Abstract
The nuclear lamins form a karyoskeleton providing structural rigidity to the nucleus. One member of the lamin family, lamin A, is first synthesized as a 74 kDa precursor, prelamin A. After the endopeptidase and methylation reactions which occur after farnesylation of the CAAX-box cysteine, there is a second endoproteolysis that occurs 15 amino acids upstream from the C-terminal farnesylated cysteine residue. Studies with knockout mice have implicated the enzyme Zmpste24 (Face-1) as a suitable candidate to perform one or both of these proteolytic reactions. Evidence has been presented elsewhere establishing that Zmpste24 possesses a zinc-dependent CAAX endopeptidase activity. In the present study, we confirm this CAAX endopeptidase activity with recombinant, membrane-reconstituted Zmpste24 and show that it can accept a prelamin A farnesylated tetrapeptide as substrate. To monitor the second upstream endoproteolytic cleavage of prelamin A, we expressed a 33 kDa prelamin A C-terminal tail in insect cells. We demonstrate that this purified substrate possesses a C-terminal farnesylated and carboxyl-methylated cysteine and, therefore, constitutes a valid substrate for assaying the second endoproteolytic step in lamin A maturation. With this substrate, we demonstrate that insect cell membranes bearing recombinant Zmpste24 can also catalyse the second upstream endoproteolytic cleavage.
Keywords: endoproteolysis, Face-1, lamin A, prelamin A, prenylation, Zmpste24
Abbreviations: CHO-K1 cells, Chinese-hamster ovary-K1 cells; E-64, trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane; FPP, farnesyl pyrophosphate; HRP, horseradish peroxidase; ICMT, isoprenyl cysteine methyltransferase; MALDI–TOF-MS, matrix-assisted laser-desorption ionization–time-of-flight MS; Ni-NTA, Ni2+-nitrilotriacetate; Opa, 1,10-orthophenanthroline; prelaminAct, the C-terminal residues 389–664 of prelamin A; Sf21, Spodoptera frugiperda
INTRODUCTION
The function, localization and binding characteristics of a large variety of cellular proteins can be modulated by post-translational lipid modification at the C-terminus with a prenyl moiety [1–3]. Proteins with a CAAX box at the C-terminus undergo a series of post-translational modifications resulting in the prenylation of the cysteine thiol through a prenyltransferase, removal of the AAX tripeptide by a CAAX endopeptidase and finally carboxyl-methylation of the newly exposed cysteinyl carboxy group via an isoprenylcysteine methyltransferase [4]. The canonical C-terminal CAAX box (where C stands for cysteine, A for an aliphatic amino acid and X for one among S, M, C, A, Q and L) can undergo modification by a farnesyl isoprenoid when X is S, M, C, A or Q or by a geranylgeranyl group when the amino acid X is a leucine.
The farnesylated nuclear lamin A precursor, prelamin A, is a unique example of a prenylated protein in mammalian cells that undergoes an additional upstream proteolysis [5] subsequent to the canonical CAAX box modifications (Figure 1). This cleavage between Tyr646 and Leu647 [6], along with the CAAX endopeptidolysis, results in the removal of 18 C-terminal amino acid residues and is required for lamin A assembly into the nuclear lamina [7]. In the genetic disease Hutchinson–Gilford progeria, deletion of this protease target site results in an aberrant lamin A molecule [8].
Figure 1. The processing pathway of prelamin A.
Prelamin A undergoes a series of post-translational modifications at the designated CAAX box, CSIM, including farnesylation, AAX endopeptidolysis, carboxyl-methylation and a second endoproteolytic cleavage between a tyrosine and a leucine residue, 15 amino acids upstream from the farnesylated cysteine residue. This results in the production of mature lamin A, along with an approx. 2 kDa released prenylated peptide.
Several lines of evidence have demonstrated that endoproteolysis of prelamin A is prenylation-dependent. When cells are incubated with lovastatin, thereby blocking farnesylation through the inhibition of isoprenoid biosynthesis, the conversion of prelamin A into lamin A is abolished, resulting in the accumulation of prelamin A into nucleoplasmic aggregates [5,6]. Experiments in which the CAAX box cysteine is mutated to abolish prenylation have also shown that prelamin A accumulates in nucleoplasmic aggregates and that conversion into lamin A is blocked [9]. Our laboratory has also reported in vitro studies with synthetic peptide substrates consistent with the conclusion that prelamin A must not only be farnesylated, but also undergo AAXing (endopeptidase-catalysed removal of the C-terminal tripeptide) and carboxyl-methylation of the CAAX box cysteine before endoproteolysis can take place [10,11]. The hexapeptide sequence (RSYLLG) around the cleavage site of prelamin A is highly conserved among prelamin A molecules from various species [6,10].
A possible candidate for the gene responsible for prelamin A maturation has arisen from genetic data obtained from both yeast and mice. Ste24 was first identified as a zinc-dependent metalloprotease in yeast whose function is to endoproteolytically process the prenylated mating factor precursor, a-factor [12]. The proteolytic maturation of a-factor parallels that of prelamin A in that it is first processed at the CAAX box by farnesylation, AAXing and carboxyl-methylation and then undergoes endoproteolysis 26 residues upstream from the prenylated cysteine residue between threonine and alanine residues [13,14]. However, unlike prelamin A, a-factor undergoes a third endoproteolytic step that cleaves 12 residues upstream from the prenylated cysteine residue, producing the mature farnesylated dodecapeptide. Ste24p catalyses both the AAXing reaction and the first endoproteolytic cleavage 26 residues N-terminal to the prenylated cysteine, apparently mediated by a canonical zinc-metalloprotease HEXXH active site [12,15–18]. The similarities between the processing of prelamin A in mammals and a-factor in yeast suggested the possibility that the human orthologue to Ste24p may play a role in prelamin A maturation. The cloned and expressed human orthologue, Zmpste24 (Face-1 or HsSte24), was shown to complement fully the yeast Ste24 by exhibiting activity in both the AAXing reaction and the first endoproteolytic cleavage, 26 residues N-terminal to the prenylated cysteine [19,20].
To elucidate the in vivo function of Zmpste24, two Zmpste24 knockout mouse lines were generated and phenotypically characterized [21,22]. It was observed that mice deficient in Zmpste24 produced a phenotype similar to those associated with defects in lamin A, known as laminopathies. On evaluating the fate of prelamin A in fibroblasts derived from these Zmpste24−/− mice, it was observed that prelamin A accumulates in these cells without conversion into mature lamin A. Furthermore, membranes from these cells were inactive, compared with wild-type, in performing the N-terminal processing of yeast a-factor, suggesting that Zmpste24, similar to Ste24p, possesses a farnesylation-dependent endoprotease activity [23]. Taken together with the in vivo accumulation of prelamin A, the hypothesis that Zmpste24 is the prelamin A endoprotease is very interesting.
In the present study, we utilize the expression of recombinant Zmpste24 and prelamin A in insect cells, as well as a synthetic CAAX endopeptidase substrate, to test further the hypothesis that Zmpste24 has endoproteolytic activity towards prelamin A. In addition to the technical advantages of expression of both putative substrate and processing enzyme in insect cells, it has been well established that insects lack lamin A [20]. Thus, it would be expected that there would be a minimum of interference from endogenously expressed substrate-specific proteases. Using this system, we present evidence that Zmpste24 is active towards prelamin A in both of the endoproteolytic cleavages that occur during lamin A maturation.
