Abstract
When the yeast protein Ypr140w was expressed in Escherichia coli, a lyso-PC [lysophosphatidylcholine (1-acylglycerophosphorylcholine)] acyltransferase activity was found associated with the membranes of the bacteria. To our knowledge, this is the first identification of a protein capable of catalysing the acylation of lyso-PC molecules to form PC. Fluorescence microscopy analysis of living yeasts revealed that the fusion protein Ypr140w–green fluorescent protein is targeted to the mitochondria. Moreover, in contrast with wild-type cells, in the absence of acyl-CoA, the yeast mutant deleted for the YPR140w gene has no lyso-PC acyltransferase activity associated with the mitochondrial fraction. When yeast cells were grown in the presence of lactate, the mutant synthesized 2-fold more triacylglycerols when compared with the wild-type. Moreover, its mitochondrial membranes contained a lesser amount of PC and cardiolipin, and the fatty acid composition of these latter was greatly changed. These modifications were accompanied by a 2-fold increase in the respiration rates (states 3 and 4) of the mitochondria. The relationship between the deletion of the YPR140w gene and the lipid composition of the ypr140wΔ cells is discussed.
Keywords: acyltransferase, cardiolipin, lysophosphatidylcholine, mitochondria, oil, tafazzin
Abbreviations: DAG, diacylglycerol; DASPMI, 2-(4-(dimethylamino)steryl)-1-methylpyridinium iodide; ER, endoplasmic reticulum; FAMES, fatty acid methyl esters; GFP, green fluorescent protein; HA, haemagglutinin; HPTLC, high-performance TLC; IPTG, isopropyl β-D-thiogalactoside; PA, phosphatidic acid; lyso-PA, 1-acylglycerophosphatidic acid; PC, phosphatidylcholine; lyso-PC, 1-acylglycerophosphorylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; TAG, triacylglycerol
INTRODUCTION
Lyso-PC [lysophosphatidylcholine (1-acylglycerophosphorylcholine] acyltransferases (EC 2.3.1.23) catalyse the acylation of lyso-PC molecules to form PC and are involved in several important physiological processes. For example, in animal cells, the acylation of lyso-PC molecules appears to be an important process of the cardiolipin remodelling occurring during cell death signalling (see [1] for a review). It has been proposed recently [2,3] that such a remodelling in human cells involves tafazzin, a protein showing some similarities to glycero-3-phosphate and lyso-PA [lyso-phosphatidic acid (1-acylglycerophosphatidic acid)] acyltransferases and linked to the Barth syndrome (an X-linked pediatric cardiomyopathy [4]. In plant cells, lyso-PC acyltransferases located in the plastid envelope [5] and in mitochondrial membranes [6] are supposed to be the key enzymes for the import of PC from the ER (endoplasmic reticulum) membranes into organelles. Briefly, on the basis of in vitro [5–7] and in vivo [8,9] studies, it has been proposed that this import involves a release of lyso-PC from endomembranes, a transfer of these molecules to chloroplasts and mitochondria and their acylation in the organelles to synthesize PC. Similarly, as noted by Nebauer et al. [10] in a recent review on the biosynthesis and cellular dynamics of lipids in yeast, mitochondria from Saccharomyces cerevisiae do not contain “a biosynthetic pathway for PC formation and all mitochondrial PC has to be imported”. Hence, as in plants, one can imagine that this import may involve a transfer of lyso-PC molecules between the ER and organelles. This would require the presence of a lyso-PC acyltransferase in mitochondria from S. cerevisiae that has not yet been evidenced. Recent results obtained by de Kroon et al. [11] suggest that a continuous equilibration of phosphatidylmonomethylethanolamine and phosphatidyldimethylethanolamine between the ER and mitochondria leads to the import of PC into these organelles. Their results do not exclude a transfer of lyso-PC between endomembranes and organelles. The exchange of these methylated phospholipids may occur through the ‘mitochondria-associated membranes’, known to be involved in the import of phosphatidylserine into mitochondria for decarboxylation to PE (phosphatidylethanolamine; see [12] for a review). In contrast with PS and PE, so far, there has been no role assigned for mitochondria-associated membranes to account for the presence of PC in mitochondrial membranes (see [10,12] for reviews).
Another interesting feature may be the involvement of the lyso-PC acyltransferases in oil synthesis in yeast and plants. In yeast, the final step of TAG (triacylglycerol) synthesis mainly occurs by the acylation of DAG (diacylglycerol) utilizing either acyl-CoAs or the acyl chain esterified to the sn-2 (stereospecific numbering for glycerol, regardless of substituents) position of PC (see [13] for a review). The first pathway catalysed by DAG acyltransferases takes place in lipid bodies and ER membranes, whereas the second pathway (catalysed by enzymes similar to lecithin:cholesterol acyltransferase) is exclusively located in the ER. Hence lipid bodies are capable of synthesizing TAG; however, as noted by Mullner and Daum [14], the ER contains the whole set of TAG-synthesizing enzymes and is the major site of TAG formation in yeast. Similarly, in various seed crops, it has been shown that the oil synthesis can occur through a transfer of the acyl chain esterified to the sn-2 position of PC to DAG molecules [15].
In S. cerevisiae, only one gene encoding a lysolipid acyltransferase (a lyso-PA acyltransferase) was assigned [16], whereas we observed a lyso-PC acyltransferase activity associated with mitochondria (see below) in addition to the acyl-CoA lyso-PC acyltransferase activity associated with the endomembranes described previously [17]. Hence we sought homologies between putative or known lysolipid acyltransferases from different organisms and open reading frames of yeast to identify at least one gene coding for a lyso-PC acyltransferase.
EXPERIMENTAL
Materials
TLC plates were HPTLC (high performance TLC) silica-gel 60 plates (Merck 60 F254). [1-14C]1-palmitoyl-sn-glycerol-3-phosphocholine (57 mCi/mmol) was purchased from New England Nuclear (Boston, MA, U.S.A.). Antibodies raised against HA (haemagglutinin) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.). All other reagents were from Sigma (St. Louis, MO, U.S.A.).
Methods
Yeast culture
The S. cerevisiae strains used in the present study are listed in Table 1. The cells were grown in a shaking incubator at 30 °C, in 500 ml Erlenmeyer flasks containing 100 ml of liquid medium YP (10 g yeast extract, 10 g peptone, 1 g potassium phosphate and 1.2 g ammonium sulphate) supplemented with 2% glucose (YPD) or 2% lactate (YPL) as the carbon substrate. The pH was set at 5.5. The cells were harvested at the exponential and stationary growth phases. Cellular ATP and ADP concentrations were determined on HClO4 extracts as described by Beauvoit et al. [18].
Table 1. S. cerevisiae strains used in the present study.
| Strain | Relevant genotype | Source |
|---|---|---|
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | EUROSCARF |
| Ypr140wΔ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YPR140W::kanMX4 | EUROSCARF |
| W303-1A | MATa ade2-1 his3-11 leu2-3 trp1Δ2 ura3-52 | |
| WKEN011A (Δcls1) | MATα ade2-1 his3-11 leu2-3 trp1Δ2 ura3-52 YDL142C::kanMX4 | EUROSCARF |
Preparation of spheroplasts and isolation of mitochondria and microsomes
Bioenergetic and biochemical analysis at the mitochondrial level requires mitochondria of good quality. The method described represents a modification of a previously published procedure [19]. Cells were grown at 30 °C aerobically under vigorous shaking (200 rev./min) in 5 litre Erlenmeyer flasks containing 1 litre of YPL medium. Cells were harvested at the mid-exponential growth phase (centrifugation for 5 min at 5000 g). The pellet was washed twice with ice-cold distilled water and then resuspended in pre-warmed buffer 1 (0.1 M Tris/HCl, 0.5 M 2-mercaptoethanol, pH 9.3, and 20 ml of buffer/g dry weight of cells). Gentle shaking of cells was maintained for 15 min at 30 °C before cells were pelleted and washed three times with ice-cold buffer 2 (10 mM Tris/HCl and 0.5 M KCl, pH 7.0). The cell pellet was resuspended in a pre-warmed buffer 3 (10 mM citrate, 10 mM disodium phosphate, 1.35 M sorbitol, 1 mM EGTA, pH 7.2, and 10 ml of buffer/g dry weight of cells) containing zymolyase (ICN Biochemicals, Cleveland, OH, U.S.A.) at 15 mg/g dry weight of cells. Digestion of cell walls was performed at 30 °C under gentle shaking. Digestion was monitored by observing the cells diluted in water under a light microscope. Spheroplasts appeared larger and of uniform hue compared with undigested cells, which appeared smaller and clear with a dark wall.
