Abstract
TraA is the DNA relaxase encoded by the broad-host-range Grampositive plasmid pIP501. It is the second relaxase to be characterized from plasmids originating from Gram-positive organisms. Full-length TraA (654 amino acids) and the N-terminal domain (246 amino acids), termed TraAN246, were expressed as 6×His-tagged fusions and purified. Small-angle X-ray scattering and chemical cross-linking proved that TraAN246 and TraA form dimers in solution. Both proteins revealed oriTpIP501 (origin of transfer of pIP501) cleavage activity on supercoiled plasmid DNA in vitro. oriT binding was demonstrated by electrophoretic mobility shift assays. Radiolabelled oligonucleotides covering different parts of oriTpIP501 were subjected to binding with TraA and TraAN246. The KD of the protein–DNA complex encompassing the inverted repeat, the nick site and an additional 7 bases was found to be 55 nM for TraA and 26 nM for TraAN246. The unfolding of both protein constructs was monitored by measuring the change in the CD signal at 220 nm upon temperature change. The unfolding transition of both proteins occurred at approx. 42 °C. CD spectra measured at 20 °C showed 30% α-helix and 13% β-sheet for TraA, and 27% α-helix and 18% β-sheet content for the truncated protein. Upon DNA binding, an enhanced secondary structure content and increased thermal stability were observed for the TraAN246 protein, suggesting an induced-fit mechanism for the formation of the specific relaxase–oriT complex.
Keywords: aggregation state, circular dichroism, DNA–protein interaction, plasmid pIP501, relaxase, small-angle X-ray scattering
Abbreviations: EMSA, electrophoretic mobility shift assay; GST, glutathione S-transferase; oriT, origin of transfer; PDDF, pair distance distribution function; SAXS, small-angle X-ray scattering; TM, transition temperature; TraAN246, protein comprising the N-terminal 246 amino acids of TraA
INTRODUCTION
Bacterial conjugation is a highly specific process whereby DNA is transferred from donor to recipient bacteria by two specialized protein complexes, namely the relaxosome and the mating-pair formation complex, which are connected via interaction with a socalled coupling protein. The relaxosome has been defined as a (multi)protein–DNA complex that is generated at the plasmid origin of transfer, oriT. The key enzyme of the relaxosome is a conjugative DNA relaxase responsible for binding and cleaving single-stranded plasmid DNA in a sequence-specific manner prior to transfer [1–3]. For Gram-positive plasmid transfer systems, little information is available on the mechanisms involved in the DNA–protein translocation process through the cell envelopes of interacting donor and recipient cells. Recently, significant similarities to type IV secretion system components involved in conjugative transfer systems in Gram-negative bacteria and protein toxin secretion systems of several Gram-negative pathogens were detected in the transfer regions of plasmids pIP501, pRE25, pSK41, pGO1 and pMRC01 originating from Gram-positive bacteria [4]. The putative type IV proteins found on the basis of amino acid identity/similarity could fulfil crucial roles in the plasmid transfer process, such as ‘coupling protein’ (VirD4 homologue), translocation energy supplier (VirB4 homologue) and lytic transglycosylase that locally opens the peptidoglycan (VirB1 homologue) (reviewed in [4]). Only two DNA relaxases originating from Gram-positive mobile elements have been purified and characterized in some detail: (i) the mobilization protein MobM, encoded by the mobilizable rolling-circle-type replicating plasmid pMV158 [5,6], and (ii) the IncQ-type relaxase TraA, encoded by pIP501. pIP501 has the broadest host range for a plasmid from Gram-positive bacteria [7]. TraA has been purified as GST (glutathione S-transferase) fusion protein and shown to cleave supercoiled plasmid DNA containing oriTpIP501. Relaxase activity is strictly dependent on the presence of Mg2+ or Mn2+ ions and is highest at temperatures between 42 and 45 °C. Transesterase activity of the TraA relaxase is located in the N-terminal region, which was demonstrated by purification of the N-terminal portion of TraA comprising the first 293 amino acids as a GST fusion protein. The truncated protein also cleaved a supercoiled pIP501 derivative, although less efficiently. Approx. 25% conversion from FI to FII forms, compared with a maximum of 55% for the full-length protein, was observed [8].
In the present study we purified the TraA relaxase (654 amino acids of the original TraA sequence) and its active N-terminal domain, TraAN246 (i.e. comprising the N-terminal 246 amino acids) as 6×His tag fusion proteins. Both proteins were shown to form dimers in solution. Far-UV CD was used to determine the secondary structure content of the pure TraA and TraAN246 proteins. By means of heat-induced unfolding, TM (the transition temperature) values for both proteins were determined to be 42 °C. EMSAs (electrophoretic mobility shift assays) proved specific binding of both TraA proteins to an imperfect inverted repeat structure located 8 bases downstream from the oriT nick site.
EXPERIMENTAL
traA cloning
The traA fragment of plasmid pIP501 (GenBank® accession no. L39769) was amplified by PCR using GenTherm™ DNA polymerase (Rapidozym, Berlin, Germany) and specific primers 5′-GAGGTGATAGGATCCGCAATCTT-3′ (forward; nucleotide positions 1382–1404) and 5′-TTTCATTTTAGGTACCTCTTGTTTTT-3′ (reverse; nucleotide positions 3383–3408) (MWG-Biotech AG, Ebersberg, Germany). The PCR product was cut with restriction enzymes BamHI and KpnI. Expression vector pQE30 (Qiagen, Hilden, Germany), which is designed to express proteins with an N-terminal 6×His tag, was also cut with BamHI and KpnI. The ligation mixture was incubated at 4 °C overnight with T4 DNA ligase (Roche Diagnostics, Mannheim, Germany) and used for transformation of Escherichia coli XL1Blue (Stratagene, Amsterdam, The Netherlands) cells. The sequence of both strands of the recombinant plasmid pQE30-traA was verified using a BigDye™ Terminators Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, U.S.A.) and an automated sequencer (ABI Prism 310; Perkin-Elmer, Wellesley, MA, U.S.A.).
traAN246 cloning
A 780 bp traAN246 fragment encoding the first 246 amino acids of TraA was also PCR-amplified. The forward primer used for cloning purposes was the same as for the traA amplification. The sequence of the reverse primer was 5′-CTCGGTACCCTAAATTTTTTCTTTTTAACTCG-3′ (nucleotide positions 2131–2162; GenBank® accession no. L39769) (MWG-Biotech AG). The PCR product was cloned into the unique BamHI and HindIII sites of pQE30. E. coli XL1Blue cells were transformed with the recombinant vector, and a positive clone, pQE30-traAN246, was sequenced in both senses of the DNA with the BigDye™ Terminators Cycle Sequencing Kit.
Expression of TraA and TraAN246
E. coli XL1 Blue (pQE30-traA) and E. coli XL1 Blue (pQE30-traAN246) were inoculated into LB (Luria–Bertani) broth containing 100 mg/l ampicillin and 40 mg/l tetracycline and incubated with shaking overnight at 37 °C. The overnight cultures were diluted 100-fold with LB broth containing the respective antibiotics, and the cultures were shaken at 37 °C until a D600 of 0.5–0.6 was reached. Transcription of the 6×His-tag fusion proteins from the T5 promoter was induced by addition of isopropyl β-D-thiogalactopyranoside (Sigma, Taufkirchen, Germany) to a final concentration of 1 mM. Incubation continued for 4 h. The cells were collected by centrifugation at 6000 g at 4 °C for 15 min.
