Abstract
In humans, a polymorphic gene encodes the drug-metabolizing enzyme NAT1 (arylamine N-acetyltransferase Type 1), which is widely expressed throughout the body. While the protein-coding region of NAT1 is contained within a single exon, examination of the human EST (expressed sequence tag) database at the NCBI revealed the presence of nine separate exons, eight of which were located in the 5′ non-coding region of NAT1. Differential splicing produced at least eight unique mRNA isoforms that could be grouped according to the location of the first exon, which suggested that NAT1 expression occurs from three alternative promoters. Using RT (reverse transcriptase)-PCR, we identified one major transcript in various epithelial cells derived from different tissues. In contrast, multiple transcripts were observed in blood-derived cell lines (CEM, THP-1 and Jurkat), with a novel variant, not identified in the EST database, found in CEM cells only. The major splice variant increased gene expression 9–11-fold in a luciferase reporter assay, while the other isoforms were similar or slightly greater than the control. We examined the upstream region of the most active splice variant in a promoter-reporter assay, and isolated a 257 bp sequence that produced maximal promoter activity. This sequence lacked a TATA box, but contained a consensus Sp1 site and a CAAT box, as well as several other putative transcription-factor-binding sites. Cell-specific expression of the different NAT1 transcripts may contribute to the variation in NAT1 activity in vivo.
Keywords: N-acetyltransferase, arylamine N-transferase (NAT), promoter, splice variant, translation, 5′ untranslated region (5′-UTR)
Abbreviations: EST, expressed sequence tag; NAT1, arylamine N-acetyltransferase 1; NAT2, arylamine N-acetyltransferase 2; RT, reverse transcriptase; uAUG, upstream AUG triplet; UTR, untranslated region
INTRODUCTION
Human NAT1 (arylamine N-acetyltransferase Type I; EC 2.3.1.5) is found in most tissues, where it catalyses the transfer of an acetyl group from acetyl-CoA to arylamine and hydrazine substrates [1–3], including the folate catabolite p-aminobenzoylglutamate [4,5]. The gene encoding NAT1 is genetically variant, and is located at 8p21.3–22, a site that is often deleted in human tumours [6–9]. A second gene (NAT2; arylamine N-acetyltransferase Type 2) that shares high sequence identity with NAT1 is located approx. 177 kb closer to the centromere of chromosome 8 [10]. Unlike NAT1, NAT2 activity appears to be confined mostly to the liver and gastrointestinal tract [11,12]. Together, the proteins encoded by these two genes are responsible for the acetylation of most exogenous arylamine, heterocyclic amine and hydrazine compounds.
The first human mRNA sequence for NAT1 was isolated from liver tissue by Ohsaka and Deguchi in 1990 [13] and was reported to lack introns. A recent study suggests that this transcript originates from an inducible promoter located approx. 250 bases upstream of the coding region [14]. The promoter consisted of an AP-1 (activator protein 1) consensus sequence flanked by two TCATT motifs. However, cDNA isolated from ovarian carcinoma and sequenced by the IMAGE Consortium (GenBank® accession number BC013732) showed identical sequence similarity to NAT1 in the coding region, but indicated the presence of multiple introns in the 5′ non-coding region. When mapped to the genomic sequence for chromosome 8, this clone revealed the presence of a second promoter for NAT1 some 12 kb upstream of the coding region. The presence of non-coding introns in the human NAT1 gene is consistent with findings in other species, such as mouse, rat and rabbit, where at least one intron upstream of the coding region has been reported [15–17].
In the human genome, approx. 18% of all genes exhibit evidence for the use of alternative promoters [18]. Splice variants that alter the 5′-UTR (untranslated region) of the message, but not the protein-coding region, often show tissue-specific expression and altered translational efficiencies. Promoters can switch during development and cell differentiation. For example, the p18(INK4c) gene utilizes one promoter in undifferentiated cells, then switches to a second promoter that generates a shorter 5′-UTR with enhanced translational efficiency once differentiation is initiated [19]. Thus the presence of splice variants that differ only in their 5′-UTR sequence can indicate differential regulation at the gene level.
In the present study, we have examined the role of NAT1 splice variants in NAT1 expression. First, we investigated the genomic organization of NAT1 using the IMAGE clone BC013732 and a set of ESTs (expressed sequence tags) that mapped to the NAT1 locus. We identified sequences deposited in the EST databases that map to different regions of 8p21.2–22, and show evidence of multiple exons in the NAT1 gene. All of the NAT1 transcripts found comprised different exon combinations located exclusively in the 5′-UTR. We then isolated RNA from a number of human cell lines and used RT (reverse transcriptase)-PCR to identify multiple transcripts for NAT1 that showed cell-specific expression. Moreover, we describe a novel transcript found only in CEM T-cells. All of the transcripts observed in the present study were generated from a single promoter located approx. 12 kb upstream of the coding sequence.
EXPERIMENTAL
Identification of NAT1 genomic organization from EST databases
A human NAT1 mRNA (GenBank® accession number BC013732) was used to search the human EST database at the NCBI using the BLAST search engine. The search identified 61 ESTs having sequence identity with the NAT1 mRNA. Sequences corresponding to NAT2 were identified by the presence of a T at +19 and a C at +42, relative to the translation start site, and were eliminated from the analysis. The ESTs were sorted into 5′ and 3′ sequences, and each was aligned against the overlapping genomic sequences (GenBank® accession numbers AB020868 and AC025062), which map to chromosome 8p21.3–22. Intron–exon boundaries were identified from the EST maps.
Cell lines and tissue culture
All cell lines (hepatocarcinoma HepG2, breast carcinoma MCF7, lung carcinoma Calu6, melanoma MM2058, T-lymphoblastoid leukaemia CEM, monocytic THP-1, colon carcinoma HT-29, cervical carcinoma HeLa and T-cell Jurkat leukaemia) were cultured at 37 °C in DMEM (Dulbecco's modified Eagle's medium), supplemented with 10% (v/v) foetal calf serum containing 80 μg/ml benzylpenicillin and 50 μg/ml gentamicin, in a humidified atmosphere of 5% CO2 in air.
