Abstract
Few cell adhesion molecules have been reported to be expressed in mature adipocytes, and the significance of cell adhesion process in adipocyte biology is also unknown. In the present study, we identified ACAM (adipocyte adhesion molecule), a novel homologue of the CTX (cortical thymocyte marker in Xenopus) gene family. ACAM cDNA was isolated during PCR-based cDNA subtraction, and its mRNA was shown to be up-regulated in WATs (white adipose tissues) of OLETF (Otsuka Long–Evans Tokushima fatty) rats, an animal model for Type II diabetes and obesity. ACAM, 372 amino acids in total, has a signal peptide, V-type (variable) and C2-type (constant) Ig domains, a single transmembrane segment and a cytoplasmic tail. The amino acid sequence in rat is highly homologous to mouse (94%) and human (87%). ACAM mRNA was predominantly expressed in WATs in OLETF rats, and increased with the development of obesity until 30 weeks of age, which is when the peak of body mass is reached. Western blot analysis revealed that ACAM protein, approx. 45 kDa, was associated with plasma membrane fractions of mature adipocytes isolated from mesenteric and subdermal adipose deposits of OLETF rats. Up-regulation of ACAM mRNAs in obesity was also shown in WATs of genetically obese db/db mice, diet-induced obese ICR mice and human obese subjects. In primary cultured mouse and human adipocytes, ACAM mRNA expression was progressively up-regulated during differentiation. Several stably transfected Chinese-hamster ovary K1 cell lines were established, and the quantification of ACAM mRNA and cell aggregation assay revealed that the degree of homophilic aggregation correlated well with ACAM mRNA expression. In summary, ACAM may be the critical adhesion molecule in adipocyte differentiation and development of obesity.
Keywords: adipose tissue, adhesion molecule, cell aggregation, CTX gene family, obesity
Abbreviations: ACAM, adipocyte adhesion molecule; BMI, body mass index; BT-IgSF, brain- and testis-specific Ig superfamily; CAR, coxsackievirus and adenovirus receptor; CHO, Chinese-hamster ovary; CLMP, CAR-like membrane protein; CTX, cortical thymocyte marker in Xenopus; CTXL, CTX-like; C-type domain, constant domain; DIO, diet-induced obesity; DMEM, Dulbecco's modified Eagle's medium; ESAM, endothelial cell-selective adhesion molecule; GPA33, glycoprotein A33; HBSS, Hanks balanced salt solution; HDM, high-density microsome; JAM, junctional adhesion molecule; LDM, low-density microsome; LETO, Long–Evans Tokushima Otsuka; OLETF, Otsuka Long–Evans Tokushima fatty; PPARγ, peroxisome-proliferator-activated receptor-γ; RACE, rapid amplification of cDNA ends; TZD, thiazolidinedione; V-type domain, variable domain; WAT, white adipose tissue
INTRODUCTION
Normal metabolic balance is maintained by a complex homoeostasis system involving multiple tissues and organs. Modern sedentary life style and excess of total lipid intake can lead to metabolic syndrome, which is pandemic in industrialized countries worldwide. Metabolic syndrome is associated with a cluster of risk factors of atherosclerosis, such as insulin resistance, hyperinsulinaemia, impaired glucose tolerance or Type II diabetes, hypertension and dyslipidaemia [1,2]. Indeed, excess body mass increases the risk of death from cardiovascular diseases in adults up to 75 years of age, and the relative risk associated with an increment of BMI (body mass index) was 1.10 for the mortality from cardiovascular diseases in 30- to 44-year-old men [3].
The regional difference in body fat is also one of the critical determinants for the vascular complications, and the accumulation of abdominal fat is the major risk factor for cardiovascular diseases both in men and women [4,5]. To give new insights into molecular mechanism of development of abdominal obesity and metabolic syndrome, we attempted to isolate genes up-regulated during the process of fat cell hypertrophy in visceral adipose tissues of OLETF (Otsuka Long–Evans Tokushima fatty) rats. OLETF rats were suitable animals for the present study, since they are characterized as having visceral fat obesity, insulin resistance, hyperinsulinaemia, hyperglycaemia, hypertension and dyslipidaemia [6]. We employed PCR-based subtractive hybridization method, i.e. representational difference analysis of cDNA, and mRNA isolated from visceral adipose tissues of 30-week-old OLETF and mRNA from 8-week-old LETO (Long–Evans Tokushima Otsuka) rats, diabetes-resistant counterparts, were used as the tester and driver respectively. In the process of cDNA subtraction, a cDNA fragment of a novel gene, OL-16, was isolated and its mRNA expression was up-regulated in the visceral adipose tissues of 30-week-old OLETF rats [7].
In the present study, we have reported cDNA cloning of the full-coding sequence of OL-16 and the gene product has been designated as ACAM (adipocyte adhesion molecule), since it is predominantly expressed in WATs (white adipose tissues) and mediates the cell–cell adhesion. Homology searches showed that ACAM is a new member of the CTX (cortical thymocyte marker in Xenopus) subfamily belonging to the Ig superfamily. ACAM shares common structural organization with other members of the CTX family: N-terminal extracellular variable (V-type) domain and constant (C2-type) Ig domain separated by J segment, a transmembrane domain and a cytoplasmic tail [8,9]. Several homologues of CTX have been identified in human: CAR (coxsackievirus and adenovirus receptor) [10–12], BT-IgSF (brain- and testis-specific Ig superfamily) [13], ESAM (endothelial cell-selective adhesion molecule) [14], GPA33 (glycoprotein A33) [15], CTXL (CTX-like) [8,9], JAM-A (junctional adhesion molecule-A) [16], JAM-B [17] and JAM-C [18–22]. The hypothetical protein MGC44287, found by the National Institutes of Health Mammalian Gene Collection (MGC) Program, also seems to be a member of the CTX family [23].
Most of the CTX family members are localized to cell–cell contacts between epithelial and endothelial cells, such as GPA33 on basolateral surfaces of intestinal epithelial cells [24], CAR at tight junctions of polarized epithelial cells in a wide range of tissues [25,26], and ESAM [14], JAM-B and JAM-C [27] in endothelial cell tight junctions. In addition, CTX family members mediate homophilic cell aggregation and cell adhesion of different cell types. JAM-A on endothelial cells is a ligand for β2 integrin LFA-1 (lymphocyte function-associated antigen 1) of T-cells [28]; JAM-C on platelets is a counter-receptor for the β2-integrin Mac-1 [18]; JAM-B on endothelial cells interacts with T-, NK (natural killer) and dendritic cells through JAM-C [21]. These molecules seem to mediate leukocyte–endothelial cell interaction during leukocyte migration through endothelial cells and platelet–leukocyte interaction in inflammatory process. Interestingly, the BT-IgSF gene was expressed in both neurons and glial cells in vitro, and was preferentially detected in pyramidal cell layers of the dentate gyrus and hippocampus and in commissure fibres of the corpus callosum [13]. This finding suggests that BT-IgSF plays a role in the development or function of the central nervous system. Thus, in various cell types and tissues, CTX family members mediate cell–cell adhesion and interactions, and are involved in various biological and pathobiological processes, such as development, inflammation, cancer metastasis and tight junctions in epithelial and endothelial cells.
In the present study, we show that ACAM is a new member of the emerging family of CTXL molecules. Unlike other CTX members, ACAM was predominantly expressed in adipocytes and was induced in WATs of rat and mouse animal models for obesity. The identification of a new adhesion molecule expressed in adipocyte lineage would open up a new field in adipocyte biology, and facilitate the elucidation of molecular mechanism of adipocyte hypertrophy and subsequent metabolic syndrome in obesity.