Purification of TraA and TraAN246
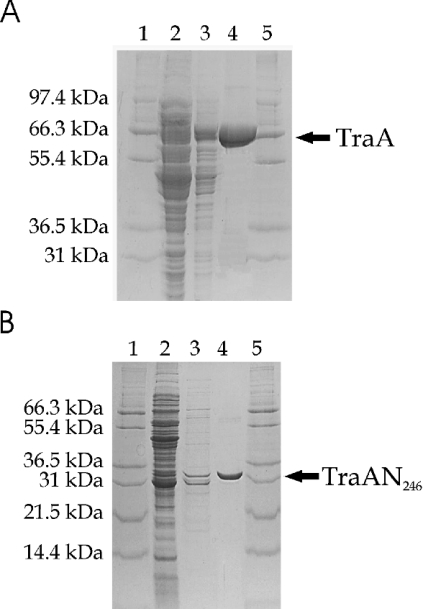
The cell pellets were suspended in BugBuster® HT buffer (Novagen, VWR International GMBH, Vienna, Austria) following the instructions provided by the supplier. The clarified supernatant was adjusted to 5 mM imidazole, 50 mM Tris/HCl, pH 8.0, and 0.5 M NaCl and loaded on to a Ni2+-charged HiTrap Chelating column (Amersham Biosciences, Uppsala, Sweden) to bind the His-tagged protein. The column was washed with several volumes of binding buffer (50 mM Tris/HCl, pH 8.0, 0.5 M NaCl, 5 mM imidazole), and finally TraA or TraAN246 was eluted with an imidazole gradient (50 mM Tris/HCl, pH 6.0, 0.5 M NaCl, 250 mM imidazole). The fractions containing the desired protein were dialysed against a buffer consisting of 20 mM Tris/HCl, pH 8.0, 300 mM NaCl, 1 mM EDTA and 0.5 mM dithiothreitol. This buffer was used as loading buffer for purification on a HiTrap Heparin column (Amersham Biosciences). The target protein was eluted with a NaCl gradient (20 mM Tris/HCl, pH 8.0, 1 M NaCl, 1 mM EDTA and 0.5 mM dithiothreitol). Samples were taken at the different purification steps and loaded on to SDS/polyacrylamide gels (Figures 1A and 1B). While the TraAN246 band migrates at 32 kDa in the denaturing PAGE according to its expected molecular mass of 31.6 kDa, the TraA band migrates at approx. 66 kDa, significantly faster than expected for its calculated molecular mass of 78.6 kDa; this behaviour may be due to the basic nature of the TraA protein and has been described previously for the HMG (high-mobility-group) proteins [9].
Figure 1. Protein purification of TraA and TraAN246.
Samples were applied to an SDS/12% (w/v) (TraA; A) or 18% (w/v) (TraAN246; B) polyacrylamide gel stained with Coomassie Brilliant Blue. Lanes 1 and 5, Mark12™ protein standard (Invitrogen, Carlsbad, CA, U.S.A.); lane 2, clarified lysate; lane 3, eluate after the HiTrap Chelating Ni2+-charged column (∼50 μg of TraA; ∼4 μg of TraAN246); lane 4, eluate after the HiTrap heparin column (∼10 μg of TraA; ∼3 μg of TraAN246).
The two-step purification procedure yielded highly pure proteins. Expression of TraA and TraAN246 resulted in 1.4 g of cells per litre of culture. The yield of purified protein was approx. 3 mg of TraA and 1.5 mg of TraAN246 per litre of culture. TraA was concentrated to 12 mg/ml and TraAN246 to 6 mg/ml. The concentration of the proteins was determined by the BCA Protein Assay procedure (Pierce, Perbio Science, Erembodegem, Belgium) using BSA as standard. The protein solutions were diluted for subsequent experiments.
CD spectroscopy
CD measurements were performed on a Jasco J-715 spectropolarimeter using 0.02 cm and 0.1 cm path-length cylindrical cells. Far-UV CD spectra were recorded from 260 to 195 nm. Each spectrum was recorded as an average of three scans taken with the following parameters: step resolution, 0.5 nm; speed, 50 nm/min; response, 1 s; bandwidth, 1 nm. The resulting spectra were baseline-corrected by subtracting the signal of the buffer. The protein samples were prepared in 50 mM Tris/HCl buffer, pH 7.5, containing 250 mM NaCl. Protein concentrations used were 46.1 μM, 20.2 μM and 57.0 μM for TraA, TraAN246 and the TraAN246–DNA complex (1:1) respectively. As DNA target, the annealed 42-mer oligonucleotide containing the inverted repeat structure, the nick site and an additional 7 bases was used. The CD signal was converted to mean residue ellipticity [θ].
Secondary structure determination and thermal stability
The secondary structure content of the two proteins was calculated using the K2d algorithm (available at http://www.embl-heidelberg.de/~andrade/k2d.html) [10,11]. CD data obtained at 20 °C were used as input for the program. Thermal denaturation curves were collected using a 0.02 cm water-jacket cylindrical cell thermostatted by an external computer-controlled water bath. The data were recorded in the temperature range 20–60 °C at 208 nm by a step scan procedure: heating rate, 20 °C/h; response, 1 s; recorded interval, 0.2 °C. The thermal denaturation curves were fitted with a sigmoidal function, and TM was determined from the point of inflection using the program Origin™ 5.0 (Microcal™, Northampton, MA, U.S.A.). For better comparability, the temperature scans were normalized according to their initial CD208 signal, as determined from the least-squares fit, and the curves were smoothed using a moving-window average algorithm with a window size of 10 points.
Sequence-based secondary structure prediction
The secondary structures of TraA and TraAN246 were calculated using the Network Protein Sequence @nalysis (NPS@; available at http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_seccons.html) [12] and the program PSIPRED (available at http://bioinf.cs.ucl.ac.uk/psipred.html) [13].
Cross-linking experiments with TraA and TraAN246
Cross-linking of the proteins was performed as described previously [14] with minor modifications. The reaction volume of 50 μl consisted of 0.5 mg/ml protein, 100 mM Bicine, pH 7.5, 300 mM NaCl, 1 mM dithiothreitol and various concentrations of glutaraldehyde (0.001%, 0.01%, 0.05% and 0.1%, v/v). The reaction was stopped after 15 min by adding 1 M glycine, pH 8.0, to final concentration of 140 mM. The samples were incubated for another 5 min. The proteins were precipitated with 400 μl of cold acetone for 2 h at −20 °C and centrifuged at 15000 g for 15 min at room temperature. Prior to loading on to an 18% (w/v) polyacrylamide gel in the presence of SDS, the pellets were dissolved in loading buffer [100 mM Tris/HCl, pH 6.8, 4% (w/v) SDS, 0.2% (w/v) Bromophenol Blue, 20% (v/v) glycerol, 200 mM β-mercaptoethanol] and heated to 100 °C for 5 min.