Identification of NAT1 transcripts by RT-PCR
Cytosolic RNA was extracted from human cell lines using an RNeasy kit (Qiagen) with on-column DNase I treatment according to the manufacturer's protocol. Oligo(dT) primer (0.5 μg) together with 1 μg of DNase-I-treated total RNA and AMV (avian myeloblastosis virus) RT (Promega) were used to generate first-strand cDNA. PCR was then performed using forward primers (Geneworks) designed for each of the exonic sequences identified from the EST database in combination with a common reverse primer located in the NAT1 coding sequence (see Supplementary Table I at http://www.BiochemJ.org/bj/387/bj3870119add.htm), and the following conditions: 95 °C for 3 min, followed by 35 cycles of 95 °C for 15 s, 50 °C for 30 s and 72 °C for 1 min. Amplified fragments were then cloned into pGEM-T Easy vector (Promega) using TA cloning. All clones were sequenced using the dideoxy terminator method.
Cloning of splice variants into pcDNA3 reporter plasmid
Each of the 5′-UTR splice variants identified in the different human cell lines was cloned into a pcDNA3 vector containing the Renilla luciferase gene inserted into the XhoI/XbaI restriction sites (pcDNA3-rLuc; a gift from Dr K. Ednie, Western Australian Institute for Medical Research) using the following strategy. First, exon 4 was amplified from genomic DNA using primers Ex4F and Ex4R (see Supplementary Table I at http://www.BiochemJ.org/bj/387/bj3870119add.htm) that contain HindIII sites, and cloned into the HindIII site in pcDNA3-rLuc. Clones with exon 4 in the correct orientation were identified by DNA sequencing. Exon 8 was then amplified using primers Ex8F and Ex8R that have KpnI and NotI sites respectively. This fragment was then cloned downstream of exon 4 using a KpnI site generated in exon 4 by Ex4R and the NotI site in pcDNA3-rLuc to generate a reporter gene with a 5′-UTR similar to the NAT1 Type IIA splice variant (pcDNA3-4/8-rLuc). To generate the Type IID 5′-UTR, the exon 6/8 fragment was amplified from CEM cDNA using primers Ex6/8F and Ex8R that contain KpnI and NotI sites respectively. This fragment was cloned into pcDNA3-rLuc containing exon 4 to generate pcDNA3-4/6/8-rLuc. Type IIB variant 5′-UTR was generated by amplifying exon 7 with primers Ex7F and Ex7R containing KpnI sites, which was then cloned into pcDNA3-4/8-rLuc to generate pcDNA3-4/7/8-rLuc. Type IIE variant 5′-UTR was made by amplifying exon 5 with primers Ex5F and Ex5R containing KpnI sites which was then cloned into pcDNA3-4/6/8-rLuc to generate pcDNA3-4/5/6/8-rLuc. All clones were sequenced to confirm the correct orientation of each exonic insert.
Cloning of NAT1 promoter II into pGL3-enhancer reporter vector
Promoter II deletion constructs were generated by cloning PCR fragments amplified from the region upstream of exon 4 with specific forward and reverse primers (see Supplementary Table I at http://www.BiochemJ.org/bj/387/bj3870119add.htm) containing KpnI and HindIII sites, respectively, into the pGL3-enhancer vector (Promega). The deletion constructs, namely pGL-2974, pGL-1861, pGL-893 and pGL-242, where the number indicates the size in bases of each fragment, were verified by DNA sequencing.
Cell transfection and dual-luciferase assay
HT-29 cells and CEM T-cells were seeded in six-well tissue culture plates at a density of 1×106 cells/well. The cells were cotransfected with 5 μg of each splice variant construct and 1 μg of pGL3-control vector (Promega) using LIPOFECTAMINE 2000 reagent (Invitrogen), according to the manufacturer's instructions. Cells were incubated at 37 °C for 48 h, and then firefly luciferase activity from the pGL3-control vector and Renilla luciferase activity from the splice variant constructs were measured using a dual-luciferase assay kit (Promega). For promoter-characterization experiments, HT-29 cells were seeded in 24-well tissue culture plates at a density of 2×105 cells/well, and were co-transfected with 1 μg of empty pGL3-enhancer vector, pGL3-2974, pGL3-1861, pGL3-893 or pGL3-257, and 0.1 μg of internal control vector pRL-SV40 (Promega). After incubation at 37 °C for 48 h, firefly luciferase activity from the pGL3-promoter constructs and Renilla luciferase activity from the internal control vector were measured.
Quantitative real-time PCR
Cytosolic RNA was extracted from HT-29 and CEM cells that had been transfected with the pcDNA3-rLuc splice variant constructs and reverse-transcribed to generate first-strand cDNA as described above. For each RNA sample, a reaction lacking RT was performed and used in real-time analysis to ensure no genomic or plasmid DNA contamination. Expression levels of Renilla luciferase from the various splice variant constructs were quantified by real-time PCR using a Rotor-Gene 3000 (Corbett Research) and SYBR Green detection. PCR conditions consisted of an initial denaturing step for 3 min at 95 °C, followed by 40 cycles of 15 s at 95 °C, 30 s at 61 °C and 30 s at 72 °C. Specific forward and reverse primers used to amplify Renilla luciferase and β-actin are shown in Supplementary Table I (available at http://www.BiochemJ.org/bj/387/bj3870119add.htm). Standard curves for Renilla luciferase and β-actin were constructed by serial dilution of the control (pcDNA3-rLuc lacking NAT1 exons) cDNA sample and then used to quantify expression levels. After normalization for β-actin, Renilla luciferase levels were expressed relative to the control. PCR products were run on 2% (w/v) agarose gels for confirmation of the correct amplicon size and identification of a single PCR product.
