Abstract
In Saccharomyces cerevisiae, Sic1, an inhibitor of Cdk (cyclin-dependent kinase), blocks the activity of S-Cdk1 (Cdk1/Clb5,6) kinase that is required for DNA replication. Deletion of Sic1 causes premature DNA replication from fewer origins, extension of the S phase and inefficient separation of sister chromatids during anaphase. Despite the well-documented relevance of Sic1 inhibition of S-Cdk1 for cell cycle control and genome instability, the molecular mechanism by which Sic1 inhibits S-Cdk1 activity remains obscure. In this paper, we show that Sic1 is functionally and structurally related to the mammalian Cki (Cdk inhibitor) p27Kip1 of the Kip/Cip family. A molecular model of the inhibitory domain of Sic1 bound to the Cdk2–cyclin A complex suggested that the yeast inhibitor might productively interface with the mammalian Cdk2–cyclin A complex. Consistent with this, Sic1 is able to bind to, and strongly inhibit the kinase activity of, the Cdk2–cyclin A complex. In addition, comparison of the different inhibitory patterns obtained using histone H1 or GST (glutathione S-transferase)–pRb (retinoblastoma protein) fusion protein as substrate (the latter of which recognizes both the docking site and the catalytic site of Cdk2–cyclin A) offers interesting suggestions for the inhibitory mechanism of Sic1. Finally, overexpression of the KIP1 gene in vivo in Saccharomyces cerevisiae, like overexpression of the related SIC1 gene, rescues the cell cycle-related phenotype of a sic1Δ strain. Taken together, these findings strongly indicate that budding yeast Sic1 and mammalian p27Kip1 are functional homologues with a structurally conserved inhibitory domain.
Keywords: cell division cycle, cyclin-dependent kinase inhibitor (Cki), molecular modelling, p27Kip1, Sic1, surface plasmon resonance
Abbreviations: Cki, cyclin-dependent kinase inhibitor; Cdk, cyclin-dependent kinase; S-Cdk1, Cdk1/Clb5,6 (Cdk activity required to start DNA replication); GST, glutathione S-transferase; pRb, retinoblastoma protein; SPR, surface plasmon resonance
INTRODUCTION
Cdks (cyclin-dependent kinases) play an essential regulatory role in cell cycle progression: it is in fact the sequential activation of Cdks by specific, unstable, regulatory subunits, named cyclins, that first triggers the onset of DNA replication and later initiates mitosis [1,2]. The evolutionary conservation of Cdks and cyclins from yeast to mammalian cells is well established [1,2]. In budding yeast a single Cdk (Cdc28; now renamed Cdk1) is involved in the control of the cell cycle. In the cell cycle there are cyclins associated with G1 (Cln1, Cln2 and Cln3 in budding yeast; cyclin D in mammals), S phase (Clb5,6 in yeast; cyclins E and A in mammals) and mitosis (Clb1,2 in yeast; cyclin B in mammals). Both cyclins and Cdks have a wide degree of redundancy, and it is currently believed that their specificity in driving the cell cycle is dependent more on their timing of expression and subcellular localization than on substrate specificity embedded in their molecular structure (reviewed in [2]).
Cdk activity is tightly regulated by various molecular mechanisms [3] that include regulatory phosphorylation, differential expression and/or localization, and interaction with regulatory proteins, such as Ckis (Cdk inhibitors), which inhibit Cdk activity by binding to Cdk–cyclin complexes.
In budding yeast, the Cki Sic1 blocks the activity of Clb-containing complexes: both Cdk1/Clb5,6 required for DNA replication, and Cdk1/Clb1,2 required in mitosis [4]. The S-Cdk1 (Cdk1/Clb5,6) kinase activity required for the initiation of DNA replication is present only when Sic1 is removed [4]. Sic1 is degraded by ubiquitin-dependent proteolysis that is regulated by multi-site phosphorylation performed by Cdk1/Cln1,2; this targets Sic1 to be specifically recognized by the F-box protein Cdc4, which recruits Sic1 for ubiquitination by the Cdc34–SCF (Skp1/Cullin/F-box) complex [5]. Deletion of Sic1 causes premature DNA replication from fewer origins, extension of the duration of S phase and inefficient separation of sister chromatids during anaphase. The ensuing double-strand breaks generate genomic instability and gross chromosomal rearrangements. Delaying S-Cdk1 activation rescues defects in both S and M phases [6].
Despite the well-documented relevance of Sic1-mediated inhibition of S-Cdk1 for cell cycle control and genomic instability, the molecular mechanism by which Sic1 inhibits S-Cdk1 activity remains obscure. Sic1 has been proposed to be a functional homologue of the mammalian Cki p21Cip1 [7], which in turn is characterized by significant sequence similarity with the Cki p27Kip1 (42% identity). Progression through S phase in mammalian cells is promoted by a Cdk2–cyclin A complex, whose activity is inhibited by p27Kip1. The X-ray structure of the inhibitor domain of p27Kip1 bound to the Cdk2–cyclin A complex has been reported [8]. The three-dimensional structure reveals that the N-terminus of p27Kip1 is extended over the surface of the Cdk2–cyclin A complex, forming hydrophobic contacts with regions on both cyclin and kinase. The inhibitory mechanism of p27Kip1 is multi-faceted: it occupies a secondary substrate recruitment site on cyclin A; it binds to the N-terminal lobe of Cdk2, flattening it out and disrupting the active site; and it inserts itself into the ATP binding pocket, blocking ATP binding to Cdk2 [8]. The fact that the inhibitory domain of Sic1 [9] does not have significant sequence identity with either p27Kip1 or p21Cip1 (as shown in this paper) has hampered a comparative analysis of the relationship between the structure and function of Cdk and Cki in budding yeast and in mammalian cells, despite remarkable similarity and functional interchangeability of their cyclins and Cdk [1,2].
