Abstract
Mass spectrometry with chemical cross-linking was used to probe the conformational changes of HSA (human serum albumin) in solution on interaction with monounsaturated OA (oleic acid) or polyunsaturated AA (arachidonic acid) or DHA (docosahexaenoic acid). Fatty acid-free or -bound HSA was modified with lysine-specific cross-linkers and digested with trypsin. Cross-linked peptides were analysed by nano-electrospray ionization MS to localize the sites of cross-linking. Our data indicated that a local conformational change involving movement of the side chains of Lys-402 of subdomain IIIA or Lys-541 of subdomain IIIB occurred upon binding of all three fatty acids. Our data also indicated that the side chains of Lys-205 (IIA) and Lys-466 (IIIA) moved closer towards each other upon binding AA or DHA, but not OA, suggesting that the conformations of HSA when bound to mono- and poly-unsaturated fatty acids are distinctively different. While these observations agreed with previous X-ray crystallographic studies, the distances between ε-amino groups of most cross-linked lysine pairs were shorter than the crystal structure predicted, possibly reflecting a discrepancy between the solution and crystal structures. This method can serve as a useful complement to X-ray crystallography, particularly in probing the structure of a protein in solution.
Keywords: arachidonic acid, conformational change, docosahexaenoic acid, MS, oleic acid, serum albumin
Abbreviations: AA, arachidonic acid; BS3, bis(sulphosuccinimidyl)suberate; 3-D, three-dimensional; DHA, docosahexaenoic acid; DSG, disuccinimidyl glutarate; DSS, disuccinimidyl suberate; ESI, electrospray ionization; HSA, human serum albumin; MS/MS, tandem MS; OA, oleic acid; Qq-TOF, quadrupole–time-of-flight
INTRODUCTION
HSA (human serum albumin) is the major protein component in human plasma. The protein consists of three homologous α-helical domains (I–III) arranged in a heart-shaped fashion. Each domain contains 10 helices, which are divided into six-helix subdomain A and four-helix subdomain B [1]. The primary function of HSA is to transport hydrophobic unesterified fatty acids that are required for the synthesis of membrane lipids and serve as the physiological energy source [2]. The tertiary structure of HSA and the interaction between albumin and fatty acids have been investigated over the past decades using X-ray crystallography [1,3–10] and NMR spectroscopy [11–13]. Great effort has been devoted to understanding the binding sites and affinities for fatty acids of different chain lengths and degrees of unsaturation. To date, a total of 11 binding sites with various affinities distributed asymmetrically across all three domains of HSA have been identified [7].
The biological properties of a protein depend on its conformation, and the protein conformation can be influenced by physical or chemical factors, for example ligand binding. Upon interacting with fatty acids, the HSA molecule undergoes dramatic conformational changes, according to X-ray crystallographic studies. The changes are described as a significant left-shifting of subdomains I and III relative to the central subdomain II, as well as rotations of side chains of amino acid residues [6]. Nevertheless, detailed mechanisms for the conformational changes, as well as the physiological implications, are still unclear, apparently because of the limited structural information obtained. It has been suggested that approaches other than X-ray crystallography and NMR are necessary in order to provide better structural elucidation pertinent to the conformational changes [14].
MS in conjunction with chemical cross-linking has emerged as a sensitive tool for probing the tertiary structure of proteins and protein–protein interactions [15–24]. Chemical cross-linkers introduce new covalent bonds into the 3-D (three-dimensional) structure of the protein [25]. The modified protein is digested and subjected to analysis by MS. Cross-linked peptides are identified by comparing the mass map of the digests from the cross-linked protein with that of the unmodified protein. Fragmentation by MS/MS (tandem MS) confirms the peptide sequence and localizes the cross-linked amino acid residues. Based on the cross-linked sites and the spacer arm lengths of the cross-linkers, low-resolution distance constraints can be obtained to assist the construction of the 3-D structure of a protein. This approach is particularly advantageous, since analysis time as well as the amount of protein sample required are significantly reduced in comparison with the traditional X-ray crystallography or NMR analysis. Moreover, there is no limitation to the molecular size of a protein of interest. Furthermore, like NMR, this method can be applied in solution, and therefore shows promise for monitoring the conformational changes of proteins due to protein–ligand or protein–protein interactions under native biological conditions.
We developed such a method to probe the unknown tertiary structure of BSA [22], which is thought to be similar to that of HSA due to the 76% sequence identity [2]. We reported valuable spatial distance information for lysine–lysine pairs in BSA, particularly for those which do not have counterparts in the crystal structure of HSA. In the present study, we applied our methodology to probe the conformational changes of HSA upon interaction with three important long-chain unsaturated fatty acids, including monounsaturated OA (oleic acid; C18:1,n−9) and polyunsaturated AA (arachidonic acid; C20:4,n−6), which are among the major fatty acids bound to HSA under normal physiological conditions [26], as well as polyunsaturated DHA (docosahexaenoic acid; C22:6,n−3), which is highly enriched in neural tissues [27] and plays an important role in neuronal survival and differentiation [28]. To introduce covalent bonds into the 3-D structure of HSA, which contains many lysine residues as well as many hydrophobic pockets, we chose three different lysine-specific homobifunctional cross-linkers with varying spacer arm lengths and hydrophobicity [22]. BS3 [bis(sulphosuccinimidyl) suberate], a water-soluble cross-linker with a spacer arm length of 11.4 Å (1.14 nm), and DSS (disuccinimidyl suberate), the water-insoluble analogue of BS3, were chosen in an attempt to generate complementary information about the lysine–lysine distance constraints for the lysine residues in either hydrophilic or hydrophobic environments [22,29]. In addition, we used DSG (disuccinimidyl glutarate), which is analogous to DSS in chemistry but contains a shorter spacer arm length (7.7 Å), to further refine the distance constraints between lysine residues. Our data indicated that all three fatty acids induced a local conformational change in domain IIIA, with little distortion in each individual domain. However, our data suggested that there is a distinctive conformational difference in the bound form of HSA when bound to polyunsaturated fatty acids, i.e. AA or DHA, in comparison with monounsaturated OA.