In vitro transcription/translation
The NAT1 open reading frame and entire 3′-UTR was amplified from the genomic DNA of a wild-type individual using NATF and NATR primers (see Supplementary Table I at http://www.BiochemJ.org/bj/387/bj3870119add.htm) and cloned into the pcDNA3-rLuc splice variant constructs described above, following excision of Renilla luciferase with NotI and ApaI. Because exon 5 contains an ApaI site, it was cloned into the pcDNA3-4/6/8 construct after NAT1. The resulting constructs, pcDNA3-NAT1 (control), pcDNA3-4/8-NAT1 (variant IIA), pcDNA3-4/6/8-NAT1 (variant IID), pcDNA3-4/7/8-NAT1 (variant IIB) and pcDNA3-4/5/6/8-NAT1 (variant IIE) were expressed using rabbit reticulocyte lysate (TNT Quick Coupled Transcription/Translation System; Promega) according to the manufacturers' instructions. Briefly, reaction mixtures contained 1 μg of plasmid DNA in a final volume of 25 μl, and were incubated for 1 h at 30 °C. Reactions were terminated by placing on ice. NAT1 activity was assayed by HPLC as described previously [20], using 2 μl of each reaction mixture. RNA was extracted from the remaining reaction mix using TRIzol® reagent (Invitrogen), separated on formaldehyde/agarose gels and stained with ethidium bromide to quantify transcript levels.
RESULTS
Identification of NAT1 splice variants from EST databases
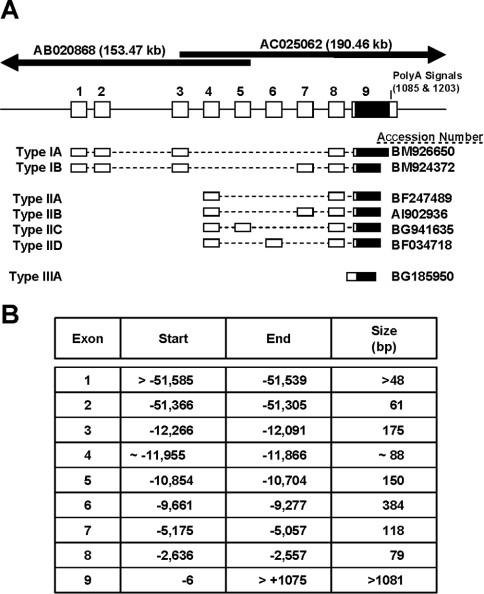
After performing a BLAST search of clone BC013732 against the NCBI human EST database, 61 potential sequences having identity with NAT1 were identified. We used the sequence differences at +19 (C in NAT1 compared with T in NAT2) and +42 (G in NAT1 compared with C in NAT2) to ensure each EST represented a sequence from the NAT1 gene. Initially, the ESTs were sorted into 5′ and 3′ fragments, and were then aligned with the overlapping genomic sequences AB020868 and AC025062, which map to more than 150 kb upstream and 180 kb downstream of the NAT1 coding sequence. We identified nine separate exons, eight of which were located upstream of the coding sequence (Figure 1A). The coding region of the gene was located entirely within exon 9 (Figure 1A, shaded box). In addition, we found two ESTs that were identical with the genomic sequence upstream of the coding region, indicating a transcript derived from a single exon (Type III). This transcript and its expression from an inducible promoter has been described recently [14], and was not investigated further in the present study. Differential splicing produced six unique mRNA isoforms that could be grouped according to the location of the first exon. Type I transcripts commenced with exon 1 and consisted of two species isolated from pooled tissues as well as mRNA from the nasopharynx. The Type IB transcript was grouped with the Type IA, because of the presence of exon 3 and absence of exon 4, although the EST only contained three bases at the end of exon 1. Type II transcripts all commence with exon 4, suggesting that they were expressed from a promoter located upstream of this sequence. Type III transcripts lacked introns, and their expression has been described elsewhere [14]. The location of each exon relative to the translational start site is shown in Figure 1(B). The largest intron in the NAT1 gene was located between exon 2 and 3, and was greater than 39 kb. Exon 8 was present in all transcripts, except in Type III. The most common transcript was Type IIA, which consists of exons 4, 8 and 9. It comprised 70% of the informative ESTs.
Figure 1. Genomic organization of the human NAT1 gene deduced from the EST database.
(A) Summary of the intron–exon organization and the seven different splice variants that were identified in the database. The mRNAs are grouped according to the first exon used. The overlapping sequences that contain the entire NAT1 gene are depicted at the top of the Figure. (B) The Table lists the first and last base for each exon relative to the translation start site. The size of each exon is also shown.
The intron–exon boundaries are shown in Table 1. All splice sites obeyed the GT-AG rule. The acceptor site for exon 4 is not shown as there was no evidence from any of the ESTs that another exon upstream of it is used for these transcripts.
Table 1. Exon–intron organization and boundary sequences of the human NAT1 gene.
Exon sequences are shown in upper case, and intron sequences are shown in lower case. The 3′-splice acceptor for exon 4 is not shown as there was no evidence from the EST databases that exons upstream of it were used.
| Exon | Exon size (bp) | 5′-Splice donor | Intron size (bp) | 3′-Splice acceptor | Exon |
|---|---|---|---|---|---|
| 1 | >31 | ACAAATCAGgtagtctct | 171 | ctttaacagATTGACCCA | 2 |
| 2 | 61 | CGCAACTGTgtaagtgcc | 39039 | ctcctctagGTTACCAGT | 3 |
| 3 | 175 | TTTTTTACGgtaagatat | 137 | ||
| 4 | 88 | AATAACCTGgtaagtgga | 1012 | caccgtcagACCGAGAGC | 5 |
| 5 | 150 | ACTACAATGgtatacttg | 1043 | tttacacagCTCTCCAAA | 6 |
| 6 | 384 | GGGATCCAGgtaagttat | 4102 | cttcaacagACGTGTACA | 7 |
| 7 | 118 | TGTTTCTGGgtcagtacc | 2421 | tatttctagAATTCAAGC | 8 |
| 8 | 79 | CTAAGAAAGgtattaagc | 2551 | cttgcttagGGGATCATG | 9 |
A list of tissues from which the NAT1 mRNA isoforms were identified was compiled and is shown in Table 2. Although some variants have only been identified in specific tissues, their distributions may be more widespread and require a more thorough investigation. Nevertheless, the presence of NAT1 mRNA in most tissues is consistent with the ubiquitous distribution of NAT1 activity [1].