Here we show that the inhibitory domain of Sic1 is functionally and structurally related to the inhibitory domain of mammalian p27Kip1 of the Kip/Cip family. Molecular modelling of the inhibitory domain of Sic1 suggested that the protein interface of the yeast inhibitor should be able to interact productively with the mammalian Cdk2–cyclin A complex. Consistently, Sic1 was shown to bind to, and strongly inhibit the kinase activity of, the Cdk2–cyclin A complex. In addition, comparison of the different inhibitory patterns obtained using as substrates histone H1 or GST (glutathione S-transferase)–pRb (retinoblastoma protein), the latter of which recognizes both the docking site and the catalytic site of Cdk2–cyclin A, offers interesting suggestions for the inhibitory mechanism of Sic1. The physiological relevance of these experimental results was confirmed by overexpressing the KIP1 gene in vivo in Saccharomyces cerevisiae. The yeast and mammalian Ckis in fact showed equivalent ability to rescue the cell cycle-related phenotype of a sic1Δ strain. Taken together, these findings strongly indicate that Sic1 from budding yeast and mammalian p27Kip1 are functional homologues with a structurally conserved inhibitory domain.
EXPERIMENTAL
Molecular modelling of Sic1 protein
Initial alignments of the Sic1 sequence (SwissProt accession number P38634) were carried out using CLUSTALW [10], and secondary structures were predicted using the PHD [11], PSIPRED [12] and JPRED [13] methods. A partial model of the Sic1 inhibitory domain (residues 215–284) was obtained by using the Swiss-Pdb Viewer 3.7 program [14] performing in silico mutagenesis of the Cdk–cyclin-interacting portion of p27Kip1, abridged from the structure of the p27Kip1–Cdk2–cyclin A complex (PDB ID 1JSU). The Sic1 model was docked on the Cdk2–cyclin A complex, and the model of the ternary complex was subsequently refined by 5000 steps of conjugate gradients energy minimization using the GROMOS force field [15].
Recombinant DNA techniques
Escherichia coli DH5α strain was used as host cell in cloning experiments, and standard recombinant DNA manipulations were performed [16]. E. coli BL21 (DE3)[pLysE] strain was used to generate recombinant Sic1–His6 protein.
Plasmid pIVEX2.4a containing the SIC1 gene was generated by cloning the entire open reading frame of the SIC1 gene obtained as a BamHI–BamHI fragment from plasmid pQE-30 Xa (Qiagen) into the BamHI restriction site of pIVEX2.4a MCS. Plasmid pEMBLyex4 containing the SIC1 gene under the control of the GAL1 promoter (pyex-SIC1) was generated by cloning the entire open reading frame of the SIC1 gene with one haemagglutinin epitope, obtained as an EcoRI–XbaI fragment from plasmid 2801 (kindly provided by E. Schwob, Institute of Molecular Genetics, CNRS UMR 5535 and Université Montpellier II, Montpellier, France), in plasmid pEMBLyex4 MCS. Plasmid pEMBLyex4 containing the KIP1 gene under the control of the GAL1 promoter (pyex-KIP1) was generated by a fusion of the SalI–BamHI GAL1 promoter fragment to the KIP1 open reading frame, cloned as an EcoRI–BamHI fragment derived from a pcDNA3-derived plasmid carrying the whole KIP1 open reading frame (Invitrogen).
The yeast strain used in this study was 4245 (MATa, ade2-1, leu2-3, ura3, trp1-1, his3-11, GAL, psi+, bar1::LEU2, sic1::HIS3, can1-100), kindly supplied by E. Schwob. The strain was subsequently transformed with empty plasmid pEMBLyex4 or with the pyex-SIC1 and pyex-KIP1 derivatives. Yeast cells were transformed by a modification of the lithium acetate procedure [17].
Yeast growth conditions
Yeast cells transformed with pEMBLyex4, pyex-SIC1 or pyex-KIP1 plasmids were grown at 30 °C in a synthetic medium containing 6.7 g/l YNB (yeast nitrogen base) without amino acids (Difco), 100 mg/l adenine, 50 mg/l leucine and tryptophan, and 2% (w/v) galactose as carbon source to induce expression of plasmid-borne genes.
The cell density of liquid cultures was determined with a Coulter counter using mildly sonicated and diluted samples. The fraction of budded cells was scored microscopically from samples of at least 300 cells after mild sonication.
Flow cytofluorimetric analysis
A total of 2×107 cells in mid-exponential growth [cell density approx. (2–8)×106 cells/ml] were harvested by filtration, mildly sonicated, fixed in 70% (v/v) ethanol and subsequently processed for FACS analysis. Cells were washed three times with PBS, resuspended in 1 ml of PBS with 1 mg/ml RNAse and incubated overnight at 37 °C. After incubation, cells were washed once with PBS, resuspended in DNA staining solution (0.46 mM propidium iodide/50 mM Tris/15 mM MgCl2, pH 7.7) and incubated in ice in the dark for 30 min. All centrifugations were performed at 18000 g for 5 min at 4 °C. Cell suspensions were transferred into FACS Falcon tubes and sonicated prior to FACS analysis. To measure cell size, forward scatter was recorded using the same cells. Analyses were performed with a FACScan (Becton Dickinson), and plots were generated with WinMDI 2.8.
Production and purification of recombinant proteins and protein complex
E. coli strain BL21 (DE3)[pLysE] was transformed with plasmid pIVEX2.4a-SIC1, cultured in Luria–Bertani broth with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol at 37 °C (A600=0.5), and induced for 4 h with 50 μM isopropyl β-D-thiogalactoside at 25 °C. Sic1–His6 protein was purified on Ni2+/nitrilotriacetate beads as described in the QIA Expressionist Handbook (Qiagen) and eluted with 100 mM imidazole. Protein concentration was measured by the Bradford method using a Bio-Rad protein assay kit. The purified protein was stored at −20 °C in 50 mM Tris/HCl, pH 7.5, and 100 mM NaCl.