EXPERIMENTAL
Isolation of a full-coding region of ACAM cDNA and nucleotide sequencing
mRNAs from visceral adipose tissues of 8-week-old LETO and 30-week-old OLETF rats were isloated, and were screened for upregulated genes in OLETF rats by using representational difference analysis of cDNA [29–31]. We isolated a partial cDNA fragment of ACAM gene described previously as OL-16 [7]. In the present study, we used Marathon cDNA Amplification Kit (Clontech, Palo Alto, CA, U.S.A.) to isolate the full-coding sequences of orthologous rat, mouse and human ACAM genes. The PCR products of 5′ and 3′ RACE (rapid amplification of cDNA ends) were subcloned into pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA, U.S.A.) and sequenced by automated DNA sequencer (ABI PRISM 310 Genetic Analyzer, Applied Biosystems, Foster City, CA, U.S.A.).
Structural and homology analyses
Kyte and Doolittle hydrophobicity/hydrophilicity plot analyses [32] were performed using GENETYX-WIN (Software Development, Tokyo, Japan). On-line homology search was performed using the BLAST program at National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple sequence alignment was performed by Clustal W (http://clustalw.genome.jp/) [33] and the dendogram was drawn by TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). The presence of signal sequence and transmembrane domain was assessed by SOSUI program (http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0.html) [34] or PSORT (http://psort.nibb.ac.jp/).
Experimental animals
For expression studies of ACAM in adipose tissues, 4-week-old OLETF (n=45) and LETO rats (n=15) (Otsuka Pharmaceutical Tokushima Research Institute, Tokushima, Japan) were individually housed under 12-h light–dark cycles and allowed free access to rodent chow (MF, Oriental Yeast, Tokyo, Japan) and water. We divided OLETF rats into two experimental groups, control group (n=15) and TZD (thiazolidinedione) group (n=15) with free access to chow containing pioglitazone (generously given by Takeda Chemical Industry, Osaka, Japan). The average food intake of each animal was measured and the TZD group received approx. 1 mg/kg of mass per day of pioglitazone during the period of the experiments. Various adipose tissues were obtained after overnight fasting.
db/db (n=10) and db/m (n=10) mice (Clea Co Ltd, Tokyo, Japan) were individually housed under 12-h light–dark cycles and allowed free access to rodent chow (MF, Oriental Yeast) and water. At 12 weeks of age, various adipose tissues were obtained after overnight fasting. ICR mice (Charles River, Tokyo, Japan) were individually housed under 12-h light–dark cycles with free access to food and water. We divided ICR mice into two experimental groups, standard chow group (corn starch chow containing 11% fat in the total calorie count; n=10) and DIO (diet-induced obesity) group (sucrose chow containing 58% fat in the total calorie count; n=10) (Research Diets, New Brunswick, NJ, U.S.A.). At 16 weeks of age, mice were killed while under deep anesthaesia after overnight fasting, and various adipose tissues were obtained.
Isolation of mature adipocytes and stromal vascular cells
Adipocytes were isolated as described previously [35]. Briefly, various adipose tissues of 30-week-old OLETF rats (approx. 2 g) were removed, rinsed with 0.9% NaCl and minced finely with scissors. Minced tissues were digested with collagenase in Krebs–Ringer/Hepes buffer (117 mM NaCl, 10 mM NaHCO3, 1 mM CaCl2, 1 mM MgSO4, 4 mM KH2PO4, 30 mM Hepes, 2.5 mM glucose, 200 nM adenosine, 4% BSA fraction V, pH 7.4) for 60 min at 37 °C. The digested tissues were filtered through a 300 mesh nylon screen and centrifuged for 2 min at 100 g at room temperature. The floating layer of adipocytes and pelleted stromal vascular cells were each collected and washed three times with Krebs–Ringer/Hepes buffer. The washed adipocytes and stromal vascular cells were then subjected to RNA isolation.
Total RNA preparation, Northern blot analyses, quantitive real-time PCR
For the expression studies of ACAM mRNA in rat and mouse adipose tissues, rat and mouse total RNAs were isolated from adipose tissues of rats and mice by guanidinium isothiocyanate–CsCl ultracentrifugation. In addition, for expression study of ACAM mRNA in human adipose tissues, human mesenteric and subdermal fats were obtained from thirteen patients who had undergone surgical resection of colon cancers. Written informed consent was given by the patients. For ACAM tissue distribution studies on human and mouse various tissues, total RNAs were obtained from Clontech. Total RNAs (20 μg) were subjected to Northern blot analyses as described previously [7,30]. For quantitative real-time PCR analysis, cDNA synthesized from 2 μg of total RNA was analysed in a Sequence Detector (model 7900; PE Applied Biosystems) with specific primers and SYBR Green PCR Master (PerkinElmer Life Sciences). The relative abundance of mRNAs was standardized with 36B4 mRNA as the invariant control. The primers used were as follows: mouse and human ACAM, 5′-AGCCGTCATGTCTACAATAACTTGA-3′ (sense) and 5′-GGGCTTGGATGGTCTCACTAGCACT-3′ (antisense).
Preparation of rabbit polyclonal anti-ACAM antibody
Antibodies were raised in rabbits against the synthetic peptide, PSQSRAFQTV, the sequence of which was derived from the C-terminal end of intracellular domain of ACAM (see Figure 1). Cysteine residue was added to the N-terminus for conjugation of the peptide to keyhole-limpet haemocyanin (Asahi Techno Glass, Tokyo, Japan). To assess the specificity of the anti-ACAM antibody, ELISA and competitive inhibition ELISA assay were performed as described previously [30,36].
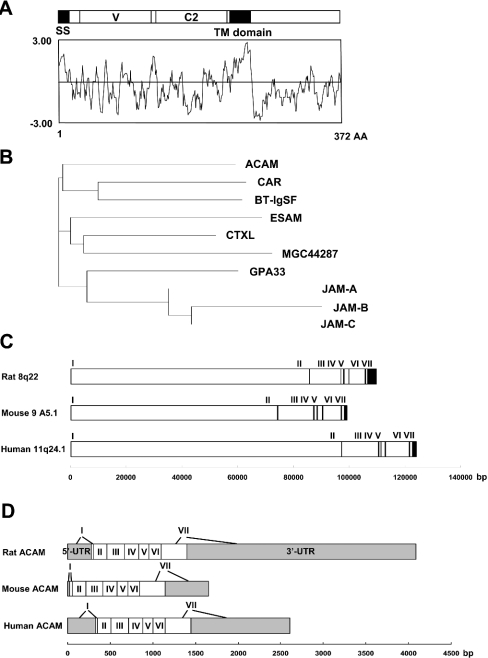
Figure 1. Structural and homology analysis of rat, mouse and human ACAM.
(A) Kyte and Doolittle hydrophobicity/hydrophilicity plot analysis of rat ACAM. Two hydrophobic domains, the signal sequence (SS) and transmembrane (TM) domain, are noted. ACAM is characterized by N-terminal V-type Ig domain (V) and C2-type Ig domain (C2). (B) Dendogram of ACAM and CTX family members. (C) Exon–intron boundaries of mouse, rat and human ACAM gene. ACAM genes are localized to chromosome 8q22 in rat (NW_044096), chromosome 9 A5.1 in mouse (NW_000352) and chromosome 11q24.1 in human (NW_024769). Exons are indicated by vertical bars and exon numbers, I to VII, are also indicated. (D) Untranslated and coding sequence of ACAM cDNAs. Exon 7, encoding cytoplasmic domains and containing 3′-untranslated region of rat, mouse and human ACAM, varies in size. UTR, untranslated regions (grey boxes).