EXPERIMENTAL
Chemicals
Fatty acid-free HSA was purchased from Sigma-Aldrich (St. Louis, MO, U.S.A.). OA, AA and DHA were purchased from Nu-Check Prep Inc. (Elysian, MN, U.S.A.). BS3, DSS and DSG were purchased from Pierce (Rockford, IL, U.S.A.). Modified trypsin was purchased from Promega (Madison, WI, U.S.A.). Potassium phosphate was purchased from Mallinckrodt (Phillipsburg, NJ, U.S.A.). Pure water was obtained from a Milli-Q UV plus ultra-pure water system (Bedford, MA, U.S.A.). H218O (99%) was purchased from Isotec (Miamisburg, OH, U.S.A.). Other chemicals were from Sigma.
Fatty acid binding, cross-linking and tryptic digestion
An aliquot of fatty acid solution in methanol was dried under nitrogen and mixed thoroughly with 10 μM HSA in pH 7.6 phosphate buffer at a 9:1 molar ratio of fatty acid to HSA. The mixture was incubated at 37 °C for 30 min. The resulting HSA/fatty acid mixture was incubated with 500 μM BS3, DSS or DSG at room temperature for 30 min. BS3 solution was prepared fresh in pH 5.0 sodium citrate buffer, whereas the hydrophobic DSS or DSG was dissolved in DMSO before use, in accordance with the manufacturer's (Pierce) instructions. Under these cross-linking conditions no intermolecular cross-linked dimers or multimers were observed by SDS/PAGE separation (results not shown). The cross-linking reaction was quenched with 1 M Tris/HCl (pH 7.4). An aliquot was subjected to CD measurement using a Jasco J-810 spectropolarimeter (JASCO Corp., Tokyo, Japan). DSS- or DSG-modified samples were dialysed against pure water using a membrane with molecular mass cut-off of 3500 Da (Waters) to remove interference due to strong UV absorption by DMSO before CD measurements. The protein was reduced with dithiothreitol (100 mM) for 1 h at 56 °C and alkylated with iodoacetamide (100 mM) for 45 min at room temperature. After dialysis against pure water using a membrane with molecular mass cut-off of 3500 Da (Waters) three times within 24 h, the sample was dried using a vacuum centrifuge and reconstituted in pure water or 99% H218O with 5% (v/v) acetonitrile [21,22], followed by tryptic digestion at 37 °C for 20 h with a trypsin/protein ratio of 1:10.
Off-line static nano-ESI (electrospray ionization) MS analysis
After desalting using a C18 ZipTip (Millipore Corp., Bedford, MA, U.S.A.), the tryptic peptides were subjected to analysis by a QSTAR pulsar Qq-TOF (quadrupole–time-of-flight) mass spectrometer (Applied Biosystems/MDS Sciex, Toronto, Canada) equipped with a nano-ESI source. The ion source voltage was set at 1100 V in the positive-ion mode. A full mass spectrum was acquired over an m/z range of 500–2000, and the ions of interest were subjected to collision-induced dissociation using high purity argon for MS/MS analysis. Accurate mass measurements were performed using internal mass calibration with known unmodified tryptic peptide ions present in the sample.
LC/nano-ESI mass spectrometric analysis
LC/nano-ESI MS analysis was performed on a QSTAR pulsar Qq-TOF mass spectrometer (Applied Biosystems/MDS Sciex, Toronto, Canada), or an Agilent ion trap mass spectrometer (XCT) equipped with an Agilent 1100 nanoflow LC system. Peptides were separated using a Zorbax 300 SB C18 column (75 μm×150 mm, 3.5 μm; Agilent, Wilmington, DE, U.S.A.) using a mobile phase that contained solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile) at a flow rate of 300 nl/min. The mobile phase composition was held initially at 3% (v/v) solvent B for 5 min, and then changed from 3% to 85% (v/v) solvent B over 70 min.
PAWS and Protein Explorer softwares
The mass values of tryptic peptides were assigned with the assistance of the Protein Analysis Work Sheet (PAWS) (Proteo-Metrics). The distances between lysine–lysine pairs determined by X-ray crystallography were obtained using Protein Explorer (Version 1.982).
RESULTS
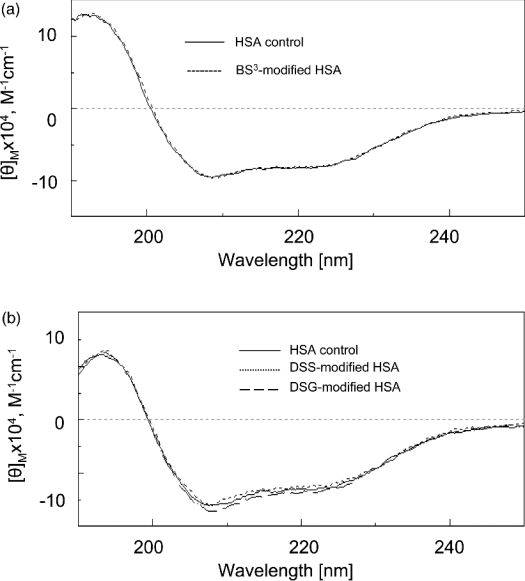
A major concern about protein cross-linking is whether the use of an excess amount of cross-linkers and the organic solvent affect the conformation of the protein [15,29]. Our cross-linking conditions included a 50:1 cross-linker/protein ratio and 5% (v/v) DMSO as the solvent for DSS and DSG. To confirm the conformation integrity of modified HSA under the experimental conditions, we recorded the far-UV CD spectra of HSA cross-linked with BS3, DSS or DSG, as shown in Figure 1. The spectra of the modified HSA were superimposable on the spectrum of the control protein. In addition, we conducted the cross-linking of HSA with BS3 in the presence or absence of 5% (v/v) DMSO, which generated virtually the same results (results not shown), further implying that no major conformational changes occur under our cross-linking conditions.
Figure 1. Far-UV CD spectra of BS3-modified (a) and DSS- or DSG-modified (b) HSA.
DSS- and DSG-modified samples were dialysed against pure water using a membrane with molecular mass cut-off of 3500 Da before CD measurements.
