Abstract
The AMBP [A1M (α1-microglobulin)/bikunin precursor] gene encodes two plasma glycoproteins: A1M, an immunosuppressive lipocalin, and bikunin, a member of plasma serine proteinase inhibitor family with prototypical Kunitz-type domain. Although previously believed to be constitutively expressed exclusively in liver, the present study demonstrates the induction of this gene by oxalate in porcine proximal tubular LLC-PK1 cells and rat kidney. In liver, the precursor protein is cleaved in the Golgi network by a furin-like enzyme to release constituent proteins, which undergo glycosylation before their export from the cell. In the renal tubular cells, A1M and bikunin co-precipitate, indicating lack of cleavage of the precursor protein. As the expression of the AMBP gene is regulated by A1M-specific cis elements and transcription factors, A1M protein was studied as a representative of AMBP gene expression in renal cells. Oxalate treatment (500 μM) resulted in a time- and dose-dependent induction of A1M protein in LLC-PK1 cells. Of the four transcription factors, HNF-4 (hepatocyte nuclear factor-4) has been reported previously to be a major regulator of AMBP gene expression in liver. Electrophoretic mobility-shift assay, supershift assay, immunoreactivity assay and transfection-based studies showed the presence of an HNF-4 or an HNF-4-like protein in the kidney, which can affect the expression of the AMBP gene. In situ hybridization and immunocytochemical studies showed that the expression of this gene in kidney was mainly restricted to cells lining the renal tubular system.
Keywords: α1-microglobulin/bikunin precursor gene, immunocytochemistry, LLC-PK cells, oxalate, renal tubular epithelial cells, transcription factor hepatocyte nuclear factor-4 (HNF-4)
Abbreviations: A1M, α1-microglobulin; AMBP, A1M/bikunin precursor; HNF, hepatocyte nuclear factor; AH4ON, AMBP-specific HNF-4-binding consensus sequence (double stranded) oligonucleotide; ApH4ON, apolipoprotein CIII-specific HNF-4-binding consensus sequence (double stranded) oligonucleotide; AP, alkaline phosphatase; DIG, digoxigenin; EMSA, electrophoretic mobility-shift assay; FastDAB, diamino-benzidine tetrahydrochloride; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; OctON, octamer transcription factor binding cis element oligonucleotide
INTRODUCTION
The AMBP [A1M (α1-microglobulin)/bikunin precursor] gene encodes two distantly related plasma glycoproteins: A1M, a lipocalin, and bikunin, a Kunitz-type serine proteinase inhibitor. The existence of this complex gene in Salmo salar (Linnaeus) [1,2] and its association with catabolism of haem [3] suggest that ancestral genes combined, during the Silurian period, may be linked to the evolution of haemoglobin and myoglobin proteins in metazoans, a hallmark in the advent of terrestrial life, approx. 500 million year ago [4]. The AMBP gene is a single-copy gene [5] clustered along with other lipocalin genes on chromosomes. AMBP gene-encoded precursor protein is cleaved into its constituent proteins in the trans-Golgi network by a furin-like enzyme [6]. Both of these proteins undergo glycosylation before their independent export from the cell. AMBP protein forms homodimers as well as heterodimers with one of its constituent proteins, A1M, but not with bikunin [6]. The AMBP gene is expressed constitutively in liver, tightly regulated by a lipocalin (A1M)-specific set of cis elements and transcription factors [7,8]. The cDNAs of A1M and bikunin are connected by a 9 nt long sequence that codes for a tripeptide, which along with the terminal arginine of A1M forms the recognition site (RXK/RR) for the furin-like enzyme that cleaves the precursor protein [9,10].
A1M is a yellowish brown-coloured 26 kDa glycoprotein [11] covalently associated with unidentified sets of chromophores. About half of the plasma A1M exists in free form [12], whereas the rest forms 1:1 complexes with other macromolecules like IgA [13], prothrombin and albumin [14] in humans, and with fibronectin [15] and α1-inhibitor-3 [16] in the rat. It has been implicated in the regulation of development [17], cell growth [18–21], metabolism [3], immune response [22–24] and in mood disorders [25–27]. Bikunin, a 28 kDa serine proteinase inhibitor belongs to a family of inter-α-inhibitors. It has been reported to prevent invasion and metastasis of cancer cells [28–30], and to modulate extracellular matrix proteins as well as levels of intracellular calcium [8].
Both of the AMBP constituent proteins have been reported in urine of all mammals (so far studied) as well as in the organic matrix of kidney stones. The urinary A1M is one of the major biomarkers of renal tubular dysfunction. Previously, A1M [31] and bikunin [32–36] proteins have been shown, individually, to prevent calcium oxalate crystal formation in vitro. AMBP gene expression is regulated by a set of four liver-specific transcription factors, known as the HNFs (hepatocyte nuclear factors)-1–4 [6,7]. In the present study, we have observed oxalate-dependent AMBP gene expression in porcine renal tubular epithelial (LLC-PK1) cells. This observation is of particular importance as oxalate concentrations in the renal tubular system are very high, especially under pathological conditions like hyperoxaluria. The present study represents a systematic analysis of induction of AMBP gene expression and de novo synthesis of its constituent proteins in renal tubular epithelial cells. It further demonstrates the existence of, probable, identical AMBP gene regulatory mechanisms in liver and kidney.
MATERIALS AND METHODS
Chemicals and reagents
Polyclonal anti-A1M antibodies were purchased from Binding Sites (Birmingham, U.K.) and from AgriSera AB (Vannas, Sweden), anti-HNF-4α polyclonal antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.), whereas anti-bikunin antibody was a gift from Dr S. R. Marengo (Case Western Reserve University, Cleveland, OH, U.S.A.). Precast polyacrylamide gels (4–20% or 8–16%) were purchased from Bio-Rad Laboratories (Hercules, CA, U.S.A.). The PCR master mix and QuantiTech SYBR® Green kit containing reagents for real-time PCR were from Applied Biosystems (Foster City, CA, U.S.A.). Non-radioactive DIG (digoxigenin)-labelling kits were purchased from Roche (Mannheim, Germany). The transfection reagent, SuperFect™, was purchased from Qiagen (Chatsworth, CA, U.S.A.).
Cells and cell culture
Primary renal cell lines LLC-PK1, generated from porcine proximal tubules were purchased from A.T.C.C. (Manassas, VA, U.S.A.). Cell cultures were maintained in DMEM/F-12 medium (where DMEM stands for Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) new born calf serum (Gibco BRL, Gaithersburg, MD, U.S.A.) and 1% antibiotic antimycotic 100×solution (final concentration 100 units/ml penicillin, 100 μg/ml streptomycin and 250 ng/ml amphotericin B). All cell cultures were kept in a humid CO2 incubator with 5% CO2/95% air at 37 °C. Cells were washed twice with serum-free medium to remove traces of serum in the medium and then incubated for at least 48 h in serum-free medium before treatment.
Immunoprecipitation and immunoblotting
A1M protein was purified by immunoprecipitation as described previously [37]. Briefly, 80% confluent serum-starved cells in 100 mm culture dishes were treated with oxalate. Proteins from cell lysates and supernatant media were immunoprecipitated using anti-A1M antibody covalently immobilized on Sepharose beads and separated under reducing and denaturing conditions on Bio-Rad precast SDS/polyacrylamide gels. After transferring the proteins on to PVDF membrane, immunoreactive bands were visualized using anti-A1M antibody, rabbit anti-sheep AP (alkaline phosphatase)-conjugated IgG and CDP Star™ (Roche) substrate. The membranes were exposed to Fuji preflashed X-ray films. Relative intensities of bands were estimated using ScanAnalysis™ software (BIOSOFT™, MO, U.S.A.). In some experiments, anti-bikunin antibody was used to determine if bikunin co-precipitated with A1M. For HNF-4α protein detection, samples were processed without immunoprecipitation and were visualized using anti-HNF-4α antibody.