SAXS (small-angle X-ray scattering) measurements
For the SAXS measurements, an evacuated high-performance SAXS instrument, ‘SAXSess’ (Anton Paar KG, Graz, Austria), was used. It is a modification of the so-called ‘Kratky compact camera’ that is described in detail elsewhere [15]. Focusing multilayer optics and a block-collimating unit (slit collimation) provided an intense monochromatic primary beam with low background. The SAXSess was attached to a conventional X-ray generator (Philips, Eindhoven, Netherlands) equipped with a sealed X-ray tube (Cu anode target type, producing Cu Kα X-rays with a wavelength of 0.154 nm) operating at 40 kV and 50 mA. The samples were transferred to standard quartz capillary tubes for the SAXSess camera (with an outer diameter of 1 mm and wall thickness of 10 μm) and placed in a thermally controlled sample holder, which was centred in the X-ray beam. The scattered X-ray intensities were measured with a BAS1800 two-dimensional imaging plate detection system (Fuji, Tokyo, Japan) and processed with SAXSQuant software (Anton Paar KG). The scattering intensity was sensed over the whole scattering range simultaneously, with the spatial resolution of 50 μm×50 μm per pixel at a sample-to-detector distance of 265 mm. The semi-transparent ‘beam stop’ (0.350 mm-thick Ni foil) attenuated the primary beam to a level that was detectable with the imaging plate. With the monochromatic primary beam of the SAXSess system, the measuring times were 60 min per sample. Scattering data were smeared, mainly because of the finite dimensions of the primary beam. For purposes of the desmearing procedure, the two primary beam cross-section profiles (beam width and beam length) were measured in the detection plane. Scattering data that were read off from the imaging plate were first corrected for the absorption of the X-rays in the sample and transformed to the q scale (program SAXSQuant; Anton Paar KG). q is the scattering vector, defined as q=4π/λ·sin(θ/2), where λ is the wavelength of the X-rays and θ is the angle between the incident beam and the scattered radiation. The SAXS measurements were corrected further for the empty capillary and pure solvent scattering, and put on the absolute scale using water as a secondary standard [16] (program PDH; PCG software, Institute of Chemistry, Graz, Austria). TraA and TraAN246 were applied at concentrations of 4.66 mg/ml and 1.2 mg/ml respectively for the SAXS measurements.
A relationship between molecular mass and particle radius [17] was developed using the values for protein volumes and masses given by Durchschlag and Zipper [18].
Cleavage assay
Plasmid pVA2241 (6.9 kb) [19] was used as a substrate for the cleavage reaction. This plasmid contains the complete oriTpIP501 region. Control reactions were performed using the mobilizable IncQ plasmid RSF1010 (8.7 kb) [20] and the oriTpIP501− plasmid pDL277 [19,21]. The reaction mixture (20 μl) contained 20 mM Tris/HCl, pH 7.8, 100 mM NaCl, 15 mM MgCl2, 0.1 mM EDTA, 0.1% Brij 58, 200 ng of BSA, 150 ng of each plasmid and a 20- or 80-fold molar excess of TraA or TraAN246. The samples were incubated at 42 °C for 60 min, reactions were stopped by adjusting the mixture to 10 mM EDTA, 0.5% SDS and 0.5 mg/ml proteinase K, and the incubation was continued at 37 °C for 30 min [20]. The mixtures were subjected to electrophoresis in 0.7% (w/v) agarose gels in TAE buffer (40 mM Tris/acetate, 1 mM EDTA, pH 8.0) at 7 V/cm for 4 h. Gels were stained with ethidium bromide and photographed. The amount of cleaved pVA2241 DNA was determined with AlphaEaseFC™ software (Biozym Scientific, Hess Oldendorf, Germany).
DNA labelling
Synthetic oligonucleotides (HPLC-grade) were purchased from VBC-GENOMICS (Vienna, Austria). Oligonucleotides were labelled with [γ-32P]ATP (1.11×1014 Bq/mmol) by T4 polynucleotide kinase (Roche Diagnostics). Unbound [γ-32P]ATP was removed by a Sephadex G-50 (Amersham Biosciences) column. The labelled oligonucleotides were annealed in the following way: oligonucleotides were diluted in TE buffer (10 mM Tris, pH 8.0, 1 mM EDTA), denatured completely (5 min at 95 °C) and annealed by quick cooling on ice. The annealed oligonucleotides were applied immediately to the EMSA or stored at 4 °C.
EMSA
Aliquots of 20 μl of binding mixtures contained 10 fmol (0.5 nM) of radiolabelled oligonucleotides and increasing TraA or TraAN246 concentrations in 20 mM Tris/HCl, pH 7.5, 0.1 mM EDTA and 200 mM NaCl. EMSA experiments with hairpin and hairpin_nic oligonucleotides were performed in the presence of 1 mg/ml BSA. All oligonucleotides used in the EMSAs are shown in Figure 6. After 30 min of incubation at 42 °C, the samples were loaded on a 10% (w/v) native polyacrylamide gel and run for 90 min at constant voltage of 80 V in ice-cold buffer containing 22.5 mM Tris/borate and 0.5 mM EDTA (pH 8.0). The amounts of bound and unbound oligonucleotides were determined in a liquid scintillation counter (Liquid Scintillation Analyser Tri-Carb 1900 CA; Packard Instruments, Groningen, The Netherlands) and/or AlphaEaseFC™ software (Biozym Scientific). For scintillation counting, each lane was cut out of the gel, and cut into 16 pieces. Each piece was solubilized with 0.5 ml of Soluene-350 (Packard Instruments) and incubated at 50 °C for 3 h. A 10 ml volume of Hionic-Fluor (Packard Instruments) was added to each vial. The samples were vortexed and allowed to adapt to room temperature before counting for radioactivity. The dissociation constant (KD) of the DNA–protein complex was estimated to be equal to the protein concentration at which half of the oligonucleotide was bound. As a negative control, a randomly chosen 42-mer oligonucleotide with no sequence identity with the pIP501 oriT (encompassing nucleotides 2179–2220 on pIP501) was used (GenBank® accession no. AJ301605). To test binding to IncP group (RP4) oriT, the oligonucleotide oriT 2 (5′-CAGCCGGGCAGGATAGGTGAAGT-3) [22] was used, and for binding to the pMV158-type oriT, the oligonucleotide 5′-ATAAAGTATAGTGTGTTATACTTTAT-3′ was used (nucleotide positions 3582–3607; GenBank® accession no. X15669, the last T was exchanged for a C). Competition assays were done similarly to other EMSA experiments: unlabelled 42-mer as specific competitor or the negative control 42-mer oligonucleotide as unspecific competitor was used. The polyacrylamide gels were exposed to Hyperfilm™-βmax (Amersham Biosciences) overnight at 4 °C.
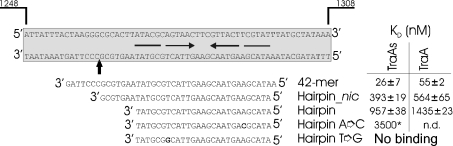
Figure 6. Oligonucleotides used for band shift experiments.
The DNA fragment of the original oriTpIP501 sequence is shown on a grey background. Position numbers refer to GenBank accession no. L39769. The experimentally determined nick site [19] is marked with a vertical arrow. Horizontal arrows indicate an inverted repeat, potentially generating a secondary structure. The exchanged base for generating a perfect hairpin is shown in bold. KD values are means±S.D. of three independent measurements (except for the hairpin oligonucleotide with TraAN246 – two measurements). KD values were determined with the liquid scintillation counter (see Experimental section). *The KD value for the TraAN246/hairpin A→C complex was calculated using AlphaEase software. n.d., not determined.