Baculovirus cells were transformed with plasmids pVL1392-GST-Cdk2 and pVL1392-GST-cyclin A separately to express Cdk2 or cyclin A proteins, or by co-expression of the two plasmids for expression of the Cdk2–cyclin A complex. GST–Cdk2 and GST–cyclin A proteins and the GST–Cdk2–cyclin A kinase complex were purified on glutathione–Sepharose 4 Fast Flow (Amersham Biosciences) and subsequently cleaved with PreScission™ Protease (Amersham Biosciences) to remove the GST tags. PreScission™ and cleaved GST tags were removed by re-chromatography on glutathione–Sepharose 4 Fast Flow. Purified proteins were kindly provided by Dr Simon Plyte (Pharmacia-Pfizer, Nerviano, Milan, Italy).
BIAcore analysis
A BIAcore X system was used to analyse molecular interactions by means of SPR (surface plasmon resonance) [18]. Sic1 protein was covalently coupled to a Sensor Chip CM5 (carboxymethylated dextran surface), using amine-coupling chemistry [19]. A surface density of 3500 resonance units was generated for Sic1 protein by using 100 μl of a 50 μg/ml protein solution in acetate buffer, pH 3.5. Solutions of each interacting protein (Cdk2, cyclin A or their complex) were injected over the surface at 25 °C in a volume of 35 μl at a flow rate of 10 μl/min in HBS running buffer [10 mM Hepes, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% (v/v) surfactant P20]. After injection, protein solutions were replaced by HBS at a continuous flow rate of 10 μl/min. Surface regeneration was accomplished by injecting successively 1 M NaCl, 100 mM NaOH and 10 mM HCl (1 min contact time). All injected solutions were run simultaneously over a control flow cell containing a blank surface (with no immobilized protein). The response observed in the control flow cell was subtracted for each sensorgram (time course of the SPR signal) and the baseline was normalized to 0 resonance units. Apparent kinetic (kon and koff) and thermodynamic (KD=koff/kon) parameters of the interactions studied were obtained by simultaneous fitting of binding curves obtained with different concentrations of each interacting protein or protein complex using the BIAevaluation 3.1 SPR kinetic software (BIAcore).
In vitro Cdk2 kinase assays
In vitro assays of the phosphorylation activity of the Cdk2–cyclin A kinase complex were run in a final volume of 30 μl of 50 mM Tris/HCl, pH 7.5, 12 mM MgCl2, 1 mM dithiothreitol, 40 nM Cdk2–cyclin A kinase complex and an appropriate concentration of substrate: histone H1 (Roche) from 1 to 30 μM or GST–pRb (residues 769–921) [Rb (769); sc-4112; Santa Cruz Biotechnology, Inc.] from 0.5 to 8 μM. When required, an appropriate Sic1 concentration (from 1.5 to 5 μM) was added. The reaction was started by adding 500 μM [γ-32P] ATP (specific radioactivity 2000 c.p.m./pmol) and stopped after a 10 min incubation at 30 °C by cooling samples on ice and adding 4×SDS sample buffer [0.25 M Tris/HCl, pH 6.8, 40% (v/v) glycerol, 9.2% (w/v) SDS, 4% (v/v) β-mercaptoethanol, 0.01% (w/v) Bromophenol Blue]. Proteins were separated by SDS/PAGE on 12% (w/v) polyacrylamide gels. Phosphorylated bands were identified by autoradiography and quantified using Scion Image software (Scion Corp.) on a 256 grey level Tiff-scanned image of the autoradiographic film.
Kinase activities of Cdk2–cyclin A towards histone H1 and GST–pRb, in the presence or absence of Sic1, were analysed with GraphPad Prism software. Kinetic parameters were calculated by fitting the data to the Michaelis–Menten equation. For histone H1 kinase assays only, the Ki value was calculated by plotting the apparent Km values obtained against the cognate Sic1 concentration.
Protein extraction and immunoblotting
A total of 2×108 exponentially growing cells were harvested by filtration and lysed using ice-cold HB1 buffer [25 mM Mops, pH 7.2, 15 mM MgCl2, 15 mM EDTA, 0.5% (v/v) Triton X-100] plus protease inhibitor mix (Complete EDTA-free Protease Inhibitor Cocktail Tablets; Roche). An equal volume of acid-washed glass beads (Sigma) was added, and cells were broken by ten vortex/ice cycles of 1 min each. Extracts were clarified by centrifugation. Protein concentration was measured by the Bradford method using a Bio-Rad protein assay kit. Typically, samples of 70–100 μg of protein were used for direct Western blotting. Protein extracts were separated using a 12% (w/v) polyacrylamide gel, and Western blots to detect Sic1 or p27Kip1 proteins were performed with purified anti-Sic1 polyclonal antibody kindly provided by Dr Oriano Marin (Dipartimento di Chimica Biologica, Università di Padova, Padova, Italy) and anti-p27Kip1 monoclonal antibody (Transduction Laboratories) respectively (1:1000 dilution). An enhanced chemiluminescence system (ECL® Western Blotting Detection Reagents; Amersham Biosciences) was used for antibody detection after immunoblotting.
