Abstract
HVS (herpesvirus saimiri) is the prototype γ-2 herpesvirus. This is a subfamily of herpesviruses gaining importance since the identification of the first human γ-2 herpesvirus, Kaposi's sarcoma-associated herpesvirus. The HVS ORF 57 (open reading frame 57) protein is a multifunctional transregulatory protein homologous with genes identified in all classes of herpesviruses. Recent work has demonstrated that ORF 57 has the ability to bind viral RNA, shuttles between the nucleus and cytoplasm and promotes the nuclear export of viral transcripts. In the present study, we show that ORF 57 shuttles between the nucleus and cytoplasm in a CRM-1 (chromosomal region maintenance 1)-independent manner. ORF 57 interacts with the mRNA export factor REF (RNA export factor) and two other components of the exon junction complex, Y14 and Magoh. The association of ORF 57 with REF stimulates recruitment of the cellular mRNA export factor TAP (Tip-associated protein), and HVS infection triggers the relocalization of REF and TAP from the nuclear speckles to several large clumps within the cell. Using a dominant-negative form of TAP and RNA interference to deplete TAP, we show that it is essential for bulk mRNA export in mammalian cells and is required for ORF 57-mediated viral RNA export. Furthermore, we show that the disruption of TAP reduces viral replication. These results indicate that HVS utilizes ORF 57 to recruit components of the exon junction complex and subsequently TAP to promote viral RNA export through the cellular mRNA export pathway.
Keywords: chromosomal region maintenance 1 (CRM-1), exon junction complex, herpesvirus, mRNA export, open reading frame 57 (ORF 57), Tip-associated protein (TAP)
Abbreviations: CMV, cytomegalovirus; CRM-1, chromosomal region maintenance 1; CTE, constitutive transport element; EJC, exon junction complex; FISH, fluorescence in situ hybridization; gB, glycoprotein B; GFP, green fluorescent protein; GST, glutathione S-transferase; HEK-293T cells, human embryonic kidney 293T cells; HSV, herpes simplex virus; HVS, herpesvirus saimiri; KSHV, Kaposi's sarcoma associated herpesvirus; LMB, leptomycin B; MOI, multiplicity of infection; REF, RNA export factor; mREF2-1, murine REF2-1; NES, nuclear export signal; NMD, nonsense-mediated decay; OMK cells, owl monkey kidney cells; ORF 57, open reading frame 57; RNAi, RNA interference; RNPS1, RNA-binding protein prevalent during S phase; RRM, RNA recognition motif; TAP, Tip-associated protein; TREX complex, transcription and export complex; YFP, yellow fluorescent protein
INTRODUCTION
The past seven years have seen significant advances in our understanding of the mechanism of mRNA export from the nucleus and how it is coupled with mRNA biogenesis and processing, catalysed in part by the identification of the Mex67 mRNA export factor in yeast [1]. It has become clear that the processes of transcription, pre-mRNA splicing, capping and polyadenylation are coupled with mRNA export. Transcription by RNA polymerase II is linked to mRNA export through the TREX complex (transcription and export complex) [2] and to splicing through the EJC (exon junction complex) [3]. The TREX complex contains two proteins directly involved in mRNA export, the RNA helicase UAP56 (Sub2 in yeast) and REF (RNA export factor)/ALY (Yra1 in yeast), and may be responsible for loading these export factors on to mRNA during transcription. The splicing-dependent recruitment of mRNA export factors involves the deposition of the EJC of proteins, approx. 20–24 bases upstream from the site of exon ligation. Components of this complex include REF, Y14, Magoh, RNPS1 (RNA-binding protein prevalent during S phase), SRm160, UPF3B and eIF4AIII [3–8]. The EJC acts as a binding site for the TAP (Tip-associated protein) mRNA export factor [9]. In yeast, the recruitment of Yra1 to mRNA is also coupled with polyadenylation, which is required for mRNA export [10].
TAP is a multidomain protein with an N-terminal substrate-binding domain (amino acids 1–372), which interacts with the retroviral CTE (constitutive transport element) RNA element and REF [11,12]. The central domain (amino acids 373–550) interacts with p15 [13] and nucleoporins [14], and the C-terminal domain (amino acids 551–619) also binds nucleoporins [14–16]. Y14 and Magoh also bind to TAP directly [5]. The TAP/p15 heterodimer provides the connection between the mRNP and the nuclear pore and shuttles between the nucleus and cytoplasm. As expected, depletion of the yeast orthologue of TAP, Mex67, leads to a block in mRNA export as does the depletion of TAP in Drosophila S2 cells and Caenorhabditis elegans [1,17,18]. There are several alternative forms of TAP in humans, which show restricted expression patterns, although the function of these is not clear at present [19,20].
Whereas the depletion of Yra1 in yeast leads to a significant blockage in mRNA export [21], an RNAi (RNA interference) knockdown of the higher eukaryotic orthologue, REF, in Drosophila S2 cells fails to block export, suggesting that there are alternative methods to recruit TAP to mRNA [22]. Evidence for alternative pathways is now emerging; for example, TAP binds to U2AF35 and is recruited to mRNA through an interaction with U2AF65 [23]. Recently, it was found that shuttling SR proteins can also act as export adaptors and promote mRNA export. 9G8, SRp20 and ASF/SF2 associate with TAP and promote the export of spliced and intronless mRNA [24]. In the context of intronless mRNAs, the SR proteins associate with a specific 22 nt element [14], whereas the site of interaction for spliced mRNAs may be through exonic splice enhancers, although SRp20 is also stably associated with the EJC [25].
Herpesviridae express a significant number of naturally intronless mRNAs that are unable to recruit mRNA export factors by splicing. In HSV (herpes simplex virus), ICP27 binds to REF directly and forms a ternary complex with TAP and these interactions promote the export of viral RNA in Xenopus oocytes. The ICP27-mediated export of viral RNA is blocked in the presence of antibodies to REF or the CTE, which binds directly to TAP, acting as a competitive inhibitor for mRNA export [26]. More recently, ICP27 has been shown to utilize the mRNA export pathway in mammalian cells [27]. Other members of the herpesvirus family appear to use the REF/TAP mRNA export pathway; for example, Epstein–Barr virus EB2 protein promotes viral mRNA export. EB2 interacts with REF and co-immunoprecipitates with TAP, although these interactions are dependent on RNA. Furthermore, a truncated form of EB2, which lacks the REF interaction domain, prevents viral mRNA export [28]. Similar results have recently been shown for the homologous ORF 57 (open reading frame 57) protein in KSHV (Kaposi's sarcoma-associated herpesvirus) [29].
The ICP27 family of proteins shuttle between the nucleus and cytoplasm as would be expected for mRNA export factors, although the pathways utilized are unclear at present. Results from previous studies suggested that the leucine-rich repeat in ICP27 provided access to the CRM-1 (chromosomal region maintenance 1)-dependent export pathway [30], although more recent results indicate that shuttling of ICP27 is not sensitive to the CRM-1 inhibitor LMB (leptomycin B). However, the leucine-rich nuclear export sequence is reported to be required for efficient export of the protein [27]. In fact, ICP27 appears to be capable of using two distinct export pathways: a viral RNA-dependent pathway, which involves TAP and is blocked by the CTE, and an RNA-independent pathway, which remains to be characterized [26]. Non-conventional, LMB-insensitive transferable nuclear export sequences have also been identified in the Epstein–Barr virus EB2 protein and the human CMV (cytomegalovirus) transactivator protein pUL69 [28,31], although their export receptors have not yet been identified.
HVS (herpesvirus saimiri) is the prototype member of the γ-2 herpesvirus family, which includes the medically important virus KSHV, which is involved in the development of Kaposi's sarcoma [32,33]. Similar to other herpesviruses, it encodes an ICP27 homologue, known as HVS ORF 57. HVS ORF 57 is a 52 kDa multifunctional transregulatory protein. Transactivation of late viral genes by ORF 57 occurs independently of target gene promoter sequences and is mediated at a post-transcriptional level [34]. In addition to its transactivation properties, ORF 57 is responsible for the repression of viral gene expression, which correlates with the presence of introns within the target gene [34]. ORF 57 also redistributes both U2 and SC-35 splicing factors during an HVS infection into intense distinct nuclear aggregations [35]. Recent studies have also demonstrated that the ORF 57 protein has the ability to bind viral RNA, shuttle between the nucleus and cytoplasm and is required for the efficient cytoplasmic accumulation of virus mRNA [36,37]. This suggests that, similar to its homologues, ORF 57 plays a pivotal role in mediating the nuclear export of viral transcripts. Analysis of functional domains encoded by HVS ORF 57 has identified a nuclear localization signal and RNA-binding domain within the N-terminal, a central leucine-rich sequence reminiscent of a CRM-1-mediated NES (nuclear export signal) and activation and repression domains within the C-terminus [36–38].
In the present study, we have characterized the mRNA export pathway used by HVS. We demonstrate that ORF 57 shuttles between the nucleus and cytoplasm in an LMB-insensitive manner and show that ORF 57 binds through its N-terminal domain to the mRNA export factor REF. The interaction with REF promotes the recruitment of TAP to form a ternary complex that promotes viral mRNA export. We show that the depletion of TAP from mammalian cells causes a block in bulk mRNA export and this blocks viral mRNA export. Finally, we demonstrate that viral replication is impaired in the presence of an excess of a dominant-negative form of TAP.
