Abstract
Localization of mRNAs provides a novel mechanism for synthesis of proteins close to their site of function. MT1 (metallothionein-1) is a small, metal-binding protein that is largely cytoplasmic but which can be found in the nucleus. The localization of rat MT1 requires the perinuclear localization of its mRNA by a mechanism dependent on the 3′-UTR (3′-untranslated region). The present study investigates the nature of this mRNA localization signal using Chinese-hamster ovary cells transfected with gene constructs in which either MT1 or the globin coding region is linked to different sequences from the MT1 3′-UTR. Deletion, mutagenesis and antisense oligonucleotide approaches indicate that nt 45–76 of the 3′-UTR, in particular nt 66–76, are required for the localization of either MT1 mRNA or chimaeric transcripts in which a β-globin coding region is linked to sequences from the MT1 3′-UTR. This section of the 3′-UTR contains a CACC repeat. Two mutations that are predicted to alter the secondary structure of this region also impair localization. Our hypothesis is that the perinuclear localization signal in MT1 mRNA is formed by a combination of the CACC repeat and its structural context.
Keywords: immunocytochemistry, in situ hybridization, localization signal, metallothionein, mRNA targeting, 3′-untranslated region (3′-UTR)
Abbreviations: CHO, Chinese-hamster ovary; MT1, metallothionein-1; ODN, oligodeoxynucleotide; 3′-UTR, 3′-untranslated region
INTRODUCTION
There is now a considerable body of evidence that in a variety of cells from yeast to Drosophila to mammals certain mRNAs are localized to different subcellular regions of the cytoplasm and/or associated with the cytoskeleton [1,2]. The number of identified localized mRNAs is steadily increasing and such mRNA targeting is believed to provide a mechanism for local synthesis of proteins close to where they function [3]; this can be important in restricting protein activity to a particular region of the cell, minimizing its damaging effects or enhancing the efficacy of protein targeting. For example, in oligodendroglia, myelin basic protein mRNA is transported down long cell processes and translation is repressed until the mRNA is localized [4]. In neurons, β-actin mRNA is transported down the axon and other mRNAs are specifically targeted to dendrites [5], whereas in fibroblasts β-actin mRNA is localized to the cell periphery under conditions when actin synthesis in this area is increased [6]. In contrast, certain mRNAs are found associated with the cytoskeleton and localized in the perinuclear cytoplasm; this includes mRNAs encoding the nuclear transcription factors MYC and FOS [7–9], and MT1 (metallothionein-1), which is normally cytoplasmic but is nuclear at the G1/S transition in the cell cycle [10,11]. Correct localization of MT1 mRNA was found necessary for subsequent nuclear localization of the protein [12,13] and we have therefore hypothesized that targeting of mRNAs to the perinuclear cytoskeleton may promote subsequent nuclear import of a range of proteins [12].
To date, all the available evidence suggests that localization of mRNAs across a range of species is due to signals within their 3′-UTRs (3′-untranslated regions) [14] and this has been shown to be the case for the targeting of c-myc, c-fos and MT1 mRNAs to the perinuclear cytoskeleton [12,13,15]. It is supposed that the localization of these mRNAs to specific sites requires proteins to bind to specific signals within the 3′-UTR, thus forming ribonucleo-protein complexes and possibly RNA transport granules [16,17]. However, the precise nature of most of these signals is not known. Deletion analysis of 3′-UTRs has shown that, in general, whole 3′-UTRs are not necessary for localization but that localization signals lie in regions of <100–200 nt. For example, the β-actin localization element lies within 45 nt [18]. However, further definition of the key features of any localization signal has been limited. Repeated motifs have been found to be present in those localization elements for Vg1 or VegT [19]. The 3′-UTR of myelin basic protein mRNA contains a 21-nt motif rich in GC [4], whereas a tandem repeat of a ACACCC motif is believed to be important in the localization element for β-actin mRNA [18]. Recently, bioinformatic analysis has suggested that repeats of CAC motifs may be a common sequence motif in localization signals [20]. If such elements are indeed common to signals which direct mRNAs to different destinations, then the context of the motifs or RNA secondary structure must also play a role in giving signal specificity.
For those mRNAs that are localized to the perinuclear cytoplasm, the localization element in c-myc lies within 86 nt [21], that of c-fos within 145 nt [9] and that of vimentin within 100 nt [22]. The localization element within the MT1 3′-UTR has not been defined, but the short length of this 3′-UTR (134 nt) means that it provides an excellent system for studying a perinuclear localization signal. The aim of the present study was to use deletion, mutagenesis and antisense approaches to narrow down the part of the 3′-UTR required for MT1 mRNA localization in transfected CHO (Chinese-hamster ovary) cells.
MATERIALS AND METHODS
Gene constructs
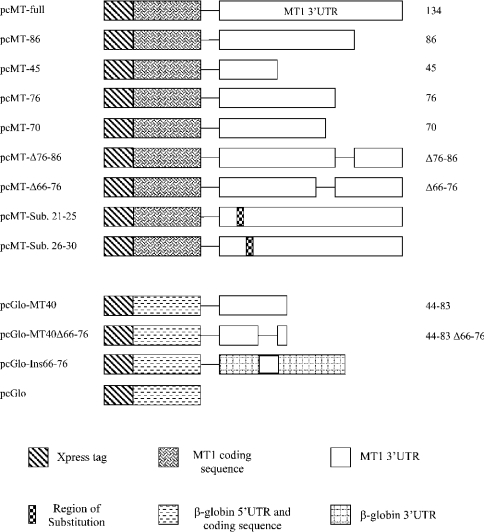
All constructs were made using cloning based on PCR and they are shown schematically in Figure 1. Primers used in cloning are shown in Table 1. pcMT-full contained the rat MT1 coding region and full 3′-UTR. It was obtained using the pcMT1 vector containing the full rat MT1 cDNA [12] as template and primers 1 and 2. The vector also contained sequences corresponding to the Xpress tag at the 5′-end of the coding sequence. Further constructs, namely pcMT-86 (nt 87–134 of the MT1 3′-UTR deleted), pcMT-45 (nt 46–134 deleted), pcMT-76 (nt 77–134 deleted) and pcMT-70 (nt 71–134 deleted) were generated by PCR combining primer 1 with either primer 3, 4, 5 or 6 respectively. All these PCR products were cloned directly into pcDNA4/HisMax-TOPO following the manufacturer's instructions (Invitrogen). Further deletion constructs (pcMT-Δ66-76 and pcMT-Δ76-86) were made in which nt 66–76 and 76–86 were deleted from the MT1 3′-UTR. These 11 nt deletions were performed by site-directed mutagenesis following the manufacturer's instructions (Quik Change®, Stratagene) using primers 7 and 8 for MT-Δ76-86 and primers 9 and 10 for MT-Δ66-76. Two substitution constructs were made in which either nt 21–25 were changed from CCCTC to GGTGG (pcMT-sub21-25) or nt 26–30 were changed from AGGTG to UCCAC (pcMT-sub26-30). The substitutions were performed by site-directed mutagenesis using primers 11 and 12 or 13 and 14 respectively. Transformation of one shot Top10 cells (Invitrogen) using post-mutagenesis PCR products was performed according to the manufacturer's instructions and plasmids containing the required deletions were selected. All constructs were verified by sequencing using the T7 forward and bovine growth hormone reverse primers.
