Abstract
Prion protein consists of an N-terminal domain containing a series of octapeptide repeats with the consensus sequence PHGGGWGQ and a C-terminal domain composed of three α-helices and two short β-strands. Several studies have shown that the N-terminal domain binds five Cu2+ ions. In the present study, we have investigated copper-catalysed oxidation of a recombinant mouse prion protein, PrP23–231. The copper-loaded PrP23–231 was found to be carbonylated by incubation with dopamine. Besides the formation of carbonyls, a cross-linked species with the dimeric size and C-terminally truncated species were generated. These reactions were retarded in the presence of Cu+- and Cu2+-specific copper chelators, catalase, and SOD (superoxide dismutase), but not in the presence of various bivalent metal ions. Together, these results indicate that the copper bound to prion protein undergoes catalytic cycling in the presence of catecholamines and causes the oxidation of the protein.
Keywords: carbonyl, copper, dopamine, metal ion, oxidative damage, prion protein
Abbreviations: BCA, 4,4′-dicarboxy-2,2′-biquinoline; BCS, 2,9-dimethyl-4,7-diphenyl-1,10-phenanthrolinedisulphonic acid; DTPA, diethylenetri-aminepenta-acetic acid; PrP, prion protein; PrP A, rabbit polyclonal anti-prion protein A antibody; PrPC, cellular PrP isoform; SOD, superoxide dismutase; TBS, Tris-buffered saline; TBS-T, TBS containing 0.1% (v/v) Tween 20
INTRODUCTION
All mammalian and avian species possess PrPC (cellular prion protein isoform), but its normal physiological function has not yet been determined. However, the property of PrPC to bind Cu2+ in vivo and in vitro suggests that its function relates to copper homoeostasis or to copper-dependent enzymatic functions [1,2]. Because Cu2+ stimulates PrPC endocytosis, it has been suggested that PrPC plays a role in shuttling Cu2+ from the synaptic space to the cell interior [3,4]. However, another study showed that cuproenzyme activity was not influenced by the degree of PrP (prion protein) expression in brain tissues [5]. The authors suggested that PrPC might act as a reversible sink or a carrier of the metal ion [5]. In addition, a recent study indicated that the expression of PrP increased copper binding to cells, but did not affect copper uptake, antioxidant enzyme activities or glutathione levels [6]. Another suggestion is that PrP might be a stress sensor for copper and might be able to initiate, following copper binding, a signal transduction process for improving cell defences by acting on the antioxidant systems [6]. An enzymatic role for copper-bound PrP was also proposed as it exhibited SOD (superoxide dismutase) activity, protecting synaptic regions from oxidative stress [7–10]. Our previous study indicated that copper ions bound to an N-terminal part of PrP (PrP23-98) catalysed oxidations of L-ascorbate and dopamine [11]. The last two studies have shown that the PrP-bound copper undergoes redox cycling in the presence of electron donors, such as superoxide ions (O2−), dopamine and L-ascorbate.
PrPC contains a C-terminal domain, residues 126–231, that has a globular fold composed of three α-helices and two short β-strands [12,13]. Under Cu2+-free conditions, the N-terminal portion of the mature PrPC, residues 23–125 of the primary translation product, is largely unstructured [13–15]. Residues 60–91 consist of an octa-peptide sequence, PHGGGWGQ, which is repeated four times. Several studies have shown that this unstructured region selectively binds Cu2+ over other bivalent metal ion species [16–24]. This octapeptide repeat region binds four Cu2+ ions co-operatively with identical co-ordination geometry [20,24–26]. The affinity of this copper binding is in the femtomolar to micromolar range [21,23]. In addition, the fifth Cu2+-binding site centred at His-96 and His-111 has also been observed [22,23,26]. In fact, affinity-purified PrPC preparations from mouse and human brain have been shown to bind three and approx. seven copper atoms respectively [10,27]. Moreover, PrPC from cultured cells was found to bind one to four copper atoms, depending on the availability of copper in the culture medium [10]. In the light of these results, it is probable that native PrP binds copper in vivo.
The evidence from several studies has suggested that the most important mechanism of oxidative damage to proteins involves catalysis by transition metals [28,29]. This process consists of reduction of Fe3+ or Cu2+ by electron donors, such as O2−, H2O2, catecholamines, L-ascorbate and mercaptans, and generation of the hydroxyl radical through reduction of H2O2 by the reduced metals. This highly reactive free radical immediately oxidizes neighbouring amino acid residues. The reaction typically results in structural alterations and loss of enzyme activity [28,29]. As regards the oxidative damage of PrP, PrP undergoes a site-specific cleavage of the octapeptide repeat region on exposure to H2O2 plus CuSO4 [30]. Also, PrP suffers aggregation and precipitation upon incubation with L-ascorbate and CuCl2, the histidine-containing octarepeat region being particularly affected by oxidation [31]. Our previous study has also indicated that carbonyl formation on copper-bound PrP23-98 and a decrease in its histidine content were induced by incubation with dopamine or L-ascorbate [11]. These studies indicate that the N-terminal domain is susceptible to copper-catalysed oxidation. However, there are conflicting data as to structural alterations in PrP produced by copper-catalysed oxidation.
Certain neurotransmitters, such as dopamine and noradrenaline, are autoxidizable and react with O2 to generate O2− and H2O2 through catalysis by heavy-metal ions [32]. Dopamine is present in various anatomical regions of the brain, its extraordinarily high concentrations being found in the striatum (37 and 53 μM in the caudate nucleus and putamen of humans respectively) [33]. Because the PrP-bound copper is reduced by electron donors, such as O2−, dopamine and L-ascorbate [7–11], it is probable that incubation of PrP with copper salts in the presence of H2O2 or L-ascorbate causes structural alterations in PrP [30,31]. However, the oxidative damage and structural alterations in copper-bound PrP, which should exist in vivo [10,27], remain largely unknown. In the present study, we have investigated catecholamine-induced oxidation of a recombinant mouse PrP23–231 to which copper has previously loaded. The results have indicated that in the presence of catecholamine, copper-bound PrP23–231 undergoes carbonylation on its own part, and partly leads to its dimerization and fragmentation.
EXPERIMENTAL
Antibodies and reagents
PrP A (rabbit polyclonal anti-prion protein A antibody) recognizing epitope 228–244 of bovine PrP (epitope 216–232 of mouse PrP) was purchased from Cosmo Bio Co. (Tokyo, Japan). Mouse monoclonal antibody SAF 32 recognizing epitope 78–91 of hamster PrP, mouse monoclonal antibody SAF 8G8 recognizing epitope 95–110 of human PrP, mouse monoclonal antibody SAF 70 recognizing epitope 142–160 of hamster PrP and mouse monoclonal antibody SAF 84 recognizing epitope 160–170 of hamster PrP were purchased from Cayman Chemical Co. (Ann Arbor, MI, U.S.A.). These antibodies exhibit cross-reactivity to mouse PrP as described in the product information. Horseradish-peroxidase-conjugated anti-mouse IgG secondary antibody was purchased from Chemicon International (Temecula, CA, U.S.A.), and horseradish-peroxidase-conjugated anti-rabbit IgG secondary antibody was from Cappel Research Products (Durham, NC, U.S.A.). Plasmid pUC 19 was purchased from Takara Bio (Tokyo, Japan). Plasmid pET-39b (+) and S-protein–agarose were purchased from Novagen (Madison, WI, U.S.A.). BSA, catecholamines, rabbit polyclonal anti-dinitrophenyl antibody and Protein G–Sepharose 4B were purchased from Sigma–Aldrich (St. Louis, MO, U.S.A.). Benzamidine–Sepharose 6B, SP Sepharose, ECL® (enhanced chemiluminescence) Western blotting detection reagents and Hybond ECL® nitrocellulose membrane were purchased from Amersham Biosciences (Piscataway, NJ, U.S.A.). EnterokinaseMax was purchased from Invitrogen (Carlsbad, CA, U.S.A.). SOD and catalase were purchased from Alexis Biochemicals (San Diego, CA, U.S.A.) and Nacalai Tesque (Kyoto, Japan) respectively. Xanthine oxidase and xanthine were purchased from Wako Pure Chemical Industries (Osaka, Japan). All other chemicals were purchased from Nacalai Tesque, unless specified otherwise. Distilled water was purified by passage through a Milli-Q Academic A10 system (Millipore, Bedford, MA, U.S.A.). The resistance of the water was 1.8×107 Ω·cm at 20 °C.
