Abstract
MT1-MMP (membrane type 1 matrix metalloproteinase) is a membrane-anchored MMP that can be shed to the extracellular milieu. In the present study we report the primary structure and activity of the major soluble form of MT1-MMP. MS analysis of the purified 50-kDa soluble MT1-MMP form shows that the enzyme extends from Tyr112 to Val524, indicating that formation of this species requires a proteolytic cleavage within the stem region. In agreement, deletion of the entire stem region of MT1-MMP inhibited shedding of the 50-kDa species. A recombinant 50-kDa species (Tyr112–Val524) expressed in cells exhibited enzymatic activity against pro-MMP-2 and galectin-3, and thus this species is a competent protease. The recombinant 50-kDa soluble form also decreased the level of surface-associated TIMP-2 (tissue inhibitor of metalloproteinase 2) when administered to cells expressing wild-type membrane-anchored MT1-MMP, suggesting that ectodomain shedding of MT1-MMP can alter the MMP/TIMP balance on the cell surface. A ∼53-kDa species of MT1-MMP was also isolated from a non-detergent extract of human breast carcinoma tissue and was found to lack the cytosolic tail, as determined with specific MT1-MMP domain antibodies. Together, these data show that MT1-MMP ectodomain shedding is a physiological process that may broaden MT1-MMP activity to the pericellular space.
Keywords: ectodomain, matrix metalloproteinase (MMP), shedding, protease, proteolysis, tissue inhibitor of metalloproteinase (TIMP)
Abbreviations: CB, collagenase buffer; Con A, concanavalin A; DMEM, Dulbecco's modified Eagle's medium; HRP, horseradish peroxidase; mAb, monoclonal antibody; MMP, matrix metalloproteinase; MS/MS, tandem MS; MT-MMP, membrane type-MMP; pAb, polyclonal antibody; TAPI-1, tumour necrosis factor-α protease inhibitor-1; TIMP, tissue inhibitor of metalloproteinase
INTRODUCTION
MT1-MMP (membrane type 1 matrix metalloproteinase) is a type I membrane-anchored MMP that has been shown to play a critical role in the pericellular degradation of connective tissue matrix by directly hydrolysing native collagen I [1–3], and indirectly by initiating a cascade of zymogen activation at the cell surface, which leads to the generation of active MMP-2 (gelatinase A) [4], MMP-13 (collagenase-3) [5,6] and MMP-9 [6,7]. MT1-MMP also degrades a variety of surface-associated proteins, including growth factors, cytokines and cell adhesion receptors [8–11], indicating that MT1-MMP is a major mediator of surface proteolysis. In agreement with this property, MT1-MMP has been shown to be involved in various pathological and physiological processes, including cancer metastasis, angiogenesis, arthritis and bone development [12,13]. Structurally, MT1-MMP possesses all of the classic MMP domains, including a pro-peptide, a zinc-containing catalytic domain, a hinge region and a haemopexin-like domain. As a membrane-anchored protein, MT1-MMP contains a transmembrane domain comprising a stretch of ∼20 hydrophobic amino acids followed by a short cytoplasmic tail [14]. Between the transmembrane domain and the haemopexin-like domain there is a short stretch of residues which constitute a so-called ‘stem’ region of unknown function [15]. Similar to all MMPs, MT1-MMP is inhibited by the TIMPs (tissue inhibitors of metalloproteinases). However, MT1-MMP discriminates among the members of the TIMP family, with TIMP-2, TIMP-3 and TIMP-4 being high-affinity inhibitors, whereas TIMP-1 is the weakest MT1-MMP inhibitor [16]. The interactions of MT1-MMP with TIMP-2 go beyond inhibition of catalytic activity, and in some instances may promote proteolysis. The complex of MT1-MMP with TIMP-2 can act as a cell surface ‘receptor’ for pro-MMP-2, which is then activated by a neighboring TIMP-2-free MT1-MMP [4]. In addition, TIMP-2 inhibits the autocatalytic turnover of MT1-MMP at the cell surface, which also promotes MT1-MMP-dependent activity [16,17].
As a membrane-anchored protein, MT1-MMP exhibits a unique mechanism of regulation involving processing and shedding, which regulate the level of mature MT1-MMP at the cell surface and generate MT1-MMP forms with different functions and cellular distribution. MT1-MMP processing is an autocatalytic event, which leads to the generation of a major 44-kDa membrane-anchored degradation product, extending from Gly285 to Val582 and thus lacking the catalytic domain and the release of an inactive 18-kDa soluble fragment, starting at Tyr112 and ending at Ala255, into the extracellular space [18–21]. The remaining membrane-anchored 44-kDa form of MT1-MMP, which contains the haemopexin-like domain, has been shown to maintain binding to collagen and thus may influence the activity of the mature MT1-MMP [22]. The soluble 18-kDa fragment is generated by two cleavage events, firstly at the Gly284–Gly285 peptide bond and then at the Ala255–Ile256 site. The latter is an amino acid upstream of the conserved methionine turn [14], and thus cleavage at this site results in inactivation of enzymatic activity and inability to bind TIMP-2. The autocatalytic processing of MT1-MMP therefore might have evolved to terminate MT1-MMP activity both at the cell surface and in the pericellular space.
Mature MT1-MMP is also shed from the cell surface via a non-autocatalytic process that results in the release of various soluble forms [21], and, as opposed to the autocatalytic processing, it generates active soluble fragments. MT1-MMP shedding has been described in cultures of human mesangial [23], breast carcinoma [21,24,25], lung fibroblast [25], endothelial [26,27] and fibrosarcoma [21] cells. Soluble MT1-MMP forms were also detected in human sputum and bronchoalveolar lavage fluid [28], suggesting that shedding occurs in vivo and may represent a physiologically relevant process. Mammalian cells that are engineered to express recombinant MT1-MMP also shed MT1-MMP into the supernatant [11,19,21]. The non-autocatalytic shedding of MT1-MMP produces a major soluble form ranging from ∼50–52 kDa and a minor form of ∼25–32 kDa. On the basis of the molecular mass, the 50–52-kDa species must represent shedding of the entire ectodomain including the catalytic domain [19,21,24–27]. In agreement, media from the cells containing the 50–52-kDa soluble species of MT1-MMP promoted processing of pro-MMP-2 [21] and cleaved fibrinogen [27]. However, the precise nature of this species is unknown. In the present study we aimed to identify and to characterize the primary structure and activity of the ∼52–50-kDa soluble form of MT1-MMP and to identify equivalent species in breast carcinoma tissues. We report the primary structure and activity of the 50-kDa soluble form of MT1-MMP, which is a fully functional enzyme that is present in vivo and that has the potential to play a role in pericellular proteolysis.
EXPERIMENTAL
Cell culture
Non-malignant monkey kidney epithelial BS-C-1 (CCL-26) and CV-1 (CCL-70) cells and human fibrosarcoma HT-1080 (CCL-121) cells were obtained from the A.T.C.C. BS-C-1, CV-1 and HT1080 cells were cultured in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% FBS (fetal bovine serum) and antibiotics (100 units/ml penicillin G sodium and 100 μg/ml streptomycin sulphate). Human HeLa S3 cells were purchased from the A.T.C.C. (CCL-2.2) and grown in suspension in MEM Spinner medium (Quality Biologicals, Gaithersburg, MD, U.S.A.) supplemented with 5% horse serum and antibiotics (100 units/ml penicillin G sodium and 100 μg/ml streptomycin sulphate).
Recombinant vaccinia viruses
The production of the recombinant vaccinia virus (vTF7-3) expressing bacteriophage T7 RNA polymerase has been described by Fuerst et al. [29]. Recombinant vaccinia viruses expressing MT1-MMP (vT7-MT1), pro-MMP-2 (vT7-GelA), TIMP-1 (vT7-T1) or TIMP-2 (vSC59-T2) were generated by homologous recombination, as described previously [29].
Recombinant proteins, antibodies and protease inhibitors
Human recombinant pro-MMP-2, TIMP-2 and TIMP-1 were expressed in HeLa S3 cells infected with the appropriate recombinant vaccinia viruses and purified to homogeneity from the medium, as described previously [30]. Human recombinant galectin-3 and the rabbit polyclonal anti-galectin-3 antibody were gifts from Dr A. Raz (Karmanos Cancer Institute, Detroit, MI, U.S.A.). The rabbit pAb (polyclonal antibody) against the haemopexin-like domain of MT1-MMP (pAb437) [20] and the mAb (monoclonal antibody) against the catalytic domain of MT1-MMP (LEM-2/15) [27], a gift from Dr A. Arroyo (Hospital de la Princesa, Madrid, Spain), have been described previously. The rabbit pAb to the cytosolic tail of MT1-MMP (pAbCT) was purchased from Triple Point Biologics (Forest Grove, OR, U.S.A.). Marimastat was obtained from British Biotech (Oxford, U.K.). TAPI-1 (tumour necrosis factor-α protease inhibitor-1) was purchased from EMD Biosciences (San Diego, CA, U.S.A.).