All subsequent steps were performed at 4 °C. The spheroplast suspension was centrifuged at 16000 g for 10 min. The pellet was washed twice with buffer 4 (10 mM Tris/maleate, 0.75 M sorbitol, 0.4 M mannitol, 2 mM EGTA and 0.1% BSA, pH 6.8) before being resuspended in 10 ml/g dry weight in buffer 5 (10 mM Tris/maleate, 0.6 M mannitol, 2 mM EGTA, 0.5 mM phosphate and 0.2% BSA, pH 6.8). The suspension was homogenized vigorously using a Potter–Elvehjem homogenizer and then washed by at least 4-fold dilution in buffer 5 and centrifuged at 500 g (low speed) for 10 min. The supernatant was carefully decanted into SS34 tubes (Sorvall, Newton, CT, U.S.A.) and centrifuged at 16000 g (high speed) for 10 min. The supernatant (supernatant A) was removed and the pellet was gently resuspended in a small volume of buffer 6 (10 mM Tris/maleate, 0.6 M mannitol, 2 mM EGTA, pH 6.8, and 0.5 mM phosphate). The suspension was diluted in buffer 6 and two further low-speed/high-speed centrifugation and resuspension cycles were performed. The mitochondrial pellet was finally resuspended in a small volume of buffer 6 to achieve a final protein concentration of 40–60 mg/ml and assayed by the biuret method using BSA as standard.
Mitochondria were frozen by dispensing the mitochondrial suspension in small droplets, through a syringe, into liquid nitrogen. The frozen beads were then stored at −80 °C. Just before use, mitochondria were diluted in a buffer (25 mM Tris and 0.4 M mannitol, pH 7.0). Microsomes were obtained by centrifugation (15 min and 150000 g using a Hitachi centrifuge) of supernatant A (see above) and pellets were then resuspended in a buffer (25 mM Tris and 0.33 M mannitol, pH 7.0). The protein amount was determined using bicinchoninic acid (Pierce), with BSA as the standard.
Cloning and ectopic expression of the gene in Escherichia coli and isolation of E. coli membranes
The YPR140w S. cerevisiae gene was amplified by PCR. The PCR product was first subcloned in pGEM T Easy vector (Promega, Charbonnieres les Bains, France). Then, we used a set of sense and antisense primers containing the appropriate restriction sites for cloning sequences of interest in the pET-15b vector (Novagen, Merck Biosciences, Badsoden, Germany). To obtain a sequence encoding the Ypr140w–HA (haemagglutinin) fusion protein, the HA tag (ATGTATCCATATGACGTGCCGGACTACGCCTCCCTC encoding the following peptide: MYPYDVPDYASL) was included in the primer. All constructs were verified by sequencing.
C41(DE3) E. coli bacteria (Avidis, Saint-Beauzire, France) were further transformed with the pET-15b containing the putative lyso-PC acyltransferase gene with or without the HA tag. Cells were grown overnight in Luria–Bertani medium with 50 μg/ml ampicillin at 37 °C. The overnight culture was diluted (1:20) and the culture was continued in 250 ml flasks at 37 °C with shaking at 280 rev./min. When the culture reached an absorbance A600 0.6, expression of the putative lyso-PC acyltransferase was induced by the addition of 1 mM IPTG (isopropyl β-D-thiogalactoside). The culture was then allowed to grow for 3 h at 30 °C. Bacterial membranes were isolated as follows: 50 ml of culture was pelleted and cells were resuspended in 2.5 ml of 0.2 M Tris/HCl (pH 8.0) buffer. First, 2.5 ml of 0.2 M Tris/HCl, 1 M sucrose and 1 mM EDTA (pH 8.0) buffer, then 25 μl of lysozyme (1 g/l) and, finally, 10 ml of water were added, and the mixture was gently stirred for 30 min at room temperature (20 °C). Membranes were then spun down by centrifugation (15 min and 150000 g, using a Hitachi centrifuge) and the pellet was suspended in a 50 mM Tris/HCl (pH 8.0) buffer; after another centrifugation, the membranes were suspended in a 50 mM Tris/HCl (pH 8.0) buffer. The amount of membrane-bound protein was determined using bicinchoninic acid (Pierce), with BSA as the standard.
Expression of the Ypr140w–GFP (green fluorescent protein) fusion protein in yeast and fluorescence microscopy measurements
The YPR140w-GFP fusion gene was obtained directly by homologous recombination in yeast. The 400 nt upstream the ATG and the entire coding sequence without the stop codon of YPR140w were amplified by PCR with the following primers: YPR140wup, ACTCACTATAGGGCGAATTGCTGATTGTGTTTGGTAGAAATGGAGCTATT; and YPR140wdw, AGCCCGGGGGATCCACTAGTATCATCCTTACCCTTTGGTTTACCCTCTGG. The PCR product was used to transform ypr140wΔ mutant cells with the plasmid pGB7 (kindly provided by Professor M. Bonneu, Plateforme de génomique fonctionelle, Bordeaux, France) derived from pRS316 [20] containing the yGFP3 gene [21]. The cells grew on a minimal medium: YNB [yeast nitrogen base without amino acids (0.175%), ammonium sulphate (0.5%), potassium phosphate (0.1%), Drop-Mix (0.2%) and auxotrophic requirements (His, Leu; 0.01%), pH 5.5] supplemented with 2% lactate as carbon source. Expression of the Ypr140w–GFP fusion protein in ypr140wΔ cells induced an increase in the cardiolipin content from 8.0±0.6 to 13.0±0.9% of total polar lipids, whereas 15.9±2.7% were found in wild-type cells expressing the fusion protein (n=3). For fluorescence microscopy measurements, 107 cells were then spun down, suspended in 50 μl of water and stained with 5 μM DASPMI [2-(4-(dimethylamino)steryl)-1-methylpyridinium iodide]. Fluorescent cells were visualized on a Leica microscope. The images were acquired with an SIS camera and processed with Corel Draw 9.0 suite software.
Western-blot analysis
Bacterial culture (1 ml) either induced with IPTG or non-induced was pelleted by centrifugation at 5000 g for 15 min and resuspended in 100 μl of SDS loading buffer. To evaluate the expression of the recombinant lyso-PC acyltransferase in the C41 (DE3) by immunoblotting, non-reduced SDS-denatured proteins of lysates (10 μg) from these recombinant bacteria were separated first by SDS/PAGE (12% polyacrylamide) and electrotransferred on to a nitrocellulose membrane. After transfer, the nitrocellulose membrane was soaked in 24 mM Trizma base, 136 mM NaCl, 2.6 mM KCl and 5% (w/v) non-fat dry milk powder (pH 8.2) for 1 h. Nitrocellulose membranes were then incubated with rabbit polyclonal anti-HA (Santa Cruz Biotechnology) diluted 1:2500 in the washing buffer [24 mM Trizma base, 136 mM NaCl, 2.6 mM KCl and 0.05% (v/v) Tween 20] for 2 h. After several washings in the same buffer, the nitrocellulose membrane was incubated with peroxidase-conjugated goat anti-rabbit IgG diluted 1:6000 (Sigma). The bound antibodies were detected using an enhanced chemiluminescence procedure (Amersham France, Les Ulis, France). Molecular-mass markers (Sigma) were electrophoresed in parallel with the bacterial proteins.