Western blotting
Mesenteric and subdermal WATs derived from OLETF and TZD rats at 30 weeks of age were subjected to subcellular fractionation of adipocytes [35]. Isolated adipocytes were suspended in TES (20 mM Tris/HCl, 1 mM EDTA, 8.7% sucrose, pH 7.4) buffer and homogenized in pre-cooled motor-driven Potter–Elvehjem grinder with 10 strokes at 1400 rev./min. After the centrifugation of homogenates with a fixed angle JA20 rotor (Beckman Coulter, Fullerton, CA) at 13200 g for 30 min (4 °C), the supernatants were centrifuged at 16000 g for 30 min (4 °C) and the pellets used as high-density microsome (HDM) fraction. Further centrifugation of the supernatant at 200000 g for 75 min (4 °C) was performed to pellet the low-density microsomes (LDMs). The pellets of the first centrifugation was resuspended in TES buffer and loaded on the sucrose cushion (20 mM Tris/HCl, 1 mM EDTA, 38.5% sucrose, pH 7.4) and centrifuged for 60 min at 100000 g (4 °C) in a Beckman model SW28 rotor and plasma membrane fraction was collected from the top of the sucrose cushion. For Western blot analyses, 20 μg of protein of each sample was subjected to SDS/PAGE under reducing conditions and the gel proteins were electroblotted on to Hybond P PVDF membrane (Amersham). Membranes were incubated with rabbit polyclonal anti-ACAM antibody (1:500 dilution) and anti-rabbit IgG conjugated with horseradish peroxidase (1:20000 dilution) at 37 °C. The blots were incubated with ECL® Plus Western Blotting Detection Reagents (Amersham), and then exposed to X-ray film [31].
Immunohistochemistry
Immunolocalization of ACAM was assessed by immunoperoxidase ABC kit (Vector Laboratories, Burlingame, CA, U.S.A.), as described previously [37]. Briefly, formalin-fixed paraffin sections (4-μm) were dewaxed, cleared and rehydrated. The sections were first incubated with rabbit anti-ACAM serum overnight at 4 °C and then incubated with biotinylated donkey anti-rabbit IgG (Chemicon, Temecula, CA, U.S.A.) for 30 min at 22 °C, and followed by treatment with 3-diamino-benzidine and H2O2.
Cell culture and adipocyte differentiation
The isolation and culture of adipose tissue-derived stromal cells were performed as below. Briefly, freshly excised subdermal fat pads from 10-week-old male ICR mice were rinsed in PBS, minced and digested for 60 min at 37 °C in DMEM (Dulbecco's modified Eagle's medium) (Sigma, St. Louis, MO, U.S.A.) with 1 mg/ml type I collagenase. The digested tissues were filtered through a 250 nylon mesh to remove undigested tissue and centrifuged at 1800 rev./min for 5 min. After washing and centrifugation steps, stromal cells were inoculated in DMEM supplemented with 10% fetal bovine serum (Invitrogen), 200 μM ascorbic acid (Sigma) and 100 units/ml penicillin and 0.1 mg/ml streptomycin (Invitrogen). After 2 days, differentiation of stromal cells was induced with 0.5 mM methyl-isobutyl-xanthine (Sigma), 0.25 μM dexamethasone (Sigma), 10 μg/ml insulin (Sigma) and 5 μM 15-deoxy-Δ12,14-prostaglandin J2 (Sigma) for 48 h and the cells were maintained in DMEM containing 10 μg/ml insulin and 5 μM 15-deoxy-Δ12,14-prostaglandin. More than 90% of the cells showed typical morphological features of adipocytes 0–6 days after the initiation of differentiation. Total RNA was isolated by using RNeasy Lipid Tissue Mini Kit (Qiagen, Hilden, Germany). In addition, total RNAs of human primary cultured adipocytes were obtained from Zen-Bio (Chapel Hill, NC, U.S.A.). For quantitative real-time PCR analysis, cDNA synthesized from 2 μg of total RNA was analysed in a Sequence Detector (model 7900; PE Applied Biosystems). The relative abundance of mRNAs was standardized with 36B4 mRNA as the invariant control.
Isolation of ACAM stably expressing CHO (Chinese-hamster ovary)-K1 cells and aggregation studies
The coding region of rat ACAM cDNA was subjected to PCR amplification and subcloned into pcDNA3.1/V5-His TOPO vector (Invitrogen) and it was designated as pcDNA3.1/ACAM. This mammalian expression vector construct was incorporated into 1.0 unit of HVJ (haemagglutinating virus of Japan) envelope vector (GenomeONE-Neo, Ishihara Sangyo, Osaka, Japan) and transfected into CHO-K1 cells (A.T.C.C.). The cells were selected in the presence of 1000 μg/ml G418 (Invitrogen). Stable transfectants were cloned, total RNAs isolated and Northern blot analysis performed. High-expressing and low-expressing CHO-K1 cell lines were selected and used for cell aggregation assay. Negative control clones were obtained by transfecting pcDNA3.1 empty vector. Transfected CHO-K1 cells were detached with PBS with 1 mM EDTA at 37 °C for 10 min. After washing with Ca2+-free HBSS (Hanks balanced salt solution) (Sigma) two times then cells were resuspended in Ca2+-free HBSS and 2% fetal calf serum, which is predialysed against Ca2+-free HBSS, by three passages through an 18-gauge needle. Single-cell suspensions, 1×106 cells/ml, were incubated in plastic wells with gentle rotation on the platform rotator for 90 min at 37 °C. The aggregation assay was stopped by the addition of 2 ml of 5% glutaraldehyde in HBSS. To measure the aggregation, the total particle numbers in cell suspensions were counted in a Coulter counter (model Z1, Beckman Coulter) with 100-μm aperture. The initial particle number (N0), identical to the total number of the cells added to the medium, and the particle number at 90 min of incubation (N90) were measured with a Coulter counter and the extent of aggregation was presented by aggregation index, (N0−N90)/N0 [38].
Statistical analysis
The results are expressed as the means±S.D. and analysed by a one-way ANOVA by Fisher's t test when multiple comparisons against the control were required. P<0.01 was regarded as statistically significant. The data were analysed with SPSS II for Windows, release 11.0.1J.
RESULTS
Identification of ACAM as a new member of the Ig superfamily
cDNA containing the full-coding sequence of rat ACAM was obtained by RACE PCR (GenBank® accession number, AF302047). The rat ACAM cDNA included 3925 bp, and it contained a 1116-bp open reading frame flanked by 5′ and 3′ untranslated regions. The open reading frame encoded 372 amino acid residues with a predicted molecular mass of approx. 41 kDa. Then, we performed real-time PCR by using primers matched to EST (expressed sequence tag) clones of both mouse and human ACAM; 1322 bp of mouse ACAM (GenBank® accession number AY326421) and 2458 bp of human ACAM (AY326422) cDNAs were also isolated. The amino acid sequence in rat is highly homologous to mouse (94%) and human (87%) and Clustal W multiple alignments indicate that the primary structure of rat, mouse and human ACAM matches throughout the entire sequence. Kyte and Doolittle hydrophobocity/hydrophilicity plot analysis predicts that the protein has two hydrophobic domains (Figure 1A). The N-terminal hydrophobic domain (amino acids 1–17 in mouse and rat; 1–18 in human) is predicted to be a signal sequence, and the second hydrophobic domain is predicted to be a transmembrane domain (amino acids 228–250 in mouse and rat; 229–251 in human) by SOSUI program. Homology searching using BLASTP program through the NCBI database revealed human ACAM has 35% identity with CAR followed by BT-IgSF, ESAM, CTXL, MGC44287, GPA33, JAM-A, JAM-B and JAM-C (Figure 1B). Homology searches of the rat, mouse and human genome sequences reveals that the ACAM genes are localized to chromosome 8q22 in rat (NW_044096), chromosome 9 A5.1 in mouse (NW_000352) and chromosome 11q24.1 in human (NW_024769) (Figure 1C). The genomic organization is conserved in rat, mouse and human, and 7 exons have been found. The exon 7 encoding cytoplasmic domains and containing 3′-untranslated region varies in size, 2988 bp in rat, 809 bp in mouse and 1463 bp in human (Figure 1D). These molecules have recently been proposed to form a novel subfamily, i.e. CTX family, within the Ig superfamily, since they share common structural features: extracellular V-type and C2-type Ig domains, with a transmembrane segment and a cytoplasmic tail. ACAM is a new member of the CTX family, since it shares the following conserved structure (Figure 2). ACAM is characterized by an N-terminal V domain with the two cysteines (denoted C1–C2 in Figure 2) forming the intra-chain disulphide bond. The V domain is terminated by motif of the J segment, VLV, which is similar to T-cell receptor J segments and also highly conserved in CTX family. The most specific characteristic in the second C2-type domain is the presence of four cysteine residues that are predicted to form two disulphide bonds (C3–C6 and C4–C5 in Figure 2). The extracellular domain contains two N-glycosylation sites in rat, mouse and human orthologues (Figure 2). The cytoplasmic tails are most divergent in CTX family members. However, at the membrane proximal portion of the cytoplasmic domain, ACAM, CAR, BT-IgSF and CTXL share the motif D/EI/LREDXXXP. In ACAM, this motif is followed by a serine-rich segment, which is not found in other members of CTX family (Figure 2). In addition, homology search of intracellular domain through the NCBI database revealed no long-ordered homology with any other intracellular domains deposited in the database.