Real-time SYBR Green PCR
Total RNA was isolated by a single-step method as described previously [38]. In brief, 80% confluent LLC-PK1 cells grown in 100 mm cell culture dishes were treated with acid guanidinium thiocyanate-phenolchloroform containing TRIzol® reagent (Invitrogen, Carlsbad, CA, U.S.A.). RNA in the aqueous phase was precipitated with 0.7 vol. of propan-2-ol in a new Eppendorf tube and centrifuged. The RNA pellets were washed with 70% ethanol. Purified RNA was air-dried and reconstituted in RNasefree distilled water. First-strand cDNA was prepared with Super-Script II kit from Invitrogen using either oligo(dT)12–18 or AMBP gene-specific primers. The quantification of PCR product was performed by using SYBR Green PCR core kit reagents and 96-well MicroAmo Optic plates (Applied Biosystems, Foster City, CA, U.S.A.). Initially, the temperature was set at 95 °C for 15 min for AmpliTaq Gold polymerase activation. A 273 bp AMBP cDNA fragment was amplified using a forward primer, 5′-GCGACGGAGAGGGAGATCA-3′ and a reverse primer, 5′-TGAACTCCTCCAGCAGGCTTT-3′ for 40 cycles consisting of the following steps: denaturation, 94 °C for 30 s; annealing, 54 °C for 30 s; and elongation, 72 °C for 45 s. The SYBR Green fluorescence was measured at the end of each elongation step. Data were analysed using SPSS software with limits set at 2. A 75 bp long GAPDH (glyceraldehyde-3-phosphate dehydrogenase) fragment was amplified using gene-specific primers (forward primer, 5′-TCGGTGTCAACGGATTTGG-3′ and reverse primer, 5′-CAATGTCCACTTTGTCACAAGAGAA-3′) to normalize the data at each end point. Values of initial amount of specific mRNA in each sample were calculated using serial dilutions of GAPDH and target gene. Briefly, SYBR Green fluorescent signals from serial dilutions of standards were plotted against the logarithm of the concentration to determine the initial relative amount of specific mRNA in each sample.
Experimental hyperoxaluric rat model
Experimental hyperoxaluria was induced in Sprague–Dawley rats following the guidelines set by Institutional Animal Care and Use Committee (University of Florida). In brief, 0.7% of ethylene glycol or 5% 4-hydroxy-L-proline was administered in the diet. The animals became hyperoxaluric within 2–3 days, showed increased crystalluria after the third day and developed calcium oxalate crystals in the kidney after the second week. For in situ hybridization and immunohistochemistry analysis, kidneys of third day hyperoxaluric rats were saline-perfused and dissected out for paraffin embedding. Urine samples collected from normal and d7 hyperoxaluric rats were used for analysis of urinary A1M protein.
Non-radioactive in situ hybridization
An AMBP cDNA template with SP6 promoter on 5′-end and T7 promoter on 3′-end was prepared (the SP6 promoter primer sequence is recognized by bacteriophage SP6-specific RNA polymerase and helps in preparing ribonucleic acid probes for hybridization). Sense and antisense riboprobes were obtained by in vitro transcription using DIG-11-UTP and appropriate RNA polymerase according to the instructions in the DIG-RNA Labeling kit (Roche).
Deparaffinized and rehydrated (5 μm thick) sections were treated with proteinase K for 10 min at 37 °C to improve permeabilization and further subjected to acetylation with 0.25% acetic anhydride in 1 M triethanolamine–HCl (pH 8.0) for 20 min. After dehydration, sections were incubated in prehybridization solution [50 mM Tris/HCl, 25 mM EDTA, 20 mM NaCl, 0.2% SDS, 250 μg/ml yeast tRNA, 250 μg/ml denatured salmon sperm DNA and 2.5×Denhardt's solution (0.02%, w/v, of Ficoll 400, 0.02% polyvinylpyrrolidone and 0.02% BSA) in 50% deionized formamide] at 42 °C for 1 h in a humidifying chamber. The prehybridization solution was then replaced by hybridization buffer [20 mM Tris/HCl, 1 mM EDTA, 300 mM NaCl, 0.2 M dithiothreitol, 500 μg/ml tRNA; 100 μg/ml denatured salmon sperm DNA, 1×Denhardt's solution and 10% (w/v) dextran sulphate in 50% (v/v) formamide] containing 50 ng/ml sense or antisense DIG-labelled riboprobe. Hybridization was performed overnight at 42 °C. After stringent washings [for 10 min with 2×SSC (where 1× SSC stands for 15 M NaCl and 0.015 M sodium citrate) and for another 10 min with 0.1×SSC], the sections were incubated for 1 h at room temperature (∼25 °C) in blocking buffer (Boehringer, Mannheim, Germany) followed by overnight incubation at 4 °C with AP-coupled sheep anti-DIG antibody (1:500 in blocking buffer). Hybridized RNA was visualized with freshly prepared Nitro Blue Tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate-4-toludine-substrate solution-mixture at room temperature in the dark. The stain-intensity was monitored under a microscope and the reaction was terminated by dipping in stop buffer (1 mM EDTA, pH 8.0, in 100 mM Tris/HCl). The specificity of the riboprobes was verified by parallel incubation with antisense and sense riboprobes on alternate sections. To detect non-specific signals, some sections were hybridized without riboprobe, whereas others were processed by omission of anti-DIG antibody. Sections were counterstained with haematoxylin to visualize cells in the background.
Immunocytochemistry
Rehydrated sections with 5 μm thickness were incubated with 0.3% H2O2 in PBS for 20 min to quench endogenous peroxidase. After blocking in 1% goat serum in PBS for 30 min at room temperature, sections were incubated with anti-A1M antibody (1:100 dilution in blocking buffer) at 4 °C overnight, followed by incubation with biotinylated anti-sheep IgG secondary antibody for 30 min at room temperature. The immunoreactivity was visualized using avidin-conjugated peroxidase (Vector Laboratories, Burlingame, CA, U.S.A.) and FastDAB (diamino-benzidine tetrahydrochloride) substrate (Sigma, St. Louis, MO, U.S.A.). Sections were counterstained with haematoxylin to visualize the cells in background. Some sections processed similarly but without primary antibody constituted controls and were helpful in determining the specificity of the primary antibody.
Gel EMSA
A double-stranded AH4ON [AMBP-specific HNF-4-binding consensus sequence (double stranded) oligonucleotide] was prepared by reannealing complementary sequences of double stranded 5′-GTCCAAGTGGCCCTTGGCAGCAT-3′ oligo, as described previously [39]. The AH4ON was DIG-labelled using DIG gel shift kit (Roche). The nuclear proteins from oxalate treated or untreated cells were prepared as described previously [40]. The reaction mixture containing 20 mM Hepes (pH 7.6), 1 mM EDTA, 10 mM ammonium sulphate, 1 mM dithiothreitol, 0.2% (v/v) Tween 20, 30 mM potassium chloride, 1 μg of poly(dI/dC), 0.1 μg of poly(L-lysine), 300 pmol of labelled oligonucleotide and 2–6 μg of nuclear proteins was incubated at 25 °C for 15 min and then shifted to 4 °C to stop the reaction.
