Abstract
Globally, the prevalence of breast cancer (BC) is increasing at an alarming level, despite early detection and technological improvements. Alkaloids are diverse chemical groups, and many within this class have been reported as potential anticancer compounds. Chabamide F (F) and chabamide G (G) are two dimeric amide alkaloids found in a traditional medicinal plant, Piper chaba, and possess significant cytotoxic effects. However, their scientific rationalization in BC remains unknown. Here, we aimed to investigate their potential and molecular mechanisms for BC through in silico approaches. From network pharmacology, we identified 64 BC-related genes as targets. GO and KEGG studies showed that they were involved in various biological processes and mostly expressed in BC-related pathways such as RAS, PI3K-AKT, estrogen, MAPK, and FoxO pathways. However, PPI analysis revealed SRC and AKT1 as hub genes, which play key roles in BC tumorigenesis and metastasis. Molecular docking revealed the strong binding affinity of F (− 10.7 kcal/mol) and G (− 9.4 and − 11.7 kcal/mol) for SRC and AKT1, respectively, as well as the acquisition of vital residues to inhibit them. Their long-term stability was evaluated using 200 ns molecular dynamics simulation. The RMSD, RMSF, Rg, and SASA analyses showed that the G-SRC and G-AKT1 complexes were excellently stable compared to the control, dasatinib, and capivasertib, respectively. Additionally, the PCA and DCCM analyses revealed a significant reduction in the residual correlation and motions. By contrast, the stability of the F-SRC complex was greater than that of the control, whereas it was moderately stable in complex with AKT1. The MMPBSA analysis demonstrated higher binding energies for both compounds than the controls. In particular, the binding energy of G for SRC and AKT1 was − 120.671 ± 16.997 and − 130.437 ± 19.111 kJ/mol, respectively, which was approximately twice as high as the control molecules. Van der Waal and polar solvation energies significantly contributed to this energy. Furthermore, both of them exhibited significant interactions with the binding site residues of both proteins. In summary, this study indicates that these two molecules could be a potential ATP-competitive inhibitor of SRC and an allosteric inhibitor of AKT1.
Keywords: Chabamide F, Chabamide G, Breast cancer, SRC, AKT1, Molecular dynamics simulation
Subject terms: Oncogene proteins, Breast cancer, Drug discovery, Molecular modelling
Introduction
Cancer is a group of diseases characterized by uncontrolled cell division, which is the consequence of various factors including mutation, viral infection, lifestyle, and so on1,2. According to GLOBOCAN estimation (2022), approximately 36 types of cancer are diagnosed in 185 countries, resulting in nearly 9.7 million deaths and 20 million new cases, which is anticipated to rise by 77% within 2050 to 35.30 million cases3. However, breast cancer (BC) represents the most commonly diagnosed malignancy in women, contributing 2.3 million cases worldwide in 2022. BC was responsible for 666,000 deaths globally, placing it as the fourth leading cause of cancer mortality3.
BC is a heterogeneous and complex disease in which each patient processes unique morphological and molecular features4. It is broadly classified into four subtypes, including estrogen receptor positive (ER +), progesterone receptor positive (PR +), human epidermal growth factor receptor positive (HER2 +), and triple-negative (TNBC)5. Primary BC can develop in any region of the breast, including the ducts, lobules, and surrounding tissues6. Although early detection improves the rate of survival, BC is frequently diagnosed after metastasis, which is a more aggressive stage7. During metastasis, tumor microenvironments, such as stromal influences or macrophages, are affected by carcinogens, resulting in the initiation and progression of angiogenesis in BC cells8. In addition, migration to lymph nodes and distant organs such as the lung, liver, and bone is a hallmark of mortality, drug resistance, and the failure of current therapies7,9. Although radiation, surgery, and immunotherapy are the most common treatments for cancer, they have many side effects, leading researchers to discover alternatives with fewer side effects10.
Alkaloids are an extensively investigated group of compounds because of their broad spectrum of biological activities, including anti-inflammatory, anti-cancer, anti-microbial, and so on11. Various alkaloids, like vinca alkaloids, taxanes (paclitaxel and docetaxel), and quinolines (camptothecin and its derivatives), are well-known cancer drugs, and a large number of plant-derived alkaloids are currently being studied for cancer therapy11. This study investigated the effects of two novel dimeric amides alkaloids containing pyrrole rings, chabamide F (F) and chabamide G (G), on BC, which have been reported to be present in the roots of Piper chaba12. Piper chaba, a member of the Piperaceae family, is a widely grown spice in the southern part of Bangladesh and is commonly named Chui Jhal13. A previous study reported that F and G induced cytotoxicity in the BC cell lines14. In addition, several dimeric alkaloids similar to F and G are also potent cytotoxic compounds15–17. However, the effect of F and G on BC, as well as their molecular mechanisms, is not yet well understood. In this study, we used an integrated in-silico approach combining network pharmacology, molecular dynamics (MD) simulations, and Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) analysis to find the effect and underlying molecular mechanisms of these two compounds against BC.
In recent years, computational algorithms have emerged as a powerful method for screening potential new lead drug candidates 18,19. It not only lowers the cost of redundant preclinical trials but also shortens the time and resources required for new drug discovery. In particular, MD simulations and MMPBSA approaches have become reliable methods for studying protein dynamics, conformational patterns, ligand binding and unbinding processes, and the free energy of macromolecular systems19–21. Network pharmacology has added a new dimension to this field and opened up new avenues to explain intricate relationships among diseases, drugs, and targets22. Utilizing these approaches, we demonstrated that F and G targeted multiple vital cancer cell signaling pathways and strongly interacted with SRC and AKT1. Proto-oncogene SRC is overexpressed in the BC and regulates tumor initiation, proliferation, and drug resistance23. AKT1, also known as protein kinase B, is a key player in the PI3K/Akt signaling pathway, which increases BC cell proliferation and reduces apoptosis24. This is the first study to comprehensively describe the molecular targets of F and G, providing new insights about their inhibitory potential as a new promising drug candidate for BC.
Methods
Determination of candidate targets
The 3D structures of F (PubChem ID: 102,480,093) and G (PubChem ID: 102,480,094) were retrieved from PubChem, and their energy was minimized by Avogadro software under the MMFF94 force field25–27.The mol2 files of each compound were uploaded to the PharmMapper (http://www.lilab-ecust.cn/pharmmapper) server to find out the predictive targets of F and G28. The parameters used were as follows: Generate Conformers: yes; Maximum Generated Conformations: 300; Target Set: Human Protein Targets Only; and Number of Reserved Matched Targets: 300. We filtered the targets based on z scores > 0, and the resulting targets were used for further analysis. The predictive targets of both compounds were submitted to the UniProtKB database (https://www.uniprot.org/) to collect the official gene symbols. The final dataset was prepared by combining the gene symbols of two datasets after removing duplicate genes.
The disease-related genes were collected from the GeneCards Database (https://www.genecards.org/) using the keyword “breast cancer,” and the top 500 genes were selected based on the relevance score29. However, targets of F and G in BC, or candidate targets, were obtained after intersecting the final dataset of both compounds and the disease related targets30.
Protein–protein interaction of candidate targets
To analyze protein–protein interaction (PPI), the candidate targets were submitted to the STRING database 11.5 (https://string-db.org/). The string database contains curated, up-to-date information on physical interactions and functional associations of protein–protein interactions31. We selected ‘Homo sapiens’ as organisms and advanced settings were set: network type: full STRING network; required score: medium confidence (0.7); FDR stringency: medium (5 percent). The network was imported into Cytoscape 3.9.1, and the CytoNCA plugin was utilized to analyze the PPI network based on degree, betweenness, closeness, and eigenvector32,33.
Gene ontology and KEGG pathway analysis
Gene ontology (GO) analysis and KEGG pathway analysis of the candidate targets were performed in DAVID (https://david.ncifcrf.gov/)34. The biological process, cellular component, molecular function, and KEGG pathway were selected, and Bonferroni correction was used to filter for P-values ≤ 0.05.
Network construction
Network analysis and network visualization were accomplished by the Cytoscape 3.9.1 (https://cytoscape.org/) software, and PPI and target-pathway networks were constructed in this study33.
Drug likeness and ADME
Drug likeness and ADME analysis were investigated in the SwissADME (www.swissadme.ch)35. According to the Lipinski rule, an oral bioavailable drug should have the following characteristics: molecular mass not more than 500 Da, hydrogen bond donors not more than 5, hydrogen bond acceptors not more than 10, and partition coefficient log P not greater than 5.
Molecular docking
The crystal structures of AKT1 (PDB: 6S9W) and SRC (PDB: 7NG7) were retrieved from the RCSB PDB database (https://www.rcsb.org/). Discovery Studio (Accelrys Software Inc., San Diego, CA) was used for removing water molecules and heteroatoms from the structures, and missing atoms were fixed by Modeller 10.436. For AKT1, capivasertib was utilized as a control, whereas dasatinib was utilized for SRC. Dasatinib is a tyrosine kinase inhibitor that inhibits SRC, and capivasertib is a potent pan-AKT inhibitor drug37,38. Both compounds were obtained from the PubChem database, followed by energy minimization, similar to the F and G preparations. To prepare the protein for docking, polar hydrogens were added and saved in pdbqt format by using AutoDock Tools 1.5.7. The ligands were also prepared by the same tool, and the final molecular docking was carried out in AutoDock Vina 1.1.2, where the exhaustiveness was set to 3239. In this study, we targeted the allosteric binding site of AKT1 and the ATP binding site of SRC. The active site of SRC and the allosteric site of AKT1 were identified from the previous studies40–44. The docking grid box (25 × 25 × 25; 1 Å) was prepared, and the grid center was assigned at X = − 11.688, Y = − 14.813, Z = 13.487 for AKT1, and X = − 16.286, Y = − 6.16, Z = − 8.498 for SRC, according to the co-crystalized ligands. Furthermore, Ligplot and Pymol were used to visualize the protein–ligand interactions45.
Molecular dynamics simulation
The MD simulation of our protein–ligand complexes was carried out in GROMACS 2022.546. The AMBER99SB-ILDN force field was utilized for generating protein topology, and ligand parameters were generated under the General Amber Force Field (GAFF) by using ACPYPE server47–49. The AMBER99SB-ILDN is an improvised and accurate force field for protein parameters with improvised side-chain torsion potentials47. GAFF contains parameters for organic molecules and pharmaceutical molecules, and it is widely used to generate ligand parameters for AMBER force fields48. A three-point (TIP3P) water model was used for the solvation of protein–ligand complexes within a cubic box at 10 Å distance from each side of the box to the protein. TIP3P is a highly efficient and frequently used computational water model in MD simulation50. However, after solvation, the system was neutralized by adding Na + and Cl − ions, followed by energy minimization by the steepest descent integrator. The complex was then equilibrated under the NVT and NPT ensembles for 1 ns, respectively, by restraining both protein and ligand positions. The system was first stabilized at the target temperature of 300 K under the NVT ensemble, and then the pressure (1 bar) was stabilized under the NPT ensemble. In these steps, we used the V-rescale thermostat and the C-rescale barostat for temperature and pressure coupling. Finally, after equilibration, the system was run for 200 ns to collect MD data, releasing restraints with 2 fs time-based integrators. During the production period, we used the V-rescale thermostat (300 K) and Parrinello-Rahman barostat (1 bar).
Trajectory and molecular mechanics poisson-boltzmann surface area binding free energy analysis
The GROMACS tools were used to calculate root mean square deviation (RMSD), root mean square fluctuations (RMSF), radius of gyration (Rg), and solvent accessibility surface area (SASA). All the average values were calculated as average ± standard deviation (SD).
The Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) binding free energy analysis of each protein–ligand complex over the 100 ns to 200 ns of the simulations were calculated by using g_mmpbsa tool, where snapshots were taken at 100 ps interval51.
Dynamic cross correlation map analysis
The Cα atoms of each ligand bound protein were used to construct the dynamic cross-correlation maps (DCCM) between two residues. The R-based Bio3D package52 was used to calculate DCCM from 100 to 200 ns and the correlation between two Cα atoms, i and j, can be defined as ,
where Δri is the displacement from i, Δrj from j, and < > denotes the average time53. A positive value indicates that the atomic movements are in the same direction, while opposite directions represent a negative value.