Table 2. Tissue distribution of NAT1 mRNA isoforms.
An asterisk denotes tumour tissue.
| Isoform | Tissue | GenBank® accession number |
|---|---|---|
| Type IA | Lung/spleen | BM926650 |
| Nasopharynx | CD702544 | |
| Type IB | Colon/kidney/stomach | BM924372 |
| Type IIA | Testis* | BG529392 |
| Kidney* | BF247489 | |
| Aorta | CD511345 | |
| Lung* | BQ231158 | |
| Retina | BM696249 | |
| Pituitary* | AV754344 | |
| Pancreas | BG655073 | |
| Prostate | T29485 | |
| Colon* | AW956027 | |
| Type IIB | Breast | AI902936 |
| Type IIC | Peripheral blood | BG941635 |
| Type IID | Ovary* | BF034718 |
| Parathyroid* | AI022585 | |
| Type III | Liver | D90041 |
| Fibrosarcoma* | BG185950 |
Distribution of NAT1 transcripts in human cell lines
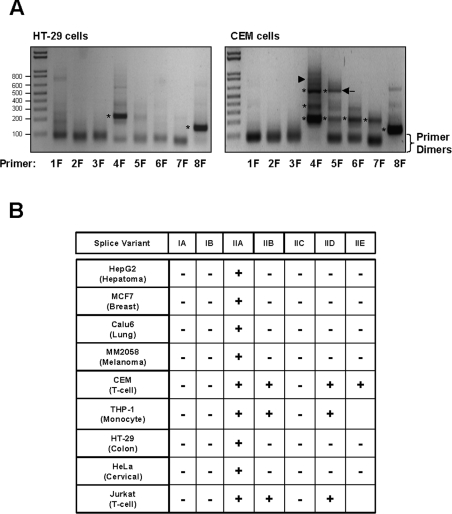
To determine the presence of the different transcripts in a number of human cell lines derived from various tissues, cytosolic RNA was isolated, reverse-transcribed and then amplified by PCR using specific primers located in each exon in combination with a common reverse primer located in exon 9. The predicted PCR products for each of the splice variants are listed in Table 3, and these were used to deduce which splice variants were expressed in each of the cell lines. Figure 2(A) shows representative gels from two of the cell lines, HT-29 cells where only exons 4 and 8 amplified (variant IIA) and CEM cells. All of the epithelial lines gave similar PCR results to those in the HT-29 cells. In contrast, the blood-derived cell lines (CEM, THP-1 and Jurkat) demonstrated more complex banding as shown in Figure 2(A, right-hand panel). Each PCR product (marked with an asterisk in Figure 2A) was cloned into pGEM-T and sequenced to confirm its identity. The results are summarized in Figure 2(B). None of the cell lines showed the presence of Type IA (exons 1, 2, 3, 8 and 9), IB (exons 2, 3, 7, 8 and 9) or IIC (exons 4, 5, 8 and 9) transcripts. Each of these variants appeared only once or twice in the EST database, suggesting that they may be rare mRNA species. Type IIA mRNA (exons 4, 8 and 9) was seen in all cell lines, confirming that this is the most common isoform. In the blood-derived cell lines (CEM, THP-1 and Jurkat), variants IIB and IID also were present.
Table 3. Predicted sizes of PCR products after amplification of cDNA for each of the transcripts identified from the EST database.
| Primer | Type IA | Type IB | Type IIA | Type IIB | Type IIC | Type IID |
|---|---|---|---|---|---|---|
| 1 | 464 | 582 | − | − | − | − |
| 2 | 409 | 527 | − | − | − | − |
| 3 | 238 | 356 | − | − | − | − |
| 4 | − | − | 230 | 348 | 380 | 614 |
| 5 | − | − | − | − | 232 | − |
| 6 | − | − | − | − | − | 230 |
| 7 | − | 230 | − | 230 | − | − |
| 8 | 154 | 154 | 154 | 154 | 154 | 154 |
Figure 2. Identification of RNA splice variants in human cell lines.
Forward primers (1F–8F specific for exons 1–8 respectively) were designed to amplify the different exons using a common reverse primer (ExonR) in the coding region. Amplification gave a series of products depending on the combination of exons. Predicted product sizes for the different RNA variants are listed in Table 3. (A) Representative electrophoretic patterns from cells that express only one variant, Type IIA (left-hand panel). A band can be seen with the 4F and 8F primers, indicating the presence of exons 4 and 8. The right-hand panel shows the electrophoretic pattern for cells expressing several variants. In this case (CEM cells), multiple bands were seen with primers 4F and 5F, while a single band was seen with 6F, 7F and 8F. From the sizes, the different RNA variants can be deduced. Bands indicated with the asterisks were cloned and sequenced to confirm their identity. In lane 5F, an unknown band of approx. 550 bp was present (arrow). When sequenced, this band represented a new variant containing exons 5, 6, 8 and 9. (B) Occurrence of the different RNA species in human cell lines. Variant IIA was present in all cells, while only the blood-derived lines showed evidence of multiple RNA species.
One PCR product obtained following amplification of CEM cell mRNA with the exon 5 primer (Figure 2A, arrow) represented a new isoform not identified in the EST database, and consisted of exons 5, 6, 8 and 9. Given that exons 5 and 6 were only observed in Type II transcripts, it is likely that exon 4 is the first exon for this variant. A faint band of the correct size (777 bp) was present in the CEM cells amplified with the primer for exon 4 (Figure 2A, lane 4F, arrowhead), but we were unsuccessful in cloning this product to confirm its identity. The novel RNA variant was named Type IIE. Interestingly, this RNA species was not observed in either the Jurkat or the THP-1 cell line (results not shown).