RESULTS
The inhibitory domain of yeast Sic1 is structurally related to that of a mammalian Cki
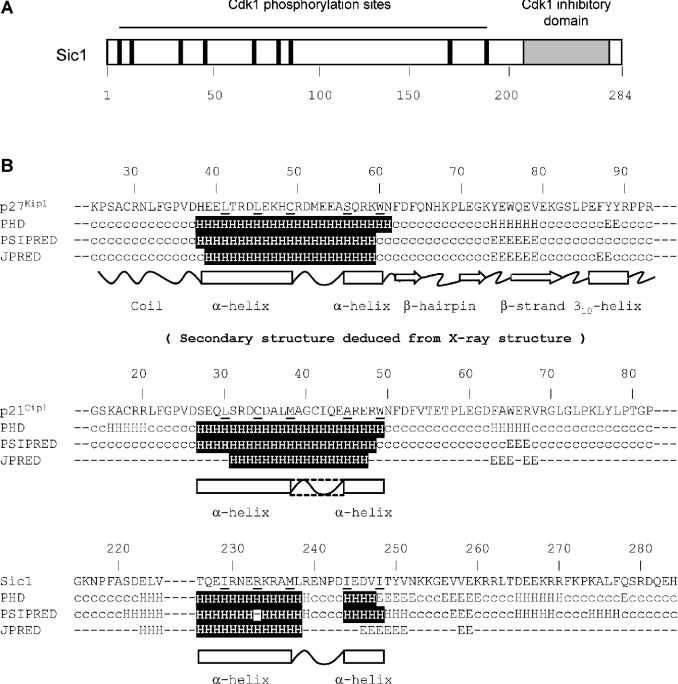
Sic1 has no obvious sequence homologues in the databanks, as indicated by the very low scores obtained using BLAST [20], PSI-BLAST [21] and FASTA [22] (results not shown). Due to its very low sequence similarity to p27Kip1, conventional homology modelling could not be applied to Sic1. Analysis of the inhibitory domain of Sic1 by fold recognition methods such as GenTHREADER [23,24] and 3D-PSSM [25,26] did not identify any suitable compatible fold (results not shown). Since it has been suggested that Sic1 may be a functional homologue of Ckis of the Kip/Cip family [7], we reasoned that local properties at the interaction surface with the Cdk–cyclin complex might be conserved. Secondary structure predictions for the sequence corresponding to the inhibitory domain of Sic1 (residues 215–284 [9]), as well as for the corresponding domains of p27Kip1 (residues 25–93 [8]) and p21Cip1 (residues 14–82 [8]), were computed as described in the Experimental section, and compared with the secondary structure deduced from the X-ray structure of p27Kip1 (Figure 1B). This analysis revealed that the secondary structures of these protein domains should be very similar. Notably, a long α-helix predicted in Sic1 (residues 226–248) shares a similar amphiphilic profile with the corresponding α-helix of p27Kip1 and p21Cip1.
Figure 1. Sequence alignment and secondary structure predictions for p27Kip1, p21Cip1 and Sic1 inhibitory domains.
(A) Schematic view of the full-length Sic1 protein, indicating the sites of phosphorylation by Cdk1 (black lines) and the Cdk1-inhibitory domain (grey). (B) Secondary structure predictions computed by PHD, PSIPRED and JPRED programs for the inhibitory domains of p27Kip1 (residues 38–60), p21Cip1 (residues 27–49) and Sic1 (residues 215–284) are shown (H, α-helix; c, coil; E, β-sheet). Residues involved in binding to either Cdk2 or cyclin A are underlined. Residues predicted to be in an α-helix within the Cdk2–cyclin A-interacting region are shown as white-on-black characters. Secondary structure deduced from X-ray crystallography is shown for p27Kip1. Corresponding expected secondary structures for p21Cip1 and Sic1 in the Cdk2–cyclin A complex are also shown.
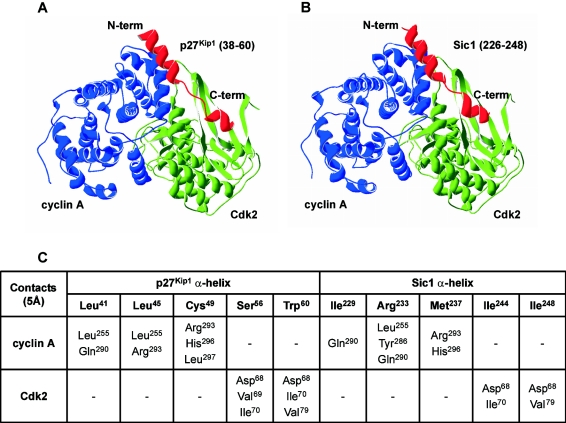
On the basis of secondary structure predictions, the inhibitory domain of Sic1 was modelled using the corresponding domain of p27Kip1 as a template, and the α-helix of p27Kip1 (residues 38–60; Figure 2A) was mutated in silico to generate the predicted α-helix of Sic1 (residues 226–248). The Sic1 model was then docked on to the Cdk2–cyclin A binary complex and optimized by energy minimization, as described in the Experimental section (Figure 2B). Analysis of the ternary complex reinforced the reliability of this structural prediction. In fact, the interface between the inhibitory domains of Sic1, Cdk2 and cyclin A is characterized by proper steric and electronic contacts that should allow the formation of a stable ternary complex. Five residues within the amphipathic α-helix of p27Kip1 establish hydrophobic contacts with either cyclin A or Cdk2 [8]; corresponding amino acids in the predicted Sic1 α-helix make contact with a significant sub-group of the interacting residues in the computed model (Figure 2C). For instance, the hydrophobic properties of residue Leu41 of p27Kip1 – positioned for specific interaction with the cyclin subunit of the Cdk2–cyclin A complex [8] – are conserved in Sic1 by residue Ile229, supporting the notion that the three Ckis share a common structural fold in the Cdk–cyclin-interacting region. Interestingly, residue Arg233 of Sic1, although not itself hydrophobic, appears able to be threaded within the cyclin A structure in the correct orientation, making use of the alkyl moiety to effect specific hydrophobic interactions.
Figure 2. Molecular visualization of surface interactions of p27Kip1 and Sic1 inhibitory domains with the Cdk2–cyclin A complex.