EXPERIMENTAL
Materials
The BacPak8 baculovirus protein expression system was obtained from Clontech and the Sf21 (Spodoptera frugiperda) insect cells used were from Invitrogen (Carlsbad, CA, U.S.A.). Sf21 cells were grown in Grace's Insect Medium (Invitrogen) containing 10% (v/v) heat-inactivated fetal bovine serum (Invitrogen), 0.1% Pluronic F-68 (Mediatech, Herndon, VA, U.S.A.), 10 μg/ml gentamicin (Invitrogen), supplemented with 500 mg/l CaCl2, 2800 mg/l KCl, L-glutamine, 3330 mg/l lactalbumin hydrolysate and 3330 mg/l yeastolate. CHO-K1 cells (Chinese-hamster ovary-K1 cells) were cultured in Ham's F-12 medium containing 5% (v/v) fetal calf serum, 100 units/ml penicillin and 100 μg/ml streptomycin (F12FC5). Antibodies used in the present study included: mouse monoclonal anti-Zmpste24 (recognizing residues 56–76; a gift from S. Young, Gladstone Institute, San Francisco, CA, U.S.A.); rabbit polyclonal anti-Zmpste24 (recognizing residues 440–455; Abgent, San Diego, CA, U.S.A.); mouse monoclonal anti-His6 (BD Biosciences); goat-anti-mouse IgG–HRP (Pierce; HRP stands for horseradish peroxidase); goat-anti-rabbit IgG–HRP (Santa Cruz Biotechnology); donkey-anti-goat IgG–HRP (Santa Cruz Biotechnology); mouse monoclonal anti-farnesyl (Sigma), goat polyclonal anti-lamin A (Santa Cruz Biotechnology), rabbit polyclonal anti-prelamin A [24] and antimouse IgG Texas Red-conjugated antibody (Biomeda, Foster City, CA, U.S.A.). Radioactive isotopes included: [35S]methionine (specific activity, 1175 Ci/mmol; ICN Biomedicals, Irvine, CA, U.S.A.); 5-R,S-[3H]mevalonate (specific activity, 60 Ci/mmol; American Radiolabeled Chemicals, St. Louis, MO, U.S.A.), [3H]methyl-S-adenosylmethionine (specific activity, 66.8 Ci/mmol; ICN Biomedicals) and [3H]FPP (where FPP stands for farnesyl pyrophosphate; specific activity, 20 Ci/mmol; American Radiolabeled Chemicals).
Cloning of prelamin A, Zmpste24 and ERp57
The template construct for prelamin A was a gift from H. J. Worman (Columbia University, New York, NY, U.S.A.) and included prelaminAct (C-terminal residues 389–664 of prelamin A) cloned into the vector pBFT4 [25]. We performed PCR-directed cloning of this insert fused to a His6 tag at the N-terminus using two separate PCRs: the first stage used the forward primer 5′-ATGGCTCATCATCATCATCATCATCTGTCCCCCAGCCCTACCTC-3′ and the reverse primer 5′-GCGAATTCTTACATGATGCTGCAGTTCT-3′. The product of this PCR was then amplified with the forward primer 5′-TAGGATCCACCATGGCTCATCATCATCATCATCATCTGT-3′ and the same reverse primer as in the first reaction. This product was directionally cloned into the (5′)-BamHI–(3′)-EcoRI restriction sites of the pBacpak8 baculovirus expression vector (Clontech). To make the RSY RLG mutant, we introduced a point mutation using the Quik Change kit (Stratagene) into this prelaminAct/pBacpak8 construct with the primer 5′-GTCACCCGCTCCTACCGCCTGGGCAACTCCAG-3′ along with its complimentary sequence. Zmpste24, fused N-terminally to a His6 tag, was cloned into the (5′)-NheI–(3′)-BamHI site of the pcDNA3.1 construct using reverse transcriptase–PCR on HeLa RNA (Stratagene). The forward primer was 5′-GCGGCTAGCATGGGGATGTGGGCATCGCT-3′ and the reverse primer was 5′-GCGGGATCCGGACATCTCAGTGTTGCTTCATAG-3′. Zmpste24 was then cloned using this as the template into pBacpak8, with the first set of primers being 5′-CATCATCATCATCATCATATGTGGGCATCGCTGGAC-3′ (forward) and 5′-GTGACGCCCGGGTCAGTGTTGCTTCATAGTTTTCAAAGC-3′ (reverse) and the second set being 5′-GCTGGCTCGAGACCATGGGCCATCATCATCATCATCATATGTGG-3′ (forward) and the same reverse primer as in the first reaction. The PCR product was directionally cloned into pBacpak8 using the (5′)-XhoI–(3′)-XmaI restriction sites. The ERp57 served as a mock-infected control to emulate the protein expression changes due to viral infection. ERp57 containing a His6 tag at the N-terminus was cloned similarly using the first set of primers, 5′-ATGGCTCATCATCATCATCATCATCGCCTCCGCCGCCTAGC-3′ (forward) and 5′-GCACGCGAATTCTACTGCTTTAGAGATCCTCCTG-3′ (reverse). The second amplification used 5′-GCTACTGGATCCATGGCTCATCATCATCATCATCATCGCCTC-3′ (forward) and the same reverse primer as in the first reaction. This was cloned into the (5′)-BamHI–(3′)-EcoRI site of pBacPak8. Each construct was verified for accuracy by sequencing. Recombinant high-titre baculovirus stock was produced for prelaminAct, Zmpste24 and ERp57 according to the manufacturer's instructions (Clontech).
Baculovirus expression method
Sf21 (Invitrogen) cells, diluted to 1×106 cells/ml, were grown in exponential phase in Grace's Insect Medium (Invitrogen) containing 10% heat-inactivated fetal bovine serum, 0.1% Pluronic F-68 and 10 μg/ml gentamicin. The cells were infected with hightitre recombinant baculovirus expressing human Zmpste24, prelaminAct or ERp57 (mock-transfected control) at a multiplicity of infection of 10. The cells were pelleted, 72 h post-infection, and then stored at −80 °C. These Sf21 cell pellets were then used for producing enzyme-enriched membranes or purified substrate.
Preparation of enzyme-enriched membranes
Sf21 cells expressing either Zmpste24 or Erp57 (mock-transfected control) were resuspended in 50 mM Tris/HCl (pH 7.0) and disrupted by either sonication or a French press at 6900 kPa. Nuclei and debris were removed by centrifugation at 500 g for 5 min and membranes were then pelleted by centrifugation at 200000 g for 1.5 h. Membranes were resuspended in 50 mM Tris/HCl and stored frozen at −80 °C in multiple aliquots [26]. Protein content was determined by the method of Lowry using the DC Protein Assay Reagent kit (Bio-Rad).
Immunoblotting
Protein samples were separated by SDS/PAGE and then electro-blotted on to a PVDF membrane (Immobilon-P; Millipore). The membranes were saturated with 5% (w/v) non-fat dry milk in TBST [50 mM Tris/HCl (pH 8.0), 138 mM NaCl, 2.7 mM KCl and 0.05% Tween 20; Sigma] and then incubated for 1 h with the primary antibody of interest. Then, the membranes were washed with TBST and incubated for 1 h with either goat-anti-mouse IgG–HRP [with anti-Zmpste24 (residues 56–76) or anti-His6 as primary], goat-anti-rabbit IgG–HRP [with anti-prelamin A or anti-Zmpste24 (residues 440–455) as primary] or anti-goat IgG–HRP (with anti-lamin A as primary). Detection was performed with the ECL® (enhanced chemiluminescence) kit (Pierce).
Metabolic labelling of prelaminAct
After infection with baculovirus (60 h post-infection), Sf21 cells were labelled either for 4 h with [35S]methionine (100 μCi/ml; specific activity, 1175 Ci/mmol) or for 16 h in growth medium containing 5-R,S-[3H]mevalonate (125 μCi/ml; specific activity, 60 Ci/mmol). During mevalonate labelling, insect cells were treated with lovastatin (10 μg/ml; Tocris Cookson, Ellisville, MO, U.S.A.). After labelling, the Sf21 cells were harvested, sonicated in lysis buffer (50 mM NaH2PO4, 300 mM NaCl and 10 mM imidazole, pH 8.0), separated into membrane and cytosolic components and purified using Ni-NTA (Ni2+-nitrilotriacetate) beads (Qiagen). The [3H]mevalonate- and [35S]methionine-labelled prelaminAct fractions were separated on a 4–12% gradient [MES (2-(N-morpholino)ethanesulphonic acid)] or 10% [MOPS (3-(N-morpholino)propanesulphonic acid)] SDS/polyacrylamide gel system (Invitrogen) respectively. Dried gels were visualized by either fluorography or a phosphoimager.