Principal component analysis
The atomic coordinates of Cα atoms in the last 100 ns of the simulation were used for the principal component analysis (PCA). The covariance matrix was constructed by the ‘gmx covar’ utility of GROMACS, followed by diagonalization to obtain eigenvalues and eigenvectors. Principal components were determined by projecting eigenvalues along eigenvectors using the 'gmx anaeig' tool. We also performed K-means clustering to analyze the PCA data based on the average silhouette approach in R.
Gibbs free energy landscape
Gibbs free energy landscape (FEL) profile was constructed from the RMSD and Rg data of each protein–ligand complex by the use of the ‘gmx sham’ tool of GROMACS. FEL was determined as,
where kB denotes Boltzmann's constant, Gi indicates Gibbs free energy, T is the temperature, Ni represents the population in a bin, and Nmax is the population of the densely populated bin. Here, we used a temperature of 300 K.
Result
Identification of candidate target genes
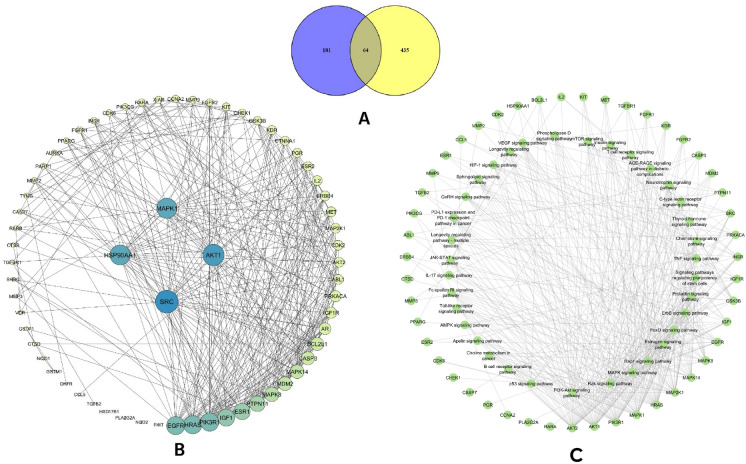
A total of 300 predictive targets were found for each compound in the PharmMapper database. However, 221 genes in F and 215 genes in G demonstrated z-scores > 0, which were further submitted to the UniProtKB database. A total of 215 official gene symbols for F and 209 for G were obtained from the UniProtKB database by considering the organism as “Homo sapiens.” The two datasets were merged, eliminating any duplicate entries, resulting in a total of 245 target genes for both compounds. Finally, the targets of both compounds were compared to the 500 BC- associated genes, and 64 genes were selected as candidate targets (Fig. 1A). The known roles of these 64 target genes are given in the Table S2.
Figure 1.
(A) The Venn diagram of the 64 candidate targets, (B) PPI interaction analysis, sorted according to the degree, and (C) Target-pathway network of the investigated candidate genes.
Analysis of PPI network
PPI analysis is a vital tool for understanding interactions among candidate targets and their functions. For this analysis, the gene symbols were presented as nodes and their interactions as edges. Our PPI network consisted of 64 nodes and 342 edges, where the mean degree was 10. The degree indicates the number of connections, and nodes with high degrees represent the hub of a network. From our analysis of degree, betweenness, closeness, and eigenvectors, we identified SRC as the most significant target gene, followed by AKT1, MAPK1, and HSP90AA1. Figure 1B depicts the hub gene network in decreasing order of nodes, and Table 1 shows the details of the top four nodes.
Table 1.
The PPI network analysis value of top four targets.
| Gene name | Degree | Eigenvector | Betweenness | Closeness |
|---|---|---|---|---|
| SRC | 32 | 0.279148 | 286.2739 | 0.180516 |
| AKT1 | 30 | 0.241861 | 389.9254 | 0.179487 |
| MAPK1 | 28 | 0.248501 | 209.7188 | 0.18 |
| HSP90AA1 | 28 | 0.228574 | 295.5486 | 0.17847 |
GO analysis of candidate genes
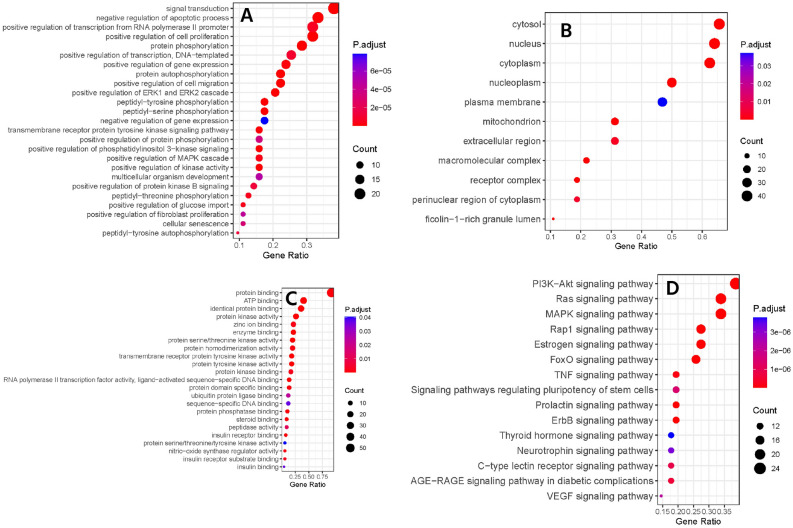
In the GO analysis, we obtained the biological processes, molecular function, and cellular components of our candidate targets. Figure 2A,B,C shows the top GO terms having P-values ≤ 0.05. The top ten biological processes regulated by the candidate targets include signal transduction, negative regulation of apoptotic process, positive regulation of cell proliferation, positive regulation of transcription from RNA polymerase II promoter, protein phosphorylation, positive regulation of DNA-templated transcription, positive regulation of gene expression, protein autophosphorylation, positive regulation of cell migration, and positive regulation of ERK1 and ERK2 cascade. Figure 2A shows the top 25 targeted biological processes. The cellular components of the candidates were the cytosol, nucleus, cytoplasm, nucleoplasm, plasma membrane, mitochondrion, extracellular region, macromolecular complex, receptor complex, perinuclear region of cytoplasm, and ficolin-1-rich granule lumen (Fig. 2B). Furthermore, the top ten molecular functions that we found were protein binding, ATP binding, identical protein binding, protein kinase activity, enzyme binding, zinc ion binding, protein serine/threonine kinase activity, protein homodimerization activity, protein tyrosine kinase activity, and transmembrane receptor protein tyrosine kinase activity (Fig. 2C).
Figure 2.
The biological process (A), cellular components (B), molecular functions (C), and KEGG pathway (D) of candidate targets.
Target pathway analysis
The KEGG pathway analysis further provides more insights about the probable molecular mechanisms of F and G. A total of 145 pathways were found, and 98 of them had P-values ≤ 0.05. However, after removing generalized pathways that focused on mostly disease terms, 53 signaling pathways were considered to be regulated by our candidate genes. Figure 2D represents the top 10 pathways, ranked in decreasing order, and Fig. 1C shows the target pathway-gene network. The most significant pathways were the Ras signaling pathway, the phosphatidylinositol 3-kinase (PI3K)-AKT signaling pathway, the estrogen signaling pathway, the mitogen-activated protein kinases (MAPK) signaling pathway, and the forkhead box O proteins (FOXO) signaling pathway. Ras is a highly mutated oncogene, and its signaling is essential for normal cell growth, proliferation, and differentiation54. Ras-dependent PI3K-AKT and MAPK pathways are upregulated in cancer stem cells, and in BC, Ras acts as a supporting actor54,55. The PI3K/AKT pathway is considered as a master regulator of cancer56. Generally, it plays a vital role in cell proliferation, survival, metabolism, and apoptosis; dysregulation of this pathway is linked to cancer progression and drug resistance in BC57. Furthermore, estrogen, MAPK, and FOXO signaling pathways are also dysregulated in BC patients58–60. Abnormal estrogen receptor signaling is a major cause of BC, where normal estrogen signaling is changed to tumor-specific signaling58. MAPK signaling pathway is involved in the development of TNBC59. FOXOs are tumor suppressor transcription factors that have a positive role in cell cycle arrest, apoptosis, and so on61. Overall, the KEGG analysis suggested that both F and G have a higher possibility of working on multiple vital cancer signaling pathways.
Drug likeness, and ADME analysis
The Lipinski rule was used to evaluate the drug likeness based on specific parameters to understand whether our compounds are good oral candidates or not. Except for the molecular weight violation with a value of 542.62, our candidate dimers, F and G, fulfilled the other three requirements, including hydrogen bond acceptors and donors, and calculated LogP. Tables 2, 3 show the details of each term of drug likeness and ADME. LogP is a gold standard for lipophilicity, and in conjugation with solubility, it explains the absorption of a drug62. Our studied compounds showed moderate solubility, calculated by using the ESOL Log S, Ali Log S, and Silicos-IT LogSw methods. Moreover, the gastrointestinal absorption was high and both were predicted to be blood–brain barrier permeable according to the boiled egg method. Furthermore, biotransformation and P-glycoprotein (P-gp) generally induce drug elimination62; both F and G were predicted as inhibitors P-gp and vital metabolizing enzymes including CYP2C19, CYP2C9, and CYP3A4.
Table 2.
Result of drug likeness and solubility of investigated ligands.
| Ligand | Molecular weight | H–bond acceptors | H–bond donors | Consensus log P | ESOL log S | ESOL class | Ali log S | Ali class | Silicos-IT LogSw | Silicos-IT class |
|---|---|---|---|---|---|---|---|---|---|---|
| F | 542.62 | 6 | 0 | 4.02 | − 5.77 | Moderately soluble | − 5.81 | Moderately soluble | − 5.35 | Moderately soluble |
| G | 542.62 | 6 | 0 | 4.08 | − 5.77 | Moderately soluble | − 5.81 | Moderately soluble | − 5.35 | Moderately soluble |
Table 3.
The ADME analysis of investigated ligands.
| Ligand | GI absorption | BBB permeant | Pgp substrate | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor |
|---|---|---|---|---|---|---|---|---|
| F | High | Yes | Yes | No | Yes | Yes | No | Yes |
| G | High | Yes | Yes | No | Yes | Yes | No | Yes |
Molecular docking analysis
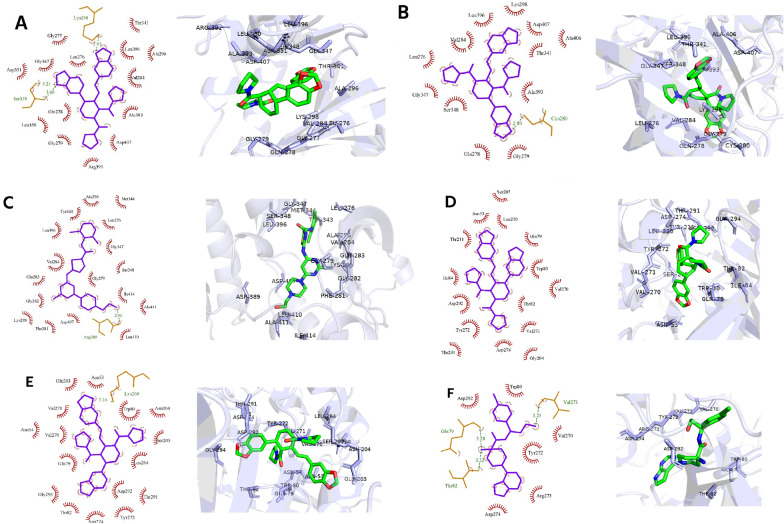
Molecular docking and further MD simulations were investigated on the top two significant proteins, SRC and AKT1. Table 4 indicates the binding energy and interacting residues of protein–ligand complexes. The kinase domain of SRC was selected, and the ATP–binding cleft formed between the small and large lobes, was targeted. Vital residues within this pocket were conserved glycine rich loop residues (Gly277—Gly282), β3-strand (Lys298), hinge residues (Glu342—Gly347), catalytic loop (His387—Asn394), and activation segment (Asp407—Glu435)40. Moreover, within the activation segment, DFGLAR residues (Asp407 – Arg412) are considered to act as a Mg2+ positioning loop. In addition, ATP-competitive inhibitors also bound with the multiple catalytic spine residues, mostly Val284, Ala296, and Leu39640,41. Here, we found the highest binding affinity against SRC for F (− 10.7 kcal/mol) followed by control, and G, with a value of − 9.4, and − 9.2 kcal/mol, respectively. Figure 3A-C denotes the molecular docking results of SRC. The F formed two hydrogen bonds with Lys298 and Ser348, and interacted hydrophobically with Leu276, Gly277, Gly347, Leu350, Asp351, Gln278, Gly279, Val284, Thr341, Ala296, Arg391, Ala393, Leu396, and Asp407 residues (Fig. 3A). For G, a hydrogen bond was found with Cys280, and hydrophobic bonds with Gln278, Leu276, Gly279, Val284, Lys298, Thr341, Gly347, Ser348, Ala393, Leu396, Ala406, and Asp407 (Fig. 3B). On the other hand, control bound with Asp389 by forming hydrogen bonds, and Leu276, Gly279, Phe281, Gly282, Glu283, Val284, Ala296, Lys298, Tyr343, Met344, Ser348, Gly347, Leu396, Asp407, Ala411, Leu410, and Ile414 residues showed hydrophobic interactions (Fig. 3C), which was almost similar to the previous studies40,63–65.