Expression of wild-type MT1-MMP
Expression of wild-type MT1-MMP in BS-C-1 and CV-1 cells was carried out by co-infecting the cells with the vTF7-3 and vTF-MT1 vaccinia viruses, as described previously [20]. In some experiments with the BS-C-1 cells, 3 h post-infection the media were replaced with serum-free DMEM supplemented with 2 μM TAPI-1 final concentration followed by overnight incubation. Then, the serum-free conditioned media of the co-infected cells were collected, clarified and concentrated on a Centricon Plus-80 concentrator for further analyses. To remove TAPI-1, the media of infected BS-C-1 cells were dialysed against collagenase buffer (CB) [50 mM Tris/HCl, pH 7.5, 150 NaCl, 5 mM CaCl2 and 0.2% Brij-35]. In some experiments, these media samples were further centrifuged (100000 g for 1 h) to remove any membrane fragments. The membrane pellet was solubilized in reducing sample buffer and resolved by SDS/PAGE followed by immunoblot analysis. The collected media and the membrane pellet were subjected to either immunoblot analyses using antibodies to various MT1-MMP domains, as described previously [20], or to isolation of soluble MT1-MMP species as described below.
MMP inhibitor affinity purification of soluble MT1-MMP forms
The hydroxamate-based broad-spectrum MMP inhibitor batimastat (BB-94) [31] was synthesized in 11 synthetic steps, and was covalently coupled to epoxy-activated Sepharose® 6B (Sigma–Aldrich, St. Louis, MO, U.S.A.) via a 10-membered linker [see Supplementary Figure 1 for the structure of the inhibitor-tethered resin (http://www.BiochemJ.org/bj/387/bj3870497add.htm)]. A manuscript describing the synthesis of the modified inhibitor used as the affinity ligand is in preparation (D. Hesek, M. Toth, S. Brown, S. O. Meroueh, H. Zhao, W. Sakr, V. Krchnak, S. Mobashery and R. Fridman, unpublished work). The resultant inhibitor-tethered matrix was equilibrated in CB and stored at 4 °C until use. Serum-free media containing soluble MT1-MMP forms or tissue extracts were incubated (12 h, 4 °C) on a rocker with the inhibitor-tethered matrix. In some experiments, the mixture of media and inhibitor-tethered matrix was supplemented with 100 nM of either TIMP-2 or TIMP-1. After a brief centrifugation, the supernatants were collected (unbound fraction) and the inhibitor-tethered matrix was washed first with CB followed by CB supplemented with 1 M NaCl. The bound proteins (bound fraction) were eluted with reducing SDS sample buffer or with 25 μM marimastat in CB. The bound and unbound fractions were resolved by reducing SDS/PAGE, followed by immunoblot analysis using domain specific anti-MT1-MMP antibodies.
Sequence of MT1-MMP ectodomain
BS-C-1 cells were co-infected to express MT1-MMP and the serum-free media were collected and subjected to the inhibitor-tethered matrix purification as described above. The bound proteins were eluted with SDS sample buffer, subjected to reducing SDS/PAGE, transferred on to a PVDF membrane and stained with Coomassie Blue R-250. An aliquot of the samples was subjected to immunoblot analysis to identify the MT1-MMP forms. The identified ∼50-kDa band was excised and sent for N-terminal microsequencing to ProSeq (Boxford, MA, U.S.A.). To determine the full sequence of the 50-kDa species, the corresponding protein band was excised from the Coomassie Blue-stained gel and sent to the Harvard Microchemistry Facility. The protein sample was digested with trypsin and AspN separately and combined for sequence analysis by microcapillary reverse-phase HPLC nano-electrospray MS/MS (tandem MS) on a Finnigan LCQ DECA XP quadropole ion trap mass spectrometer. The resulting MS/MS spectra of the peptides were correlated with database using the algorithm SEQUEST and programs developed in the Facility [32]. The MS/MS peptide sequences were reviewed for consensus with known proteins and the result manually confirmed for fidelity. The peptides covered 72% of the active MT1-MMP species by amino acid count.
Generation and expression of MT1-MMP mutants
To generate the 50-kDa (Tyr112–Val524) soluble species of MT1-MMP, Ile525 was substituted with a termination codon using the QuikChange® site-directed mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.) in the pTF7EMCV-1-MT1 expression vector containing the full-length human pro-MT1-MMP [20]. Deletion of the entire stem region (ΔPro509–Gly535) to generate Δstem-MT1 was carried out by mutagenic PCR using the same kit. The MT1-MMP mutants containing a single Ile525→Phe or a double Val524→Glu/Ile525→Glu substitution within the stem region were also generated with the QuikChange® site-directed mutagenesis kit and pTF7EMCV-1-MT1 as a template. The fidelity of the constructs was verified by DNA sequencing in both directions. The pTF7EMCV-1-MT1 expression vectors encoding wild-type and mutant MT1-MMP forms were expressed in CV-1 cells using the infection/transfection procedure of the vaccinia expression system, as described previously [20,33]. After infection/transfection (18 h), the serum-free media were collected, clarified and concentrated with a Microcon YM-10 centrifugal filter device. The cells were solubilized in cold lysis buffer (25 mM Tris/HCl, pH 7.5, 1% Nonidet P40 and 100 mM NaCl with protease inhibitors), centrifuged at 4 °C (15000 g, 30 min) and the supernatant (lysate) was collected. The lysate and concentrated media samples were subjected to immunoblot analyses using domain-specific anti-MT1-MMP antibodies.
MT1-MMP activity assays
To examine the activity of the soluble recombinant 50-kDa (Tyr112–Val524) species, CV-1 cells in six-well plates were infected/transfected with vTF7-3 vaccinia virus and pTF7EMCV-1-MT1-50 to express the soluble 50-kDa (Tyr112–Val524) as described previously [20,33]. Control cells were infected with same amounts of vTF7-3, but received no plasmid DNA. After 18 h, the serum-free media were clarified by centrifugation, concentrated (10×) with a Microcon YM-10 centrifugal filter device and analysed for MT1-MMP activity by the MMP-14 Biotrak activity assay system (RPN 2637, Amersham Biosciences, Piscataway, NJ, U.S.A.) according to the manufacturer's intructions. Briefly, MT1-MMP species in biological samples are captured by a specific MT1-MMP antibody immobilized on a microtitre plate. The immobilized MT1-MMP is then incubated with a modified pro-urokinase detection enzyme, which can only be activated by active MT1-MMP. The activated detection enzyme then cleaves a chromogenic peptide substrate (S-2444™) and the resultant colour was monitored at 405 nm in a spectrophotometer. The concentration of active MT1-MMP in the sample is then determined by interpolation from a standard curve of a soluble recombinant MT1-MMP, provided in the kit, and is reported as ng/ml of active MT1-MMP. In some experiments, CV-1 cells infected/transfected to express 50-kDa (Tyr112–Val524) species or wild-type MT1-MMP, received either 10 nM pro-MMP-2 or 20 nM galectin-3 in serum-free media, 4 h after infection/transfection. After overnight incubation at 37 °C, aliquots of the conditioned media were analysed by gelatin zymography, as described [20], or immunoblotting using an antibody to galectin-3.
Analysis of MT1-MMP forms shed by HT1080 cells
HT1080 cells grown in 150 mm tissue culture dishes were untreated or treated overnight with either 10 μg/ml Con A (concanavalin A) or 100 nM phorbol ester (PMA) in serum-free media. The conditioned media were collected and clarified by centrifugation and subjected to purification by the inhibitor-tethered matrix as described above. The bound proteins were eluted with SDS sample buffer and subjected to immunoblot analysis. To label cell surface proteins, confluent HT1080 cells in 150 mm dishes were untreated or treated with Con A (10 μg/ml) in serum-free media overnight and then surface biotinylated with 0.5 mg/ml sulpho-N-hydroxysuccinimido-biotin as described previously [21]. The surface-biotinylated cells were cultured overnight in serum-free media to allow MT1-MMP shedding. The conditioned media were collected, clarified by centrifugation and subjected to inhibitor-tethered affinity purification as described above. Aliquots of the eluates were subjected to SDS/PAGE followed by transfer on to a nitrocellulose membrane and detection of biotinylated proteins with streptavidin/HRP (horseradish peroxidase) and enhanced chemiluminescence. The positive bands were further identified with anti-MT1-MMP antibodies by immunoblot analysis.