MATERIALS AND METHODS
Plasmid constructs
Details of the plasmid constructs are given in the Supplementary Methods (available at http://www.BiochemJ.org/bj/387/bj3870295add.htm).
Interspecies heterokaryon assays
Cos-7 cells, seeded at 2×105 cells per 35 mm-diameter Petri dish, were transiently transfected with 2 μg of pRSVORF 57 [34,36,38]. After 18 h, mouse 3T3 cells (5×105 cells/well) were plated on to the Cos-7 cells in a medium containing 50 μg/ml cycloheximide and 25 nM LMB, when required. After 4 h, the cells were washed with PBS and fused by the addition of 2 ml of 50% (w/v) poly(ethylene glycol) in PBS. After 2 min, the cells were washed extensively in PBS. After washing, the cells were returned to a medium containing 50 μg/ml cycloheximide for 60 min. Cells were then fixed with 4% (v/v) formaldehyde in PBS, washed three times with PBS and permeabilized in 0.5% (v/v) Triton X-100 for 5 min. The cells were rinsed in PBS and blocked by preincubation with 1% (w/v) non-fat milk powder for 1 h at 37 °C. A 1:100 dilution of anti-ORF 57 antibody [34,36,38] was layered over the cells and incubated for 1 h at 37 °C. Fluorescein-conjugated anti-mouse immunoglobulin (Dako) at 1:50 dilution and a co-stain of 0.5 μg/ml Hoechst dye (Sigma) were added for 1 h at 37 °C. After each incubation step, cells were washed extensively with PBS. The immunofluorescence slides were observed using a Zeiss Axiovert 135TV inverted microscope with a Neofluar ×40 oil immersion lens.
Northern-blot analysis
Northern-blot analysis was performed as described previously [34]. Total, nuclear and cytoplasmic RNAs were isolated from transfected cells. Briefly, total RNA was isolated using TRIzol® reagent (Life Technologies) according to the manufacturer's instructions. In contrast, to isolate nuclear and cytoplasmic RNA, the cellular pellet was resuspended in 200 μl of RNA extraction buffer [0.14 M NaCl, 1.5 mM MgCl2, 10 mM Tris/HCl (pH 8.6), 0.5% Triton X-100, 1 mM dithiothreitol and 1000 units/ml RNase inhibitor], vortex-mixed for 15 s and then left on ice for 5 min, and the nuclear fraction was pelleted. The supernatant contained the cytoplasmic fraction. Protease digestion buffer [200 μl; 0.2 M Tris/HCl (pH 8.0), 25 mM EDTA, 0.3 M NaCl and 2% (w/v) SDS] was added to the supernatant fraction and vortex-mixed before incubating for 30 min at 37 °C. Proteinase K was added to a final concentration of 50 μg/ml. An equal volume of phenol/chloroform/isoamyl alcohol (25:24:1, by vol.) solution was added, mixed vigorously and centrifuged. Ice-cold propan-2-ol (400 μl) was then added to the aqueous phase, mixed and chilled on ice for 30 min. RNA was precipitated and air-dried. The pellet containing the nuclear fraction was redissolved in 200 μl of 50 mM Tris/HCl (pH 8.0) and 1 mM EDTA, and MgCl2 and dithiothreitol were added to final concentrations of 10 and 0.1 mM respectively. RNase-free pancreatic DNase I was then added to a final concentration of 2 μg/ml and incubated for 60 min at 37 °C. EDTA and SDS were then added to final concentrations of 10 mM and 0.2% (w/v) respectively, followed by extraction using phenol/chloroform. After centrifugation, the RNA was precipitated and allowed to air-dry. Finally, the pellets were redissolved in 200 μl of Tris/EDTA buffer (pH 7.6). Total, nuclear and cytoplasmic RNAs were separated by elecrophoresis on 1% denaturing formaldehyde–agarose gel. The RNA was transferred on to Hybond-N membranes and hybridized with 32P-radiolabelled random-primed probes specific for gB (glycoprotein B) and actin-coding sequences.
Yeast two-hybrid screen for ORF 57-interacting proteins
The GAL4-based yeast two-hybrid system screening technique [39] was used to identify ORF 57-interacting proteins as described previously [37]. Briefly, a human kidney cDNA-GAL4 activation domain fusion library in the vector pACT2 (ClonTech) was utilized to identify ORF 57-interacting proteins. The bait plasmid was transformed into the Saccharomyces cerevisiae strain, HF7c (ClonTech). Clones were selected on minimal synthetic dropout medium in the absence of tryptophan. Yeast clones harbouring the bait plasmids were then sequentially transformed with the ‘prey’ library. Positive clones potentially harbouring ORF 57-interacting species were identified both by their ability to grow on media without tryptophan, leucine and histidine and by detection of β-galactosidase activity. Plasmids were isolated from positive yeast clones, selecting for pACT2 cDNA library plasmids by transformation of the leucine auxotroph into the Escherichia coli strain, HB101.
To confirm the specificity of the interactions, pACT-2 library plasmids were transformed into yeast strains harbouring no plasmid, yeast containing pGBT9 vector only, yeast containing pLAM5 (a GAL4 human lamin C fusion) or pDBD57. Only those library plasmids demonstrating a requirement of the pDBD57 plasmid for induced expression of HIS3 or lacZ reporter genes were considered further and selected for DNA sequencing.
Viruses, cell culture and transfections
HVS (strain A11) was propagated in OMK cells (owl monkey kidney cells) that were maintained in Dulbecco's modified Eagle's medium (Life Technologies), supplemented with 10% (v/v) foetal calf serum. Cos-7, HeLa and HEK-293T (human embryonic kidney 293T) cells were also maintained in Dulbecco's modified Eagle's medium supplemented with 10% foetal calf serum. The plasmids used in the transfections were prepared using Qiagen plasmid kits according to the manufacturer's instructions. Transfections were performed using LIPOFECTAMINE™ or LIPOFECTAMINE™ 2000 (Invitrogen) according to the manufacturer's instructions. For the RNA export assays using pLUCSALRRE, 100 ng of pLUCSALRRE was co-transfected with 200 ng of the REV expression plasmids and 100 ng of a CMV-driven β-galactosidase expression vector. Transfections were performed in triplicate, and luciferase and β-galactosidase activities were measured in duplicate. The luciferase activities were averaged and normalized to the β-galactosidase levels, which acted as a transfection rate control.
Co-immunoprecipitation assays and GST (glutathione S-transferase) pull downs
HEK-293T cells were transfected using 2 μg of the appropriate DNAs, or infected with HVS at an MOI (multiplicity of infection) of 1. After 24 h, cells were harvested and lysed with lysis buffer (0.3 M NaCl, 1% Triton X-100 and 50 mM Hepes buffer, pH 8.0) containing protease inhibitors (leupeptin and PMSF). For each immunoprecipitation, 10 μl of the anti-ORF 57 polyclonal antibody [36] prebound to Protein A–Sepharose beads (Amersham Biosciences) or 40 μl of anti-myc affinity agarose (Sigma) was incubated with the respective cell lysate for 3 h at 4 °C. The beads were then pelleted, washed and the precipitated polypeptides were subjected to SDS/PAGE (12% gel) and immunoblot analysis. GST pull-down assays were performed as described previously [26] and RNase was added at the beginning of all the pull-down experiments together with the reticulocyte lysate. To confirm that RNase treatments were effective, an in vitro transcription/translation reaction for ORF 57 was spiked with [32P]UTP. After a GST–TAP pull down, RNA was extracted from the total reaction, resolved on a denaturing polyacrylamide gel and visualized using a phosphoimager. His6-ORF 57 (8–120) was expressed from the pET14b derivative expression vector in BL21(DE3) cells and purified using Talon resin. His6-mREF2-1 was purified as described previously [26]. For MS analysis of ORF 57 8–120, the sample (50 μg at 0.5 mg/ml in 40 mM ammonium acetate) was analysed in methanol/1% aqueous formic acid (1:1, v/v) by positive electrospray ionization MS using a Platform II mass spectrometer (Micromass, Atlas Park, Manchester, U.K.). A capillary voltage of 3.5 kV and a sampling cone voltage of 40 V were used. Mass accuracy was ensured by calibrating the data with a reference spectrum obtained from horse heart myoglobin (16951.49 Da). A mass accuracy of 0.01% is thus expected.
Immunoblot analysis
Polypeptides were subjected to SDS/PAGE (9 or 12% gel) and soaked for 10 min in transfer buffer [25 mM Tris, 192 mM glycine and 20% (v/v) methanol]. The proteins were transferred on to nitrocellulose membranes by electroblotting for 3 h at 250 mA. After transfer, the membranes were soaked in PBS and blocked by preincubation with 2% non-fat milk powder for 2 h at 37 °C. Membranes were incubated with a 1:500 dilution of anti-myc (Sigma), 1:1000 dilution of anti-TAP or a 1:1000 dilution of anti-GFP antibodies (ClonTech; GFP stands for green fluorescent protein), washed with PBS and then incubated for 1 h at 37 °C with a 1:1000 dilution of secondary immunoglobulin conjugated with horseradish peroxidase (Dako) in blocking buffer. His-tagged proteins were identified using a monoclonal antibody to His6 directly coupled with horseradish peroxidase (Sigma). After five washes with PBS, the nitrocellulose membranes were developed using ECL® (Pierce).