Table 4.
Molecular docking results of all ligands in complex with SRC, and AKT1.
| Protein | Ligand | Binding energy (kcal/mol) | Hydrogen bonds | Hydrophobic interactions |
|---|---|---|---|---|
| SRC | F | − 10.7 | Lys298, Ser348 | Leu276, Gly277, Gly347, Leu350, Asp351, Gln278, Gly279, Val284, Thr341, Ala296, Arg391, Ala393, Leu396, Asp407 |
| G | − 9.2 | Cys280 | Gln278, Leu276, Gly279, Val284, Lys298, Thr341, Gly347, Ser348, Ala393, Leu396, Ala406, Asp407 | |
| Control | − 9.4 | Asp389 | Leu276, Gly279, Phe281, Gly282, Glu283, Val284, Ala296, Lys298, Tyr343, Met344, Ser348, Gly347, Leu396, Asp407, Ala411, Leu410, Ile414 | |
| AKT1 | F | − 10.7 | N/A | Asn53, Gln79, Trp80, Thr82, Ile84, Ser205, Leu210, Thr211, Val270, Val271, Tyr272, Asp274, Thr291, Asp292, Gly294 |
| G | − 11.7 | Lys268 | Asn53, Asn54, Gln79, Trp80, Thr82, Gln203, Asn204, Ser205, Leu264, Val270, Val271, Tyr272, Asp274, Thr291, Asp292, Gly294 | |
| Control | − 9.7 | Gln79, Thr82, Val271 | Trp80, Val270, Tyr272, Arg273, Asp274, Asp292 |
Figure 3.
The 2D and 3D interactions of molecular docking of (A) F, (B) G, and (C) control with SRC protein and F (D), G (E), and control (F) with AKT1 protein.
Regarding the allosteric binding pocket of AKT1, previous studies reported that the inhibitory effect of allosteric inhibitors is critically dependent on the hydrophobic interaction with the pleckstrin-homology (PH) domain residues, Trp80 and Gln7942,43. Other reported interacting residues are Thr82, Ile84, Glu85, Ser205, Leu210, Leu264, Lys268, Val270, Tyr272, Arg273, Ile290, and Cys31043,44. Here, high negative binding energy was seen for G, followed by F, and control, with values of − 11.7, − 10.7, and − 9.7 kcal/mol. Figure 3D,E, and F indicates the two-dimensional interacting pattern of ligands with AKT1. The F showed only hydrophobic interactions and bound with Asn53, Gln79, Trp80, Thr82, Ile84, Ser205, Leu210, Thr211, Val270, Val271, Tyr272, Asp274, Thr291, Asp292, and Gly294 (Fig. 3D). The G formed one hydrogen bond with Lys268, and demonstrated hydrophobic interactions with Asn53, Asn54, Gln79, Trp80, Thr82, Gln203, Asn204, Ser205, Leu264, Val270, Val271, Tyr272, Asp274, Thr291, Asp292, and Gly294 (Fig. 3E). On the contrary, three hydrogen bonds with Gln79, Thr82, and Val271, and hydrophobic interactions with Trp80, Val270, Tyr272, Arg273, Asp274, and Asp292 were noticed for the control-AKT1 complex (Fig. 3F). The interactions of the control-AKT1 complex were similar to the previously reported AKT1 allosteric inhibitor VIII43.
MD simulations analysis of SRC-ligand complex
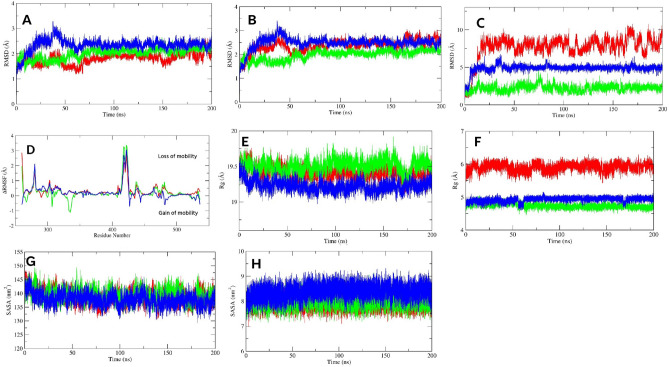
To understand the protein–ligand complex stability, first we analyzed the RMSD over 200 ns, which describes the average structural deviation between two structures. Here, we analyzed Cα RMSD of ligand bound proteins, and RMSD of backbone-ligand complexes (termed as protein–ligand complexes), and ligand RMSD over the simulation period. Figure 4A,B indicate the Cα-RMSD, and protein–ligand complex RMSD, respectively. Interestingly, we found lower structural deviations for F and G compared to the control. The Cα-RMSD of F bound protein was less stable from the starting to 25 ns and fluctuated within 1.2–2.9 Å. However, after 25 ns to 50 ns, the protein exhibited a stable behavior (mean, 1.7 ± 0.10 Å) that again stabilized after 66 ns to 150 ns (average, 2.1 ± 0.10 Å). The F bound protein moderately fluctuated from 150 to 180 ns, followed by a stable behavior until last. For the G-bound protein, the Cα-RMSD was gradually increased from the beginning, mean 1.9 Å ± 0.38 Å, to 22 ns, followed by a steady fall till 62 ns. However, after 70 ns, a complete convergence was observed until the end of the simulation study, where the mean was 2.4 ± 0.11 Å. In the protein–ligand complex state, both showed identical fluctuations to Cα-RMSD, indicating stability after F and G binding. On the other hand, the stability of the control bound protein was comparatively lower than our investigated compounds; the control bound protein demonstrated high fluctuations in multiple areas of the first 50 ns, including from the beginning to 14 ns, and 33 to 48 ns, with a mean of 1.7 ± 0.20, and 1.8 ± 0.17 Å, respectively. The Cα-RMSD gradually increased with lower fluctuations within 67 to 100 ns, followed by a convergence state until 147 ns (mean, 1.8 ± 0.08 Å). However, stability decreased after 148 ns, and unstable behavior was dominant till the end of the study (mean, 1.9 ± 0.20 Å). The control-protein complex sate showed more unstable behavior with no convergences and fluctuated with a mean of 2.5 ± 0.26 Å over the simulation.
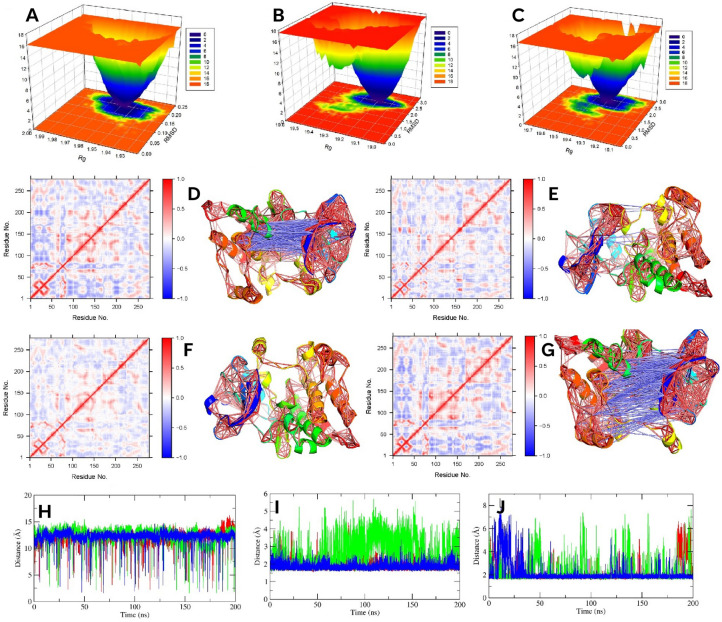
Figure 4.
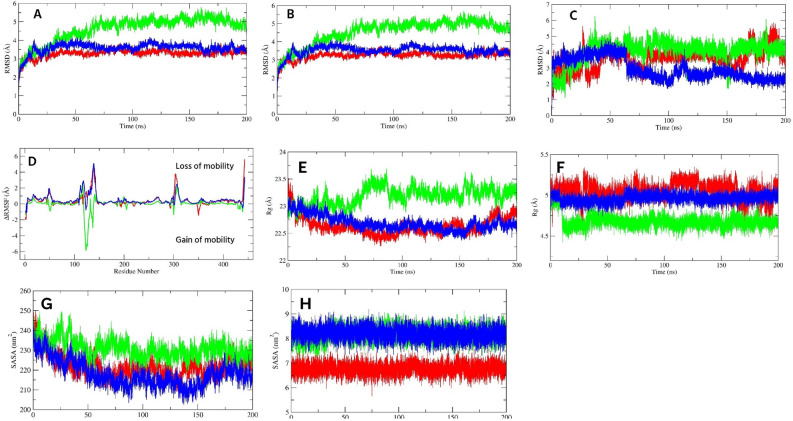
The Cα-RMSD (A), protein–ligand complex RMSD (B), ligand RMSD (C), and ligand bound ΔRMSF (D) of SRC protein. (E, F, G, and H) indicate the protein–ligand complex Rg (E), ligand Rg (F), protein–ligand complex SASA (G), and ligand SASA (H) of SRC protein. The green, blue, and red color represents F, G, and control, respectively.
The ligand RMSD further showed that the F and G were well-equipped within the binding pocket than the control (Fig. 4C). Over the simulation, their structural deviations were lower; the G showed more stability. The G demonstrated a very convergent state and lower fluctuations, particularly after 45 ns to the end of the study, near a mean of 4.9 ± 0.25 Å. However, it reached a stable state after 5 ns, and before 45 ns, this ligand demonstrated moderate RMSD values. Notably, from 30 to 37 ns, the RMSD was higher (mean, 5.4 ± 0.46 Å). The RMSD value of F was lower than the G (average, 2.0 ± 0.44 Å), and over the simulation, no abrupt high fluctuations were observed. However, the F was more stable after 78 ns to 200 ns (mean, 2.2 ± 0.38 Å) than the first 77 ns (mean, 2.2 ± 0.51 Å). By contrast, the control showed higher variability over the simulation, thereby lower stability. The RMSD rose gradually from the beginning that stabilized after 15 ns. However, after 15 ns to the end of the simulation, no convergence was noticed, and the mean fluctuations were 7.6 ± 1.15 Å, which was an indication of higher conformational changes.