Figure 2. Amino acid sequences of rat, mouse, and human ACAM and sequence alignment with other CTX family members in human.
Multiple sequence alignment was performed by Clustal W, using amino acid sequences of rat ACAM (rACAM; GenBank® accession number, AF302047), mouse ACAM (mACAM; AY326421), human ACAM (hACAM; AY326422), CAR (Hs.473417), BT-IgSF (Hs.112873), ESAM (Hs.173840), CTXL (Hs.112377), MGC44287 (Hs.177164), GPA33 (Hs.437229), JAM-A (Hs.414880), JAM-B (Hs.436494) and JAM-C (Hs.419149). Identical and similar amino acids are boxed in black and grey respectively. Signal sequences are indicated by broken lines. Transmembrane segments are underlined. Rabbit polyclonal serum was raised against a synthetic peptide at the C-terminal end (bold and underlined). N-glycosylation sites are indicated by closed circles. ACAM, CAR, BT-IgSF and CTXL share the motif D/EI/LREDXXXP (indicated by asterisks). The six cysteine residues, C1 to C6, forming disulphide bonds in V- and C2-type Ig domains, are indicated. The J-segment motif (VLV) is indicated by J.
ACAM mRNA is predominantly expressed in WATs
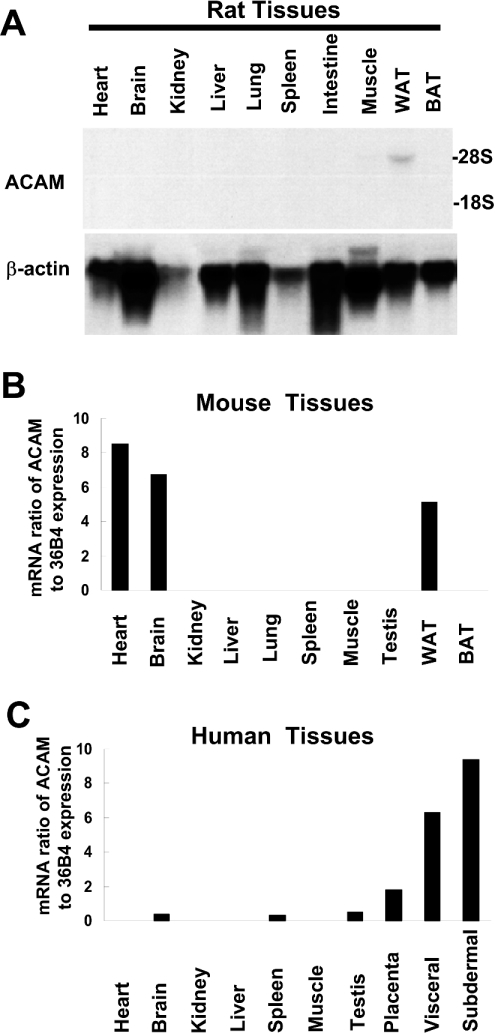
To survey gene expression of ACAM mRNA in adipose tissues and various non-adipose tissues, we performed Northern blot analyses and quantitative real-time PCR using various tissues from rat, mouse and human. In OLETF rats, a single transcript of approx. 5.0 kb was found exclusively in WATs and mRNA signals were not found in other tissues (Figure 3A). In mouse, the high expression level of ACAM mRNA was not restricted to WATs and was also detected in heart and brain (Figure 3B). In human, ACAM mRNA was predominantly expressed in WATs, including visceral and subdermal fat pads (Figure 3C).
Figure 3. Northern blot analyses and real-time PCR of ACAM.
(A) In OLETF rat, single transcripts, approx. 5.0 kb, are found in WATs and absent in other non-adipose tissues. BAT, brown adipose tissue. (B) In mouse, the high expression level of ACAM mRNA is not restricted to WATs and is also detected in heart and brain, as revealed by quantitative real-time PCR. (C) In human, ACAM mRNA is predominantly expressed in WATs, including visceral and subdermal fat pads. In (B) and (C), the relative abundance of mRNAs is standardized with 36B4 mRNA as the invariant control.
ACAM gene expression is up-regulated in mature adipocytes in OLETF rats
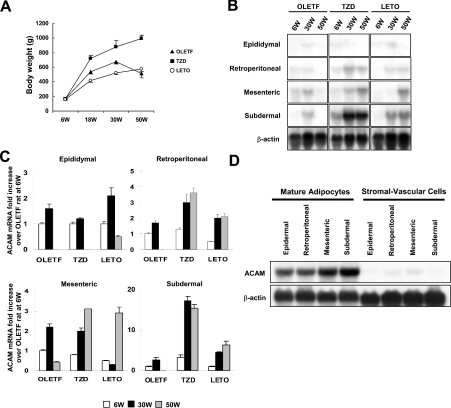
Since the fragment of rat ACAM cDNA was isolated from visceral adipose tissues of OLETF rats, an animal model of Type II diabetes characterized by abdominal obesity, hypertension and dyslipidaemia, Northern blot analyses of ACAM were performed using OLETF rats and their diabetes-resistant counterparts, LETO rats. The body mass of OLETF rats reached a peak at 30 weeks of age (674.8±32.3 g), and then their body mass was decreased to 513.0±55.9 g at 50 weeks of age, because of impaired secretion of insulin and hyperglycaemia. LETO rats did not develop diabetes and they gradually gained weight due to the aging process. TZD rats administered with pioglitazone showed sustained insulin secretion during the experimental period and continuously gained weight and reached 996.0±44.3 g at 50 weeks (Figure 4A). In OLETF rats at 6 weeks of age, ACAM mRNA was detected in mesenteric and retroperitoneal fat pads, and then mRNA expression prominently increased in mesenteric (2.2±0.16-fold) and subdermal (2.6±0.59-fold) adipose tissues at 30 weeks of age. At 50 weeks, OLETF rat body mass declined and ACAM mRNA signals were barely detectable in all WATs, and thus ACAM mRNA expression is closely linked to the status of obesity. The administration of pioglitazone resulted in continuous increase in body mass and, in parallel, ACAM mRNA expression increased in retroperitoneal (3.0±0.51-fold) and mesenteric (2.0±0.14-fold) adipose tissues compared with OLETF rats at 6 weeks of age. Interestingly, ACAM mRNA expression was remarkably accentuated by pioglitazone treatment in subdermal adipose tissues both at 30 (17.3±1.04-fold) and 50 (15.4±0.92-fold) weeks (Figures 4B and 4C). In LETO rats, ACAM mRNA expression also paralleled with body mass during the aging process and reached their peak at 50 weeks (Figures 4B and 4C).
Figure 4. Northern blot analyses of ACAM in WATs of OLETF rats.
(A) The body mass of OLETF rats, TZD rats [OLETF rats with free access to chow containing pioglitazone (approx. 1 mg/kg of body mass per day] and LETO rats are shown. (B) Total RNAs (20 μg) isolated from various WATs of 6-week (6W), 30-week (30W) and 50-week (50W)-old rats were subjected to Northern blot analyses. (C) For each adipose tissue, the fold increase in the ACAM mRNA/β-actin ratio compared with OLETF rats at 6 weeks of age is indicated. Each value is expressed as the mean±S.D. (D) Mature adipocytes and stromal vascular cells were isolated by collagenase digestion and ACAM mRNA expression was analysed with Northern blot analysis. ACAM mRNA is predominantly expressed in mature adipocyte fractions and barely detected in stromal vascular cells.
