=5).
Localization of CTR1, CTR2 and OPR mRNA
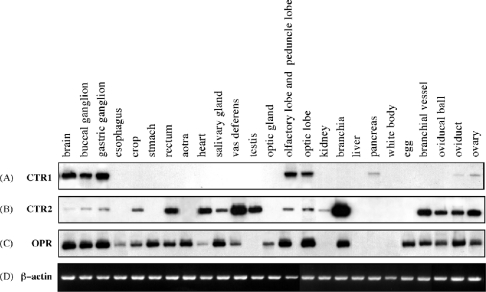
Southern blot analysis of RT-PCR products for CTR1, CTR2 and OPR was performed to detect the tissue distribution of CTR1, CTR2 and OPR mRNAs in the central and peripheral nervous systems, and in several peripheral tissues of Octopus (Figures 4A–4D). The CTR1 transcripts were highly expressed in central and peripheral nervous systems, whereas relatively low expression was seen in pancreas, oviduct and ovary. CTR2 transcripts were present abundantly in the various peripheral tissues, but faintly in the nervous systems. OPR transcripts were widely expressed in both the nervous system and the peripheral tissues.
Figure 4. Southern blot analysis of RT-PCR products for CTR1 (A), CTR2 (B), OPR (C) and β-actin (D) transcripts isolated from octopus tissues.
DISCUSSION
Previously, we showed that the common octopus, Octopus vulgaris, has two members of the OT/VP superfamily, OP and CT [12]. The distinct biological activities of OP and CT [12] were indicative of the presence of receptors specific for each peptide. Indeed, the octopus cephalotocin receptor, CTR1, exhibited binding specificity for CT, but not for OP. Moreover, the presence of a subtype of CTR1 was supported by the fact that mammalian VP receptors and Lymnaea LSCPRs constitute the subfamily [7–10,16,17]. In the present study, we have cloned two novel receptors of OP and CT from the octopus. Comparative sequence analysis revealed that CTR2 and OPR share high similarity to the OT/VP receptors. OPR, CTR1, and CTR2 genes, similar to all the vertebrate receptors, have an intron between the sixth and seventh TM domains (Figure 2). Furthermore, functional analysis of CTR2 and OPR expression in the Xenopus oocytes provided evidence that CTR2 and OPR exerted specific activation by CT and OP respectively (Figure 3). These results lead to the conclusion that CTR2 and OPR are receptors specific for CT and OP respectively, and that these octopus receptors share a common ancestor with the vertebrate OT/VP receptors.
Although the differences in the polarity of amino acid residues at position 8 are believed to confer on OT and VP families the binding selectivity for their respective receptors [3], both annetocin (threonine at position 8) and Lys-conopressin (lysine at position 8) induced egg-laying-like behaviour in the leech, Whitmania pigra, to a similar degree [20], and activated annetocin receptor [21]. In the present study, we found unique structure–activity relationships between OP and CT. Amino acid residues at positions 2–5 are completely different, and the isoleucine at position 8 is conserved between OP and CT (Table 1). Nevertheless, CTR1, CTR2 and OPR exerted high specific activation by CT and OP respectively (Figure 3) [14]. These data revealed that the difference in the polarity of the amino acid at position 8 is not responsible for the binding specificity of OP and CT for these receptors. Five glutamine residues in TM domains 2, 3, 4 and 6 and the lysine residue localized in TM3, which are requisite for the interaction of OT and VP with their receptors [18], are highly conserved in the OT/VP receptors, CTR1, and CTR2, whereas three glutamine and lysine residues are substituted by different residues in OPR. The residue at position 3 is a phenylalanine or isoleucine, position 4 is a glutamine or arginine, and position 5 is an asparagine residue in most OT/VP family peptides, whereas they are tryptophan, threoine and serine residues in OP. The substitution of amino acid residues suggests that the ligand–receptor binding mode for OP and OPR differs from other OT/VP superfamily peptides and receptors. The sequences Phe-Gln-Val-Leu-Pro-Gln-Leu, Gly-Pro-Asp, Asp-Cys-Trp-Ala and Pro-Trp-Gly in the vertebrate OT/VP receptors (Figure 1), which are proposed to play an important role in ligand binding [19], are substituted by Phe-Asn-Ile-Leu-Pro-Gln-Leu, Ala-Gly-Asp, Asp-Cys-Trp-Val and Ala-Trp-Val, and Phe-His-Ile-Leu-Pro-Thr-Ile, Gly-Asp-Ile, Leu-Cys-Arg-Pro and Glu-Leu-Ala sequences in the corresponding regions of CTR2 and OPR respectively. Such amino acid substitutions were also found in other invertebrate OT/VP receptors, including CTR1, annetocin receptor, LSCPR1 and LSCPR2. Moreover, these invertebrate receptors are not activated by vertebrate OT/VP family peptides [14,16,17,21]. Consequently, it is suggested that the substitution of amino acid residues in those receptors is consistent with the binding specificity of octopus OT/VP superfamily peptides to their receptors, which is dependent on amino acid residues of the peptide between position 2 and 5 of OP and CT, unlike those of vertebrate counterparts, and binding specificity of invertebrate OT/VP superfamily peptides to their receptors has been established in the evolutionary lineage distinct from those of vertebrates.