Analysis of TIMP-2 cellular distribution
CV-1 cells in six-well plates were infected/transfected to express wild-type MT1-MMP as described previously [33]. As a control, some cells were infected with the vaccinia virus, but received no plasmid DNA. After 4 h, the media was changed and replaced with serum-free media containing 3.4 nM of recombinant human TIMP-2 in the presence or absence of media containing ∼50 ng of recombinant 50-kDa (Tyr112–Val524) MT1-MMP, which was previously produced as described above. The amount of soluble 50-kDa (Tyr112–Val524) MT1-MMP species was determined by the MMP-14 Biotrak activity assay system. After 16 h media were collected and clarified by centrifugation. The cells were rinsed twice with PBS, solubilized with cold lysis buffer and centrifuged. The lysate and media samples were analysed for TIMP-2 concentration by ELISA (QIA40, Oncogene Research Products, San Diego, CA, U.S.A.) as described by the manufacturer.
Identification of MT1-MMP in benign and malignant human breast tissue
Samples of benign and malignant breast tissue obtained from invasive breast carcinomas were collected from excess tissue sent for histopathological diagnosis to the Department of Pathology at Harper Hospital (Detroit, MI, U.S.A.). A total of six tumour samples of invasive breast carcinoma tissue, each from a different patient, were collected. Benign tissue was also collected from adjacent areas of the same tumour samples. The six malignant and benign tissue samples were pooled and homogenized in cold homogenization buffer (25 mM Tris/HCl pH 7.5, 8.5% sucrose, 50 mM NaCl and protease inhibitors) and the extracts were centrifuged several times. To reduce lipid contamination, the supernatants were filtered through a 0.45 μm membrane filter and ultracentrifuged to pellet all insoluble particles. The total protein concentration was measured and adjusted with homogenization buffer. Equal protein amounts of benign and tumour tissue extracts were incubated with 100 μl of the inhibitor-tethered matrix with constant rotation at 4 °C overnight. The bound proteins were eluted with 25 μM marimastat in CB as described above. Aliquots of the eluates were subjected to SDS/PAGE and the MT1-MMP forms were detected by immunoblot analysis using domain-specific anti-MT1-MMP antibodies.
RESULTS
Soluble MT1-MMP forms
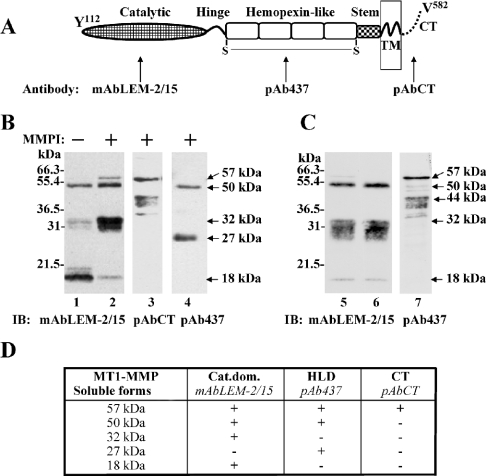
BS-C-1 cells were infected to express human MT1-MMP and then treated or untreated with TAPI-1, a metalloproteinase inhibitor. The media were then analysed for the presence of MT1-MMP soluble forms by immunoblot analysis using anti-MT1-MMP antibodies recognizing different enzyme domains (Figure 1A). As shown in Figure 1(B), media of untreated cells analysed with a mAb to the catalytic domain (mAbLEM-2/15) contained three soluble MT1-MMP species of ∼50, 32 and 18 kDa (Figure 1B, lane 1). TAPI-1 treatment of the cells inhibited the shedding of the 18-kDa form, which is in agreement with this species being the product of autocatalytic processing [18,21] (Figure 1B, lane 2). Also, in the presence of TAPI-1, higher amounts of the 32-kDa fragment, which sometimes appeared as a smear, were detected in the media (Figure 1B, lane 2). The amount of the 50-kDa species also slightly increased in the presence of TAPI-1 when compared with untreated cells. A ∼57-kDa species was also detected in the presence of TAPI-1 (Figure 1B, lane 2). A pAb against the cytosolic tail (pAbCT) of MT1-MMP recognized the 57-kDa species and a series of ∼44– 40-kDa fragments (Figure 1B, lane 3), but not the 50-, 32- and 18-kDa species, suggesting that the presence of 57- and 44– 40-kDa fragments in the media represent membrane-anchored fragments shed in membrane vesicles. In agreement with this assertion, ultracentrifugation of the media (Figure 1C, lane 5) derived from TAPI-1-treated cells (to detect the 32-kDa species) showed that the 57- and 44– 40-kDa forms were found in the pellet fraction (Figure 1C, lane 7), whereas the 50-, 32- and 18-kDa species were detected in the supernatant (Figure 1C, lane 6). Note that the pellet fraction (Figure 1C, lane 7) was examined with the pAb against the haemopexin-like domain of MT1-MMP (pAb437) to identify the membrane-bound 44–40-kDa species lacking the catalytic domain [20], whereas the total medium sample (Figure 1C, lane 5) was examined with mAbLEM-2/15 against the catalytic domain and thus the 44–40-kDa species is not recognized in the total media sample (Figure 1C, lane 5). However, a small amount of the 57-kDa species was readily detectable in the total sample under these conditions (Figure 1C, lane 5). That the 57- and 44– 40-kDa detected in the pellet fraction are indeed present in the medium is clearly shown with the antibody against the cytosolic tail (Figure 1B, lane 3). Taken together, the ultracentrifugation data indicated that the 50-, 32- and 18-kDa species are truly soluble species of MT1-MMP.
Figure 1. Characterization of soluble MT1-MMP forms.
(A) Domain structure of active MT1-MMP with the domain specific antibodies used in the present study. (B) Media from BS-C-1 cells expressing MT1-MMP in the absence (lane 1) or in the presence of 2 μM TAPI-1 (lanes 2–4) were concentrated and subjected to SDS/PAGE (15% reducing gel) followed by immunoblot analysis using the mAbLEM-2/15 to the catalytic domain (lanes 1 and 2), pAbCT to cytoplasmic tail (lane 3) and pAb437 to the haemopexinlike domain (lane 4) of MT1-MMP. (C) Media of MT1-MMP expressing BS-C-1 cells obtained in the presence of 2 μM TAPI-1 were collected (lane 5) and subjected to ultracentrifugation (supernatant, lane 6; pellet, lane 7). The samples were subjected to SDS/PAGE (15% reducing gel) followed by immunoblot analysis using the mAbLEM-2/15 to the catalytic domain (lanes 5 and 6) and pAb437 to the haemopexin-like domain (lane 7). (D) Summary of the species of soluble forms of MT1-MMP detected in the media of BS-C-1 cells expressing MT1-MMP and their recognition by various anti-MT1-MMP antibodies. Cat.dom., catalytic domain; CT, C-terminus; HLD, haemopexin-like domain; TM, transmembrane.
The antibody pAb437 recognized the 50-kDa species and thus this form represents shedding of the entire ectodomain (Figure 1B, lane 4) in medium from TAPI-1-treated cells. This antibody also detected a minor soluble fragment of ∼27 kDa, which was present in the media regardless of TAPI-1 treatment (Figure 1B, lane 4). Since this form contains the haemopexin-like domain, the 27-kDa fragment is likely to be generated from shedding of the 44-kDa membrane bound form of MT1-MMP formed by autocatalytic processing of the 57-kDa active form [18,20]. Figure 1(D) summarizes all the species of MT1-MMP detected in the media and their recognition by various domain antibodies.