RESULTS
The relaxase contains a mixed α/β-fold
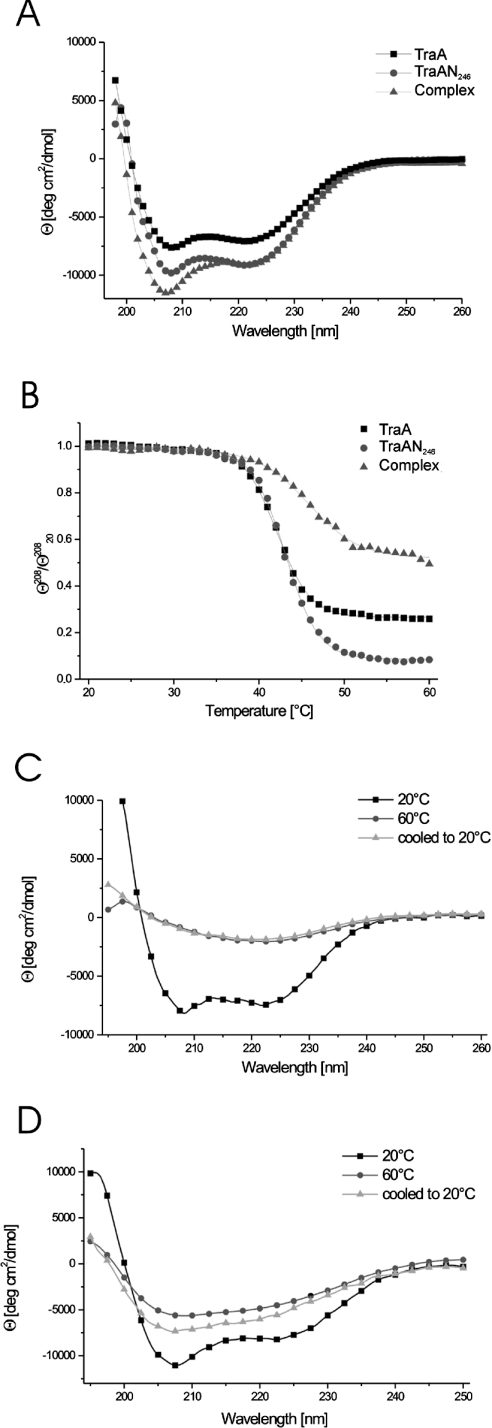
In order to check the structural integrity and to compare the fold content of the full-length and the C-terminally truncated TraA proteins, the far-UV CD spectra of purified TraA and TraAN246 at room temperature were recorded. The spectra were normalized for concentration and residue number and superimposed (Figure 2A), showing mainly α-helical features for both proteins and a higher fold content for the TraAN246 protein. To quantify the secondary structure fractions in the two proteins, we used the K2d program [10]. The results obtained for TraA revealed 30% α-helix, 13% β-sheet and 57% random coil, whereas TraAN246 showed a higher β-sheet content (27% α-helix and 18% β-sheet). This distribution indicates that the β-sheet elements are concentrated mainly in the N-terminal relaxase domain and that the C-terminal domain of TraA constitutes a predominantly α-helical fold. These results are in very good agreement with secondary structure predictions using the program PSIPRED [12], which yield a mixed α/β-fold for the relaxase domain (41% α, 15% β) and an almost entirely α-helical prediction for the C-terminal part (residues 247–654: 70% α, 3% β).
Figure 2. CD analysis.
Far-UV CD spectra of the proteins were recorded in a 0.02 cm cylindrical cell with a scan rate of 50 nm/min at 20 °C. The spectra represent the accumulation of three repetitive scans and are normalized for mean residue weight and concentration of the respective protein. (A) TraA, TraAN246 and complex (TraAN246+42-mer) spectra recorded at 20 °C. (B) Heat-induced unfolding of TraA, TraAN246 and the complex: [θ]208 values recorded during the scan were normalized for the room temperature values ([θ]20820) in order to yield comparable unfolding curves. (C, D) Heat denaturation of TraA (C) and TraAN246 (D). Spectra of the samples were taken at 20 °C, and following heating to 60 °C, and then cooling to 20 °C.
TraA and TraAN246 unfold at the same temperature
To answer the question of whether the N- and C-terminal domains of TraA are independent fold entities, we investigated the thermal stability of TraA and TraAN246. We performed temperature scans on the CD spectropolarimeter following a step-scan procedure, where the CD spectra of the proteins were recorded at 10° intervals upon heating and cooling. In addition, the CD signal at 208 nm was recorded every 0.2° over the whole temperature range (Figure 2B). The TM for both proteins was 42 °C and the unfolding curves were very similar in shape, indicating a single domain transition or at least a highly co-operative unfolding procedure.
TraA precipitated in the measuring cell when the temperature scan approached 60 °C, and the corresponding CD spectrum showed a major reduction of the CD signal. No refolding was observed upon cooling to room temperature (Figure 2C). In contrast, the TraAN246 protein retained some secondary structure when heated to 60 °C, but the minimum of the spectrum was shifted towards shorter wavelengths, as expected for a higher random coil fraction (Figure 2D). Only a slight refolding effect could be observed for TraAN246 upon cooling, which means that the heat denaturation is essentially irreversible for both TraA and TraAN246.
The relaxase domain is stabilized through DNA binding
DNA-binding proteins often undergo conformational rearrangements upon DNA binding [23,24]. We investigated the effect of oriT–TraAN246 complex formation by CD spectroscopy. At room temperature, the CD spectrum of the 1:1 stoichiometric TraAN246–DNA complex showed a 20% increase in the CD signal at 208 nm (Figure 2A). In order to analyse the effect of DNA binding on the thermodynamic behaviour of TraAN246 we performed a temperature scan of the complex from 20 to 65 °C (Figure 2B). The TM of the complex, as determined by non-linear least-squares fit using a Boltzmann function, was 48.8 °C, indicating a significant stabilization of the relaxase domain. On approaching the high-temperature end of the scan the protein precipitated, and the unfolding procedure was irreversible (results not shown).
TraA and TraAN246 form dimers in solution
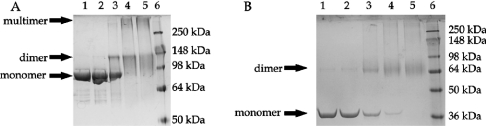
The quaternary structures of the full-length TraA protein and of TraAN246 were investigated by chemical cross-linking experiments. Using increasing amounts of glutaraldehyde as cross-linking reagent, both full-length TraA and TraAN246 formed dimers. In lane 1 of Figures 3(A) and 3(B), the proteins without cross-linking reagent were loaded on an SDS/18%-polyacrylamide gel, showing the expected positions for the monomeric proteins. With the lowest glutaraldehyde concentration (0.001%; lane 2 in Figures 3A and 3B) a faint band appeared at the expected position corresponding to double the molecular mass of the monomers (expected molecular mass 157.1 kDa for TraA and 63.3 kDa for TraAN246). With increasing glutaraldehyde concentrations (0.01%, 0.05% and 0.1% in lanes 3–5 respectively) the monomer bands gradually vanished and the dimer bands became the predominant species. Only in the case of TraA did the intensity of the dimer bands decrease at high cross-linker concentrations (Figure 3A, lanes 4 and 5), and a high molecular mass species appeared, indicative of unspecific cross-linking.
Figure 3. Glutaraldehyde cross-linking of TraA and TraAN246.
Samples of 0.5 mg/ml TraA (A) or TraAN246 (B) were incubated with increasing glutaraldehyde concentrations, then the products were loaded on to SDS/18%-polyacrylamide gels, electrophoresed at a constant voltage of 180 V and stained with Coomassie Brilliant Blue. Lane 1, no glutaraldehyde; lane 2, 0.001% glutaraldehyde; lane 3, 0.01%; lane 4, 0.05%; lane 5, 0.1%; lane 6, SeeBlue® plus2 prestained protein standards (Invitrogen).