The HNF-4–AH4ON complex was separated on precast 5% polyacrylamide/0.5×TBE gel (1×TBE stands for 100 mM Tris, pH 7.9, 100 mM boric acid and 2 mM EDTA). In some experiments, 100×unlabelled AH4ON was used as a competitor for binding HNF-4 along with labelled oligonucleotide. The protein–DNA complexes were transferred to positively charged nylon membranes and visualized using an anti-DIG-AP conjugate and a CSPD chemiluminescent substrate. Membranes were then exposed to X-ray films and scanned using ScanAnalysis software (BIOSOFT™).
To identify the nuclear factor, the reaction mixture was incubated for 15 min at 25 °C, followed by the addition of anti-HNF-4α antibody, and incubated further for 20 min. The whole complex was then separated on 5% polyacrylamide/0.5×TBE gel. The band supershift was visualized following the method described above.
Oligonucleotide transfection in LLC-PK1 cells
To determine if sequestration of endogenous HNF-4 will abrogate AMBP gene expression, 80% confluent LLC-PK1 cells were transfected with reannealed ApH4ON [apolipoprotein CIII-specific HNF-4-binding consensus sequence (double stranded) oligonucleotide] 5′-CTCAGCTTGTACTTTGGTACAACTA-3′. ApH4ON or a mock cis element corresponding to OctON (octamer transcription factor binding cis element oligonucleotide) were transfected using SuperFect™ reagent following the manufacturer's instructions (Qiagen). In brief, 1 μg of DNA was mixed with 50 μl of SuperFect™ reagent in 300 μl of serum-free medium. The reaction was incubated for 10 min at room temperature. After adding 3 ml of medium containing serum and antibiotics, the contents were immediately laid over the cells. Control cells were laid over with a medium containing vehicle (transfection reagent SuperFect™) alone. After 6 h, the cells were treated with oxalate and incubated further for 3 h. Cell lysate and media from each dish were pooled, and A1M protein was estimated as described above in immunoprecipitation and immunoblotting experiments.
RESULTS
Induction of the AMBP gene products by oxalate
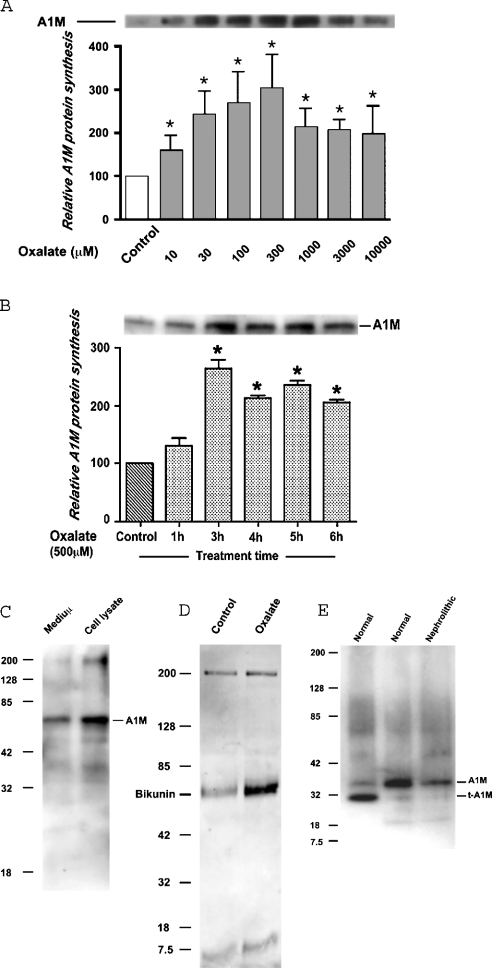
AMBP gene expression is regulated by A1M-specific cis elements/transcription factors and A1M is located proximal to bikunin. In the present study, only A1M was selected as the protein of choice. In liver, both AMBP gene products, A1M and bikunin are excretory proteins; therefore protein estimation was made by pooling the supernatant medium and cell lysate from culture plates. A1M protein was purified by immunoprecipitation and separated on polyacrylamide gel under reducing and denaturing conditions. A1M protein was seen as a 60–65 kDa band. Serum-starved LLC-PK1 cells in 100 mm culture plates were challenged with different concentrations of oxalate for 3 h. As seen in Figure 1(A), a dose as low as 30 μM was capable of inducing A1M protein in LLC-PK1 cells. Oxalate (300 μM) showed maximal induction, which declined with higher concentrations. In time-dependent response experiments, cells were treated with 500 μM oxalate for different periods of time. Figure 1(B) shows that treatment for 3 h resulted in peak induction of A1M protein, which declined on further exposure. Oxalate treatment at 500 μM for 3 h did not show any overt sign of cell membrane disruption, as measured by a lactate dehydrogenase release assay (results not shown).
Figure 1. Oxalate-inducible A1M protein synthesis.
(A) Semi-confluent LLC-PK1 cells were treated with different concentrations of oxalate (10 μM to 10 mM) for 3 h. The medium and cell lysate from each culture dish were pooled to estimate the total amount of A1M produced. Immunoprecipitation and immunoblotting were performed as described in the Materials and methods section. Maximal induction of A1M protein was seen for 300 μM oxalate. (B) Semi-confluent cells were treated with 500 μM oxalate for different time periods from 1–6 h. At 3 h treatment, A1M production reached its maximal peak and declined afterwards. Results represent means±S.E.M. (n=3). *P<0.01 versus basal levels in controls. (C) Semi-confluent LLC-PK1 cells were treated with 500 μM oxalate for 3 h. The medium and cell lysate were processed separately for immunoprecipitation and immunoblotting under reducing and denaturing conditions using anti-A1M antibody. The presence of A1M protein in a medium indicated its secretion from cells. (D) Semi-confluent serum-starved cells were treated with 500 μM oxalate. The medium and cell lysate from each dish were pooled and processed for immunoprecipitation using anti-A1M antibody and immunoblotted with anti-bikunin antibody. Immunoreactivity of a band to anti-bikunin antibody indicates co-precipitation of bikunin with A1M. (E) Urine samples from normal and experimental hyperoxaluric rats were analysed for the presence of A1M. Lanes 1 and 2 show striking difference in degree of A1M truncation in urine samples from two normal rats. Rat with crystalluria or nephrolithic condition showed decreased levels of urinary A1M (lane 3).
To find out if A1M protein was exported out of cell, A1M protein was detected separately in both supernatant medium and cell lysate. As shown in Figure 1(C) A1M appeared both in the medium and cell lysate.
To determine if the anti-A1M antibody-immunoreactive band size (60–65 kDa) was a result of bikunin co-precipitation, immunoreactivity of A1M-immunoprecipitated protein was determined by using anti-bikunin antibody. Figure 1(D) shows immunoreactivity of a band of same molecular mass, suggesting co-precipitation of bikunin. This may be due to non-cleavage of AMBP protein into its constituent proteins in these cells. Cellular A1M also formed a complex with other proteins at a molecular mass of 200 kDa.
A1M from normal rat urine was (Figure 1E) separated as untruncated (∼36 kDa) and/or truncated bands (∼30 kDa). Hyperoxaluric rats with crystalluria and/or calcium oxalate crystals in the kidney showed a decreased level of urinary A1M protein, with little or no truncation.