Isolation of the 50-kDa MT1-MMP soluble form
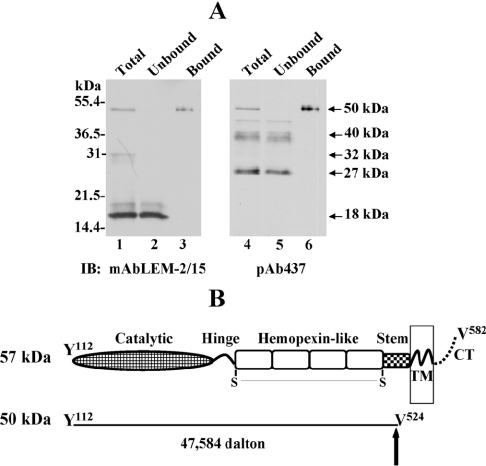
The characterization of the shed MT1-MMP forms with the domain antibodies suggested the possibility that both the 50- and the 32-kDa species represent catalytically competent enzymes, which upon shedding may act in the extracellular milieu. To establish the precise nature of these species, it was necessary to purify them and determine their N- and C-terminal ends. To this end, we have developed an MMP-inhibitor based affinity purification method in which the broad-spectrum inhibitor batimastat (BB-94) is tethered to a resin [see Supplementary Figure 1 (http://www.BiochemJ.org/bj/387/bj3870497add.htm)]. The inhibitor resin was designed to specifically bind active MMPs in biological fluids. The effectiveness of the affinity approach was first tested with media conditioned by surface-biotinylated HT1080 cells untreated or treated with PMA or Con A, two agents previously shown to promote MT1-MMP shedding [21,24]. After elution, bound biotinylated MT1-MMP forms were identified by streptavidin/HRP and mAb LEM-2/15 to the catalytic domain. The specificity of the avidin detection was determined using media from non-biotinylated HT1080 cells, whereas the specificity of the binding was established by competition with exogenous TIMPs (TIMP-2 and TIMP-1) using conditioned medium from BS-C-1 cells expressing MT1-MMP. As shown in Figure 2(A), the inhibitor-tethered matrix captured a ∼50–52-kDa species of MT1-MMP present in the media of PMA- or Con A-treated HT1080 cells, which was detected in the eluate by immunoblot analysis with the catalytic domain antibody (Figure 2A, lanes 1 and 2). Streptavidin/HRP detection of the fraction that had bound to the inhibitor-tethered resin showed the presence of a major biotinylated ∼50–52-kDa species (Figure 2B, lane 3), which was also identified with the LEM-2/15 mAb, consistent with being it MT1-MMP (Figure 2B, lane 8). The media from Con A-treated cells contained higher levels of the biotinylated ∼50–52-kDa species and two minor biotinylated species of ∼40 and ∼32 kDa, which bound to the inhibitor resin (Figure 2B, lane 4). Whereas the biotinylated ∼50–52-kDa species was recognized with LEM-2/15 antibody, the smaller species were not (Figure 2B, lane 8). The biotinylated ∼32-kDa species may be the 32-kDa soluble fragment of MT1-MMP (shown in Figures 1B and 1C), which due to its low amount and the difference in sensitivity of the detection methods could only be detected by streptavidin/HRP. The fractions derived from media of non-biotinylated HT1080 cells that had bound to the resin were not detected by streptavidin/HRP (Figure 2B, lanes 1 and 2), but the bound 50-kDa MT1-MMP species was identified by immunoblot analysis (Figure 2B, lane 6). demonstrating the specificity of the biotinylation and avidin detection procedures. Addition of recombinant TIMP-2 to the medium of BS-C-1 cells expressing MT1-MMP prior to the affinity purification blocked binding of the 50-kDa species to the inhibitor resin and the bulk of this form was recovered in the unbound fraction (Figure 2C, upper panel). In contrast, in the absence of TIMPs or in the presence of TIMP-1, the 50-kDa species was recovered in the bound fraction (Figure 2C, lower panel), indicating that the binding of MT1-MMP to the inhibitor-tethered matrix is specifically mediated by interactions with the active site. Taken together, these studies demonstrate that soluble active MT1-MMP forms, which are shed from the cell surface, can be captured with an immobilized MMP inhibitor and that they represent enzyme species with intact active sites.
Figure 2. MT1-MMP forms shed by surface biotinylated HT1080 cells.
(A) Serum-free conditioned media from PMA (‘TPA’)- (lane 1) or Con A-treated (lane 2) HT1080 cells were incubated with the MMP inhibitor-tethered matrix and the bound proteins were detected by immunoblot (IB) analysis using mAbLEM-2/15. (B) Untreated (lanes 1, 3, 5 and 7) or Con A-treated (lanes 2, 4, 6 and 8) HT1080 cells were surface biotinylated (lanes 3, 4, 7 and 8) or non-biotinylated (lanes 1, 2, 5 and 6). The serum-free conditioned media were collected, incubated with the inhibitor-tethered matrix and the bound fractions were subjected to SDS/PAGE (12% reducing gel) and transferred on to a nitrocellulose membrane. The same membrane was developed with streptavidin/HRP (lanes 1–4) and with mAbLEM-2/15 (lanes 5–8). (C) Serum-free media of MT1-MMP expressing BS-C-1 cells were incubated with the inhibitor-tethered matrix in the absence or presence of recombinant TIMP-2 or TIMP-1. The 50 kDa form of MT1-MMP in the unbound and bound fractions were detected by immunoblot analysis using the mAbLEM-2/15.
Next, we set out to purify sufficient soluble active MT1-MMP species for sequencing as described in the Experimental section. To this end, we used media of BS-C-1 cells infected to express MT1-MMP [21]. The immunoblot of Figure 3(A) shows the results of a representative purification procedure with the inhibitor-tethered matrix. The 50-kDa soluble species was recovered in the resin-bound fractions (Figure 3A, lanes 3 and 6), whereas the fragments containing a truncated (18 kDa) [21] or lacking (∼40 and ∼27 kDa) catalytic domain were recovered in the unbound fraction (Figure 3A, lanes 2 and 5 respectively). The 50-kDa species was isolated and first subjected to N-terminal sequencing by 11 cycles of Edman degradation. This study indicated an N-terminus starting at Tyr112, which is the consensus start of active MT1-MMP [20]. For determination of the C-terminal end of the soluble 50-kDa species, the purified enzyme was excised from the gel and sent for ion-trap MS/MS peptide sequencing. Two enzymes (trypsin and AspN) were used to digest the protein and the resulting peptides were analysed as described in the Experimental section. These analyses showed that the last detected amino acid, according to the spectra defining the C-terminal end of the 50-kDa form, was Val524, which is a non-specific site for both enzymes used for sample preparation indicating that the cleavage site in ectodomain shedding is at the V524–Ile525 site within the stem region [see Supplementary Figure 2 (http://www.BiochemJ.org/bj/387/bj3870497add.htm) for MS/MS data]. The soluble character of this protein, its electrophoretic mobility on SDS/PAGE and the lack of the cytosolic tail, as shown by immunoblot analysis with an antibody against the cytosolic tail of MT1-MMP (Figure 1B), also support this finding. Taken together, these data indicated that the 50-kDa soluble species of MT1-MMP starts at Tyr112 and extends up to Val524 in the stem region (Figure 3B).
Figure 3. Isolation and structure of the 50-kDa soluble form of MT1-MMP.
(A) Concentrated medium of MT1-MMP expressing BS-C-1 cells (lane 1 and 4) was incubated with the inhibitor-tethered matrix and the unbound (lanes 2 and 5) and bound fractions (lanes 3 and 6) were subjected to SDS/PAGE (12% reducing gel) followed by immunoblot (IB) analysis using mAbLEM-2/15 to the catalytic domain (lanes 1–3) or pAb437 to the haemopexin-like domain (lanes 4–6). (B) Structure of the 50-kDa soluble species of MT1-MMP as determined by MS/MS sequencing. The arrow indicates the putative site of cleavage at the stem region.
Shedding of the 50-kDa species requires the stem region
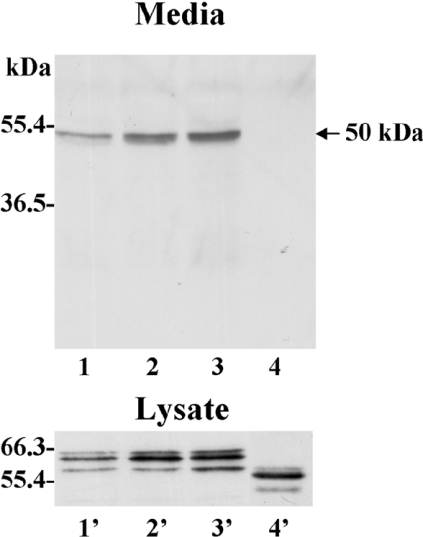
The sequencing data suggested that shedding of the 50-kDa species of MT1-MMP occurs at the Val524–Ile525 peptide bond within the stem region, which is located between the haemopexin-like domain and the transmembrane domain [15]. Therefore, we examined the effects of specific amino acid substitutions at the Val524–Ile525 site (Ile525→Phe and Val524→Glu/Ile525→Glu) and of a deletion of the entire stem region (Δstem-MT1, which lacks Pro509 to Gly535) on shedding of the 50-kDa species. Ile525 was replaced by phenylalanine to maintain the hydrophobic nature of the residue, but to introduce a bulky aromatic side chain to deter cleavage. The glutamate substitutions at both Val524 and Ile525 (double mutant) were generated in an attempt to mask the hydrophobicity of the cleavage site, which is flanked by several acidic residues [15]. These studies showed that the substitutions Ile525→Phe (Figure 4, lane 2) and Val524→Glu/Ile525→Glu (Figure 4, lane 3) at the Val524–Ile525 cleavage site had no effect on shedding of the 50-kDa species when compared with the wild-type enzyme (Figure 4, lane 1). In contrast, deletion of the entire stem region completely abrogated shedding of the 50-kDa species (Figure 4, lane 4) consistent with the cleavage being located within the stem region. Expression of these mutants was confirmed in the lysates by immunoblot analysis, showing that all forms were expressed (Figure 4, lysates). The reduced molecular mass of the Δstem-MT1 (Figure 4, lane 4′) compared with the wild-type enzyme (Figure 4, lane 1′) is consistent with the deletion of the entire stem region.