Expression and purification of PrP23–231 and preparation of copper-loaded PrP23–231
The DNA encoding PrP23–231 was amplified via PCR from mouse genomic DNA under the standard conditions using primers 5′-CGGGATCCCGAAAAAGCGGCCAAAGCCTGGAGGG-3′ (sense primer which includes an artificial BamHI site) and 5′-CCGGAATTCCTAGGATCTTCTCCCGTCGTAATAGGC-3′ (antisense primer which includes an artificial EcoRI site). The PCR product was ligated to the BamHI and EcoRI sites of plasmid pUC19, and the sequence of the insert was verified by DNA sequence analysis. A HindIII/BamHI adapter containing an SrfI site was ligated to the HindIII and BamHI sites of the resulting construct. After digestion of pUC19 with SrfI and EcoRI, the insert was ligated to the SrfI and EcoRI sites of plasmid pET-39b (+). Escherichia coli cells of the strain BL21 (DE3) pLysS were transformed with the construct pET-39b (+)/PrP23–231, and the resulting recombinant clone was cultured at 37 °C in LB (Luria–Bertani) broth containing 30 μg/ml kanamycin. When the A600 of the culture reached approx. 0.8, IPTG (isopropyl β-D-thiogalactoside) was added to a concentration of 1 mM, and the cells were cultured at 37 °C for 2 h. Then the cells were spun down at 5500 g for 20 min, washed with PBS and suspended in 20 mM Tris buffer (pH 7.5) containing 0.15 M NaCl, 0.1% Triton X-100 and 1 mM 2-mercaptoethanol. The cell suspension was stored at −20 °C until use.
The cells were disrupted on ice with a sonicator (Digital Sonifier S-250D; Branson Ultrasonics, Danbury CT, USA) at 4 °C for 10 min, and then the lysate was centrifuged at 35000 g for 20 min at 4 °C. The supernatant was applied to an S-protein–agarose column equilibrated with 20 mM Tris buffer (pH 7.5) containing 0.15 M NaCl and 0.1% (v/v) Triton X-100. After the column was washed with 25 mM TBS (Tris-buffered saline), pH 7.5 (Tris buffer containing 0.136 M NaCl and 2.68 mM KCl), the Dsb A/S·Tag fusion protein was eluted with 0.2 M sodium citrate buffer (pH 2), dialysed against 20 mM Mes buffer (pH 6.0) containing 1 mM CaCl2 at 4 °C for ~18 h, and centrifuged at 19000 g at 4 °C for 10 min. The supernatant was treated with Entero-kinaseMax at room temperature (25 °C) for 2 h, and after addition of NaCl to a final concentration of 0.15 M and adjustment of pH to 7.5 with 1 M Tris/HCl, the EnterokinaseMax was removed by chromatography on a benzamidine–Sepharose 6B column equilibrated with TBS. After addition of Tween 20 to a final concentration of 0.1%, the pass-through fraction containing recombinant PrP23–231 and Dsb A/S·Tag protein was applied to an S-protein–agarose column equilibrated with TBS-T [TBS containing 0.1% (v/v) Tween 20]. The eluate containing the recombinant PrP23–231 was applied to an SP Sepharose column equilibrated with TBS. PrP23–231 was eluted with stepwise NaCl gradients (0.2, 0.3, 0.35 and 0.4 M). Fractions containing PrP23–231 were dialysed against 5 mM Mes buffer (pH 7.5) at 4 °C and stored at −80 °C until use. The concentration of PrP23–231 was determined spectrophotometrically by using a molar absorption coefficient at 280 nm of 62160 M−1·cm−1, which was deduced from the contents of tryptophan and tyrosine (molar absorption coefficient, 5690 M−1·cm−1 for tryptophan and 1280 M−1·cm−1 for tyrosine) [34].
Copper-loaded PrP23–231 was prepared by incubation of the recombinant PrP23–231 (2 μM) with CuCl2 (10 μM) and urea (6 M) at 37 °C for 30 min, followed by dialysis at 4 °C for ~18 h, first against a 10 μM CuCl2 solution to remove urea, and secondly against 5 mM Mes buffer (pH 7.5) to remove free copper. The copper content of the resulting copper-loaded PrP23–231 was measured with an atomic absorption spectrophotometer (AA-6800, Shimadzu, Kyoto, Japan).
Western blotting
Mixtures (100 μl) containing 50 mM Mes (pH 7.5), 1 μM copper-loaded PrP23–231 and catecholamine were incubated at 37 °C for 15–60 min, before 2 μl of 500 mM EDTA was added to stop the reaction. The reaction mixtures were mixed with gel-loading buffer containing dithiothreitol, and then heated at 95 °C for 5 min. For detection of PrP and carbonyl groups, the samples were analysed by SDS/PAGE followed by immunoblotting.
The membranes were blocked with 0.5% (w/v) non-fat dried milk in TBS-T at room temperature for 1 h, and then incubated with primary antibody (0.2 μg/ml SAF 32, 0.6 μg/ml 8G8, 0.6 μg/ml SAF 70, 0.2 μg/ml SAF 84, PrP A diluted 2500-fold or rabbit polyclonal anti-dinitrophenyl antibody diluted 2000-fold) in TBS-T containing 1% (w/v) BSA for 1 h with shaking at room temperature. The membranes were washed with TBS-T and incubated with horseradish-peroxidase-conjugated anti-mouse IgG secondary antibody (diluted 1:5000 in TBS-T containing 1% BSA) or horseradish-peroxidase-conjugated anti-rabbit IgG secondary antibody (diluted 1:5000 in TBS-T containing 1% BSA) at room temperature for 1 h. The blots were developed by ECL® Western blotting detection reagents, and their signals were recorded using a CCD (charge-coupled device) camera (Lumino Capture Af-650; ATTO Co., Tokyo, Japan). In the case of the carbonyl assay, a replicate blot was stained for protein with Coomassie Blue. Relative chemiluminescence intensities of bands were determined using ATTO Densitograph Software Library, version 4.0. The results were subjected to statistical evaluation using an unpaired Student's t test.
Immunoprecipitation
Mixtures containing 50 mM Mes (pH 7.5) and 1 μM copper-loaded PrP23–231 were incubated in the absence or presence of 10 μM dopamine at 37 °C for 30 min, before 2 μl of 500 mM EDTA was added to stop the reaction, and then mixtures were dialysed at 4 °C for 8 h against 5 mM Mes buffer (pH 7.5). Anti-PrP monoclonal antibody SAF 32 (3 μg) was added to the reaction mixtures (0.5 ml). After 1 h of incubation at room temperature, Protein G–Sepharose 4B was added. After further incubation for 1 h, the Sepharose beads were pelleted and washed extensively with 50 mM Mes buffer (pH 7.5) containing 0.15 M NaCl. The washed beads were mixed with 2× gel-loading buffer containing dithiothreitol, and then heated at 95 °C for 5 min. PrP was detected by SDS/PAGE, followed by immunoblotting with antibodies SAF 84 and PrP A (0.2 μg/ml SAF 84 and 2500-fold diluted PrP A in TBS-T containing 1% BSA).