Purification of His6-tagged prelamin A and Zmpste24
Sf21 cells, expressing recombinant protein, were subject to lysis by sonication in a lysis buffer [50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole (pH 8.0) and 1 mg/ml E-64, where E-64 stands for trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane]. Membrane and cytosolic fractions were separated by centrifugation (100000 g for 1 h). Membrane-bound proteins were solubilized in 1% (v/v) Nonidet P40 (prelamin A) or 1.5% (v/v) octyl glucoside (Zmpste24), homogenized, and the insoluble material was removed by centrifugation at 100000 g. Cytosolic and membrane fractions were incubated for 2 h at room temperature (25 °C) with Ni-NTA beads in lysis buffer. After incubation, the beads were washed with 8 vol. of lysis buffer containing 20 mM imidazole and then eluted with 1 vol. of lysis buffer containing 250 mM imidazole for 15 min. Samples were analysed by SDS/PAGE and immunoblotting.
Immunoprecipitation with anti-farnesyl
Ni-purified [35S]methionine-labelled prelaminAct was immunoprecipitated by incubation with an anti-farnesyl antibody in 30 mM Hepes (pH 7.5), 10 mM NaCl, 5 mM MgCl2, 25 mM NaF, 1 mM EDTA and 5% Nonidet P40 for 16 h at 4 °C. The complexes were isolated on Protein A–Sepharose beads. The beads were washed and the immunoprecipitate was resuspended in 0.1 M NaOH and incubated for 30 min at 30 °C to release the precipitated protein. The pH of the eluted sample was neutralized by the addition of 0.1 M Hepes-free acid. The products were separated by SDS/PAGE (10% gel; Invitrogen), transferred on to a PVDF membrane and then visualized with a FUJIFILM FLA-5000 PhosphoImager (FUJIFILM Medical Systems USA, Stamford, CT, U.S.A.).
MALDI–TOF-MS (matrix-assisted laser-desorption ionization–time-of-flight MS)
Proteins were purified by SDS/PAGE, visualized by Coomassie Blue staining and the appropriate bands were excised. The isolated proteins were treated with iodoacetamide and digested with trypsin. MALDI–TOF was with a PerSeptive Voyager DE-RP mass spectrometer in the linear or reflector mode. This was performed by M. A. Gawinowicz at the Protein Chemistry Core Facility at Columbia University.
Mammalian cell culture and transfection
For transient transfections, CHO-K1 cells were seeded at 3× 105 cells/well in a 6-well culture plate and transfected with the Zmpste24/pcDNA3.1 construct using the Stratagene Gene-Jammer transfection reagent according to the manufacturer's instructions.
Indirect immunofluorescence
Transiently transfected CHO-K1 cells on glass coverslips were briefly rinsed in PBS and then fixed with 4% (v/v) formaldehyde in PBS (pH 7.4) for 15 min at 20 °C. After three washes with PBS, the cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min on ice, quickly washed and blocked with 10% (w/v) BSA in PBS for 5 min on ice. The coverslips were incubated for 1 h at room temperature with a mouse monoclonal anti-Zmpste24 antibody (diluted 1:500) developed against residues 56–76. Cells were then washed and incubated at room temperature for 1 h with anti-mouse IgG Texas Red-conjugated antibody (diluted 1:500). Images were obtained by digital deconvolution of 10-slice stacks acquired on a Nikon Diaphot 200 microscope equipped with a Photometrics Sensys cooled charge-coupled-device digital camera or Nikon D100 Oncor Z-Drive using Oncor Image Software.
Farnesylation of CSIM tetrapeptide
Each reaction mixture contained, in a total volume of 100 μl, 50 mM Tris chloride (pH 7.5), 50 μM ZnCl2, 3 mM MgCl2, 20 mM KCl, 1 mM dithiothreitol and 0.2% octyl β-D-glucoside [27], with 10 μM FPP, 100 μM Ac-CSIM-OH (Sigma-GenoSys) and 0.3 μg of farnesyltransferase (Sigma). When the radiolabelled substrate was prepared, unlabelled FPP was replaced by [3H]FPP (20 Ci/mmol) in the reaction above (final concentration of 1 μM). After 1 h incubation at 37 °C, the reaction was terminated by the addition of 0.1 μl of concentrated HCl. Ac-(farnesyl)CSIM-OH was extracted with ethyl acetate (2×100 μl), dried under N2 and dissolved in ethyl acetate. To confirm purity, reaction products were separated by TLC on plastic-backed Silica Gel G thin layer sheets (20 cm×20 cm; Whatman) developed with chloroform/acetone/methanol/acetic acid (70:15:8:2, by vol.) [28]. The farnesylated product was visualized by brief staining with iodine vapour (unlabelled substrate) or by fluorography ([3H]FPP-labelled substrate). Fluorography was accomplished by spraying the TLC plates with Enhance (PerkinElmer) and exposing the film to a Kodak-AR film in the presence of an intensifying screen.
Base release assay for AAXing activity
In a total volume of 20 μl [250 μM zinc acetate, 200 mM NaCl and 200 mM Hepes (pH 7.5)], CAAX proteolysis mixtures contained either 2 μM farnesylated a-factor or 2 μM Ac-(farnesyl)CSIM-OH (see above) and 30 μg of membrane protein from insect cells expressing either ZmpSte24 or Erp57. After incubation at 30 °C for 25 min, reactions were terminated by heating to 95 °C for 1 min [10]. Methylation reactions were then initiated by the addition of premixed 100 μg of Escherichia coli lipid and 0.13 μg of purified Ste14p (isoprenyl cysteine methyltransferase) and [3H]methyl-S-adenosylmethionine (55 μCi/ml; specific activity, 66.8 Ci/mmol) to the reaction mixtures [29]. After incubating for 60 min at 30 °C, the reactions were stopped by the addition of 1 M NaOH/1% SDS, which initiated base hydrolysis of the incorporated [3H]methyl group. Then, 50 μl of each mixture was immediately spotted on to a heavy filter paper lodged into the neck of a scintillation vial containing 10 ml of scintillation fluid (ScintiSafe Econo 2; Fisher Scientific, Fair Lawn, NJ, U.S.A.). After 2–3 h, the filters were removed, and the radioactivity released was determined by liquid-scintillation counting.
CAAX proteolysis reaction on [3H]farnesylated CSIM tetrapeptide using purified and membrane-reconstituted Zmpste24
Zmpste24 was expressed in insect cells and Ni-purified as described above. The activity of purified Zmpste24 was reconstituted by diluting the purified Zmpste24 five times into heat-inactivated insect membranes in 50 mM Tris (pH 7.0) and then forming vesicles by sonication for 3 min. In a total volume of 20 μl, CAAX proteolysis mixtures contained Ac-*fCSIM-OH (20000 c.p.m.), 30 μg of either reconstituted Zmpste24 or heat-inactivated membranes and a buffer containing 200 mM NaCl, 200 mM Hepes (pH 7.5) and 30 mM zinc acetate. In the samples preincubated for 15 min at 30 °C with 2 mM Opa (1,10-orthophenanthroline; a Zn2+ chelator), zinc acetate was not used in the buffer. After incubation at 37 °C for 20 min, the reactions were terminated by the addition of 100 μl of chloroform/methanol (1:1) followed by 100 μl of 1 M citric acid to achieve phase separation. The (lower) organic layer was collected, dried under N2, dissolved in ethyl acetate and spotted on to plastic TLC plates which were developed in chloroform/acetone/methanol/acetic acid (70:15:8:2, by vol.). Substrate (retention factor Rf=0.3) and product (Rf=0.58) were localized by autoradiography and counted using a liquid-scintillation counter. For scintillation counting, the spots were cut and counted in 10 ml of scintillation fluid (Fisher Scientific).
Co-transfection of Zmpste24 and prelaminAct into Sf21 cells
The recipient cells used for co-transfection were Sf21. Cells were infected (baculovirus), 8 h after plating, with either prelaminAct or both prelaminAct and Zmpste24 in the presence or absence of lovastatin (10 μg/ml; Tocris Cookson). Cells were collected, 64 h post-infection, by scraping and centrifugation at 1000 g for 5 min. The cells were washed with PBS, resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl and 10 mM imidazole, pH 8.0) and prelaminAct was purified with Ni-NTA beads. The purified products were analysed by SDS/PAGE and immunoblotting with anti-lamin A or anti-prelamin A.