Effect of NAT1 5′-UTR on gene expression
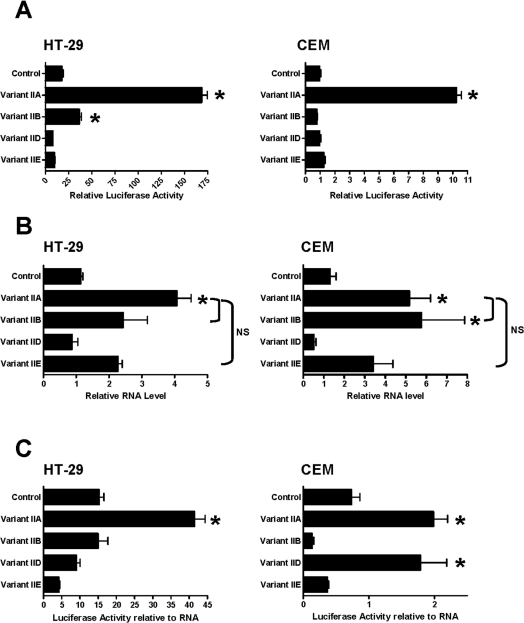
Each of the 5′-UTR sequences identified in the human cell lines (summarized in Figure 2B) was cloned into pcDNA3 upstream of the Renilla luciferase reporter gene. The resulting constructs were transiently transfected into HT-29 cells (representative of cells with a single NAT1 mRNA species) or CEM cells (representative of cells with various NAT1 mRNA species), and gene expression was determined 48 h later. In both cell lines, the Type IIA splice variant showed a 9–11-fold increase in luciferase expression relative to the pcDNA3-rLuc control plasmid (Figure 3A). The Type IIB variant also showed enhanced expression in HT-29 cells, but to a lesser extent. All other constructs were similar to the control. Expression of all constructs was greater in the HT-29 cells compared with CEM cells. This difference in expression was not due to different transfection efficiencies, since Renilla luciferase activity is reported relative to firefly luciferase activity. The data may reflect differences in transcriptional/translational efficiency for the constructs in the HT-29 cells.
Figure 3. Effect of the variant 5′-UTR sequences on gene expression.
(A) Each of the variants identified in Figure 2 was cloned upstream of the Renilla luciferase reporter gene and then transiently transfected into either HT-29 cells (left-hand panel) or CEM cells (right-hand panel), along with the firefly luciferase gene as a control. Results are the means±S.E.M. (n=3) of Renilla luciferase activity relative to firefly luciferase activity. Asterisks indicate results significantly greater than control vector (one-way ANOVA, followed by Dunnett's test; P<0.05). (B) RNA levels for each of the variants shown in (A) for HT-29 cells (left-hand panel) and CEM cells (right-hand panel). Real-time PCR was used to quantify the amount of RNA in cells following transfection with the different RNA splice variants. Results are means±S.E.M. (n=3–6), presented as the ratio of luciferase RNA to β-actin RNA. Asterisks indicate data significantly greater than control vector (one-way ANOVA, followed by Dunnett's test; P<0.05). When variant IIA was compared with IIB or IIE, no significant difference (NS) was seen. (C) Luciferase activity was normalized to RNA levels. Results are means±S.E.M. (n=3). Asterisks indicate results significantly greater than control vector (one-way ANOVA, followed by Dunnett's test; P<0.05).
To determine whether the different levels of expression shown in Figure 3(A) were due to different amounts of mRNA, real-time PCR was employed to quantify the levels of each message relative to β-actin mRNA (Figure 3B). The Type IIA variant produced significantly higher levels of mRNA compared with the control vector in both cell lines (one-way ANOVA; P<0.05). However, the amount of mRNA from variants IIA, IIB and IIE were not different (one-way ANOVA; P>0.05). When luciferase activities were normalized for RNA levels, variant IIA still showed greater expression compared with the control (Figure 3C). However, unlike that seen in HT-29 cells, variant IID also showed significantly greater expression than control in CEM cells.
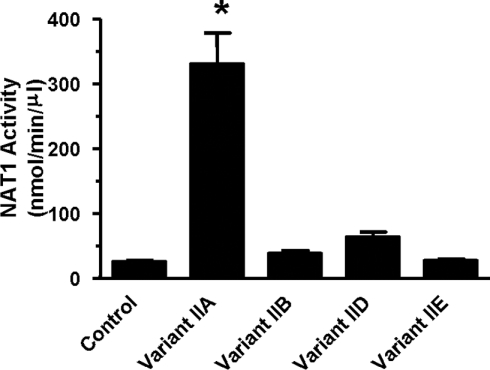
To examine further the effect of the different splice variants on translational efficiency, each 5′-UTR, along with the entire NAT1 coding sequence and the 3′-UTR, was cloned into pcDNA3 and expressed in an in vitro transcription/translation system. mRNA levels were determined from ethidium-bromide-stained agarose gels, and were not different between variants (one-way ANOVA; P>0.05; results not shown). Expressed NAT1 activity is shown in Figure 4. Variant IIA showed a 13-fold increase in translational efficiency compared with the control. None of the other variants were different from the control.
Figure 4. Expression of NAT1 splice variants in an in vitro transcription/translation system.
Each splice variant was cloned into pcDNA3 upstream of the NAT1 coding sequence and the entire 3′-UTR, and was used as template for in vitro transcription/translation. After 1 h of incubation at 30 °C, 2 μl was used to measure NAT1 activity, and RNA was extracted from the remainder, electrophoresed through agarose and stained with ethidium bromide to quantify mRNA levels. Results are means±S.E.M. (n=3). Asterisks indicate results significantly greater than control vector (one-way ANOVA, followed by Dunnett's test; P<0.05).
Since the translational efficiency of genes can by affected by both the number of upstream AUG triplets (uAUGs), as well as the folding complexity of the 5′-UTR, the different NAT1 variants were examined for each of these features. Table 4 shows the number of uAUGs and the predicted free energy of folding for each of the 5′-UTR sequences investigated in Figure 3. The Type IIA variant, which demonstrated the highest level of expression, has only one uAUG which is defined as a ‘weak’ initiation codon (does not contain an A or G at −3, or a G at −4). In contrast, the Type IIB, IID and IIE variants have 4, 5 and 10 uAUGs respectively, with many partially or completely resembling a Kozak sequence. The predicted free energy decreased with 5′-UTR size, being greatest for Type IIE, which has 708 bases.