Figure 1. Details of gene constructs.
The coding region of MT1 was linked to its own entire 3′-UTR or with increasingly large deletions so that pcMT-86, -76, -70 and -45 contained bases 0–86, 0–76, 0–70 and 0–45 respectively of MT1 3′-UTR. Site-directed mutagenesis was used to delete different 11 nt regions to create pcMT-Δ66-76 and pcMT-Δ76-86, as well as substitutions of nt 21–25 and 26–30. The coding region of β-globin was used as a reporter gene and linked either to the 3′-UTR of the pcDNA3 vector (pcGlo) or to nt 44–83 of MT1 3′-UTR (pcGlo-MT40). Site-directed mutagenesis was used to create a deletion of bases 66–76 within the MT1 3′-UTR sequences (pcGlo-MT40Δ66–76). An insert of 11 nt, corresponding to region 66–76 of the MT1 3′-UTR, was introduced into the globin 3′-UTR in pcKGG to produce pcGlo-Ins66–76.
Table 1. Sequences of primers used in PCR cloning and site-directed mutagenesis.
| Primer number | Sequence |
|---|---|
| MT1 deletion | |
| 1 | 5′-AGTACTTTTCCCGACTTCAGCAG-3′ (forward) |
| 2 | 5′-CCCCGAGAATAAACAGGCTT-3′ (reverse) |
| 3 | 5′-CCGGAAATTATTTACACCTGAGGGC-3′ (reverse) |
| 4 | 5′-TTTAGTCTGGGTGGAGGTGTAC-3′ (reverse) |
| 5 | 5′-GGTGGAGGTGTACGGCAAGACT-3′ (reverse) |
| 6 | 5′-GGTGTACGGCAAGACTCTGAGTT-3′ (reverse) |
| 7 | 5′-TTGCCGTACACCTCCACAACCCCGTTTTCTACCG-3′ (forward) |
| 8 | 5′-CGGTAGAAAACGGGGTTGTGGAGGTGTACGGCAA-3′ (reverse) |
| 9 | 5′-ACTCAGAGTCTTGCCGTCAGTTTACTAAACCCCG-3′ (forward) |
| 10 | 5′-CGGGGTTTAGTAAACTGACGGCAAGACTCTGAGT-3′ (reverse) |
| 11 | 5′-GTGACGAACAGTGCTGCTGGGTGGAGGTGTAAATAATTTCCGGACC-3′ (forward) |
| 12 | 5′-GGTCCGGAAATTATTATTTACACCTCCACCCAGCAGCACTGTTCGTCAC-3′ (reverse) |
| 13 | 5′-GACGAACAGTGCTGCTGCCCTCTCCACTAAATAATTTCCGGACCAACTCA-3′ (forward) |
| 14 | 5′-GAGTTGGTCCGGAAATTATTTAGTGGAGAGGGCAGCAGCACTGTTCGTC-3′ (reverse) |
| β-Globin/MT1 construct | |
| 15 | 5′-GGGGTACCCCAAACAGACA-3′ (forward) |
| 16 | 5′-CAAGACTCTGAGTTGGTCAGTGGTATTTGTGAGC-3′ (reverse) |
| 17 | 5′-GCTCACAAATACCACTGACCAACTCAGAGTCTTG-3′ (forward) |
| 18 | 5′-TAGTAAACTGGGTGGAGGTGTACGG-3′ (reverse) |
| 19 | 5′-CTCAGAGTCTTGCCGTCCAGTTTAAAAGGGCC-3′ (forward) |
| 20 | 5′-GGCCCTTTTAAACTGGACGGCAAGACTCTGAG-3′ (reverse) |
| 21 | 5′-CTCTGCCAAAAATTATGGGCACCTCCACCGACATCATGAAGCCCCT-3′ (forward) |
| 22 | 5′-AGGGGCTTCATGATGTCGGTGGAGGTGCCCATAATTTTTGGCAGAG-3′ (reverse) |
Overlapping PCR was used to make a construct containing the rabbit β-globin coding region and an appropriate fragment of MT1 3′-UTR. The coding region of β-globin was amplified from pcKGG [12] with specific primers, where the 3′-end of the reverse primer annealed to the end of the coding region and the 5′-end was complementary to the region 45–55 of MT1 3′-UTR (primers 15 and 16). The 40 nt sequence from MT1 3′-UTR was amplified from pcMT1 using specific primers, where the 5′-end of the forward primer was complementary to the end of the β-globin coding region (primers 17 and 18). Both PCR products were extracted from the gel and subjected to annealing PCR. The full overlapped product was cloned into pcDNA4/HisMax-TOPO. Sequencing showed the construct to consist of the β-globin coding region linked to nt 44–83 from the MT1 3′-UTR. This construct was then used as a template for another round of mutagenesis to produce pcGlo-MT-Δ66-76 in which the sequence 66–76 from the inserted 40 nt of MT1 3′-UTR was deleted using primers 19 and 20. A further construct was made in which the nt 66–76 from MT1 3′-UTR were inserted into the β-globin 3′-UTR at nt 30 by site-directed mutagenesis using pcKGG as template and primers 21 and 22. A control plasmid (pcGlo) was produced by cloning the 5′-UTR and coding sequence of β-globin into pcDNA4/HisMax-TOPO.