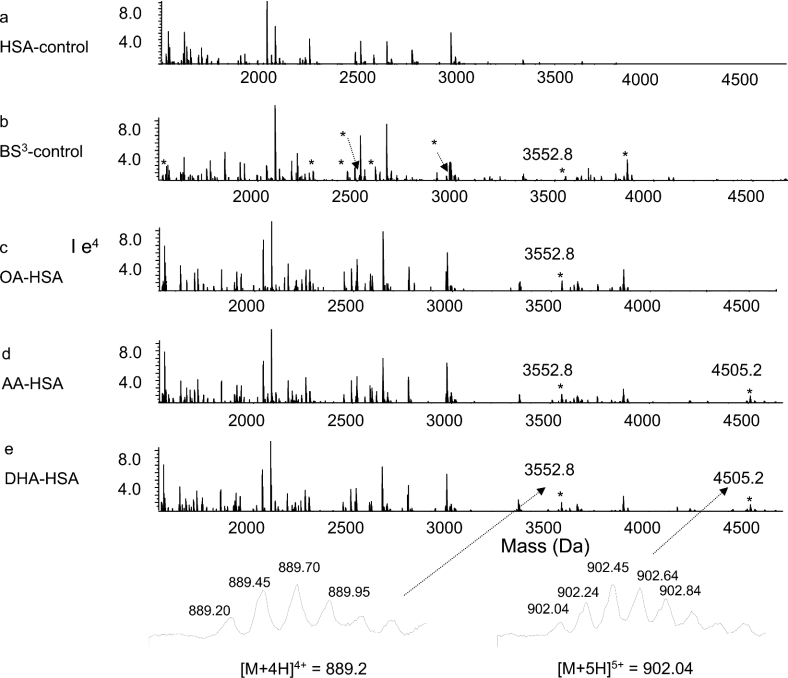
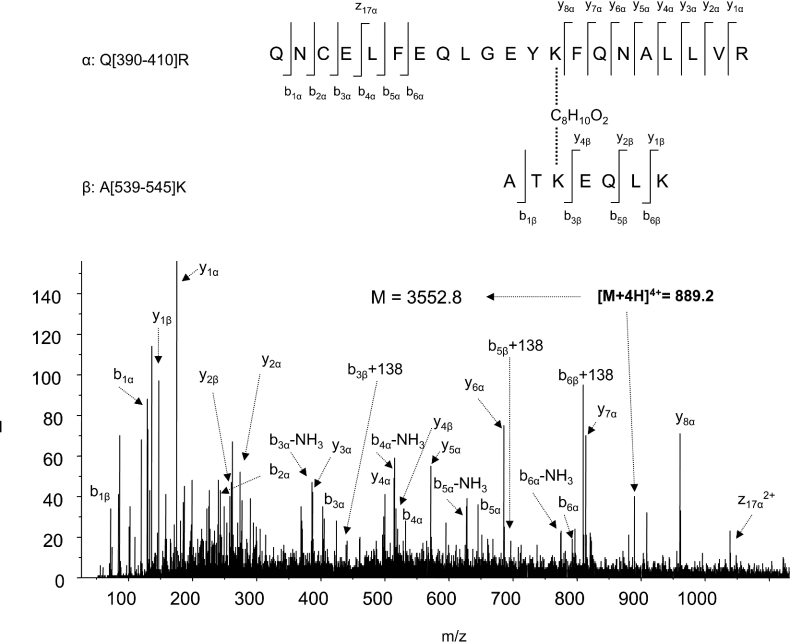
The ε-amino groups of two lysine residues reacted with BS3, DSS and DSG, resulting in a net mass increase of 138, 138 and 96 Da respectively, whereas modification of a single lysine residue (end-capping) increased the mass by 156, 156 and 114 Da respectively [15,22]. The cross-linked peptides were identified by comparing the mass spectra of tryptic digests obtained from unmodified and cross-linker-modified HSA. Furthermore, tryptic digestion in H218O confirmed a through-space cross-linked pair between two peptide segments by a characteristic 8 Da mass shift, due to the complete incorporation of two 18O atoms for each C-teminus [21,22]. Cross-linking with BS3 generated eight new peaks, which are marked with asterisks (*) in Figure 2(b). For example, a cross-linked peptide with a mass value of 3552.8 Da, which was reconstructed from a quadruply charged ion at m/z 889.2 (bottom left of Figure 2), emerged in BS3-modified sample (Figure 2b). The mass of the peptide shifted to 3560.8 Da when digested in H218O (results not shown), indicating involvement of a through-space cross-linking. With non-cross-linked control HSA, the peak was absent (Figure 2a). The identity of the cross-linked peptide was revealed by MS/MS, as shown in Figure 3. The data clearly revealed that this peak resulted from through-space cross-linking between two peptide segments, Gln-390–Arg-410 and Ala-539–Lys-545, with Lys-402 located in subdomain IIIA cross-linked to Lys-541 from subdomain IIIB, via C8H10O2 (derived from C8H12O2, the cross-linking arm of BS3, with two hydrogens subtracted due to cross-linking). The lysines participating in the cross-linking reaction were not hydrolysed by trypsin. The N-terminal b ions and C-terminal y ions resulting from the amide bond cleavages are labelled in Figure 3, in accordance with the nomenclature suggested by Schilling et al. [30]. In addition to b and y ions, the neutral loss of NH3 (−17 Da) from b ions, probably from side chains of Gln-390 or Asn-391, was also observed. Ions b5 and b6 included C8H10O2 with a net m/z increase of 138. The accurate mass measurement showed that the mass of the peptide was 3552.798 Da, which represented an error within 8 p.p.m., further confirming the assignment of the cross-linking pair.
Figure 2. Nano-ESI-QqTOF reconstructed mass spectra of tryptic digests from unmodified (a), and BS3-modified control (b), OA-bound (c), AA-bound (d) and DHA-bound (e) HSA.
The peak with a mass of 3552.8 Da emerged from BS3-modified samples (b–e). The peak with a mass of 4505 Da emerged from AA- or DHA-bound HSA samples (d, e), but was absent from OA-bound HSA (c). Cross-linked peptides in the fatty acid-free control (b) are marked with an asterisk (*).