Table 4. Number of AUG triplets and secondary-structure free energy for NAT1 5′-UTR sequences.
| uAUG* | |||||
|---|---|---|---|---|---|
| Variant | Length | Strong | Adequate | Weak | Free energy (kcal/mol)† |
| Type IIA | 166 | 1 | −47.2 | ||
| Type IIB | 284 | 2 | 2 | −80.5 | |
| Type IID | 550 | 1 | 4 | −167.7 | |
| Type IIE | 700 | 2 | 2 | 6 | −215.6 |
* The context of the uAUG was defined according to Meijer and Thomas [30] as ‘strong’ if it contained both an A, or G, at −3 and a G at +4, ‘adequate’ if it contained either of these nucleotides, and ‘weak’ if it contained neither.
† Calculated by the method of Mathews et al. [31] using the RNAstructure software. 1 kcal=4.184 kJ.
Identification of a promoter sequence upstream of exon 4
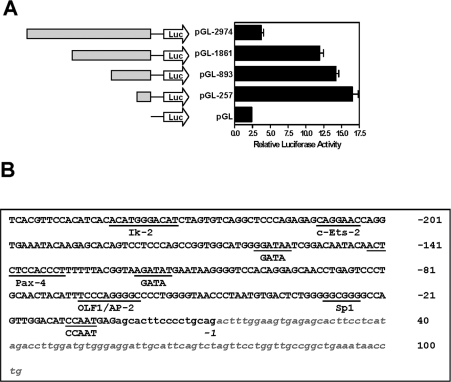
Since every cell line investigated demonstrated one or more splice variants from the Type II group, we cloned the region upstream of exon 4 (2974 bp) into the pGL-3 enhancer vector to investigate potential promoter activity. Deletion mutants were also constructed in order to identify the approximate region of the minimal promoter sequence. Figure 5(A) indicates a significant increase in reporter activity when the promoter was truncated from 2974 bp to 1861 bp, suggesting the presence of silencing sequences within this region. Further deletion of the promoter to as little as 257 bp did not result in a loss of promoter activity (Figure 5A, right-hand panel), suggesting that the main regulatory sequences resided in this region, which is shown in Figure 5(B). Analysis of this sequence using MatInspector identified a conserved Sp1 site approx. 40 bases upstream of the start of exon 4. There was no evidence of a TATA box. Several other putative transcription factor binding sites also were identified, including c-Ets-2, GATA and Olf-1 (olfactory neuronal transcription factor-1).
Figure 5. Promoter activity of the sequence upstream of exon 4.
(A) Various promoter lengths were inserted into the pGL3-enhancer vector and were transiently transfected into HT-29 cells, together with the Renilla luciferase reporter vector as a control. Results are means±S.E.M. (n=3). (B) The sequence of the 257 bases (upper case) upstream of exon 4 that showed maximal promoter activity. The lower case sequence indicates bases downstream of the promoter, and include the start of exon 4 deduced from the clone BG655073. The promoter sequence was examined using MatInspector [32] to identify putative transcription factor binding sites (underlined).
DISCUSSION
NAT1 is widely expressed in humans, although activity varies between individuals, and between tissues within an individual. It has been identified in early development [21], which is not characteristic of most drug-metabolizing enzymes, including NAT2. Interindividual variation in activity is the result of a number of genetic and functional polymorphisms [1,3]. However, other genetic and environmental factors clearly contribute to the overall activity of the enzyme in vivo. NAT1 activity is subject to substrate-dependent down-regulation, where the protein is polyubiquitinated and targeted to the proteasome for degradation [20]. Moreover, NAT1 is readily and irreversibly inactivated by substrate intermediates such as the N-hydroxy metabolite of p-aminobenzoic acid and other hydroxamic acids that attack the active site cysteine residue [22,23]. More recently, it was demonstrated that NAT1 activity could be modulated by radicals including NO intermediates and H2O2 [24–26]. Thus the level of NAT1 activity within specific tissues of an individual is the net effect of genetic and environmental factors. In the present study, we have described a further cause of variation in NAT1 activity, multiple cell-specific mRNA splice variants that have differing translational efficiencies.
From an investigation of the EST database at the NCBI, as well as direct sequencing of cDNA from several human cell lines, at least eight different splice variants for human NAT1 were identified, including a novel variant Type IIE. A total of nine exons were identified with the entire gene encompassing over 52 kb. Each of the mRNA variants differed according to the combination of non-coding exons present in the 5′-UTR of the message. The Type IIA variant was present in all human cell lines investigated and was the only species identified in the epithelial cell lines. In contrast, multiple splice variants were present in the three blood-derived cell lines CEM, THP-1 and Jurkat. When the different NAT1 5′-UTR sequences identified in the human cell lines were cloned upstream of the luciferase reporter gene, the Type IIA variant showed the greatest gene expression compared with the control vector or the other NAT1 5′-UTR sequences. The only exception was the Type IID variant in CEM cells. This difference in expression could not be accounted for by a difference in RNA levels, since the amount of Type IIA RNA was similar to that for Type IIB and IIE. The data suggested that the Type IIA splice variant was more efficiently translated. To investigate this possibility further, each of the variants, along with the entire NAT1-coding sequence and the 3′-UTR, were examined in an in vitro transcription/translation system. mRNA levels were similar for each variant, but the Type IIA variant showed a much greater level of expression, supporting the more efficient translation of this mRNA. Translational efficiency is affected by both the secondary structure of the 5′-UTR and the number of uAUGs, both of which can hamper ribosomal scanning of the mRNA for the initiation codon. The Type IIA variant not only has the least number of uAUGs, but also the highest free energy level. Secondary structures, such as that predicted for the other variants (Table 4), with a free energy less than −50 kcal/mol (1 kcal≡4.184 kJ) have been shown to be sufficient to impede scanning [27].