Cell culture
CHO cells (ECACC 85050302) with undetectable expression of MT were maintained in a mixture of F12/Ham's medium supplemented with 10% FCS and 1% penicillin/streptomycin in an atmosphere of 5% CO2. Cells (80% confluent) growing in a monolayer were transfected using LIPOFECTAMINE™ Plus (Invitrogen) according to the manufacturer's instructions. Cells, 48 h after transfection, were selected using zeocin. Subsequently, for immunocytochemistry, cells were seeded at a density of 4×103 cells/well in eight-well culture slides and left to attach for 48 h; for in situ hybridization, cells were seeded at the higher density of 8×103 cells/well.
Antisense ODNs (oligodeoxynucleotides)
ODNs were designed so as to be complementary to different 15–20 nt sections of the MT1 3′-UTR (Table 2). Phosphorothioated ODNs were chemically synthesized (MWG-Biotech) and introduced into CHO cells using LIPOFECTAMINE™ Plus. Cells were incubated with ODNs at a concentration of 600 nM for 48 h before analysis of their effect on perinuclear localization of MT protein, as monitored by immunocytochemistry.
Table 2. Sequences of ODNs complementary to the MT1 3′-UTR and quantification of their effects on MT1 localization.
A series of ODNs was designed so as to be complementary to different regions of the MT1 3′-UTR. Cells expressing the full MT1 construct were incubated with each ODN (600 nM) before immunostaining with anti-XPress antibody and FITC-conjugated goat anti-mouse secondary IgG. Distribution of MT1 was assessed in at least 100 cells and classified as either showing perinuclear localization or no localization. Results shown are means±S.E.M. for 5–6 experiments.
| ODN | 5′–3′ Sequence | Region of 3′-UTR to which complementary | Cells with perinuclear localization (%) |
|---|---|---|---|
| ODN-1 | ACGGCAAGACTCTGAGTTGG | 45–65 | 90±2 |
| ODN-2 | AGTAAACTGGGTGGAGGTGT | 66–86 | 8±7 |
| ODN-2.1 | GTGGAGGTGTACGGC | 61–75 | 70±10 |
| ODN-2.2 | GGGGTTTAGTAAACTGG | 76–92 | 6±2 |
| ODN-2.3 | TAGTAAACTGGGTGGAG | 70–86 | 30±9 |
| ODN-2.4 | GGTGTACGGCAAGACTC | 54–70 | 70±7 |
| Control | ATTCACATGCTCGGTAGAA | 95–115 | 91±4 |
| Sense | TGTGGAGGTGGGTCAAATGA | 86–66 | 93±2 |
Immunocytochemistry and in situ hybridization
Studies of both protein and mRNA distribution were performed in cells grown in multiwell chamber slides so that the different cell lines could be studied under identical conditions and the quantification of staining be directly comparable.
Immunocytochemistry was performed using an anti-Xpress antibody (Invitrogen) directed against the Xpress peptide epitope tag from the pcDNA4/HisMax-TOPO plasmid. Cells were washed three times with PBS and fixed with 1% (w/v) paraformaldehyde in PBS for 20 min, before being permeabilized with 1% paraformaldehyde/0.05% Triton X-100 in PBS for 10 min. The cells were washed in PBS and incubated with 10% (v/v) normal sheep serum in PBS for 30 min to allow saturation of non-specific antibody binding sites. After a brief wash in PBS containing 1% BSA, cells were incubated with anti-Xpress antibody diluted 1/400 in PBS/1% BSA for 45 min at room temperature (∼20 °C). After four washes in PBS/1% BSA, cells were incubated with FITC-conjugated goat anti-mouse IgG (Sigma) at a dilution of 1/150 in PBS/1% BSA. Finally, cells were rinsed four times in PBS/1% BSA and mounted in Citifluor. Two controls were performed routinely: untransfected CHO cells taken through the whole procedure described above and transfected cells that were not incubated with the primary antibody but with PBS/1% BSA.
In situ hybridization was performed using digoxigenin-labelled riboprobes as described previously [9,23]. Cells were washed with PBS, fixed in 4% paraformaldehyde in PBS for 10 min at 4 °C, dehydrated in 70% (v/v) ethanol and permeabilized with 4% paraformaldehyde/0.2% Triton X-100 in PBS for 10 min. After prehybridization in 2×SSC (0.15 M NaCl/0.015 M sodium citrate) containing 50% (v/v) formamide for 10 min at room temperature, hybridization was performed overnight at 55 °C with 200 ng of digoxigenin-labelled antisense riboprobe. The antisense MT1 probe was a 520 nt ApaI–BamHI fragment generated using SP6 RNA polymerase. The antisense globin probe was a 511 nt ApaI–BamHI fragment generated using SP6 RNA polymerase as described previously [23]. In addition, control sense probes were generated from the same fragments using T7 RNA polymerase and other controls were performed with either no probe in the hybridization mixture or with RNase A treatment of the samples. After hybridization, cells were washed in 5×SSC for 30 min at room temperature, and then for 30 min in 2×SSC/50% formamide at 55 °C, before treatment with 20 μg/ml RNase A in wash buffer (10 mM Tris/HCl, pH 7.5, containing 0.4 M NaCl and 5 mM EDTA) for 30 min at 37 °C to remove non-specifically bound probe. After a brief wash in wash buffer and two further washes in 2×SSC, the bound probe was detected by incubation with alkaline phosphate-linked anti-digoxigenin IgG (Roche, Lewes, East Sussex, U.K.) and then with 4-Nitro Blue Tetrazolium for 16 h (Roche).
Standard microscopy was performed using an Olympus BX51 microscope and digital images of the cells were captured under the ×100 oil immersion lens using an Olympus DP50 digital camera and Analysis Viewfinder Lite SIS Software. Staining was quantified by examining fields of view at random and assigning localization characteristics (presence or absence of a distinct ring of perinuclear staining) of approx. 100 cells in each of at least three separate experiments. Cells were classified as exhibiting perinuclear localization of protein or mRNA or no localization. Profiles were obtained by drawing an arbitrary line across the image of the cell, and Image Gauge V4.0 software was used to display the staining intensity along this line.
RESULTS
Previous studies [12,13] have shown that the 3′-UTR of MT1 is necessary for perinuclear localization of MT1 mRNA, its association with the cytoskeleton and the localization of MT1 protein in the perinuclear cytoplasm during G1 and in the nucleus during the G1–S transition of the cell cycle. To define which part(s) of the MT1 3′-UTR is responsible for mRNA localization, two approaches were used in the present study: first, CHO cells, which express undetectable levels of endogenous MT1, were transfected with gene constructs with deletions within the 3′-UTR; secondly, the effects of ODNs complementary to different MT1 3′-UTR sequences were investigated in transfected cells expressing the MT1 coding region linked to the full 3′-UTR. The gene constructs made with different parts of the 3′-UTR deleted are shown in Figure 1 and the 3′-UTRs to which the ODNs are complementary are indicated in Table 2.