Lipid analysis
Cells from 40 ml of culture were harvested by centrifugation when cultures had reached an A600 of ∼8–9 (end of the exponential phase) or ∼15–17 (stationary phase). The resulting pellets were then washed twice with 40 ml of water and resuspended in 2 ml of water. To extract yeast lipids from whole cells, 500 μl of the cell suspensions were vigorously shaken for 10 min with glass beads in the presence of 2 ml of chloroform/methanol (2:1, v/v). After centrifugation, the organic phase was isolated and the remaining lipids were further extracted twice by the addition of 2 ml of chloroform to the aqueous phase and by shaking (in the presence of the glass beads) for 10 min. The organic phases were then pooled and evaporated to dryness. Next, the lipids were redissolved in 50 μl of chloroform/methanol (2:1, v/v). Alternatively, they were extracted from the subcellular fraction by adding microsomal membranes or mitochondrial proteins to 2 ml of chloroform/methanol (2:1, v/v) plus 500 μl of water. After phase separation, the organic phase was isolated and 2 ml of chloroform was added to the remaining aqueous phase. After a new phase separation, the organic phases were pooled and evaporated to dryness. The lipids were then suspended in 50 μl of chloroform/methanol (2:1, v/v).
Neutral and polar lipids were purified from the extracts by one-dimensional TLC using hexane/diethyl ether/acetic acid (90:15:2, by vol.) and chloroform/methanol/1-propanol/methyl acetate/0.25% KCl (10:4:10:10:3.6, by vol.) as the solvent respectively [22,23]. The lipids were then located by spraying the plates with a solution of 0.001% (w/v) primuline in 80% acetone, followed by visualization under UV light. The silica gel zones corresponding to the various lipids were then scraped from the plates and added to 1 ml of methanol/2.5% H2SO4 containing 5 μg of heptadecanoic acid methyl ester. After maintaining the lipids in the mixture at 80 °C for 1 h, 1.5 ml of water was added and FAMES (fatty acid methyl esters) were extracted using 0.75 ml of hexane. Separation of FAMES was performed by GC (Hewlett–Packard 5890 series II) on a 15 m×0.53 mm Carbowax column (Alltech Associates, Deerfield, IL, U.S.A.) by flame-ionization detection. The oven temperature was programmed to be 160 °C for 1 min, followed by a 20 °C·min−1 ramp to 190 °C and a second ramp of 5 °C·min−1 to 210 °C, and maintained at this temperature for a further 6 min. The retention times of FAMES were determined by comparison with standards, and they were quantified using heptadecanoic acid methyl ester as standard.
Enzymatic assays
Lyso-PC acyltransferase reactions were conducted in 100 μl of assay mixtures containing various amounts of labelled lyso-PC (see the Figure legends), 0.4 M mannitol, 25 mM Tris/HCl (pH 7.0), various amounts of mitochondrial proteins (see the Figure legends) and, when added, 5 nmol of oleoyl-CoA. Incubations were performed at 30 °C for 10 and 30 min in the presence and absence of oleoyl-CoA respectively (30 min when bacterial membranes were used). Reactions were stopped by the addition of 2 ml of chloroform/methanol (2:1, v/v) and 500 μl of water. The organic phase was isolated and the aqueous phase was re-extracted with 2 ml of chloroform. These combined lipid extracts were dried, redissolved in 50 μl of chloroform/methanol (2:1, v/v), and the lipids were separated by HPTLC as described above. The radioactivity incorporated into PC was quantified using a Phosphor-Imager (Amersham Biosciences–Molecular Dynamics).
Respiration rate determination
Oxygen consumption was measured in a thermostatically controlled chamber (maintained at 28 °C) containing a Clarke electrode (Gilson or similar), which was connected to a recording device monitoring the changes of oxygen content in the medium over time. Mitochondria were added to 0.5 mg of protein/ml in an assay buffer (1.5 ml) containing 0.65 M mannitol, 0.36 mM EGTA, 10 mM Tris/maleate (pH 6.8) and 3 mM phosphate (as Tris phosphoric acid). To start reaction-inducing state 4 respiration (absence of phosphorylation), the respiratory substrate NADH was added to 4 mM (NADH as such can be used as a respiration substrate because yeast mitochondria contain an NADH dehydrogenase located on the cytoplasmic side of the inner membrane). State 4 respiration was maintained for a period of time sufficient to measure a constant rate of oxygen consumption. State 3 respiration (maximal phosphorylation rate) was started by adding ADP to 1 mM.
RESULTS
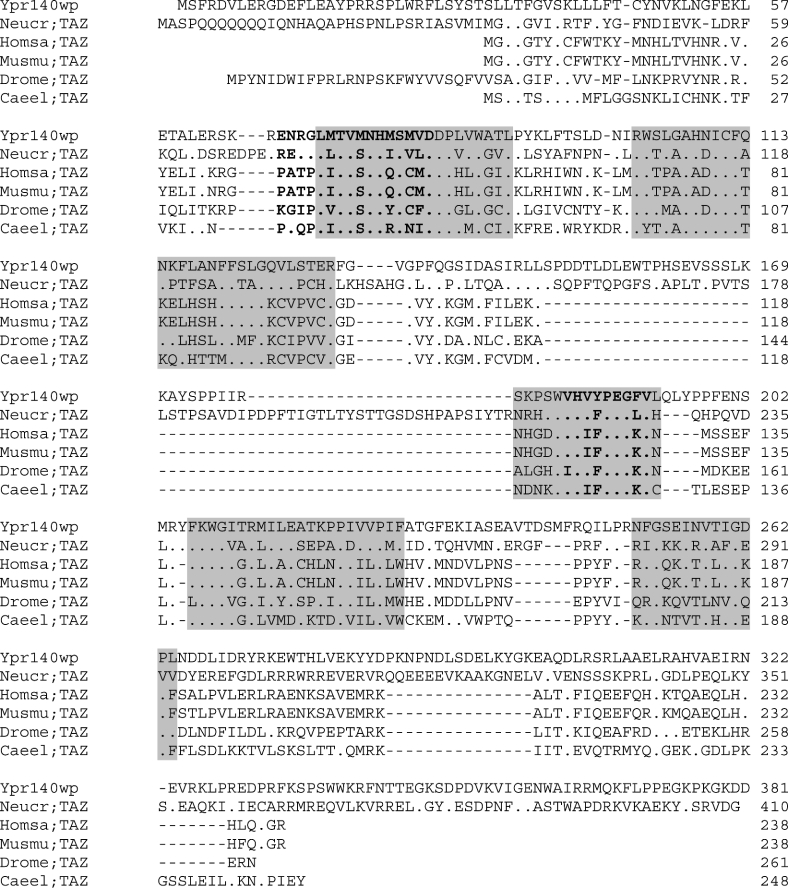
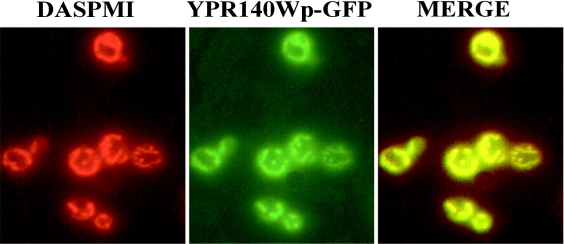
‘Tafazzin’, a human protein linked to the Barth syndrome is considered to be a lysolipid acyltransferase [4], and we used its sequence to identify some open reading frames of interest in S. cerevisiae. From our analysis, it appeared that only one yeast gene (YPR140w) has a significant homology with this protein. Interestingly, a similar analysis performed by using the coding sequence of the sole lysolipid acyltransferase identified so far in S. cerevisiae (Ydl052cp, a lyso-PA acyltransferase) did not lead to identifying the YPR140w sequence. The Ypr140wp (Figure 1) is a protein containing 381 amino acids, with the five boxes corresponding to the tafazzin signature. In addition, its sequence contains two of the four homology blocks (blocks I and III) previously identified by the alignment sequence of various other acyltransferases [4]. The sequence analysis of Ypr140wp strongly suggested the presence of one transmembrane helix from amino acid 26 to amino acid 47 (TMpred program, EMB.net.org). Further analysis of this sequence underlined the absence of peroxisomal targeting signal and vacuolar targeting motif, and did not suggest a mitochondrial location of this protein. In contrast, the use of the Psort II server indicated the presence of an XXRR-like motif in the N-terminus (SFRD) and a KKXX-like motif in the C-terminus (KGKD) that might be ER membrane retention peptide signals. Such a finding appeared puzzling because it was shown previously that when the yeast tafazzin gene is disrupted, an abnormal cardiolipin (a lipid exclusively associated with mitochondria) profile is observed [3,24]. In addition, a recent analysis [25] of the proteome of S. cerevisiae mitochondria indicated the presence of Ypr140wp in this organelle. Nevertheless, of the 750 proteins identified in the present study, 106 have been reported to be located in other cellular compartments. Hence, because of the lack of a clear-cut result concerning the location of this protein in yeast, a construction encoding the sequence of Ypr140wp fused upstream with the GFP was introduced into the Δypr140w strain. Fluorescence microscopy analysis of living cells grown on a lactate-supplemented medium revealed that the Ypr140w–GFP fusion protein is targeted to the mitochondria (well-differentiated network), as shown by the co-staining with the fluorescent mitochondrial marker DASPMI (Figure 2).