RNAi and immunofluorescence analysis
For heterokaryon analysis, the cells were fixed with 100% ice-cold methanol for 10 min. The cells were rinsed in PBS and blocked by preincubation with 1% non-fat milk powder for 1 h at 37 °C. A 1:100 dilution of anti-ORF 57 polyclonal antiserum [34,36,38] or 1:50 dilution of anti-myc monoclonal antibody was layered over the cells and incubated for 1 h at 37 °C. Texas Red-conjugated immunoglobulin (Dako) at 1:200 dilution was added for 1 h at 37 °C. After each incubation step, cells were washed extensively with PBS. The immunofluorescence slides were examined using a Zeiss Axiovert 135TV inverted microscope with a Neofluar ×40 oil immersion lens.
To deplete endogenous TAP, RNAi was performed in HeLa cells by transfecting pSUPERTAP with Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's instructions. Cells were co-transfected with YFP–tubulin (ClonTech; YFP stands for yellow fluorescent protein) as a marker for the transfection. Immunofluorescence of TAP was detected at 96 h post-transfection using the anti-TAP-C antibody at 1:300 dilution [40] and a TRITC-labelled anti-rabbit secondary antibody at 1:100 dilution. To detect mRNA, FISH (fluorescence in situ hybridization) was performed using Cy3-labelled oligo(dT) as described previously [41]. Images were collected using a Zeiss LSM510Meta confocal microscope.
RESULTS
ORF 57 shuttles between the nucleus and cytoplasm and mediates viral mRNA export through a CRM-1-independent pathway
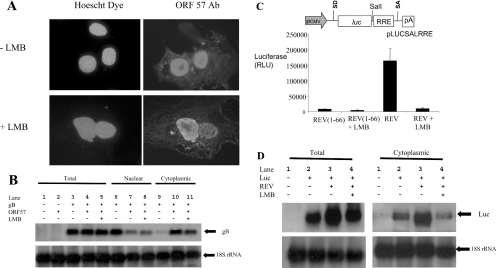
To determine whether the ORF 57 protein shuttles between the nucleus and cytoplasm through a CRM-1-dependent or -independent pathway, an interspecies heterokaryon assay was utilized [30,36,42]. Monkey Cos-7 cells transiently transfected with pRSVORF 57, a eukaryotic expression vector encoding the complete coding region of ORF 57 [34,36,38], were fused with mouse 3T3 cells in a medium with or without LMB. Cells were then incubated with an anti-ORF 57 monoclonal antibody and co-stained with Hoechst dye. Hoechst dye allowed the differentiation between monkey and mouse nuclei. As reported previously [36,42], monkey cells stained diffusely throughout the nuclei, whereas mouse nuclei stained with a distinctive speckled pattern. Analysis of the interspecies heterokaryons in the absence of LMB demonstrated, as described previously, that ORF 57 was detected in both monkey and mouse cell nuclei, indicating that ORF 57 has the ability to shuttle (Figure 1A). In addition, similar results were observed in the presence of LMB, suggesting that ORF 57 can shuttle between the nucleus and cytoplasm through a CRM-1-independent pathway.
Figure 1. ORF 57 shuttles in the presence of LMB.
(A) Cos-7 cells were transiently transfected with 2 μg of pRSVORF 57. After 18 h, mouse 3T3 cells were plated on to the Cos-7 cells in a medium containing cycloheximide in the absence or presence of LMB. After 4 h, the cells were washed with PBS and fused by the addition of 2 ml of 50% poly(ethylene glycol) in PBS. After washing, the cells were returned to a medium containing cycloheximide in the absence or presence of LMB for 60 min. Cells were then fixed and incubated with a 1:100 dilution of anti-ORF 57 antibody and then co-stained with 0.5 μg/ml Hoechst dye and fluorescein-conjugated anti-mouse immunoglobulin. (B) HEK-293T cells remained untransfected (lanes 1) or were transfected with pUCgB in the absence (lanes 3, 6 and 9) and presence of pRSVORF 57 (lanes 2, 4, 5, and 7, 8 and 10) or incubated in the presence of LMB (lanes 5, 8 and 11). Total (lanes 1–5), nuclear (lanes 6–8) and cytoplasmic (lanes 9–11) RNAs were then isolated and separated by elecrophoresis on a 1% denaturing formaldehyde–agarose gel. The RNA was transferred on to Hybond-N membranes and hybridized with 32P-radiolabelled random-primed probes specific for the HVS gB and 18 S rRNA-coding sequences. (C) HEK-293T cells were transiently transfected with pLUCSALRRE together with the indicated plasmids and a β-galactosidase control. Luciferase activities [RLU (relative light units)] were measured in triplicate on three separate occasions and averaged. Data were normalized for transfection efficiency by assaying for β-galactosidase activity. A schematic representation of pLUCSALRRE is shown above the graph; pCMV, CMV immediate early promoter; SD, splice donor; luc, luciferase gene; SalI, unique SalI restriction site; RRE, REV response element; SA, splice acceptor; pA, polyadenylation signal. (D) HEK-293T cells remained untransfected (lane 1) or were transfected with pLUCSALRRE in the absence (lane 2) and presence of pREV (lane 3) or incubated in the presence of LMB (lane 4). Total and cytoplasmic RNA was then isolated and separated by elecrophoresis on a 1% denaturing formaldehyde–agarose gel. The RNA was transferred on to Hybond-N membranes and hybridized with 32P-radiolabelled random-primed probes specific for the luciferase and 18 S rRNA-coding sequences.
To determine whether ORF 57 mediates nuclear export of viral RNA through a CRM-1-independent pathway, Northern-blot analysis was performed. Total, nuclear and cytoplasmic RNAs were isolated separately from HEK-293T cells transfected with pUCgB, a transfer vector containing the full-length coding region and promoter of the HVS late gB gene, in the absence and presence of pRSVORF 57 and LMB (Figure 1B). In the absence of ORF 57, the gB transcript accumulates in the nucleus and very little is observed in the cytoplasm (lanes 6 and 9); in contrast, the presence of ORF 57 leads to efficient export and accumulation of the gB transcript in the cytoplasm (lanes 7 and 10). The export of the gB transcript is not significantly decreased in the presence of LMB (lanes 8 and 11), indicating that ORF 57 can function through a CRM-1-independent pathway. To confirm that the LMB treatment was effective in these experiments, we utilized an RNA export assay based on HIV-1 REV that utilizes the CRM-1 pathway (Figure 1C). In this assay, a luciferase gene is placed within an inefficiently spliced intron that contains the REV response element. This reporter plasmid is similar to the pDM128 plasmids and derivatives widely used to monitor RNA export in vivo [43,44]. The pre-mRNA is normally retained in the nucleus and the exported, spliced mRNA lacks the luciferase gene. In the presence of a truncated form of REV (amino acids 1–66) that lacks the NES, background luciferase activity was observed. In the presence of functional REV, the pre-mRNA was actively exported through the CRM-1 pathway and luciferase activity was readily detected. The presence of LMB effectively inhibited the REV-dependent mRNA export, confirming that LMB treatment of cells was effectively blocking the CRM-1-dependent export pathway. Moreover, to confirm this result, Northern-blot analysis was performed. Total and cytoplasmic RNAs were isolated from cells transfected with the luciferase reporter gene in the absence and presence of REV and LMB. Northern-blot analysis demonstrated that LMB blocked CRM-1-dependent nuclear export of the mRNA in a manner similar to the export assay (Figure 1D).
The cellular protein REF/ALY interacts with HVS ORF 57
To identify additional ORF 57-interacting cellular proteins, 1.5×106 independent cDNA clones of a human kidney cDNA library fused to the GAL4 activation domain were screened. Of them, 24 clones were identified as capable of activating histidine and β-galactosidase reporter gene expression in the presence of the ORF 57–DBD fusion protein. The specificity of the interaction was confirmed by transforming the putative ORF 57-interacting cellular clones into yeast strains harbouring no plasmid, yeast containing pGBT9 vector only, yeast containing pLAM5 (a GAL4 human lamin C fusion) or pDBD57. Only those library plasmids demonstrating a requirement of pDBD57 for induced expression of histidine and β-galactosidase reporter genes were considered further.
Clones which fulfilled all these criteria were sequenced and BLAST-searched against the EMBL/GenBank® database. Analysis revealed that 14 ORF 57-interacting clones corresponded to known cellular proteins. Nine clones encoded importin alpha isoforms, which we have reported previously [37], and five clones encoded the cellular protein REF/Aly.