To understand the residual flexibility during simulations, we calculated the ΔRMSF by subtracting the RMSF value of Cα atoms of the ligand bound protein from the Cα-RMSF of the apo protein (Fig. 4D). In the present study, a ΔRMSF > 0.3 Å indicated a significant decrease in mobility66. All investigated ligand bound proteins showed a higher loss of mobilities within 409–425 residues, which is a part of the activation segment that includes short α-helices (αAL1 and αAL2), important for autoinhibition40. Additional lower flexible regions were the glycine-rich loop and its nearby residues of the N-lobe (276–282). The other regions of the N-lobe also experienced lower movements. Among the investigated ligands, the G bound protein reduced significant flexibilities of Cα atoms. Residues 266–270, and 274–320 of the N-lobe demonstrated a ΔRMSF of 0.4 and 0.5 Å, respectively. Other lower movement regions were 437–441, and 464–465; both showed a mean value of 0.4, and 0.5 Å, respectively. By contrast, higher flexibility was distinguishable within 260–261, 427, 481–482, and 536 residues. For F bound protein, 260–261, 276–289, 294–296, 303–305, and 315–320 residues of the N-lobe demonstrated lower mobility with a value of 1.4, 0.6, 0.3, 0.6, and 0.3 Å, respectively. In other regions, 436–443 (0.6 Å), and 464–484 (0.4 Å) exhibited significantly lower mobilities, while 267, 307, and 332–337 (-0.8 Å) residues showed higher flexibilities. On the other hand, for control bound protein, no significant flexibilities were noticed, while a considerable decrease in mobility was observed within 260–262, 266–269, 276–286, and 300–320 of the N-lobe, where the mean was 1.6, 0.4, 0.6, and 0.5 Å, respectively. Other significant residues were 437–439, 464–470, 477–485, and 533–535, which demonstrated a mean of 0.6, 0.6, 0.5, and 0.4 Å, respectively.
The radius of gyration is an indication of protein compactness and folding rate67. The higher Rg values leads to higher flexibilities of the protein, indicating instability during ligand binding. The Rg of all protein–ligand complex are given in the Fig. 4E. In the Rg analysis of protein–ligand complexes, we found very stable trajectories for F and G-protein complexes. The average Rg value of the G-protein complex (19.22 ± 0.07 Å) was the lowest compared to F and control. The mean values for both F and control-protein complexes throughout the simulation were 19.50 ± 0.09 Å and 19.41 ± 0.08 Å, respectively. The G-protein complex showed no abrupt fluctuations, whereas the F-protein complex had a sharp fall from 160 to 180 ns. On the other hand, the control-protein complex demonstrated a convergence state from 65 to 165 ns and lower compactness from 65 to 160 ns. Overall, all three ligands increased protein compactness after binding.
Regarding ligand Rg, both F and G were highly compact and fluctuated near a mean of 4.74 ± 0.07 and 4.92 ± 0.07 Å, respectively (Fig. 4F). The Rg gradually decreased over time after 58 ns to the end of the study. On the other hand, control manifested a higher value of Rg than our investigated ligands and fluctuated near a mean of 5.91 ± 0.12 Å. In addition, it exhibited fluctuations over the whole study, indicating lesser compactness.
SASA is another important parameter to understand protein stability, as it is the measure of the surface area that is available for the solvent to interact with and often varies when the ligand binds. The higher the SASA value, the more solvent access to the protein and protein expansion, thereby lowering its stability. In this study, the G-protein complex showed the lowest average SASA value of 137.80 ± 2.32 nm2, followed by the control and F-protein complex with values of 138.84 ± 2.42 nm2 and 139.50 ± 2.32 nm2, respectively (Fig. 4G). This indicated that the binding of G decreased interaction with solvents, and reduced protein folding, thereby increasing protein stability compared to control. In addition, all of them showed a similar pattern of fluctuation, having almost the same SASA values, indicating identical protein expansion behavior.
In the case of ligand SASA, all investigated ligands showed no higher fluctuations and fluctuated near a mean of 7.92 ± 0.22, 8.03 ± 0.26, and 8.38 ± 0.28 nm2 for control, F, and G, respectively (Fig. 4H). This result indicated that all ligands were well occupied in the binding pocket and less interacted with solvents.
To get further insights about the global energy minima and thermodynamic stability, we analyzed the FEL from the RMSD and Rg values of protein–ligand complexes. Figure 5A,B,C denotes the FEL of each protein–ligand complex; a lower FEL value is indicated by more blue colors. Binding of F induced more thermodynamically stable sates than other protein–ligand complexes; formed a broad, deep, well-defined valley. It suggested that the interaction of F with SRC was more favorable in the simulation, which indicated higher stability. By contrast, for G and control-protein complexes, one less deep valley was also noticed, which was much deeper in the case of the control-protein complex, along with a large valley. It indicated the presence of partially folded states in addition to a higher percentage of native states. The partially folded states represent moderate FEL values, thereby indicating the moderate stability of multiple conformations of a protein–ligand complex. However, drug efficacy depends not only on the thermodynamics of a system but also on the binding kinetics between protein and ligand68. Overall, our investigated complexes decreased entropy and formed thermodynamically stable conformations.
Figure 5.
The FEL plot of F (A), G (B), and control (C) bound protein. (D, E, F and G) denote 2D DCCM plot and 3D highly correlated (≥ ± 0.4) residues of apo, F, G, and control bound protein, respectively. (H, I, and J) denotes distance between 298–313, 298–407, and 313–412 resides, respectively. Green, blue, and red color represents F and G, and control, respectively.
Further DCCM analysis was carried out to understand the correlated residues during the simulation. Figure 5D–G denote the 2D DCCM plot, where more blue indicates negative correlations and more red indicates positive correlations. In addition, the 3D plot of highly correlated residues (≥ ± 0.4) is also added to Fig. 5. It was noted that both F and G bound protein decreased the overall positive and negative correlation compared to the apo and control bound protein. Notably, the G decreased the correlated motions more than the F; showed no higher anticorrelation. Furthermore, both also demonstrated lower positive and no negative correlations near the ligand binding pocket, indicating both stabilized the protein significantly. On the other hand, the control reduced correlated motions, although not as much as F and G bound proteins. Additionally, near the binding pocket, it showed a higher anti-correlation.
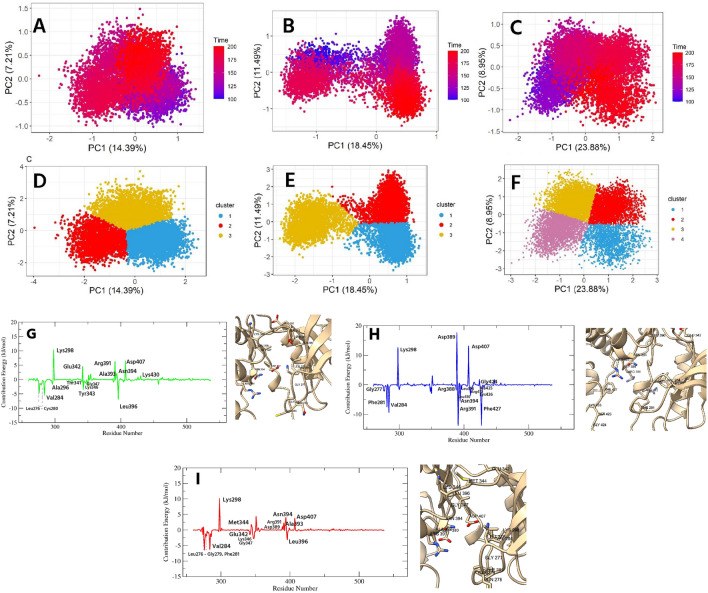
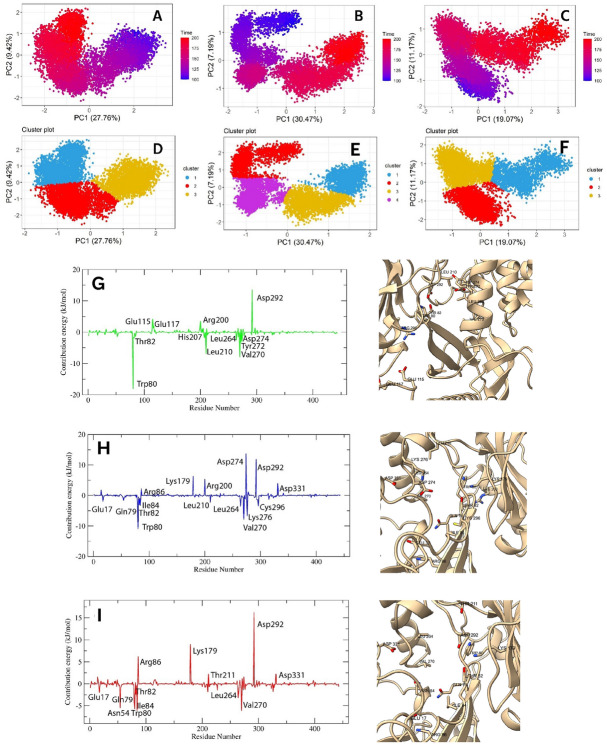
To understand the protein motion after ligand binding, we analyzed the PCA of Cα coordinates from 100 to 200 ns of the simulation period. In this study, the first 15 eigenvalues of F, G, and control represent 61.51, 64.12, and 67.03% of the total motion. Fig. S1 indicates the eigenvalue vs. eigenvector index plot. From this figure, it was evident that the first two eigenvalues represent most of the variability, thereby, these two eigenvalues were projected along the eigenvector direction for obtaining the first and second principal components, termed PC1 and PC2, respectively. The 2D scatter and cluster plots PC1 and PC2 are presented in Fig. 6A–F, where each dot indicates correlated motions. Both F and G bound proteins showed compacted motion within a smaller subspace than the control. The motion of the G bound protein was more compact than that of the F; however, both F and G formed three well-defined clusters, indicating the decreased motion after ligand binding. Compacted motions indicated that the F and G reduced protein movement, as expected for an inhibitor. In addition, F and G-induced lower movements indicated that the protein remained in a compacted state during simulation, thereby preventing its effect. On the other hand, control bound protein showed a more spreading nature, occupied a larger space, and formed four clusters that indicated higher motion of the protein compared to our investigated ligands.
Figure 6.
The PCA plot of F (A), G (B), and control (C) bound protein. The (D, E, F) denotes the clusters of F, G, and control bound protein, respectively. The MMPBSA plot of interacting residues for SRC in complex with F, G, and control denoted by (G), (H), and (I), respectively. The 3D images indicate the binding residues of ligands.
Hydrogen bonds (H-bonds) are important for the stability of the protein and ligand interaction. It determines the binding specificity of drugs and also influences the drug pharmacokinetics69,70. Here, we considered a distance and angle of 3.5 Å and 120°, respectively, for the H-bond analysis66. For this study, H-bonds with an occupancy ≥ 60% were classified as strong, intermediate (40–59%), and lower (25–39%). Table 5 denotes the H-bonds occupancy level of the ligands. Both F and G-protein complexes formed several strong to moderate hydrogen bonds by acting as a donor or acceptor. The F formed strong hydrogen bonds with Tyr343 of the hinge as an acceptor and with Gln278 of the glycine-rich loop and Glu342 of the hinge as donors. Moderate occupancy was shown by Leu396, Met344, and Ser348 residues as donors and Ser348, Tyr343, and Met344 as acceptors. Furthermore, Leu276, Lys298, Gly347, and Leu396 residues exhibited low H-bond occupancy. The G as a donor formed strong H-bonds with the catalytic loop residue Ala393 and Gly424 of the activation segment, and while as an acceptor formed bonds with Leu276 of the glycine rich loop, and Asn394 of the catalytic. In addition, Phe427 and Arg391 acted as acceptors, and Phe281, Phe427, and Arg391 acted as donors and showed moderate H-bonds. Furthermore, low occupancy was observed for Phe281, GLy282, and Tyr343 when they acted as acceptors. By contrast, for the control-SRC complex, hydrogen bond occupancy was lower than our investigated ligands. A strong bond was seen with Gly347 (donor), while multiple residues, including Tyr343 and Ser348, Met344 and Gln278, demonstrated moderate H-bonds as donors and acceptors, respectively. In addition, one lower interaction, with Ala393, was found.
Table 5.