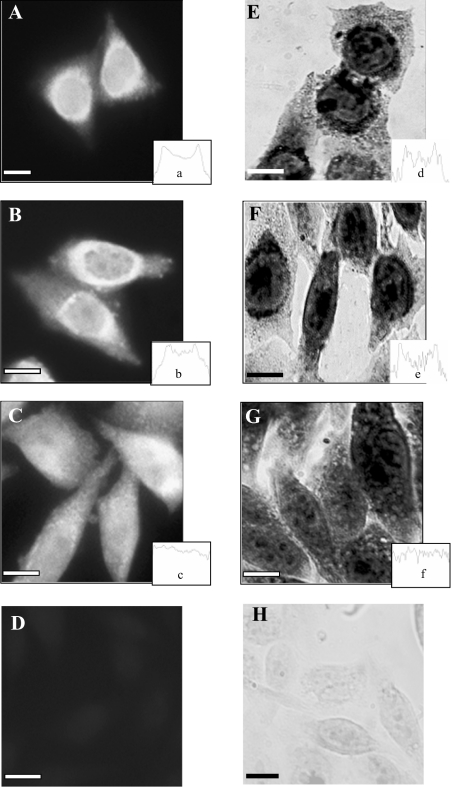
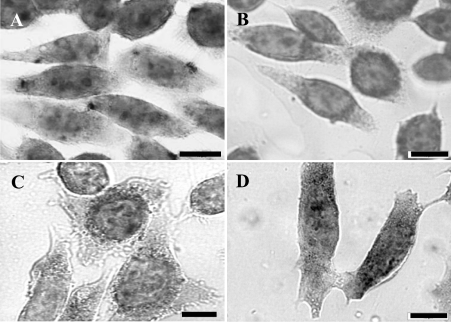
Immunocytochemistry of cells expressing the full MT1 construct showed staining as a ring around the nucleus and there was little or no staining in the cytoplasm towards the cell periphery. These results indicated that the tagged MT1 protein was concentrated in the perinuclear cytoplasm (Figure 2A), as we have observed previously in unsynchronized cells expressing untagged MT1 [12]. Initial deletion analysis showed that in cells expressing MT1 with nt 87–134 of the 3′-UTR removed (Figure 2B) MT1 was also perinuclear, but removal of nt 46–134 caused a change in staining pattern such that the ring of perinuclear staining was absent or much less intense and staining was seen throughout the cytoplasm into the cell periphery: this change in staining indicates a loss of MT1 localization (Figure 2C). In situ hybridization showed that in cells expressing the full MT1 (Figure 2E) the staining pattern corresponding to mRNA distribution was a distinct, dark ring around the nucleus with little or no staining in the cytoplasm towards the cell periphery. In addition, there was staining within the nucleus, often in discrete areas. In cells expressing MT1, with nt 87–134 of the 3′-UTR removed, the staining pattern was similar, with the mRNA localized in the perinuclear cytoplasm (Figure 2F). However, in cells expressing MT1, with nt 46–134 removed to produce a truncated 0–45 nt 3′-UTR, the staining pattern was significantly different in that the ring of perinuclear staining was absent or much less marked and diffuse staining was seen throughout the cytoplasm into the cell periphery (Figure 2G) as well as in the nucleus. Thus in these cells the perinuclear mRNA localization was lost. Controls of either untransfected cells incubated with specific antibodies (Figure 2D) or sense riboprobe (Figure 2H) showed no staining. Quantification of the staining as illustrated by the profiles shown in Figures 2(A)–2(H) confirmed these observations. The results indicate that part, or all, of the region required for the localization of the MT1 mRNA resides in the 41 nt region between nt 45 and 86.
Figure 2. Immunocytochemistry and in situ hybridization of transfected CHO cells expressing MT1 gene constructs with deletions in the 3′-UTR.
Results show either immunostaining of cells after incubation with anti-XPress antibody and FITC-conjugated goat anti-mouse secondary IgG (A–C) or in situ hybridization with digoxigenin-labelled antisense riboprobes after incubation with anti-digoxigenin antibodies and secondary antibody linked to alkaline phosphatase (E–G). There was a distinct perinuclear localization of MT1 protein and mRNA in cells transfected with either the pcMT-full (A, E) or pcMT-86 (B, F) constructs, whereas the protein and the mRNA did not show any specific localization in cells transfected with pcMT-45 (C, G). Controls of untransfected cells incubated with specific antibodies (D) or sense riboprobe (H) showed no staining. Scale bars, 10 μm. Quantification of the staining is illustrated in typical staining profiles across individual cells from the different transfected cell populations (A–F). Note that in the profiles from in situ hybridization there is a peak of staining that corresponds to consistent, circular areas of intra-nuclear staining, as found previously with c-myc transcripts [8].
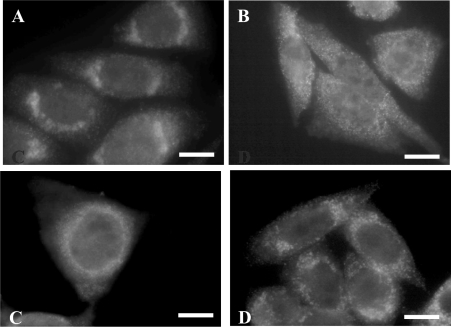
Further studies have concentrated on examining the role of this region of the 3′-UTR. Our first approach was to use antisense technology, since this methodology has been used successfully to disrupt localization of β-actin transcripts by its 3′-UTR [18]. The ODNs used were 15–20 nt in length and complementary to different regions of the 41 nt region of interest (see Table 2). These ODNs were incubated with CHO cells expressing tagged MT1 containing the full 3′-UTR, and MT1 distribution was assessed by immunocytochemistry. Initially, two 20-mer ODNs were used, complementary to either the proximal or distal end of the 41 nt region. Incubation with ODN-1 (complementary to nt 45–66) had no effect on localization of MT1 (Figure 3A), whereas, in contrast, ODN-2 (complementary to nt 66–86) caused a loss of perinuclear localization (Figure 3B). Quantification of the data showed that more than 90% of cells exhibited delocalized MT1 after treatment with ODN-2 (Table 2). Further experiments were then performed with shorter ODNs (15-mer or 17-mer) complementary to different sequences around the 66–86 region. Both ODN-2.2 and -2.3 (complementary to nt 76–92 and 70–86 respectively) caused a loss of the perinuclear localization of MT1 (Table 2), with more than 90 and 70% of cells showing delocalization with ODN-2.2 and -2.3 respectively. The ODN-2.1 (complementary to nt 61–75) and ODN-2.4 (complementary to nt 54–70) had much smaller effects on localization, with quantification showing only 25–30% of the cells exhibiting delocalization. Control ODNs, either complementary to a region outside the 41 nt region (nt 95–115) or sense sequences, had no effect on localization. Overall, these experiments indicate that the most effective ODNs were those complementary to sequences within or close to, the 66–86 nt region of the MT1 3′-UTR.