ORF 57 interacts with the N- and C-terminal domains of REF
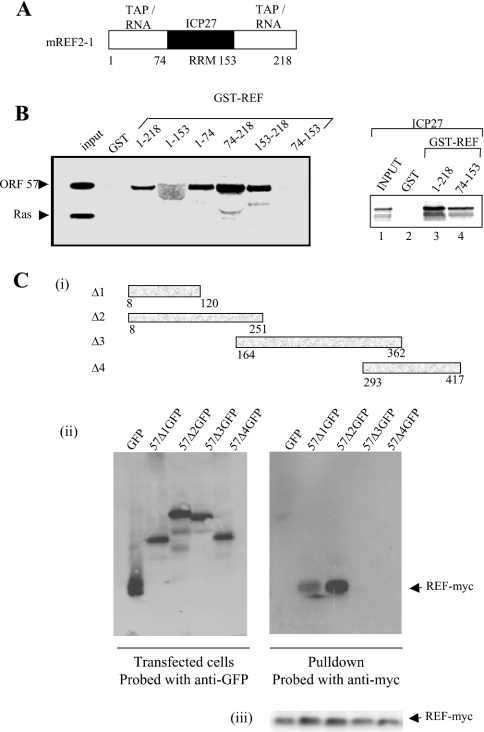
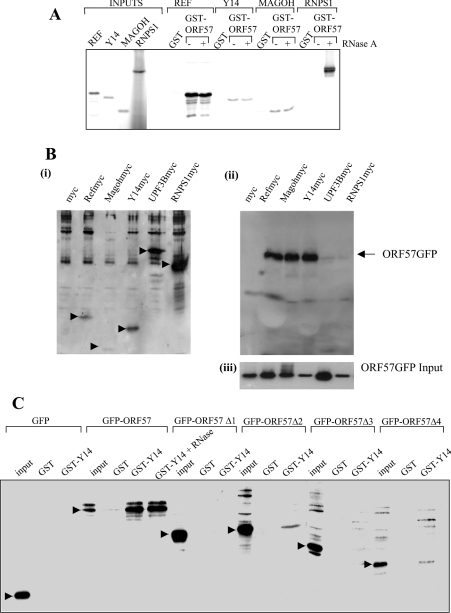
To confirm that REF interacts with HVS ORF 57 in vitro, we used a GST pull-down assay in conjunction with ORF 57 produced by in vitro transcription/translation. The mREF2-1 used in these studies can be divided into three domains of approx. 70 amino acids each. The N- and C-terminal domains mediate dimerization or oligomerization of REF and interactions with TAP and RNA, whereas the central RRM (RNA recognition motif) domain mediates interactions with HSV-1 ICP27 [26] (Figure 2A). When the interactions of ICP27 and ORF 57 with mREF2-1 are compared, we find that ICP27 interacts specifically with full-length mREF2-1 and the central RRM domain, albeit more weakly, as reported previously ([26] and Figure 2B, right panel). In contrast, ORF 57 interacted with the N- and C-terminal domains of mREF2-1 and failed to interact with the central RRM domain (Figure 2B, left panel). The human Ras protein control failed to interact with any of the GST–REF fusion proteins in these experiments (Figure 2B), thus indicating that truncation of REF had not created non-specific protein-binding surfaces, as shown previously by the finding that ICP27 failed to interact with the isolated N- or C-terminal domains of REF [26].
Figure 2. The N-terminal domain of ORF 57 binds to the N- and C-terminal domains of REF.
(A) The domain structure for mREF2-1 is shown schematically. The central RNA recognition motif spans residues 74–153 approximately. Interactions with other proteins and RNA are shown above each domain of the protein. (B) ORF 57 binds to the N- and C-terminal domains of REF. [35S]Methionine-labelled ICP27 (right panel), ORF 57 and human Ras (left panel), synthesized in rabbit reticulocyte lysates, were incubated with beads coated with the indicated GST fusion proteins. Then, 5% of the input and 50% of the bound material were loaded on to the gel. The diffuse ORF 57 band seen in lane 4 (left panel) was caused by co-migration of ORF 57 with the GST–REF(1–153) fusion protein. (C) REF binds to the N-terminal domain of ORF 57. (i) Schematic representation of the ORF 57–GFP deletion series. (ii) Cell lysates of p57GFP, p57Δ1–4GFP and pREF-myc transfected cells were immunoprecipitated using polyclonal ORF 57 antiserum. Bound proteins were resolved by SDS/PAGE and the presence of ORF 57 was detected using anti-GFP antiserum (left panel) and REF–myc was detected using anti-myc antiserum (right panel). (iii) As a control, Western-blot analysis was performed on the supernatants before immunoprecipitation using an anti-myc antibody to confirm that REF was expressed in all samples.
REF interacts with the N-terminal domain of HVS ORF 57
To delineate the domain within ORF 57 required for the specific interaction with mREF2-1, a series of ORF 57–GFP truncations were utilized in GST pull-down experiments. Protein extracts prepared from cells transfected with the ORF 57–GFP deletions were incubated with GST–REF bound to glutathione beads and the bound proteins were detected using an anti-GFP monoclonal antibody, since the anti-ORF 57 polyclonal antibody does not recognize ORF 57 on Western blots [34] (Figure 2C). The results demonstrated that GFP failed to interact with REF, whereas ORF 57 Δ1 (amino acids 8–120) and Δ2 (amino acids 8–251) specifically bound to GST–REF (Figure 2C). These results indicate that, whereas the minimal REF interaction domain for ORF 57 encompasses amino acids 8–120, the interaction with REF appears to be strengthened with ORF 57 (amino acids 8–251). As a suitable control, Western-blot analysis was performed to confirm that REF–myc was expressed in all samples [Figure 2C (iii)].
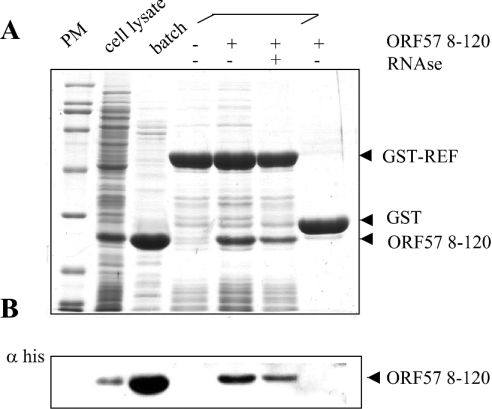
To confirm that the interaction between ORF 57 and mREF2-1 was not bridged by other eukaryotic proteins, we utilized a GST-pull-down assay with GST–REF and His6-tagged ORF 57 (8–120) expressed in E. coli (Figure 3). To verify the expression of ORF 57, a sample of the E. coli lysate (lane 2) was incubated with a cobalt affinity resin and found to yield purified ORF 57 (8–120) (lane 3). The fragment of ORF 57 encompassing amino acids 8–120 used in these experiments has a predicted mass of 15.077 kDa; however, the purified ORF 57 (8–120) protein migrated anomalously after SDS/PAGE with a mass of approx. 26 kDa. Therefore we confirmed its mass using MS. This analysis gave an experimental mass of 14.945 kDa for ORF 57 8–120, which agrees well with the predicted molecular mass for this fragment (14.946 kDa), if the first methionine is removed in E. coli. A cell lysate of E. coli expressing His6-tagged ORF 57 (8–120) (lane 2) was incubated with GST or GST–REF bound to glutathione–Sepharose. GST–REF selectively pulled down the ORF 57 (8–120) from the cell lysate, whereas GST alone did not. The identity of the ORF 57 bound to GST–REF was confirmed by Western blotting using an anti-His antibody (Figure 3B). The presence of RNase in the binding reaction moderately decreased the interaction between ORF 57 and GST–REF, suggesting that RNA may be stabilizing the interaction in the pull-down assays, although the ORF 57–GST–REF interaction was still readily detectable after RNase treatment.
Figure 3. ORF 57 (8–120) binds to REF without additional eukaryotic proteins.
An aliquot of a cell lysate from E. coli expressing His6-tagged ORF 57 (8–120) is shown in lane 2 (numbering from the left), incubation of an aliquot of this lysate with Talon resin leads to the purification of ORF 57 (8–120) (lane 3). Beads coated with the indicated GST fusions were incubated with the same E. coli cell lysate containing His-tagged ORF 57 in lanes 4–7 in the presence or absence of RNase as indicated. Bound proteins were analysed by SDS/PAGE followed by Coomassie Blue staining. An aliquot of each sample was also analysed by Western blotting with an antibody specific for the His6 tag and the results are shown in (B).
ORF 57 recruits TAP through REF
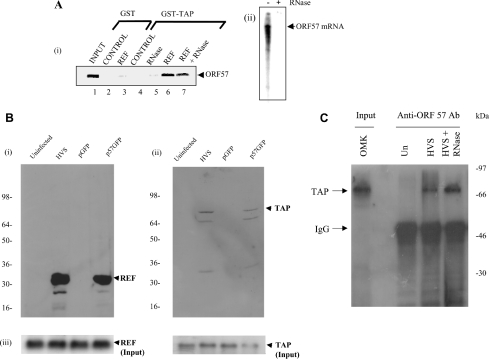
Since ORF 57 associates with the TAP interaction domains of REF, we examined whether the ORF 57–REF interaction might be mutually exclusive with the TAP–REF interaction or whether a ternary complex could form between ORF 57, REF and TAP. We immobilized GST or GST–TAP on Sepharose beads and looked for an interaction with in vitro translated ORF 57 [Figure 4A (i)]. No interaction was seen between ORF 57 and GST beads, even when supplemented with purified REF protein. Similarly, we did not detect an interaction between ORF 57 and GST–TAP [Figure 4A (i), lanes 4 and 5]. However, when the binding reaction was supplemented with purified REF protein, we did detect the formation of a ternary complex between GST–TAP, REF and ORF 57, and this interaction was only moderately decreased with RNase treatment [Figure 4A (i), lanes 6 and 7]. To confirm that the RNase treatment was effective in these experiments, an in vitro transcription/translation reaction with ORF 57 was performed and it was spiked with radiolabelled UTP. The RNA was subsequently extracted from pull-down assays in the presence and absence of RNase and analysed by denaturing PAGE (Figure 4A, ii). RNase treatment effectively degraded the RNA in the binding reaction, suggesting that, although the presence of RNA appears to enhance the formation of the binary and ternary complexes between GST–REF/ORF 57 and GST–TAP/REF/ORF 57, it may not be a prerequisite for these interactions. Taken together, these results indicate that REF, TAP and ORF 57 can form a ternary complex and that the interaction of TAP with ORF 57 is promoted by REF.