Figure 3. Nano-ESI-QqTOF-MS/MS analysis of the cross-linked peptide with mass of 3552.8 Da depicted in Figure 2.
The sequence of the peptide is shown using the single-letter code, based on the fragmentation observed in the spectrum. The fragments in the spectra indicated two peptide segments, Q[390–410]R (Gln-390–Arg-410) and A[539–545]K (Ala-539–Lys-545), cross-linked at Lys-402/Lys-541. The peptide segments are labelled α and β respectively according to the nomenclature suggested by Schilling et al. [30]. N-terminal b ions and C-terminal y ions resulting from the amide bond cleavages are labelled.
The spectra of tryptic digests obtained from modified HSA before and after fatty acid binding were different from each other, as shown in Figures 2(c)–2(e). In addition to the peaks observed in the BS3 control (Figure 2b), a new peak with a mass of 4505.2 Da, reconstructed from the quadruply charged ion at m/z 1127.3 and the quintuply charged ion at m/z 902.0 (bottom right of Figure 2), were detected upon binding of AA (Figure 2d) or DHA (Figure 2e) to HSA. MS/MS analysis indicated that this peak originated from the through-space cross-linking between Cys-200–Arg-209 and Met-446–Arg-472, with Lys-205 of subdomain IIA linked to Lys-466 of subdomain IIIA. The peak was not seen in the spectrum obtained from OA-bound HSA (Figure 2c). In addition, a considerable increase in the intensity of the cross-linking peptide between Lys-402 of IIIA and Lys-541 of IIIB with mass of 3552.8 Da, discussed above, was observed in all three fatty acid-bound HSA samples.
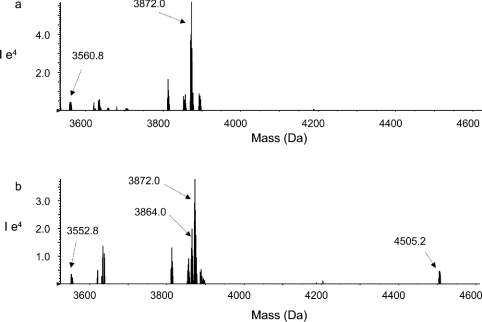
The changes in the peak intensities were further confirmed by the 18O labelling technique. The BS3-modified fatty acid-free HSA sample digested in H218O was mixed with the fatty acid-bound HSA sample digested in 16O water. The mixture was subjected to mass spectrometric analysis. For example, Figure 4(a) shows the reconstructed mass spectrum of the 18O-labelled fatty acid-free sample at a mass range of 3500–4600 Da. A cross-linked peptide peak with a mass of 3872.0 Da resulting from the cross-linking between Lys-402 and Lys-525 was identified. In addition, a weak peak with mass of 3560.8 Da due to the cross-linking of Lys-402 to Lys-541 was detected. Figure 4(b) shows the spectrum obtained from the mixture of DHA-bound sample digested in 16O water and the 18O-labelled fatty acid-free sample (mixed at a ratio of 1:2). The peak height ratio of 3864.0 Da (corresponding to the tryptic peptide cross-linked between Lys-402 and Lys-525) over 3872.0 Da was approx. 1:2. Even though the 18O-labelled fatty acid-free sample was twice concentrated, the peak with mass of 3560.8 Da was hardly detected in the mixture because of the dilution. However, the presence of the peak at 3552.8 Da was obvious. The peak with a mass of 4505.2 Da was also identified only in the mixture, whereas the corresponding 18O-labelled peak at 4513.2 Da was not present in the 18O-labelled control sample (Figure 4a).
Figure 4. Nano-ESI-QqTOF reconstructed mass spectra of BS3-modified HSA samples.
(a) Fatty acid-free sample digested in H218O; (b) mixture of the DHA-bound sample digested in normal H216O and the fatty acid-free sample digested in H18O (at a ratio of 1:2).
Modification by DSS generated similar results. A total of 12 cross-linked pairs were identified in the DSS-modified control. Eight of them were identical to the pairs seen with BS3. Although DSS generated more cross-linked pairs, no further changes in cross-linked pairs were observed due to fatty acid binding other than the ones observed with BS3.
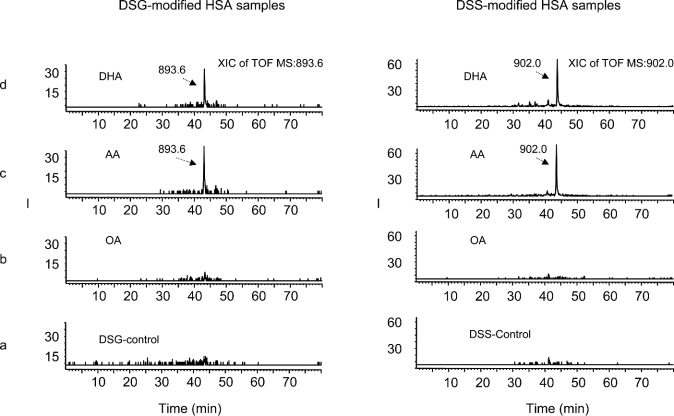
DSG was used to further refine the distance constraints for the cross-linked lysine pairs. Although three of the cross-linked pairs found in DSS-modified samples were not detected in the DSG-modified samples, the rest of the cross-linked peptides were consistent with those obtained from the DSS-modified counterparts. Figure 5 shows the confirmation of the cross-linked pair of Lys-205/Lys-466 induced upon AA or DHA binding, by LC/MS/MS, which allowed more reliable identification of the cross-linked peptides due to the separation of tryptic peptides prior to the mass spectrometric detection. The ion chromatograms for the peptide pair Lys-205/Lys-466 cross-linked with DSG (Figure 5, left) or DSS (Figure 5, right) are shown using quintuply charged ions of m/z 893.6 or 902.0 for a molecular mass of 4463 or 4505 Da respectively. The chromatographic peak eluting at approx. 43.5 min was observed in the digests obtained from AA- or DHA-bound HSA (Figures 5c and 5d), whereas it was not present in the digests obtained from OA-bound (Figure 5b) or control HSA (Figure 5a). The MS/MS spectrum obtained from the ion at m/z 902.0 was virtually identical to that from the ion at m/z 893.6. The fragments detected in the spectrum (results not shown) revealed that this peptide resulted from the through-space cross-linking between the peptide segments Met-446–Arg-472 and Cys-200–Arg-209, with Lys-205 (subdomain IIA) linked to Lys-466 (subdomain IIIA), via C5H4O2, derived from the cross-linking arm of DSG. The DSS-modified peptide showed a mass of 4505 Da (as seen from BS3-modified samples in Figure 2), an increase of 42 Da compared with its DSG-modified counterpart, because DSS and BS3 contain longer spacer arms (C8H12O2) than DSG (C5H6O2).