Figure 4. Mutations and truncations at the stem region of MT1-MMP: effect on shedding.
Concentrated media and cell lysates of CV-1 cells infected/transfected to express wild-type (lanes 1 and 1′), Ile525→Phe (lanes 2 and 2′), Val524→Glu/Ile525→Glu (lanes 3 and 3′) or Δstem (lanes 4 and 4′) MT1-MMP were subjected to immunoblot analysis using mAbLEM-2/15 to the catalytic domain of MT1-MMP.
Enzymatic activity of the 50-kDa MT1-MMP species
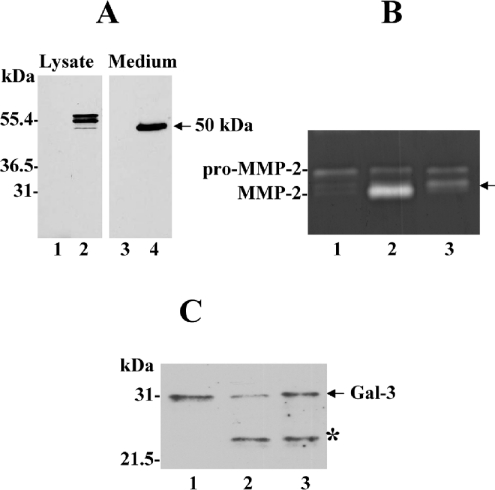
A recombinant 50-kDa (Tyr112–Val524) MT1-MMP was expressed in cells for functional studies as described in the Experimental section. As shown in Figure 5(A), the cell lysate showed presence of three MT1-MMP forms, which are likely to be the pre-pro-form, the zymogen and the active form of the recombinant enzyme (Figure 5A, lane 2). In the media, we detected the recombinant 50-kDa species, as expected (Figure 5A, lane 4). The recombinant 50-kDa species appeared to be a stable form, since lower-molecular-mass fragments were not detected in the media, even after prolonged incubations, suggesting that the 32-kDa species is not likely to be a degradation product of the soluble form (50 kDa) of MT1-MMP. The 50-kDa-containing medium was examined for enzymatic activity using a peptide-based substrate assay as described in the Experimental section, and found to be active when compared with control media. The amount of active 50-kDa enzyme detected in the media was approx. 17±0.5 ng/ml based on a standard curve using a soluble recombinant MT1-MMP. Also, cells expressing the recombinant 50-kDa (Tyr112–Val524) enzyme converted pro-MMP-2 into the intermediate form (Figure 5B, lane 3), which is consistent with the ability of MT1-MMP to cleave within the prodomain of pro-MMP-2 at the Asn37–Leu38 peptide bond, as reported previously [34,35]. As expected, cells expressing the wild-type membrane-bound active MT1-MMP (Figure 5B, lane 2) accomplished full pro-MMP-2 activation [20]. The activity studies also showed that galectin-3, a 31-kDa member of the β-galactoside-binding proteins, was cleaved to the 22-kDa degradation product [36] when exposed to cells expressing membrane-anchored wild-type MT1-MMP or the recombinant 50-kDa enzyme form (Figure 5C, lanes 2 and 3 respectively). This activity was inhibited by TIMP-2, but not by TIMP-1, indicating that the galectin-3 cleavage was mediated by MT1-MMP (results not shown). Thus the shed ectodomain of MT1-MMP is a fully functional protease that can promote pericellular proteolysis.
Figure 5. Expression and activity of recombinant soluble 50-kDa MT1-MMP.
(A) CV-1 cells were infected/transfected with (lanes 2 and 4) or without (lanes 1 and 3) a plasmid encoding the 50-kDa MT1-MMP as described in the Experimental section. The media and lysates were analysed by immunoblot using mAbLEM-2/15 to the catalytic domain. (B and C) CV-1 cells were infected/transfected to express wild-type MT1-MMP (lane 2) or the recombinant 50-kDa (Tyr112–Val524) soluble form (lane 3). Post-transfection (4 h), the media were replaced with serum-free media containing 10 nM pro-MMP-2 (B) or 20 nM galectin-3 (C) and incubated for an additional 18 h. An aliquot of the media was analysed by gelatin zymography (B) or by immunoblot analysis using an anti-galectin-3 antibody (C). The arrow in (B) indicates the intermediate form of MMP-2 and the asterisk in (C) indicates the cleaved product of galectin-3. Lanes 1 in (B) and (C) show the control pro-MMP-2 and control galectin-3 respectively.
50-kDa ectodomain binds TIMP-2 in the pericellular space
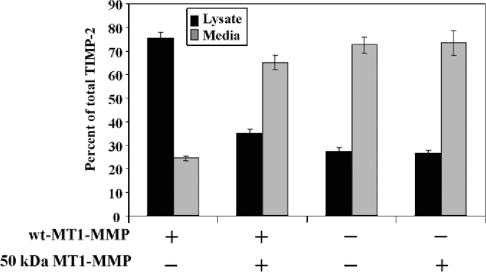
It has been previously shown that expression of the active MT1-MMP (57 kDa) promotes binding of TIMP-2 to the cell surface [20,26]. Since the 50-kDa species of MT1-MMP also binds TIMP-2 (Figure 2C), we wished to examine whether excess of the soluble 50-kDa species in the pericellular space of cells expressing membrane-bound active MT1-MMP (57 kDa) could alter the distribution of TIMP-2 between the cell surface and the media. To this end, CV-1 cells were infected/transfected to express the wild-type MT1-MMP and then were incubated with TIMP-2 in the presence or absence of the recombinant 50-kDa species. Addition of the exogenous soluble 50-kDa species to the cells was preferred over co-expression of wild-type and 50-kDa MT1-MMP to avoid potential problems of intracellular trafficking and activation. The level of TIMP-2 associated with the cells and in the media was then measured by ELISA. In the absence of the 50-kDa enzyme, most of the TIMP-2 (75%) was found in the cell lysate fraction consistent with binding of inhibitor to active MT1-MMP on the cell surface (Figure 6). Addition of the 50-kDa species to the cells expressing MT1-MMP caused a significant decrease in cell-associated TIMP-2, which was then mostly localized in the supernatant fraction. The controls showed that in cells without membrane-anchored MT1-MMP and with soluble 50-kDa species alone, most of the TIMP-2 was detected in the media, demonstrating the specificity of the inhibitor binding. These studies suggest that shedding of MT1-MMP ectodomain may alter the balance of TIMP-2 between the cell surface and the pericellular space.
Figure 6. Cellular distribution of TIMP-2 in the presence of membrane-anchored and soluble MT1-MMP.
CV-1 cells were infected/transfected to express or not wild-type (wt) MT1-MMP. Post-transfection (4 h), the cells received 3.4 nM TIMP-2 alone or with ∼50 ng of recombinant 50-kDa MT1-MMP in serum-free media for 16 h. Then, the cell lysate and media were harvested and analysed for TIMP-2 levels by ELISA.