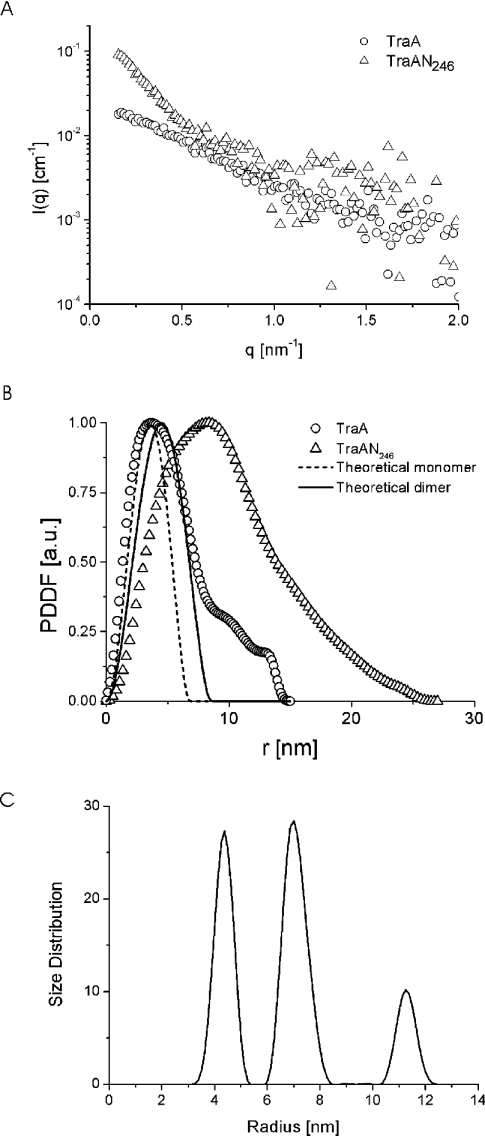
To confirm further the dimeric state in solution of the TraA protein as well as of the relaxase domain TraAN246, SAXS experiments were performed. The scattering curves of the two proteins are shown in Figure 4(A). From these, PDDFs (pair distance distribution functions) [25] were derived, showing a symmetrical peak for TraA with a maximum at 3.7 nm (Figure 4B). The shape of the PDDF curve is indicative of a homogeneous species of approximately spherical particles with a maximum dimension of 9.5 nm and a small percentage of bigger structures (impurities or dust), resulting in a shoulder at larger radii.
Figure 4. SAXS data for TraA and TraAN246 solutions.
(A) Experimental scattering data for TraA (○) and TraAN246 (△) in Tris/NaCl buffer (see the Experimental section). (B) PDDFs of TraA (○) and TraAN246 (△) solutions derived from the scattering functions; the theoretical PDDFs are shown for a sphere corresponding to the TraA monomer and the TraA dimer. (C) Size distribution function of TraAN246 solution, weighted by particle volume.
Calculating the theoretical scattering functions of protein molecular mass standards and approximating them to homogeneous spheres results in a relationship of VS≈(2.12±0.16)MM between the molecular mass MM (kDa) and the volume of the sphere VS (nm3) [17]. Assuming a spherical shape of the TraA protein and taking the molecular mass as 78.6 kDa, one can calculate VS and, via the corresponding radius, the theoretical PDDFs of the monomeric and dimeric forms of the TraA protein. The resulting maximum dimensions are 6.8 nm for the monomeric and 8.6 nm for the dimeric form, which is in good agreement with the maximum dimension of 9.5 nm obtained in the SAXS experiment. The peak shape of the theoretical PDDF corresponding to the dimeric form fits well with the curve obtained experimentally for TraA. The main differences between the experimental and theoretical PDDFs of the dimer occur at small distances (<3 nm). They are due mainly to the inhomogeneities of the protein, in contrast with the constant density of the sphere used for the calculation of the theoretical curve. The possibility cannot be ruled out that a small amount of monomer might be present in the sample, but the main component consists of dimers. A quantitative separation is not feasible, since the size difference is not large enough, and deviations from perfect spherical shape [26] might lead to incorrect estimates of small amounts of monomers.
In contrast, the PDDF of the TraAN246 protein exhibited pronounced asymmetry, with its maximum located at 8.4 nm and a shoulder at longer distances. Calculating the size distribution function of the TraAN246 sample yielded three well separated peaks denoting three species with radii of 4.3, 7 and 11 nm (Figure 4C). In order to determine the accurate molecular masses of the three species of TraAN246, the SAXS measurements were performed on absolute scale and water was used as a secondary standard [16]. In the case of TraAN246 we assumed that the peaks at 4.3 and 7 nm in the distribution function correspond to protein species of defined size, and that the peak at 11 nm is generated by small amounts of impurities or unspecific protein aggregates. In order to attribute molecular mass values to each peak, the scattering intensity for each species has to be calculated (G. Fritz and A. Bergmann, unpublished work; details available on request from G.F. (email gerhard.fritz@uni-graz.at). For the first peak (r=4.3 nm) this calculation yielded a molecular mass of 73.5±15 kDa, corresponding to the dimeric form of TraAN246, and for the second peak (r=7 nm) we obtained a value of 300±30 kDa, indicative of a TraAN246 aggregate of 9–11 monomers.
TraA and TraAN246 show nicking activity in vitro
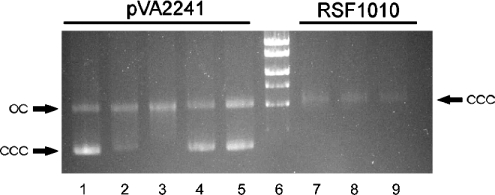
The relaxation activities of TraA and TraAN246 on supercoiled plasmid DNA were tested. The oriTpIP501-containing plasmid pVA2241 (6.9 kb) was used as substrate for the reaction. The plasmid DNA subjected to the reaction contained ∼73% supercoiled form (ccc). TraA and TraAN246 caused an increase in the amount of relaxed form (oc). Applying the conditions of the cleavage assay optimized for GST–TraA and GST–TraA* (N-terminal 293 amino acids of TraA) [8], the maximum conversion of the supercoiled plasmid DNA into the relaxed form obtained by incubation with TraA for 60 min was approx. 78% oc form (Figure 5, lane 3), and with TraAN246 it was approx. 49% oc form (Figure 5, lane 5). No relaxation was observed for the IncQ plasmid RSF1010 (8.7 kb) belonging to the same oriT nick region family as pIP501 [3] and the oriTpIP501− plasmid pDL277 (6.6 kb) [19,21] (results not shown). In comparison with the data of Kurenbach and co-workers [8], who showed that the first 293 amino acids of TraA are sufficient for oriTpIP501 cleavage, TraAN246 (N-terminal 246 amino acids of TraA) now represents the smallest TraA portion exhibiting specific transesterase activity.
Figure 5. Cleavage assay.
A 0.7% (w/v) agarose gel was run in TAE buffer (40 mM Tris/acetate, 1 mM EDTA, pH 8.0), and stained with ethidium bromide. Lane 1, pVA2241 without protein; lane 2, pVA2241 with 20-fold molar excess TraA; lane 3, pVA2241 with 80-fold excess TraA; lane 4, pVA2241 with 20-fold excess TraAN246; lane 5, pVA2241 with 80-fold excess TraAN246; lane 6, MassRuler™ DNA Ladder [high range: 10, 8, 6, 5, 4 and 3 kb (MBI Fermentas, St. Leon-Rot, Germany)]; lane 7, RSF1010 with 80-fold excess TraA; lane 8, RSF1010 with 80-fold excess TraAN246; lane 9, RSF1010 without protein. oc, open circular plasmid DNA; ccc, covalently closed circular plasmid DNA.