In vitro endoprotease assay of [3H]mevalonate-labelled prelaminAct
PrelaminAct, purified from insect cells and metabolically labelled with [3H]mevalonate (see above), was incubated with membranes from insect cells expressing either Zmpste24 or Erp57 (mock-transfected). The reaction mixture was separated by SDS/PAGE (4–12% MES) and visualized by fluorography. Reactions were also performed with [3H]mevalonate-labelled prelamin A immobilized on Ni-NTA beads. The amount of [3H]mevalonate-labelled material released was determined at various times by liquid-scintillation counting.
In vitro endoprotease assay using [35S]methionine-labelled prelaminAct as a substrate
Purified [35S]methionine-labelled prelaminAct was incubated with 30 μg of insect membranes in 50 mM Tris/HCl (pH 7.0) in a final volume of 20 μl at 37 °C for various time periods. Each reaction contained the thiol protease inhibitor E-64 (Roche Molecular Biochemicals) to decrease background protease activity from the Sf21 insect cells. A prelaminActΔCAAX-pBFT4 construct (a gift from H. J. Worman), which contained residues 389–660 (a stop codon after the terminal cysteine residue), was in vitro translated in the presence of [35S]methionine with the Quick TnT system obtained from Promega [25]. The product from this reaction was diluted to 400:1 and incubated with membranes as above. Products were separated on 10% MOPS SDS/polyacrylamide gels, then visualized using a phosphoimager. Densitometry was performed using the provided software.
RESULTS
The insect-cell-expressed and purified prelaminAct is fully processed
In designing a suitable substrate for assaying endoproteolytic activity, a number of factors were taken into consideration. Most importantly, in order for the prelamin A endoprotease to cleave the prelamin A substrate, it is necessary for the substrate to be fully post-translationally modified at the CAAX motif (Figure 1) [10]. In addition, since these assays were conducted in vitro, the substrate had to be soluble and, therefore, the N-terminal coiled-coil domain of lamin A was omitted. In the light of these considerations, we expressed and purified a C-terminal fragment of prelamin A (residues 389–664 including the CAAX motif, CSIM) tagged with His6 at the N-terminus, using baculovirus expression in insect cells, and then verified that the substrate was properly post-translationally modified. This C-terminal fragment of prelamin A, prelaminAct, has previously been demonstrated to be water-soluble and, therefore, is suitable for in vitro studies [25].
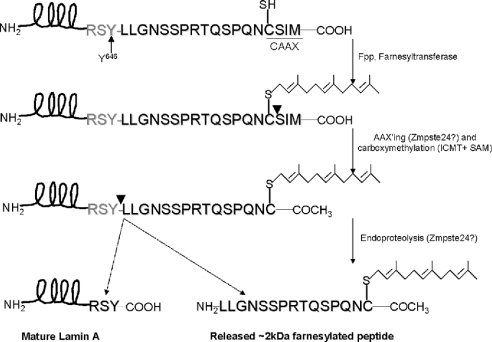
The purified substrate is recognized in immunoblots by a prelamin A-specific antibody (Figure 2A). The prelamin A antibody was raised against the 14-amino acid sequence of prelamin A (LLGNSSPRTQSPQN), residues 647–660 [24], which is removed during endoproteolytic maturation (Figure 1). Immunoblots with an anti-lamin A-specific antibody were also positive (results not shown). The lamin A antibody recognizes a portion of lamin A excluding the prelamin A-specific C-terminal 15 residues. These results indicate that the recombinantly expressed substrate contains both a prelamin A-specific sequence at the C-terminal end and a lamin A-specific sequence.
Figure 2. CAAX box modification of membrane-associated prelaminAct expressed in insect cells.
PrelaminAct fused to His6 at the N-terminus was cloned and expressed in insect cells by means of baculovirus. The expressed prelaminAct is solubilized from membranes and purified by binding and elution from Ni-agarose beads as described in the Experimental section. (A) SDS/PAGE and immunoblotting with anti-prelamin A of purified prelaminAct cytosolic fraction (lane 1), membrane fraction (lane 2) and mock-transfected purification (lane 3). (B) Metabolic labelling of prelaminAct with [3H]mevalonate. Prelamin A was isolated by SDS/PAGE and visualized by fluorography. (C) Immunoprecipitation with anti-farnesyl antibody of purified cytosolic and membrane-associated prelaminAct metabolically labelled with [35S]methionine. Immunoprecipitates (lanes 1 and 3) and supernatants (lanes 2 and 4) of membrane (lanes 1 and 2) and cytosolic fractions (lanes 3 and 4) were analysed by SDS/PAGE and visualized by fluorography. (D) MALDI–TOF analysis of Ni-agarose- and SDS/PAGE-purified membrane-associated prelaminAct. The 33 kDa band was excised from the gel and digested with trypsin. The C-terminal peptide (depicted in the inset) modified by AAXing, farnesylation and carboxyl-methylation (molecular mass, 995.13 Da) was observed.
To confirm that the prelamin A substrate is prenylated, we metabolically labelled prelaminAct expressed in insect cells with [3H]mevalonate, a precursor in the biosynthetic pathway of isoprenoids. After labelling, the prelaminAct substrate was affinity-purified, separated by SDS/PAGE and visualized using fluorography (Figure 2B). The single 3H-labelled band at 33 kDa indicates that the insect cell host is competent for prenylating the specific CAAX box sequence associated with prelamin A, CSIM.
It is possible for CAAX box proteins to be modified with a geranylgeranyl moiety rather than a farnesyl. The above mevalonate labelling experiment did not distinguish between these two potential forms of modification. To verify that the substrate is farnesylated, we immunoprecipitated [35S]methionine-labelled prelaminAct with an anti-farnesyl cysteine-specific antibody (Figure 2C). The membrane-associated prelaminAct was precipitated by this antibody, consistent with the conclusion that prelaminAct is properly farnesylated. In contrast, only a small percentage of the cytosolic fraction of prelaminAct was recognized by this antibody (Figure 2C, lanes 3 and 4), consistent with the conclusion that the cytosolically localized protein is not farnesylated. It has been observed that expression of farnesylated proteins, such as Ras, in insect cells results in only a portion of the expressed proteins being lipidated. This is due to limitations on the capacity of insect cells for isoprenoid biosynthesis [30]. Therefore this result is in agreement with the observation that prenylation targets proteins to membranes. On the basis of these observations, the membrane fraction from insect cells expressing prelaminAct was used as the source of prenylated substrate.
Finally, to demonstrate directly that the substrate is farnesylated, AAXed and carboxyl-methylated, we performed MALDI–TOF-MS on trypsin-digested, purified, membrane-associated prelaminAct, isolated as a band from an SDS/polyacrylamide gel. The C-terminal farnesylated, AAXed and carboxyl-methylated peptide, TQSPQNC (molecular mass, 995.13 Da), was observed (Figure 2D). Furthermore, incompletely processed intermediates, as well as the nascent precursor peaks, were absent from the spectra. This leads us to conclude that the membrane-bound purified substrate is completely modified with the correct post-translational modifications at the CAAX box and, therefore, can be employed as a valid substrate for assaying the second upstream endoproteolytic conversion in the processing of prelamin A.
Zmpste24 expressed in insect cells has a CAAX endopeptidase activity in vitro
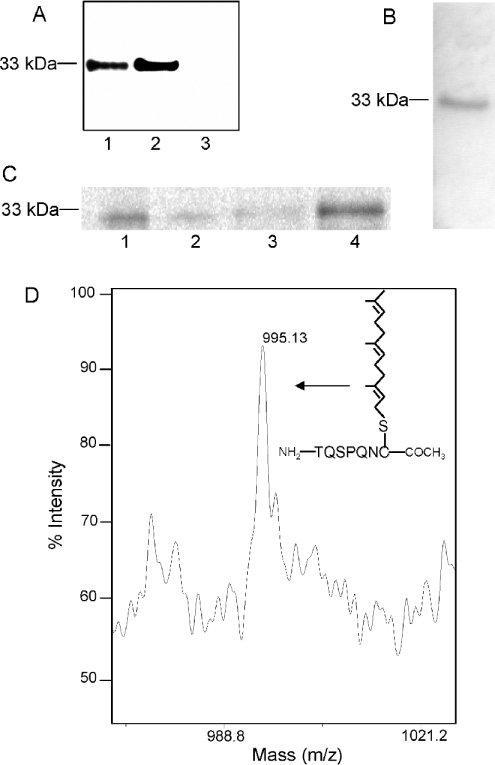
Cloning of the full-length cDNA for human Zmpste24 was accomplished by reverse transcriptase–PCR from a HeLa cDNA library. The results obtained by expressing this construct in CHO-K1 cells and then performing indirect immunofluorescence with a Zmpste24-specific antibody (Figure 3A) are consistent with Zmpste24 localization to the endoplasmic reticulum (reticular staining throughout the cell) and to the nuclear envelope (perinuclear staining). Partial localization of Zmpste24 to the nuclear envelope is consistent with our previous studies on the subcellular localization of prelamin A endoproteolytic processing [10,19].