To confirm ACAM expression in mature adipocytes, we separated mature adipocytes and stromal vascular cells by collagenase digestion and analysed ACAM mRNA expression in both fractions by using Northern blotting. The expression of ACAM mRNA in mature adipocytes was predominantly expressed in mature adipocyte fraction and barely detected in stromal vascular cells (Figure 4D).
ACAM is a plasma membrane bound protein of adipocytes
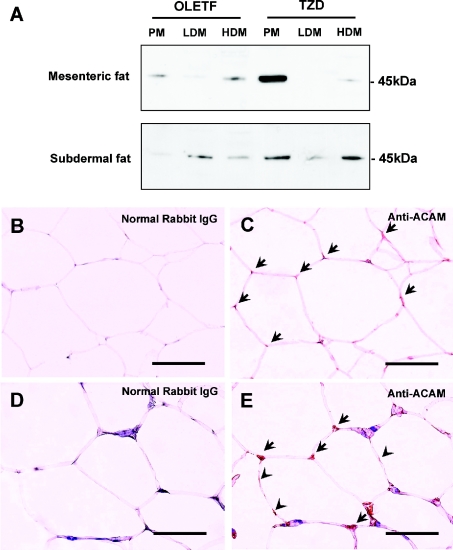
To examine the subcellular localization of ACAM protein in mature adipocytes, mature adipocytes were isolated by collagenase digestion and subjected to subcellular fractionation. Since mRNA expression was prominent in mesenteric and subdermal fats, Western blot analyses were performed using these WATs. Anti-ACAM antibody revealed major bands at approx. 45 kDa in the plasma membrane fraction of mesenteric and subdermal adipose tissues of OLETF rats. Pioglitazone up-regulated the ACAM protein expression in mesenteric and subdermal adipose tissues of OLETF rats (Figure 5A). In the HDM and LDM fractions, faint bands at approx. 45 kDa were detected (Figure 5A). Immunohistochemistry of subdermal adipose tissues in TZD rats revealed faint immunoreactivity of ACAM on the surface of adipocytes (arrowheads in Figure 5E), and somehow more intense staining was obtained at corners of polygonal-shaped adipocytes (arrows in Figure 5E). In OLETF rats, immunoreactivity on the cell surface was weak and almost indistinguishable from the background staining (Figures 5B and 5C), and ACAM protein bands were also weak in plasma membrane fractions (Figure 5A). No immunoreactivity was found on endothelial cells in both OLETF and TZD rats (results not shown).
Figure 5. Western blot and immunohistochemistry of ACAM in WATs of OLETF rats.
(A) Mesenteric and subdermal WATs derived from OLETF and TZD rats at 30 weeks of age were subjected to subcellular fractionation of adipocytes: plasma membrane (PM), HDM and LDM fractions. Anti-ACAM antibody reveals approx. 45 kDa major bands in plasma membrane fraction of mesenteric and subdermal adipose tissues of OLETF rats. Pioglitazone up-regulates the ACAM protein expression in mesenteric and subdermal adipose tissues of OLETF rats. In HDM and LDM fractions, faint bands at approx. 45 kDa were detected. (B–E) Immunostaining of ACAM in subdermal adipose tissues of 30-week-old OLETF rats (C) and TZD rats (E). Normal rabbit IgG was incubated with subdermal adipose tissues of OLETF rats (B) and TZD rats (D) as a negative control. In subdermal adipose tissues in TZD rats, faint immunoreactivity of ACAM is detected on the surface of adipocytes [arrowheads in (E)] and somehow more intense staining is observed at corners of polygonal-shaped adipocytes [arrows in (E)]. Scale bars, 100 μm.
Expression level of ACAM mRNA is up-regulated in obesity model mice and human obesity state
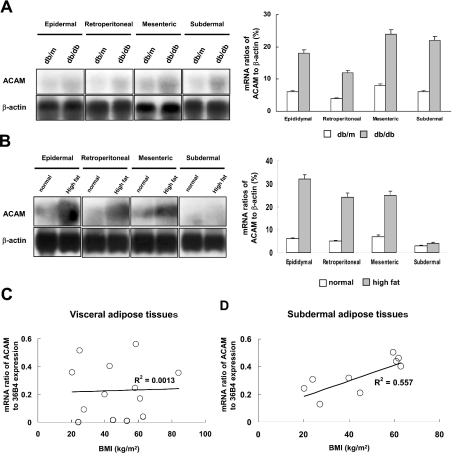
The db/db mouse, lacking hypothalamic leptin receptors, is a model of Type II diabetes and obesity, and db/m mouse is a nondiabetic control animal. To confirm whether ACAM mRNA expression regulation in adipose tissues is similar in other species, Northern blot analyses were performed by using RNAs extracted form db/db and db/m mice at 12 weeks of age, where obesity was prominent in db/db mice. ACAM mRNA expression increased, particularly in mesenteric (24±1.4%, mRNA densitometric ratio to β-actin) and subdermal fat pads (22±1.3%, mRNA densitometric ratio to β-actin) of db/db mice, and thus similar regulation of ACAM expression found in OLETF rats was also observed in db/db mice (Figure 6A). We next investigated changes of ACAM mRNA expression in DIO mice. Total RNAs were extracted from WATs of DIO mice and control lean mice at 16 weeks of age and Northern blot analyses were performed. ACAM mRNA expression increased in visceral adipose tissues of DIO mice compared with lean control mice (Figure 6B). Finally, we examined the relationship between ACAM mRNA expression in human WATs and BMI. A significant positive correlation was observed between BMI and ACAM mRNA expression level in human subdermal adipose tissues (R2=0.557, P<0.01, n=9). There was no significant correlation between BMI and ACAM mRNA expression level in human visceral adipose tissues (Figure 6C). These results indicated that ACAM mRNA expression is strongly correlated with accumulation of WATs in human and animal obesity state.
Figure 6. ACAM mRNA expression in obese animal models and human subjects.
(A) Northern blot analyses were performed by using RNA extracted from db/db and db/m mice at 12 weeks of age, and mRNA ratios of ACAM/β-actin (%), measured with densitometer, are indicated. (B) Total RNAs were extracted from WATs of DIO ICR mice and control lean mice at 16 weeks of age, and Northern blot analyses show the ACAM mRNA expression increases in visceral adipose tissues of DIO mice compared with lean control mice. (C) There is no significant correlation between BMI and ACAM mRNA expression level in human visceral adipose tissues. (D) A significant positive correlation is observed between BMI and ACAM mRNA expression level in human subdermal adipose tissues (R2=0.557, P<0.01, n=9). In (C) and (D), the relative abundance of the mRNAs is standardized with 36B4 mRNA as the invariant control.
ACAM mRNA expression is regulated during the differentiation of adipocytes
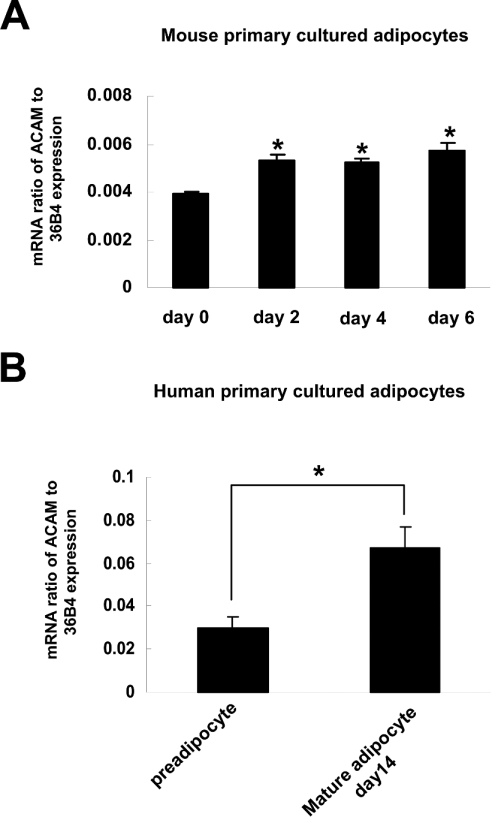
To investigate the regulation of expression of ACAM during adipocyte differentiation of mice and human primary cultured adipocytes, we analysed the time course of ACAM mRNA expression by quantitative real-time PCR. In mice primary cultured adipocytes, ACAM mRNA expression was rather low prior to hormonal induction, and it progressively increased during the adipocyte differentiation. In human primary cultured adipocytes, ACAM mRNA expression in day 14 mature adipocytes was higher than that in day 0 pre-adipocytes (Figures 7A and 7B). These results indicated that up-regulation of ACAM mRNA expression might be associated with adipocyte maturation.