OPR widely distributed in both the nervous systems and peripheral tissues (Figure 4C). The distribution of OPR in peripheral tissues is in agreement with our previous study, given that OP evoked rhythmic contractions with increased tonus in the rectum, oviduct, and efferent branchial vessel, rhythmic contractions in the spermatophoric gland, and tonic contractions in the ring slice of the anterior aorta in a previous study [12]. Therefore, a major physiological role for OP is contractile action of various tissues though OPR.
Expression of CTR1 mRNA was limited in the central and peripheral nervous systems and tissues associated with reproduction [14]. On the other hand, CTR2 mRNA was mainly distributed in peripheral tissues, although CT did not induce contractile activities on the smooth muscles of several Octopus tissues (Figures 4A and 4B). These expression profiles indicate two major physiological roles of CTR2-medicated CT, namely, ammonia excretion and production/secretion of steroid hormones. Arg-vasotocin caused prominent transient increases in urea excretion from branchia of toadfish, and Arg-vasotocin receptor mRNA was expressed in the branchia [22,23]. Furthermore, ammonia is excreted from the branchia of octopus [24]. Consequently, it can be presumed that CT is involved in the control of ammonia excretion via CTR2 expressed in the branchia. In mammals, V1aR has been identified in the epididymis, vas deferens, Leydig cells and Leydig-derived cell lines, and the presence of VP has been reported in the Leydig cells of several mammals, suggesting that VP regulates testosterone production [25–29]. These findings, combined with the abundant expression of CTR2 in the vas deferens and testis, support a notion that CT participates in production and/or secretion of the steroid hormones via CTR2.
The OP and CT mRNA were distributed in different regions of the octopus brain [12,13], and OP stimulated contractions in various tissues for which CT is devoid of any contractile activity [12]. Such distinct localization and physiological action suggest that OP and CT gene expressions are controlled by different transcriptional factors. Therefore also of interest is the transcriptional regulation of the OP and CT genes. Transcription of OT and VP gene is positively and negatively regulated by distinct factors: EREs (oestrogen-responsive elements) and HREs (hormone-response elements) are involved in the promoter regions of the OT gene, whereas CRE (cAMP-response element), AP-2 (activator protein-2) and GRE (glucocorticoid-response element) are found in the promoter regions of the VP gene [2,30]. We found that the promoter regions of OP and CT genes have several putative regulatory elements, such as GRE-, CRE- and AP-2-binding sites located in the VP promoter, whereas ERE and HRE sites, which are typical for the OT gene promoter, are absent in both promoters (A. Kanda, H. Satake, T. Kawada and H. Minakata, unpublished work). Whether only these VP gene-like or additional novel elements are involved in the distinct transcriptional regulation is now being examined.
In conclusion, we have cloned octopus OP and CT receptors specific for OP and CT respectively, and noted the differences in the tissue distribution of CTR1, CTR2 and OPR mRNA, suggesting that octopus established the advanced physiological functions through regulation by OP and CT. The octopus OT/VP gene also evolved through duplication, whereas the ligand–receptor selectivities were established through different evolutionary lineages to those of their vertebrate counterparts. The analysis of not only the sequences of peptides and receptors, but also their ligand–receptor binding mode, revealed unprecedented molecular and functional evolution of the OT/VP superfamily in invertebrates.
Online data
Acknowledgments
This work was funded in part by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science to H.M. (12640669, 15207007).