Figure 3. Cloning and expression of Zmpste24.
(A) Subcellular localization of Zmpste24 expressed in mammalian cells. Zmpste24 was cloned from a HeLa cDNA library into the mammalian expression vector pcDNA3.1 and transiently transfected into CHO-K1 cells. Subcellular localization was determined by indirect immunofluorescence with anti-Zmpste24 (residues 56–76). Note both the cytosolic and perinuclear distribution of Zmpste24. (B) Immunoblot with anti-Zmpste24 (residues 56–76; lanes 1–3), anti-Zmpste24 (residues 440–455; lane 4) or anti-His6 (lane 5) of expressed and Ni-agarose-purified Zmpste24 from insect cells. Lane 1, Ni-agarose-purified elution of mock-transfected cells; lane 2, total cell extract before purification; lanes 3–5, Ni-agarose-purified elutions of Zmpste24.
To produce recombinant enzyme, we cloned and expressed human Zmpste24 N-terminally fused to a His6 tag in the baculovirus system. Ni-bead-purified Zmpste24 runs on SDS/PAGE at 54 kDa, the predicted molecular mass of the protein, although it appears to undergo proteolysis during expression and/or purification (Figure 3). Since the His6 tag is at the N-terminus, the N-terminus of Zmpste24 is intact. The presence of an intact N-terminus is further confirmed by immunoblotting the purified material with anti-Zmpste24 (residues 56–76) and anti-His6 (Figure 3B, lanes 3 and 5 respectively). MALDI–TOF mass spectral analysis of a tryptic digest of the 54 kDa band indicated sequences in this protein predicted to be found in Zmpste24 with 34% sequence coverage spanning residues 50–481 (results not shown). This included a sequence corresponding to the predicted C-terminal peptide of Zmpste24. A band appearing at 54 kDa in the immunoblot with anti-Zmpste24 (residues 440–455) is also consistent with the purified Zmpste24 having an intact C-terminus (Figure 3B, lane 4). The above results are consistent with producing an appropriately constructed Zmpste24 enzyme.
Tam et al. [17] have reported that the human enzyme Zmpste24, similar to its yeast homologue Ste24p, possesses endopeptidase activity towards the farnesylated yeast a-factor CAAX box, resulting in the removal of the C-terminal tripeptide adjacent to the prenylated cysteine residue, i.e. AAXing [17]. Therefore, to ascertain that the insect cell-expressed Zmpste24 construct is enzymically active, we assayed Zmpste24 membranes for AAXing activity using farnesylated a-factor as substrate with the standard base release assay. Our results showed a 5.9±1.0-fold increase in the activity of membranes from cells expressing Zmpste24 relative to the activity of membranes from mock-transfected cells. These results demonstrate that the expressed Zmpste24 has CAAX endopeptidase activity towards the a-factor substrate.
We also confirmed that Zmpste24 can process the prelamin A-specific CAAX sequence, CSIM, utilizing a synthetic N-(Ac)-Cys-(farnesyl)-Ser-Ile-Met tetrapeptide as the substrate in the base release assay. The results show a 9.2±2.9-fold increase in the activity of membranes from cells expressing Zmpste24 relative to the activity of membranes from mock-transfected cells. Therefore Zmpste24 has CAAX endopeptidase activity towards the prelamin A-specific CAAX box sequence, CSIM.
To determine if the AAXing activity being measured is due directly to Zmpste24, we purified Zmpste24 and reconstituted the enzyme into heat-inactivated membranes. We also employed a TLC system for measuring the formation of product to increase the sensitivity of the assay. Our results demonstrate that purified and reconstituted Zmpste24 can AAX the prelamin A model substrate, farnesylated CSIM (Table 1). There is an 8-fold increase in activity with Zmpste24-transfected cells, and a decrease in activity on the addition of the Zn2+ chelator Opa that is not statistically distinguishable from the level observed in mock-transfected cells (Table 1). Inhibition by Opa is in agreement with the idea that the AAXing activity of Zmpste24 is due to a Zn2+-dependent active site.
Table 1. AAXing activity on purified and reconstituted Zmpste24 is Zn2+-sensitive.
Zmpste24 was purified using Ni-agarose chromatography and reconstituted into heat-inactivated insect membranes. N-(Ac)-CSIM was in vitro farnesylated with [3H]FPP (20 Ci/mmol) and farnesyltransferase and incubated with either mock-infected membranes (i) or reconstituted Zmpste24 preincubated with no inhibitor (ii) or 2 mM Opa (iii). Reactions were repeated in triplicate. Products were extracted, separated on TLC, visualized by fluorography, and the N-(Ac)-S-farnesyl-L-cysteine product was identified by co-migration with a commercial standard and counted. Results are expressed as means±S.D. When comparing the results between non-inhibited (ii) and Opa-inhibited (iii) Zmpste24 or the results between non-inhibited (ii) and mock-transfected (i) Zmpste24, the difference is significant with P<0.05 (t test: for two samples assuming equal variances). However, the difference between the mock-transfected (i) and Opa-inhibited (iii) Zmpste24 results is not significant (P=0.08).
| Sample | Activity [pmol of product·(mg of protein)−1·h−1] |
|---|---|
| (i) Mock | 0.02±0.02 |
| (ii) Zmspte24 | 0.16±0.02 |
| (iii) Zmpste24+Opa | 0.08±0.03 |
Zmpste24 can endoproteolytically process prelamin A to lamin A
Previous experiments in which the mouse orthologue to Zmpste24 was knocked out in mice demonstrated that Zmpste24−/− cells could not process prelamin A to mature lamin A [21,22]. This leads to the hypothesis that Zmpste24 may be performing the first, second or both of the endoproteolytic steps during the processing of prelamin A. The experiments described above confirm that Zmpste24 indeed possesses the first endoproteolytic AAXing activity towards the substrate prelamin A. It is, of course, possible that Zmpste24 may also catalyse the second endoproteolytic cleavage, converting prelamin A into the mature lamin A.
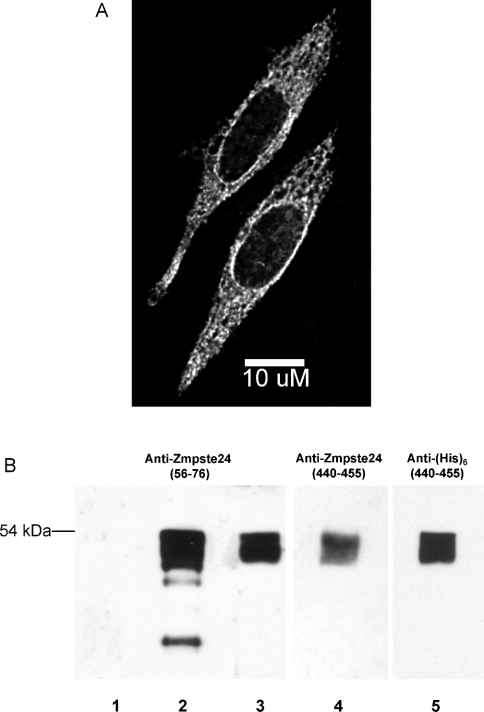
To test this hypothesis in vivo, we expressed either prelaminAct or both prelaminAct and Zmpste24 in insect cells and then monitored the conversion of prelamin A into lamin A using immunoblotting. Co-expression of Zmpste24 and prelamin A results in a product (with a molecular mass 2 kDa lower than that for prelaminAct) that is recognized by a lamin A antibody, indicating that conversion of prelamin A into lamin A is mediated by Zmpste24 expression (Figure 4). This conversion into lamin A is blocked by incubation of the cells with lovastatin, an inhibitor of isoprenoid biosynthesis, which we have previously demonstrated to block prelamin A maturation in mammalian cells by blocking prenylation [5]. We also observed that the prelamin A antibody only recognized the putative 33 kDa prelaminAct band (results not shown), whereas the lamin A antibody recognizes both the top prelaminAct band and the bottom lamin A band (Figure 4, lane 3). This result demonstrates that the change in migration is due to a cleavage that removes the C-terminal prelamin A sequence.