α-Helices of p27Kip1 (residues 38–60) (A) and Sic1 (residues 226–248) (B) are shown in red; Cdk2 and cyclin A are visualized in green and blue respectively. (C) Analysis of the interface contacts of p27Kip1 and Sic1 long amphipathic α-helixes with the Cdk2–cyclin A complex were conducted within the 5 Å range. The indicated residues of the Sic1 α-helix are involved in the same contacts as the corresponding residues of the p27Kip1 α-helix (see Figure 1B) with regard to the Cdk2–cyclin A complex.
In summary, these findings indicate that, despite their low sequence similarity, the inhibitory domains of yeast Sic1 and mammalian p27Kip1 may be structurally related.
The yeast Sic1 protein binds to the mammalian Cdk2–cyclin A complex
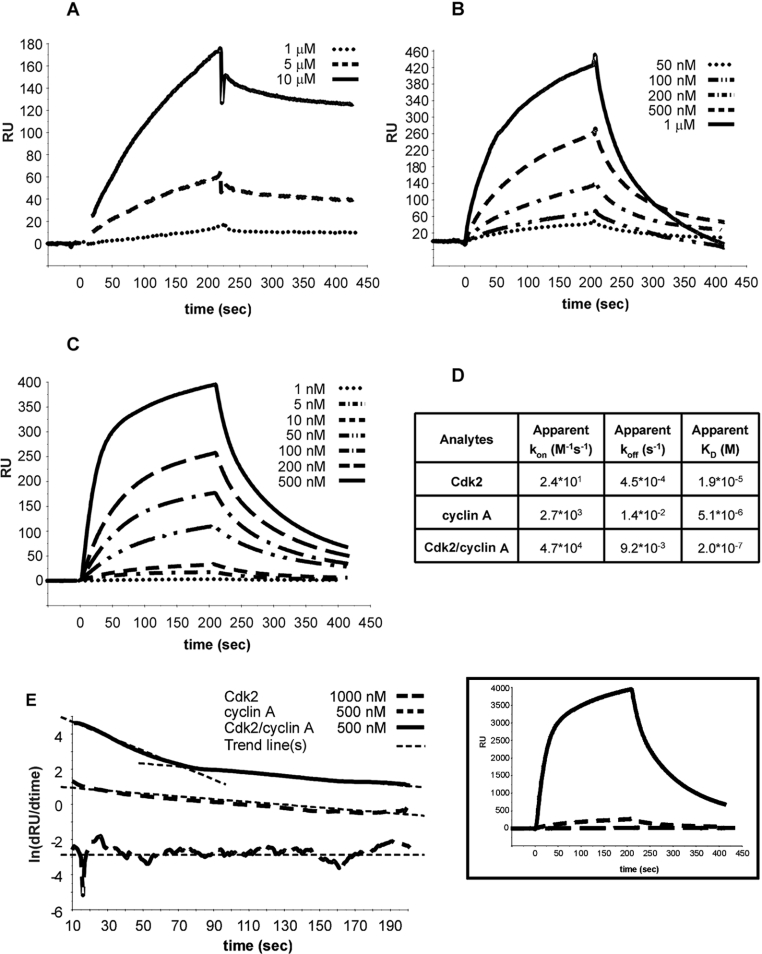
The results reported above predict that the inhibitory domain of Sic1 has the ability to interact with the mammalian Cdk2–cyclin A complex. To test this hypothesis directly, we tested the interactions of Sic1 with mammalian Cdk2 and cyclin A (alone or in complex) by SPR using a BIAcore X. In all experiments described, Sic1 was immobilized on the sensor chip and apparent kinetic (kon and koff) and thermodynamic (KD=koff/kon) parameters of the studied interactions were determined by fluxing several concentrations of each chosen interactor. Typical sensorgrams obtained using Cdk2, cyclin A or the Cdk2–cyclin A complex are reported in Figures 3(A)–3(C) respectively. Apparent kinetic and thermodynamic parameters obtained after fitting sensorgrams are summarized in Figure 3(D). These data indicate that the affinity of Sic1 for Cdk2 is very low, while binding of Sic1 to cyclin A and, much more so, to the Cdk–cyclin complex is more favourable. These findings suggest that Sic1 could realize its inhibitory function by interacting first with the cyclin and then extending on the surface of the kinase complex to reach the binding site on the kinase. A two-step mechanism of binding of Sic1 to the Cdk2–cyclin A complex is supported by the secondary plot of ln[d(RU)/d(time)] against time (where RU=response) derived by sensorgrams shown for clarity on the side of the secondary plots (Figure 3E). The interactions of Sic1 with cyclin A and Cdk2 individually show a curve with a unique slope (indicative of a single binding site), whereas the interaction with the Cdk2–cyclin A complex is better described by a curve with two slopes, indicative of Sic1 making contact with two sites on the complex.
Figure 3. Analysis of the binding of Sic1 to Cdk2, cyclin A and the Cdk2–cyclin A complex by BIAcore.
Sic1 was immobilized on the sensor chip and the binding curves were recorded by flowing the analytes Cdk2 (A), cyclin A (B) and the Cdk2–cyclin A complex (C) on the chip surface. (D) Values for apparent association rate (kon), dissociation rate (koff) and affinity constant (KD) were obtained by fitting of data in (A)–(C), as detailed in the Experimental section. (E) Analysis of stoichiometry of binding between Sic1 and the analytes, obtained by plotting ln[d(RU)/d(time)] against time. Trend line(s) approximating each curve are also shown. The original graph is shown to the right.
Inhibition by Sic1 of Cdk2–cyclin A kinase activity
To investigate the mechanism of inhibition of Cdk2–cyclin A activity by Sic1, a kinetic analysis using the purified proteins was conducted using either histone H1 or GST–pRb as substrate, as described in the Experimental section. Substrate recruitment to Cdk2 has been shown to rely upon interactions with a docking site located on cyclin A for substrates such as E2F1, p21Cip1, p27Kip1, p107 and pRb [27], while histone H1, which is also phosphorylated by Cdk2–cyclin A, binds only to the catalytic site on Cdk2 and not to the docking site [28]. It was therefore of interest to compare the effects of adding Sic1 to two assays of Cdk2–cyclin A activity, one utilizing pRb as substrate and the other using histone H1.