The hydrogen bond analysis of ligands in complex with SRC protein.
| Ligand | Ligand as H bond acceptor | Ligand as H bond acceptor | ||||
|---|---|---|---|---|---|---|
| Donor | Acceptor | % of occupancy | Donor | Acceptor | % of occupancy | |
| F | Tyr343 | F | 66.63 | F | Gln278 | 70.94 |
| Leu396 | 54.49 | Glu342 | 66.26 | |||
| Met344 | 50.66 | Ser348 | 57.08 | |||
| Ser348 | 49.07 | Tyr343 | 43.09 | |||
| Leu276 | 34.54 | Met344 | 40.96 | |||
| Lys298 | 33.03 | Leu276 | 37.09 | |||
| Gly347 | 29.55 | Leu396 | 25.67 | |||
| Gly347 | 25.33 | |||||
| G | Leu276 | G | 120.32 | G | Ala393 | 71.48 |
| Asn394 | 82.44 | Gly424 | 61.58 | |||
| Phe281 | 54.06 | Phe427 | 51.40 | |||
| Phe427 | 52.63 | Arg391 | 49.52 | |||
| Arg391 | 46.14 | Phe281 | 37.44 | |||
| Gly282 | 35.00 | |||||
| Tyr343 | 30.13 | |||||
| Control | Gly347 | Control | 61.34 | Control | Met344 | 46.67 |
| Tyr343 | 53.54 | Gln278 | 43.50 | |||
| Ser348 | 53.06 | Ala393 | 35.03 | |||
Further binding free energy of the protein–ligand complex was analyzed by the MMPBSA approach, which is a reliable method to assess the protein–ligand interaction71. Table 6 denotes each term of MMPBSA for all protein–ligand complexes. In our study, the highest binding energy (ΔE Bind) was observed for the G-protein complex (− 120.671 ± 16.997 kJ/mol), followed by the F-protein complex (− 86.363 ± 17.878 kJ/mol) and the control (− 65.042 ± 18.419 kJ/mol). The Van Der Waal’s and SASA energies for all complexes, which are two elements of binding energy, followed a similar trend as binding energy. On the other hand, the electrostatic energy for the F-protein complex was the highest, − 43.661 ± 11.113 kJ/mol, followed by the control and the G-protein complex. Moreover, F-protein complex also showed a high polar solvation energy of 183.821 ± 26.903 kJ/mol. By contrast, this energy for the G and control-protein complex was almost similar, with values of 170.754 ± 17.704 kJ/mol and 174.734 ± 23.403 kJ/mol, respectively.
Table 6.
The MMPBSA binding free energy analysis of each ligand-SRC complex.
| Ligand | Energy terms (kJ/mol ± SD) | ||||
|---|---|---|---|---|---|
| ΔE Van der Waal | ΔE Electrostatic | ΔE Solvation; Polar | ΔE Solvation; SASA | ΔE Bind | |
| F | − 203.475 ± 15.515 | − 43.661 ± 11.113 | 183.821 ± 26.903 | − 23.047 ± 1.113 | − 86.363 ± 17.878 |
| G | − 245.731 ± 13.724 | − 19.832 ± 7.105 | 170.754 ± 17.704 | − 25.863 ± 1.105 | − 120.671 ± 16.997 |
| Control | − 185.922 ± 16.917 | − 32.705 ± 12.601 | 174.734 ± 23.403 | − 21.150 ± 1.690 | − 65.042 ± 18.419 |
To understand the contributed residues, we decomposed the binding energy to each residue. Energy value ≥ ± 1 kJ/mol was considered a significant contribution. Figure 6G–I represents the interacting residues of F, G, and control with the SRC. Each compound showed strong binding affinity for Lys298; the interaction energy of G was the highest, followed by F and control, with a value of 12.6839, 10.4582, and 10.187 kJ/mol, respectively. The G also strongly interacted with catalytic loop residues, including Asp389, Arg391, Asn394, and Arg388, with a value of 17.7453, − 13.8728, − 5.622, and − 1.9266 kJ/mol, respectively (Fig. 6H). In addition, the Mg2+ positioning part of the active segment residues, including Asp407, Leu410, and Arg412 showed an energy value of 13.2121, − 3.7387, and − 1.4984 kJ/mol, respectively. Another residue of the active segment, Phe427, also contributed significant energy (− 13.8282 kJ/mol) while other residues of this segment, withinGly424—Lys426, also demonstrated moderate energy. Furthermore, the catalytic spine residues, Val284, and Leu396 also interacted and showed an energy of − 9.473, and − 1.5639 kJ/mol, respectively. Apart from these, Gly277 and Phe281 of the conserved glycine loop of the N-lobe exhibited strong binding affinity of − 2.2146, and − 7.5838 kJ/mol, respectively, while Trp285, Leu300, Ser348, and Leu350-Asp351 were the remaining interacting residues. The F-protein complex demonstrated strong interactions with the catalytic spine residues including Leu396, Val284, and Ala296 with a value of − 6.9862, − 5.596, and − 1.7137 kJ/mol, respectively (Fig. 6G). Aside from Lys298, catalytic loop residues including Arg391, Ala393, and Asn394 showed energies of 6.385, 1.598, and 2.1723 kJ/mol, respectively. Moreover, F also bound with the five residues of the conserved glycine rich loop, from Leu276 to Cys280, within − 4.8 to − 1.4 kJ/mol, while hinge residues including Glu342—Tyr343, and Lys346—Gly347 also showed significant energy, within -3 to 4 kJ/mol. Furthermore, the first residue of DFG active segment Asp407 (6.5905 kJ/mol) and gatekeeper residues Thr341 (− 1.0015 kJ/mol) were also interacted. The other binding residues were Val326, Leu350—Asp351, Lys430, and Glu457. On the other hand, the contribution of interacting residues for control was lower than our investigated ligands. In addition to Lys298, the catalytic loop residues, Asp389, Arg391, and Ala393-Asn394 showed binding energies within 1–4 kJ/mol (Fig. 6I). The conserved glycine rich loop residues, Leu276—Gly279, and Phe281 exhibited energy values within − 6.5 to − 1.0 kJ/mol. Moreover, Asp407 of the active segment and Val284 and Leu396 of the catalytic spine exhibited contributions of 3.8211, − 6.3903, and − 3.2682 kJ/mol, respectively. Furthermore, hinge region residues (Glu342, Met344, Lys346-Gly347) demonstrated interactions within − 2.7 to − 1.6 kJ/mol and the other residues were, Val274—Lys275, Glu283, Trp285—Met286, Ser348, and Asp351 (− 1 to − 2.7 kJ/mol).
We also analyzed the distance of multiple residues vital for receptor inactivation. In the SRC, the salt bridge between invariant residue Lys298 and Glu313 is considered very crucial for receptor activation40,72. In the inactivate state, this salt bridge is broken; Lys298 interacts with Asp407, and Glu313 of αC interacts with Arg412 of the activation loops. Figure 5H,I,J denotes the distance analysis over the simulation. The distance between Lys298-Glu313 was nearly 12 Å in all three protein–ligand complexes, which indicated the breakage of the bonds (Fig. 5H). On the other hand, a lower distance between Lys298-Asp407, and Lys298-Arg412 was visible in all three cases, indicating their interactions over the simulation period (Fig. 5I,J).
MD simulation of AKT1-ligand complex
In search of the effect of F and G on the allosteric site of AKT1, first we analyzed the Cα-RMSD of ligand bound proteins, followed by protein–ligand complexes, and ligand RMSD. Figure 7A,B represent the Cα-RMSD, and protein–ligand complex RMSD, respectively. The F-bound protein exhibited late stabilization; from 0 to 87 ns, the Cα-RMSD value increased steadily, with a peak value of 5.2 Å at 79 ns. However, stability gained following 87 ns where fluctuations were observed near a mean of 5.0 ± 0.19 Å till 185 ns, followed by a lower RMSD until the end of the study. The F-protein complex also demonstrated similar RMSD profile as Cα-RMSD. Overall, no abrupt fluctuations were distinguished after stabilization, that indicated its stability. In the G bound protein, first convergence was noted after 31 ns which continued till 98 ns (mean, 3.7 ± 0.14 Å). Following 99 to 115 ns, the RMSD gradually increased (within 3.2–4.1 Å), followed by another converged state, from 124 to 160 ns (average, 3.6 ± 0.09 Å). The Cα-RMSD decreased after 160 ns; however, fluctuations were moderate till the end of the study, with a mean of 3.5 ± 0.14 Å. In the complex state, we also found identical fluctuations to Cα-RMSD, and over the simulations, the average fluctuation was lower, which was an indication of higher stability. The control bound protein also showed such lower deviations and high stability; initially, this protein showed high Cα-RMSD that remained until 38 ns (within 0.9–3.6 Å) after which it stabilized. However, this convergence period continued until 56 ns with a mean of 3.3 ± 0.08 Å. Following 68 ns, the Cα atoms fluctuated, within 3.1–3.8 Å, that stabilized again for a short period, from 100 to 112 ns. However, apart from the two more convergences, from 134–172, and 188–200 ns, the protein fluctuated with a mean of 3.4 ± 0.12 Å. The complex state also manifested a similar fluctuations pattern.
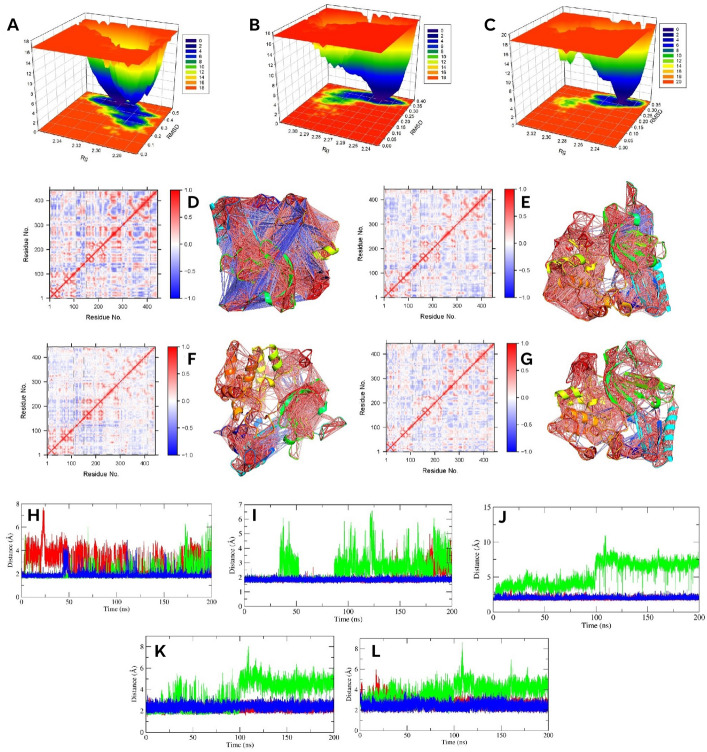
Figure 7.
The Cα-RMSD (A), protein–ligand complex RMSD (B), ligand RMSD (C), and ligand bound ΔRMSF (D) of AKT1 protein. The protein-AKT1 complex Rg, ligand Rg, protein–ligand complex SASA, and ligand SASA are denoted by (E), (F), (G), and (H), respectively. The green, blue, and red color represents F, G, and control, respectively.
Regarding ligand RMSD, both F and G showed lower conformational changes (Fig. 7C). The F demonstrated a stable RMSD until 12 ns, after which the RMSD value increased with a peak of 5.0 Å that stabilized after 32 ns. From 32 to 60 ns, a stable profile was noticed, followed by fluctuations with a mean of 4.4 ± 0.39 Å from 61 to 92 ns. The F achieved stability after 93 ns, and except from 149–153 and 179–185 ns, it lasted till the end of the simulation (mean 4.2 ± 0.43 Å), indicating stable and minimal conformational changes. The RMSD of G followed a rising pattern from beginning with lower fluctuations and reached convergence at 44 ns. This convergence period sustained till 64 ns (mean 3.9 ± 0.34 Å), after which the RMSD steadily fell till 102 ns within 1.7–4.2 Å. However, no abrupt fluctuations were noted after 116 ns and till the end of the simulations, the RMSD pattern was stable, decreased, and fluctuated near a mean of 2.4 ± 0.26 Å, suggesting a stable profile. On the other hand, the control showed higher fluctuations in the first 40 ns and last 44 ns, with a mean of 2.8 ± 0.59, and 4.1 ± 0.66 Å, respectively. However, within 41–66 ns, fluctuations were comparatively lower, average 3.8 ± 0.48 Å, that reduced after 67 ns to 80 ns. The control demonstrated convergence from 81 to 153 ns, where the mean was 3.7 ± 0.24 Å. Overall, the control showed a moderate to lower stability depending on the simulation time.