Figure 3. Immunocytochemistry of transfected CHO cells expressing MT1 gene constructs (pcMT-full) after incubation with antisense oligonucleotides.
Results show immunostaining of cells after incubation with anti-XPress antibody and FITC-conjugated goat anti-mouse secondary IgG. There was a distinct perinuclear localization of MT1 in cells incubated with ODN-1 (A) but not ODN-2 (B). Controls in which no ODNs were applied (C) or incubation was with a sense ODN (D) showed perinuclear localization of MT1. Scale bars, 10 μm.
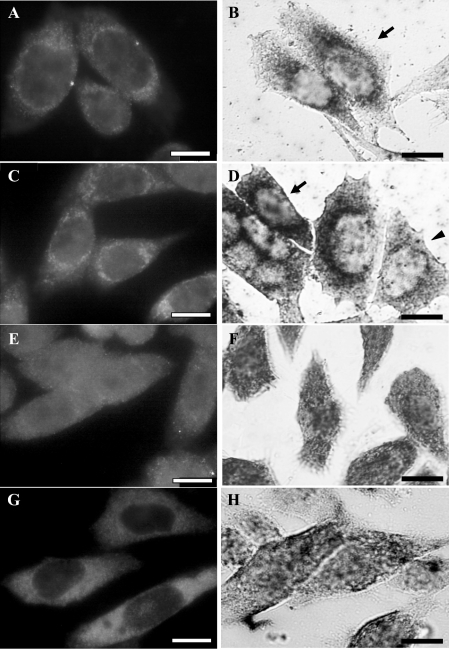
To investigate this region in more detail, further deletion analysis was performed using constructs made by PCR cloning and site-directed mutagenesis. Site-directed mutagenesis was used to delete two different 11 nt sequences corresponding to regions 76–86 and 66–76 (pcMT-Δ76-86 and pcMT-Δ66-76 respectively; see Figure 1). In parallel, further constructs were made in which nt 70–134 and 76–134 were deleted from the full MT1 construct so as to produce constructs expressing the tagged MT1 coding region linked to nt 0–70 and 0–76 of the 3′-UTR respectively (pcMT-70 and pcMT-76; Figure 1). Immunocytochemisty and in situ hybridization analysis of cells expressing constructs with deletion of nt 76–86 (Figures 4A and 4B) or nt 76–134 (Figures 4C and 4D) showed distinct rings of dense staining around the nucleus that were considerably darker than in other parts of the cytoplasm; there was only very little, weak staining in the cytoplasm towards the cell periphery. As shown in Figures 4(B) and 4(D) the ring of staining was present regardless of whether the cells adopted an elongated (arrows) or less extended, polygonal (arrowhead) morphology. Cells expressing constructs in which nt 71–134 or 66–76 were deleted showed a different staining pattern produced by immunocytochemistry (Figures 4E and 4G) or in situ hybridization (Figures 4F and 4H). In these cells, the immunocytochemical staining spread out from the nucleus so that the distinct perinuclear ring was absent and the staining appeared diffuse and present throughout most of the cytoplasm. Similarly, after in situ hybridization of these cells the distinct perinuclear ring of staining was absent. Using the presence of a ring of perinuclear staining as indicative of perinuclear localization, the staining pattern was assessed in at least 100 cells at random, so as to quantify the extent of localization (Table 3). Such quantification confirmed visual inspection of the staining pattern in indicating that 76–90% of cells expressing constructs with nt 76–86 or 76–134 deleted showed perinuclear localization of MT1 mRNA and protein, whereas only 13–29% of the cells expressing constructs in which nt 71–134 or 66–76 were deleted showed a perinuclear mRNA or protein distribution (Table 3). These results indicate that removal of bases 70–76 or 66–76 causes loss of localization and suggest that these form part of the localization signal, consistent with the data from the antisense experiments.
Figure 4. Immunocytochemistry and in situ hybridization of transfected CHO cells expressing MT1 gene constructs to investigate the role of the 66–86 nt region of the 3′-UTR.
Cells were subjected to either immunostaining with anti-XPress antibody and FITC-conjugated goat anti-mouse secondary IgG (A, C, E, G) or in situ hybridization with digoxigenin-labelled riboprobes, incubation with anti-digoxigenin antibody and secondary antibody linked to alkaline phosphatase (B, D, F, H). There was a distinct perinuclear localization of MT1 protein and mRNA in cells transfected with either pcMT-Δ76-86 (A, B) or pcMT-76 (C, D), whereas cells expressing pcMT-70 (E, F) or pcMT-Δ66-76 (G, H) showed no perinuclear localization of either MT1 protein or mRNA. CHO cells adopt elongated (arrows), oval or polygonal morphology (arrowhead) and the perinuclear ring of staining was observed regardless of overall morphology. For example, in (B) and (D), after in situ hybridization, the ring of staining is seen in cells with elongated (arrows) or polygonal (arrowhead) morphology. Scale bars, 10 μm.
Table 3. Quantification of perinuclear localization of MT1 in CHO cells transfected with constructs to investigate the role of the 66–86 nt region of the 3′-UTR.