Figure 4. ORF 57 recruits TAP through REF.
(A) (i) [35S]Methionine-labelled ORF 57 was synthesized in vitro and incubated with beads coated with the indicated GST or GST fusion proteins. In the control lanes, 5 μg of purified thioredoxin was added to the binding reactions, and in some binding reactions, 5 μg of purified REF was added as indicated. RNase was included in some binding reactions as indicated. The bound proteins were eluted and one-tenth of the input (lane 1) and one-third of the bound fractions (lanes 2–7) were analysed by SDS/PAGE. (ii) Radiolabelled ORF 57 mRNA generated from an in vitro transcription/translation reaction was subjected to phenol extraction of a complete GST–TAP pull-down assay in the presence or absence of RNase as indicated. The recovered RNA was analysed by denaturing PAGE and visualized using a phosphoimager. (B) HEK-293T cells were transfected with pREF-myc or pTAP-myc and then studied for three cases: uninfected, superinfected with HVS (MOI of 1) or transfected with pGFP or p57GFP. After 24 h, cell lysates were immunoprecipitated using ORF 57 antiserum. The bound proteins were resolved by SDS/PAGE and the presence of (i) REF–myc and (ii) TAP–myc were detected by Western-blot analysis. (iii) As a suitable control, Western-blot analysis was performed using REF–myc and TAP–myc on whole cell extracts to confirm that TAP and REF were expressed in all samples. (C) Western-blot analysis, using an anti-TAP Ab (antibody), of immunoprecipitates obtained with anti-ORF 57 Ab from HVS mock and infected cell extracts (24 h post-infection), after separation by SDS/PAGE: input OMK cellular extract (lane 1, numbering from the left), uninfected extract+anti-ORF 57 Ab (lane 2), infected extract+anti-ORF 57 Ab (lane 3) and infected extract+anti-ORF 57 Ab+RNase (lane 4).
To confirm that the ORF 57–TAP interaction could also be observed in vivo, co-immunoprecipitation studies were performed. Control untransfected HEK-293T cells were compared with cells transfected with pRSVORF 57 or HVS-infected cells (MOI of 1) and expression vectors for myc-tagged REF and TAP. After 24 h, the cells were harvested and cell lysates were utilized in co-immunoprecipitation analysis using an anti-ORF 57 polyclonal antiserum. Immunoblot detection was performed using myc-specific antiserum. The results demonstrate that, in cells transfected with an ORF 57 expression vector and HVS-infected cells, ORF 57 immunoprecipitates with both REF and TAP (Figure 4B). As a suitable control, Western-blot analysis was performed to confirm that TAP and REF were expressed in all samples (Figure 4B, lower panel). To confirm that ORF 57 forms a complex containing endogenous TAP, ORF 57 was immunoprecipitated from HVS-infected cell lysates using an anti-ORF 57 polyclonal antiserum, and TAP was detected using an anti-TAP polyclonal antiserum. The results demonstrate that in HVS-infected cells, ORF 57 co-immunoprecipitates with endogenous TAP (Figure 4C).
HVS ORF 57 interacts with other members of the EJC
Since REF forms part of the EJC deposited on mRNA during splicing, we decided to investigate whether ORF 57 might interact with additional components of this complex. We screened for interactions with Y14, Magoh and RNPS1 (Figure 5). We could not synthesize full-length SRm160 in a reticulocyte lysate to test this EJC component. We observed a strong interaction with REF, which was insensitive to RNase treatment. A weaker but reproducible interaction was detected with Y14 and Magoh. In this assay, we also observed an interaction with RNPS1, but this was seen only after RNase treatment. A similar RNase-dependent interaction between Y14 and RNPS1 has been reported previously [5].
Figure 5. ORF 57 associates with other components of the EJC.
(A) ORF 57 interacts with components of the EJC in vitro. [35S]Methionine-labelled components of the EJC were synthesized in vitro and incubated with beads coated with the indicated GST or GST fusion proteins for REF, YI4, Mogoh and RNSP1. RNase was added where indicated. Bound proteins were eluted and one-tenth of the input (left-hand side of the gel) and one-third of the bound fractions were analysed by SDS/PAGE. (B) HEK-293T cells were transfected with myc-tagged expression constructs for REF, YI4, Magoh, RNSP1 and UPF3B (i) and in the presence of pRSVORF 57 (ii). After 24 h, cell lysates were utilized in co-immunoprecipitation analysis using an anti-myc affinity resin. Bound proteins were resolved by SDS/PAGE and the presence of ORF 57–GFP was detected using a GFP-specific antiserum. (iii) As a suitable control, Western-blot analysis was performed using anti-GFP on whole cell extracts to confirm that ORF 57–GFP was expressed in all samples. (C) Y14 associates with the N- and C-terminal domains of ORF 57. GST pull-downs were performed with GST or GST–Y14 and cell extracts from HEK-293T cells were transfected with the indicated GFP–ORF 57 fusions. RNase was added to the binding reactions where indicated. The bound proteins were detected by Western blotting using a GFP-specific antibody. The specific band corresponding to the expected size for the GFP–ORF 57 fusion is indicated in each case by an arrow.
To confirm that ORF 57 interacts with additional members of the EJC in vivo, co-immunoprecipitation studies were performed. Control untransfected HEK-293T cells were compared with cells transfected with pRSVORF 57 and myc-tagged expression constructs for REF, Y14, Magoh, RNSP1 and UPF3B. After 24 h, the cells were harvested and cell lysates were utilized in co-immunoprecipitation analysis using an anti-myc affinity resin. Polypeptides precipitated from cellular extracts were then analysed by Western blotting with a GFP-specific antiserum. The results demonstrate that, in ORF 57-transfected cells, ORF 57 specifically interacts with the EJC components REF, Y14 and Magoh. In contrast, RNPS1 and UPF3B failed to co-immunoprecipitate with GFP–ORF 57 in significant amounts (Figure 5B). To define the Y14 interaction domain in ORF 57, we utilized a GST pull-down assay with GST–Y14 which was incubated with cell extracts containing truncations of ORF 57 fused to GFP as described in Figure 2(C). A strong interaction between full-length GFP–ORF 57 and GST–Y14 was seen and it was insensitive to RNase treatment. No interaction was seen with ORF 57 (8–120). Weak interactions were seen with ORF 57 (8–240) and the C-terminal domain of ORF 57 (293–417); however the efficiency of the pull down of these truncated forms of ORF 57 was significantly compromised, suggesting that both domains are required for optimal interaction with Y14.
ORF 57 co-localizes with REF and TAP in the nucleus
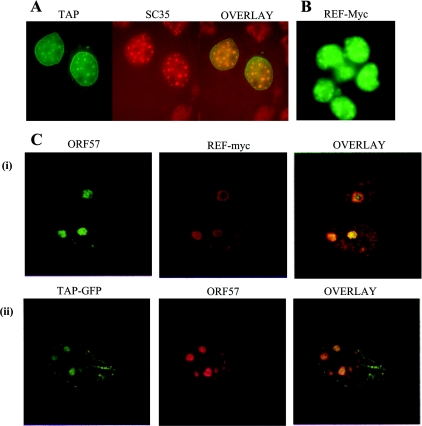
Many of the components of the EJC have been found to localize to the nuclear speckles, which are reorganized after infection with herpesviruses [35,45,46]. We initially determined whether TAP might also be localized in the nuclear speckles. To do this, we examined the localization of TAP–GFP in Cos-7 cells (Figure 6A). TAP shows a nuclear-rim staining as reported previously and a diffuse intranuclear staining. TAP also co-localized with the splicing factor SC35 to the nuclear speckles. The Myc-tagged REF expression vector also localized to the nuclear speckles as reported previously (Figure 6B) [47]. The effects of ORF 57 expression on the localization of myc-tagged REF and TAP–GFP were investigated in Cos-7 cells. In contrast with the speckled localization seen for REF and TAP with 10–30 small speckles per nucleus, ORF 57 localizes to 2–5 larger speckles that resemble nucleoli. When the localization of REF and TAP in cells expressing ORF 57 was examined, we found that REF and TAP were redistributed from the nuclear speckles to the larger structures and co-localized with ORF 57 (Figure 6C). Similar results were observed in HVS-infected cells (results not shown). Co-localization of ORF 57 with importin-α has also been observed in the nucleoli [37]; however, the implications of this subcellular compartmentalization have not been elucidated so far.
Figure 6. ORF 57 co-localizes REF and TAP in the nucleus.
(A) Cos-7 cells were transfected with TAP–GFP, fixed and then immunostained with a primary antibody specific for SC35 followed by a TRITC-labelled secondary antibody. (B) HeLa cells were transfected with a myc-tagged REF expression vector and then immunostained with the anti-myc monoclonal antibody 9E10 followed by an anti-mouse FITC-labelled secondary antibody. (C) Cos-7 cells were transfected with pRSVORF 57 in the presence of (i) pREF-myc or (ii) TAP–GFP. ORF 57 was detected using an anti-ORF 57 monoclonal antiserum and (i) anti-mouse FITC-labelled secondary antibody or (ii) anti-mouse Texas Red conjugate. REF–myc was detected using a primary antibody specific for myc followed by a TRITC-labelled secondary antibody. TAP–GFP was directly visualized using fluorescence microscopy.