Figure 1. Amino acid sequence alignment of S. cerevisiae Ypr140wp with various tafazzin proteins.
Proteins other than Ypr140wp are tafazzins from Neurospora crassa (Neucr;TAZ; EAA29034), human (Homsa;TAZ; AAO84344), drosophila (Drome;TAZ; AAD48409), mouse (Musmu;TAZ; AAH15305) and Caenorhabditis elegans (Caeel;TAZ; CAA92638). Identical amino acids are represented by dots. Sequences were compared using the multiple alignment program CLUSTAL W version 1.7. The tafazzin signature is boxed and the phospholipid and glycerol acyltransferase motifs are in boldface letters.
Figure 2. Mitochondrial location of Ypr140wp.
ypr140wΔ cells expressing the Ypr140w–GFP fusion protein were grown in YNB medium supplemented with 2% lactate. A total of 107 cells were then stained with 5 μM DASPMI and examined under a fluorescence microscope as described in the Experimental section.
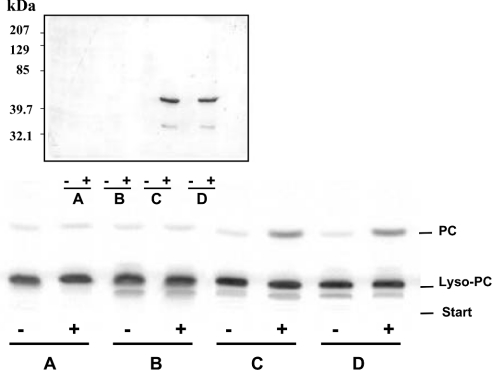
To determine whether Ypr140wp displays a lyso-PC acyltransferase activity, we first expressed its full-length sequence in C41 (DE3) E. coli bacteria (pET-15b was used as a vector). After IPTG induction, the amount of Ypr140wp synthesized by such bacteria was too low to be revealed by a mere electrophoresis staining (results not shown). However, when the cells were transformed with the plasmid containing the sequence encoding the Ypr140w–HA fusion protein, induction of the protein synthesis could be observed by Western blotting, using an antibody to HA (Figure 3, upper panel). Unfortunately, no significant lyso-PC acyltransferase activity [0.16 and 0.19 nmol of PC·(30 min)−1·mg−1] was associated with the membrane of bacteria expressing the Ypr140w–HA fusion protein. In contrast, when C41 (DE3) E. coli bacteria expressed Ypr140wp, their membranes were capable of catalysing the acylation of labelled lyso-PC to form PC. Figure 3 (lower panel) demonstrates the synthesis of 1.92±0.16 nmol of PC·(30 min)−1·(mg of membrane-bound proteins)−1 (n=3; two different clones). This activity was (almost) undetectable in the absence of IPTG [0.17±0.01 nmol·(30 min)−1·mg−1] (n=3; two different clones) or when bacteria were transformed with the empty vector [0.15 and 0.20 nmol of PC synthesis·(30 min)−1·mg−1]. Moreover, the rate of PC synthesis from labelled lyso-PC in membranes of E. coli expressing Ypr140wp was the same in the absence or presence of exogenous acyl-CoAs. Therefore this activity appeared to be acyl-CoA-independent. In agreement with this, when labelled acyl-CoA and unlabelled lyso-PC were used as substrates, no synthesis of labelled PC occurred (results not shown).
Figure 3. Lyso-PC acyltransferase activity associated with membranes of E. coli expressing Ypr140wp and induction by IPTG of Ypr140w–HA fusion protein synthesis.
Upper panel, immunoblot experiments were performed by using rabbit polyclonal antibody to HA as described in the Experimental section (10 μg of lysates per lane). Experimental details are as given in the legend to Figure 2, except that pET-15b contained the coding sequence of the Ypr140w–HA fusion protein instead of Ypr140wp. Lower panel, lyso-PC acyltransferase activities were determined using 1 nmol of labelled lyso-PC and 100 μg of membrane proteins as described in the Experimental section. Membrane proteins were obtained from C41 (DE3) E. coli (A), C41 (DE3) E. coli transformed with pET-15b (B), E. coli transformed with pET-15b containing the coding sequence of Ypr140wp [clone 1 (C) and clone 2 (D)]. The protein synthesis was induced (+) or not (−) by IPTG.
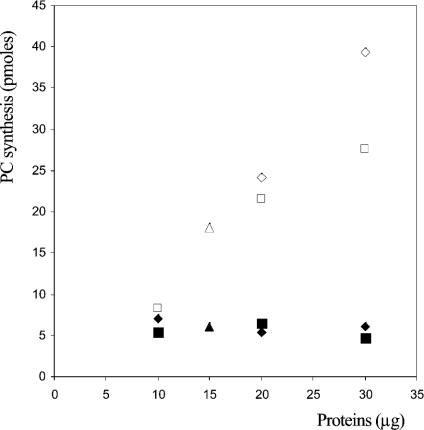
To confirm the lyso-PC acyltransferase activity of Ypr140wp, we compared the rate of acylation of lyso-PC molecules in mitochondria purified from yeast deleted for YPR140w (mutant obtained from the EUROSCARF library) to the activity associated with the mitochondria purified from wild-type cells. When experiments were performed in the absence of acyl-CoA, it clearly appeared that mitochondria from wild-type cells were capable of catalysing the synthesis of labelled PC from labelled lyso-PC, whereas no activity was detected when mitochondria purified from the ypr140wΔ mutant cells were used (Figure 4). This result clearly confirmed that Ypr140wp is capable of catalysing the acylation of lyso-PC molecules in the absence of acyl-CoA. In the presence of acyl-CoA, a much higher activity was measured and no greater difference in the acylation of lyso-PC molecules by mitochondria from wild-type and mutant cells appeared (results not shown). Because of the lack of several commercially available labelled lysolipids, to determine the specificity of the enzyme towards various lysolipids, we first determined the Km value of the enzyme for lyso-PC (7 μM, results not shown). We further performed a set of experiments to determine (for [S]≫Km) the effect of isotopic dilution of this labelled substrate by various unlabelled lysolipids. Under these conditions, the addition of an unlabelled lysolipid with a higher affinity for the enzyme compared with lyso-PC would induce a higher ‘isotopic dilution’ effect than that observed with unlabelled lyso-PC. It is shown in Table 2 that the addition of other lysolipids did not induce such an ‘isotopic dilution’ effect. The same results (i.e. no ‘isotopic dilution’ effect) were obtained when 3 nmol of DAG was added to 3 nmol of labelled lyso-PC (in the presence of 1 mM CHAPS; results not shown). These results clearly showed that, in comparison with lyso-PC, these other lipids were not used [or poorly used for lyso-PE (1-acylglycerophosphorylethanolamine)] by the enzyme under study.