Ultrafiltration experiment
The aliquots of the above reaction mixtures prepared for Western blotting were used for measurement of copper released from the protein during the incubation. The reaction mixtures (400 μl) were dispensed into cups of Ultrafree-MC centrifugal filter units (Biomax-5; Millipore) and centrifuged at 5000 g for 15 min at 4 °C, and the copper concentration of the resulting filtrates was measured with an atomic absorption spectrophotometer.
SOD assay
SOD activity was measured by the xanthine oxidase/Nitro Blue Tetrazolium method [35]. The reaction mixtures contained 80 μM Nitro Blue Tetrazolium, 100 μM xanthine, 0.00375 unit/ml xanthine oxidase and 50 mM Mes buffer (pH 7.5), with or without copper-loaded PrP23–231 at room temperature. The Nitro Blue Tetrazolium reduction was followed spectrophotometrically at 560 nm.
RESULTS
Copper-loaded PrP23–231 undergoes structural damage in the presence of catecholamine
Because the Cu2+ ions bound to PrP are reducible by electron donors, such as O2−, catecholamines and L-ascorbate, as shown in previous studies [7–11], we aimed to examine oxidative damage to the PrP molecule by the copper in the presence of electron donors. We prepared copper-loaded PrP23–231 by incubation of recombinant mouse PrP23–231 and CuCl2 in the presence of urea, followed by dialysis essentially as described by Brown et al. [7,8]. The refolded PrP23–231 contained 5.0–5.2 mol of copper per mol of protein, and exhibited SOD activity as measured by the formazan formation assay [35], like the copper-loaded PrP prepared by Brown's group [7–10] (results not shown). Because the interaction of PrP and dopamine is feasible in the brain, we studied the effect of dopamine on copper-loaded PrP23–231. The copper-loaded PrP23–231 was incubated with 10 μM dopamine at 37 °C for 30 min. After centrifugation (at 10000 g for 10 min at 4 °C) of the reaction mixture, the supernatants contained the same amounts of protein as a control with dopamine omitted (results not shown). This is in contrast with the report by Requena et al. [31], who indicated that PrP formed an aggregate in its incubation with L-ascorbate and CuCl2. When the sample of the dopamine treatment was analysed by SDS/PAGE, followed by staining with Coomassie Blue, the sample of the dopamine treatment showed a significantly decreased staining for the PrP23–231 band as compared with the control, indicating the decomposition of the PrP23–231 molecule (results not shown).
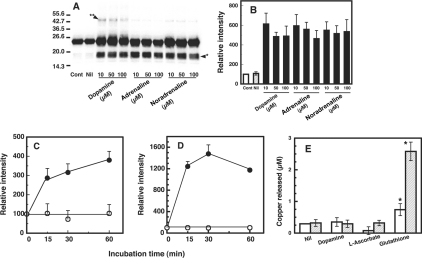
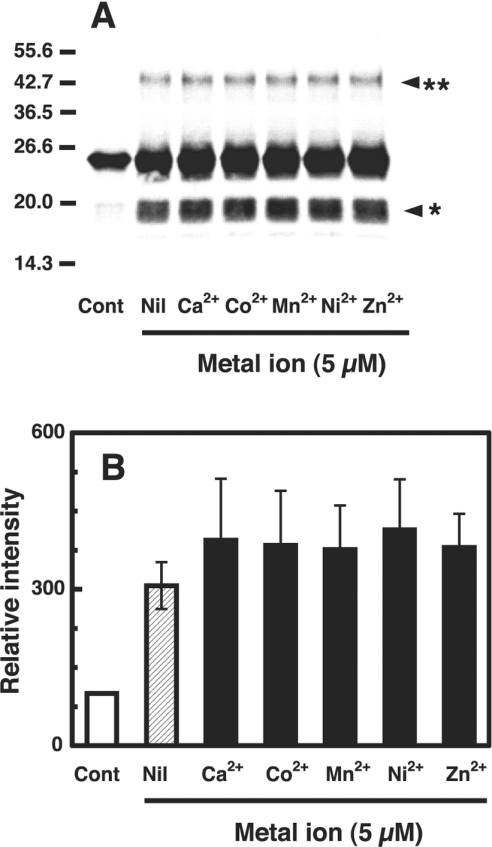
To examine further the oxidative damage to copper-loaded PrP23–231, the PrP23–231 preparation (1 μM, containing 5.2 μM copper) was treated with varying concentrations (10, 50 and 100 μM) of dopamine, adrenaline or noradrenaline, and analysed by Western blot analysis using SAF 84 antibody that recognizes epitope 160–170 of hamster PrP. The PrP23–231 remained as a major band at 23 kDa (Figure 1A, Cont and Nil). Additionally, a strong, broad band and a faint band were observed at positions of 17–19 kDa (Figure 1A, arrowhead*) and 46 kDa (Figure 1A, arrowhead**) respectively, indicating the formation of fragments and dimerization of PrP23–231. The degrees of fragmentation did not change upon increasing the concentrations of catecholamines (Figure 1B). The time-course study with dopamine showed a dramatic increase in the signal of the fragments (Figure 1C) and the dimer (Figure 1D) occurred within 15 min. The signal of fragments gradually increased until 60 min; the signal of the dimer increased until 30 min, then decreased.
Figure 1. Fragmentation and dimerization of the copper-loaded PrP23–231 in the presence of catecholamine.
(A) Western blot immunoassay for copper-loaded PrP23–231. Mixtures containing 50 mM Mes (pH 7.5) and 1 μM copper-loaded PrP23–231 (containing 5.2 μM copper) were incubated with or without (Nil) the indicated concentrations of dopamine, adrenaline or noradrenaline at 37 °C for 30 min. As a control, copper-loaded PrP23–231 was kept on ice was used (Cont). PrP was detected by SDS/PAGE, followed by immunoblotting with antibody SAF 84. Molecular masses in kDa are indicated on the left. The positions of the fragments at 17–19 kDa and the dimer at 46 kDa are marked by arrowhead* and arrowhead** respectively. (B) Quantification of the Western blot data. Relative chemiluminescence intensities of the bands of the fragments (A, arrowhead*) were determined using ATTO Densitograph Software Library. Because the copper-loaded PrP23–231 preparation contained a trace of the 17–19 kDa fragments, their signal was taken as 100% for expression of intensities of the test samples. Data are means±S.D. (n=3). (C) and (D) Time course of formation of 17–19 kDa fragments and dimer (46 kDa). A mixture containing 50 mM Mes (pH 7.5) and 1 μM copper-loaded PrP23–231 (containing 5.2 μM copper) was incubated without (○) or with 10 μM dopamine (●). PrP was detected by SDS/PAGE followed by immunoblotting with antibody SAF 84. Signals of 17–19 kDa fragments (C) and dimer (46 kDa) (D) were determined using ATTO Densitograph Software Library and plotted as a percentage of the control (zero time). Data are means±S.D. (n=3). (E) Release of copper from copper-loaded PrP23–231 by reducing substances. Copper-loaded PrP23–231 (1 μM, containing 5.2 μM copper) was incubated with dopamine, L-ascorbate or glutathione at 10 μM, mixtures were centrifuged immediately (open bars) or after 30 min (hatched bars) in ultrafiltration cups, and copper concentrations of the filtrates were measured by atomic absorption spectrophotometry. The measured concentrations were corrected for the copper concentration of the buffer used. Data are means±S.D. (n=3). *Significant difference (P≤0.01) from the experiment with no additions (Nil).