Oxalate-inducible AMBP gene expression in renal tubular cells
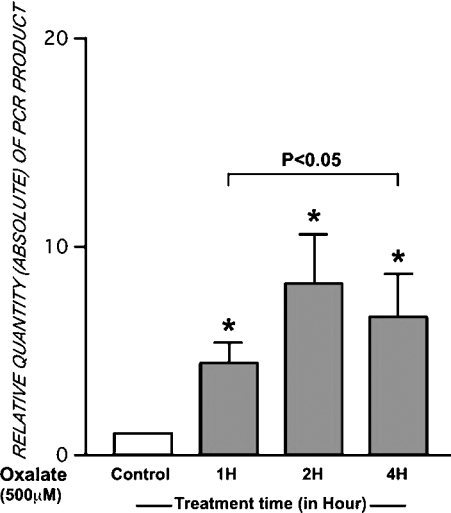
SYBR® Green dye binds to the minor groove of double-stranded DNA and in bound form it fluoresces. This property of SYBR Green dye is utilized to monitor the PCR product amplification in real time. First-strand cDNA was prepared from LLC-PK1 cells treated with 500 μM potassium oxalate for 0–4 h. Figure 2 shows oxalate-induced expression of the AMBP gene in LLC-PK1 cells. Potassium oxalate (500 μM) treatment for 2 h showed maximal AMBP transcription. These results demonstrate oxalate-inducible AMBP gene expression in renal cells. Treatment of LLC-PK1 cells with oxalate for different time periods showed statistically significant increases in AMBP gene expression. Data were normalized with that of GAPDH, a housekeeping gene.
Figure 2. Oxalate-inducible AMBP gene expression.
RNA was isolated from LLC-PK1 cells with or without oxalate treatment. A reverse transcriptase product served as a template for SYBR Green dye-facilitated quantification of real-time PCR amplicons. PCR was performed in 96-well MicroAmo optic plates using GeneAmp 5700 sequence detection system. AmpliTaq Gold was activated at 95 °C for 15 min followed by amplification for 40 cycles (94 °C for 30 s; 54 °C for 30 s; 72 °C for 45 s). GAPDH gene was used as a control standard for quantification of the initial amount of mRNA. The results were analysed using two-way ANOVA. The values are the means±S.E.M. (n=4). *P<0.05 versus basal levels in controls.
Localization of AMBP gene expression site in kidney by in situ hybridization
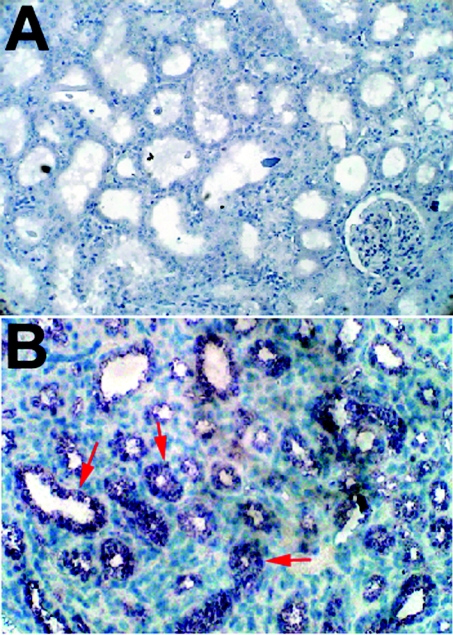
Paraffin embedded 5 μm thick sections of kidneys from normal and day 3 hyperoxaluric rats were hybridized with sense or antisense A1M riboprobe. Figure 3(B) shows staining of hybridized mRNA in cells lining the lumen of proximal tubules as well as those of distal tubules, whereas a section from normal rat kidney (Figure 3A) showed little or no staining. The specificity of antisense AMBP riboprobes was confirmed by staining some sections with sense riboprobe. This finding indicates localized expression of the AMBP gene in kidney cells besides liver and pancreas [41].
Figure 3. Localization of AMBP gene expression in kidney.
Sections of kidney with 5 μm thickness from normal and experimental hyperoxaluric rats were hybridized with DIG-labelled sense or antisense riboprobe. Hybridized riboprobe was detected using AP-coupled sheep anti-DIG antibody and freshly prepared Nitro Blue Tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate-4-toludine substrate. An antisense riboprobe-hybridized section showed dark blue staining (B) in epithelial cells lining the renal tubular system. The specificity of the riboprobe was confirmed by the absence of any staining with sense riboprobe. Sections from normal rat kidney showed little or no staining (A).
Localization of A1M protein by immunocytochemistry
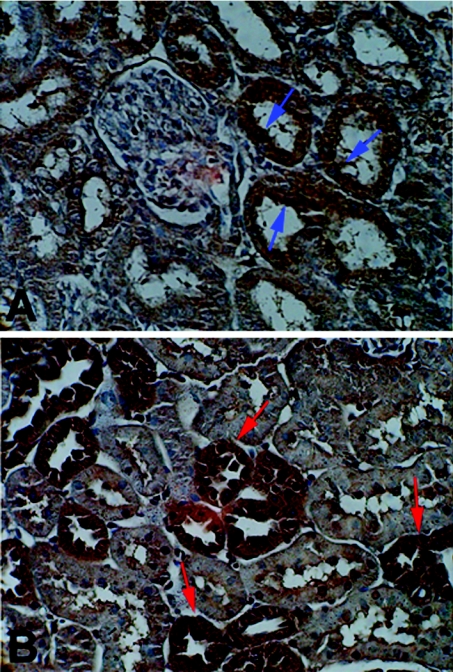
To determine the distribution of A1M proteins, sections of kidney from normal and d3 experimental hyperoxaluric rats were treated with primary antibody against A1M and visualized with avidin-conjugated peroxidase and FastDAB substrate. As shown in Figure 4, the site of A1M protein immunoreactivity, as expected, corresponded with the site of AMBP gene expression. These results further supported the observation that renal tubular cells in response to oxalate do locally synthesize AMBP gene-encoded proteins. In normal rats, a little immunoreactivity was seen on the apical surface of renal tubular cells. It was most probably due to adsorption of A1M proteins present in the glomerular filtrate. The proximal tubular cells do possess receptors, like megalin and cubulin, which mediate the uptake of various proteins from the glomerular filtrate.
Figure 4. Immunocytological detection of A1M protein.
Sections from perfused kidney of normal and hyperoxaluric rats were labelled with anti-A1M antibody and visualized using biotin–avidin conjugated peroxidase and FastDAB substrate system. The dark brown staining of A1M protein (B) in renal tubular epithelial cells indicates the local synthesis of A1M protein and its site of synthesis tallies with the site of AMBP gene expression by in situ hybridization analysis. Slides were counterstained with haematoxylin to visualize other cells in the background. Slides from control animals also showed a little dark brown staining (A). This staining was detected on the cell surface facing the lumen of the tubules, indicating possible absorption/adsorption of A1M from glomerular filtrate.