Figure 4. Acyl-CoA-independent lyso-PC-acyltransferase activity associated with mitochondria purified from ypr140wΔ mutant and wild-type cells.
Lyso-PC acyltransferase activities were determined for 30 min using 0.5 nmol of labelled lyso-PC and various amounts of mitochondria purified from mutant (closed symbols) or wild-type (open symbols) cells. Results are representative of three independent experiments.
Table 2. Isotopic dilution of the labelled substrate (lyso-PC) by various lysolipids.
Lyso-PC acyltransferase activities were determined by incubating 20 μg of mitochondria for 30 min with 3 nmol of labelled lyso-PC and 3 nmol each of various unlabelled lysolipids. Results represent the means±S.D. for three determinations. The isotopic dilution factor was determined as VlpcD/Vlpx, where Vlpc and Vlpx represent the rates of incorporation of labelled lyso-PC (3 nmol) in the presence of 3 nmol of unlabelled lyso-PC and in the presence of 3 nmol of lyso-PX (where PX=PC, PA, PE, PI or PG) respectively; D (=2) is the ‘isotopic dilution factor’ in the presence of 3 nmol of unlabelled lyso-PC. lyso-PG, 1-acylglycerophosphorylglycerol; lyso-PI, 1-acylglycerophosphorylinositol; MAG, monoacylglycerol.
| Unlabelled lysolipid added (3 nmol) | Labelled PC synthesized (nmol·mg−1·h−1) | ‘Isotopic dilution factor’ of the labelled substrate |
|---|---|---|
| Lyso-PC | 2.3±0.03 | 2 |
| Lyso-PA | 4.4±1 | 1.04 |
| Lyso-PE | 3.7±0.07 | 1.24 |
| Lyso-PI | 5.4±1.8 | 0.85 |
| Lyso-PG | 4.7±1 | 0.98 |
| MAG | 5.5±0.6 | 0.84 |
Ypr140wp appears to be an enzyme involved in lipid metabolism. We therefore determined the lipid composition of the ypr140wΔ mutant and the wild-type cells grown in media supplemented with 2% (w/v) glucose or 2% lactate (whatever culture medium was used, no significant difference in growth rate was observed; results not shown). We first determined the total phospholipid, DAG, TAG, non-esterified fatty acid and steryl ester content. After culturing in a medium supplemented with 2% glucose, no significant differences in lipid amounts were observed, irrespective of the phase chosen to harvest cells (exponential or stationary phase). In contrast, when cells were grown in the presence of 2% lactate (i.e. under strict respiratory conditions), the mutant cells harvested during the stationary phase contained approx. 2-fold more TAGs compared with the wild-type cells (0.46±0.08 μg·absorbance unit−1·ml−1 and 0.25±0.03 μg·absorbance unit−1·ml−1 respectively; n=4, two separate cultures). As expected (see e.g. [26]), the synthesis of TAGs was much lower during the exponential phase, so differences in the oil content of mutant and wild-type cells were more difficult to highlight. When the results were expressed as a percentage of the total lipids, the cells harvested during the stationary phase contained 25.8% of TAGs for the mutant but only 15.4% for the wild-type (Table 3). The percentage of steryl esters was also slightly higher in the mutant, whereas correlatively the percentage of all other lipids was lower. This repeatedly observed increase in the amount of TAGs and steryl esters in the mutant cells was not accompanied by significant changes in the fatty acid composition of the various lipids, except for a slight increase in the percentage of 16:1 fatty acid esterified to neutral lipids (Table 3). Nevertheless, by itself, this increase could not account for the increase in the amount of TAGs. In other words, the deletion of YPR140w induced an increase in the oil synthesis in yeast but did not seem to affect greatly the ‘specificity’ of the overall process towards the various acyl chains. Moreover, no variation in the fatty acid composition of the various lipids was observed when cells were harvested during the exponential phase. Similar results (i.e. no change in the fatty acid composition) were obtained when cells were harvested during the stationary or exponential phase from a medium supplemented with 2% glucose (results not shown).
Table 3. Distribution and fatty acid composition of lipids from ypr140wΔ mutant and wild-type cells grown in the presence of 2% lactate and harvested during the stationary phase.
Lipids were purified and quantified as described in the Experimental section. Results represent the means±S.D. for four analyses performed in two separate cultures.
| Fatty acid composition | |||||
|---|---|---|---|---|---|
| Lipids | Percentage of total lipids | 16:0 (%) | 16:1 (%) | 18:0 (%) | 18:1 (%) |
| Polar lipids | |||||
| Wild-type | 71.1±0.75 | 8.8±0.16 | 49.1±0.24 | 3.5±0.03 | 38.5±0.29 |
| Mutant | 63.6±4.7 | 11.2±0.62 | 48.7±1.62 | 4.5±2.19 | 35.7±0.03 |
| Diacylglycerols | |||||
| Wild-type | 8.1±0.49 | 16.7±0.51 | 36.4±0.63 | 7.0±0.16 | 39.9±0.05 |
| Mutant | 6.6±0.70 | 15.5±0.09 | 42.3±0.3 | 5.6±0.04 | 36.6±0.37 |
| Non-esterified fatty acids | |||||
| Wild-type | 3.5±0.64 | 17.2±1.47 | 47.1±1.3 | 6.4±0.12 | 29.3±0.25 |
| Mutant | 2.4±0.28 | 19.8±2.83 | 46.8±2.81 | 7.0±0.42 | 26.5±0.43 |
| TAGs | |||||
| Wild-type | 15.4±0.03 | 14.9±0.24 | 42.3±0.32 | 7.3±0.7 | 35.5±0.1 |
| Mutant | 25.8±2.22 | 12.7±0.2 | 50.2±0.35 | 5.4±0.06 | 31.8±0.1 |
| Steryl esters | |||||
| Wild-type | 1.71±0.19 | 16.8±0.63 | 41.0±1.1 | 7.0±0.45 | 35.1±0.6 |
| Mutant | 2.50±0.46 | 17.2±0.32 | 53±0.44 | 5.1±0.2 | 24.6±0.6 |
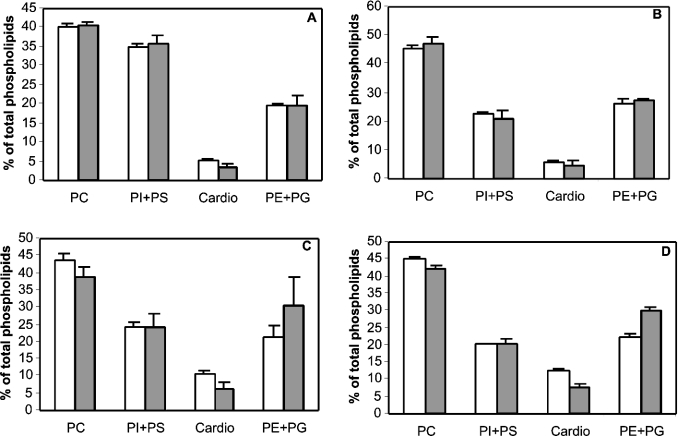
A further set of experiments was performed to determine the distribution of various phospholipids in the mutant and wild-type cells. Figure 5 shows the phospholipid compositions of cells grown on a medium supplemented with 2% glucose or 2% lactate. In the presence of glucose, no significant difference was observed in the phospholipid composition of wild-type and ypr140wΔ cells. These results perfectly fit the lipid compositions of wild-type and ypr140wΔ cells deposited by G. Daum in the ‘Lipid profile database’ (http://mips.gsf.de/proj/eurofan/eurofan_2/n4/datasheets/YPR140Wtab.html), i.e. PC, 45 and 42.5%; PS, 5.8 and 6.8%; PA, 2.5 and 1.8%; PI (phosphatidylinositol), 16 and 15.9%; cardiolipin, 2.5 and 3.2%; PE, 24 and 25.5% for the wild-type and mutant respectively. In contrast, when cells were grown in the presence of lactate, a significant decrease in the percentage of cardiolipin + PC was observed in mutant cells harvested during the exponential phase (54.6±2.6% in wild-type cells compared with 45.2±4.7% for the mutant) and the stationary phase (57.5±1.2% compared with 49.9±2.3%). These differences in phospholipid distribution were accompanied by a change in the composition of fatty acids esterified to cardiolipins. Table 4 clearly shows that cardiolipins from mutant cells contained much less oleic acid (18:1) when compared with wild-type cells, this decrease being compensated by an increase in the percentage of palmitic acid (16:0). This result was observed with cells harvested both during the exponential and stationary phases. The fatty acid composition of other phospholipids purified from mutant and wild-type cells did not significantly vary whatever the culture medium used (glucose or lactate) and whatever the phase chosen to harvest cells (results not shown).