TAP is required for bulk mRNA export in mammalian cells
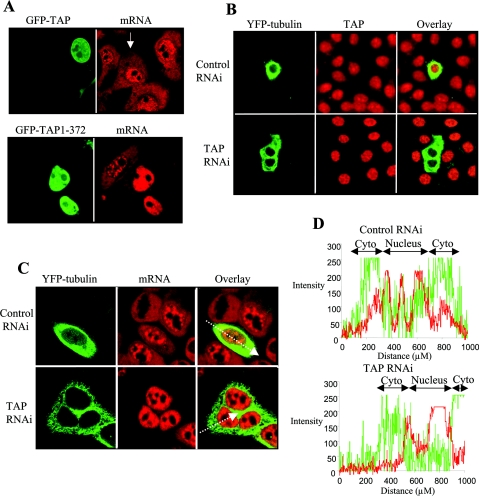
The essential role for TAP in bulk mRNA export has been demonstrated in Drosophila S2 cells by RNAi [22]. Furthermore, mRNA export can be blocked in Xenopus oocytes by injection of the retroviral CTE, which specifically associates with TAP through its N-terminal domain and acts as a competitive inhibitor for interaction with proteins involved in bulk mRNA export. The CTE has also been used to confirm that ICP27 utilizes TAP to export viral RNA in Xenopus oocytes [26]. It is conceivable that there are differences in the mRNA export pathways used in Xenopus oocytes and mammalian cells in which Herpesviridae replicate. Therefore we sought to disrupt TAP function in mammalian cells to analyse the effects on bulk mRNA and viral RNA export. TAP has a modular domain structure and the N-terminal domain is the substrate-interacting domain, associating with a variety of proteins including REF, Magoh, Y14 and SR proteins [5,12,24]. We anticipated that deletion of the central and C-terminal domains would render the substrate-binding domain incapable of docking at the nuclear pore and would act as a dominant-negative inhibitor of mRNA export when overexpressed (Figure 7). The distribution of bulk mRNA in OMK cells was analysed by FISH. In untransfected cells (indicated by the arrow in Figure 7A) and in cells overexpressing wild-type GFP–TAP, mRNA was readily detected in both the nucleus and cytoplasm. In contrast, cells overexpressing GFP–TAP (1–372) demonstrated significant nuclear accumulation and little cytoplasm staining for mRNA, indicating that bulk mRNA export had been compromised in these cells.
Figure 7. TAP is required for bulk mRNA export in mammalian cells.
(A) TAP (1–372) blocks mRNA export. OMK cells were transfected with the GFP–TAP or GFP–TAP (1–372). At 48 h post-transfection, the cells were fixed, permeabilized and total mRNA was detected by FISH using a Cy3-labelled oligo(dT) probe. The arrow indicates an untransfected cell. (B) TAP can be depleted by RNAi. HeLa cells were transfected with 700 ng of pSUPERLUC (control RNAi) or 700 ng of pSUPERTAP (TAP RNAi), together with 100 ng of a YFP–tubulin expression vector. Cells were fixed and permeabilized at 96 h post-transfection. TAP was detected using a rabbit polyclonal antibody and a TRITC-labelled secondary antibody. (C) mRNA export is blocked in cells depleted for TAP. HeLa cells were transfected with 700 ng of pSUPERLUC (control RNAi) or 700 ng of pSUPERTAP (TAP RNAi) together with 100 ng of a YFP–tubulin expression vector. Cells were fixed and permeabilized 96 h post-transfection. Bulk mRNA was detected by FISH using a Cy3-labelled oligo(dT) probe. The lines through the two cells in the right panel correspond to the path for the x-axis of the fluorescence intensity profiles shown in (D). (D) Fluorescence intensity profiles for the control and TAP-depleted cells shown in (C); Cyto, cytoplasm. The red line corresponds to the fluorescence from the mRNA detection by FISH and the green line corresponds to the fluorescence from the YFP–tubulin.
To provide more direct evidence for the essential role of TAP in mammalian mRNA export, we depleted the protein using RNAi in HeLa cells. HeLa cells were transfected with a control pSUPERLUC or pSUPERTAP, which expresses a short hairpin RNA active against TAP, together with YFP–tubulin, which acted as a marker for transfection. Depletion of endogenous TAP was confirmed using immunofluorescence with an anti-TAP antibody. At 96 h post-transfection, TAP protein was no longer detected in cells transfected with pSUPERTAP. In control transfected and untransfected HeLa cells, mRNA was readily detected in the nucleus and cytoplasm (Figures 7C and 7D). In contrast, cells depleted for TAP displayed a robust nuclear accumulation of mRNA and cytoplasmic mRNA was barely detected (Figures 7C and 7D). These effects were seen as early as 72 h (results not shown), but were most pronounced at 96 h post-transfection. These results indicate that TAP is essential for bulk mRNA export in mammalian cells. The YFP–tubulin was detected at 96 h post-transfection in these experiments, despite a gradually increasing block in mRNA export as TAP was depleted, and this probably represents YFP–tubulin mRNA, which escaped the export block earlier during the experiment and then went on to produce protein that was not turned over during the course of the experiment.
Functional TAP is required for ORF 57-mediated viral mRNA nuclear export
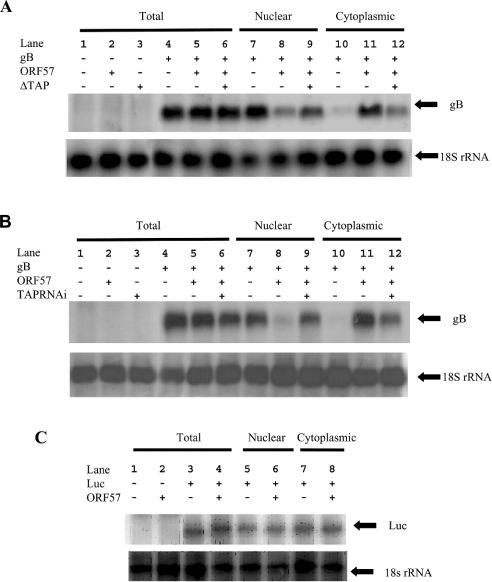
We have demonstrated previously that ORF 57 is a nucleocytoplasmic shuttling protein that mediates the nuclear export of viral mRNAs [36]. To determine whether TAP is required for viral RNA export, Northern-blot analysis was performed. Total, nuclear and cytoplasmic RNAs were isolated separately from HEK-293T cells transfected with pUCgB, in the absence and presence of pRSVORF 57 and pTAPΔGFP or pSUPERTAP and assayed by Northern-blot analysis (Figures 8A and 8B). In the presence of ORF 57, the gB mRNA efficiently accumulates in the cytoplasm (Figures 8A and 8B, compare lanes 10 and 11). However, when TAP activity is compromised in the cell either by the dominant-negative form (Figure 8A) or through depletion of TAP by RNAi (Figure 8B), the nuclear export of gB mRNA is retarded and there is a significant nuclear accumulation of the mRNA (Figures 8A and 8B, compare lanes 8 and 9 with lanes 11 and 12). These results suggest that the ORF 57–TAP interaction is required for the efficient nuclear export of late viral transcripts and viral transcripts use the cellular mRNA export pathway. In these experiments, we cannot exclude the possibility that the block in viral RNA export is an indirect effect caused by the depletion of another cellular factor required for viral RNA export. However, with the dominant-negative form of TAP, the viral RNA export block is seen at 48 h post-transfection, suggesting that the effects may be directly caused by inhibiting TAP function. In addition, to demonstrate that the ORF 57-mediated nuclear export is specific for viral transcripts, Northern-blot analysis was repeated using an ORF 57-independent RNA. Total, nuclear and cytoplasmic RNAs were isolated separately from HEK-293T cells transfected with pGL3 (Promega), a plasmid containing the luciferase reporter gene under the control of the simian virus 40 promoter, in the absence and presence of pRSVORF 57 and assayed by Northern-blot analysis (Figure 8C). The results demonstrate that ORF 57 had little effect on the nuclear–cytoplasmic distribution of the ORF 57-independent transcript.
Figure 8. Functional TAP is required for viral mRNA export.
(A) HEK-293T cells remained untransfected (lane 1) or were transfected with pUCgB in the absence (lanes 4, 7 and 10) and presence of pRSVORF 57 (lanes 5, 6, 8, 9, 11 and 12) or presence of pGFP-TAP (1–372) (lanes 3, 6, 9 and 12). (B) HeLa cells remained untransfected (lane 1) or were transfected with pUCgB in the absence (lanes 4, 7 and 10) and presence of pRSVORF 57 (lanes 5, 6, 8, 9, 11 and 12) or presence of pSUPERTAP (lanes 3, 6, 9 and 12). Total (lanes 1–6), nuclear (lanes 7–9) and cytoplasmic (lanes 10–12) RNAs were then isolated and separated by electrophoresis on a 1% denaturing formaldehyde–agarose gel. The RNA was transferred on to Hybond-N membranes and hybridized with 32P-radiolabelled random-primed probes specific for the HVS gB and 18 S rRNA-coding sequences. (C) HEK-293T cells remained untransfected (lane 1) or were transfected with pGL-3 in the absence (lanes 3, 5 and 7) and presence of pRSVORF 57 (lanes 4, 6 and 8). Total (lanes 1–4), nuclear (lanes 5–6) and cytoplasmic (lanes 7–8) RNAs were then isolated and separated by electrophoresis on a 1% denaturing formaldehyde–agarose gel. The RNA was transferred on to Hybond-N membranes and hybridized with 32P-radiolabelled random-primed probes specific for the Luc and 18 S rRNA-coding sequences.