The sequences of the different human NAT1 mRNAs suggested that at least three different promoters are used for NAT1 expression. The first (Promoter I) is located approx. 245 bases upstream of the coding region, and has been described elsewhere in detail [14]. This promoter gives rise to mRNA that is identical with the genomic sequence (intronless). However, the lack of transcripts in the public databases originating from this promoter suggests that it may not be important for the constitutive expression of NAT1. From the present study, two other promoters are probably used to generate NAT1 transcripts. The first (Promoter II) is upstream of exon 4 and gives rise to the Type II transcripts. We identified a 257 bp sequence from this region that was active in a promoter-reporter assay. Promoter II is a TATA-less sequence with a consensus Sp1 site and a CAAT box. This region of the human NAT1 gene bears no resemblance to the recently defined promoter sequence for the mouse homologue gene Nat2 [28]. Indeed, the organization of the mouse Nat2 gene is less complex than that of the human NAT1 gene, and appears to more closely resemble that of the human NAT2 gene. Further studies are required to define the exact sequences in this region of the human NAT1 gene that are involved in gene expression.
The third promoter (Promoter III), not investigated in the present study, is located upstream of exon 1 approx. 51 kb from the coding region, and results in Type I transcripts. Since Type I transcripts were not identified in any of the cell lines in the present study, their expression may be restricted to particular tissues or cells, or they may be expressed only under as yet unknown conditions. A recent study by Husain et al. [29] also failed to identify transcripts from Promoter III in MCF-7 cells. However, they reported another possible promoter for NAT1 located in an internal exon (Exon 6) [29], although no clones in the public databases appear to originate from this sequence. The reason why the NAT1 gene might utilize several different promoters is unclear. Using a luciferase reporter assay, Husain et al. [29] showed that the strength of Promoter II was considerably greater than that of Promoter I in unstimulated MCF-7 cells. Thus Promoter II is most likely to be responsible for the constitutive expression of NAT1.
In summary, we have identified several NAT1 splice variants that differ in their 5′-UTR, cell-specificity and translational efficiency. These novel transcripts may contribute to the variation in NAT1 activity in vivo.
Online data
Acknowledgments
This work was supported by the National Health and Medical Research Council of Australia (grant number 212066).
References
- 1.Butcher N. J., Boukouvala S., Sim E., Minchin R. F. Pharmacogenetics of the arylamine N-acetyltransferases. Pharmacogenomics J. 2002;2:30–42. doi: 10.1038/sj.tpj.6500053. [DOI] [PubMed] [Google Scholar]
- 2.Dupret J.-M., Goodfellow G. H., Janezic S. A., Grant D. M. Structure–function studies of human arylamine N-acetyltransferases NAT1 and NAT2: functional analysis of recombinant NAT1/NAT2 chimeras expressed in Escherichia coli. J. Biol. Chem. 1994;269:26830–26835. [PubMed] [Google Scholar]
- 3.Hein D. W., Doll M. A., Fretland A. J., Leff M. A., Webb S. J., Xiao G. H., Devanaboyina U. S., Nangju N. A., Feng Y. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol. Biomarkers Prev. 2000;9:29–42. [PubMed] [Google Scholar]
- 4.Minchin R. F. Acetylation of p-aminobenzoylglutamate, a folic acid catabolite, by recombinant human arylamine N-acetyltransferase and U937 cells. Biochem. J. 1995;307:1–3. doi: 10.1042/bj3070001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward A., Summers M. J., Sim E. Purification of recombinant human N-acetyltransferase type 1 (NAT1) expressed in. E coli and characterization of its potential role in folate metabolism. Biochem. Pharmacol. 1995;49:1759–1767. doi: 10.1016/0006-2952(95)00087-g. [DOI] [PubMed] [Google Scholar]
- 6.Brown M. R., Chuaqui R., Vocke C. D., Berchuck A., Middleton L. P., Emmert-Buck M. R., Kohn E. C. Allelic loss on chromosome arm 8p: analysis of sporadic epithelial ovarian tumors. Gynecol. Oncol. 1999;74:98–102. doi: 10.1006/gyno.1999.5439. [DOI] [PubMed] [Google Scholar]
- 7.Akiyama Y., Arai T., Nagasaki H., Yagi O. K., Nakahata A., Nakajima T., Ohkura Y., Iwai T., Saitoh K., Yuasa Y. Frequent allelic imbalance on chromosome 18q21 in early superficial colorectal cancers. Jpn. J. Cancer Res. 1999;90:1329–1337. doi: 10.1111/j.1349-7006.1999.tb00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lutchman M., Pack S., Kim A. C., Azim A., Emmert-Buck M., van Huffel C., Zhuang Z., Chishti A. H. Loss of heterozygosity on 8p in prostate cancer implicates a role for dematin in tumor progression. Cancer Genet. Cytogenet. 1999;115:65–69. doi: 10.1016/s0165-4608(99)00081-3. [DOI] [PubMed] [Google Scholar]
- 9.Wistuba I. I., Behrens C., Virmani A. K., Milchgrub S., Syed S., Lam S., Mackay B., Minna J. D., Gazdar A. F. Allelic losses at chromosome 8p21–23 are early and frequent events in the pathogenesis of lung cancer. Cancer Res. 1999;59:1973–1979. [PubMed] [Google Scholar]
- 10.Sim E., Payton M., Noble M., Minchin R. An update on genetic, structural and functional studies of arylamine N-acetyltransferases in eucaryotes and procaryotes. Hum. Mol. Genet. 2000;9:2435–2441. doi: 10.1093/hmg/9.16.2435. [DOI] [PubMed] [Google Scholar]