We next examined whether the addition of dopamine, L-ascorbate or glutathione releases copper from the copper-bound PrP23–231, because they are known to interact with copper [11,36,37]. These compounds (a final concentration of 10 μM) were added to copper-loaded PrP23–231, and the amount of copper filterable by ultrafiltration was measured immediately and at 30 min after mixing. In the experiment with dopamine and L-ascorbate, the filterable copper was not detected (Figure 1E), indicating that the binding of copper to PrP23–231 was not affected by these compounds. On the other hand, in the experiment with glutathione, 9% of the PrP23–231-bound copper was released even immediately after mixing, and the released copper increased to 44% after 30 min of incubation. The copper-loaded PrP23–231 produced the dimer and fragments by incubation with L-ascorbate, whereas the effect by glutathione was not observed (results not shown).
Inhibitory effect of metal chelators, catalase and SOD on carbonyl formation of copper-loaded PrP23–231
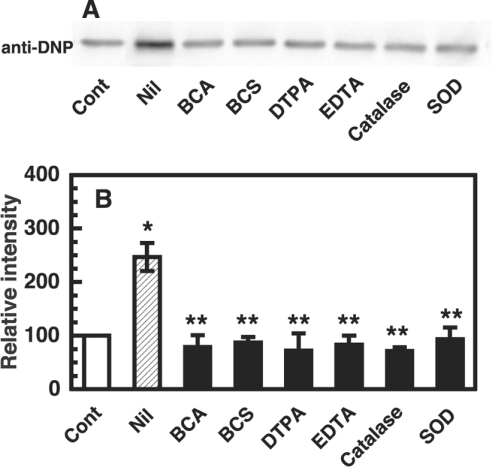
To investigate the oxidative protein damage on copper-loaded PrP, we used the formation of carbonyls as a marker. Copper-loaded PrP23–231 was incubated with 10 μM dopamine at 37 °C for 30 min, and the protein was analysed by Western blotting using rabbit polyclonal anti-dinitrophenyl antibody. A major band was observed at the position of PrP23–231 (Figure 2A). To ascertain the participation of copper and reactive oxygen species in the formation of carbonyls on copper-loaded PrP23–231, the effects of various copper chelators, catalase and SOD were tested. The metal chelators, BCA (4,4′-dicarboxy-2,2′-biquinoline) and BCS (2,9-dimethyl-4,7-diphenyl-1,10-phenanthrolinedisulphonic acid) as Cu+ chelators, DTPA (diethylenetriaminepenta-acetic acid) and EDTA as Cu2+ chelators, were used at 100 μM in the present study. The inhibition was totally complete with either of the copper chelators (Figure 2B). Catalase and SOD also completely inhibited the formation of carbonyls on copper-loaded PrP23–231 (Figure 2B).
Figure 2. Inhibitory effects of metal chelators, catalase and SOD on carbonyl formation of copper-loaded PrP23–231.
(A) Western blot immunoassay for carbonyl on copper-loaded PrP23–231. Copper-loaded PrP23–231 (1 μM, containing 5.2 μM copper) and 10 μM dopamine were incubated at 37 °C for 30 min without (Nil) or with either of the indicated metal chelators (100 μM) or enzyme (catalase 1000 units/ml or SOD 1000 units/ml). Copper-loaded PrP23-23 without the incubation was used as a control (Cont). For the detection of carbonyl groups, the samples were analysed by SDS/PAGE followed by immunoblotting. (B) Quantification of the Western blot data. Signals of specific bands were determined using ATTO Densitograph Software Library and indicated as a percentage of the control value (copper-loaded PrP23–231 incubated without dopamine, Cont). Data are means±S.D. (n=4–5). *Significant difference (P≤0.01) from the experiment with control. **Significant difference (P≤0.01) from the experiment with no additions (Nil).
Inhibitory effect of metal chelators, catalase and SOD on fragmentation and dimerization of the copper-loaded PrP23–231
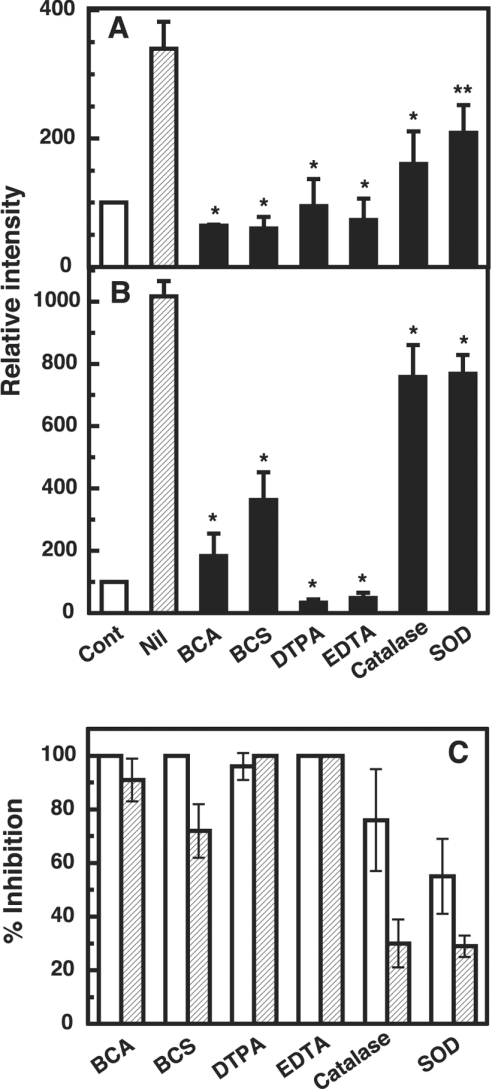
To examine the participation of copper and reactive oxygen species in the formation of the dimer (46 kDa species; see Figure 1A, arrowhead**) and fragments (17–19 kDa; see Figure 1A, arrow-head*) from copper-loaded PrP23–231, the effects of various copper chelators, catalase and SOD were tested in the same experimental setting as in Figure 2 (Figure 3). The formation of fragments was totally or nearly totally inhibited, depending on the kind of the copper chelators (BCA, BCS, DTPA or EDTA) (Figures 3A and 3C). Catalase and SOD also reduced the formation of fragments, albeit to lesser extents (Figures 3A and 3C). For the dimer formation, BCA, DTPA and EDTA were completely inhibitory, while BCS was less effective (Figures 3B and 3C). The inhibitory effect of catalase and SOD on the dimer formation was very weak (Figures 3B and 3C).
Figure 3. Inhibitory effects of metal chelators, catalase and SOD on fragmentation and dimerization of the copper-loaded PrP23–231.
(A) and (B) Quantification of the Western blot data. Copper-loaded PrP23–231 (1 μM, containing 5.2 μM copper) and 10 μM dopamine were incubated for 30 min at 37 °C without (Nil) or with either of the indicated metal chelators (100 μM) or enzyme (catalase, 1000 units/ml, or SOD, 1000 units/ml). PrP was detected by SDS/PAGE followed by immunoblotting with antibody SAF 84. Signals of 17–19 kDa fragments (A) and a dimer (46 kDa) (B) were determined using ATTO Densitograph Software Library and plotted as a percentage of the control (copper-loaded PrP23–231 incubated without dopamine, Cont). Data are means±S.D. (n=3). Significant difference (*P≤0.01 and **P≤0.05) from the experiment with no additions (Nil). (C) Percentage inhibition for fragmentation (open bars) and dimerization (hatched bars) are expressed as follows: inhibition (%)=[(relative intensity for Nil−relative intensity for inhibitor)/(relative intensity for Nil−100)]×100.