Functional TAP is required for HVS replication
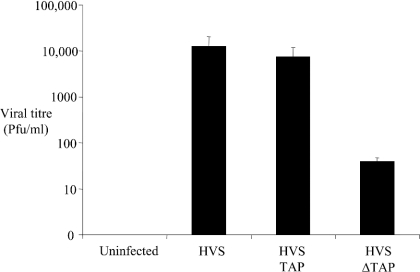
Infection of OMK cells with HVS leads to expression of viral lytic proteins and production of infectious virus, which can be easily monitored with a plaque assay. We investigated whether inhibition of viral RNA export might block viral replication by expressing the dominant-negative form of TAP in OMK cells and then challenging these cells with HVS. OMK cells were mock-transfected or transfected with GFP–TAP or GFP–TAP (1–372). The OMK cells were subsequently infected with HVS–GFP at an MOI of 1, and incubated at 37 °C for 5 days until destruction of the cell sheet had occurred. The supernatants were then harvested from each well and the viral titres measured by plaque assay (Figure 9). The results show that overexpression of TAP does not influence viral titre (column 2 versus column 3). However, viral titre from GFP–TAP (1–372) pretransfected cells is decreased by approx. 100-fold (column 4). These results demonstrate that the disruption of the cellular mRNA export pathway with the dominant-negative form of TAP can have a profound effect on viral replication.
Figure 9. A dominant-negative form of TAP reduces viral replication.
OMK cells remained untransfected or were transfected with pTAP-GFP or transfected with pGFP-TAP (1–372). After 24 h, the cells were infected with HVS–GFP at an MOI of 1. The supernatants were harvested 5 days post-infection and viral titres were determined using plaque assays. The variations between three replicated assays are indicated.
DISCUSSION
We have shown that ORF 57 shuttles between the nucleus and cytoplasm and transports viral RNA by a CRM-1-independent pathway. This is despite ORF 57 encoding a leucine-rich motif that resembles an HIV REV NES [36]. However, we believe that the ORF 57 leucine-rich NES plays some role in nuclear export since it enables the nuclear export of a heterologous protein and mutational analysis of the conserved leucine residues abrogates the ability of the ORF 57 protein to shuttle between the nucleus and cytoplasm. However, this sequence is unlikely to function as a conventional leucine-rich NES since it is LMB-insensitive and does not interact with CRM-1 (D. J. Goodwin and A. Whitehouse, unpublished work). This is consistent with recent observations for HSV ICP27, which appears to require a leucine-rich nuclear export sequence for export but does not associate with or use the CRM-1 pathway [26,27,30]. The leucine-rich NES found in ORF 57 and ICP27 may represent a motif important for interaction with other proteins or may simply be required for the overall structural integrity of the protein.
We have demonstrated that REF associates directly with the N-terminal domain of ORF 57 and this region has been narrowed down to amino acids 8–120, although a stronger interaction is seen with amino acids 8–251, suggesting that sequences between amino acids 121 and 253 either contribute directly to the interaction with REF or to the folding of the N-terminal domain. The former explanation seems more plausible since it has been shown that amino acids 1–215 from KSHV ORF 57 interact strongly with REF and a weaker interaction is seen with amino acids 181–328, indicating that there are two potential binding sites for REF on KSHV ORF 57 [29]. Furthermore, an REF interaction domain in EB2 has been mapped between amino acids 218 and 236 [28]. However, no direct interaction between EB2 amino acids 218–236 and REF was presented [28] and deletion of this domain may have altered the overall structural integrity of EB2, impairing its interaction with REF. The identification of the N-terminus of HVS ORF 57 as a minimal interaction domain for REF is consistent with the interaction domain identified for REF in HSV ICP27, which, by combining results from two separate mapping experiments [26,27,29], appears to lie between amino acids 110 and 138.
The results presented here indicate that HVS ORF 57 interacts with both the N-terminal (amino acids 1–74) and C-terminal domains (amino acids 153–218) of REF2-1 with equal affinity and does not interact with the central RNA recognition motif domain under the assay conditions used. KSHV ORF 57 also interacts with the N- and C-terminal domains of REF [29]. However, this mode of interaction with REF contrasts with that for ICP27, which binds to the central RRM domain only [26] and is more reminiscent of the interaction between EBV EB2 and REF, which involves the C-terminal domain of REF. The N- and C-terminal domains of REF are also used for interaction with RNA and TAP [47–49], although we have found that ORF 57 can still form a ternary complex with REF and TAP in vitro. Therefore the interaction domains for ORF 57 and TAP may not directly overlap within the N- and C-terminal domains. Alternatively, REF and TAP might interact with either domain of REF in such a way that, in ternary complexes, ORF 57 is associated with the N-terminus and TAP with the C-terminus or vice versa. A structural analysis of these interactions is probably required to resolve these possibilities.
REF forms part of the EJC on spliced mRNA and interacts with Y14 in this complex [5]. We have now shown that ORF 57 interacts with Y14 and Magoh and co-immunoprecipitates with these proteins. Y14 and Magoh are involved in the process of NMD (nonsense-mediated decay) [6,48,49] and, in Drosophila, they are implicated in the localization of oskar mRNA in the developing oocyte [50]. Y14 co-immunoprecipitates with Upf3B, a component of the NMD machinery, and when it is artificially tethered to an mRNA, it can trigger NMD, as can another component of the EJC, RNPS1 [6,7,49]. We have screened an HVS mRNA with a premature stop codon for ORF 57-mediated NMD but found no evidence for this (results not shown). Consistent with this, we fail to observe RNPS1 and UPF3B in ORF 57 immunoprecipitates. These results suggest that ORF 57 may recruit a stripped-down version of the EJC to its mRNA, which fails to recruit the NMD factor Upf3B, or these proteins are not stably associated with the complex. This may be due to the manner in which ORF 57 associates with REF, Y14 and Magoh, precluding the interactions with proteins involved in NMD. The role of Y14 and Magoh in ORF 57-stimulated gene expression remains unclear, although they may be responsible for localizing certain transcripts in the cytoplasm or improving the stability of the transport complex formed with TAP, p15 and REF since Y14 and Magoh both interact with TAP [5]. Recent results indicate that components of the EJC including Y14 and Magoh can stimulate polysome loading of mRNAs and increase their translation [51]. It may be the case that ORF 57 enhances viral mRNA translation in this manner by recruiting Y14 and Magoh.
We have confirmed that HVS mRNA transport occurs through the mRNA export pathway using RNAi on TAP and a dominant-negative mutant of TAP to block the mRNA export pathway. On the basis of these experiments, we have also demonstrated that TAP is the major mRNA exporter in mammalian cells, since we see very little cytoplasmic mRNA in cells where TAP has been depleted by RNAi. Similarly, the export of an intronless HVS transcript, gB, is inhibited when TAP function is compromised in the cell. However, the inhibition of viral RNA export is not complete, probably because we have failed to inhibit completely TAP function in the cell. Alternatively, the viral RNA may use other RNA export pathways albeit less efficiently, when the major mRNA export pathway is compromised. The block in viral RNA export has the side effect of blocking viral replication, as expected. However, we cannot exclude the possibility that viral replication and RNA export are impeded because other cellular proteins are no longer being produced due to the mRNA export block when TAP function is inhibited in the cell.
Herpesviridae appear to modify the gene expression pathway to suit their needs during infection, for example splicing factors are redistributed after infection and splicing is inhibited. We have shown previously that HVS ORF 57 is responsible for the redistribution of splicing factors into distinct nuclear aggregations [35]. We now show that, by expression of ORF 57, the mRNA export factors are also relocalized to large clumps within the cell. It will be interesting to determine whether viral transcription occurs within or around these structures and whether they represent specialized viral gene expression machines dedicated to transcription, processing and export of viral mRNAs at the expense of cellular mRNAs.
From these results, a simple model for HVS mRNA export can be predicted in which ORF 57 initially associates with viral mRNA. This triggers the recruitment of REF, Y14 and Magoh, which, in turn, leads to the recruitment of TAP and subsequent mRNA export. Whether ORF 57 has specific viral mRNA-binding sites and where these might reside in the mRNA is unclear at present. It is interesting to note that in transient transfection assays, KSHV ORF 57 synergistically enhances the transactivation capabilities of the major transcriptional protein, ORF 50 [52]. This suggests that ORF 57 may act at several levels to regulate viral gene expression. REF forms part of the TREX complex [2] and can be recruited to intronless mRNAs during transcription. It is possible that ORF 57 is also recruited to mRNA during the transcription process and associates with the TREX complex or a virally modified form of the TREX complex. Further experiments are now required to determine whether ORF 57 functions as a sequence-specific mRNA-binding protein and whether transcription plays a role in HVS mRNA export.
Online data
Acknowledgments
S.A.W. was supported by a grant from the BBSRC, A.W. by grants from the Yorkshire Cancer Research, BBSRC and Royal Society and L.R. by a BBSRC studentship. We thank J. Katahira for providing the anti-TAP-C antibody and B. Clements for a critical reading of the paper.