The ΔRMSF analysis further provides insights about the residual mobility. Figure 7D denotes the ΔRMSF of the ligand bound AKT1 protein. In the G and control bound proteins, most of the residues of AKT1 demonstrated significant lower mobility; the G bound protein showed a higher loss of residual flexibility than the control. For the F bound protein, mobility was also reduced; however, lower than that of the other two ligands. Here, we also noted that F, G, and control decreased the flexibilities of 303–313 residues that lie within the activation loop, where the mean was 1.17, 1.84 and 1.64 Å. Interestingly, the PH-kinase linker and its nearby regions showed dynamic behavior; it (within 110–145 residues) demonstrated significant loss of mobility in the G, and control bound proteins with a value of 1.5 and 2.05 Å, respectively. By contrast, in the F bound protein, the flexibility of 119–139 residues in this region increased near a mean of − 2.58 Å. Apart from these, the G followed by control and F also significantly decreased the motion PH domain.
In the Rg analysis, we found lower Rg values for G and control-protein complexes, while the F-protein complex manifested a higher Rg value. Figure 7E represents the Rg of our investigated ligands complexed with AKT1. The fluctuations of the F-protein complex of last 100 ns (23.24 ± 0.10 Å) were comparatively lower than the first 100 ns (23.12 ± 0.18 Å), demonstrating increased compactness after the first 100 ns of the study. This Rg profile of the first 100 ns might be associated with late stabilization, as indicated by the high RMSD at the beginning. On the other hand, for the G-protein complex, compactness was higher compared to the control. A gradual decrease in Rg value with lower fluctuations was noted until 154 ns (mean, 22.70 ± 0.14 Å). After 155 to 200 ns, this complex fluctuated near an average of 22.64 ± 0.07 Å. The control-protein complex also exhibited a decreasing pattern of Rg till 94 ns where the mean was 22.62 ± 0.18 Å. Following 94 ns, the Rg value increased slightly, which was higher in magnitude than the periods of 135–152 and 170–200 ns.
Regarding ligand Rg, both F and G demonstrated lower mean Rg values, 4.67 ± 0.07, and 4.96 ± 0.05, respectively, than the control (5.05 ± 0.10 Å) (Fig. 7F). Over the course of the simulation, the G showed nearly a stable line except for a higher Rg from 61–65 ns than any other investigated ligand, while the F showed lower Rg values after 10 ns to the end of the simulation, suggesting their compactness in the binding site. On the other hand, high fluctuations were noticed for control, particularly after 148 ns to the end of the simulation, indicating its less compactness than our investigated compounds.
From the SASA analysis, we found that both F and G demonstrated a decreasing pattern of SASA values until 155 ns (Fig. 7G). After 155 ns to the end of the simulation, fluctuations were found near a mean of 229.59 ± 3.33, and 218.05 ± 3.23 nm2 for the F and G-protein complexes, respectively. For the control-protein complex, the SASA decreased sharply till 66 ns, and after 67 to 200 ns, the mean fluctuation was 222.30 ± 3.78 nm2. However, over the study, the G-protein complex demonstrated a lower mean SASA value (217.94 ± 6.42 nm2) followed by the control and F-protein complex, 222.08 ± 5.53, and 230.79 ± 4.60 nm2, respectively, suggesting that the G efficiently decreased the solvent accessibility area of the protein surface, thereby resulting in a significant decrease in expansion and increased stability.
In the case of the ligand SASA, no significant variations were observed in the investigated ligands. Figure 7H denotes the ligand SASA profile over the simulation. The control demonstrated a lower mean SASA value, 6.76 ± 0.22 nm2, than the F and G, 8.17 ± 0.26, and 8.19 ± 0.25 nm2, respectively. However, SASA changes depending on the molecular size and polarity. Both of our investigated compounds fluctuated near a similar mean value, indicating that both were well-equipped in the binding pocket.
FEL analysis further provides more insights about the thermodynamically stable states. Similar to SRC, FEL was generated from the RMSD and Rg data. Figure 8A,B,C represents the plot of FEL for each protein–ligand complex. The blue color was more enriched in the F-protein complex and formed a well-defined valley. However, the less blue color also spread over a certain area with less deep peaks. Overall, it indicated that the interaction of F with AKT1 was more thermodynamically stable, despite some intermediately stable states. On the other hand, both the G and control-protein complexes manifested a similar, well-defined valley. The binding of G formed a much wider valley enriched with more blue color than the the control which was a demonstration of more stable states and lower entropy. However, it was worth noting that these two complexes also showed multiple partially folded states, indicating moderate stability of some conformations.
Figure 8.
The FEL plot of F (A), G (B), and control (C) bound protein of AKT1. The 2D DCCM plots and 3D highly correlated (≥ ± 0.4) residues of apo, F, G, and control bound AKT1 are denoted by (D, E, F, and G), respectively. The H, I, J, K, and L indicate the distance between 17–273, 23–323, 25–322, 25–323, and 25–324, respectively.
The DCCM analysis further gives more insights about the corrected residues. The Fig. 8D–G represents the DCCM, where higher positive correlation is indicated by more red color, while more blue color represents negative correlation. Compared to the apo protein, we found decreased correlated and anticorrelated motion after binding with ligands. This decrease was higher in G bound protein, followed by control and F bound protein. However, multiple residues of the PH-kinase linker region (~ 119–125 residues) showed higher anticorrelated motion in the F and G bound proteins than the control bound protein.
To analyze the motion after ligand binding, we performed PCA over the last 100 ns of the simulation. Fig. S1 indicates the eigenvectors vs. eigenvalues plot, and Fig. 9A–F denotes the plot of PC1/PC2 and cluster analysis. A total of 1329 eigenvalues with their corresponding eigenvectors were obtained for each ligand bound protein. However, the first two eigenvectors explained most of the variability, 70.19, 70.29, and 63.65% for F, G, and control bound proteins, respectively. From Fig. 9, it was evident that the motion of the G bound protein confined within a smaller subspace than any other investigated ligands, denoting higher stability. However, these motions were confined in four clusters; cluster 2 was less compacted and contained motions from 100 to 125 ns (Fig. 9B,E). The motion of F bound protein was also confined within a smaller space, which formed three compacted clusters (Fig. 9A,D), however, less than control bound protein (Fig. 9C,F). On the other hand, the flexibilities of the control bound protein also low and formed three well-defined clusters; cluster-1 was less compacted (Fig. 9F). Overall, G, followed by F and decreased atomic movements and showed higher stability after binding with AKT1.
Figure 9.
The PCA plot of F (A), G (B), and control (C) bound AKT1 protein. The (D, E, and F) denotes the clusters of F, G, and control bound protein, respectively. The interactions of F (G), G (H), and control (I) with AKT1, analyzed by MMPBSA plot having an energy value ≥ ± 2 kJ/mol.
Further H-bonds analysis (Table 7) showed that F formed strong H-bonds with Trp80 (as both a donor and an acceptor) and Asp53 (as an acceptor). In addition, F demonstrated moderate occupancy with Asn53 and Val271 as donors and with Asn54 as an acceptor. Other residues, including Leu210, Gln203, and Asp292, exhibited lower hydrogen bonds. The H-bond interactions of G were higher; Tyr272, Gln79, and Asn53 as acceptors and Trp80, Asn53, and Leu295 as donors showed higher occupancy. Trp80, Asn54, Gly294, and Leu295 also demonstrated moderate bonds as acceptors, while Asp274, Thr82, and Tyr272 contributed as donors. Furthermore, lower bond occupancy was found with Lys112, Asp274, Asp292, Val271, Asn54, and Gly294. By contrast, the control strongly interacted with Gln79, Asn54, and Trp80 as donors, whereas as acceptors, it bound with Arg86 and Trp80. Furthermore, moderated H-bonds were distinguished with Arg273, whereas Val271, Tyr326, Asp292, Tyr326, and Gln79 showed lower interactions.
Table 7.
The hydrogen bond analysis of ligands in complex with AKT1 protein.
| Ligands | Ligand as H bond acceptor | Ligand as H bond acceptor | ||||
|---|---|---|---|---|---|---|
| Donor | Acceptor | % of occupancy | Donor | Acceptor | % of Occupancy | |
| F | Trp80 | F | 110.58 | F | Trp80 | 78.40 |
| Asn53 | 64.32 | Asn53 | 44.24 | |||
| Asn54 | 41.34 | Val271 | 41.35 | |||
| Leu210 | 33.99 | Leu210 | 27.67 | |||
| Gln203 | 27.72 | Asp292 | 27.17 | |||
| G | Trp80 | G | 98.45 | G | Tyr272 | 111.14 |
| Gln79 | 81.68 | Gln79 | 73.40 | |||
| Leu295 | 73.40 | Asn53 | 62.56 | |||
| Asp274 | 52.32 | Gly294 | 57.48 | |||
| Thr82 | 52.30 | Trp80 | 48.30 | |||
| Tyr272 | 45.11 | Leu295 | 47.20 | |||
| Asn54 | 31.69 | Asn54 | 43.83 | |||
| Gly294 | 26.81 | Lys112 | 36.35 | |||
| Asp274 | 35.43 | |||||
| Asp292 | 27.38 | |||||
| Val271 | 26.81 | |||||
| Control | Arg86 | Control | 73.34 | Control | Gln79 | 92.31 |
| Trp80 | 71.80 | Asn54 | 79.62 | |||
| Tyr326 | 29.12 | Trp80 | 67.63 | |||
| Gln79 | 27.93 | Arg273 | 49.61 | |||
| Val271 | 36.92 | |||||
| Tyr326 | 29.10 | |||||
| Asp292 | 28.82 | |||||
Further analysis was carried out to find the binding energy through MMPBSA analysis. The details of each term are shown in Table 8. The G-protein complex demonstrated the highest ΔEBind, − 130.437 ± 19.111 kJ/mol, which was almost twice than the control-protein complex, -68.129 ± 24.043 kJ/mol. The F-protein complex also exhibited high binding energy, with a value of − 98.249 ± 19.884 kJ/mol. For the G-protein complex, energy terms including Van der Waal, electrostatic, polar solvation, and SASA were − 302.438 ± 10.980, − 57.744 ± 9.634, 258.571 ± 22.001, − 28.826 ± 0.973 kJ/mol, respectively, which was higher or nearly similar than any other investigated ligands. In the F-protein complex, high van der Waal, and SASA energies were dominant, whereas electrostatic and polar solvation energies were comparatively lower than the control. For the binding of control, Van der Waal, electrostatic, and polar solvation energies play key roles.
Table 8.