Since it has been shown that pRb contains in the C-terminal region (beginning at residue 870) the motif able to bind the docking site, a commercially available GST–pRb fusion protein (spanning residues 769–921 of pRb), which contains both the cyclin A-binding motif and Cdk/cyclin phospho-acceptor sites [27], was used in the assay.
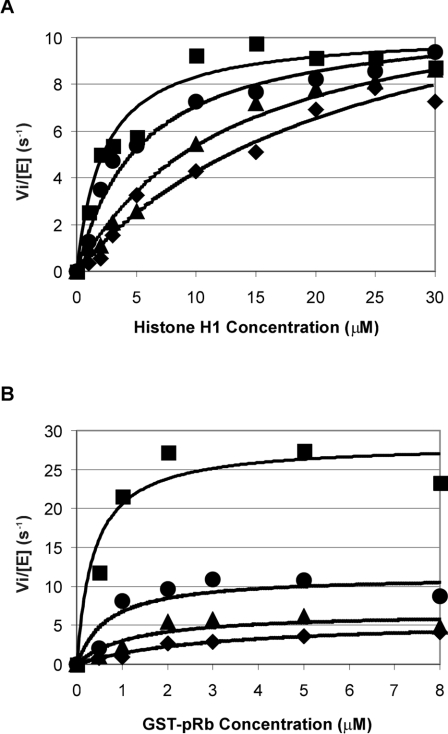
The kinase activity of Cdk2–cyclin A towards histone H1 and GST–pRb was determined by in vitro kinase assays in the presence of [γ-32P]ATP. The initial velocities of histone H1 and GST–pRb phosphorylation, obtained in the presence of a fixed ATP concentration (500 μM) at different Sic1 concentrations, were calculated by densitometry and plotted as a function of substrate concentration. Experimental data for the uninhibited and inhibited reactions are reported in Figures 4(A) (histone H1 as substrate) and 4(B) (GST–pRb as substrate), together with the corresponding Michaelis–Menten curves. The histone H1 kinase assay indicated that, under our experimental conditions, Sic1 is acting as a competitive inhibitor. The apparent Km values of the Cdk2–cyclin A complex were used to calculate the Ki in a secondary plot of apparent Km against Sic1 concentration. The obtained Ki value for Sic1 (500 nM) was found to be of the same order of magnitude as the KD value obtained by BIAcore analysis (Figure 3D). In contrast, the GST–pRb kinase assay showed that both kinetic parameters (Vmax and Km) changed in the presence of Sic1, with a very marked decrease in the Vmax value (Figure 4B).
Figure 4. Inhibition of Cdk2–cyclin A kinase activity by Sic1.
Cdk2–cyclin A kinase activity was measured with various histone H1 (A) or GST–pRb (B) concentrations at several fixed Sic1 concentrations: 0 μM (■), 1.5 μM (●), 3.5 μM (▲) and 5 μM (◆). The ATP concentration used in the assay was 500 μM, and the Cdk2–cyclin A concentration was 40 nM.
In summary, our analysis shows that Sic1 presents different inhibitory patterns with regard to the Cdk2–cyclin A complex depending on whether histone H1 or GST–pRb protein is used as substrate.
Overexpression of KIP1 in yeast rescues the cell cycle-related phenotype of a sic1Δ strain
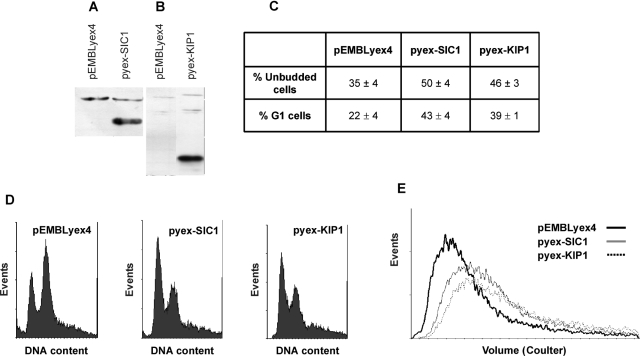
To ascertain the physiological significance of promiscuous interactions among yeast and mammalian Ckis, Cdk and cyclins, the ability of the yeast and mammalian Ckis to rescue the cell cycle-related phenotype of yeast cells deleted for the SIC1 gene was examined. Cells carrying disruption or deletion of SIC1 are viable [29], but display phenotypes indicative of aberrant cell cycle progression. In fact, cultures of sic1Δ cells have a low fraction of G1 unbudded cells and display an altered cell size ([30] and Figure 5). The effects of Sic1 and p27Kip1 overexpression, driven by a GAL1 promoter (Figures 5A and 5B), on cell cycle-related parameters were thus investigated in cells growing exponentially in galactose-supplemented media.
Figure 5. Overexpression of KIP1 rescues the cell cycle-related phenotype in a sic1Δ strain.
sic1Δ strains transformed with the URA3-based plasmid pEMBLyex4 or the same plasmid expressing the SIC1 and KIP1 genes under the control of a galactose-inducible promoter were grown in a synthetic medium containing 2% (w/v) galactose, collected in mid-exponential phase (approx. 4×106 cells/ml) and scored for expression of Sic1 (A) or p27Kip1 (B) protein by immunoblots with antibodies as described in the Experimental section, for the fraction of unbudded cells (C) by direct microscopic examination, for the fraction of cells in G1 by FACS analysis (C and D), and for cell size by forward scatter (E). Data in (C) are means±S.D. of at least three independent experiments. Representative DNA and size distributions and immunoblots are shown in the other panels.