Figure 5. Extracted ion chromatograms of a cross-linked peptide obtained from DSG-modified (left) and DSS-modified (right) HSA samples analysed by HPLC/nano-ESI/MS/MS.
(a) Fatty acid-free control; (b) OA-bound HSA; (c) AA-bound HSA; (d) DHA-bound HSA. A new peak with a mass of 4463 Da (left) or 4505 Da (right), derived from the quintuply charged ion at m/z of 893.6 or 902.0 respectively, eluted at around 43.5 min with AA- or DHA-bound HSA (c, d).
All 13 cross-linked peptides that were identified are listed in Table 1. Among them, 11 pairs were commonly detected with or without fatty acid binding. Four pairs resulting from the cross-linking of Lys-212/Lys-240, Lys-276/Lys-286, Lys-64/Lys-73 and Lys-313/Lys-317 were not observed with the hydrophilic cross-linker BS3, indicating that at least an amino group of each pair is in the hydrophobic environment. In addition, cross-linked pairs Lys-212/Lys-351, Lys-414/Lys-541 and Lys-276/Lys-286, which were detected with DSS, were not observed with DSG, indicating that the distance between the ε-amino groups of lysine residues cross-linked in each pair is less than 11.4 Å but greater than 7.7 Å. The rest of the cross-linked pairs were observed with DSG, indicating that the distance between their ε-amino groups was within 7.7 Å. These distance constraints obtained by cross-linking were not consistent with the corresponding distances determined by X-ray crystallography for fatty acid-free HSA (pdb entry 1ao6) shown in Table 1. Except for the cross-linked pairs Lys-205/Lys-466 and Lys-402/Lys-541, all cross-linked peptides were detected with similar intensities in both fatty acid-free and fatty acid-bound samples. The α-carbon distances in the crystal structure of OA-bound (PDB 1gni) or AA-bound (PDB 1gnj) HSA molecules are comparable with those from fatty acid-free HSA. For those fatty acid-bound forms, the distances between the ε-amino groups of many lysine pairs (Lys-212/Lys-240, Lys-402/Lys-525, Lys-276/Lys-286, Lys-541/Lys-545, Lys-402/Lys-541 and Lys-205/Lys-466) are not available due to the absence of at least one ε-amino group of the lysine pairs. The three-dimensional structure of DHA-bound HSA has not been reported.
Table 1. Cross-linked peptides identified in HSA.
The distances between the α-carbons of the cross-linked lysine pair in fatty acid-free HSA are taken from X-ray crystallography results (PDB 1ao6). N.A., not applicable, because Lys-4 is missing in the crystal structure. Values in parentheses represent the distance between ε-amino groups.
| Observed mass of cross-linked peptide (Da) | Cross-linked peptide assigned by MS/MS analysis | Cross-linked lysines | Calculated mass of cross-linked peptide (Da) | Error (p.p.m.) | HSA control | HSA–OA | HSA–AA | HSA–DHA | Distance between α-carbons of lysine pair in HSA (Å) |
|---|---|---|---|---|---|---|---|---|---|
| 4012.900* | Ala-210–Arg-218∼Leu-234–Arg-257 | Lys-212 (IIA)/Lys-240 (IIA) | 4012.923 | 6 | + | + | + | + | 10.2 (12.0) |
| 3864.013 | Gln-390–Arg-410∼Lys-525–Lys-534 | Lys402 (IIIA)/Lys-525 (IIIB) | 3864.049 | 9 | + | + | + | + | 11.6 (11.6) |
| 2452.302† | Ala-210–Arg-218∼Leu-349–Lys-359 | Lys-212 (IIA)/Lys-351 (IIB) | 2452.336 | 14 | + | + | + | + | 13.4 (16.4) |
| 2593.483*† | Lys-414–Arg-428∼Ala-539–Lys-545 | Lys-414 (IIIA)/Lys-541 (IIIB) | 2593.468 | 6 | + | + | + | + | 15.3 (16.4) |
| 2512.193 | Asp-1–Arg-10∼Phe-11–Lys-20 | Lys-4 (before IA)/Lys-12 (IA) | 2512.235 | 17 | + | + | + | + | N.A. |
| 4639.246*† | Leu-275–Lys-313 | Lys-276 (IIA)/Lys-286 (IIA) | 4639.184 | 13 | + | + | + | + | 10.0 (14.9) |
| 2967.327 | Leu-349–Lys-372 | Lys-351 (IIB)/Lys-359 (IIB) | 2967.345 | 6 | + | + | + | + | 12.8 (14.8) |
| 3548.657* | Thr-52–Arg-81 | Lys-64 (IA)/Lys-73 (IA) | 3548.658 | 0.3 | + | + | + | + | 13.2 (10.3) |
| 4289.936* | Ser-287–Lys-323 | Lys-313 (IIB)/Lys-317 (IIB) | 4289.944 | 2 | + | + | + | + | 8.6 (14.1) |
| 2278.170 | Ala-539–Lys-557 | Lys-541 (IIIB)/Lys-545 (IIIB) | 2278.155 | 7 | + | + | + | + | 7.8 (13.6) |
| 1537.756 | Val-433–Lys-444 | Lys-436 (IIIA)/Lys-439 (IIIA) | 1537.738 | 12 | + | + | + | + | 6.4 (16.1) |
| 3552.798 | Gln-390–Arg-410∼Ala-539–Lys-545 | Lys-402 (IIIA)/Lys-541 (IIIB) | 3552.828 | 8 | ‡ | + | + | + | 14.6 (20.1) |
| 4505.181 | Cys-200–Arg-209∼Met-446–Arg-472 | Lys-205 (IIA)/Lys-466 (IIIA) | 4505.186 | 1 | − | − | + | + | 14.1 (15.2) |
* Peaks not observed with BS3.