The MMPBSA binding free energy analysis of AKT1-ligand complex.
| Ligand | Energy terms (kJ/mol ± SD) | ||||
|---|---|---|---|---|---|
| ΔE Van der Waal | ΔE Electrostatic | ΔE Solvation; Polar | ΔE Solvation; SASA | ΔE Bind | |
| F | − 221.942 ± 13.117 | − 35.940 ± 11.839 | 183.796 ± 24.936 | − 24.163 ± 1.100 | − 98.249 ± 19.884 |
| G | − 302.438 ± 10.980 | − 57.744 ± 9.634 | 258.571 ± 22.001 | − 28.826 ± 0.973 | − 130.437 ± 19.111 |
| Control | − 214.685 ± 13.974 | − 58.669 ± 22.339 | 226.931 ± 42.575 | − 21.705 ± 1.140 | − 68.129 ± 24.043 |
Decomposing the ΔEBind revealed the contribution of residues to binding energy, providing more insights about the interacting residues. Both F and G interact with a significant number of residues. Figure 9G–I denotes the interaction plot of the MMPBSA analysis, where residues are marked as having an energy value ≥ ± 2 kJ/mol. Interestingly, our examined F and G bound very strongly to Trp80, two times stronger than the control, with values of -18.1683, -11.034, and -5.7412 kJ/mol, respectively. In addition, with the first residues of the DFG loop, Asp292 also exhibited high affinity, with values of 13.5926, 11.925, and 16.283 kJ/mol for F, G, and control, respectively. Apart from these, the F also interacted strongly with multiple kinase domain residues, including Val270, Leu210, Tyr272, Asp274, and Leu264, where the energy contribution ranged between − 2 and − 7.9 kJ/mol (Fig. 9G). Moreover, the pH domain residue, Thr82, showed a binding energy of -2.0353 kJ/mol, while other residues, including Glu17, Asp32, Glu49, Leu52, Thr81, and Ile84, demonstrated energy within ± 1.0 to ± 1.9 kJ/mol. By contrast, strong positive interactions were found for Glu115, Arg200, and Glu117 (2.05–4.24 kJ/mol), while the other kinase domain residues including Glu116, Lys179, Asn199, Gln203, Ser205, His207, Thr211, Ala212, Lys276, Glu267, Lys268, Glu278, and Lys297, showed lower energy, within ± 1.0 to ± 1.9 kJ/mol. For the G-protein complex, Asp274 also played a key contribution (13.7828 kJ/mol) (Fig. 9H). The kinase domain residues including Leu264, Val270, Lys276, Cys296, Leu210, Lys179, Arg200, and Asp331 exhibited strong interaction within ± 2 to ± 8 kJ/mol. The G was also bound to multiple PH domain residues, including Glu17, Gln79, Thr82, Ile84, and Arg86 (− 3.3 to − 2.0 kJ/mol). Additionally, other residues that demonstrated binding energy within ± 1.0 to ± 1.9 kJ/mol were Lys14, Asn54, His265, Tyr272, Arg273, Glu278, Phe293, Gly294, Leu295, and Tyr315. On the other hand, the control bound with the PH domain residues, including Glu17, Asn54, Thr82, Gln79, and Ile84, within − 2.0 to 5.7 kJ/mol and with Arg86 (6.2488 kJ/mol) (Fig. 9I). In the kinase domain, Val270, Leu264, Asp331, Thr211 and Lys179 manifested energy value ≥ ± 2 kJ/mol. Lower contributing residues were Lys14, Asp32, Leu210, Met227, Val271, Arg273, Asp274, Lys276, Asp323, and Tyr326.
We further analyzed the distance between Glu17-Arg273, Arg23-Asp323, and Arg25 with Glu322, Asp323, and Asn324, reported by Wu et al., 2010 in the inactivate state of AKT143. Figure 8H–L represents the distance between each investigated pair. The salt bridge between Glu17-Arg273 gives additional stability, and in our study, the G-protein complex demonstrated the lowest distance between them, followed by the F and control-protein complex (Fig. 8H). The distances in the reaming interacting residues were also highly stable and lower than the control in the G-protein complex (Fig. 8I, J, K, and L). On the other hand, the F-protein complex showed comparatively higher distances for the remaining residues; notably, distances between Arg25-Glu322 and Arg25-Asp323 (Fig. 8J, and K) were increased after 100 ns, indicating different residual interactions might be responsible for its stabilization.
Discussions
BC is a prevalent form of neoplastic malignancy that affects the mammary glands in women6. Since BC metastasis is a complicated process that involves numerous signaling pathways and is a leading cause of death in women, it is imperative to find potent drugs to combat BC7. The investigated dimer amides in our study, chabamide F and chabamide G, have been reported as cytotoxic against multiple cancer cell lines; notably, the G was found as highly potent against colon cancer cell lines14. However, their effect on BC, along with their molecular mechanisms, remains poorly understood. In the present study, we comprehensively uncover their biological targets, probable mechanisms, key target proteins, drug likeness and ADME properties as new lead compounds. Figure 10 represents the chemical structure of these two compounds.
Figure 10.
Chemical structure of chabamide F and chabamide G.
Network pharmacology is a promising approach to describing relationships among biological systems, drugs, and diseases12,22. Both F and G are dimer, moderately soluble, highly GI absorbable, and follow the Lipinski rule except for a violation of molecular weight. From 245 drug targets and 500 BC targets, we identified 64 BC-related genes as candidate targets for F and G. The GO analysis revealed that these target genes were linked to various biological processes, including signal transduction, cell apoptosis and proliferation, transcriptional activity from RNA polymerase II, and so on. The KEGG pathway investigation showed that they were enriched mostly in the RAS, PI3K-AKT, estrogen, MAPK, and FoxO signaling pathways. Among them, PI3K/AKT and MAPK signaling pathways are mostly reported in BC and are important signaling pathways, particularly in TNBC, which is an aggressive subtype of BC with a poor prognosis59,73. Furthermore, the crosstalk between estrogen signaling and metabolic regulators allows BC cells to rearrange their metabolism pathways74. RAS signaling has been shown to activate in luminal breast cancer, lowering survival rates75. Furthermore, PI3K-AKT-FOXO signaling has been reported in the hormone-independent growth of many breast cancers76.
The PPI network analysis is a valuable approach to identifying key target proteins in a hub gene network. From the STRING database to network analysis, we found a network of 64 nodes with 342 edges. Further degree, betweenness, closeness, and eigenvector analyses revealed SRC, AKT1, MAPK1, and HSP90AA1 as key targets of F and G. However, SRC and AKT1 had higher degree values compared to the rest, indicating more interactions with other proteins. Therefore, it was expected that they played a vital role in regulating the network, and we conducted further analyses on these two proteins. SRC proto-oncogene is a non-receptor tyrosine kinase that promotes malignant transformation and is typically overexpressed in a variety of BC subtypes23,77. Generally, activated estrogen or progesterone receptors can bind directly with SRC kinase, and activated SRC kinase can trigger the Ras/Raf/MAPK cascade, causing BC cell proliferation23. On the other hand, AKT overactivation is a frequent event in malignancies, which is a key player in the PI3K/AKT/mTOR pathway78. AKT1 expression is upregulated in 40% of cases of BC and ovarian cancer and associated with tumor initiation, proliferation, and reducing apoptosis78. Therefore, these two proteins are vital targets for new drug discovery against BC.
Molecular docking, MD simulations, and MMPBSA are popular approaches in modern drug discovery. It can accurately evaluate the kinetics, thermodynamics, and absolute ligand affinities of a drug-receptor complex71,79. Therefore, it has become easier to find promising active compounds against a molecular target for further preclinical tests. For further experiments, we targeted the ATP biding site for SRC kinase and the allosteric site of AKT1, which lies between the PH and kinase domains. Binding of ATP triggers activation of the SRC kinase domain. Ligands that bind to the ATP binding pocket and also interact with respective amino acids, such as asparagine located within the catalytic loop and aspartate at the start of the activation segment, are a new class of promising kinase inhibitors40. On the other hand, allosteric inhibition of AKT1 offers several advantages compared to ATP-competitive inhibitors. It has shown greater selectivity, reduced toxicity, and fewer adverse reactions80. The MD simulation of SRC revealed that F and G stabilized the protein better than the control, dasatinib. The G-protein complex showed a more stable fluctuation pattern, higher stability, higher compactness, lower motion, and decreased cross-correlated residues in the RMSF, RMSD, Rg, PCA, and DCCM than the F and control protein complexes. The F also showed similar stable pattern; notably showed more thermodynamically stable states than the investigated ligands. Further MMPBSA analysis revealed that the binding energy of both F and G was higher than that of dasatinib; in particular, G showed a two-fold higher binding energy than dasatinib. All investigated compounds broke the key salt bridge between Lys298 and Glu313, and F and G interacted strongly with Lys298 compared to the control. Interestingly, the asparagine (Asn394) within the catalytic loop and the aspartate (Asp407) at the start of the activation segment demonstrated strong contributing residues for G binding, followed by F and control. Additionally, G also strongly interacted with catalytic loop residues (Asp389, Arg391, and Arg388), active segment residues (Leu410, Arg412, Phe427, and Gly424-Lys426), catalytic spine residues (Val284, and Leu396), and conserved glycine loops (Gly277 and Phe281) of the N-lobe. The F showed a similar interaction profile; however, it showed interaction with hinge and gatekeeper residues (Glu342-Tyr343, Lys346-Gly347, and Thr341), whereas lower interactions with active segments were found. Overall, these interactions are important for inhibition of SRC kinase40 and our investigated compounds maintained such interactions of ligands over the 100 ns period which indicates both F and G could potentially interfere with SRC activation and be a highly effective inhibitors of SRC. Previous studies have reported that oncogenic activation of SRC activates various downstream pathways, such as PI3K/AKT, MAPK, interleukin 8 (IL8), signal transducer and activator of transcription 3 (STAT3), vascular endothelial growth factor (VEGF), and cytoskeletal formation81. Recent studies on BC mentioned that SRC phosphorylation activates the β-catenin pathway in TNBC and increases resistance to trastuzumab for HER2 + breast cancer82,83. Therefore, it might be possible that interference with SRC activation caused by F and G could potentially interfere with the downstream signaling pathways of SRC.
In the case of AKT1, we found higher stability after binding with G compared to F and capivasertib. The F showed comparatively moderately stable behavior, as indicated by RMSD, RMSF, Rg, and SASA values. However, the G reduced the motion of correlated residues of the PH and kinase domains more significantly than the F, and both compounds also showed stronger hydrogen bond occupancy with Trp80. Interaction with Trp80 is considered a vital residue for allosteric inhibition of AKT1/243. MMPBSA analysis also further confirmed the strong binding of both compounds to Trp80. Moreover, the binding energy of G was the highest, followed by F and control. The MMPBSA analysis showed that both interacted with multiple reported residues of PH and kinase domains, which are vital for allosteric inhibition43,44. Notably, PH domain residues (Glu17, Gln79, Thr82, Ile84, and Arg86) and kinase domain residues (Asp274, Leu264, Val270, Lys276, Cys296, Leu210, Lys179, Arg200, and Asp331) showed significant contributions to the G-protein complex. For stabilizing F at the binding pocket, Thr82 of the PH domain and kinase residues such as Val270, Leu210, Tyr272, Asp274, Leu264, Glu115, Arg200, and Glu117 played vital roles. The control molecule also demonstrated significant interactions with both PH and kinase domain. Furthermore, the G also demonstrated a lower distance between Glu17-Arg273, Arg23-Asp323, and Arg25 with Glu322, Asp323, and Asn324 compared to capivasertib and F over the 200 ns simulation period, which are generally found in the inactive state43. Overall, G could be a strong inhibitor of AKT1, and F could be a moderate inhibitor. Activated AKT modulates the functions of various downstream proteins, including mammalian target of rapamycin (mTOR), glucose-like transporter 4 (GLUT4), FOXO1, Glycogen synthase kinase-3 beta (GSK3β), cyclin D1, S6, and WEE1 in cancer cells24,78,84. In breast cancer, AKT1 plays roles in tumor initiation in ErbB-2- and PyMT-mediated BC, cell proliferation in BRCA1-associated BC, VEGF-mediated angiogenesis, drug resistance, changing the phenotype of stem cells, and more24. Overall, AKT1 has a broader effect on BC; thus, both F and G might have broader implications for AKT-mediated cancer pathways.
Conclusion
In this study, we investigated the effects and molecular mechanisms of two dimeric amides, chabamides F and chabamide G, in BC. Using the network pharmacology approach, we identified 64 target genes for F and G that were strongly related to the RAS, PI3K-AKT, estrogen, MAPK, and FoxO signaling pathways. The PPI analysis highlighted SRC and AKT1 as significant targets, and subsequent molecular docking and MD simulations revealed that both F and G were highly stable at the binding pocket of SRC. In addition, they had a greater binding affinity in the MMPBSA study than the control, dasatinib. On the other hand, after binding with AKT1, the G-protein complex was more stable than the control, capivasertib, followed by F. However, MMPBSA analysis demonstrated a higher binding energy for G and F than capivasertib. Overall, both investigated compounds in complex with SRC and AKT1 demonstrated stability over a 200 ns simulation period and kept interactions with the active site residues for a longer period of time in the MMPBSA analysis. In summary, both F and G would be potential drug candidates for SRC and AKT1 inhibition.