AMBP gene regulatory transcription factor
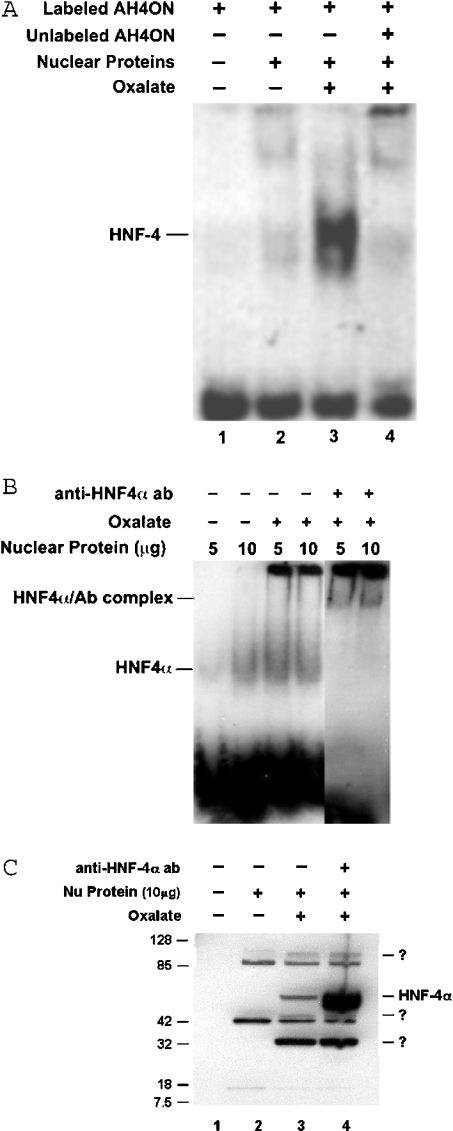
To determine the mechanism of AMBP gene expression regulation in the kidney, we studied the presence of a major AMBP regulatory HNF-4 transcription factor in LLC-PK1 cells. The transcription factor HNF-4 has previously been reported to promote AMBP gene transcription [42] in HepG2 hepatocytes. Nuclear extracts from cells with or without oxalate treatment were incubated with DIG-labelled AH4ON. The DNA–protein complex was run on a 5% polyacrylamide/0.5×TBE gel to observe mobility shift in oligonucleotide migration. As shown in Figure 5(A), AH4ON specifically bound to nuclear protein and this binding was competed out by unlabelled AH4ON. As cis element oligonucleotide specifically binds to HNF-4, these observations indicate that LLC-PK1 cells do express HNF-4 or an HNF-4-like transcription factor and that its specific binding to AH4ON increased with the oxalate treatment of the cells.
Figure 5. AMBP gene regulatory transcription factor.
Liver-specific transcription factor, HNF-4-binding cis element oligonucleotide was DIG-labelled. (A) AH4ON was incubated with nuclear proteins with or without oxalate treatment. HNF-4–DNA complex was run on 5% polyacrylamide/0.5×TBE gel. In lane 1, labelled oligonucleotide was run alone, lanes 2 and 3 had labelled AH4ON and nuclear proteins from untreated and oxalatetreated cells respectively. In lane 4, nuclear proteins were preincubated with 100×unlabelled AH4ON as a competitor before adding labelled AH4OH to determine the specificity of AH4ON. Unlabelled AH4ON completely sequestered HNF-4, indicating that HNF-4–DNA binding was specific. (B) In some experiments, anti-HNF-4α antibody was added to the reaction mixture and incubated for an additional 20 min at 25 °C, before the separation of complexes on 5% polyacrylamide/0.5×TBE gel. Lanes 5 and 6 showed supershift due to oligo–HNF-4α–antibody complex formation. (C) Samples (DNA–protein complexes) from a supershift assay experiment were separated on precast 4–20% polyacrylamide gel under reducing conditions and HNF-4α protein was detected by using anti-HNF-4α antibody. Lanes 3 and 4 show oxalate-specific induction of HNF-4α protein of molecular mass approx. 55 kDa in nuclear extract when compared with that in control (lane 2).
To confirm the identity of HNF-4, anti-HNF-4α antibody was added to the binding reaction mixture and incubated further for 20 min. On separation of oligo complexes on 5% polyacrylamide/0.5×TBE gel, an oligo–protein–antibody complex showed supershift (Figure 5B) due to the presence of the HNF-4α factor.
These results were further supported with experiment demonstrating an oxalate specific increase in the activity of HNF-4α. In brief, DNA–protein complexes from an EMSA-supershift assay experiment were separated on 4–20% precast polyacrylamide gel under reducing conditions and tested for their immunoreactivity with anti-HNF-4α antibody. As shown in Figure 5(C), a band of molecular mass of approx. 55 kDa was observed corresponding to HNF-4α. Interestingly, oxalate-treated samples (lanes 3 and 4) showed a specific induction of HNF-4α when compared with that in control cells (lane 2). Three other bands that immunoreacted were also observed, one (110 kDa) possibly a dimer of HNF-4α.
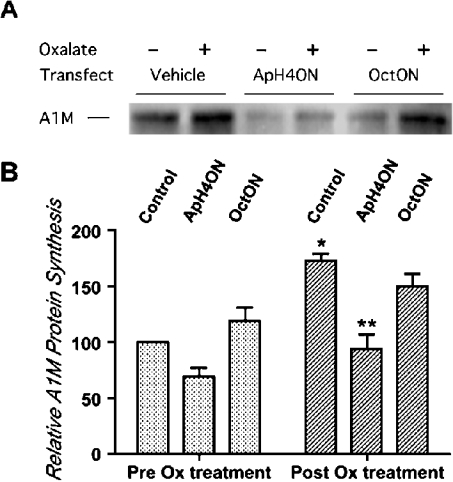
These results were further confirmed by transfection of ApH4ON in LLC-PK1 cells. The transfected oligonucleotide competed with the internal AMBP gene cis element by sequestering HNF-4, thereby attenuating the AMBP gene expression. A non-specific cis element for OctON was used as mock transfection. As shown in Figures 6(A) and 6(B), cells transfected with ApH4ON showed significantly decreased expression of the AMBP gene in untreated as well as in oxalate-treated cells. In control cells vehicle, the transfection reagent SuperFect™ alone induced a small increase in AMBP gene expression. The oligonucleotide corresponding to the octamer-binding site did not alter AMBP gene expression. These results demonstrate that AH4ON specifically bound to HNF-4 and that HNF-4 was responsible for increase in the AMBP gene expression in renal tubular epithelial cells.
Figure 6. Sequestering of HNF-4 by ApH4ON decreases endogenous A1M synthesis in LLC-PK1 cells.
Semi-confluent LLC-PK1 cells were transfected with double-stranded oligonucleotide ApH4ON or OctON (a non-specific oligonucleotide) using SuperFect™ reagent following the manufacturer's instructions (Qiagen). Cells transfected with ApH4ON showed substantially lower endogenous production of A1M, whereas those transfected with OctON did not. (A) A1M protein in cells treated with vehicle alone and cells transfected with ApH4ON and OctON on exposure to oxalate for 3 h. (B) Graphical representation of results shown in (A). Results represent two-way ANOVA with means±S.E.M. (n=3). *P<0.01 when compared with basal levels in cells without oxalate treatment.
DISCUSSION
The organic matrix of kidney stones consists of more than twenty different proteins. Little information is available about their exact role in bio-mineralization. The proteins, which have been shown to inhibit calcium oxalate crystal formation in vitro, are also integral parts of the organic matrix of kidney stones. The association of AMBP gene-regulated proteins with kidney stones and their unfailing presence in the urine formed the basis of this study. We were interested in investigating if AMBP gene was locally expressed in renal cells, and if so, could its expression be affected by high oxalate concentration in the renal tubular system?