References
- 1.Cunningham E. T., Jr, Sawchenko P. E. Reflex control of magnocellular vasopressin and oxytocin secretion. Trends Neurosci. 1991;14:406–411. doi: 10.1016/0166-2236(91)90032-p. [DOI] [PubMed] [Google Scholar]
- 2.Gimpl G., Fahrenholz F. The oxytocin receptor system: structure, function, and regulation. Physiol. Rev. 2001;81:629–683. doi: 10.1152/physrev.2001.81.2.629. [DOI] [PubMed] [Google Scholar]
- 3.Barberis C., Mouillac B., Durroux T. Structural bases of vasopressin/oxytocin receptor function. J. Endocrinol. 1998;156:223–229. doi: 10.1677/joe.0.1560223. [DOI] [PubMed] [Google Scholar]
- 4.Lane T. F., Sower S. A., Kawauchi H. Arginine vasotocin from the pituitary gland of the lamprey (Petromyzon marinus): isolation and amino acid sequence. Gen. Comp. Endocrinol. 1988;70:152–157. doi: 10.1016/0016-6480(88)90104-9. [DOI] [PubMed] [Google Scholar]
- 5.Heierhorst J., Lederis K., Richter D. Presence of a member of the Tc1-like transposon family from nematodes and Drosophila within the vasotocin gene of a primitive vertebrate, the Pacific hagfish Eptatretus stouti. Proc. Natl. Acad. Sci. U.S.A. 1992;89:6798–6802. doi: 10.1073/pnas.89.15.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoyle C. H. Neuropeptide families: evolutionary perspectives. Regul. Pept. 1998;73:1–33. doi: 10.1016/s0167-0115(97)01073-2. [DOI] [PubMed] [Google Scholar]
- 7.Rozen F., Russo C., Banville D., Zingg H. H. Structure, characterization, and expression of the rat oxytocin receptor gene. Proc. Natl. Acad. Sci. U.S.A. 1995;92:200–204. doi: 10.1073/pnas.92.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morel A., O'Carroll A. M., Brownstein M. J., Lolait S. J. Molecular cloning and expression of a rat V1a arginine vasopressin receptor. Nature (London) 1992;356:523–526. doi: 10.1038/356523a0. [DOI] [PubMed] [Google Scholar]
- 9.Saito M., Sugimoto T., Tahara A., Kawashima H. Molecular cloning and characterization of rat V1b vasopressin receptor: evidence for its expression in extra-pituitary tissues. Biochem. Biophys. Res. Commun. 1995;212:751–757. doi: 10.1006/bbrc.1995.2033. [DOI] [PubMed] [Google Scholar]
- 10.Lolait S. J., O'Carroll A. M., McBride O. W., Konig M., Morel A., Brownstein M. J. Cloning and characterization of a vasopressin V2 receptor and possible link to nephrogenic diabetes insipidus. Nature (London) 1992;357:336–339. doi: 10.1038/357336a0. [DOI] [PubMed] [Google Scholar]
- 11.Young J. Z. Multiple matrices in the memory system of Octopus. In: Abbott N. J., Williamson R., Maddock L., editors. Cephalopod Neurobiology. Oxford: Oxford University Press; 1995. [Google Scholar]
- 12.Takuwa-Kuroda K., Iwakoshi-Ukena E., Kanda A., Minakata H. Octopus, which owns the most advanced brain in invertebrates, has two members of vasopressin/oxytocin superfamily as in vertebrates. Regul. Pept. 2003;115:139–149. doi: 10.1016/s0167-0115(03)00151-4. [DOI] [PubMed] [Google Scholar]
- 13.Kanda A., Takuwa-Kuroda K., Iwakoshi-Ukena E., Minakata H. Single exon structures of the oxytocin/vasopressin superfamily peptides of octopus. Biochem. Biophys. Res. Commun. 2003;309:743–748. doi: 10.1016/j.bbrc.2003.08.061. [DOI] [PubMed] [Google Scholar]
- 14.Kanda A., Takuwa-Kuroda K., Iwakoshi-Ukena E., Furukawa Y., Matsushima O., Minakata H. Cloning of Octopus cephalotocin receptor, a member of the oxytocin/vasopressin superfamily. J. Endocrinol. 2003;179:281–291. doi: 10.1677/joe.0.1790281. [DOI] [PubMed] [Google Scholar]
- 15.Bockaert J., Pin J. P. Molecular tinkering of G-protein-coupled receptors: an evolutionary success. EMBO J. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Kesteren R. E., Tensen C. P., Smit A. B., van Minnen J., van Soest P. F., Kits K. S., Meyerhof W., Richter D., van Heerikhuizen H., Vreugdenhil E., Geraerts W. P. M. A novel G-protein-coupled receptor mediating both vasopressin- and oxytocin-like functions of Lys-conopressin in Lymnaea stagnalis. Neuron. 1995;15:897–908. doi: 10.1016/0896-6273(95)90180-9. [DOI] [PubMed] [Google Scholar]