Cells expressing the different MT1 deletion constructs were subjected either to immunostaining with anti-XPress antibody and FITC-conjugated goat anti-mouse secondary IgG or to in situ hybridization with digoxigenin-labelled riboprobes specific to MT1 transcripts. At least 100 cells were examined for perinuclear localization or lack of localization. Experiments were repeated at least three times and the percentage of localization is expressed as means±S.E.M.
| Construct | Perinuclear localization of MT1 by immunocytochemistry (%) | Perinuclear localization of MT1 mRNA by in situ hybridization (%) |
|---|---|---|
| MT-Δ76-86 | 89±3 | 88±4 |
| MT-Δ66-76 | 13±8 | 15±3 |
| MT-76 | 76±7 | 80±2 |
| MT-70 | 29±6 | 26±1 |
The results from the deletion experiments indicate that nt 45–86 or regions within this sequence, are required for localization of MT1 transcripts by its 3′-UTR. Further experiments were performed to investigate if this region was sufficient for localization and to do this we used β-globin coding sequences as a reporter. We have found previously that β-globin transcripts containing the coding region linked to its own 3′-UTR or to a vector 3′-UTR are not localized but that chimaeric transcripts containing β-globin coding region linked to a 3′-UTR containing a localization signal (e.g. MT1) are localized [12]. Thus this strategy was used with the region of MT1 3′-UTR implicated in localization, and a construct was made in which the β-globin coding region was linked to nt 44–83 of the MT1 3′-UTR (pcGlo-MT40; Figure 1). In situ hybridization analysis of cells expressing β-globin with no 3′-UTR showed, as found previously [12], a pattern of diffuse staining both within the nucleus and throughout the cytoplasm (Figure 5A), and quantification of the staining pattern showed very few (13%) cells exhibiting a perinuclear ring of staining (Table 4). In contrast, cells expressing transcripts in which β-globin coding region was linked to nt 44–83 of the MT1 3′-UTR showed evidence of localized staining. As shown in Figure 5(B) many of these cells showed a distinct ring of perinuclear staining with little or no staining in the remaining cytoplasm. Quantification showed that 43% of the cells exhibited such a perinuclear ring of staining (Table 4), indicating that nt 44–83 from the MT1 3′-UTR were capable of directing localization of the β-globin reporter transcripts to some extent. Using a similar quantitative approach to localization, previous deletion and mutation analysis of the β-actin 3′-UTR has also described occurrence of localization in a limited proportion of cells [18], and this presumably reflects a situation where the fully effective localization signal can direct localization in a high proportion of cells, but a less effective, partial signal directs mRNA localization in some cells but not others depending on the cells’ precise physiological state. Therefore the present results suggest that this 40 nt sequence is a critical feature of the perinuclear localization signal present in the MT1 3′-UTR, but that it is less efficient when not present in the context of the whole MT1 3′-UTR.
Figure 5. In situ hybridization showing effect of part of the MT1 3′-UTR on localization of reporter β-globin transcripts.
Results show in situ hybridization of CHO cells expressing chimaeric gene constructs in which the β-globin coding region was linked either to MT1 3′-UTR sequences or to the vector 3′-UTR (pcGlo). In situ hybridization was performed with digoxigenin-labelled riboprobes after incubation with anti-digoxigenin antibody and secondary antibody linked to alkaline phosphatase. Cells expressing pcGlo showed diffuse nuclear and cytoplasmic staining with no perinuclear localization of transcripts (A). Cells expressing pcGlo-MT40 (B) showed distinct rings of perinuclear staining in many cells, but those expressing pcGlo-MT40Δ66–76 (D) in which nt 66–76 were deleted showed diffuse cytoplasmic and nuclear staining with no evidence of localization of transcripts. Cells expressing pcGlo-Ins66–76 showed distinct rings of perinuclear staining in many cells and an example is shown in (C). Scale bars, 10 μm.
Table 4. Quantification of the effect of MT1 3′-UTR on perinuclear localization of chimaeric β-globin transcripts.
Cells expressing constructs with the β-globin coding sequences linked either to the vector 3′-UTR (pcGlo), to a 40 nt section of the MT1 3′-UTR (pc-Glo-MT40) or the same 40 nt section with an 11 nt deletion (pcGlo-MT40-Δ66-76), were subjected to in situ hybridization. At least 100 cells were examined for perinuclear localization or lack of localization. Experiments were repeated at least three times and the percentage of localization is expressed as means±S.E.M.
| Construct | Cells showing perinuclear localization by in situ hybridization (%) |
|---|---|
| PcGlo | 13±5 |
| pcGlo-MT40 | 43±7 |
| pcGlo-MT40-Δ66-76 | 10±3 |
| pcGlo-Ins66-76 | 33±6 |
Site-directed mutagenesis was used to produce a chimaeric β-globin/MT1 3′-UTR construct in which the sequence of nt 66–76 was deleted (pcGlo-MT40Δ66-76; see Figure 1). In situ hybridization of cells expressing this construct showed diffuse staining in the nucleus and throughout the cytoplasm, with no evidence of a ring of perinuclear staining (Figure 5D). Quantification of the staining by assessing the proportion of cells exhibiting a ring of perinuclear staining confirmed the loss of β-globin transcript localization in these cells (see Table 4), again showing the importance of this 11 nt sequence in the determination of perinuclear localization. To address whether this 11 nt sequence was sufficient to localize β-globin transcripts, site-directed mutagenesis was used to produce a construct in which nt 66–76 from the MT1 3′-UTR were inserted into the β-globin 3′-UTR (pcGlo-Ins66-76; see Figure 1). In situ hybridization using a probe to β-globin showed that many of the cells expressing this construct exhibited a distinct ring of perinuclear staining characteristic of perinuclear mRNA localization; an example is shown in Figure 5(C). Quantification of the staining distribution showed that only 33% of the cells exhibited localization of β-globin transcripts (Table 4), indicating that nt 66–76 from the MT1 3′-UTR are capable of directing localization of the β-globin reporter transcripts to some extent, but not as effectively as the full 3′-UTR.
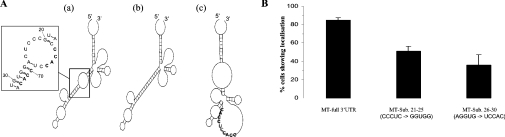
The 66–76 sequence contains a CACC repeat. The observation that only 33% of the cells expressing pcGlo-Ins66-76 and 43% of cells expressing pcGlo-MT40 showed localization suggests that the context of this sequence and nt 44–86, possibly due to secondary structure, is important. Computer simulation of the secondary structure of the MT1 3′-UTR (Figure 6) suggests that the region 66–76 is part of a stem and internal loop structure formed with region 21–30. Further constructs were made to test whether the structural context of nt 66–76 was important for the localization. As shown in Figure 6(A), mutation of nt 21–25 from CCCUC to GGUGG is predicted to result in part of the CACC repeat no longer being single-stranded but forming a double-stranded structure. In addition, the mutation of nt 26–30 from AGGUG to UCCAC is also predicted to alter the secondary structure in this region (Figure 6A). The staining pattern obtained from immunocytochemistry of cells expressing these constructs was quantified using the presence of a ring of perinuclear staining as indicative of localization. The results obtained showed that, after the mutation of nt 21–25, only 50% of cells exhibited perinuclear localization of MT1, and similarly, after the mutation of nt 26–30, only 40% of cells exhibited localization (Figure 6B). The results from cells expressing these two constructs indicate that both mutations impair, but do not destroy, the localization activity of the MT1 3′-UTR, and they are consistent with a model in which the secondary structure contributes to the effectiveness of the localization signal.