A ∼53-kDa MT1-MMP form is detected in breast carcinoma tissue
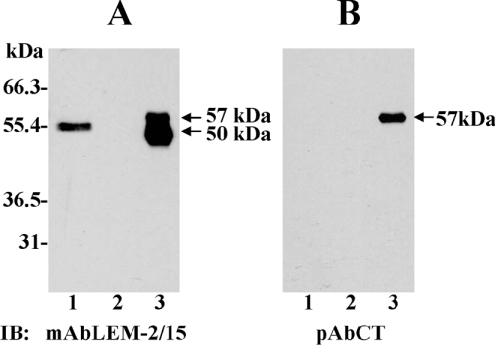
MT1-MMP is highly expressed in human breast carcinomas [37]. Therefore, we examined whether the MT1-MMP ectodomain could be detected in tissue extracts of benign and invasive breast carcinoma tissues obtained from fresh biopsies after surgery. To ensure that we would detect the soluble ectodomain (without the transmembrane domain), the tissues (benign and malignant) were homogenized in a sucrose-containing buffer without detergents. The extracts were then subjected to purification on the BB-94 resin to isolate the ectodomain of MT1-MMP, as described in the Experimental section. The MT1-MMP species in the bound fraction were detected by immunoblot analysis. As shown in Figure 7(A), the LEM-2/15 mAb detected a ∼53-kDa species in the bound fraction derived from extracts of tumour (Figure 7A, lane 1), but not from benign (Figure 7A, lane 2) tissues, indicative of the high level of expression of MT1-MMP in breast carcinomas [37]. Interestingly, the molecular mass of this species (∼53 kDa) was slightly higher from that exhibited by the soluble 50-kDa MT1-MMP form found in cells infected to express MT1-MMP. The ∼53-kDa form was not detected when the same bound fractions were tested with the pAb to the cytosolic tail of MT1-MMP (Figure 7B, lane 1), suggesting that this enzyme form represents a species of MT1-MMP lacking the cytosolic tail and possibly the transmembrane domain too, which is present in invasive breast carcinoma tissues.
Figure 7. MMP inhibitor-tethered affinity isolation of MT1-MMP in extracts of benign and tumour breast tissues.
Tumour and benign breast tissues isolated from six different human invasive breast carcinomas were homogenized, as described in the Experimental section, and incubated with the MMP inhibitor-tethered matrix. The bound protein fraction from tumour (lane 1) and benign (lane 2) tissue were eluted from the immobilized inhibitor, and subjected to immunoblot (IB) analysis of the same blot with either the mAbLEM-2/15 to the catalytic domain (A) or the pAbCT to the cytosolic tail (B) of MT1-MMP. Lane 3 shows the membrane-anchored (57 kDa) and soluble (50 kDa) MT1-MMP found in BS-C-1 cells as a control. Note that the ∼53-kDa form in the tumour tissue shown in (A) (lane 1) is only recognized by the antibody to the catalytic domain.
DISCUSSION
MT1-MMP by the virtue of being a type I transmembrane protein undergoes a series of regulatory processes at the cell surface that tightly control enzyme level on the plasma membrane. MT1-MMP can be autocatalytically processed to an inactive species [18–20], endocytosed from and/or recycled back to the cell surface [38–43] and shed from the surface into the extracellular space [11,21,24]. We previously showed that ectodomain shedding of MT1-MMP is a complex process involving both autocatalytic and non-autocatalytic proteolysis that results in the release of a series of soluble MT1-MMP forms from the cell surface [21]. Herein we have purified and expressed the major shed form of MT1-MMP produced by non-autocatalytic shedding and determined its primary structure and catalytic competence. The soluble ∼52–50-kDa species isolated from the media of MT1-MMP expressing cells was sequenced and determined to extend from Tyr112 to Val524. Thus, a cleavage at the Val524–Ile525 site in the stem region is involved in the formation of the 50-kDa soluble form of MT1-MMP. A previous study showed that MT5-MMP is also shed by cleavage at the stem region. However, as opposed to MT1-MMP, MT5-MMP shedding is mediated by furin, which cleaves the ectodomain of MT5-MMP in the trans-Golgi network [44]. It has been proposed that the stem region of MT5-MMP contains a cryptic furin-type convertase motif that is absent from the stem of MT1-MMP [44]. Thus, although both shedding processes are non-autocatalytic, they appear to involve the action of different proteases (discussed below).
Structurally, the Val524–Ile525 site within the stem region of MT1-MMP is surrounded by a glutamic acid-rich environment that is present in MT1-MMP, but not in other type I transmembrane MT-MMPs [14]. Although there is no structural information on the stem region, often the portions of proteins that connect different domains are surface flexible regions, which is consistent with the size and the location of the stem region in MT1-MMP. This likely flexibility for this region of MT1-MMP would predispose it to proteolysis, such as documented in the present report. A hallmark of such flexible structural regions is that modification of the identified site of proteolysis by mutagenesis is not likely to eliminate the proteolytic fragmentation. Even if mutagenesis would alter the specific cleavage site for a given enzyme such that MT1-MMP would no longer serve as a substrate at that site, the flexibility of the region would present an adjacent site for cleavage by the same enzyme or another. This may explain the lack of effect of various amino acid substitutions at the Val524–Ile525 site.
Although the MS analysis of the purified 50-kDa species expressed in infected BS-C-1 cells indicates a C-terminal end at Val524, we found slight differences in the molecular mass of soluble MT1-MMP (∼50–53 kDa) from various sources (HT1080 cells, fibroblasts, breast carcinomas). These small differences in mass may be related to the existence of additional cleavage sites and/or to cell specific glycosylation of MT1-MMP [45]. Nevertheless, our data are consistent with the shedding of the 50-kDa species requiring proteolysis within the stem region, since Δstem-MT1 was not shed. Consistently, we also found that mouse embryonic fibroblasts deficient in both presenilin 1 and 2 and transfected to express human MT1-MMP shed the soluble 50-kDa species (results not shown), indicating that presenilin-dependent intra-membrane proteolysis is not involved in this process. Although the nature of the protease(s) involved in MT1-MMP ectodomain shedding remains unknown, various protease inhibitors including metalloproteinase inhibitors, such as TIMP-2, TIMP-1, marimastat [21], and TAPI-1 (Figure 1A), had no inhibitory effect, indicating that a metalloproteinase is not likely to be involved. In fact, metalloproteinase inhibitors caused a noticeable increase in the amount of the 50-kDa species detected in the media. Since metalloproteinase inhibitors inhibit the autocatalytic processing of MT1-MMP, this observation suggests that the rate of ectodomain shedding is influenced by the level of MT1-MMP on the cell surface, which is regulated by diverse processes such as endocytosis and processing [46].
The sequencing information allowed us to express a soluble recombinant 50-kDa species (Tyr112–Val524) that showed catalytic activity against two distinct extracellular MMP substrates, namely pro-MMP-2 and galectin-3. This is consistent with previous studies using various MT1-MMP soluble forms [3,34,35,47,48]. Thus, ectodomain shedding of MT1-MMP has the potential to directly contribute to pericellular proteolysis. However, previous studies showed that cell invasion of a collagen matrix required the membrane-anchored form of MT1-MMP [2]. While this may be the case for collagen degradation, membrane-anchoring of MT1-MMP may not be an essential requirement for activity against other substrates. Recent evidence shows that MT1-MMP can also degrade important signalling molecules [11]. Thus we can propose that cellular activities other than collagen invasion may be equally or better served by shed forms of MT1-MMP. It will be interesting to determine the relative contribution of soluble versus membrane-tethered MT1-MMP to the cellular degradome, and whether there are preferred substrates (extracellular matrix versus non-extracellular matrix) for each enzyme form, which may influence different cellular activities.
We have found that the ectodomain of MT1-MMP (50 kDa), when added to cells expressing the membrane-anchored form of MT1-MMP (57 kDa), caused a significant decrease in cell associated TIMP-2. These data suggest that, under certain conditions, a soluble ectodomain of MT1-MMP may withdraw TIMP-2 away from the membrane-anchored form and thus alter the delicate MMP/TIMP balance on the cell surface. TIMP-2 binding to the membrane-tethered active MT1-MMP has a major impact on MT1-MMP activity [4] and on autocatalytic processing [18,20]. Therefore, soluble active forms of MT1-MMP may indirectly influence the extent of pericellular proteolysis mediated by the membrane-anchored enzyme by modulating the availability of local TIMPs [49].
The use of the inhibitor-tethered matrix also allowed us to identify, for the first time, a ∼53-kDa species of MT1-MMP in extracts of invasive breast carcinomas, which was recognized by an antibody to the catalytic domain, but not by an antibody to the cytosolic tail. Although the precise location of the cleavage site(s) (intra-stem or intra-transmembrane) involved in the formation of the ∼53-kDa species is unknown, the method used to homogenize the tissue for the affinity step is not suitable for solubilization of membrane-anchored proteins. This strongly suggests that the ∼53-kDa species is not anchored to the membrane and thus it may be a soluble form of MT1-MMP. The identification of this species in breast tumours suggests that ectodomain shedding of MT1-MMP occurs in vivo. In summary, the present report provides structural and functional information on the major soluble species of MT1-MMP produced by ectodomain shedding and underscores the complexity of MT1-MMP processing on the cell surface, which may impact on MT1-MMP-dependent proteolysis in the pericellular space. Furthermore, from the present work and previous studies with MT5-MMP [44] and MT3-MMP [33], the emerging view is that ectodomain shedding by cleavage at the stem region is a novel mechanism of MT-MMP regulation of enzymatic activity.