Overexpression of either Sic1 or p27Kip1 significantly increased the fraction of unbudded cells (Figure 5C) and cells with a G1 DNA content (Figures 5C and 5D), as one may expect for a Cki that expresses its function at the G1/S transition. Analysis of forward scatter (a measure of cell size) showed that overexpression of either Sic1 or p27Kip1 resulted in a shift in the size distribution. Figure 5(E) shows that sic1Δ cells expressing either the yeast or the mammalian Cki had size distributions shifted towards larger cells compared with pEMBLyex4-transformed sic1Δ cells.
DISCUSSION
In budding yeast, Sic1 blocks the activity of the S-Cdk1 kinase required for the initiation of DNA replication, the onset of which requires removal of Sic1 [4]. A relevant role for Sic1 in the control of the G1-to-S transition is indicated by the fact that deletion of Sic1 causes premature DNA replication from fewer origins (sparse DNA firing), extension of S phase duration and inefficient separation of sister chromatids during anaphase. Genomic instability is generated by the ensuing double-strand breaks and gross chromosomal rearrangements [6].
The novelty of our results lies in the indication that Sic1 is structurally and functionally related to mammalian p27Kip1, a member of the Kip/Cip family of Ckis, sharing a conserved inhibitory domain. Moreover, we have collected evidence showing that Sic1, like p27Kip1, interacts with both the docking site and the catalytic site of the Cdk2–cyclin A complex.
The lack of sufficient sequence identity between Sic1 and p27Kip1, and the failure of fold recognition methods to identify any suitable compatible fold, hampered the possibility of using conventional modelling techniques. In order to ascertain the degree of evolutionary conservation of the inhibitory domain of yeast and mammalian Ckis, we used in silico mutagenesis – guided by the results of secondary structure prediction experiments – to build the putative Cdk–cyclin-interacting helix of Sic1 and later to dock it to the mammalian Cdk2–cyclin A complex. Although our docking experiment does not have a resolution high enough to unambiguously assign inter-residue contacts, inspection of the computed ternary complex (in a form after energy minimization by molecular mechanics) suggested that the inhibitory domain of Sic1 has a surface that is able to interact with both subunits of the mammalian Cdk2–cyclin A complex.
A sequential mechanism for the inhibition of Cdk2–cyclin complexes has been proposed for Kip/Cip inhibitors on a structural basis [8,31], and more recently confirmed by studies of binding of p27Kip1 to Cdk2–cyclin A by SPR and NMR [32]. The latter study showed that the intrinsic flexibility of the mammalian inhibitor in solution affects the kinetics of its interaction with Cdk2, cyclin A and the Cdk2–cyclin A complex. In particular, the different affinities observed for binding of p27Kip1 to Cdk2 and cyclin A strongly suggest that p27Kip1 recognizes and binds to the cyclin A subunit of Cdk2–cyclin A complex as the first step in a sequential mechanism, and then folds into an elongated structure that extends to the kinase subunit [32].
Our computational data suggest that Sic1 may bind to the mammalian Cdk2–cyclin A complex with a similar mechanism and thus share a similar mode of inhibition with Kip/Cip proteins. Consistently, the interaction of Sic1 with mammalian Cdk2 and cyclin A (alone or in complex), tested by BIAcore, indicated that the affinity of Sic1 for Cdk2 is quite low, while binding of Sic1 to cyclin A and (to a much greater extent) to the Cdk2–cyclin A complex is strong. In addition, secondary plots derived by BIAcore sensorgrams indicate that Sic1 makes contact with two sites on the complex.
A two-site mechanism for binding of Sic1 to the Cdk–cyclin complex is further supported by the results of experiments testing the inhibition of pRb phosphorylation by Sic1. In fact, while histone H1 (which lacks a cyclin A binding motif) at high concentrations overcomes the inhibitory activity of Sic1, pRb at high concentrations is not able to reverse the inhibition by Sic1, the major effect of which is thus a consistent decrease in the apparent Vmax of the Cdk2–cyclin A complex with regard to the pRb substrate (Figure 4B). Proteins that have a cyclin A binding motif, such as pRb, have to interact first with the docking site on cyclin A in order to be phosphorylated; in fact, both site-directed mutagenesis of the pRb cyclin binding motif and competition with a short peptide spanning the Cdk2–cyclin A binding motif present in the E2F1 substrate prevents its phosphorylation by Cdk2 [27]. Thus binding of pRb to the catalytic site of the kinase may displace the phosphorylatable Sic1 domain from the Cdk2 catalytic site, without resulting in significant pRb phosphorylation, so explaining the observed decrease in apparent Vmax.
Cross and Jacobson [33] showed previously that Clb5 and p27Kip1 interact in vivo in yeast in two-hybrid experiments, and proposed that a conserved region of cyclins may provide interactions with their targets. Our experiments go one step further by showing that the p27Kip1 protein can functionally substitute in vivo for Sic1, since overexpression of the KIP1 gene in Saccharomyces cerevisiae was able to rescue the cell cycle-related phenotype of a sic1Δ strain, in a manner indistinguishable from overexpression of the homologous yeast Cki gene. It should also be remembered that Sic1 has been shown previously to be a functional homologue of the Schizosaccharomyces pombe Cdk inhibitor Rum1, despite the very limited sequence identity between the Ckis from the two yeasts [34].
Taken together, the results presented in this paper support the notion that Sic1, although not showing significant sequence identity with p27Kip1, shares a conserved inhibitory domain, interacts with the Cdk–cyclin complex by a similar two-site mechanism and can be functionally replaced in vivo by p27Kip1.
Acknowledgments
We particularly thank Professor Lorenzo A. Pinna for suggestions and critical reading of the manuscript. We are grateful to Dr Oriano Marin for purified anti-Sic1 polyclonal antibody from rabbit, and to Dr Simon Plyte (Pharmacia-Pfizer) for the kind supply of Cdk2–cyclin A kinase complex purified from baculovirus. This work was supported by research grants AIRC, FIRB 2001 and PSO CNR-MIUR to L.A., and COFIN 2002 to M.V.