Figure 6. Sequence substitutions that are predicted to affect the secondary structure of the 66–76 region cause loss of localization.
(A) Prediction using the Mfold program of the secondary structure of the wild-type MT1 3′-UTR (a) and after substitution of nt 21–25 (b) or 26–30 (c). (B) Quantification of immunocytochemistry. At least 100 cells were examined for perinuclear localization of MT1 and scored as showing localization or not. Results are shown from four separate experiments and the percentage of cells exhibiting localization is shown as means±S.E.M.
DISCUSSION
In mammalian cells, 3′-UTR signals are responsible for sorting of mRNAs such that β-actin mRNA is localized to the cell periphery and c-myc, c-fos, vimentin and MT1 mRNAs to the perinuclear cytoskeleton [9,12,13,22]. The present study provides the first information on the nature of the localization signal within the MT1 3′-UTR. The data implicating 11 nt within a 40 nt region of MT1 3′-UTR give a more detailed picture of a perinuclear localization signal than that available from studies of the c-myc, c-fos and vimentin 3′-UTRs in which the signals appear present within 86, 145 or 100 nt but in which the precise signals have not been defined.
Four approaches were used to narrow down the region of the MT1 3′-UTR responsible for perinuclear localization: deletion analysis, mutagenesis, antisense ODNs and reporter studies. Since MT1 protein localization has been shown to depend on mRNA localization [13,15], protein distribution as assessed by immunocytochemistry was used as an indirect measure of mRNA localization. In addition, effects on mRNA localization were confirmed by in situ hybridization. Truncation of the MT1 3′-UTR to the first 86 nt had no effect on mRNA or protein distribution, but deletion of a further 41 nt showed that the deletion of the region between nt 45 and 86 led to delocalization of MT1 protein and mRNA. These results indicate that the localization signal lies partly or wholly within this region. Using β-globin as a reporter, in situ hybridization studies showed that the addition of nt 44–83 from the MT1 3′-UTR were sufficient to localize these transcripts to the perinuclear cytoplasm. Together, these results indicate that the localization signal lies within the region between nt 44 and 83.
Analysis of cells transfected with MT1 constructs with different truncated 3′-UTRs or with deletions created by mutagenesis further narrowed down the region containing a perinuclear localization element. Truncation of the 3′-UTR to the first 76 nt had no effect on MT1 mRNA or protein localization, but truncation to the first 70 nt led to a very large loss in localization. Similarly, deletion of nt 66–76 led to a loss of localization of either MT1 mRNA or the β-globin reporter driven by the MT1 3′-UTR, whereas the deletion of nt 76–86 had essentially no effect. Thus these further deletion studies point to the region between 66 and 76 as being particularly important in the localization signal. They may not represent the whole signal but they are essential for localization.
Further analysis of this region was investigated by using antisense ODNs. Such an approach is based on the idea that ODNs complementary to the region involved in localization will hybridize with that part of the transcript and prevent localization by disrupting the localization signal or protein binding to it. Incubation of cells with a 20-mer ODN complementary to nt 66–86 led to loss of MT1 localization, but incubation with an ODN complementary to nt 45–66 had no effect. Using shorter ODNs, those complementary to 70–86 caused almost total loss of localization and those complementary to 54–70 or 61–75 had a smaller but significant effect. These results are consistent with the region 66–76 being important in localization. However, an ODN complementary to 76–92 also caused delocalization and, at present, we have no clear explanation for this effect, since deletion studies show region 0–76 to be sufficient for localization and removal of 76–86 to have no effect. One explanation may be that binding of this particular ODN complementary to nt 76–92 has secondary effects, such as an induction of a change in RNA structure, which interferes with the RNA signal. Overall, with this one exception, the results from the ODN experiments are consistent with the deletion studies and support them in indicating the importance of the 44–76 region, particularly 66–76, in the perinuclear localization signal.
The sequence between 66 and 76 is ACACCUCCACC, containing a CACC repeat. A recent bioinformatic analysis has suggested that CAC repeats may be a common feature of localization signals, particularly in mRNAs localized in Xenopus oocytes but also including that of β-actin mRNA [20]. In situ hybridization studies showed that the insertion of ACACCUCCACC, corresponding to nt 66–76 from the MT1 3′-UTR, into the β-globin 3′-UTR was sufficient to cause localization of the β-globin reporter transcripts to the perinuclear cytoplasm. This observation, together with the other deletion, mutagenesis and ODN data, indicates that the 11 nt containing the CACC repeat are a critical part of the localization signal. However, quantification of the in situ hybridization data showed that either these 11 nt or the region 44–83 produces localization in only 33–43% of cells, indicating that these sections of the 3′-UTR are not as effective as when present in the context of the full MT1 3′-UTR. In turn, this suggests that another factor, such as secondary structure, contributes to the localization signal. Computer simulation of the secondary structure using the Mfold program suggests a complex structure for the rat MT1 3′-UTR with nt 44–83 being part of stems, loops and bulges (Figure 6A): on such a model, the first CACC motif is predicted to be in a stem and the second to form part of an internal loop (Figure 6A), a configuration comparable with that proposed for the β-actin mRNA in which the Zipcode-binding protein ZBP1 binds the CAC-containing motif present in the loop [17]. Mutagenesis of the regions 21–25 and 26–30 in the MT1 3′-UTR, which are predicted to alter the secondary structures containing the CACC repeat, was found to reduce the proportion of cells exhibiting perinuclear localization of MT1 (Figure 6B). These results would suggest that these mutations affect the effectiveness of the localization signal such that it can only direct localization in cells under a more limited range of cell conditions, and as a result localization is observed in a smaller proportion of cells. In turn therefore, the results suggest that the structural context of the CACC repeat contributes to the localization signal. Our hypothesis is that the perinuclear localization of MT1 mRNA is driven by a localization element in which the CACC repeat between nt 66–76 and secondary structure are both critical features. This is currently being tested using a combination of chemical and enzymic cleavage analysis of the secondary structure of MT1 3′-UTR and deletion/mutagenesis.