- 11.Windmill K. F., Gaedigk A., Hall P. M., Samaratunga H., Grant D. M., McManus M. E. Localization of N-acetyltransferases NAT1 and NAT2 in human tissues. Toxicol. Sci. 2000;54:19–29. doi: 10.1093/toxsci/54.1.19. [DOI] [PubMed] [Google Scholar]
- 12.Ilett K. F., Ingram D. M., Carpenter D. S., Teitel C. H., Lang N. P., Kadlubar F. F., Minchin R. F. Expression of monomorphic and polymorphic N-acetyltransferases in human colon. Biochem. Pharmacol. 1994;47:914–917. doi: 10.1016/0006-2952(94)90493-6. [DOI] [PubMed] [Google Scholar]
- 13.Ohsako S., Deguchi T. Cloning and expression of cDNAs for polymorphic and monomorphic arylamine N-acetyltransferases from human liver. J. Biol. Chem. 1990;265:4630–4634. [PubMed] [Google Scholar]
- 14.Butcher N. J., Arulpragasam A., Pope C., Minchin R. F. Identification of a minimal promoter sequence for the human N-acetyltransferase Type I gene that binds AP-1 (activator protein 1) and YY-1 (Yin and Yang 1) Biochem. J. 2003;376:441–448. doi: 10.1042/BJ20030650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blum M., Heim M., Meyer U. A. Nucleotide sequence of rabbit NAT1 encoding monomorphic arylamine N-acetyltransferase. Nucleic Acids Res. 1990;18:5287. doi: 10.1093/nar/18.17.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebisawa T., Deguchi T. Structure and restriction fragment length polymorphism of genes for human liver arylamine N-acetyltransferases. Biochem. Biophys. Res. Commun. 1991;177:1252–1257. doi: 10.1016/0006-291x(91)90676-x. [DOI] [PubMed] [Google Scholar]
- 17.Fakis G., Boukouvala S., Buckle V., Payton M., Denning C., Sim E. Chromosome mapping of the genes for murine arylamine N-acetyltransferases (NATs), enzymes involved in the metabolism of carcinogens: identification of a novel upstream noncoding exon for murine Nat2. Cytogenet. Cell Genet. 2000;90:134–138. doi: 10.1159/000015648. [DOI] [PubMed] [Google Scholar]
- 18.Landry J. R., Mager D. L., Wilhelm B. T. Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet. 2003;19:640–648. doi: 10.1016/j.tig.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Phelps D. E., Hsiao K. M., Li Y., Hu N., Franklin D. S., Westphal E., Lee E. Y., Xiong Y. Coupled transcriptional and translational control of cyclin-dependent kinase inhibitor p18INK4c expression during myogenesis. Mol. Cell. Biol. 1998;18:2334–2343. doi: 10.1128/mcb.18.4.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butcher N. J., Arulpragasam A., Minchin R. F. Proteasomal degradation of N-acetyltransferase 1 is prevented by acetylation of the active site cysteine: a mechanism for the slow acetylator phenotype and substrate-dependent down-regulation. J. Biol. Chem. 2004;279:22131–22137. doi: 10.1074/jbc.M312858200. [DOI] [PubMed] [Google Scholar]
- 21.Pacifici G. M., Bencini C., Rane A. Acetyltransferase in humans: development and tissue distribution. Pharmacology. 1986;32:283–291. doi: 10.1159/000138181. [DOI] [PubMed] [Google Scholar]
- 22.Butcher N. J., Ilett K. F., Minchin R. F. Inactivation of human arylamine N-acetyltransferase 1 by the hydroxylamine of p-aminobenzoic acid. Biochem. Pharmacol. 2000;60:1829–1836. doi: 10.1016/s0006-2952(00)00501-3. [DOI] [PubMed] [Google Scholar]
- 23.Sticha K. R., Bergstrom C. P., Wagner C. R., Hanna P. E. Characterization of hamster recombinant monomorphic and polymorphic arylamine N-acetyltransferases: bioactivation and mechanism-based inactivation studies with N-hydroxy-2-acetylaminofluorene. Biochem. Pharmacol. 1998;56:47–59. doi: 10.1016/s0006-2952(98)00101-4. [DOI] [PubMed] [Google Scholar]
- 24.Dairou J., Atmane N., Rodrigues-Lima F., Dupret J.-M. Peroxynitrite irreversibly inactivates the human xenobiotic-metabolizing enzyme arylamine N-acetyltransferase 1 (NAT1) in human breast cancer cells: a cellular and mechanistic study. J. Biol. Chem. 2004;279:7708–7714. doi: 10.1074/jbc.M311469200. [DOI] [PubMed] [Google Scholar]
- 25.Dairou J., Atmane N., Dupret J.-M., Rodrigues-Lima F. Reversible inhibition of the human xenobiotic-metabolizing enzyme arylamine N-acetyltransferase 1 by S-nitrosothiols. Biochem. Biophys. Res. Commun. 2003;307:1059–1065. doi: 10.1016/s0006-291x(03)01311-1. [DOI] [PubMed] [Google Scholar]
- 26.Atmane N., Dairou J., Paul A., Dupret J.-M., Rodrigues-Lima F. Redox regulation of the human xenobiotic metabolizing enzyme arylamine N-acetyltransferase 1 (NAT1): reversible inactivation by hydrogen peroxide. J. Biol. Chem. 2003;278:35086–35092. doi: 10.1074/jbc.M303813200. [DOI] [PubMed] [Google Scholar]
- 27.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boukouvala S., Price N., Plant K. E., Sim E. Structure and transcriptional regulation of the Nat2 gene encoding for the drug-metabolizing enzyme arylamine N-acetyltransferase type 2 in mice. Biochem. J. 2003;375:593–602. doi: 10.1042/BJ20030812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Husain A., Barker D. F., States J. C., Doll M. A., Hein D. W. Identification of the major promoter and non-coding exons of the human arylamine N-acetyltransferase 1 gene (NAT1) Pharmacogenetics. 2004;14:397–406. doi: 10.1097/01.fpc.0000114755.08559.6e. [DOI] [PubMed] [Google Scholar]
- 30.Meijer H. A., Thomas A. A. Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem. J. 2002;367:1–11. doi: 10.1042/BJ20011706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathews D. H., Sabina J., Zuker M., Turner D. H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 32.Quandt K., Frech K., Karas H., Wingender E., Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.