Copper-loaded PrP23–231 undergoes structural damage in the presence of H2O2 and O2−
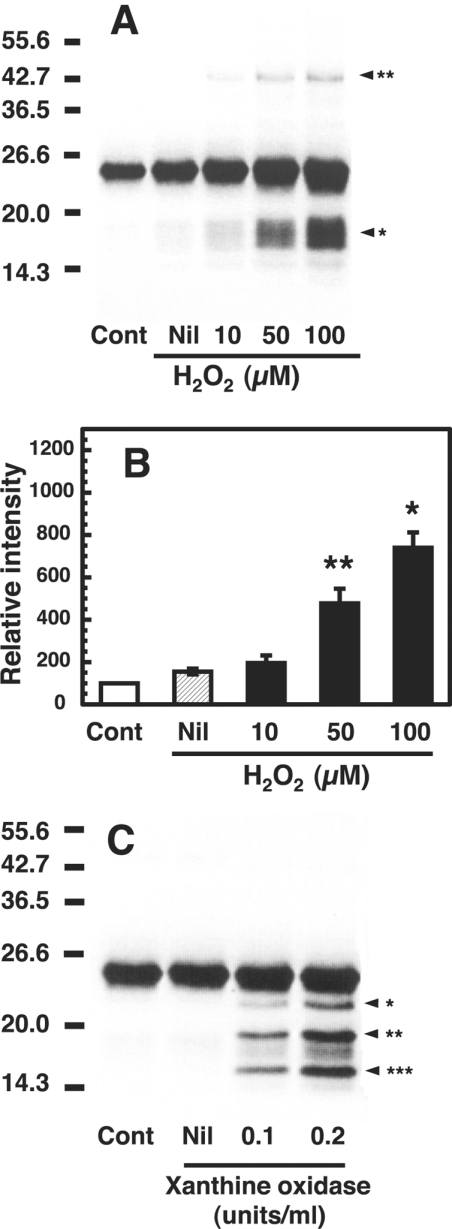
Because the inhibitory effect of SOD and catalase on the formation of the dimer and fragments was observed, we examined the effect of H2O2 and O2− on copper-loaded PrP23–231. When copper-loaded PrP23–231 was incubated with H2O2 (0–100 μM) at 37 °C for 30 min, Western blots of copper-loaded PrP23–231 showed formations of fragments at 17–19 kDa (Figure 4A, arrowhead*) and a dimer at approx. 46 kDa (Figure 4A, arrowhead**). The formation of the fragments was dependent on the H2O2 concentration (Figure 4B). As a control, PrP23–231 without copper loading did not induce any oxidative damage to the protein upon incubation with H2O2 (50 μM) (results not shown).
Figure 4. Fragmentation and dimerization of the copper-loaded PrP23–231 in the presence of H2O2 and O2−.
(A) Western blot immunoassay of copper-loaded PrP23–231 treated with H2O2. Mixtures containing 50 mM Mes (pH 7.5) and 1 μM copper-loaded PrP23–231 (containing 5.0 μM copper) were incubated without (Nil) or with the indicated concentrations of H2O2 at 37 °C for 30 min. Copper-loaded PrP23–231 without the incubation was used as a control (Cont). PrP was detected by SDS/PAGE followed by immunoblotting with antibody SAF 84. Molecular masses in kDa are indicated on the left. The positions of the fragments at 17–19 kDa and the dimer at 46 kDa are marked by arrowhead* and arrowhead** respectively. (B) Quantification of the Western blot data. Signals of the 17–19 kDa fragments were determined using ATTO Densitograph Software Library and plotted as a percentage of the control (zero time, Cont). Data are means±S.D. (n=3). Significant difference (*P≤0.01 and **P≤0.05) from the experiment with no additions (Nil). (C) Western blot immunoassay of copper-loaded PrP23–231 treated with O2−. Mixtures containing 50 mM Mes (pH 7.5), 0.1 mM xanthine and 1 μM copper-loaded PrP23–231 (containing 5.0 μM copper) were incubated without (Nil) or with the indicated concentrations of xanthine oxidase at 37 °C for 30 min. As a control, copper-loaded PrP23–231 was incubated without xanthine (Cont). PrP was detected by SDS/PAGE, followed by immunoblotting with antibody SAF 84. Molecular masses in kDa are indicated on the left.
Next, copper-loaded PrP23–231 was incubated with xanthine (100 μM) and xanthine oxidase (O2− producer; 0.1 and 0.2 unit/ml) at 37 °C for 30 min. Western blots of copper-loaded PrP23–231 showed major three discrete bands at 14.3–23 kDa besides the band of PrP23–231, but no signal at 46 kDa (Figure 4C).
Effect of metal ions on fragmentation and dimerization of the copper-loaded PrP23–231
As it has been reported that PrP may interact with metal ions other than copper [16,23,38], we examined the effect of various metal ions (5 μM) on dimerization and fragmentation of the copper-loaded PrP23–231 (containing 5 μM copper) in the presence of 10 μM dopamine at 37 °C for 30 min. All metal ions had no significant effect (Figures 5A and 5B). This is different from the observation that Ca2+, Mn2+ and Zn2+ had a significant protective effect on the cleavage of PrP by CuSO4 plus H2O2 [30].
Figure 5. Effect of metal ions on fragmentation and dimerization of the copper-loaded PrP23–231 in the presence of dopamine.
(A) Western blot immunoassay for copper-loaded PrP23–231. Mixtures containing 50 mM Mes (pH 7.5), 10 μM dopamine and 1 μM copper-loaded PrP23–231 (containing 5.0 μM copper) were incubated without (Nil) or with one of the indicated metal ions (5 μM) at 37 °C for 30 min. As a control, copper-loaded PrP23–231 was incubated with 10 μM dopamine only (Cont). PrP was detected by SDS/PAGE, followed by immunoblotting with antibody SAF 84. Molecular masses in kDa are indicated on the left. (B) Quantification of the Western blot data. Signals of fragments of PrP at 17–19 kDa were determined using ATTO Densitograph Software Library and plotted as a percentage of the control (zero time, Cont). Data are means±S.D. (n=3).
Generation of C-terminally truncated species from copper-loaded PrP23–231 in the presence of dopamine
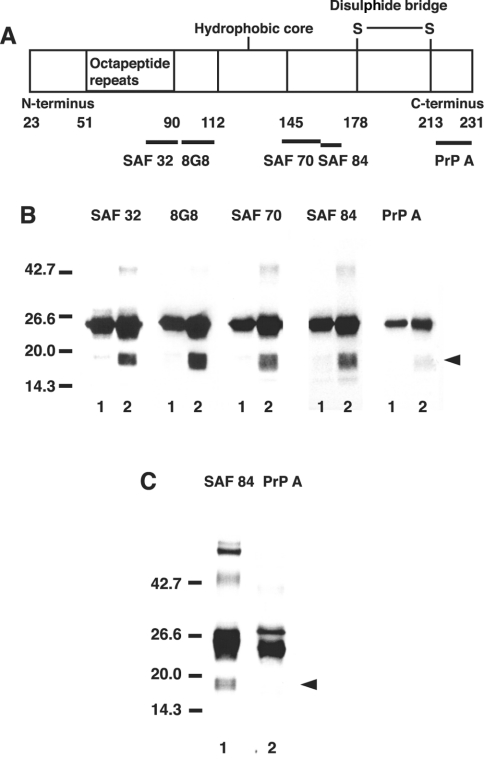
We examined which part of the PrP23–231 molecule the 17–19 kDa fragments were derived from. The epitopes of PrP that are recognized by the antibodies used in the present study are diagrammatically presented in Figure 6(A). Western blots showed a major band at 23 kDa that reacted with all the antibodies, SAF 32, 8G8, SAF 70, SAF 84 and PrP A (Figure 6B). In the presence of dopamine, fragments with molecular masses of 17–19 kDa were recognized by all the antibodies, except PrP A that binds to epitope 216–232 of mouse PrP (epitope 228–244 of bovine PrP) (Figure 6B). We examined further whether the fragment of PrP immunoprecipitated with SAF 32 includes the epitopes recognized by SAF 84 and PrP A. The 17–19 kDa fragments in the immunoprecipitates immunoreacted with SAF 84, but not with PrP A antibody (Figure 6C). These results indicate that the C-terminal part was cleaved off in the 17–19 kDa fragments.