- 17.van Kesteren R. E., Tensen C. P., Smit A. B., van Minnen J., Kolakowski L. F., Meyerhof W., Richter D., van Heerikhuizen H., Vreugdenhil E., Geraerts W. P. M. Co-evolution of ligand–receptor pairs in the vasopressin/oxytocin superfamily of bioactive peptides. J. Biol. Chem. 1996;271:3619–3626. doi: 10.1074/jbc.271.7.3619. [DOI] [PubMed] [Google Scholar]
- 18.Mouillac B., Chini B., Balestre M. N., Elands J., Trumpp-Kallmeyer S., Hoflack J., Hibert M., Jard S., Barberis C. The binding site of neuropeptide vasopressin V1a receptor. Evidence for a major localization within transmembrane regions. J. Biol. Chem. 1995;270:25771–25777. doi: 10.1074/jbc.270.43.25771. [DOI] [PubMed] [Google Scholar]
- 19.Sharif M., Hanley M. R. Peptide receptors. Stepping up the pressure. Nature (London) 1992;357:279–280. doi: 10.1038/357279a0. [DOI] [PubMed] [Google Scholar]
- 20.Fujino Y., Nagahama T., Oumi T., Ukena K., Morishita F., Furukawa Y., Matsushima O., Ando M., Takahama H., Satake H., et al. Possible functions of oxytocin/vasopressin-superfamily peptides in annelids with special reference to reproduction and osmoregulation. J. Exp. Zool. 1999;284:401–406. doi: 10.1002/(sici)1097-010x(19990901)284:4<401::aid-jez6>3.3.co;2-l. [DOI] [PubMed] [Google Scholar]
- 21.Kawada T., Kanda A., Minakata H., Matsushima O., Satake H. Identification of a novel receptor for an invertebrate oxytocin/vasopressin superfamily peptide: molecular and functional evolution of the oxytocin/vasopressin superfamily. Biochem. J. 2004;382:231–237. doi: 10.1042/BJ20040555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahlmann S., Meyerhof W., Hausmann H., Heierhorst J., Schonrock C., Zwiers H., Lederis K., Richter D. Structure, function, and phylogeny of [Arg8]vasotocin receptors from teleost fish and toad. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1342–1345. doi: 10.1073/pnas.91.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry S. F., Montpetit C. J., McKendry J., Desforges P. R., Gilmour K. M., Wood C. M., Olson K. R. The effects of endothelin-1 on the cardiorespiratory physiology of the freshwater trout (Oncorhynchus mykiss) and the marine dogfish (Squalus acanthias) J. Comp. Physiol. 2001;171:623–634. doi: 10.1007/s003600100213. [DOI] [PubMed] [Google Scholar]
- 24.Potts W. T. Ammonia extraction in octopus defleini. Comp. Biochem. Physiol. 1965;14:339–355. doi: 10.1016/0010-406x(65)90209-4. [DOI] [PubMed] [Google Scholar]
- 25.Ivell R., Hunt N., Hardy M., Nicholson H., Pickering B. Vasopressin biosynthesis in rodent Leydig cells. Mol. Cell. Endocrinol. 1992;89:59–66. doi: 10.1016/0303-7207(92)90211-n. [DOI] [PubMed] [Google Scholar]
- 26.Maggi M., Malozowski S., Kassis S., Guardabasso V., Rodbard D. Identification and characterization of two classes of receptors for oxytocin and vasopressin in porcine tunica albuginea, epididymis, and vas deferens. Endocrinology. 1987;120:986–994. doi: 10.1210/endo-120-3-986. [DOI] [PubMed] [Google Scholar]
- 27.Meidan R., Hsueh A. J. Identification and characterization of arginine vasopressin receptors in the rat testis. Endocrinology. 1985;116:416–423. doi: 10.1210/endo-116-1-416. [DOI] [PubMed] [Google Scholar]
- 28.Ascoli M., Pignataro O. P., Segaloff D. L. The inositol phosphate/diacylglycerol pathway in MA-10 Leydig tumor cells. Activation by arginine vasopressin and lack of effect of epidermal growth factor and human choriogonadotropin. J. Biol. Chem. 1989;264:6674–6681. [PubMed] [Google Scholar]
- 29.Adashi E. Y., Hsueh A. J. Direct inhibition of testicular androgen biosynthesis revealing antigonadal activity of neurohypophysial hormones. Nature (London) 1981;293:650–652. doi: 10.1038/293650a0. [DOI] [PubMed] [Google Scholar]
- 30.Burbach J. P. Regulation of gene promoters of hypothalamic peptides. Front. Neuroendocrinol. 2002;4:342–369. doi: 10.1016/s0091-3022(02)00005-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.