Figure 4. Zmpste24 processing of prelaminAct in transfected insect cells.
Zmpste24 was virally transfected into insect cells expressing N-terminally His6-tagged prelaminAct or prelaminAct mutated at the endoproteolytic cleavage site (L647R). The cells were then incubated for 72 h in the presence or absence of lovastatin. PrelaminAct and laminAct were purified using Ni-agarose chromatography, separated by SDS/PAGE and visualized by immunoblotting with anti-lamin A.
To ascertain the specific site of cleavage at the C-terminus of prelamin A, we repeated this co-expression experiment with a prelaminAct construct bearing a point mutation (L647R, Leu647→Arg) at the p1 site of the conserved hexapeptide sequence RSYRLG, instead of the wild-type sequence RSYLLG. This mutation of prelamin A cannot be processed to lamin A when expressed in mammalian cells [9]. Expression of Zmpste24 did not result in cleavage of this prelaminAct mutant (Figure 4, lane 5), thereby supporting the conclusion that Zmpste24 hydrolyses the peptide bond in prelamin A between Tyr646 and Leu647, as expected for the bona fide prelamin A endoprotease. This level of specificity is particularly supportive of the conclusion that the processing is governed by the expression of Zmpste24, considering the absence of lamin A in insect cells and, therefore, the probable deficiency of comparable endogenous protease activities.
In vitro studies on the prelamin A endoprotease activity of Zmpste24
To determine if the prelamin A endoproteolytic cleavage can be reconstituted in vitro, we first utilized a prelaminAct substrate that was metabolically labelled with [3H]mevalonate. This particular substrate allowed us to follow the endoproteolysis reaction simply by measuring the counts associated with the C-terminal farnesylated fragment. When incubated in vitro with Zmpste24-enriched membranes, this substrate released an approx. 2 kDa 3H-labelled fragment, having the approximate size of the last 15 residues of prelamin A, as the sole labelled product (Figure 5A).
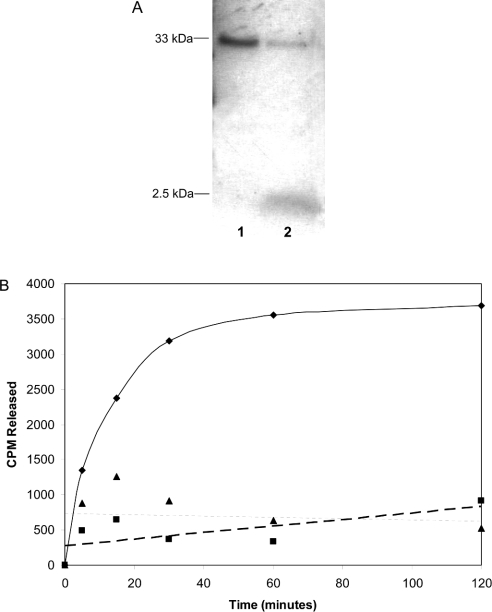
Figure 5. Zmpste24-catalysed cleavage of a 2 kDa fragment from prelaminAct.
(A) PrelaminAct was metabolically labelled for 16 h with [3H]mevalonate, 48 h after infecting with the prelaminAct construct. PrelaminAct was then purified with Ni-agarose. Products formed after incubation with either mock-transfected (lane 1) or Zmpste24 (lane 2) membranes were separated by SDS/PAGE and visualized by fluorography. (B) Purified prelaminAct from insect cell cytosol was in vitro farnesylated with [3H]FPP and farnesyltransferase and immobilized on Ni-agarose beads. The beads were incubated with membranes from Zmpste24 or mock-transfected cells or no enzyme. At various times, aliquots of the supernatant were counted to determine release of the farnesylated C-terminus of prelamin A (◆, Zmpste24 membranes; ■, mock-infected; ▲, no membranes).
To evaluate the kinetics of the release of this 2 kDa prenylated fragment, we performed cleavage reactions on immobilized [3H]mevalonate-labelled prelaminAct and then monitored tritium counts in the supernatant at various times. Formation of the product increases with time and goes to completion between 1 and 2 h (Figure 5B). Notably, membranes from mock-infected cells did not give rise to a measurable product, consistent with the conclusion that product formation arises from the expressed Zmpste24 rather than an endogenous activity of the membranes of insect cells.
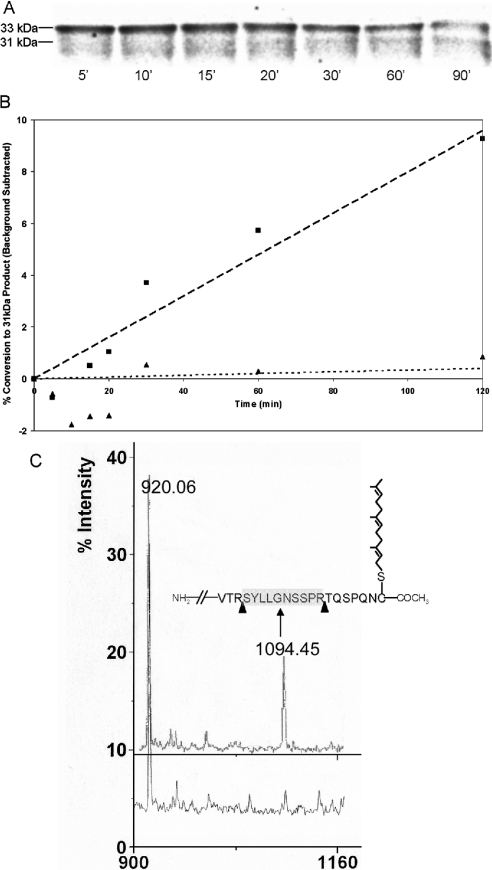
These results demonstrated that the prenylated C-terminal fragment of prelaminAct is being released on incubation with Zmpste24. However, to confirm that these results reflect the physiological processing pathway of prelamin A, we sought to determine whether the parent 33 kDa substrate was being converted into a 31 kDa product. To do this, we expressed and purified a 33 kDa prelaminAct that was metabolically labelled with [35S]methionine and then monitored the processing of this substrate on incubation with membranes from cells expressing Zmpste24. The results reveal that the 33 kDa prelaminAct band was converted into a 31 kDa form in a time-dependent fashion (Figure 6A). In the controls with membranes from mock-transfected cells incubated with wild-type prelaminAct substrate, we observed a non-specific proteolytic activity that degraded the substrate over time. Preincubation of membranes with the thiol protease inhibitor E-64 decreased this non-specific background degradation of prelamin A and allowed the measurement of enriched Zmpste24 activity, but did not completely eliminate the background. The extent to which endoproteolytic conversion occurred was quantified by densitometry and expressed as percentage conversion into the 31 kDa form (Figure 6B). The nonspecific activity seen in mock-transfected membranes was subtracted from these results as background. The RSYRLG mutant of prelaminAct substrate showed little or no conversion above background in this assay. It is clear that the expression of Zmpste24 is resulting in proteolytic activity that is consistent with the expected substrate specificity of the prelamin A endoprotease.
Figure 6. Zmpste24 can endoproteolytically process prelaminAct at the sequence RSYLLG.