References
- 1.Vermeulen K., Van Bockstaele D. R., Berneman Z. N. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolifer. 2003;36:131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray A. W. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–234. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- 3.Obaya A. J., Sedivy J. M. Regulation of cyclin-Cdk activity in mammalian cells. Cell. Mol. Life Sci. 2002;59:126–142. doi: 10.1007/s00018-002-8410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwob E., Bohm T., Mendenhall M. D., Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 5.Nash P., Tang X., Orlicky S., Chen Q., Gertler F. B., Mendenhall M. D., Sicheri F., Pawson T., Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature (London) 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 6.Lengronne A., Schwob E. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1) Mol. Cell. 2002;9:1067–1078. doi: 10.1016/s1097-2765(02)00513-0. [DOI] [PubMed] [Google Scholar]
- 7.Peter M., Herskovitz I. Joining the complex: cyclin-dependent kinase inhibitory proteins and the cell cycle. Cell. 1994;79:181–184. doi: 10.1016/0092-8674(94)90186-4. [DOI] [PubMed] [Google Scholar]
- 8.Russo A. A., Jeffrey P. D., Patten A. K., Massaguè J., Pavletich N. P. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature (London) 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 9.Hodge A., Mendenhall M. The cyclin-dependent kinase inhibitory domain of the yeast Sic1 protein is contained within the C-terminal 70 amino acids. Mol. Gen. Genet. 1999;262:55–64. doi: 10.1007/s004380051059. [DOI] [PubMed] [Google Scholar]
- 10.Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rost B., Sander C., Schneider R. PHD – an automatic mail server for protein secondary structure prediction. Comput. Appl. Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 12.Jones D. T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 13.Cuff J. A., Barton G. J. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins Struct. Funct. Genet. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 14.Guex N., Peitsch M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 15.Heiner A. P., Berendsen H. J., van Gunsteren W. F. MD simulation of subtilisin BPN′ in a crystal environment. Proteins Struct. Funct. Genet. 1992;14:451–464. doi: 10.1002/prot.340140406. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J., Fritsch E. F., Maniatis T. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 17.Schiestl R. H., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 18.Malmqvist M. BIACORE: an affinity biosensor system for characterization of biomolecular interactions. Biochem. Soc. Trans. 1999;27:335–340. doi: 10.1042/bst0270335. [DOI] [PubMed] [Google Scholar]
- 19.Johnsson B., Lofas S., Lindquist G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem. 1991;198:268–277. doi: 10.1016/0003-2697(91)90424-r. [DOI] [PubMed] [Google Scholar]
- 20.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U.S.A. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones D. T. GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol. 1999;287:797–815. doi: 10.1006/jmbi.1999.2583. [DOI] [PubMed] [Google Scholar]
- 24.McGuffin L. J., Jones D. T. Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics. 2003;19:874–881. doi: 10.1093/bioinformatics/btg097. [DOI] [PubMed] [Google Scholar]
- 25.Fischer D., Barret C., Bryson K., Elofsson A., Godzik A., Jones D., Karplus K. J., Kelley L. A., Maccallum R. M., Pawowski K., et al. CAFASP-1: critical assessment of fully automated structure prediction methods. Proteins Struct. Funct. Genet. Suppl. 1999;3:209–217. doi: 10.1002/(sici)1097-0134(1999)37:3+<209::aid-prot27>3.3.co;2-p. [DOI] [PubMed] [Google Scholar]
- 26.Kelley L. A., MacCallum R. M., Sternberg M. J. E. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 27.Adams P. D., Li X., Sellers W. R., Baker K. B., Leng X., Harper J. W., Taya Y., Kaelin W. G. Retinoblastoma protein contains a C-terminal motif that targets it for phosphorylation by cyclin-cdk complexes. Mol. Cell. Biol. 1999;19:1068–1080. doi: 10.1128/mcb.19.2.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulman B. A., Lindstrom D. L., Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. U.S.A. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendenhall M. D., Al-jumaily W., Nugroho T. T. The Cdc28 inhibitor p40SIC1. Prog. Cell Cycle Res. 1995;1:173–185. doi: 10.1007/978-1-4615-1809-9_14. [DOI] [PubMed] [Google Scholar]
- 30.Nugroho T. T., Mendenhall M. D. An inhibitor of yeast cyclin-dependent protein kinase plays an important role in ensuring the genomic integrity of daughter cells. Mol. Cell. Biol. 1994;14:3320–3328. doi: 10.1128/mcb.14.5.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan D. O. Under arrest at atomic resolution. Nature (London) 1996;382:295–296. doi: 10.1038/382295a0. [DOI] [PubMed] [Google Scholar]
- 32.Lacy E. R., Filippov I., Lewis W. S., Otieno S., Xiao L., Weiss S., Hengst L., Kriwacki R. W. p27 binds cyclin-CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat. Struct. Mol. Biol. 2004;11:358–364. doi: 10.1038/nsmb746. [DOI] [PubMed] [Google Scholar]
- 33.Cross F. R., Jacobson M. D. Conservation and function of a potential substrate-binding domain in the yeast Clb5 B-type cyclin. Mol. Cell. Biol. 2000;20:4782–4790. doi: 10.1128/mcb.20.13.4782-4790.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez-Diaz A., Gonzalez I., Arellano M., Moreno S. The Cdk inhibitors p25rum1 and p40SIC1 are functional homologues that play similar roles in the regulation of the cell cycle in fission and budding yeast. J. Cell Sci. 1998;111:843–851. doi: 10.1242/jcs.111.6.843. [DOI] [PubMed] [Google Scholar]