Figure 5. Phospholipid composition of ypr140wΔ mutant and wild-type cells.
Mutant (closed bars) or wild-type (open bars) cells were grown in a medium supplemented with either 2% glucose (A, B) or 2% lactate (C, D), and harvested during the exponential (A, C) or stationary (B, D) phase. Lipids were purified and quantified as described in the Experimental section. The error bars represent the means±S.D. for three determinations. PS, phosphatidylserine; Cardio, cardiolipin.
Table 4. Fatty acid composition of cardiolipin purified from ypr140wΔ mutant and wild-type cells.
Cultures were performed and lipids were purified and quantified as described in the Experimental section. Results represent the means±S.D. for three determinations.
| Fatty acid composition | |||||
|---|---|---|---|---|---|
| Phase | 16:0 (%) | 16:1 (%) | 18:0 (%) | 18:1 (%) | |
| 2% glucose | |||||
| Exponential | Wild-type | 25.3±0.6 | 39.1±0.4 | 3.9±0.1 | 31.8±0.4 |
| Mutant | 24.9±1 | 41.9±1.8 | 2.9±0.3 | 30.3±0.6 | |
| Stationary | Wild-type | 29.4±4.4 | 35.1±3.8 | 4.1±1.7 | 31.5±2.3 |
| Mutant | 30.1±2.1 | 37.5±1.4 | 2.8±0.8 | 29.6±0.5 | |
| 2% lactate | |||||
| Exponential | Wild-type | 7.6±1.8 | 42.2±0.6 | 1.7±0.4 | 48.5±1.5 |
| Mutant | 19.6±1.6 | 37.4±2.7 | 4.1±1.5 | 38.9±0.4 | |
| Stationary | Wild-type | 7.6±1.1 | 42.4±0.9 | 1.4±0.5 | 48.6±1 |
| Mutant | 15.6±0.9 | 42.4±1.2 | 2.2±0.3 | 39.8±0.2 | |
To confirm the decrease in cardiolipin content and the changes observed in their fatty acid composition in mutant cells, we further analysed the phospholipid composition of mitochondria purified from mutant and wild-type cells. The data in Table 5 confirm those obtained after the purification of lipids from the whole cells, i.e. there was a decrease in the PC and cardiolipin contents in purified mitochondria from the mutant, and the main effect on the fatty acid composition of individual lipids was a decrease in the percentage of 18:1 associated with cardiolipins. Moreover, as observed with whole cells, this decrease was compensated mainly by an increase in the percentage of 16:0 fatty acid. In contrast, when lipids from the microsomal membranes of mutant and wild-type cells were analysed, no clear-cut differences appeared, either in the phospholipid distribution or in their fatty acid composition.
Table 5. Fatty acid compositions of phospholipids extracted from microsomes and from purified mitochondria of ypr140wΔ mutant and wild-type cells.
Results represent the means±S.D. for three determinations. Note that cardiolipin was not detected in the microsomal fraction.
| Fatty acid composition | ||||||
|---|---|---|---|---|---|---|
| Percentage of the total phospholipids | 16:0 (%) | 16:1 (%) | 18:0 (%) | 18:1 (%) | ||
| Microsomes | ||||||
| PC | Wild-type | 44.1±2 | 13.5±0.7 | 45.9±0.5 | 7.5±0.3 | 33.1±0.5 |
| Mutant | 46.4±1.4 | 12.5±0.5 | 45.4±0.2 | 7.7±0.05 | 34.3±0.3 | |
| PI+PS | Wild-type | 14.3±0.1 | 23.9±0.6 | 31.8±0.8 | 7.9±0.1 | 36.4±1.2 |
| Mutant | 12.8±0.7 | 27.1±2.9 | 28.2±1 | 12.9±3.5 | 31.9±5.3 | |
| PI | Wild-type | 24.4±2.4 | 27±0.7 | 21.7±1.7 | 15.6±1.3 | 35.7±0.2 |
| Mutant | 24.8±2.2 | 27.1±0.6 | 19.6±0.9 | 18.1±0.8 | 35.3±0.5 | |
| PE+PG | Wild-type | 17.3±1.9 | 15.6±1.2 | 37.6±5.1 | 11.1±2.5 | 35.7±2.1 |
| Mutant | 16±1.7 | 15.8±2.1 | 42.7±2.6 | 7.8±1.6 | 33.7±1.2 | |
| Mitochondria | ||||||
| PC | Wild-type | 37.1±0.6 | 9.2±0 | 57±0.2 | 4.1±0 | 29.8±0.1 |
| Mutant | 33.7±1.1 | 8.4±0.3 | 55.3±0.7 | 4.0±0 | 32.9±0.4 | |
| PI+PS | Wild-type | 18.7±0.4 | 22.2±1.1 | 28.6±2.4 | 9.9±0.9 | 39.4±0.5 |
| Mutant | 26.2±1.4 | 22.1±0.1 | 25.9±1 | 10.6±0.8 | 41.4±0.4 | |
| Cardiolipin | Wild-type | 16.8±0.2 | 5.8±0.6 | 40.9±3.2 | 2±0.9 | 51.3±1.6 |
| Mutant | 9.8±3 | 16.9±1.1 | 42.6±2.1 | 4±1.8 | 36.4±1.5 | |
| PE+PG | Wild-type | 27.3±0.3 | 6.9±1.1 | 51.2±0.4 | 0.7±0.2 | 41.2±1 |
| Mutant | 30.3±3 | 4.9±0.4 | 53.3±0.2 | 0.4±0 | 41.4±0.5 | |
The deletion of YPR140w induced changes in the neutral and polar lipid compositions of yeast only when cells were grown in a medium supplemented with lactate, and the main effects of the mutation on the phospholipid and fatty acid compositions were observed with mitochondrial lipids. We therefore compared the respiration rates of purified mitochondria from mutant and wild-type cells. Higher respiration rates were found for mitochondria from ypr140wΔ cells (Table 6). This increase in the respiration rate was observed both at state 4 (i.e. in the absence of ADP) and state 3 (i.e. in the presence of 1 mM ADP). The coupling efficiencies between respiration and the ATP synthesis determined with mitochondria purified from both mutant and wild-type cells appeared similar. In other words, whereas the mutation induced a change in the lipid composition of the mitochondrial membranes and an increase in the respiration rates, the proton permeability of the inner mitochondrial membrane did not seem to be altered in the mutant cells. Moreover, as expected from the fact that the respiratory control ratio was the same, the ATP and ADP contents of wild-type and mutant cells did not differ (Table 6). ATP/ADP ratios in wild-type and mutant cells were similar to values previously reported for aerobic growing cells [27]. These results show that the differences in the amount of TAG in wild-type and mutant cells do not result from differences in the ‘energy charge’ of wild-type and mutant cells.
Table 6. Respiration rates of mitochondria purified from ypr140wΔ mutant and wild-type cells and ATP and ADP content of these cells.