PrelaminAct or its mutant (L647R) were metabolically labelled with [35S]methionine in insect cells and purified with Ni-agarose. The proteins were incubated with either Zmpste24 or mock-transfected membranes in the presence of E-64 for various times. (A) WT (wild-type) prelaminAct substrate and endoproteolytic products were separated by SDS/PAGE and visualized by phosphoimager analysis. (B) Relative formation of the 31 kDa product ([31 kDa counts]/[33 kDa+31 kDa counts]–background) as a function in time. Background is defined as the counts observed for mock-transfected membranes incubated with WT substrate. Products continue to be formed for at least 2 h (■, Zmpste24 membranes with WT prelaminAct; ▲, Zmpste24 membranes with RSYRLG mutant prelaminAct). (C) MALDI–TOF analysis of tryptic digest of 33 kDa (upper spectrum) and 31 kDa (lower spectra) gel excised bands, indicating the disappearance of the peak corresponding to the peptide SYLLGNSSPR (molecular mass 1094.21 Da). The peak appearing at 920.06 Da corresponds to the predicted peptide from the prelaminAct sequence between residues 428 and 435. The inset depicts the predicted C-terminal sequence of prelaminAct along with the appropriate CAAX modifications. Arrows represent tryptic cleavage sites. The released peptide (SYLLGNSSPR) has a predicted molecular mass of 1094.45 Da.
The 33 kDa band is recognized by prelamin A antibody, whereas both the 33 and 31 kDa bands are recognized by the lamin A antibody (results were independently verified on unlabelled substrate; results not shown). Confirmation of the identity of these two bands by mass spectral analysis showed that the 31 kDa band is missing in the C-terminal peak corresponding to the peptide SYLLGNSSPR (molecular mass, 1094.21 Da). This is consistent with the conclusion that Zmpste24 cleaves between Tyr646 and Lys647, as would be the case for lamin A maturation in whole mammalian cells (Figure 6C).
As mentioned earlier, the membrane-bound substrate is fully processed in that it is farnesylated, AAXed and carboxyl-methylated. Our previous studies, assaying the activity of nuclear envelope fractions on a model peptide substrate, have shown that it is necessary for prelamin A to be fully processed at the CAAX box before it can act as a substrate for the endoprotease [10]. To assess whether the dependence on processing of prelamin A by Zmpste24 is contingent on full processing at the CAAX box, we performed in vitro translation of a [35S]methionine-labelled prelaminAct construct containing a stop codon after the cysteine residue, designated prelaminAct ΔCAAX [25]. Deleting the CAAX box in this manner ensured that the substrate would be completely unprocessed. In the absence of a C-terminal, farnesylated and carboxyl-methylated cysteine residue, prelaminAct substrate is not cleaved by Zmpste24 to any measurable extent (results not shown). Comparing this to the results when a fully processed substrate was utilized (Figure 6A), we are led to the conclusion that cleavage of prelamin A to lamin A by the endoprotease is, indeed, dependent on processing at the CAAX box. This also corroborates the findings of the co-transfection experiment, which demonstrated that lovastatin treatment abolishes the conversion of prelamin A into lamin A (Figure 4).
DISCUSSION
Studies from other laboratories on Zmpste24−/− mice have clearly demonstrated that prelamin A proteolytic maturation is defective in such animals and that the phenotype of these animals is consistent with pathologies caused by lamin A/C mutations [21,22]. These mouse genetic data, in conjunction with the knowledge of the parallel processing pathways of a-factor in yeast and lamin A in mammals, generated the obvious hypothesis that Zmpste24 is the prelamin A endoprotease. Further support for this hypothesis is given by the observation that patients with mutations in Zmpste24 that abolish or diminish complementation of a-factor production in yeast exhibit some of the same pathology as patients with a genetic laminopathy referred to as mandibuloacral dysplasia [31].
However, direct demonstration of prelamin A endoprotease activity of Zmpste24 has not been reported previously. An understanding of how Zmpste24 endoproteolytically cleaves prelamin A is complicated by several technical hurdles. On the basis of its activity in processing yeast a-factor, Zmpste24, similar to Ste24p, is believed to have two enzymic activities: a CAAX endopeptidase activity as well as a prelamin A endoprotease activity. The first activity is partially redundant with at least one other enzyme in mammalian cells (Ras-converting enzyme-1), making it difficult to design and interpret specific AAXing assays on crude systems. Assaying these two activities independent of one another requires a very careful design of physiologically relevant substrates. Still, the design of a bona fide substrate is complicated by the insolubility of full-length prelamin A and the possible presence of non-specific proteases within the system being studied. For these reasons, as well as other technical advantages, the most definitive biochemical characterization of the endoproteolytic activities of Zmpste24 have been accomplished previously with the yeast a-factor as substrate. Results of previous studies have shown that human Zmpste24 can catalyse both the AAXing and the N-terminal cleavage of a-factor 26 residues upstream from the prenylated cysteine [17,23,32].
The present study extends these observations to prelamin A as a substrate, providing substantial additional support to the hypothesis that Zmpste24 is the prelamin A endoprotease.
This is important since the a-factor substrate differs from prelamin A in the nature of the N-terminal cleavage. The cleavage of a-factor by Ste24 is between a threonine and an alanine residue, 26 amino acid residues upstream from the farnesylated cysteine. In contrast, the cleavage of prelamin A occurs between a tyrosine and a leucine residue, 15 amino acid residues upstream from the farnesylated cysteine. Furthermore, in prelamin A, there appears to be a highly conserved hexapeptide domain (RSYLLG), which is the actual recognition site of the prelamin A endoprotease [9]. Indeed, this sequence is recognized to be unique among mammalian proteins to A-type lamins in the PIR (Protein Information Resource) database. The amino acid residues around the yeast a-factor cleavage site are not homologous with prelamin A, suggesting the possibility that, although Zmpste24 may be capable of cleaving both substrates, it may not do so by the same mechanism.
The possible significance of the differences between prelamin A and a-factor in determining the specificity of endoproteolytic processing argues for the importance of using a suitable substrate. We, therefore, tried to prepare a prelamin A substrate to determine the functionality in prelamin A processing. The construct chosen has previously been reported to function in vitro much like full-length prelamin A but has the advantage of being water-soluble [25]. Mass spectral characterization of the membrane-bound, expressed prelaminAct protein confirmed that the proper CAAX modifications are taking place during insect expression. Endoproteolysis of this substrate by Zmpste24-enriched membranes, in vitro, gives rise to the expected 31 and 2 kDa products, indicating that prelamin A is being converted into lamin A. The lack of endoproteolysis, in vitro, of prelamin A mutant proteins altered either at the cleavage site or at the farnesylation site, mutations that have previously been shown not to undergo processing in mammalian expression studies [9], strongly support the conclusion that this particular system is accurately reconstituting the in vivo reactions.
In the earlier studies, the zinc dependence of the endoproteolytic activity of Zmpste24 was demonstrated for both steps by showing that the AAXing and N-terminal processing reactions for a-factor were both inhibited by the zinc chelator Opa [23,32]. These observations are consistent with both the cleavages of a-factor being catalysed by the zinc metalloprotease activity of Zmpste24. We also find inhibition of AAXing by Opa (Table 1), but, in preliminary studies, do not find this inhibitor to be active in blocking the second endoproteolytic cleavage (results not shown). The significance of this observation is currently under investigation.
The in vitro reconstitution of prelamin A endoproteolysis should be useful in the analysis of possible defects in prelamin A endoproteolytic maturation in the pathologies associated with various laminopathies [33]. For example, in the autosomal dominant Hutchinson–Gilford progeria, the endoprotease site is missing due to variant splicing of the lamin A mRNA [8]. Evidence has been presented that this mutation can act as a dominant-negative interfering with proper lamina assembly and the maturation of lamin A encoded by the wild-type allele [34]. The reconstituted system described here would permit the direct testing of that hypothesis.
Acknowledgments
This work was supported by NIH grant R01GM059578 to M.S.S. We thank Dr S. Young for helpful discussions.