Figure 7. ACAM mRNA expression in mice and human primary cultured adipocytes.
(A) In mice primary cultured adipocytes, ACAM mRNA expression is rather low prior to hormonal induction and it progressively increases during adipocyte differentiation. (B) In human primary cultured adipocytes, ACAM mRNA expression in day 14 mature adipocytes is higher than that in day 0 pre-adipocytes. The relative abundance of mRNAs is standardized with 36B4 mRNA as the invariant control.
ACAM mediates cell–cell adhesion
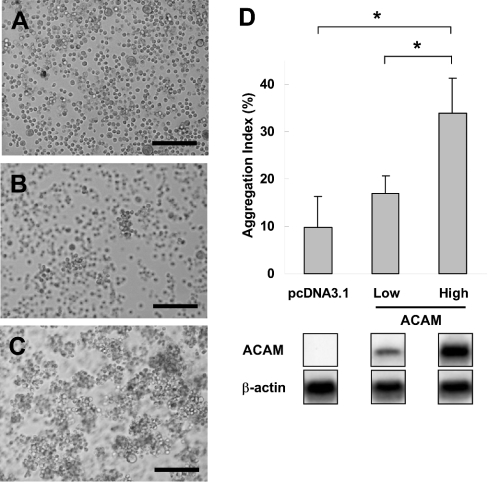
Since CTX family members mediate the cell–cell adhesion in epithelial cells or endothelial cells, we attempt to verify whether ACAM also mediates cell–cell adhesion. We transfected pcDNA3.1/ACAM into CHO-K1 cells and we established 30 stable cell lines after G418 selection. We further performed Northern blot analyses of ACAM mRNA expression and selected low expression (CHO-L2) and high expression (CHO-H11) cell lines. pcDNA3.1-transfected CHO-K1 cells showed scattered single cell suspension (Figure 8A). In the low expression cell line, small cell aggregates comprising 10–15 cells were observed, whereas larger aggregates were found in the high expression cell line after 90 min of incubation (Figures 8B and 8C). The aggregation index was significantly higher in the high expression cell line (33.8±7.4%) than in the pcDNA3.1-transfected CHO-K1 cells (9.8±6.4%) (Figure 8D).
Figure 8. Aggregation assay using ACAM stably transfected CHO-K1 cells.
(A) pcDNA3.1 transfected CHO-K1 cells, as control, show scattered single cell suspension. (B) In low expression cell line, small cell aggregates consisting of 10-15 cells are observed. (C) Larger aggregates are found in high expression cell line after 90 min of incubation. (D) The initial particle number (N0), identical to the total number of the cells added to the medium, and the particle number at 90 min of incubation (N90) was measured with Coulter counter. The aggregation index, (N0−N90)/N0, is significantly higher in the high expression cell line (33.8±7.4%) than pcDNA3.1-transfected CHO-K1 cells (9.8±6.4%).The results are expressed as the means±S.D; *P<0.001. Scale bars, 200 μm.
DISCUSSION
We have identified a type 1 transmembrane glycoprotein with V-type and C2-type Ig domains, which is a novel member of the CTX gene family. We have designated this new gene as ACAM, because this gene has several original features, including: (i) ACAM is a novel member of CTX gene family, (ii) ACAM is predominantly expressed in WATs, (iii) ACAM expression is strongly correlated with WAT mass of human and rodents with obesity, (iv) ACAM expression is differentially regulated during maturation of human and mouse primary cultured adipocytes, and (v) ACAM is a plasma membrane-bound protein found on mature adipocytes and mediates homophilic cell aggregation.
The orthologous genes in rat, mouse and human revealed striking homology, and the genomic organization in seven exons is also well conserved. X-ray structural analysis of JAMs in the CTX family shows that V-type and C2-type Ig-like domains in the extracellular domain are separated by a J segment or short linker, VLV, which connects N- and C-terminal Ig domains in a rather extended conformation, leading to an estimated elbow angle of approx. 125° [39]. The VLV motif is well conserved in the CTX family and is also found in ACAM. It was also demonstrated that an R(V/I/L)EW motif is required in homophilic dimerization of N-terminal V-like domains in JAMs by mutagenesis analysis [39]. In ACAM, CAR and BT-IgSF, the DIEW motif located in the N-terminal domain suggests that the dimerization of N-terminal domains exerts homophilic adhesive activities in these molecules. Unlike structural similarities in extracellular domains of the CTX family, the cytoplasmic tail revealed divergent amino acid sequences, and ACAM was characterized by a serine-rich segment, SSSSSGSRSSRS, suggesting possible interaction with adaptor molecules involved in signal transduction or cytoskeletal proteins.
We originally isolated rat ACAM cDNA (GenBank® accession number AF302047) from visceral adipose tissues of OLETF rats by using a PCR-based subtraction method as reported previously [7]. Mouse ACAM was independently isolated from 3T3-L1 cells, as adipocyte-specific protein 5 (AK033766), by using signal sequence trap by a retrovirus-mediated expression screening method [40]. Raschperger et al. [41] have found human and mouse orthologues of ACAM by bioinformatics approaches, and they named it CLMP (CAR-like membrane protein), and also detected CLMP mRNA expression in various non-adipose tissues, such as small intestine, placenta, heart and brain. In addition, CLMP was a component of the tight junction of epithelial cells, co-localized with ZO-1 (zonula occludens-1) and occludin. However, we found that the ACAM expression in WATs of obese animals was prominent compared with other tissues, at least in human and rats.
The expression of ACAM in WATs of OLETF rats increased during the development of obesity at 30 weeks of age, it decreased along with the reduction of body mass due to hyperglycaemia and impaired secretion of insulin, and finally mRNA expression was barely detectable at 50 weeks. mRNA expression of ACAM was not related to diabetes, since non-diabetic LETO rats also revealed continuous increases in body mass, as well as ACAM mRNA expression, in WATs up to 50 weeks of age. Further evidence for a correlation between ACAM and development of obesity is provided by experiments with genetically obese db/db mice, DIO mice and obese human subjects. ACAM mRNA expression level is high in WATs of human and rodents with obesity. Furthermore, ACAM mRNA level was markedly up-regulated during the course of the differentiation of human and mouse primary cultured adipocytes. This evidence strongly supports the suggestion that ACAM expression may be associated with obesity and adipocyte maturation. The PPARγ (peroxisome-proliferator-activated receptor-γ) agonist pioglitazone prominently increased the expression of ACAM in OLETF rats. Interestingly, the regulation by the PPARγ agonist differed in various WATs, and it induced prominent mRNA expression in subdermal adipose tissues of OLETF rats. Since we did not find PPARγ-responsive elements in the region of −500 to 100 bp around the transcription start sites in rat, mouse and human, ACAM mRNA may be indirectly induced via up-regulation of other transcriptional factors.