Figure 6. Production of C-terminally truncated species from copper-loaded PrP23–231 in the presence of dopamine.
(A) Diagrammatic presentation of the PrP regions recognized by the antibodies employed in the present study. The numbers represent the position of amino acid residue of the primary translation product. (B) Western blot immunoassay for copper-loaded PrP23–231. Mixtures containing 50 mM Mes (pH 7.5) and 1 μM copper-loaded PrP23–231 (containing 5.0 μM copper) were incubated in the absence (lanes 1) or presence of 10 μM dopamine (lanes 2) at 37 °C for 30 min. PrP was detected by SDS/PAGE, followed by immunoblotting with antibodies SAF 32, 8G8, SAF 70, SAF 84 and PrP A. Molecular masses in kDa are indicated on the left. (C) Immunoprecipitation of PrP. The complete reaction mixture was incubated as above, and anti-PrP monoclonal antibody (SAF 32) was added and allowed to stand for 1 h at room temperature. After incubation for 1 h with Protein G–Sepharose 4B, the beads were pelleted and washed extensively with 50 mM Mes buffer (pH 7.5) containing 0.15 M NaCl. The washed beads were mixed with 2× gel-loading buffer and heated at 95 °C for 5 min. PrP was detected by SDS/PAGE, followed by immunoblotting with antibodies SAF 84 (lane 1) and PrP A (lane 2). Molecular masses in kDa are indicated on the left side.
DISCUSSION
As regards copper-catalysed oxidation of PrP, McMahon et al. [30] reported that an N-terminally truncated PrPC with a molecular mass of 28.5 kDa was generated when the conditioned medium containing soluble full-length glycosylated PrPC (33 kDa) lacking its glycosylphosphatidylinositol anchor released from Chinese-hamster ovarian cells expressing wild-type mouse PrP was treated with H2O2 and CuSO4. Requena et al. [31] have indicated that Syrian hamster recombinant SHa29–231 prion protein suffers aggregation and precipitation concomitant with copper-catalysed oxidation using L-ascorbate and CuCl2, and that the histidine-containing octarepeat region is particularly affected by the oxidation. In the present study, we have investigated copper-catalysed oxidation of copper-loaded recombinant PrP23–231, in the presence of catecholamine. In the presence of dopamine, the copper-loaded PrP23–231 underwent carbonylation, and dimerization and degradation of the protein occurred concomitantly, but aggregates were not formed. All the antibodies, except PrP A that binds to epitope 216–231 of mouse PrP, recognized the degradation products. This result indicates that the degradation products hold residues 77–169 of PrP, and suggests that C-terminal part containing epitope 216–231 was cleaved off in the 17–19 kDa fragments. The discrepancy between the results of McMahon et al. [30], Requena et al. [31] and the present study may be due to a difference of redox activity of the copper between the copper-loaded PrP23–231 and the copper–PrP complex formed by mixing the protein with free copper ions (CuCl2 or CuSO4). It is also possible that the discrepancy is caused by the differences in the composition of the reaction mixtures, such as the difference of electron donors (25 mM L-ascorbate, 0.1–5 mM H2O2 and 10 μM dopamine for the respective studies) and/or pH (Hepes buffer at 7.2, Hepes buffer at 7.0 and Mes buffer at 7.5 for the respective studies). In general, these studies have indicated clearly that copper-catalysed oxidation of PrP results in structural alterations, which might be associated with the pathology of prion disease.
The oxidative damage of copper-loaded PrP23–231 induced by dopamine was copper-dependent because various copper chelators inhibited carbonyl formation, dimerization and fragmentation. The data also indicate that Cu+ is generated during the reaction, because Cu+ chelators were effective. Copper-bound PrP is known to have SOD activity [7–10]. The dismutation of O2− requires redox cycling of the copper. Although Cu2+ readily co-ordinates to deprotonated amide bonds, as shown in previous studies [20,26], the proposed binding sites will probably not co-ordinate Cu+ [26,39]. The copper-binding sites would significantly restructure to do so. Also catalase and SOD inhibited carbonyl formation, dimerization and fragmentation, albeit to lesser extents. Furthermore, we examined the effect of H2O2 and O2− on copper-loaded PrP23–231. Similar to the effect of dopamine, H2O2 induced dimerization and fragmentation of copper-loaded PrP23–231, while O2− induced its fragmentation only. These results apparently indicate the participation of PrP23–231-bound copper (Cu2+ and Cu+) and the reactive oxygen species in the formation of dimer and fragments.
Reactive oxygen species levels are known to increase with age [28], and under conditions of oxidative stress: the H2O2 concentration reaches 26–160 μM in the brain [40]. It is feasible from the results of the present study that the interaction of H2O2 at these levels causes the oxidative damage in vivo to copper-loaded PrP23–231, because the damage was observed at 50–100 μM H2O2. In contrast with copper-loaded PrP23–231, PrP23–231 without copper loading did not induced any oxidative damage to the protein upon incubation with H2O2 (results not shown). Also a study has indicated that a site-specific cleavage of PrPC was observed at 2.5 mM H2O2 with CuSO4, but not without copper [30]. These data suggest that copper-bound PrP could be susceptible to oxidative damage.
We examined whether the addition of dopamine, L-ascorbate or glutathione affects the binding of copper to PrP23–231. In the experiment with dopamine and L-ascorbate, no copper was released from the copper-loaded PrP23–231, whereas glutathione released a large amount (44%) of copper after 30 min of incubation. We also examined whether oxidative damage of copper-loaded PrP23–231 was induced by L-ascorbate and glutathione. Similar to the effect of dopamine, L-ascorbate induced dimerization and fragmentation of copper-loaded PrP23–231, but glutathione did not. Glutathione forms a stable complex with copper because of its high affinity to Cu(I), and thus thereby diminished the oxidation on the protein.
It has been reported that PrPC may interact with metal ions other than Cu2+ [16,23,38]. In addition, a study has indicated that metal ions such as Zn2+, Mn2+ or Ca2+ have a significant protective effect on H2O2 cleavage when added in combination with Cu2+ [30]. Thus PrPC could interact with more than one metal ion at a time, or the presence of additional metal ions could interfere with Cu2+ binding. From a more recent spectroscopic study of Mn2+ binding to PrP, however, Mn2+ is indicated not to bind to the octarepeat region of PrP [41]. We examined the effect of metal ions on the oxidative modification of copper-loaded PrP23–231 in the presence of dopamine. All metal ions tested (Co2+, Zn2+, Mn2+ or Ca2+) had no significant effect on the oxidative modification of copper-loaded PrP23–231. From these data, we presumed that these metal ions do not share the binding site with copper.