References
- 1.Glomset J. A., Gelb M. H., Farnsworth C. C. Prenyl proteins in eukaryotic cells: a new type of membrane anchor. Trends Biochem. Sci. 1990;15:139–142. doi: 10.1016/0968-0004(90)90213-u. [DOI] [PubMed] [Google Scholar]
- 2.Rando R. R. Chemical biology of protein isoprenylation/methylation. Biochim. Biophys. Acta. 1996;1300:5–16. doi: 10.1016/0005-2760(95)00233-2. [DOI] [PubMed] [Google Scholar]
- 3.Zhang F. L., Casey P. J. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 4.Sinensky M. Recent advances in the study of prenylated proteins. Biochim. Biophys. Acta. 2000;1484:93–106. doi: 10.1016/s1388-1981(00)00009-3. [DOI] [PubMed] [Google Scholar]
- 5.Beck L. A., Hosick T. J., Sinensky M. Isoprenylation is required for the processing of the lamin A precursor. J. Cell Biol. 1990;110:1489–1499. doi: 10.1083/jcb.110.5.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber K., Plessmann U., Traub P. Maturation of nuclear lamin A involves a specific carboxy-terminal trimming, which removes the polyisoprenylation site from the precursor; implications for the structure of the nuclear lamina. FEBS Lett. 1989;257:411–414. doi: 10.1016/0014-5793(89)81584-4. [DOI] [PubMed] [Google Scholar]
- 7.Lutz R. J., Trujillo M. A., Denham K. S., Wenger L., Sinensky M. Nucleoplasmic localization of prelamin A: implications for prenylation-dependent lamin A assembly into the nuclear lamina. Proc. Natl. Acad. Sci. U.S.A. 1992;89:3000–3004. doi: 10.1073/pnas.89.7.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eriksson M., Brown W. T., Gordon L. B., Glynn M. W., Singer J., Scott L., Erdos M. R., Robbins C. M., Moses T. Y., Berglund P., et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature (London) 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hennekes H., Nigg E. A. The role of isoprenylation in membrane attachment of nuclear lamins. A single point mutation prevents proteolytic cleavage of the lamin A precursor and confers membrane binding properties. J. Cell Sci. 1994;107:1019–1029. doi: 10.1242/jcs.107.4.1019. [DOI] [PubMed] [Google Scholar]
- 10.Kilic F., Dalton M. B., Burrell S. K., Mayer J. P., Patterson S. D., Sinensky M. In vitro assay and characterization of the farnesylation-dependent prelamin A endoprotease. J. Biol. Chem. 1997;272:5298–5304. doi: 10.1074/jbc.272.8.5298. [DOI] [PubMed] [Google Scholar]
- 11.Sinensky M., Fantle K., Trujillo M., McLain T., Kupfer A., Dalton M. The processing pathway of prelamin A. J. Cell Sci. 1994;107:61–67. doi: 10.1242/jcs.107.1.61. [DOI] [PubMed] [Google Scholar]
- 12.Fujimura-Kamada K., Nouvet F. J., Michaelis S. A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast a-factor precursor. J. Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderegg R. J., Betz R., Carr S. A., Crabb J. W., Duntze W. Structure of Saccharomyces cerevisiae mating hormone a-factor. Identification of S-farnesyl cysteine as a structural component. J. Biol. Chem. 1988;263:18236–18240. [PubMed] [Google Scholar]
- 14.Chen P., Sapperstein S. K., Choi J. D., Michaelis S. Biogenesis of the Saccharomyces cerevisiae mating pheromone a-factor. J. Cell Biol. 1997;136:251–269. doi: 10.1083/jcb.136.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyartchuk V. L., Rine J. Roles of prenyl protein proteases in maturation of Saccharomyces cerevisiae a-factor. Genetics. 1998;150:95–101. doi: 10.1093/genetics/150.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyartchuk V. L., Ashby M. N., Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- 17.Tam A., Nouvet F. J., Fujimura-Kamada K., Slunt H., Sisodia S. S., Michaelis S. Dual roles for Ste24p in yeast a-factor maturation: NH2-terminal proteolysis and COOH-terminal CAAX processing. J. Cell Biol. 1998;142:635–649. doi: 10.1083/jcb.142.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tam A., Schmidt W. K., Michaelis S. The multispanning membrane protein Ste24p catalyzes CAAX proteolysis and NH2-terminal processing of the yeast a-factor precursor. J. Biol. Chem. 2001;276:46798–46806. doi: 10.1074/jbc.M106150200. [DOI] [PubMed] [Google Scholar]
- 19.Kilic F., Johnson D. A., Sinensky M. Subcellular localization and partial purification of prelamin A endoprotease: an enzyme which catalyzes the conversion of farnesylated prelamin A to mature lamin A. FEBS Lett. 1999;450:61–65. doi: 10.1016/s0014-5793(99)00482-2. [DOI] [PubMed] [Google Scholar]
- 20.Khosravi-Far R., Der C. J. Prenylation analysis of bacterially expressed and insect cell-expressed Ras and Ras-related proteins. Methods Enzymol. 1995;255:46–60. doi: 10.1016/s0076-6879(95)55008-9. [DOI] [PubMed] [Google Scholar]
- 21.Pendas A. M., Zhou Z., Cadinanos J., Freije J. M., Wang J., Hultenby K., Astudillo A., Wernerson A., Rodriguez F., Tryggvason K., et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat. Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- 22.Bergo M. O., Gavino B., Ross J., Schmidt W. K., Hong C., Kendall L. V., Mohr A., Meta M., Genant H., Jiang Y., et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13049–13054. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leung G. K., Schmidt W. K., Bergo M. O., Gavino B., Wong D. H., Tam A., Ashby M. N., Michaelis S., Young S. G. Biochemical studies of Zmpste24-deficient mice. J. Biol. Chem. 2001;276:29051–29058. doi: 10.1074/jbc.M102908200. [DOI] [PubMed] [Google Scholar]
- 24.Sinensky M., Fantle K., Dalton M. An antibody which specifically recognizes prelamin A but not mature lamin A: application to detection of blocks in farnesylation-dependent protein processing. Cancer Res. 1994;54:3229–3232. [PubMed] [Google Scholar]
- 25.Barton R. M., Worman H. J. Prenylated prelamin A interacts with Narf, a novel nuclear protein. J. Biol. Chem. 1999;274:30008–30018. doi: 10.1074/jbc.274.42.30008. [DOI] [PubMed] [Google Scholar]
- 26.Otto J. C., Kim E., Young S. G., Casey P. J. Cloning and characterization of a mammalian prenyl protein-specific protease. J. Biol. Chem. 1999;274:8379–8382. doi: 10.1074/jbc.274.13.8379. [DOI] [PubMed] [Google Scholar]
- 27.Goldstein J. L., Brown M. S., Stradley S. J., Reiss Y., Gierasch L. M. Nonfarnesylated tetrapeptide inhibitors of protein farnesyltransferase. J. Biol. Chem. 1991;266:15575–15578. [PubMed] [Google Scholar]
- 28.Georgopapadakou N. H., Hall C. C., Lambros T., Liu W., Watkins J. D. A radiometric assay for Ras-processing peptidase using an enzymatically radiolabeled peptide. Anal. Biochem. 1994;218:273–277. doi: 10.1006/abio.1994.1178. [DOI] [PubMed] [Google Scholar]
- 29.Hrycyna C. A., Wait S. J., Backlund P. S., Jr, Michaelis S. Yeast STE14 methyltransferase, expressed as TrpE-STE14 fusion protein in Escherichia coli, for in vitro carboxylmethylation of prenylated polypeptides. Methods Enzymol. 1995;250:251–266. doi: 10.1016/0076-6879(95)50077-4. [DOI] [PubMed] [Google Scholar]
- 30.Khosravi-Far R., Der C. J. Prenylation analysis of bacterially expressed and insect cell-expressed Ras and Ras-related proteins. Methods Enzymol. 1995;255:46–60. doi: 10.1016/s0076-6879(95)55008-9. [DOI] [PubMed] [Google Scholar]
- 31.Agarwal A. K., Fryns J. P., Auchus R. J., Garg A. Zinc metalloproteinase, ZMPSTE24, is mutated in mandibuloacral dysplasia. Hum. Mol. Genet. 2003;12:1995–2001. doi: 10.1093/hmg/ddg213. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt W. K., Tam A., Michaelis S. Reconstitution of the Ste24p-dependent N-terminal proteolytic step in yeast a-factor biogenesis. J. Biol. Chem. 2000;275:6227–6233. doi: 10.1074/jbc.275.9.6227. [DOI] [PubMed] [Google Scholar]
- 33.Mounkes L., Kozlov S., Burke B., Stewart C. L. The laminopathies: nuclear structure meets disease. Curr. Opin. Genet. Dev. 2003;13:223–230. doi: 10.1016/s0959-437x(03)00058-3. [DOI] [PubMed] [Google Scholar]
- 34.Goldman R. D., Shumaker D. K., Erdos M. R., Eriksson M., Goldman A. E., Gordon L. B., Gruenbaum Y., Khuon S., Mendez M., Varga R., et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]