Online data
Acknowledgments
This work was supported by National Institutes of Health grant (NCI-CA61986) to R. F. We are grateful to Dr Alicia Arroyo (Hospital de la Princesa, Madrid, Spain) for providing the mAbLEM-2/15.
References
- 1.Hotary K. B., Allen E. D., Brooks P. C., Datta N. S., Long M. W., Weiss S. J. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell. 2003;114:33–45. doi: 10.1016/s0092-8674(03)00513-0. [DOI] [PubMed] [Google Scholar]
- 2.Hotary K., Allen E., Punturieri A., Yana I., Weiss S. J. Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J. Cell Biol. 2000;149:1309–1323. doi: 10.1083/jcb.149.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohuchi E., Imai K., Fujii Y., Sato H., Seiki M., Okada Y. Membrane type 1 matrix metalloproteinase digests interstitial collagens and other extracellular matrix macromolecules. J. Biol. Chem. 1997;272:2446–2451. doi: 10.1074/jbc.272.4.2446. [DOI] [PubMed] [Google Scholar]
- 4.Strongin A. Y., Collier I., Bannikov G., Marmer B. L., Grant G. A., Goldberg G. I. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J. Biol. Chem. 1995;270:5331–5338. doi: 10.1074/jbc.270.10.5331. [DOI] [PubMed] [Google Scholar]
- 5.Knauper V., Will H., Lopez-Otin C., Smith B., Atkinson S. J., Stanton H., Hembry R. M., Murphy G. Cellular mechanisms for human procollagenase-3 (MMP-13) activation. Evidence that MT1-MMP (MMP-14) and gelatinase a (MMP-2) are able to generate active enzyme. J. Biol. Chem. 1996;271:17124–17131. doi: 10.1074/jbc.271.29.17124. [DOI] [PubMed] [Google Scholar]
- 6.Cowell S., Knauper V., Stewart M. L., D'Ortho M. P., Stanton H., Hembry R. M., Lopez-Otin C., Reynolds J. J., Murphy G. Induction of matrix metalloproteinase activation cascades based on membrane-type 1 matrix metalloproteinase: associated activation of gelatinase A, gelatinase B and collagenase 3. Biochem. J. 1998;331:453–458. doi: 10.1042/bj3310453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toth M., Chvyrkova I., Bernardo M. M., Hernandez-Barrantes S., Fridman R. Pro-MMP-9 activation by the MT1-MMP/MMP-2 axis and MMP-3: role of TIMP-2 and plasma membranes. Biochem. Biophys. Res. Commun. 2003;308:386–395. doi: 10.1016/s0006-291x(03)01405-0. [DOI] [PubMed] [Google Scholar]
- 8.Kajita M., Itoh Y., Chiba T., Mori H., Okada A., Kinoh H., Seiki M. Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J. Cell Biol. 2001;153:893–904. doi: 10.1083/jcb.153.5.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deryugina E. I., Ratnikov B. I., Postnova T. I., Rozanov D. V., Strongin A. Y. Processing of integrin αv subunit by membrane type 1 matrix metalloproteinase stimulates migration of breast carcinoma cells on vitronectin and enhances tyrosine phosphorylation of focal adhesion kinase. J. Biol. Chem. 2002;277:9749–9756. doi: 10.1074/jbc.M110269200. [DOI] [PubMed] [Google Scholar]
- 10.Endo K., Takino T., Miyamori H., Kinsen H., Yoshizaki T., Furukawa M., Sato H. Cleavage of syndecan-1 by membrane type matrix metalloproteinase-1 stimulates cell migration. J. Biol. Chem. 2003;278:40764–40770. doi: 10.1074/jbc.M306736200. [DOI] [PubMed] [Google Scholar]
- 11.Tam E. M., Morrison C. J., Wu Y. I., Stack M. S., Overall C. M. Membrane protease proteomics: Isotope-coded affinity tag MS identification of undescribed MT1-matrix metalloproteinase substrates. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6917–6922. doi: 10.1073/pnas.0305862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmbeck K., Bianco P., Caterina J., Yamada S., Kromer M., Kuznetsov S. A., Mankani M., Robey P. G., Poole A. R., Pidoux I., et al. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell. 1999;99:81–92. doi: 10.1016/s0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- 13.Seiki M. Membrane-type 1 matrix metalloproteinase: a key enzyme for tumor invasion. Cancer Lett. 2003;194:1–11. doi: 10.1016/s0304-3835(02)00699-7. [DOI] [PubMed] [Google Scholar]
- 14.Massova I., Kotra L. P., Fridman R., Mobashery S. Matrix metalloproteinases: structures, evolution, and diversification. FASEB J. 1998;12:1075-1095. [PubMed] [Google Scholar]
- 15.Pei D. Identification and characterization of the fifth membrane-type matrix metalloproteinase MT5-MMP. J. Biol. Chem. 1999;274:8925–8932. doi: 10.1074/jbc.274.13.8925. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez-Barrantes S., Bernardo M., Toth M., Fridman R. Regulation of membrane type-matrix metalloproteinases. Semin. Cancer Biol. 2002;12:131–138. doi: 10.1006/scbi.2001.0421. [DOI] [PubMed] [Google Scholar]
- 17.Toth M., Bernardo M. M., Gervasi D. C., Soloway P. D., Wang Z., Bigg H. F., Overall C. M., DeClerck Y. A., Tschesche H., Cher M. L., et al. Tissue inhibitor of metalloproteinase (TIMP)-2 acts synergistically with synthetic matrix metalloproteinase (MMP) inhibitors but not with TIMP-4 to enhance the (membrane type 1)-MMP-dependent activation of pro-MMP-2. J. Biol. Chem. 2000;275:41415–41423. doi: 10.1074/jbc.M006871200. [DOI] [PubMed] [Google Scholar]
- 18.Lehti K., Lohi J., Valtanen H., Keski-Oja J. Proteolytic processing of membrane-type-1 matrix metalloproteinase is associated with gelatinase A activation at the cell surface. Biochem. J. 1998;334:345–353. doi: 10.1042/bj3340345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehti K., Valtanen H., Wickstrom S., Lohi J., Keski-Oja J. Regulation of membrane-type-1 matrix metalloproteinase activity by its cytoplasmic domain. J. Biol. Chem. 2000;275:15006–15013. doi: 10.1074/jbc.M910220199. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez-Barrantes S., Toth M., Bernardo M. M., Yurkova M., Gervasi D. C., Raz Y., Sang Q. A., Fridman R. Binding of active (57 kDa) membrane type 1-matrix metalloproteinase (MT1-MMP) to tissue inhibitor of metalloproteinase (TIMP)-2 regulates MT1-MMP processing and pro-MMP-2 activation. J. Biol. Chem. 2000;275:12080–12089. doi: 10.1074/jbc.275.16.12080. [DOI] [PubMed] [Google Scholar]
- 21.Toth M., Hernandez-Barrantes S., Osenkowski P., Bernardo M. M., Gervasi D. C., Shimura Y., Meroueh O., Kotra L. P., Galvez B. G., Arroyo A. G., et al. Complex pattern of membrane type-1 matrix metalloproteinase shedding. Regulation by autocatalytic cell surface inactivation of active enzyme. J. Biol. Chem. 2002;277:26340–26350. doi: 10.1074/jbc.M200655200. [DOI] [PubMed] [Google Scholar]
- 22.Tam E. M., Wu Y. I., Butler G. S., Stack M. S., Overall C. M. Collagen binding properties of the membrane type-1 matrix metalloproteinase (MT1-MMP) haemopexin C domain. The ectodomain of the 44-kDa autocatalytic product of MT1-MMP inhibits cell invasion by disrupting native type I collagen cleavage. J. Biol. Chem. 2002;277:39005–39014. doi: 10.1074/jbc.M206874200. [DOI] [PubMed] [Google Scholar]
- 23.Kazes I., Delarue F., Hagege J., Bouzhir-Sima L., Rondeau E., Sraer J. D., Nguyen G. Soluble latent membrane-type 1 matrix metalloprotease secreted by human mesangial cells is activated by urokinase. Kidney Int. 1998;54:1976–1984. doi: 10.1046/j.1523-1755.1998.00216.x. [DOI] [PubMed] [Google Scholar]