We showed previously that copper bound to the N-terminal part of PrP, PrP23-98, catalyses the oxidation of dopamine [11]. In the case of copper-loaded PrP23–231, too, the dopamine oxidation should occur in association with carbonyl formation and other oxidative damage. The mechanism of the dopamine-induced oxidative damage of copper-loaded PrP23–231 may be delineated as follows [42,43]. The copper catalysis of the oxidation of catechol to o-quinone is mediated through the formation of a PrP23–231–Cu2+–catechol complex, which favours the transfer of electrons from catechol to PrP23–231–Cu2+ and the formation of PrP23–231–CuO2+ from the resulting PrP23–231–Cu+ and O2. The PrP23–231–CuO2+ species effectively oxidizes catechol or can be cleaved to O2− and PrP23–231–Cu2+. Generation of O2− always leads to the formation of H2O2, which is reduced by PrP23–231–Cu+ to form the hydroxyl radical. This highly reactive radical attacks the protein backbone and side chains of amino acid residues directly [28]. This mechanism is consistent with the results that a copper chelator (Cu2+ or Cu+ chelator), catalase and SOD inhibited the oxidative damage induced by dopamine, and that the copper remains bound to PrP23–231 during the course of this copper reaction, because the copper bound to PrP23–231 was shown not to be released.
As regards the reduction potentials of copper complex, Bandy et al. [44] investigated the reactivity of copper complexes with a range of abilities to catalyse the reaction with O2 in autoxidation of 6-hydroxydopamine, and showed that the autoxidation of 6-hydroxydopamine was accelerated in the presence of Cu2+–1,10-phenanthroline2 (standard reduction potential at E′0=170 mV) and Cu2+–2,2-dipyridyl2 (E′0=120 mV), whereas Cu2+–His2 (E′0=−50 mV) and Cu2+–EDTA (E′0=−520 mV) were not the effective catalyst. From these data, it was suggested that the reduction potentials of the most effective catalysts fell between the one-electron reduction potential of 6-hydroxydopamine (E′0 QH./QH2 between 530 and 810 mV) and that of O2 (E′0 O2/O2−=−160 mV). The same consideration for autoxidation of dopamine suggests that the range of the effective reduction potential will be between 530 mV (the one-electron reduction potentials of catechol, the functional part of dopamine) [45] and −160 mV. Because the PrP-bound copper undergoes redox cycling in the presence of dopamine as demonstrated in our previous study [11] and the present study, the reduction potential of at least one copper of the copper-loaded PrP is assumed to fall in this range. As for the reduction potential of copper-loaded PrP, the reduction potentials of prion octapeptide PHGGGWGQ–Cu2+ and HGGG–Cu2+ complexes were determined (E′0=−311 and −289 mV respectively) [46]. However, these values do not predict that the autoxidation of dopamine is thermodynamically feasible. This discrepancy remains to be elucidated.
Acknowledgments
This work was supported by Grant-in-Aid for Scientific Research (C) (2) 14570128 (to N. S.) from Japan Society for the Promotion of Science.
References
- 1.Brockes J. P. Topics in prion cell biology. Curr. Opin. Neurobiol. 1999;9:571–577. doi: 10.1016/S0959-4388(99)00016-1. [DOI] [PubMed] [Google Scholar]
- 2.Brown D. R. Prion and prejudice: normal protein and the synapse. Trends Neurosci. 2001;24:85–90. doi: 10.1016/s0166-2236(00)01689-1. [DOI] [PubMed] [Google Scholar]
- 3.Pauly P. C., Harris D. A. Copper stimulates endocytosis of the prion protein. J. Biol. Chem. 1998;273:33107–33110. doi: 10.1074/jbc.273.50.33107. [DOI] [PubMed] [Google Scholar]
- 4.Brown L. R., Harris D. A. Copper and zinc cause delivery of the prion protein from the plasma membrane to a subset of early endosomes and the Golgi. J. Neurochem. 2003;87:353–363. doi: 10.1046/j.1471-4159.2003.01996.x. [DOI] [PubMed] [Google Scholar]
- 5.Waggoner D. J., Drisaldi B., Bartnikas T. B., Casareno R. L., Prohaska J. R., Gitlin J. D., Harris D. A. Brain copper content and cuproenzyme activity do not vary with prion protein expression level. J. Biol. Chem. 2000;275:7455–7458. doi: 10.1074/jbc.275.11.7455. [DOI] [PubMed] [Google Scholar]
- 6.Rachidi W., Vilette D., Guiraud P., Arlotto M., Riondel J., Laude H., Lehmann S., Favier A. Expression of prion protein increases cellular copper binding and antioxidant enzyme activities but not copper delivery. J. Biol. Chem. 2003;278:9064–9072. doi: 10.1074/jbc.M211830200. [DOI] [PubMed] [Google Scholar]
- 7.Brown D. R., Wong B. S., Hafiz F., Clive C., Haswell S. J., Jones I. M. Normal prion protein has an activity like that of superoxide dismutase. Biochem. J. 1999;344:1–5. [PMC free article] [PubMed] [Google Scholar]
- 8.Brown D. R., Hafiz F., Glasssmith L. L., Wong B. S., Jones I. M., Clive C., Haswell S. J. Consequences of manganese replacement of copper for prion protein function and proteinase resistance. EMBO J. 2000;19:1180–1186. doi: 10.1093/emboj/19.6.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong B. S., Pan T., Liu T., Li R., Petersen R. B., Jones I. M., Gambetti P., Brown D. R., Sy M. S. Prion disease: a loss of antioxidant function? Biochem. Biophys. Res. Commun. 2000;275:249–252. doi: 10.1006/bbrc.2000.3158. [DOI] [PubMed] [Google Scholar]
- 10.Brown D. R., Clive C., Haswell S. J. Antioxidant activity related to copper binding of native prion protein. J. Neurochem. 2001;76:69–76. doi: 10.1046/j.1471-4159.2001.00009.x. [DOI] [PubMed] [Google Scholar]
- 11.Shiraishi N., Nishikimi M. Carbonyl formation on a copper-bound prion protein fragment, PrP23–98, associated with its dopamine oxidase activity. FEBS Lett. 2002;511:118–122. doi: 10.1016/s0014-5793(01)03324-5. [DOI] [PubMed] [Google Scholar]
- 12.Riek R., Hornemann S., Wider G., Billeter M., Glockshuber R., Wüthrich K. NMR structure of the mouse prion protein domain PrP121–321. Nature (London) 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 13.Zahn R., Liu A., Luhrs T., Riek R., von Schroetter C., Lopez Garcia F., Billeter M., Calzolai L., Wider G., Wüthrich K. NMR solution structure of the human prion protein. Proc. Natl. Acad. Sci. U.S.A. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donne D. G., Viles J. H., Groth D., Mehlhorn I., James T. L., Cohen F. E., Prusiner S. B., Wright P. E., Dyson H. J. Structure of the recombinant full-length hamster prion protein PrP29–231: the N terminus is highly flexible. Proc. Natl. Acad. Sci. U.S.A. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riek R., Hornemann S., Wider G., Glockshuber R., Wüthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP23–231. FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 16.Hornshaw M. P., McDermott J. R., Candy J. M., Lakey J. H. Copper binding to the N-terminal tandem repeat region of mammalian and avian prion protein: structural studies using synthetic peptides. Biochem. Biophys. Res. Commun. 1995;214:993–999. doi: 10.1006/bbrc.1995.2384. [DOI] [PubMed] [Google Scholar]
- 17.Brown D. R., Qin K., Herms J. W., Madlung A., Manson J., Strome R., Fraser P. E., Kruck T., von Bohlen A., Schulz-Schaeffer W., et al. The cellular prion protein binds copper in vivo. Nature (London) 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 18.Stockel J., Safar J., Wallace A. C., Cohen F. E., Prusiner S. B. Prion protein selectively binds copper(II) ions. Biochemistry. 1998;37:7185–7193. doi: 10.1021/bi972827k. [DOI] [PubMed] [Google Scholar]