The functional role of ACAM in mature adipocytes remains under speculation at this point. Immunohistochemical and Western blot analyses indicated that ACAM is mainly associated with plasma membranes of mature adipocytes. The extracellular domain of ACAM is characterized by the presence of V-type and C2-type Ig-like domains, and its structure suggests that it mediates homotypic or heterotypic interactions with other Ig-like adhesion molecules. Until now we have not found any reports of adhesion molecules and the CTX gene family expressed in mature adipocytes. Cadherins are integral membrane proteins with a large extracellular domain that mediates homophilic cell–cell adhesion, a single transmembrane domain and short intracellular tail, and are a key molecule involved in embryogenesis and tissue differentiation. Although N-cadherin and cadherin-11 are expressed in immature adipocytes, after hormonal induction they are rapidly down-regulated during adipocyte differentiation and cannot be detected in mature adipocytes [42]. During the growth phase, pre-adipocytes are morphologically similar to fibroblasts, and at confluence the induction of differentiation by appropriate treatment leads to drastic cell shape changes, i.e. conversion to a spherical shape and accumulation of lipid droplets [43]. In such conversion, cell adhesion molecules and extracellular matrix components modulate the interaction of cells with their environment in a manner that influences cell differentiation [44]. Similarly, it can be speculated that a physical connection or cell–cell contact via cell adhesion molecules influences the morphology and differentiation of the cells via alterations in signal transduction or cytoskeletal organization. The up-regulation of ACAM mRNA in adipocytes after hormonal induction suggests that ACAM is one of the critical molecules, which initiates the conversion of pre-adipocytes to mature adipocytes.
In conclusion, the possible role of ACAM in adipocyte maturation and development of obesity is suggested by its specific up-regulated expression in mature adipocytes and adipose tissues of obese rodents and human. The structural analysis and aggregation assay revealed that the biological action of ACAM seems to be mediated by homophilic adhesive activity of the extracellular domain and intracellular signalling events yet uncharacterized. It is possible that the intervening ACAM expression or activities may alter the adipocyte differentiation status and contribute to therapy of obesity and metabolic syndrome.
Acknowledgments
We thank Dr Yasushi Matsuki and Dr Eijiro Watanabe for their valuable suggestions and discussion. We also thank Dr Junichiro Futami (Department of Bioscience and Biotechnology, Faculty of Engineering, Okayama University) for helpful advice regarding the Western blot analysis. This work was supported by Uehara Memorial Foundation, The Naito Foundation, ONO Medical Foundation, Japan Heart Foundation/Pfizer Grant for Research on Hypertension, Hyperlipidemia and Vascular Metabolism, Grant-in-Aid for Scientific Research (C), Ministry of Education, Science and Culture, Japan (14571025) to J.W., and Grant-in-Aid for Scientific Research (B), Ministry of Education, Science and Culture, Japan (14370319) to H.M. H.Z. was supported by International Society of Nephrology/Kirin Fellowship Award.
References
- 1.Lyon C. J., Law R. E., Hsueh W. A. Minireview: adiposity, inflammation, and atherogenesis. Endocrinology. 2003;144:2195–2200. doi: 10.1210/en.2003-0285. [DOI] [PubMed] [Google Scholar]
- 2.Matsuzawa Y., Funahashi T., Nakamura T. Molecular mechanism of metabolic syndrome X: contribution of adipocytokines adipocyte-derived bioactive substances. Ann. N.Y. Acad. Sci. 1999;892:146–154. doi: 10.1111/j.1749-6632.1999.tb07793.x. [DOI] [PubMed] [Google Scholar]
- 3.Stevens J., Cai J., Pamuk E. R., Williamson D. F., Thun M. J., Wood J. L. The effect of age on the association between body-mass index and mortality. N. Engl. J. Med. 1998;338:1–7. doi: 10.1056/NEJM199801013380101. [DOI] [PubMed] [Google Scholar]
- 4.Rexrode K. M., Carey V. J., Hennekens C. H., Walters E. E., Colditz G. A., Stampfer M. J., Willett W. C., Manson J. E. Abdominal adiposity and coronary heart disease in women. JAMA. 1998;280:1843–1848. doi: 10.1001/jama.280.21.1843. [DOI] [PubMed] [Google Scholar]
- 5.Kannel W. B., Cupples L. A., Ramaswami R., Stokes J., 3rd, Kreger B. E., Higgins M. Regional obesity and risk of cardiovascular disease: the Framingham Study. J. Clin. Epidemiol. 1991;44:183–190. doi: 10.1016/0895-4356(91)90265-b. [DOI] [PubMed] [Google Scholar]
- 6.Kawano K., Hirashima T., Mori S., Saitoh Y., Kurosumi M., Natori T. Spontaneous long-term hyperglycemic rat with diabetic complications: Otsuka Long–Evans Tokushima Fatty (OLETF) strain. Diabetes. 1992;41:1422–1428. doi: 10.2337/diab.41.11.1422. [DOI] [PubMed] [Google Scholar]
- 7.Hida K., Wada J., Zhang H., Hiragushi K., Tsuchiyama Y., Shikata K., Makino H. Identification of genes specifically expressed in the accumulated visceral adipose tissue of OLETF rats. J. Lipid Res. 2000;41:1615–1622. [PubMed] [Google Scholar]
- 8.Chretien I., Robert J., Marcuz A., Garcia-Sanz J. A., Courtet M., Du Pasquier L. CTX, a novel molecule specifically expressed on the surface of cortical thymocytes in Xenopus. Eur. J. Immunol. 1996;26:780–791. doi: 10.1002/eji.1830260409. [DOI] [PubMed] [Google Scholar]
- 9.Chretien I., Marcuz A., Courtet M., Katevuo K., Vainio O., Heath J. K., White S. J., Du Pasquier L. CTX, a Xenopus thymocyte receptor, defines a molecular family conserved throughout vertebrates. Eur. J. Immunol. 1998;28:4094–4104. doi: 10.1002/(SICI)1521-4141(199812)28:12<4094::AID-IMMU4094>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Tomko R. P., Xu R., Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freimuth P., Springer K., Berard C., Hainfeld J., Bewley M., Flanagan J. Coxsackievirus and adenovirus receptor amino-terminal immunoglobulin V-related domain binds adenovirus type 2 and fiber knob from adenovirus type 12. J. Virol. 1999;73:1392–1398. doi: 10.1128/jvi.73.2.1392-1398.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergelson J. M., Cunningham J. A., Droguett G., Kurt-Jones E. A., Krithivas A., Hong J. S., Horwitz M. S., Crowell R. L., Finberg R. W. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 13.Suzu S., Hayashi Y., Harumi T., Nomaguchi K., Yamada M., Hayasawa H., Motoyoshi K. Molecular cloning of a novel immunoglobulin superfamily gene preferentially expressed by brain and testis. Biochem. Biophys. Res. Commun. 2002;296:1215–1221. doi: 10.1016/s0006-291x(02)02025-9. [DOI] [PubMed] [Google Scholar]
- 14.Hirata K., Ishida T., Penta K., Rezaee M., Yang E., Wohlgemuth J., Quertermous T. Cloning of an immunoglobulin family adhesion molecule selectively expressed by endothelial cells. J. Biol. Chem. 2001;276:16223–16231. doi: 10.1074/jbc.M100630200. [DOI] [PubMed] [Google Scholar]
- 15.Heath J. K., White S. J., Johnstone C. N., Catimel B., Simpson R. J., Moritz R. L., Tu G. F., Ji H., Whitehead R. H., Groenen L. C., et al. The human A33 antigen is a transmembrane glycoprotein and a novel member of the immunoglobulin superfamily. Proc. Natl. Acad. Sci U.S.A. 1997;94:469–474. doi: 10.1073/pnas.94.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin-Padura I., Lostaglio S., Schneemann M., Williams L., Romano M., Fruscella P., Panzeri C., Stoppacciaro A., Ruco L., Villa A., et al. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 1998;142:117–127. doi: 10.1083/jcb.142.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmeri D., van Zante A., Huang C. C., Hemmerich S., Rosen S. D. Vascular endothelial junction-associated molecule, a novel member of the immunoglobulin superfamily, is localized to intercellular boundaries of endothelial cells. J. Biol. Chem. 2000;275:19139–19145. doi: 10.1074/jbc.M003189200. [DOI] [PubMed] [Google Scholar]