† Peaks not observed with DSG.
‡ Weak peak.
DISCUSSION
The mechanism of the high-affinity binding of HSA to fatty acids has been described by a combination of hydrophobic and electrostatic interactions [3]. The hydrophobic amino residues in HSA that form hydrophobic cavities in each domain interact with the alkyl chain of the fatty acids, whereas two or three basic amino acid residues at the entrance of the hydrophobic pocket interact with the carboxy group of fatty acids. It has been suggested that hydrophobic interactions are the dominant mechanism [3]. In addition, the hydrophobic pockets accommodate different lengths of fatty acids, based on the depth of the pockets [7].
The hydrophobic environment may prevent nearby lysine residues from reaching the hydrophilic BS3 for cross-linking. On the other hand, the hydrophobic DSS can access lysine residues in hydrophobic surroundings. However, all surface lysine residues that are exposed to solvents should be available to both DSS and BS3. This view is consistent with the cross-linking results shown in Table 1. For example, Lys-73 and Lys-240 are surrounded by a hydrophobic environment [22], explaining the absence of Lys-64/Lys-73 and Lys-212/Lys-240 pairs when BS3 was used as the cross-linker. Eight cross-linked lysine pairs observed in both DSS- and BS3-modified control (fatty acid-free) HSA suggested that these lysine residues are exposed to the surface. The cross-linking results suggested that the distances between all cross-linked lysine pairs in fatty acid-free HSA were within 11.4 Å (between ε-amino groups) or 24 Å (between α-carbons, considering the maximum distance derived from the two side-chain lengths plus the spacer arm length of BS3 or DSS). Using DSG, nine cross-linked lysine pairs were identified; further refining 7.7 Å (between ε-amino groups) or 20 Å (between α-carbons) distance constraints for these lysine pairs. The results were consistent with the distances between α-carbons of lysine pairs determined by X-ray crystallography in most cases (Table 1). However, our results concerning the distance constraints of the ε-amino groups were not consistent with X-ray crystallographic data.
The cited maximum span lengths of the cross-linkers are based on a fully extended conformation of both the cross-linking arm of a cross-linker and the reactive side chains of two lysine residues in a perfect co-operative orientation [31]. In addition to the lysine–lysine distance from each α-carbon, the cross-linking would depend critically on the orientation of the lysine side chains in the most energetically favourable conformation in solution. Our data showed that three cross-linked peptides, including Lys-212/Lys-351, Lys-414/Lys-541 and Lys-276/Lys-286, were observed with DSS but not with DSG, even though the crystal structure indicated that the distances between their α-carbons were within the maximum distance allowed by DSG (20 Å). These data suggested that the orientation of the lysine side chains was such that the distance between the two ε-amino groups in the cross-linked lysine pair is within 11.4 Å but beyond 7.7 Å in solution. However, the distances between the ε-amino groups of these cross-linked pairs were determined to be 16.4, 16.4 and 14.9 Å respectively by X-crystallography (PDB 1ao6) (Table 1). The distance constraints for most ε-amino groups deduced from our data also differed from those obtained from X-ray crystallographic studies. Since the side chains are much more flexible than the bonds between α-carbons, it is possible that the cross-linkers could instantaneously trap the protein in a short-lived, less populated conformational state. Consequently, the most energetically favoured conformation of side chains may not always be represented. Alternatively, the discrepancy in the spatial distances may reflect a different conformational state of HSA in solution in comparison with the crystal structure.
The 11 cross-linked peptides derived from fatty acid-free HSA were cross-linked between lysine pairs within an individual domain. Included were intra-subdomain cross-linking pairs Lys-212/Lys-240 in subdomain IIA, Lys-276/Lys-286 in IIA, Lys-351/Lys-359 in IIB, Lys-64/Lys-73 in IA, Lys-313/Lys-317 in IIB, Lys-541/Lys-545 in IIIB and Lys-436/Lys-439 in IIIA, and the inter-subdomain cross-linking of Lys-402 of IIIA to Lys-525 of IIIB, Lys-212 of IIA to Lys-351 of IIB, and Lys-414 of IIIA to Lys-541 of IIIB. These cross-linkings were also observed when OA, AA or DHA was bound to HSA, with intensities similar to those observed for the fatty acid-free form, suggesting that no major distortion occurs in general within an individual domain upon fatty acid binding. In this regard, the cross-linking data appear to be consistent with the results obtained from X-ray crystallographic studies [14].
The inter-domain cross-linked peptide involving Lys-205 of subdomain IIA and Lys-466 of subdomain IIIA was identified only upon binding of AA or DHA to HSA. The fact that the cross-linking was observed using DSG indicated that the distance between the reactive amino groups in the side chains of the lysine pair was within 7.7 Å. In the fatty acid-free HSA molecule, the cross-linking was not detected even with DSS or BS3, despite the single modification (end-capping) for both Lys-205 and Lys-466, implying that the side chains of the lysine pair were separated by more than 11.4 Å, which is consistent with the distance determined by X-ray crystallography. It was therefore concluded that the side chains of Lys-205 and Lys-466 moved towards each other upon AA or DHA binding due to a conformational change of HSA after binding to the long-chain polyunsaturated fatty acids, as depicted in Figure 6. In the crystal structures of HSA complexed with OA or AA (PDB 1gni and 1gnj respectively), the distances between the α-carbons of Lys-205 and Lys-466 are 16.6 and 17.7 Å respectively, whereas the distances between the ε-amino groups of the lysine residues are not available due to the lack of the ε-amino group in Lys-466 in the crystal structure. Our data indicating that the cross-linking between Lys-205 and Lys-466 was not detected in OA-bound HSA molecules suggested a distinct difference in the method of binding of monounsaturated fatty acid to HSA in comparison with polyunsaturated fatty acids.