- 19.Miura T., Hori-i A., Mototani H., Takeuchi H. Raman spectroscopic study on the copper(II) binding mode of prion octapeptide and its pH dependence. Biochemistry. 1999;38:11560–11569. doi: 10.1021/bi9909389. [DOI] [PubMed] [Google Scholar]
- 20.Viles J. H., Cohen F. E., Prusiner S. B., Goodin D. B., Wright P. E., Dyson H. J. Copper binding to the prion protein: structural implications of four identical cooperative binding sites. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2042–2047. doi: 10.1073/pnas.96.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whittal R. M., Ball H. L., Cohen F. E., Burlingame A. L., Prusiner S. B., Baldwin M. A. Copper binding to octarepeat peptides of the prion protein monitored by mass spectrometry. Protein Sci. 2000;9:332–343. doi: 10.1110/ps.9.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasnain S. S., Murphy L. M., Strange R. W., Grossmann J. G., Clarke A. R., Jackson G. S., Collinge J. XAFS study of the high-affinity copper-binding site of human PrP91–231 and its low-resolution structure in solution. J. Mol. Biol. 2001;311:467–473. doi: 10.1006/jmbi.2001.4795. [DOI] [PubMed] [Google Scholar]
- 23.Jackson G. S., Murray I., Hosszu L. L., Gibbs N., Waltho J. P., Clarke A. R., Collinge J. Location and properties of metal-binding sites on the human prion protein. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8531–8535. doi: 10.1073/pnas.151038498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burns C. S., Aronoff-Spencer E., Dunham C. M., Lario P., Avdievich N. I., Antholine W. E., Olmstead M. M., Vrielink A., Gerfen G. J., Peisach J., et al. Molecular features of the copper binding sites in the octarepeat domain of the prion protein. Biochemistry. 2002;41:3991–4001. doi: 10.1021/bi011922x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aronoff-Spencer E., Burns C. S., Avdievich N. I., Gerfen G. J., Peisach J., Antholine W. E., Ball H. L., Cohen F. E., Prusiner S. B., Millhauser G. L. Identification of the Cu2+ binding sites in the N-terminal domain of the prion protein by EPR and CD spectroscopy. Biochemistry. 2000;39:13760–13771. doi: 10.1021/bi001472t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burns C. S., Aronoff-Spencer E., Legname G., Prusiner S. B., Antholine W. E., Gerfen G. J., Peisach J., Millhauser G. L. Copper coordination in the full-length, recombinant prion protein. Biochemistry. 2003;42:6794–6803. doi: 10.1021/bi027138+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong B. S., Chen S. G., Colucci M., Xie Z., Pan T., Liu T., Li R., Gambetti P., Sy M. S., Brown D. R. Aberrant metal binding by prion protein in human prion disease. J. Neurochem. 2001;78:1400–1408. doi: 10.1046/j.1471-4159.2001.00522.x. [DOI] [PubMed] [Google Scholar]
- 28.Berlett B. S., Stadtman E. R. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 29.Butterfield D. A., Kanski J. Brain protein oxidation in age-related neurodegenerative disorders that are associated with aggregated proteins. Mech. Ageing Dev. 2001;122:945–962. doi: 10.1016/s0047-6374(01)00249-4. [DOI] [PubMed] [Google Scholar]
- 30.McMahon H. E., Mange A., Nishida N., Creminon C., Casanova D., Lehmann S. Cleavage of the amino terminus of the prion protein by reactive oxygen species. J. Biol. Chem. 2001;276:2286–2291. doi: 10.1074/jbc.M007243200. [DOI] [PubMed] [Google Scholar]
- 31.Requena J. R., Groth D., Legname G., Stadtman E. R., Prusiner S. B., Levine R. L. Copper-catalyzed oxidation of the recombinant SHa29–231 prion protein. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7170–7175. doi: 10.1073/pnas.121190898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halliwell B., Gutteridge J. M. C. New York: Oxford University Press; 1999. Free Radicals in Biology and Medicine. [Google Scholar]
- 33.Sano I., Gamo T., Kakimoto Y., Taniguchi K., Takesada M., Nishinuma K. Distribution of catechol compounds in human brain. Biochim. Biophys. Acta. 1959;32:586–587. doi: 10.1016/0006-3002(59)90652-3. [DOI] [PubMed] [Google Scholar]
- 34.Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 35.Beauchamp C., Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 36.Walaas E., Walaas O., Haavaldsen S. Spectrophotometric and electron-spin resonance studies of complexes of catecholamines with Cu(II) ions and the interaction of ceruloplasmin with catecholamines. Arch. Biochem. Biophys. 1963;100:97–109. doi: 10.1016/0003-9861(63)90040-7. [DOI] [PubMed] [Google Scholar]
- 37.Corazza A., Harvey I., Sadler P. J. 1H,13C-NMR and X-ray absorption studies of copper(I) glutathione complexes. Eur. J. Biochem. 1996;236:697–705. doi: 10.1111/j.1432-1033.1996.0697d.x. [DOI] [PubMed] [Google Scholar]
- 38.Qin K., Yang Y., Mastrangelo P., Westaway D. Mapping Cu(II) binding sites in prion proteins by diethyl pyrocarbonate modification and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometric footprinting. J. Biol. Chem. 2002;277:1981–1990. doi: 10.1074/jbc.M108744200. [DOI] [PubMed] [Google Scholar]
- 39.Kroneck P. M., Vortisch V., Hemmerich P. Model studies on the coordination of copper in biological systems: the deprotonated peptide nitrogen as a potential binding site for copper(II) Eur. J. Biochem. 1980;109:603–612. doi: 10.1111/j.1432-1033.1980.tb04833.x. [DOI] [PubMed] [Google Scholar]
- 40.Hyslop P. A., Zhang Z., Pearson D. V., Phebus L. A. Measurement of striatal H2O2 by microdialysis following global forebrain ischemia and reperfusion in the rat: correlation with the cytotoxic potential of H2O2 in vitro. Brain Res. 1995;671:181–186. doi: 10.1016/0006-8993(94)01291-o. [DOI] [PubMed] [Google Scholar]
- 41.Garnett A. P., Viles J. H. Copper binding to the octarepeats of the prion protein. Affinity, specificity, folding, and cooperativity: insights from circular dichroism. J. Biol. Chem. 2003;278:6795–6802. doi: 10.1074/jbc.M209280200. [DOI] [PubMed] [Google Scholar]
- 42.Balla J., Kiss T., Jameson R. F. Copper(II)-catalyzed oxidation of catechol by molecular oxygen in aqueous solution. Inorg. Chem. 1992;31:58–62. [Google Scholar]
- 43.Bindoli A., Rigobello M. P., Deeble D. J. Biochemical and toxicological properties of the oxidation products of catecholamines. Free Radical Biol. Med. 1992;13:391–405. doi: 10.1016/0891-5849(92)90182-g. [DOI] [PubMed] [Google Scholar]
- 44.Bandy B., Walter P. B., Moon J., Davison A. J. Reaction of oxygen with 6-hydroxydopamine catalyzed by Cu, Fe, Mn, and V complexes: identification of a thermodynamic window for effective metal catalysis. Arch. Bioch. Biophys. 2001;389:22–30. doi: 10.1006/abbi.2001.2285. [DOI] [PubMed] [Google Scholar]
- 45.Steenken S., Neta P. One-electron redox potentials of phenols: hydroxy- and aminophenols and related compounds of biological interest. J. Phys. Chem. 1982;86:3661–3667. [Google Scholar]
- 46.Bonomo R. P., Impellizzeri G., Pappalardo G., Rizzarelli E., Tabbi G. Copper(II) binding modes in the prion octapeptide PHGGGWGQ: a spectroscopic and voltammetric study. Chemistry. 2000;6:4195–4202. doi: 10.1002/1521-3765(20001117)6:22<4195::aid-chem4195>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]