- 18.Santoso S., Sachs U. J., Kroll H., Linder M., Ruf A., Preissner K. T., Chavakis T. The junctional adhesion molecule 3 (JAM-3) on human platelets is a counterreceptor for the leukocyte integrin Mac-1. J. Exp. Med. 2002;196:679–691. doi: 10.1084/jem.20020267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cunningham S. A., Rodriguez J. M., Arrate M. P., Tran T. M., Brock T. A. JAM2 interacts with alpha4beta1. Facilitation by JAM3. J. Biol. Chem. 2002;277:27589–27592. doi: 10.1074/jbc.C200331200. [DOI] [PubMed] [Google Scholar]
- 20.Phillips H. M., Renforth G. L., Spalluto C., Hearn T., Curtis A. R., Craven L., Havarani B., Clement-Jones M., English C., Stumper O., et al. Narrowing the critical region within 11q24-qter for hypoplastic left heart and identification of a candidate gene, JAM3, expressed during cardiogenesis. Genomics. 2002;79:475–478. doi: 10.1006/geno.2002.6742. [DOI] [PubMed] [Google Scholar]
- 21.Liang T. W., Chiu H. H., Gurney A., Sidle A., Tumas D. B., Schow P., Foster J., Klassen T., Dennis K., DeMarco R. A., et al. Vascular endothelial-junctional adhesion molecule (VE-JAM)/JAM 2 interacts with T, NK, and dendritic cells through JAM 3. J. Immunol. 2002;168:1618–1626. doi: 10.4049/jimmunol.168.4.1618. [DOI] [PubMed] [Google Scholar]
- 22.Arrate M. P., Rodriguez J. M., Tran T. M., Brock T. A., Cunningham S. A. Cloning of human junctional adhesion molecule 3 (JAM3) and its identification as the JAM2 counter-receptor. J. Biol. Chem. 2001;276:45826–45832. doi: 10.1074/jbc.M105972200. [DOI] [PubMed] [Google Scholar]
- 23.Strausberg R. L., Feingold E. A., Grouse L. H., Derge J. G., Klausner R. D., Collins F. S., Wagner L., Shenmen C. M., Schuler G. D., Altschul S. F., et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnstone C. N., Tebbutt N. C., Abud H. E., White S. J., Stenvers K. L., Hall N. E., Cody S. H., Whitehead R. H., Catimel B., Nice E. C., et al. Characterization of mouse A33 antigen, a definitive marker for basolateral surfaces of intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2000;279:G500–G510. doi: 10.1152/ajpgi.2000.279.3.G500. [DOI] [PubMed] [Google Scholar]
- 25.Cohen C. J., Shieh J. T., Pickles R. J., Okegawa T., Hsieh J. T., Bergelson J. M. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc. Natl. Acad. Sci. U.S.A. 2001;98:15191–15196. doi: 10.1073/pnas.261452898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagai M., Yaoita E., Yoshida Y., Kuwano R., Nameta M., Ohshiro K., Isome M., Fujinaka H., Suzuki S., Suzuki J., et al. Coxsackievirus and adenovirus receptor, a tight junction membrane protein, is expressed in glomerular podocytes in the kidney. Lab. Invest. 2003;83:901–911. doi: 10.1097/01.lab.0000073307.82991.cc. [DOI] [PubMed] [Google Scholar]
- 27.Ebnet K., Aurrand-Lions M., Kuhn A., Kiefer F., Butz S., Zander K., Meyer zu Brickwedde M. K., Suzuki A., Imhof B. A., Vestweber D. The junctional adhesion molecule (JAM) family members JAM-2 and JAM-3 associate with the cell polarity protein PAR-3: a possible role for JAMs in endothelial cell polarity. J. Cell Sci. 2003;116:3879–3891. doi: 10.1242/jcs.00704. [DOI] [PubMed] [Google Scholar]
- 28.Ostermann G., Weber K. S., Zernecke A., Schroder A., Weber C. JAM-1 is a ligand of the β(2) integrin LFA-1 involved in transendothelial migration of leukocytes. Nat. Immunol. 2002;3:151–158. doi: 10.1038/ni755. [DOI] [PubMed] [Google Scholar]
- 29.Wada J., Kanwar Y. S. Characterization of mammalian translocase of inner mitochondrial membrane (Tim44) isolated from diabetic newborn mouse kidney. Proc. Natl. Acad. Sci. U.S.A. 1998;95:144–149. doi: 10.1073/pnas.95.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H., Wada J., Hida K., Tsuchiyama Y., Hiragushi K., Shikata K., Wang H., Lin S., Kanwar Y. S., Makino H. Collectrin, a collecting duct-specific transmembrane glycoprotein, is a novel homolog of ACE2 and is developmentally regulated in embryonic kidneys. J. Biol. Chem. 2001;276:17132–17139. doi: 10.1074/jbc.M006723200. [DOI] [PubMed] [Google Scholar]
- 31.Yang Q., Dixit B., Wada J., Tian Y., Wallner E. I., Srivastva S. K., Kanwar Y. S. Identification of a renal-specific oxido-reductase in newborn diabetic mice. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9896–9901. doi: 10.1073/pnas.160266197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 33.Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitaku S., Hirokawa T., Tsuji T. Amphiphilicity index of polar amino acids as an aid in the characterization of amino acid preference at membrane-water interfaces. Bioinformatics. 2002;18:608–616. doi: 10.1093/bioinformatics/18.4.608. [DOI] [PubMed] [Google Scholar]
- 35.Joost H. G., Schurmann A. Subcellular fractionation of adipocytes and 3T3-L1 cells. Methods Mol. Biol. 2001;155:77–82. doi: 10.1385/1-59259-231-7:077. [DOI] [PubMed] [Google Scholar]
- 36.Wada J., Kanwar Y. S. Identification and characterization of galectin-9, a novel β-galactoside-binding mammalian lectin. J. Biol. Chem. 1997;272:6078–6086. doi: 10.1074/jbc.272.9.6078. [DOI] [PubMed] [Google Scholar]
- 37.Tsuchiyama Y., Wada J., Zhang H., Morita Y., Hiragushi K., Hida K., Shikata K., Yamamura M., Kanwar Y. S., Makino H. Efficacy of galectins in the amelioration of nephrotoxic serum nephritis in Wistar Kyoto rats. Kidney Int. 2000;58:1941–1952. doi: 10.1111/j.1523-1755.2000.00366.x. [DOI] [PubMed] [Google Scholar]
- 38.Takeichi M. Functional correlation between cell adhesive properties and some cell surface proteins. J. Cell Biol. 1977;75:464–474. doi: 10.1083/jcb.75.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kostrewa D., Brockhaus M., D'Arcy A., Dale G. E., Nelboeck P., Schmid G., Mueller F., Bazzoni G., Dejana E., Bartfai T., et al. X-ray structure of junctional adhesion molecule: structural basis for homophilic adhesion via a novel dimerization motif. EMBO J. 2001;20:4391–4398. doi: 10.1093/emboj/20.16.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsuruga H., Kumagai H., Kojima T., Kitamura T. Identification of novel membrane and secreted proteins upregulated during adipocyte differentiation. Biochem. Biophys. Res. Commun. 2000;272:293–297. doi: 10.1006/bbrc.2000.2759. [DOI] [PubMed] [Google Scholar]
- 41.Raschperger E., Engstrom U., Pettersson R. F., Fuxe J. CLMP, a novel member of the CTX family and a new component of epithelial tight junctions. J. Biol. Chem. 2003;279:796–804. doi: 10.1074/jbc.M308249200. [DOI] [PubMed] [Google Scholar]
- 42.Shin C. S., Lecanda F., Sheikh S., Weitzmann L., Cheng S. L., Civitelli R. Relative abundance of different cadherins defines differentiation of mesenchymal precursors into osteogenic, myogenic, or adipogenic pathways. J. Cell. Biochem. 2000;78:566–577. [PubMed] [Google Scholar]
- 43.Gregoire F. M., Smas C. M., Sul H. S. Understanding adipocyte differentiation. Physiol. Rev. 1998;78:783–809. doi: 10.1152/physrev.1998.78.3.783. [DOI] [PubMed] [Google Scholar]
- 44.Jones P. L., Schmidhauser C., Bissell M. J. Regulation of gene expression and cell function by extracellular matrix. Crit. Rev. Eukaryot. Gene Expr. 1993;3:137–154. [PubMed] [Google Scholar]