The aetiology of urolithiasis (kidney stones) in the majority of cases is linked to a high concentration of oxalate in the urine, a condition known as hyperoxaluria. In mammals, in spite of heavy doses of dietary intake, the concentration of oxalate in serum remains low; however, in glomerular filtrate, oxalate concentrations can become fairly high. With the reabsorption of water, the oxalate concentration in a tubular system rises above the saturation point (supersaturation) and oxalate forms calcium oxalate crystals that are excreted in the urine (crystalluria). Crystalluria has been reported in normal subjects as well as in patients with kidney stone disease, although in the latter case crystals are usually bigger and form larger aggregates [43,44]. Do these fluctuations in oxalate concentrations have implications for AMBP gene expression in the renal tubular system? The real-time PCR analyses clearly demonstrated (Figure 2) that AMBP gene expression in renal tubular cells is sensitive to oxalate concentrations. Similarly, immunoprecipitation and immunoblotting assays demonstrate de novo synthesis of A1M protein in LLC-PK1 cells.
Owing to its covalent binding with a variety of unknown chromophores, the mass and charge of A1M protein vary considerably. The mass of A1M isolated under reducing and denaturing conditions was approx. 60–65 kDa in LLC-PK1 cells. Is this mass due to the absence of cleavage of precursor AMBP protein and that A1M and bikunin co-precipitate? An anti-bikunin antibody did show immunoreactivity for bikunin, thus indicating co-precipitation of bikunin with A1M. The renal proximal tubular cells perhaps lack furin-like enzyme required for cleavage of the precursor protein. Evidently, the co-precipitation of A1M and bikunin accounts for the molecular mass of the immunoreactive band. A1M also formed a complex (∼200 kDa) with an unidentified protein that did not resolve even on boiling with 2-mercaptoethanol. Under reducing conditions alone it formed complexes with many other proteins with molecular masses ranging between 30 and 200 kDa (results not shown). The variation in mass of A1M in different tissues and among different species has also been reported previously by other workers [6,31,41,45].
In rat, the mass of urinary A1M was approx. 36 kDa. In normal rats it showed different extents of truncation. The mass of truncated A1M was approx. 30 kDa (Figure 3B). Allhorn et al. [3], have claimed that removal of C-terminal LIPR tetrapeptide from mature A1M was necessary for its biological activity [3]. The striking variation in levels of truncated A1M among normal rats (n=9) was difficult to explain. A1M isolated from LLC-PK1 cell lysate or from supernatant media showed no truncation (Figure 1E).
The in situ hybridization (Figure 3B) and immunocytochemical (Figure 4B) staining of A1M protein demonstrated that local transcription and translation of the AMBP gene was restricted to cells lining the proximal tubule and collecting ducts. Hyperoxaluric rats with crystalluria or calcium oxalate crystals in their kidneys showed decreased levels of urinary A1M (Figure 1C). This observation tallies with similar observations made previously in human calcium oxalate kidney stone formers [31]. Whether this decrease in urinary A1M was due to its binding to calcium oxalate crystals or due to its increased catabolism in proximal tubular cells, or both, is not certain.
In liver, the AMBP gene has previously been reported to be expressed constitutively [5–7,46], although the complex nature of the AMBP gene core-enhancer and four transcription factors [7,8], which can exert both positive and negative influence in different combinations, indicate its inducible nature. Its core-enhancer sequence consists of six clustered boxes (1–6), whereas three other boxes 7, 8 and 9 are localized within boxes 2, 1 and 6 respectively [7]. These enhancer boxes are binding sites for four liver-specific nuclear factors (HNF-1–4). The transcription factor HNF-4 has the major AMBP-enhancing effect, except for a combination of HNF-4-box 9, which has repressive activities [8]. Studies on the AMBP cis element oligonucleotide, which specifically binds to HNF-4, demonstrated the existence of HNF-4 or HNF-4-like nuclear factor in LLC-PK1 cells. The EMSA as well as oligonucleotide transfection assays (Figures 5 and 6) showed that binding of HNF-4 to its corresponding AMBP cis element was increased by oxalate treatment. The supershift assay and immunoreactivity of anti-HNF-4α antibody confirmed the identity of the transcription factor as HNF-4α. These observations indicate the existence of identical AMBP gene-regulatory mechanisms in the kidney and liver, although further study is required to understand the mechanism in detail.
Besides their association with kidney stone, A1M and bikunin show many other properties. A1M possesses generic immunosuppressive [47] properties and inhibits antigen-stimulated proliferation and secretion of interleukin-2 [48] in leukocytes, generation of free radicals and interleukins in human peripheral monocytes [49], and attachment of monocytes to endothelial cells [50]. It has a strong mitogenic effect on guinea pig and human lymphocytes; however, in serum its mitogenic activities are suppressed by other proteins [51]. Bikunin contains two tandemly repeated Kunitz-type homologies. It complexes with one or more of the three heavy-chain proteins (H1, H2 and H3) through chondroitin sulphate to form different inter-α-inhibitors. Inter-α-inhibitors are extracellular proteins and are important modulators of extracellular matrix composition [52]. Bikunin also blocks constitutive expression of a major inflammatory protein, TGF-β1, in cancer cells [28–30].
The exact significance of local synthesis of these proteins, namely A1M and bikunin [53,54], in renal tubule cells is not clear, but they do have immunosuppressive properties and both are inhibitors of calcium oxalate crystal formation in vitro. In conclusion, AMBP gene expression in renal tubular cells may be important in imparting general immunoprotection against oxalate toxicity.
Acknowledgments
We are thankful to P. Glenton, J. Cornelius and H. Hatch for their valuable technical help. We are also thankful to Department of Pathology, Immunology and Laboratory Medicine at University of Florida and National Institutes of Health (ID# RO1DK53962 to S.R.K.) for providing financial support for this work.