The ATP and ADP contents [nmol·(absorbance unit)−1·ml−1] of the cell were determined as described in the Experimental section. Results represent the means±S.D. for six determinations in two separate cultures. Mitochondria were purified and the respiration rates were determined as described in the Experimental section. Respiration rates [(nanoatom O2)·mg−1·min−1] of states 4 and 3 were determined in the presence of 4 mM NADH and 4 mM NADH+1 mM ADP respectively.
| Respiratory rate of mitochondria | |||
|---|---|---|---|
| State 4 | State 3 | Ratio (state 3/state 4) | |
| Wild-type | 177±6 | 378±18 | 2.1 |
| Mutant | 397±20 | 730±25 | 1.9 |
| ATP/ADP content of mitochondria | |||
| ATP | ADP | ||
| Exponential phase | |||
| Wild-type | 15.7±0.4 | 3.9±0.7 | |
| Mutant | 14.6±0.2 | 3.8±0.4 | |
| Stationary phase | |||
| Wild-type | 11.0±0.2 | 7.0±0.6 | |
| Mutant | 11.5±0.4 | 7.3±0.6 | |
DISCUSSION
We provide evidence in the present study that Ypr140wp is located in mitochondria, and we show that, in contrast with what was observed by using mitochondria purified from wild-type cells, mitochondria purified from yeast deleted for YPR140w were not capable of catalysing the acylation of lyso-PC in the absence of acyl-CoA. In agreement with this, when Ypr140wp was expressed in E. coli, an acyl-CoA-independent lyso-PC acyltransferase activity was found associated with the membranes of the bacteria. To our knowledge, this is the first report of a coding sequence of a protein (Ypr140wp) with a lyso-PC acyltransferase activity. We set up several experiments to determine the acyl-donor, but without success. Among these experiments, we purified membranes containing labelled lipids [PE, PG (phosphatidylglycerol), cardiolipin, but no PC in E. coli membranes] from the bacteria expressing Ypr140wp and grown in the presence of [14C]acetate. When exogenous lyso-PC molecules were added to these membranes, synthesis of labelled PC occurred as expected, but we did not notice any significant decrease in any of the labelled E. coli lipids. Interestingly, synthesis of labelled PC was observed even when the lyso-PC molecules added were unlabelled. This clearly shows that the PC formation did not result from the transfer of an acyl chain from a lyso-PC molecule to another one. Another set of experiments was performed by incubating various exogenous lipids with labelled lyso-PC and mitochondria purified from wild-type cells. None of the lipids added (PE, PG, DAG, TAG and cardiolipin) induced an increase in PC synthesis. When the ‘mitochondria/exogenous lipids’ mixture was sonicated before the incubation or when CHAPS (4 mM) was added to the incubation mixture, the same results were obtained. This clearly suggests that the absence of an effect was not because the substrate did not reach the enzyme. In addition, it is also possible that the amount of lipid present in the mitochondrial membranes was sufficient to insure a ‘saturating concentration’ of the putative acyl-donor. Nevertheless, the absence of an increase in lyso-PC acyltransferase activity when exogenous TAGs were added clearly suggests that the enzyme under study does not catalyse the reverse reaction of oil synthesis from DAG and PC. To confirm that cardiolipin is not the acyl-donor, we purified mitochondria from a yeast strain unable to synthesize this lipid (see Table 1). The acyl-CoA-independent lyso-PC acyltransferase activity associated with these mitochondria devoid of cardiolipin (checked by HPTLC analysis) was the same as the activity associated with mitochondria purified from the corresponding wild-type strain (results not shown). Hence it appears that cardiolipin is not the acyl-donor and, therefore, that the changes in the cardiolipin composition of ypr140wΔ cells did not result from a block in the transfer of an acyl chain from cardiolipin to lyso-PC molecules. Nevertheless, it remains possible that PC molecules synthesized by Ypr140wp are further used for the metabolism (remodelling) of cardiolipid. In agreement with this, a remodelling of cardiolipin by PC was recently observed in mitochondria purified from rat liver and human lymphoblasts [2]. This remodelling (synthesis) of cardiolipid also occurred in lymphoblast mitochondria from patients with tafazzin deletion, but to a lesser extent. It can be hypothesized that there are some changes in the lipid compositions, and also perhaps in the protein composition, of the mitochondria, which induce a decrease in the efficiency of the transfer of an acyl chain from PC to cardiolipin.
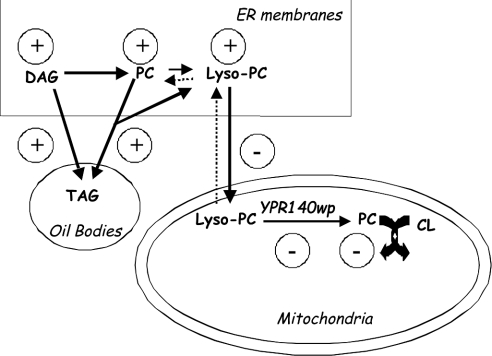
Deletion of the YPR140w gene also induced an increase in the oil content of the cell. As expected and as generally observed (see for example, [26]), by staining cells with oil red, we found that these TAGs were located in lipid droplets present in the cytosol (results not shown). In good agreement with this, no or few neutral lipids were found associated with purified mitochondria or microsomal membranes. The reasons for such an increase in the oil content of the mutant cells that grew in the presence of lactate remain to be established clearly, but the following hypothesis is possible (Scheme 1): we proposed some years ago [5] that lyso-PC might transfer from ER membranes to chloroplasts and mitochondria can be used as a substrate for organelle lipid synthesis. As previously proposed by Stymne and Stobart [28], we assumed that lyso-PC synthesis in ER results from the reverse reaction catalysed by the lyso-PC acyltransferase located in these membranes (see also [29]). Therefore, since less lyso-PC is used to form PC in ypr140wΔ mitochondria, it could be assumed that the amount of lyso-PC tends to increase in ER membranes and, therefore, that the rate of the reverse reaction catalysed by the lyso-PC acyltransferase in these membranes tends to be reduced. Such a lower synthesis of lyso-PC molecules might tend to increase the amount of precursors of this compound in the ER membranes, namely DAG and PC. Interestingly, it is well known that these lipids are both substrates of oil synthesis in yeast: DAG can be acylated by acyl-CoAs to form TAG, but also by fatty acids esterified to the sn-2 position of PC located in the ER ([30] and see [13,14] for reviews). In addition, since the latter reaction leads to the synthesis of lyso-PC molecules, it might tend to decrease the rate of the reverse reaction catalysed by the lyso-PC acyltransferase in ER membranes and to increase the amount of PC and DAG in these membranes and therefore the oil synthesis. Hence, especially when cells were grown in the presence of lactate (i.e. under conditions requiring an optimal biogenesis of mitochondria), a decrease in the amount of lyso-PC acylated through the mitochondrial pathway might lead to a higher TAG content in the mutant cells. Such an assumption is in agreement with the present findings.
Scheme 1. Possible explanation for the lipid phenotype of ypr140wΔ cells.
For more clarity, the TAG synthesis pathway occurring in lipid bodies is not shown. + (−) represents the rates that might increase (decrease) or tend to increase (decrease) in ypr140wΔ cells. CL, cardiolipin.
Acknowledgments
Our Special thanks to Dr S. Mongrand (UMR 5200 CNRS) for his helpful comments throughout this project. We also thank Professor M. Bonneu and D. Lapaillerie (Plateforme de Génomique Fonctionelle, Bordeaux, France) for their contribution to expression of the Ypr140w–GFP fusion protein in yeast, and Professor M. Rigoulet (IBGC-CNRS) and Dr J. Joubes (UMR 5200). We are grateful to Dr F. Ichas (IECB-CNRS) for her contribution to the visualization of the lipid droplets by microscopy. This work was partially supported by the Conseil Régional d'Aquitaine (France).