TraA and TraAN246 bind specifically to oriTpIP501
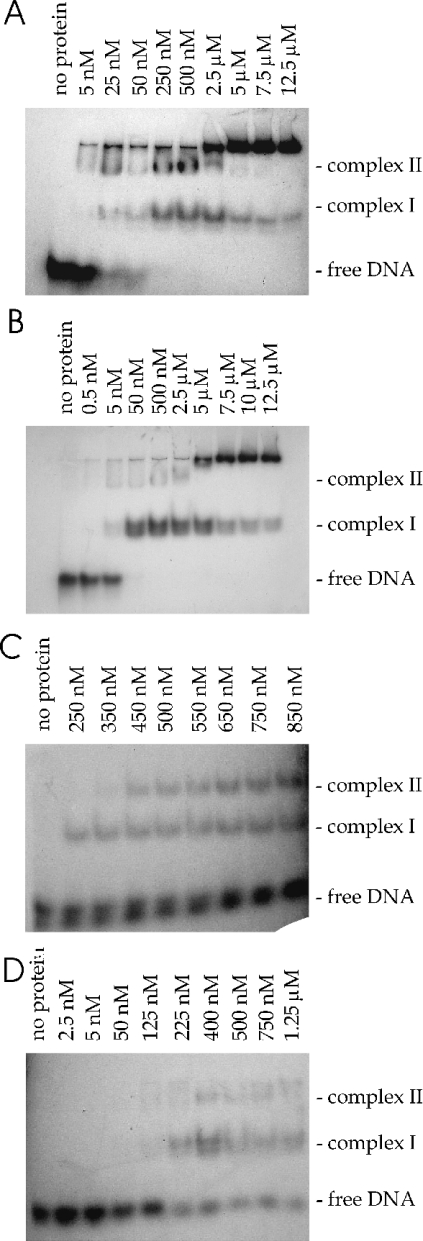
The oligonucleotides used for the band-shift experiments are shown in Figure 6. The 42-mer oligonucleotide comprises the sequence of the DNA strand which is nicked in vivo by the TraA protein [19]. It contains an inverted repeat with the potential to build a secondary structure, a stem–loop structure of 11 bp and a bulge in the middle. Its annealing temperature is TM=66.2 °C, and ΔG=−22.7 kJ/mol (−5.42 kcal/mol) at 42 °C (calculated with mFold [27,28]). At the 3′ terminus there is a single-stranded region comprising the nick site and an additional 7 bases (nucleotides 1256–1297 on pIP501; accession no. L39769). The oligonucleotide–protein binding reactions produced more than one shifted species. Low concentrations of protein TraA favoured the formation of a slowly migrating complex (complex II in Figure 7A), whereas higher concentrations resulted in an increasing fraction of the faster migrating complex (complex I). For TraAN246, predominantly complex I was formed at low protein concentrations (Figure 7B). At high protein concentrations both proteins formed large aggregates that were unable to enter the gel. In order to identify the nature of the shifted complexes, band shift experiments were repeated with chemically cross-linked protein preparations, where the main fraction of the protein formed dimers. Using the cross-linked TraA protein, the complex II in Figure 7(A) was identified as the TraA-dimer–DNA complex. When using the cross-linked TraAN246 sample, however, the shifted band coincided with the complex I band in Figure 7B (results not shown; see supplementary material at http://www.BiochemJ.org/bj/387/bj3870401add.htm). Complex II in Figure 7(B) therefore represents a higher oligomer of TraAN246, which was also found in solution in the SAXS experiment with this protein, or unspecific binding of more than one TraAN246 dimer to one oligonucleotide.
Figure 7. TraA and TraAN246 bind to oriTpIP501.
Increasing amounts of TraA (A and C) or TraAN246 (B and D) were incubated with 0.5 nM DNA at 42 °C for 30 min and then loaded on to a 10% (w/v) native polyacrylamide gel. The EMSAs were performed with the 42-mer oligonucleotide (A and B) and the hairpin_nic oligonucleotide (C and D).
The 42-mer oligonucleotide was bound by TraA and TraAN246 with KD values of 55 and 26 nM respectively. TraA and TraAN246 bound the hairpin_nic oligonucleotide, generating both retarded complexes (Figures 7C and 7D), but with a lower affinity than the 42-mer, resulting in KD values of 564 nM for TraA and 393 nM for TraAN246. KD increased approximately another 3-fold when only the hairpin was available. Since the wild-type hairpin contains a mismatch (see Figure 6), perfect hairpins were generated and also used in the EMSA experiments. The A→C perfect hairpin bound to TraAN246 approximately 4 times more weakly (KD=3.5 μM) than the wild-type hairpin (KD=953 nM). Applying high protein concentrations to oligonucleotides containing the hairpin without flanking sequences (original bulged hairpin and hairpin with A→C exchange) generated complex I and a complex that did not enter the gel. Incubation of TraA or TraAN246 with the perfect hairpin (G→T exchange in the left half repeat of the inverted repeat) resulted exclusively in a high-molecular-mass DNA–protein complex that did not enter the gel (results not shown).
Putative cross-reactivity of TraA and TraAN246 on oriTs of different oriT nick region families [3] was investigated by applying oligonucleotides containing oriTpMV158 or oriTRP4 to the EMSAs. Even using a protein/DNA ratio of 35000, the pIP501 DNA relaxase did not bind to the heterologous origins (results not shown). The specificity of TraA and TraAN246 binding was tested further by EMSAs with a randomly selected 42-mer oligonucleotide. The oligonucleotide was incubated with increasing TraA or TraAN246 concentrations up to 5 μM. No binding was observed. In competition assays the labelled 42-mer oligonucleotide (oriT 42-mer or random 42-mer) was incubated first with TraA or TraAN246 and then with different concentrations of the respective unlabelled 42-mer oligonucleotide. A 250-fold excess of the specific competitor completely inhibited binding, whereas a 1000-fold excess of unspecific competitor DNA (random 42-mer) did not affect binding of the TraA and TraAN246 proteins (results not shown). This demonstrates that the pIP501 TraA relaxase binds specifically to its cognate oriT region.
DISCUSSION
TraA is the second relaxase to be characterized from plasmids originating from Gram-positive organisms. The Gram-positive conjugative transfer machinery might require fewer components than the type IV secretion system present in Gram-negative bacteria, presumably because it is easier to cross the barrier of the Gram-positive cell wall (inner membrane only, compared with inner and outer membranes separated by the periplasm in Gram-negative bacteria). Nevertheless, the relaxase, the key enzyme initiating DNA transfer, seems to follow a quite highly conserved mechanism in Gram-positive as well as Gram-negative bacteria. The 654-amino-acid TraA protein from plasmid pIP501 belongs to the IncQ-type family of conjugative DNA relaxases. TraA exhibits a two-domain structure like other relaxases, where the relaxase function has been proposed to reside in the N-terminal domain. The function of the C-terminal domain is unknown. It has been shown previously that the N-terminal 293 amino acids of the TraA protein are sufficient for cleavage activity in vitro [8]. The results of in vitro cleavage experiments performed in the present study show that an even shorter construct, the C-terminal truncation mutant TraAN246, contains the functional relaxase domain. In addition, TraAN246 exhibits a similar fold and slightly higher secondary structure content than full-length TraA. The analysis of secondary structure content yields a mixed α-helix/β-sheet fold for both TraA proteins, but with higher β-sheet content for the relaxase domain TraAN246. This is in good agreement with the results of sequence-based secondary structure predictions, which suggest an α/β fold for the N-terminal domain, but a predominantly α-helical C-terminal domain.
Thermal denaturation curves are very similar for TraA and TraAN246 and the transition temperature was nearly identical for the two proteins (TM=42 °C and 43 °C respectively). Interestingly, a temperature range of 42–45 °C was found to be optimal for the transesterase activity of the GST–TraA fusion protein [8]. Temperatures below 37 °C and above 50 °C resulted in a significant decrease in cleavage activity [29]. The optimization of the gel retardation assays also yielded 42 °C as the optimal temperature. Incubation at 30 °C resulted in no visible binding, and less DNA was bound by the TraA proteins at 37 °C than at 42 °C. Presumably, the elevated temperatures, which trigger partial unfolding of the relaxase, are necessary to enable the DNA substrate to enter the active site.