References
- 1.Hanley S., Powell R. Sequence of a cDNA clone encoding the Atlantic salmon alpha-1-microglobulin/bikunin protein. Gene. 1994;147:297–298. doi: 10.1016/0378-1119(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 2.Hanley S., McIneney J. O., Powell R. Sequence analysis and evolutionary aspects of piscine alpha-1-microglobulin/bikunin mRNA transcripts. Mol. Mar. Biol. Biotechnol. 1995;4:105–109. [PubMed] [Google Scholar]
- 3.Allhorn M., Berggard J., Nordberg J., Olsson M. L., Akerstrom B. Processing of the lipocalin α1-microglobulin by hemoglobin induces heme-binding and heme-degradation properties. Blood. 2002;99:1894–1901. doi: 10.1182/blood.v99.6.1894. [DOI] [PubMed] [Google Scholar]
- 4.Hardison R. The evolution of hemoglobin: studies of a very ancient protein suggest that changes in gene regulation are an important part of the evolutionary story. American Scientist. 1999;87:126–137. [Google Scholar]
- 5.Diarra-Mehrpour M., Bourguignon J., Sesboue R., Salier J. P., Leveillard T., Martin J. P. Structural analysis of the human inter-alpha-trypsin inhibitor light-chain gene. Eur. J. Biochem. 1990;191:131–139. doi: 10.1111/j.1432-1033.1990.tb19102.x. [DOI] [PubMed] [Google Scholar]
- 6.Tyagi S., Salier J. P., Lal S. K. The liver specific human α1-microglobulin/bikunin precursor (AMBP) is capable of self association. Arch. Biochem. Biophys. 2002;399:66–72. doi: 10.1006/abbi.2001.2745. [DOI] [PubMed] [Google Scholar]
- 7.Rouet P., Raguenez G., Tronche F., Mfou'ou V., Salier J. P. Hierarchy and positive/negative interplays of the hepatocyte nuclear factors HNF1, -3 and -4 in the liver-specific enhancer for the human alpha-1-microglobulin/bikunin precursor. Nucleic Acids Res. 1995;23:395–404. doi: 10.1093/nar/23.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rouet P., Raguenez G., Ruminy P., Salier J. P. An array of binding sites for hepatocyte nuclear factor 4 of high and low affinities modulates the liver-specific enhancer for the human alpha1-microglobulin/bikunin precursor. Biochem. J. 1998;334:577–584. doi: 10.1042/bj3340577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molloy S. S., Bresnahan P. A., Leppla S., Klimpel K. R., Thomas G. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J. Biol. Chem. 1992;267:16396–16402. [PubMed] [Google Scholar]
- 10.Bratt T., Olsson H., Sjoberg E. M., Jergil B., Akerstrom B. Cleavage of the 1-microglobulin-bikunin precursor is localized to the Golgi apparatus of rat liver cells. Biochim. Biophys. Acta. 1993;1157:147–154. doi: 10.1016/0304-4165(93)90058-g. [DOI] [PubMed] [Google Scholar]
- 11.Ekstrom B., Berggard I. Human alpha-1-microglobulin. Purification procedure, chemical and physiochemical properties. J. Biol. Chem. 1977;252:8048–8057. [PubMed] [Google Scholar]
- 12.Flower D. R. Experimentally determined lipocalin structures. Biochim. Biophys. Acta. 2000;1482:46–56. doi: 10.1016/s0167-4838(00)00147-3. [DOI] [PubMed] [Google Scholar]
- 13.Grubb A., Mendez E., Fernandez-Luna J. L., Lopez C., Mihaesco E., Vaerman J. P. The molecular organization of the protein HC-IgA complex. J. Biol. Chem. 1986;261:14313–14320. [PubMed] [Google Scholar]
- 14.Berggard T., Thelin N., Falkenberg C., Enghild J. J., Akerstrom B. Prothrombin, albumin and immunoglobulin A form covalent complexes with alpha-1-microglobulin in human plasma. Eur. J. Biochem. 1997;245:676–683. doi: 10.1111/j.1432-1033.1997.00676.x. [DOI] [PubMed] [Google Scholar]
- 15.Falkenberg C., Enghild J. J., Thogersen I. B., Salvesen G., Akerstrom B. Isolation and characterization of fibronectin-alpha-1-microglobulin complex in rat plasma. Biochem. J. 1994;301:745–751. doi: 10.1042/bj3010745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falkenberg C., Allhorn M., Thogersen I. B., Valnickova Z., Pizzo S. V., Salvesen G., Akerstrom B., Enghild J. J. Alpha-1-microglobulin destroys the proteinase inhibitory activity of alpha-1-inhibitor-3 by complex formation. J. Biol. Chem. 1995;270:4478–4483. doi: 10.1074/jbc.270.9.4478. [DOI] [PubMed] [Google Scholar]
- 17.Logdberg L. E., Akerstrom B., Badve S. Tissue distribution of the lipocalin alpha-1 microglobulin in the developing human fetus. J. Histochem. Cytochem. 2000;48:1545–1552. doi: 10.1177/002215540004801111. [DOI] [PubMed] [Google Scholar]
- 18.Logdberg L., Akerstrom B. Immunosuppressive properties of alpha 1-microglobulin. Scand. J. Immunol. 1981;13:383–390. doi: 10.1111/j.1365-3083.1981.tb00148.x. [DOI] [PubMed] [Google Scholar]
- 19.Logdberg L., Akerstrom B., Shevach E. Alpha 1-microglobulin is mitogenic for guinea pig lymphocytes. Scand. J. Immunol. 1986;24:575–581. doi: 10.1111/j.1365-3083.1986.tb02173.x. [DOI] [PubMed] [Google Scholar]
- 20.Babiker-Mohamed H., Akerstrom B., Logdberg L. Mitogenic effect of alpha 1-microglobulin on mouse lymphocytes. Evidence of T- and B-cell cooperation, B-cell proliferation, and a low-affinity receptor on mononuclear cells. Scand. J. Immunol. 1990;32:37–44. doi: 10.1111/j.1365-3083.1990.tb02889.x. [DOI] [PubMed] [Google Scholar]
- 21.Babiker-Mohamed H., Olsson M. L., Boketoft A., Logdberg L., Akerstrom B. Alpha 1-microglobulin is mitogenic to human peripheral blood lymphocytes. Regulation by both enhancing and suppressive serum factors. Immunobiology. 1990;180:221–234. doi: 10.1016/S0171-2985(11)80330-X. [DOI] [PubMed] [Google Scholar]
- 22.Wester L., Michaelsson E., Holmdahl R., Olofsson T., Akerstrom B. Receptor for alpha1-microglobulin on T lymphocytes: inhibition of antigen-induced interleukin-2 production. Scand. J. Immunol. 1998;48:1–7. doi: 10.1046/j.1365-3083.1998.00378.x. [DOI] [PubMed] [Google Scholar]
- 23.Wester L., Fast J., Labuda T., Cedervall T., Wingardh K., Olofsson T., Akerstrom B. Carbohydrate groups of alpha1-microglobulin are important for secretion and tissue localization but not for immunological properties. Glycobiology. 2000;10:891–900. doi: 10.1093/glycob/10.9.891. [DOI] [PubMed] [Google Scholar]
- 24.Santin M., Cannas M. Collagen-bound alpha1-microglobulin in normal and healed tissues and its effect on immunocompetent cells. Scand. J. Immunol. 1999;50:289–295. doi: 10.1046/j.1365-3083.1999.00597.x. [DOI] [PubMed] [Google Scholar]
- 25.Shikimi T., Uegaki J., Inagaki T., Mitsuoka S. Changes in the regression slope correlating between urinary contents of alpha-1-microglobulin and ulinastatin and its relation to severity in mood disorders. Nurs. Health Sci. 2001;3:95–100. doi: 10.1046/j.1442-2018.2001.00078.x. [DOI] [PubMed] [Google Scholar]
- 26.Shikimia T., Kakub K., Uegakib J., Inagakib T., Senob H., Ishinoc H., Takaoria S. Serum contents of the free forms of alpha1-microglobulin and ulinastatin: relation to diseased states in patients with mood disorders. Neuropsychobiology. 2001;43:145–149. doi: 10.1159/000054883. [DOI] [PubMed] [Google Scholar]