References
- 1.Esposti M. D. The mitochondrial battlefield and membrane lipids during cell death signalling. Ital. J. Biochem. 2003;52:43–50. [PubMed] [Google Scholar]
- 2.Xu Y., Kelley R. I., Blank T. J. J., Schlame M. Remodeling of cardiolipin by phospholipid transacylation. J. Biol. Chem. 2003;278:51380–51385. doi: 10.1074/jbc.M307382200. [DOI] [PubMed] [Google Scholar]
- 3.Vaz F. M., Houtkooper R. H., Valianpour F., Barth P. G., Wanders R. J. Only one spice variant of the human TAZ gene encodes a functional protein with a role in cardiolipin metabolism. J. Biol. Chem. 2003;278:43089–43094. doi: 10.1074/jbc.M305956200. [DOI] [PubMed] [Google Scholar]
- 4.Neuwald A. F. Barth syndrome may be due to an acyltransferase deficiency. Curr. Biol. 1997;7:465–466. doi: 10.1016/s0960-9822(06)00237-5. [DOI] [PubMed] [Google Scholar]
- 5.Bessoule J.-J., Testet E., Cassagne C. Synthesis of phosphatidylcholine in the chloroplast envelope after import of lysophosphatidylcholine from endoplasmic reticulum membranes. Eur. J. Biochem. 1995;228:490–497. [PubMed] [Google Scholar]
- 6.Testet E., Bessoule J.-J., Mongrand S., Guillot-Salomon T., Cantrel C., Cassagne C. Occurrence of an acyl-CoA:1-acylglycerophosphorylcholine acyltransferase in plant mitochondria. FEBS Lett. 1996;399:87–91. doi: 10.1016/s0014-5793(96)01293-8. [DOI] [PubMed] [Google Scholar]
- 7.Testet E., Verdoni N., Cassagne C., Bessoule J.-J. Transfer and subsequent metabolism of lysolipids studied by immobilizing subcellular compartments in alginate beads. Biochim. Biophys. Acta. 1999;1440:73–80. doi: 10.1016/s1388-1981(99)00118-3. [DOI] [PubMed] [Google Scholar]
- 8.Mongrand S., Bessoule J.-J., Cassagne C. A re-examination in vivo of the phosphatidylcholine-galactolipid metabolic relationship during plant lipid biosynthesis. Biochem. J. 1997;327:853–858. doi: 10.1042/bj3270853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mongrand S., Cassagne C., Bessoule J.-J. Import of lyso-PC into chloroplasts likely at the origin of eukaryotic plastidial lipids. Plant Physiol. 2000;122:845–852. doi: 10.1104/pp.122.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nebauer R., Birner-Grünberg R., Daum G. Biogenesis and cellular dynamics of glycerophospholipids in the yeast Saccharomyces cerevisiae. In: Daum G., editor. Topics in Current Genetics Lipid Metabolism and Membrane Biogenesis. Berlin and Heidelberg: Springer-Verlag; 2004. pp. 125–168. [Google Scholar]
- 11.de Kroon A. I., Koorengevel M. C., Vromans T. A., de Kruijff B. Continuous equilibration of phosphatidylcholine and its precursors between endoplasmic reticulum and mitochondria in yeast. Mol. Biol. Cell. 2003;14:2142–2150. doi: 10.1091/mbc.E02-08-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voelker D. R. Interorganelle transport of aminoglycerophospholipids. Biochim. Biophys. Acta. 2000;1486:97–107. doi: 10.1016/s1388-1981(00)00051-2. [DOI] [PubMed] [Google Scholar]
- 13.Sorger S., Daum G. Triacylglycerol biosynthesis in yeast. Appl. Microbiol. Biotechnol. 2003;61:289–299. doi: 10.1007/s00253-002-1212-4. [DOI] [PubMed] [Google Scholar]
- 14.Mullner H., Daum G. Dynamics of neutral lipid storage in yeast. Acta Biochim. Pol. 2004;51:323–347. [PubMed] [Google Scholar]
- 15.Dahlqvist A., Stahl U., Lenman M., Banas A., Lee M., Sandager L., Ronne H., Stymne S. Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6487–6492. doi: 10.1073/pnas.120067297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagiec M. M., Wells G. B., Lester R. L., Dickson R. C. A suppressor gene that enables Saccharomyces cerevisiae to grow without making sphingolipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J. Biol. Chem. 1993;268:22156–22163. [PubMed] [Google Scholar]
- 17.Richard M. G., McMaster C. R. Lysophosphatidylcholine acyltransferase activity in Saccharomyces cerevisiae: regulation by a high-affinity Zn2+binding site. Lipids. 1998;33:1229–1234. doi: 10.1007/s11745-998-0328-1. [DOI] [PubMed] [Google Scholar]
- 18.Beauvoit B., Rigoulet M., Bunoust O., Raffard G., Canioni P., Guérin B. Interactions between glucose metabolism and oxidative phosphorylations on respiratory-competent Saccharomyces cerevisiae cells. Eur. J. Biochem. 1993;214:163–172. doi: 10.1111/j.1432-1033.1993.tb17909.x. [DOI] [PubMed] [Google Scholar]
- 19.Law R. H., Manon S., Devenish R. J., Nagley P. ATP synthase from Saccharomyces cerevisiae. Methods Enzymol. 1995;260:133–163. doi: 10.1016/0076-6879(95)60135-x. [DOI] [PubMed] [Google Scholar]
- 20.Sikorski R. S., Hierter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cormack B. P., Bertram G., Egerton M., Gow N. A. R., Falkow S., Brown A. J. P. Yeast-enhanced green fluorescent protein (yEGFP) a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 22.Juguelin H., Heape M. A., Boiron F., Cassagne C. A quantitative developmental study of neutral lipids during myelinogenesis in the peripheral nervous system of normal and Trembler mice. Dev. Brain. Res. 1986;25:249–252. doi: 10.1016/s0006-8993(86)80233-5. [DOI] [PubMed] [Google Scholar]
- 23.Vitiello F., Zanetta J. P. Thin layer chromatography of phospholipids. J. Chromatogr. 1978;166:637–640. doi: 10.1016/s0021-9673(00)95654-1. [DOI] [PubMed] [Google Scholar]
- 24.Gu Z., Valianpour F., Chen S., Vaz F. M., Hakkaart G. A., Wanders R. J., Greenberg M. L. Aberrant cardiolipin metabolism in the yeast taz1 mutant: a model for Barth syndrome. Mol. Microbiol. 2004;51:149–158. doi: 10.1046/j.1365-2958.2003.03802.x. [DOI] [PubMed] [Google Scholar]
- 25.Sickmann A., Reinders J., Wagner Y., Joppich C., Zahedi R., Meyer H. E., Schonfisch B., Perschil I., Chacinska A., Guiard B., et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandager L., Gustavsson M. H., Stahl U., Dahlqvist A., Banas A., Lenman M., Ronne H., Stymne S. Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 2002;277:6478–6482. doi: 10.1074/jbc.M109109200. [DOI] [PubMed] [Google Scholar]
- 27.Larsson C., Nilsson A., Blomberg A., Gustafsson L. Glycolytic flux is conditionally correlated with ATP concentration in Saccharomyces cerevisiae: a chemostat study under carbon or nitrogen-limiting conditions. J. Bacteriol. 1997;179:7243–7250. doi: 10.1128/jb.179.23.7243-7250.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stymne S., Stobart A. K. Evidence for the reversibility of the acyl-CoA:lysophosphatidylcholine acyltransferase in microsomal preparations from developing safflower (Carthamus tinctorius L.) cotyledons and rat liver. Biochem. J. 1984;223:305–314. doi: 10.1042/bj2230305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Domergue F., Abbadi A., Ott C., Zank T. K., Zahringer U., Heinz E. Acyl carriers used as substrates by the desaturases and elongases involved in very long chain polyunsaturated fatty acid biosynthesis reconstituted in yeast. J. Biol. Chem. 2003;278:35115–35126. doi: 10.1074/jbc.M305990200. [DOI] [PubMed] [Google Scholar]
- 30.Oelkers P., Tinkelenberg A., Erdeniz N., Cromley D., Billheimer J. T., Sturley S. L. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J. Biol. Chem. 2000;275:15609–15612. doi: 10.1074/jbc.C000144200. [DOI] [PubMed] [Google Scholar]