Upon binding of the 42-mer oligonucleotide comprising the inverted repeat and the nick site, the relaxase domain exhibited an increased CD signal at 208 nm and the thermal unfolding transition was shifted by 6 °C, indicating stabilization of the relaxase domain. The first observation suggests a conformational rearrangement of TraAN246 upon binding the oriT site DNA, reminiscent of an induced-fit mechanism. This has been described for several DNA-binding proteins, e.g. bZIP transcription factors, where the basic domain is unfolded in the unbound state and only acquires its α-helical conformation upon binding to the DNA [23,24], and the conjugation protein TraM, which features an N-terminal recognition helix that is unfolded in the free state [14]. The second observation, i.e. the increased thermal stability of the complex, is also a commonly observed phenomenon of DNA-binding proteins [30]. The higher transition temperature of the complex as well as the broadening of its unfolding transition (Figure 2B) indicates an intimate contact of the 42-mer oligonucleotide with the putative binding pocket of the relaxase domain.
Comparison of the binding of TraA and TraAN246 to the 42-mer and the hairpin_nic oligonucleotides shows that both proteins formed two distinct complexes (complexes I and II, Figure 7). The highest binding affinity was observed for the 42-mer oligonucleotide (Figures 7A and 7B). Deletion of the seven bases proximal to the nick site resulted in a 15-fold reduction in the TraAN246 binding affinity, suggesting that this region comprises a part of the oriT that is crucial for high-affinity relaxase binding (Figure 7D). Deletion of another seven bases (5′ to the nick site) generates the hairpin oligonucleotide, which showed a further ∼2.5-fold decrease in affinity for the relaxase protein. Similar results were published for the relaxase domain of TrwC [31], whose binding affinities for the hairpin and a corresponding hairpin_nic oligonucleotide differed by ∼7-fold.
Gel retardation experiments with idealized hairpins showed that the bulge in the original imperfect hairpin is required for efficient specific binding of the protein: the A→C exchange in the right half repeat resulted in a 4-fold decrease in TraAN246 binding affinity. High-molecular-mass DNA–protein complexes were formed. At high protein concentrations, complex I was visible. An even more drastic effect on protein binding was exerted by the G→T exchange in the left half repeat. Only applying high protein concentrations (≥2.5 μM TraA and ≥3.75 μM TraAN246) to this idealized hairpin resulted in visible DNA binding. However, exclusively high-molecular-mass protein–DNA complexes were formed that did not enter the gel. Since neither complex I nor II occurred, the guanine in the left half repeat of the inverted repeat might play an important role in specific binding.
Unexpectedly, the relaxase domain TraAN246 showed a higher binding affinity than the full-length TraA protein with all specific oligonucleotides tested in the present study (Figures 7B and 7D). The reason for this difference may be either the reduced accessibility of the binding site in the relaxase domain due to steric interference by the C-terminal domain of TraA or a conformational change in the binding site induced by the larger C-terminal domain.
Investigation of the aggregation state of the relaxase domain and the whole TraA protein by chemical cross-linking and SAXS showed that TraAN246 as well as TraA forms dimers in solution at the concentrations used for the experiments (16–39 μM in the case of TraAN246 and 6.4–59 μM in the case of TraA). Provided that the dimerization domain and relaxase activity domain do not overlap, these results could explain the occurrence of two complex bands in the EMSA experiments, which were performed at much lower protein concentrations than the dimerization experiments. A monomer–dimer equilibrium is likely to occur, but it is not clear a priori whether both species are able to form a specific DNA complex. Therefore the nature of the complexes observed in the EMSAs (Figure 7) was investigated using cross-linked proteins in preliminary EMSA experiments. In the case of the full-length TraA protein, complex II was identified as the TraA-dimer–DNA complex and complex I as the TraA-monomer complex. In the case of TraAN246, complex I co-migrated with the complex of the cross-linked dimer (see supplementary material at http://www.BiochemJ.org/bj/387/bj3870401add.htm), and no faster migrating complex could be identified. Thus TraAN246 apparently does not form a stable monomer–DNA complex.
For both TraA and TraAN246, the dimer–DNA complex appeared to form the predominant species (Figures 7A and 7B), which has important implications for the mechanism of this enzyme, especially for its proposed activity of re-ligation of the nicked plasmid. The product of the cleavage reaction is a covalent adduct of the relaxase bound to the 5′-phospho group at the nick site [32]. Genetic evidence for a rolling-circle-replication-like mechanism was presented for the transfer of the cleaved strand in the IncF and IncQ systems [33,34]. This mechanism requires a second cleavage reaction in order to generate a free 3′-hydroxy group for re-ligation [35]. Relaxases of the IncF and IncW type feature a tandem repeat of tyrosines which are both active in the DNA cleavage reaction [3]. For the relaxase TrwC of plasmid R388, de la Cruz and co-workers [36] have shown that two out of the four tyrosine residues are actively involved in cleavage and strand transfer of linear oligonucleotides, but only the first (Tyr-18) is able to cleave supercoiled DNA containing the R388 oriT. From these data the authors have developed a model in which the first active tyrosine (Tyr-18) catalyses the initial cleavage reaction, whereas the other (Tyr-26) is responsible for the second cleavage and thus termination of conjugation. According to this model, relaxases of the IncF and IncW types are active in initiation as well as termination of conjugation as monomers.
The relaxases TraI (RP4) and MobA (RSF1010) of the IncP and IncQ families, on the other hand, have been shown to carry only one active-site tyrosine for the DNA strand transfer reaction [37,38]. Therefore the monomeric form of these relaxases is insufficient for the second cleavage, and it was postulated for TraI of RP4 that it acts as a dimer in the termination of transfer DNA replication [32]. The tendency of full-length TraA and of the TraA relaxase domain to form stable dimers is consistent with the requirements of the second cleavage to be performed in close proximity to the site of the first cleavage/transesterification reaction in order to enable efficient DNA re-ligation at termination of strand transfer. For a precise description of the spatial arrangement of the active sites within the dimer and the elucidation of the reaction mechanism, three-dimensional structural information is needed. Using the data obtained from the present work, crystallization trials of the relaxase domain alone and its specific DNA complexes have been initiated.
Online data
Acknowledgments
J.K. received a NaFöG Ph.D. scholarship and a scholarship from Berliner Programm zur Förderung der Chancengleichheit für Frauen in Forschung und Lehre, as well as a travel grant from the Dr Heinrich Jörg-Stiftung and the Deutscher Akademischer Austauschdienst (DAAD). This work was supported by the Austrian Science Foundation – FWF (projects P15040 and F01805). G.F. thanks the Zukunftsfonds des Landes Steiermark for financial support. Generous support from the Environmental Microbiology Group and Institute of Ecology of the TU Berlin is acknowledged.