References
- 1.Segref A., Sharma K., Doye V., Hellwig A., Huber J., Luhrmann R., Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strasser K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondon A. G., Aguilera A., Struhl K., Reed R., et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature (London) 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 3.Le Hir H., Izaurralde E., Maquat L. E., Moore M. J. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan C. C., Dostie J., Diem M. D., Feng W., Mann M., Rappsilber J., Dreyfuss G. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10:200–209. doi: 10.1261/rna.5230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kataoka N., Diem M. D., Kim V. N., Yong J., Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J. 2001;20:6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim V. N., Kataoka N., Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science. 2001;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- 7.Lykke-Andersen J., Shu M. D., Steitz J. A. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- 8.Palacios I. M., Gatfield D., St Johnston D., Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature (London) 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 9.Le Hir H., Gatfield D., Izaurralde E., Moore M. J. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei E. P., Silver P. A. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes Dev. 2002;16:2761–2766. doi: 10.1101/gad.1032902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gruter P., Tabernero C., von Kobbe C., Schmitt C., Saavedra C., Bachi A., Wilm M., Felber B. K., Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 12.Stutz F., Bachi A., Doerks T., Braun I. C., Seraphin B., Wilm M., Bork P., Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fribourg S., Braun I. C., Izaurralde E., Conti E. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol. Cell. 2001;8:645–656. doi: 10.1016/s1097-2765(01)00348-3. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y., Steitz J. A. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 15.Bachi A., Braun I. C., Rodrigues J. P., Pante N., Ribbeck K., von Kobbe C., Kutay U., Wilm M., Gorlich D., Carmo-Fonseca M., et al. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant R. P., Hurt E., Neuhaus D., Stewart M. Structure of the C-terminal FG-nucleoporin binding domain of Tap/NXF1. Nat. Struct. Biol. 2002;9:247–251. doi: 10.1038/nsb773. [DOI] [PubMed] [Google Scholar]
- 17.Herold A., Klymenko T., Izaurralde E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA. 2001;7:1768–1780. [PMC free article] [PubMed] [Google Scholar]
- 18.Tan W., Zolotukhin A. S., Bear J., Patenaude D. J., Felber B. K. The mRNA export in Caenorhabditis elegans is mediated by Ce-NXF-1, an ortholog of human TAP/NXF and Saccharomyces cerevisiae Mex67p. RNA. 2000;6:1762–1772. doi: 10.1017/s1355838200000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jun L., Frints S., Duhamel H., Herold A., Abad-Rodrigues J., Dotti C., Izaurralde E., Marynen P., Froyen G. NXF5, a novel member of the nuclear RNA export factor family, is lost in a male patient with a syndromic form of mental retardation. Curr. Biol. 2001;11:1381–1391. doi: 10.1016/s0960-9822(01)00419-5. [DOI] [PubMed] [Google Scholar]
- 20.Yang J., Bogerd H. P., Wang P. J., Page D. C., Cullen B. R. Two closely related human nuclear export factors utilize entirely distinct export pathways. Mol. Cell. 2001;8:397–406. doi: 10.1016/s1097-2765(01)00303-3. [DOI] [PubMed] [Google Scholar]
- 21.Strasser K., Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herold A., Klymenko T., Izaurralde E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA. 2001;7:1768–1780. [PMC free article] [PubMed] [Google Scholar]
- 23.Zolotukhin A. S., Tan W., Bear J., Smulevitch S., Felber B. K. U2AF participates in the binding of TAP (NXF1) to mRNA. J. Biol. Chem. 2002;277:3935–3942. doi: 10.1074/jbc.M107598200. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y., Gattoni R., Stevenin J., Steitz J. A. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 25.Reichert V. L., Le Hir H., Jurica M. S., Moore M. J. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes Dev. 2002;16:2778–2791. doi: 10.1101/gad.1030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koffa M. D., Clements J. B., Izaurralde E., Wadd S., Wilson S. A., Mattaj I. W., Kuersten S. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 2001;20:5769–5778. doi: 10.1093/emboj/20.20.5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen I. H., Sciabica K. S., Sandri-Goldin R. M. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 2002;76:12877–12889. doi: 10.1128/JVI.76.24.12877-12889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiriart E., Bardouillet L., Manet E., Gruffat H., Penin F., Montserret R., Farjot G., Sergeant A. A region of the Epstein-Barr virus (EBV) mRNA export factor EB2 containing an arginine-rich motif mediates direct binding to RNA. J. Biol. Chem. 2003;278:37790–37798. doi: 10.1074/jbc.M305925200. [DOI] [PubMed] [Google Scholar]
- 29.Malik P., Blackbourn D. J., Clements J. B. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 2004;279:33001–33011. doi: 10.1074/jbc.M313008200. [DOI] [PubMed] [Google Scholar]
- 30.Soliman T. M., Silverstein S. J. Identification of an export control sequence and a requirement for the KH domains in ICP27 from herpes simplex virus type 1. J. Virol. 2000;74:7600–7609. doi: 10.1128/jvi.74.16.7600-7609.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lischka P., Rosorius O., Trommer E., Stamminger T. A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69. EMBO J. 2001;20:7271–7283. doi: 10.1093/emboj/20.24.7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boshoff C., Weiss R. A. Epidemiology and pathogenesis of Kaposi's sarcoma-associated herpesvirus. Philos. Trans. R. Soc. London Ser. B. 2001;356:517–534. doi: 10.1098/rstb.2000.0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schulz T. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) J. Gen. Virol. 1998;79:1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- 34.Whitehouse A., Cooper M., Meredith D. M. The immediate-early gene product encoded by open reading frame 57 of herpesvirus saimiri modulates gene expression at a posttranscriptional level. J. Virol. 1998;72:857–861. doi: 10.1128/jvi.72.1.857-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper M., Goodwin D. J., Hall K. T., Stevenson A. J., Meredith D. M., Markham A. F., Whitehouse A. The gene product encoded by ORF 57 of herpesvirus saimiri regulates the redistribution of the splicing factor SC-35. J. Gen. Virol. 1999;80:1311–1316. doi: 10.1099/0022-1317-80-5-1311. [DOI] [PubMed] [Google Scholar]
- 36.Goodwin D. J., Hall K. T., Stevenson A. J., Markham A. F., Whitehouse A. The open reading frame 57 gene product of herpesvirus saimiri shuttles between the nucleus and cytoplasm and is involved in viral RNA nuclear export. J. Virol. 1999;73:10519–10524. doi: 10.1128/jvi.73.12.10519-10524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodwin D. J., Whitehouse A. A gamma-2 herpesvirus nucleocytoplasmic shuttle protein interacts with importin alpha 1 and alpha 5. J. Biol. Chem. 2001;276:19905–19912. doi: 10.1074/jbc.M009513200. [DOI] [PubMed] [Google Scholar]
- 38.Goodwin D. J., Hall K. T., Giles M. S., Calderwood M. A., Markham A. F., Whitehouse A. The carboxy terminus of the herpesvirus saimiri ORF 57 gene contains domains that are required for transactivation and transrepression. J. Gen. Virol. 2000;81:2253–2265. doi: 10.1099/0022-1317-81-9-2253. [DOI] [PubMed] [Google Scholar]
- 39.Chien C. T., Bartel P. L., Sternglanz R., Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. U.S.A. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katahira J., Strasser K., Podtelejnikov A., Mann M., Jung J. U., Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang G., Taneja K. L., Singer R. H., Green M. R. Localization of pre-mRNA splicing in mammalian nuclei. Nature (London) 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
- 42.Mears W. E., Rice S. A. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology. 1998;242:128–137. doi: 10.1006/viro.1997.9006. [DOI] [PubMed] [Google Scholar]
- 43.Kotsopoulou E., Kim V. N., Kingsman A. J., Kingsman S. M., Mitrophanous K. A. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J. Virol. 2000;74:4839–4852. doi: 10.1128/jvi.74.10.4839-4852.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiegand H. L., Lu S., Cullen B. R. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11327–11332. doi: 10.1073/pnas.1934877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin T. E., Barghusen S. C., Leser G. P., Spear P. G. Redistribution of nuclear ribonucleoprotein antigens during herpes simplex virus infection. J. Cell Biol. 1987;105:2069–2082. doi: 10.1083/jcb.105.5.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phelan A., Carmo-Fonseca M., McLaughlan J., Lamond A. I., Clements J. B. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc. Natl. Acad. Sci. U.S.A. 1993;90:9056–9060. doi: 10.1073/pnas.90.19.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Z., Luo M. J., Straesser K., Katahira J., Hurt E., Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature (London) 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- 48.Fribourg S., Gatfield D., Izaurralde E., Conti E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat. Struct. Biol. 2003;10:433–439. doi: 10.1038/nsb926. [DOI] [PubMed] [Google Scholar]
- 49.Rodrigues J. P., Rode M., Gatfield D., Blencowe B. J., Carmo-Fonseca M., Izaurralde E. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. U.S.A. 2001;98:1030–1035. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hachet O., Ephrussi A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 2001;11:1666–1674. doi: 10.1016/s0960-9822(01)00508-5. [DOI] [PubMed] [Google Scholar]
- 51.Nott A., Le Hir H., Moore M. J. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirshner J. R., Lukac D. M., Chang J., Ganem D. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 2000;74:3586–3597. doi: 10.1128/jvi.74.8.3586-3597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.