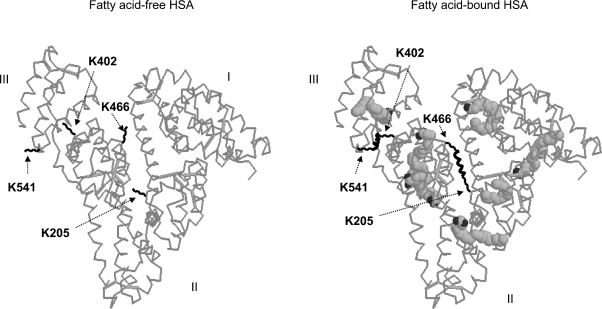
Figure 6. Conformational changes of HSA induced by fatty acid binding depicted based on the crystal structure of HSA complexed with AA.
The rotation of the side chains of Lys-402 (domain IIIA) and Lys-541 (domain IIIB) towards each other upon binding to a fatty acid is suggested. In addition, depicted is the movement of the side chains of Lys-205 from domain IIA and Lys-466 from domain IIIA closer towards each other upon binding of polyunsaturated AA or DHA. The cross-linking resulting from these movements is indicated. Fatty acid molecules are represented in space-filling form, and their binding sites shown in the right panel are based on the PDB entry 1gnj.
NMR studies have shown that polyunsaturated fatty acids exhibit binding behaviour quite distinct from that of saturated and monounsaturated fatty acids when interacting with HSA [10,32]. Similarly, in an investigation using X-ray crystallography, Peptipsa et al. [8] reported that AA and OA showed different patterns of binding to HSA. It was suggested that, at least at two sites, the presence of four double bonds in AA can cause a notable difference in the bound conformations of the fatty acid methylene tail. One site is located in the interface between subdomains IA and IIA, and the other is located in subdomain IIIA [6]. This conformational change induced by the multiple double bonds in AA or DHA is consistent with the relative movement of Lys-205 located at the beginning of IIA closer to Lys-466 in IIIA suggested by our data.
The observation of the significant enhancement of cross-linking between Lys-402 (IIIA) and Lys-541 (IIIB) upon HSA binding to all three fatty acids suggested that a local conformational change occurred in domain III. It is likely that the side chains of the lysine residues rotated upon accommodating incoming fatty acids, resulting in a conformation in which the side chains of the lysines pointed in the same direction, allowing the cross-linking to occur (Figure 6).
In conclusion, we have demonstrated in the present study that chemical cross-linking with mass spectrometric detection can be used to monitor the conformational changes of HSA associated with fatty acid binding in a solution state. Changes in conformation upon binding to various fatty acids were deduced from the changes in cross-linked peptides characterized by MS/MS. A local conformational change in domain III involving the movement of the side chains at Lys-402 and Lys-541 was detected upon interaction of HSA with all three unsaturated fatty acids (OA, AA and DHA). Our results also appear to be in agreement with the data obtained by X-ray crystallography, in that fatty acids did not introduce major distortions into each of three individual domains. In particular, the conformational change involving the movement of the side chains of Lys-205 from domain IIA and Lys-466 from domain IIIA towards each other occurred upon binding of AA or DHA, but not OA, suggesting a distinctively different conformation of HSA bound to polyunsaturated AA or DHA in comparison with the monounsaturated OA-bound counterpart. Given enough cross-linking data using more cross-linkers with various spacer arm lengths and chemistry, detailed structural information concerning conformational changes of proteins in solution after protein–ligand or protein–protein interactions could be achieved. While X-ray crystallography provides detailed 3-D structural information, the present method should be a useful complement for probing the conformation of proteins in biological media.
References
- 1.Carter D. C., He X., Munson S. H., Twigg P. D., Gernert K. M., Broom M. B., Miller T. Y. Three-dimensional structure of human serum albumin. Science. 1989;244:1195–1198. doi: 10.1126/science.2727704. [DOI] [PubMed] [Google Scholar]
- 2.Carter D. C., Ho J. X. Structure of serum albumin. Adv. Protein Chem. 1994;45:153–203. doi: 10.1016/s0065-3233(08)60640-3. [DOI] [PubMed] [Google Scholar]
- 3.Spector A. Fatty acid binding to plasma albumin. J. Lipid Res. 1975;16:165–179. [PubMed] [Google Scholar]
- 4.He X. M., Carter D. C. Atomic structure and chemistry of human serum albumin. Nature (London) 1992;358:209–215. doi: 10.1038/358209a0. [DOI] [PubMed] [Google Scholar]
- 5.Sugio S., Kashima A., Mochizuki S., Noda M., Kobayashi K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 1999;12:439–446. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 6.Curry S., Mandelkow H., Brick P., Franks N. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat. Struct. Biol. 1998;5:827–835. doi: 10.1038/1869. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharya A. A., Grune T., Curry S. Crystallographic analysis reveals modes of binding of medium and long-chain fatty acids to human serum albumin. J. Mol. Biol. 2000;303:721–732. doi: 10.1006/jmbi.2000.4158. [DOI] [PubMed] [Google Scholar]
- 8.Peptipsa I., Grune T., Bhattacharya A. A., Curry S. Crystal structure of human serum albumin complexed with monounsaturated and polyunsaturated fatty acids. J. Mol. Biol. 2001;314:955–960. doi: 10.1006/jmbi.2000.5208. [DOI] [PubMed] [Google Scholar]
- 9.Zunszain P., Ghuman J., Komatsu T., Tsuchida E., Curry S. Crystal structure analysis of human serum albumin complexed with hemin and fatty acid. BMC Struct. Biol. 2003;3:1–9. doi: 10.1186/1472-6807-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cistola D. P., Small D. M., Hamilton J. A. Carbon 13 NMR studies of saturated fatty acids bound to bovine serum albumin, I. The filling of individual fatty acid binding sites. J. Biol. Chem. 1987;262:10971–10979. [PubMed] [Google Scholar]