- 24.Harayama T., Ohuchi E., Aoki T., Sato H., Seiki M., Okada Y. Shedding of membrane type 1 matrix metalloproteinase in a human breast carcinoma cell line. Jpn. J. Cancer Res. 1999;90:942–950. doi: 10.1111/j.1349-7006.1999.tb00839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H., Bauzon D. E., Xu X., Tschesche H., Cao J., Sang Q. A. Immunological characterization of cell-surface and soluble forms of membrane type 1 matrix metalloproteinase in human breast cancer cells and in fibroblasts. Mol. Carcinog. 1998;22:84–94. [PubMed] [Google Scholar]
- 26.Zucker S., Drews M., Conner C., Foda H. D., DeClerck Y. A., Langley K. E., Bahou W. F., Docherty A. J., Cao J. Tissue inhibitor of metalloproteinase-2 (TIMP-2) binds to the catalytic domain of the cell surface receptor, membrane type 1-matrix metalloproteinase 1 (MT1-MMP) J. Biol. Chem. 1998;273:1216–1222. doi: 10.1074/jbc.273.2.1216. [DOI] [PubMed] [Google Scholar]
- 27.Galvez B. G., Matias-Roman S., Albar J. P., Sanchez-Madrid F., Arroyo A. G. Membrane type 1-matrix metalloproteinase is activated during migration of human endothelial cells and modulates endothelial motility and matrix remodeling. J. Biol. Chem. 2001;276:37491–37500. doi: 10.1074/jbc.M104094200. [DOI] [PubMed] [Google Scholar]
- 28.Maisi P., Prikk K., Sepper R., Pirila E., Salo T., Hietanen J., Sorsa T. Soluble membrane-type 1 matrix metalloproteinase (MT1-MMP) and gelatinase A (MMP-2) in induced sputum and bronchoalveolar lavage fluid of human bronchial asthma and bronchiectasis. APMIS. 2002;110:771–782. doi: 10.1034/j.1600-0463.2002.1101102.x. [DOI] [PubMed] [Google Scholar]
- 29.Fuerst T. R., Earl P. L., Moss B. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol. Cell. Biol. 1987;7:2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson M. W., Gervasi D. C., Mobashery S., Fridman R. Kinetic analysis of the binding of human matrix metalloproteinase-2 and -9 to tissue inhibitor of metalloproteinase (TIMP)-1 and TIMP-2. J. Biol. Chem. 1997;272:29975–29983. doi: 10.1074/jbc.272.47.29975. [DOI] [PubMed] [Google Scholar]
- 31.Brown P. D. Matrix metalloproteinase inhibitors. Breast Cancer Res. Treat. 1998;52:125–136. doi: 10.1023/a:1006119319695. [DOI] [PubMed] [Google Scholar]
- 32.Chittum H. S., Lane W. S., Carlson B. A., Roller P. P., Lung F. D., Lee B. J., Hatfield D. L. Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry. 1998;37:10866–10870. doi: 10.1021/bi981042r. [DOI] [PubMed] [Google Scholar]
- 33.Zhao H., Bernardo M. M., Osenkowski P., Sohail A., Pei D., Nagase H., Kashiwagi M., Soloway P. D., DeClerck Y. A., Fridman R. Differential inhibition of membrane type 3 (MT3)-matrix metalloproteinase (MMP) and MT1-MMP by tissue inhibitor of metalloproteinase (TIMP)-2 and TIMP-3 regulates pro-MMP-2 activation. J. Biol. Chem. 2004;279:8592–8601. doi: 10.1074/jbc.M308708200. [DOI] [PubMed] [Google Scholar]
- 34.Will H., Atkinson S. J., Butler G. S., Smith B., Murphy G. The soluble catalytic domain of membrane type 1 matrix metalloproteinase cleaves the propeptide of progelatinase A and initiates autoproteolytic activation. Regulation by TIMP-2 and TIMP-3. J. Biol. Chem. 1996;271:17119–17123. doi: 10.1074/jbc.271.29.17119. [DOI] [PubMed] [Google Scholar]
- 35.Pei D., Weiss S. J. Transmembrane-deletion mutants of the membrane-type matrix metalloproteinase-1 process progelatinase A and express intrinsic matrix-degrading activity. J. Biol. Chem. 1996;271:9135–9140. doi: 10.1074/jbc.271.15.9135. [DOI] [PubMed] [Google Scholar]
- 36.Ochieng J., Fridman R., Nangia-Makker P., Kleiner D. E., Liotta L. A., Stetler-Stevenson W. G., Raz A. Galectin-3 is a novel substrate for human matrix metalloproteinases-2 and -9. Biochemistry. 1994;33:14109–14114. doi: 10.1021/bi00251a020. [DOI] [PubMed] [Google Scholar]
- 37.Ueno H., Nakamura H., Inoue M., Imai K., Noguchi M., Sato H., Seiki M., Okada Y. Expression and tissue localization of membrane-types 1, 2, and 3 matrix metalloproteinases in human invasive breast carcinomas. Cancer Res. 1997;57:2055–2060. [PubMed] [Google Scholar]
- 38.Galvez B. G., Matias-Roman S., Yanez-Mo M., Vicente-Manzanares M., Sanchez-Madrid F., Arroyo A. G. Caveolae are a novel pathway for membrane-type 1 matrix metalloproteinase traffic in human endothelial cells. Mol. Biol. Cell. 2004;15:678–687. doi: 10.1091/mbc.E03-07-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang A., Lehti K., Wang X., Weiss S. J., Keski-Oja J., Pei D. Regulation of membrane-type matrix metalloproteinase 1 activity by dynamin-mediated endocytosis. Proc. Natl. Acad. Sci. U.S.A. 2001;98:13693–13698. doi: 10.1073/pnas.241293698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uekita T., Itoh Y., Yana I., Ohno H., Seiki M. Cytoplasmic tail-dependent internalization of membrane-type 1 matrix metalloproteinase is important for its invasion-promoting activity. J. Cell Biol. 2001;155:1345–1356. doi: 10.1083/jcb.200108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozanov D. V., Deryugina E. I., Monosov E. Z., Marchenko N. D., Strongin A. Y. Aberrant, persistent inclusion into lipid rafts limits the tumorigenic function of membrane type-1 matrix metalloproteinase in malignant cells. Exp. Cell Res. 2004;293:81–95. doi: 10.1016/j.yexcr.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Remacle A., Murphy G., Roghi C. Membrane type I-matrix metalloproteinase (MT1-MMP) is internalised by two different pathways and is recycled to the cell surface. J. Cell Sci. 2003;116:3905–3916. doi: 10.1242/jcs.00710. [DOI] [PubMed] [Google Scholar]
- 43.Wang X., Ma D., Keski-Oja J., Pei D. Co-recycling of MT1-MMP and MT3-MMP through the trans-Golgi network. Identification of DKV582 as a recycling signal. J. Biol. Chem. 2004;279:9331–9336. doi: 10.1074/jbc.M312369200. [DOI] [PubMed] [Google Scholar]
- 44.Wang X., Pei D. Shedding of membrane type matrix metalloproteinase 5 by a furin-type convertase: a potential mechanism for down-regulation. J. Biol. Chem. 2001;276:35953–35960. doi: 10.1074/jbc.M103680200. [DOI] [PubMed] [Google Scholar]
- 45.Wu Y. I., Munshi H. G., Sen R., Snipas S. J., Salvesen G. S., Fridman R., Stack M. S. Glycosylation broadens the substrate profile of membrane type 1 matrix metalloproteinase. J. Biol. Chem. 2004;279:8278–8289. doi: 10.1074/jbc.M311870200. [DOI] [PubMed] [Google Scholar]
- 46.Osenkowski P., Toth M., Fridman R. Processing, shedding, and endocytosis of membrane type 1-matrix metalloproteinase (MT1-MMP) J. Cell. Physiol. 2004;200:2–10. doi: 10.1002/jcp.20064. [DOI] [PubMed] [Google Scholar]
- 47.d'Ortho M. P., Will H., Atkinson S., Butler G., Messent A., Gavrilovic J., Smith B., Timpl R., Zardi L., Murphy G. Membrane-type matrix metalloproteinases 1 and 2 exhibit broad-spectrum proteolytic capacities comparable to many matrix metalloproteinases. Eur. J. Biochem. 1997;250:751–757. doi: 10.1111/j.1432-1033.1997.00751.x. [DOI] [PubMed] [Google Scholar]
- 48.Kinoshita T., Sato H., Takino T., Itoh M., Akizawa T., Seiki M. Processing of a precursor of 72-kilodalton type IV collagenase/gelatinase A by a recombinant membrane-type 1 matrix metalloproteinase. Cancer Res. 1996;56:2535–2538. [PubMed] [Google Scholar]
- 49.Karagiannis E. D., Popel A. S. A theoretical model of type I collagen proteolysis by matrix metalloproteinase (MMP) 2 and membrane type 1 MMP in the presence of tissue inhibitor of metalloproteinase 2. J. Biol. Chem. 2004;279:39105–39114. doi: 10.1074/jbc.M403627200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.