Acknowledgments
D. N. received a Faculty Research Studentship from University of Newcastle, and M. L.-M. and H. C. were supported by University of Newcastle and BBSRC (Biotechnology and Biological Sciences Research Council) respectively. We thank Dr H. Vasconcelos for advice on the experiments with oligonucleotides. The work was supported by BBSRC (grant 13/C14236).
References
- 1.Jansen R.-P. RNA-cytoskeletal associations. FASEB J. 1999;13:455–466. [PubMed] [Google Scholar]
- 2.Tekotte H., Davis I. Intracellular mRNA localization: motors move messages. Trends Genet. 2002;18:636–642. doi: 10.1016/s0168-9525(02)02819-6. [DOI] [PubMed] [Google Scholar]
- 3.Hesketh J. Translation and the cytoskeleton: a mechanism for targeted protein synthesis. Mol. Biol. Rep. 1994;19:233–243. doi: 10.1007/BF00986965. [DOI] [PubMed] [Google Scholar]
- 4.Ainger K., Avossa D., Morgan F., Hill S. J., Barry C., Barbarese E., Carson J. H. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J. Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassell G. J., Zhang H., Byrd A. L., Femino A. M., Singer R. H., Taneja K. L., Lifshitz L. M., Herman I. M., Kosik K. S. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J. Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kislauskis E. H., Li Z., Singer R. H., Taneja K. L. Isoform-specific 3′-untranslated sequences sort alpha-cardiac and beta-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J. Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hesketh J. E., Campbell G. P., Whitelaw P. F. c-myc mRNA in cytoskeletal-bound polysomes in fibroblasts. Biochem. J. 1991;274:607–609. doi: 10.1042/bj2740607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hesketh J., Campbell G., Piechaczyk M., Blanchard J.-M. Targeting of c-myc and beta-globin coding sequences to cytoskeletal-bound polysomes by c-myc 3′ untranslated region. Biochem. J. 1994;298:143–148. doi: 10.1042/bj2980143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalgleish G., Veyrune J.-L., Blanchard J.-M., Hesketh J. mRNA localization by a 145-nucleotide region of the c-fos 3′-untranslated region. Links to translation but not stability. J. Biol. Chem. 2001;276:13593–13599. doi: 10.1074/jbc.M001141200. [DOI] [PubMed] [Google Scholar]
- 10.Mahon P., Beattie J., Glover L. A., Hesketh J. Localisation of metallothionein isoform mRNAs in rat hepatoma (H4) cells. FEBS Lett. 1995;373:76–80. doi: 10.1016/0014-5793(95)01000-5. [DOI] [PubMed] [Google Scholar]
- 11.Tsujikawa K., Imai T., Kakutani M., Kayamori Y., Mimura Y., Otaki N., Kimura M., Fukuyama R., Shimizu N. Localization of metallothionein in nuclei of growing primary cultured adult rat hepatocytes. FEBS Lett. 1991;283:239–242. doi: 10.1016/0014-5793(91)80597-v. [DOI] [PubMed] [Google Scholar]
- 12.Mahon P., Partridge K., Beattie J. H., Glover L. A., Hesketh J. E. The 3′ untranslated region plays a role in the targeting of metallothionein-I mRNA to the perinuclear cytoplasm and cytoskeletal-bound polysomes. Biochim. Biophys. Acta. 1997;1358:153–162. doi: 10.1016/s0167-4889(97)00058-x. [DOI] [PubMed] [Google Scholar]
- 13.Levadoux M., Mahon C., Beattie J. H., Wallace H. M., Hesketh J. E. Nuclear import of metallothionein requires its mRNA to be associated with the perinuclear cytoskeleton. J. Biol. Chem. 1999;274:34961–34966. doi: 10.1074/jbc.274.49.34961. [DOI] [PubMed] [Google Scholar]
- 14.Oleynikov Y., Singer R. H. RNA localization: different zipcodes, same postman? Trends Cell Biol. 1998;8:381–383. doi: 10.1016/s0962-8924(98)01348-8. [DOI] [PubMed] [Google Scholar]
- 15.Levadoux-Martin M., Hesketh J. E., Beattie J. H., Wallace H. M. Influence of metallothionein-1 localization on its function. Biochem. J. 2001;355:473–479. doi: 10.1042/0264-6021:3550473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farina K. L., Huttelmaier S., Musunuru K., Darnell R., Singer R. H. Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment. J. Cell Biol. 2003;160:77–87. doi: 10.1083/jcb.200206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross A. F., Oleynikov Y., Kislauskis E. H., Taneja K. L., Singer R. H. Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kislauskis E. H., Zhu X., Singer R. H. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J. Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bubunenko M., Kress T. L., Vempati U. D., Mowry K. L., King M. L. A consensus RNA signal that directs germ layer determinants to the vegetal cortex of Xenopus oocytes. Dev. Biol. 2002;248:82–92. doi: 10.1006/dbio.2002.0719. [DOI] [PubMed] [Google Scholar]
- 20.Betley J. N., Frith M. C., Graber J. H., Choo S., Deshler J. O. A ubiquitous and conserved signal for RNA localization in chordates. Curr. Biol. 2002;12:1756–1761. doi: 10.1016/s0960-9822(02)01220-4. [DOI] [PubMed] [Google Scholar]
- 21.Veyrune J.-L., Campbell G. P., Wiseman J., Blanchard J.-M., Hesketh J. E. A localisation signal in the 3′ untranslated region of c-myc mRNA targets c-myc mRNA and beta-globin reporter sequences to the perinuclear cytoplasm and cytoskeletal-bound polysomes. J. Cell Sci. 1996;109:1185–1194. doi: 10.1242/jcs.109.6.1185. [DOI] [PubMed] [Google Scholar]
- 22.Bermano G., Shepherd R. K., Zehner Z. E., Hesketh J. E. Perinuclear mRNA localisation by vimentin 3′-untranslated region requires a 100 nucleotide sequence and intermediate filaments. FEBS Lett. 2001;497:77–81. doi: 10.1016/s0014-5793(01)02438-3. [DOI] [PubMed] [Google Scholar]
- 23.Bermano G., Hesketh J. E. The study of mRNA-cytoskeleton interactions and mRNA sorting in mammalian cells. In: Carraway K. L., Carraway C. A. C., editors. Cytoskeleton Signalling and Cell Regulation: A Practical Approach. Oxford University Press; 1999. pp. 209–244. [Google Scholar]