- 11.Cistola D. P., Small D. M., Hamilton J. A. Carbon-13 NMR studies of saturated fatty acids bound to bovine serum albumin, II. Electrostatic interactions in individual fatty acid binding sites. J. Biol. Chem. 1987;262:10980–10985. [PubMed] [Google Scholar]
- 12.Choi J. K., Curry S., Qin D., Bittman R., Hamilton J. A. Interactions of very long-chain saturated fatty acids with serum albumin. J. Lipid Res. 2002;43:1000–1010. doi: 10.1194/jlr.m200041-jlr200. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton J. A., Era S., Bhamidipati S. P., Reed R. G. Location of the three primary binding sites for long-chain fatty acids on bovine serum albumin. Proc. Natl. Acad. Sci. U.S.A. 1991;88:2051–2054. doi: 10.1073/pnas.88.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curry S., Brick P., Franks N. P. Fatty acid binding to human serum albumin: New insights from crystallographic studies. Biochim. Biophys. Acta. 1999;1441:131–140. doi: 10.1016/s1388-1981(99)00148-1. [DOI] [PubMed] [Google Scholar]
- 15.Young M. M., Tang N., Hempel J. C., Oshiro C. M., Taylor E. W., Kuntz I. D., Gilson B. W., Dollinger G. High throughout protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5802–5806. doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rappsiber J., Siniossoglou S., Hurt E. C., Mann M. A generic strategy to analyze the spatial organization of multi-protein complexes by cross-linking and mass spectrometry. Anal. Chem. 2000;72:267–275. doi: 10.1021/ac991081o. [DOI] [PubMed] [Google Scholar]
- 17.Muller D. R., Schindler P., Towbin H., Wirth U., Voshol H., Hoving S., Steinmetz M. O. Isotope-tagged cross-linking reagents. A new tool in mass spectrometric protein interaction analysis. Anal. Chem. 2001;73:1927–1934. doi: 10.1021/ac001379a. [DOI] [PubMed] [Google Scholar]
- 18.Bennett K. L., Kussmann M., Bjork P., Godzwon M., Mikkelsen M., Sorensen P., Roepstorff P. Chemical cross-linking with thiol-cleavable reagents combined with differential mass spectrometric peptide mapping – A novel approach to assess intermolecular protein contacts. Protein Sci. 2000;9:1503–1518. doi: 10.1110/ps.9.8.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taverner T., Hall N. E., Ohair R. A. J., Simpson R. J. Characterization of an antagonist interleukin-6 dimer by stable isotope labeling, cross-linking and mass spectrometry. J. Biol. Chem. 2002;277:46487–46492. doi: 10.1074/jbc.M207370200. [DOI] [PubMed] [Google Scholar]
- 20.Pearson K. M., Pannell L. K., Fales H. M. Intramolecular cross-linking experiments on cytochrome c and ribonuclease A using an isotope multiplet method. Rapid Commun. Mass Spectrom. 2002;16:149–159. doi: 10.1002/rcm.554. [DOI] [PubMed] [Google Scholar]
- 21.Back J. W., Notenboom V., Koning L. J., Muijsers A. O., Sixma T. K., Koster C. G., Jong L. Identification of cross-linked peptides for protein interaction studies using mass spectrometry and 18O labeling. Anal. Chem. 2002;74:4417–4422. doi: 10.1021/ac0257492. [DOI] [PubMed] [Google Scholar]
- 22.Huang B. X., Dass C., Kim H.-Y. Probing three-dimensional structure of bovine serum albumin by chemical cross-linking and mass spectrometry. J. Am. Soc. Mass Spectrom. 2004;15:1237–1247. doi: 10.1016/j.jasms.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Sinz A. Chemical cross-linking and mass spectrometry for mapping three-dimensional structures of proteins and protein complexes. J. Mass Spectrom. 2003;38:1225–1237. doi: 10.1002/jms.559. [DOI] [PubMed] [Google Scholar]
- 24.Back J. W., de Jong L., Muijsers A. O., de Koster C. G. Chemical cross-linking and mass spectrometry for protein structural modeling. J. Mol. Biol. 2003;331:303–313. doi: 10.1016/s0022-2836(03)00721-6. [DOI] [PubMed] [Google Scholar]
- 25.Wong S. S. Boca Raton, FL: CRC Press; 1991. Chemistry of Protein Conjugation and Cross-linking. [Google Scholar]
- 26.Saifer A., Goldman L. The free fatty acids bound to human serum albumin. J. Lipid Res. 1961;2:268–270. [Google Scholar]
- 27.Salem N., Jr, Kim H. Y., Yergey J. A. Docosahexaenoic acid: membrane function and metabolism. In: Simopoulos A. P., Kifer R. R., editors. Health Effects of Polyunsaturated Fatty Acids in Seafoods. New York: Academic Press; 1986. pp. 263–317. [Google Scholar]
- 28.Kim H. Y., Akbar M., Lau A., Edsall L. Inhibition of neuronal apoptosis by docosahexaenoic acid (22:6n-3) J. Biol. Chem. 2000;275:35215–35223. doi: 10.1074/jbc.M004446200. [DOI] [PubMed] [Google Scholar]
- 29.Haniu M., Narhi L. O., Arakawa T., Elliott S., Rohde M. F. Recombinant human erythropoietin (rHuEPO): Cross-linking with disuccinimidyl esters and identification of the interfacing domains in EPO. Protein Sci. 1993;2:1441–1451. doi: 10.1002/pro.5560020908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schilling B., Row R. H., Gibson B. W., Guo X., Young M. M. MS2assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 2003;14:834–850. doi: 10.1016/S1044-0305(03)00327-1. [DOI] [PubMed] [Google Scholar]
- 31.Green N. S., Reisler E., Houk K. N. Quantitative evaluation of the lengths of homobifunctional protein cross-linking reagents used as molecular rulers. Protein Sci. 2001;10:1293–1304. doi: 10.1110/ps.51201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richieri G. V., Anel A., Kleinfeld A. M. Interactions of long-chain fatty acids and albumin: determination of the free fatty acid levels using the fluorescent probe ADIFAB. Biochemistry. 1993;32:7574–7580. doi: 10.1021/bi00080a032. [DOI] [PubMed] [Google Scholar]