References
- 1.Lanka E., Wilkins B. M. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 2.Pansegrau W., Lanka E. Enzymology of DNA transfer by conjugative mechanisms. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 3.Zechner E. L., de la Cruz F., Eisenbrandt R., Grahn A. M., Koraimann G., Lanka E., Muth G., Pansegrau W., Thomas C. M., Wilkins B. M., Zatyka M. Conjugative-DNA transfer processes. In: Thomas C. M., editor. The Horizontal Gene Pool. Bacterial Plasmids and Gene Spread. Amsterdam: Harwood Academic Publishers; 2000. pp. 87–174. [Google Scholar]
- 4.Grohmann E., Muth G., Espinosa M. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grohmann E., Guzmán L., Espinosa M. Mobilisation of the streptococcal plasmid pMV158: interactions of MobM protein with its cognate oriT DNA region. Mol. Gen. Genet. 1999;261:707–715. doi: 10.1007/s004380050014. [DOI] [PubMed] [Google Scholar]
- 6.Guzmán L. M., Espinosa M. The mobilization protein, MobM, of the streptococcal plasmid pMV158 specifically cleaves supercoiled DNA at the plasmid oriT. J. Mol. Biol. 1997;266:688–702. doi: 10.1006/jmbi.1996.0824. [DOI] [PubMed] [Google Scholar]
- 7.Kurenbach B., Bohn C., Prabhu J., Abudukerim M., Szewzyk U., Grohmann E. Intergeneric transfer of the Enterococcus faecalis plasmid pIP501 to Escherichia coli and Streptomyces lividans and sequence analysis of its tra region. Plasmid. 2003;50:86–93. doi: 10.1016/s0147-619x(03)00044-1. [DOI] [PubMed] [Google Scholar]
- 8.Kurenbach B., Grothe D., Farías M. E., Szewzyk U., Grohmann E. The tra region of the conjugative plasmid pIP501 is organized in an operon with the first gene encoding the relaxase. J. Bacteriol. 2002;184:1801–1805. doi: 10.1128/JB.184.6.1801-1805.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johns E. W. History, definitions and problems. In: Johns E. W., editor. The HMG Chromosomal Proteins. London and New York: Academic Press; 1982. pp. 1–9. [Google Scholar]
- 10.Merelo J. J., Andrade M. A., Prieto A., Morán F. Proteinotopic feature maps. Neurocomputing. 1994;6:443–454. [Google Scholar]
- 11.Andrade M. A., Chacón P., Merelo J. J., Morán F. Evaluation of secondary structure of proteins from UV circular dichroism using an unsupervised learning neural network. Protein Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- 12.Combet C., Blanchet C., Geourjon C., Deléage G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 2000;291:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 13.McGuffin L. J., Bryson K., Jones D. T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 14.Verdino P., Keller W., Strohmaier H., Bischof K., Lindner H., Koraimann G. The essential transfer protein TraM binds to DNA as a tetramer. J. Biol. Chem. 1999;274:37421–37428. doi: 10.1074/jbc.274.52.37421. [DOI] [PubMed] [Google Scholar]
- 15.Bergmann A., Orthaber D., Scherf G., Glatter O. Improvement of SAXS measurements on Kratky slit systems by Göbel mirrors and imaging-plate detectors. J. Appl. Crystallogr. 2000;33:869–875. [Google Scholar]
- 16.Orthaber D., Bergmann A., Glatter O. SAXS experiments on absolute scale with Kratky systems using water as a secondary standard. J. Appl. Crystallogr. 2000;33:218–225. [Google Scholar]
- 17.Teller D. C. Accessible area, packing volumes and interaction surfaces of globular proteins. Nature (London) 1976;260:729–731. doi: 10.1038/260729a0. [DOI] [PubMed] [Google Scholar]
- 18.Durchschlag H., Zipper P. Prediction of hydrodynamic parameters of biopolymers from small-angle scattering data. J. Appl. Crystallogr. 1997;30:1112–1124. [Google Scholar]
- 19.Wang A., Macrina F. L. Streptococcal plasmid pIP501 has a functional oriT site. J. Bacteriol. 1995;177:4199–4206. doi: 10.1128/jb.177.15.4199-4206.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scherzinger E., Lurz R., Otto S., Dobrinski B. In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res. 1992;20:41–48. doi: 10.1093/nar/20.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krah E. R., III, Macrina F. L. Genetic analysis of the conjugal transfer determinants encoded by the streptococcal broad-host-range plasmid pIP501. J. Bacteriol. 1989;171:6005–6012. doi: 10.1128/jb.171.11.6005-6012.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Götz A., Pukall R., Smit E., Tietze E., Prager R., Tschäpe H., van Elsas J. D., Smalla K. Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl. Environ. Microbiol. 1996;62:2621–2628. doi: 10.1128/aem.62.7.2621-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiss M. A., Ellenberger T., Wobbe C. R., Lee J. P., Harrison S. C., Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature (London) 1990;347:575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]
- 24.Patel L., Abate C., Curran T. Altered protein conformation on DNA binding by Fos and Jun. Nature (London) 1990;347:572–575. doi: 10.1038/347572a0. [DOI] [PubMed] [Google Scholar]
- 25.Glatter O. A new method for the evaluation of small-angle scattering data. J. Appl. Crystallogr. 1977;10:415–421. [Google Scholar]
- 26.Mittelbach P., Porod G. Zur Röntgenkleinwinkelstreuung verdünnter kolloidaler Systeme VII. Die Berechnung der Streukurven von dreiachsigen Ellipsoiden. Acta Phys. Austriaca. 1962;XV:122–147. [Google Scholar]
- 27.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurenbach B. Ph.D. thesis. Technische Universität Berlin; 2004. Konjugativer DNA Transfer zwischen Gram-positiven und Gram-negativen Bakterien: Transferkomponenten des Multiresistenzplasmids pIP501 aus Streptococcus agalactiae. [Google Scholar]
- 30.Greenfield N., Vijayanathan V., Thomas T. J., Gallo M. A., Thomas T. Increase in the stability and helical content of estrogen receptor alpha in the presence of the estrogen response element: analysis by circular dichroism spectroscopy. Biochemistry. 2001;22:6646–6652. doi: 10.1021/bi002846l. [DOI] [PubMed] [Google Scholar]
- 31.Guasch A., Lucas M., Moncalián G., Cabezas M., Pérez-Luque R., Gomis-Ruth F. X., de la Cruz F., Coll M. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat. Struct. Biol. 2003;10:1002–1010. doi: 10.1038/nsb1017. [DOI] [PubMed] [Google Scholar]
- 32.Pansegrau W., Lanka E. Mechanisms of initiation and termination reactions in conjugative DNA processing. Independence of tight substrate binding and catalytic activity of relaxase (TraI) of IncP alpha plasmid RP4. J. Biol. Chem. 1996;271:13068–13076. doi: 10.1074/jbc.271.22.13068. [DOI] [PubMed] [Google Scholar]
- 33.Gao Q., Luo Y., Deonier R. C. Initiation and termination of DNA transfer at F plasmid oriT. Mol. Microbiol. 1994;11:449–458. doi: 10.1111/j.1365-2958.1994.tb00326.x. [DOI] [PubMed] [Google Scholar]
- 34.Rao X. M., Meyer R. J. Conjugal mobilization of plasmid DNA: termination frequency at the origin of transfer of plasmid R1162. J. Bacteriol. 1994;176:5958–5961. doi: 10.1128/jb.176.19.5958-5961.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkins B. M., Lanka E. DNA processing and replication during plasmid transfer between Gram-negative bacteria. In: Clewell D. B., editor. Bacterial Conjugation. New York: Plenum Publishing Corp.; 1993. pp. 105–136. [Google Scholar]
- 36.Grandoso G., Avila P., Cayon A., Hernando M. A., Llosa M., de la Cruz F. Two active-site tyrosyl residues of protein TrwC act sequentially at the origin of transfer during plasmid R388 conjugation. J. Mol. Biol. 2000;295:1163–1172. doi: 10.1006/jmbi.1999.3425. [DOI] [PubMed] [Google Scholar]
- 37.Pansegrau W., Schroder W., Lanka E. Relaxase (TraI) of IncP alpha plasmid RP4 catalyzes a site-specific cleaving-joining reaction of single-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 1993;90:2925–2929. doi: 10.1073/pnas.90.7.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scherzinger E., Kruft V., Otto S. Purification of the large mobilization protein of plasmid RSF1010 and characterization of its site-specific DNA-cleaving/DNA-joining activity. Eur. J. Biochem. 1993;217:929–938. doi: 10.1111/j.1432-1033.1993.tb18323.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.