- 27.Kaku K., Shikimi T., Kamisaki Y., Inagaki T., Ishino H., Okunishi H., Takaori S. Nullification of a positive correlation between urinary contents of alpha1-microglobulin and ulinastatin with intracerebroventricularly administered 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and 1-methyl-4-phenylpyridinium (MPP+) in mice. Psychopharmacology. 1998;136:374–378. doi: 10.1007/s002130050580. [DOI] [PubMed] [Google Scholar]
- 28.Hirashima Y., Kobayashi H., Suzuki M., Tanaka Y., Kanayama N., Terao T. Transforming growth factor-beta1 produced by ovarian cancer cell line HRA stimulates attachment and invasion through an up-regulation of plasminogen activator inhibitor type-1 in human peritoneal mesothelial cells. J. Biol. Chem. 2003;278:26793–26802. doi: 10.1074/jbc.M212187200. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi H., Suzuki M., Tanaka Y., Kanayama N., Terao T. A Kunitz-type protease inhibitor, bikunin, inhibits ovarian cancer cell invasion by blocking the calcium-dependent transforming growth factor-beta 1 signaling cascade. J. Biol. Chem. 2003;278:7790–7799. doi: 10.1074/jbc.M210407200. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki M., Kobayashi H., Tanaka Y., Hirashima Y., Kanayama N., Takei Y., Saga Y., Suzuki M., Itoh H., Terao T. Suppression of invasion and peritoneal carcinomatosis of ovarian cancer cell line by overexpression of bikunin. Int. J. Cancer. 2003;104:289–302. doi: 10.1002/ijc.10950. [DOI] [PubMed] [Google Scholar]
- 31.Tardivel S., Medetognon J., Randoux C., Kebede M., Drueke T., Daudon M., Hennequin C., Lacour B. Alpha-1-microglobulin: inhibitory effect on calcium oxalate crystallization in vitro and diseased urinary concentration in calcium oxalate stone formers. Urol. Res. 1999;27:243–249. doi: 10.1007/s002400050117. [DOI] [PubMed] [Google Scholar]
- 32.Atmani F., Mizon J., Khan S. R. Inter-a-inhibitor: a protein family involved in the inhibition of calcium oxalate crystallization. Scanning Microsc. 1996;10:425–433. [PubMed] [Google Scholar]
- 33.Atmani F., Khan S. R. Inter-a-inhibitor, another serum protein with potential involvement in calcium oxalate nephrolithiasis. J. Am. Soc. Nephrol. 1996;7:1798A. [Google Scholar]
- 34.Atmani F., Lacour B., Strecker G., Parvy P., Drueke T., Daudon M. Molecular characteristics of uronic-acid-rich protein, a strong inhibitor of calcium oxalate crystallization in vitro. Biochem. Biophys. Res. Commun. 1993;19:1158–1165. doi: 10.1006/bbrc.1993.1338. [DOI] [PubMed] [Google Scholar]
- 35.Atmani F., Mizon J., Khan S. R. Identification of uronic acid-rich protein as urinary bikunin, the light chain of inter-a-inhibitor. Eur. J. Biochem. 1996;236:984–990. doi: 10.1111/j.1432-1033.1996.00984.x. [DOI] [PubMed] [Google Scholar]
- 36.Atmani F., Khan S. R. Role of urinary bikunin in the inhibition of calcium oxalate crystallization. J. Am. Soc. Nephrol. 1999;10:S385–S388. [PubMed] [Google Scholar]
- 37.Akerstrom B., Bratt T., Enghild J. J. Formation of the α1-microglobulin chromophore in mammalian and insect cells: a novel post-translational mechanism? FEBS Lett. 1995;362:50–54. doi: 10.1016/0014-5793(95)00206-o. [DOI] [PubMed] [Google Scholar]
- 38.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 39.Chouard T., Blumenfeld M., Bach I., Vandekerckhove J., Cereghini S., Yaniv M. A distal dimerization domain is essential for DNA-binding by the atypical HNF1 homeodomain. Nucleic Acid Res. 1990;18:5853–5863. doi: 10.1093/nar/18.19.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grewal J. S., Bag J. Slow troponin C gene expression in chicken heart and liver is regulated by similar enhancers. FEBS Lett. 1996;383:267–272. doi: 10.1016/0014-5793(96)00247-5. [DOI] [PubMed] [Google Scholar]
- 41.Berggard T., Oury T. D., Thergersen I. B., Akerstrom B., Enghild J. J. α1-Microglobulin is found both in blood and in most tissues. J. Histochem. Cytochem. 1998;46:887–893. doi: 10.1177/002215549804600803. [DOI] [PubMed] [Google Scholar]
- 42.Rouet P., Raguenez G., Tronche F., Yaniv M., N'Guyen C., Saleir J. P. A potent enhancer made of clustered liver-specific elements in the transcription control sequences of human alpha 1-microglobulin/bikunin gene. J. Biol. Chem. 1992;267:20765–20773. [PubMed] [Google Scholar]
- 43.Robertson W. G., Peacock M., Nordin B. E. C. Calcium crystalluria in recurrent renal stone formers. Lancet. 1969;2:21–24. doi: 10.1016/s0140-6736(69)92598-7. [DOI] [PubMed] [Google Scholar]
- 44.Crassweller P. O., Brandes L., Katirtzoglou A., Oreopoulos D. G. Studies on crystalluria in recurrent calcium lithiasis. Can. J. Surg. 1979;22:527–529. [PubMed] [Google Scholar]
- 45.Akerstrom B., Bratt T., Enghild J. J. Formation of the α1-microglobulin chromophores in mammalian and insect cells: a novel post-translational mechanism. FEBS Lett. 1995;362:50–54. doi: 10.1016/0014-5793(95)00206-o. [DOI] [PubMed] [Google Scholar]
- 46.Berggard T., Enghild J. J., Badve S., Salafia C. M., Logdberg L., Akerstrom B. Histologic distribution and biochemical properties of alpha 1-microglobulin in human placenta. Am. J. Reprod. Immunol. 1999;41:52–60. doi: 10.1111/j.1600-0897.1999.tb00075.x. [DOI] [PubMed] [Google Scholar]
- 47.Logdberg L., Wester L. Immunocalins: a lipocalin subfamily that modulates immune and inflammatory responses. Biochim. Biophys. Acta. 2000;1482:284–297. doi: 10.1016/s0167-4838(00)00164-3. [DOI] [PubMed] [Google Scholar]
- 48.Wester L., Michaelsson E., Holmdahl R., Olofsson T., Akerstrom B. Receptor for alpha1-microglobulin on T lymphocytes: inhibition of antigen-induced interleukin-2 production. Scand. J. Immunol. 1998;48:1–7. doi: 10.1046/j.1365-3083.1998.00378.x. [DOI] [PubMed] [Google Scholar]
- 49.Takagi K., Itoh Y., Enomoto H., Koyamaishi Y., Maeda K., Kawai T. A comparative study of serum alpha 1-microglobulin and beta 2-microglobulin levels in cancerous and other diseases. Clin. Chim. Acta. 1980;108:277–283. doi: 10.1016/0009-8981(80)90014-5. [DOI] [PubMed] [Google Scholar]
- 50.Logdberg L., Akerstrom B. Immunosuppressive properties of alpha-1-microglobulin. Scand. J. Immunol. 1981;13:383–390. doi: 10.1111/j.1365-3083.1981.tb00148.x. [DOI] [PubMed] [Google Scholar]
- 51.Babiker M. H., Olsson M. L., Boketoft A., Logdberg L., Akerstrom B. Alpha-1 microglobulin is mitogenic to human peripheral blood lymphocytes regulation by both enhancing and suppressive serum factors. Immunobiology. 1990;180:221–234. doi: 10.1016/S0171-2985(11)80330-X. [DOI] [PubMed] [Google Scholar]
- 52.Chen L., Mao S. J. T., Larsen W. J. Identification of a factor in fetal bovine serum that stabilizes the cumulus extracellular matrix, a role for a member of the inter-alpha-inhibitor family. J. Biol. Chem. 1992;267:12380–12386. [PubMed] [Google Scholar]
- 53.Iida S., Peck A. B., Johnson-Tardieu J., Moriyama M., Byer K. J., Khan S. R. Temporal changes in mRNA expression for bikunin in the kidneys of rats during calcium oxalate nephrolithiasis. J. Am. Soc. Nephrol. 1999;10:986–996. doi: 10.1681/ASN.V105986. [DOI] [PubMed] [Google Scholar]
- 54.Iida S., Peck A. B., Byer K. J., Khan S. R. Expression of bikunin mRNA in renal epithelial cells after oxalate exposure. J. Urol. 1999;162:1480–1486. [PubMed] [Google Scholar]