Supplementary Information
Abbreviations
- BC
Breast cancer
- ΔEBind
Binding free energy
- F
Chabamide F
- G
Chabamide G
- DCCM
Dynamic cross correlation maps
- FEL
Free energy landscape
- FOXO
Forkhead box O proteins
- GO
Gene ontology
- MAPK
Mitogen-activated protein kinases
- MD
Molecular dynamics
- MMPBSA
Molecular Mechanics Poisson-Boltzmann Surface Area
- H-bond
Hydrogen bond
- PCA
Principal component analysis
- PPI
Protein-protein interaction
- PI3K
Phosphatidylinositol 3-kinase
- RMSD
Root means square deviation
- RMSF
Root means square fluctuation
- Rg
Radius of gyration
- SASA
Solvent accessibility surface area
- SD
Standard deviation
- TNBC
Triple-negative
Author contributions
Conceptualization: P.R, and D.K.S; Methodology: P.R, and D.K.S; Software: P.R, and D.K.S; Validation: P.R; Formal analysis: P.R, and D.K.S; Investigation: P.R, and D.K.S; Resources: P.R, D.K.S, and S.J.U; Data curation: P.R, and S.J.U; Writing—original draft preparation: P.R, and D.K.S; Writing—review and editing: P.R, D.K.S, and S.J.U; Visualization: P.R; Supervision: S.J.U; Project administration: P.R, D.K.S, and S.J.U.
Funding
This research received no financial grant from any funding agencies in the commercial, public, or not-for-profit sectors.
Data availability
All software is freely available, and the manuscript and supporting information file contain the data used to reach the conclusion.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-70854-0.
References
- 1.Diori Karidio, I. & Sanlier, S. H. Reviewing cancer’s biology: an eclectic approach. J. Egypt. Natl. Cancer Inst.33, 1–7 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Upadhyay, A. Cancer: an unknown territory; rethinking before going ahead. Genes Dis.8, 655–661 (2021). 10.1016/j.gendis.2020.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J. Clin.74, 229–263 (2024). [DOI] [PubMed] [Google Scholar]
- 4.Luque-Bolivar, A., Pérez-Mora, E., Villegas, V. E. & Rondón-Lagos, M. Resistance and overcoming resistance in breast cancer. Breast Cancer: Targets Therapy12, 211–229 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winters, S., Martin, C., Murphy, D. & Shokar, N. K. Breast cancer epidemiology, prevention, and screening. Progr. Mol. Biol. Transl. Sci.1(151), 1–32 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Feng, Y. et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis.5, 77–106 (2018). 10.1016/j.gendis.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park, M. et al. Breast cancer metastasis: mechanisms and therapeutic implications. Int. J. Mol. Sci.23, 6806 (2022). 10.3390/ijms23126806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo, S. & Deng, C.-X. Effect of stromal cells in tumor microenvironment on metastasis initiation. Int. J. Biol. Sci.14, 2083 (2018). 10.7150/ijbs.25720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, W., Hoffmann, A. D., Liu, H. & Liu, X. Organotropism: new insights into molecular mechanisms of breast cancer metastasis. NPJ Precision Oncol.2, 4 (2018). 10.1038/s41698-018-0047-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy, A., Datta, S., Bhatia, K. S., Jha, P. & Prasad, R. Role of plant derived bioactive compounds against cancer. S. Afr. J. Bot.149, 1017–1028 (2022). 10.1016/j.sajb.2021.10.015 [DOI] [Google Scholar]
- 11.Di Dalmazi, G., Giuliani, C., Bucci, I., Mascitti, M. & Napolitano, G. Promising role of alkaloids in the prevention and treatment of thyroid cancer and autoimmune thyroid disease: a comprehensive review of the current evidence. Int. J. Mol. Sci.25, 5395 (2024). 10.3390/ijms25105395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandran, U., Mehendale, N., Patil, S., Chaguturu, R. & Patwardhan, B. Network pharmacology. Innovative approaches in drug discovery, 127 (2017).
- 13.Islam, M. T. et al. Chemical profile, traditional uses, and biological activities of Piper chaba Hunter: a review. J. Ethnopharmacol.257, 112853 (2020). 10.1016/j.jep.2020.112853 [DOI] [PubMed] [Google Scholar]
- 14.Rao, V. R. S. et al. Novel dimeric amide alkaloids from Piper chaba Hunter: isolation, cytotoxic activity, and their biomimetic synthesis. Tetrahedron67, 1885–1892 (2011). 10.1016/j.tet.2011.01.015 [DOI] [Google Scholar]
- 15.Ngo, Q. M. T. et al. Cytotoxic activity of alkaloids from the fruits of Piper nigrum. Nat. Product Commun.13, 1934578X1801301114 (2018). [Google Scholar]
- 16.Ren, J. et al. Chabamide induces cell cycle arrest and apoptosis by the Akt/MAPK pathway and inhibition of P-glycoprotein in K562/ADR cells. Anticancer Drugs26, 498–507 (2015). 10.1097/CAD.0000000000000209 [DOI] [PubMed] [Google Scholar]
- 17.Muharini, R., Liu, Z., Lin, W. & Proksch, P. New amides from the fruits of Piper retrofractum. Tetrahedron Lett.56, 2521–2525 (2015). 10.1016/j.tetlet.2015.03.116 [DOI] [Google Scholar]
- 18.Singh, R., Manna, S., Nandanwar, H. & Purohit, R. Bioactives from medicinal herb against bedaquiline resistant tuberculosis: removing the dark clouds from the horizon. Microbes Infect.26, 105279. 10.1016/j.micinf.2023.105279 (2024). 10.1016/j.micinf.2023.105279 [DOI] [PubMed] [Google Scholar]
- 19.Ahmed, M., Maldonado, A. M. & Durrant, J. D. From byte to bench to bedside: molecular dynamics simulations and drug discovery. BMC Biol.21, 299. 10.1186/s12915-023-01791-z (2023). 10.1186/s12915-023-01791-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta, A. & Purohit, R. Identification of potent BRD4-BD1 inhibitors using classical and steered molecular dynamics based free energy analysis. J. Cell Biochem.125, e30532. 10.1002/jcb.30532 (2024). 10.1002/jcb.30532 [DOI] [PubMed] [Google Scholar]
- 21.Kumar, A., Rajendran, V., Sethumadhavan, R. & Purohit, R. Evidence of colorectal cancer-associated mutation in MCAK: a computational report. Cell Biochem. Biophys.67, 837–851. 10.1007/s12013-013-9572-1 (2013). 10.1007/s12013-013-9572-1 [DOI] [PubMed] [Google Scholar]
- 22.Sakle, N. S., More, S. A. & Mokale, S. N. A network pharmacology-based approach to explore potential targets of Caesalpinia pulcherima: an updated prototype in drug discovery. Sci. Rep.10, 17217 (2020). 10.1038/s41598-020-74251-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo, J. et al. SRC kinase-mediated signaling pathways and targeted therapies in breast cancer. Breast Cancer Res.24, 99 (2022). 10.1186/s13058-022-01596-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinz, N. & Jücker, M. Distinct functions of AKT isoforms in breast cancer: a comprehensive review. Cell Commun. Signal.17, 1–29 (2019). 10.1186/s12964-019-0450-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanwell, M. D. et al. Avogadro: an open-source molecular builder and visualization tool. J. Cheminform.4, 17 (2012). 10.1186/1758-2946-4-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanwell, M. D. et al. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform.4, 1–17 (2012). 10.1186/1758-2946-4-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halgren, T. A. Merck molecular force field I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem.17, 490–519 (1996). [DOI] [Google Scholar]
- 28.Liu, X. et al. PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucl. Acids Res.38, W609–W614 (2010). 10.1093/nar/gkq300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stelzer, G. et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protocols Bioinform.54(1), 1–30 (2016). 10.1002/cpbi.5 [DOI] [PubMed] [Google Scholar]
- 30.Oliveros, J. C. VENNY. An interactive tool for comparing lists with Venn Diagrams. (2007).
- 31.Szklarczyk, D. et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucl. Acids Res.51, D638–D646 (2023). 10.1093/nar/gkac1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang, Y., Li, M., Wang, J., Pan, Y. & Wu, F.-X. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems127, 67–72 (2015). 10.1016/j.biosystems.2014.11.005 [DOI] [PubMed] [Google Scholar]
- 33.Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13, 2498–2504 (2003). 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherman, B. T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucl. Acids Res.50, W216–W221 (2022). 10.1093/nar/gkac194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daina, A., Michielin, O. & Zoete, V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep.7, 42717 (2017). 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webb, B. & Sali, A. Comparative protein structure modeling using Modeller. Curr. Protocols Bioinform.54(1), 5–6 (2016). 10.1002/cpbi.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montero, J. C., Seoane, S., Ocaña, A. & Pandiella, A. Inhibition of SRC family kinases and receptor tyrosine kinases by dasatinib: possible combinations in solid tumors. Clin. Cancer Res.17, 5546–5552 (2011). 10.1158/1078-0432.CCR-10-2616 [DOI] [PubMed] [Google Scholar]
- 38.Mullard, A. FDA approves first-in-class AKT inhibitor. Nat. Rev. Drug Discov.23, 9. 10.1038/d41573-023-00202-w (2024). 10.1038/d41573-023-00202-w [DOI] [PubMed] [Google Scholar]
- 39.Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem.31, 455–461 (2010). 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roskoski, R. Jr. Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol. Res.94, 9–25 (2015). 10.1016/j.phrs.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 41.Roskoski, R. Jr. Src protein–tyrosine kinase structure and regulation. Biochem. Biophys. Res. Commun.324, 1155–1164 (2004). 10.1016/j.bbrc.2004.09.171 [DOI] [PubMed] [Google Scholar]
- 42.Truebestein, L. et al. Structure of autoinhibited Akt1 reveals mechanism of PIP3-mediated activation. Proc. Natl. Acad. Sci.118, e2101496118 (2021). 10.1073/pnas.2101496118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, W.-I. et al. Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition. PloS ONE5, e12913 (2010). 10.1371/journal.pone.0012913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quambusch, L. et al. Cellular model system to dissect the isoform-selectivity of Akt inhibitors. Nat. Commun.12, 5297 (2021). 10.1038/s41467-021-25512-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laskowski, R. A. & Swindells, M. B. (ACS Publications, 2011).
- 46.Van Der Spoel, D. et al. GROMACS: fast, flexible, and free. J. Comput. Chem.26, 1701–1718 (2005). 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- 47.Lindorff-Larsen, K. et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinform.78, 1950–1958 (2010). 10.1002/prot.22711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. Development and testing of a general amber force field. J. Comput. Chem.25, 1157–1174 (2004). 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- 49.Sousa da Silva, A. W. & Vranken, W. F. ACPYPE-Antechamber python parser interface. BMC Res. Notes5, 1–8 (2012). 10.1186/1756-0500-5-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Florová, P., Sklenovsky, P., Banas, P. & Otyepka, M. Explicit water models affect the specific solvation and dynamics of unfolded peptides while the conformational behavior and flexibility of folded peptides remain intact. J. Chem. Theory Comput.6, 3569–3579 (2010). 10.1021/ct1003687 [DOI] [PubMed] [Google Scholar]
- 51.Kumari, R., Kumar, R., Consortium, O. S. D. D. & Lynn, A. g_mmpbsa A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inform. Model.54, 1951–1962 (2014). 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- 52.Grant, B. J., Skjaerven, L. & Yao, X. Q. The Bio3D packages for structural bioinformatics. Protein Sci.30, 20–30 (2021). 10.1002/pro.3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma, M., Sharma, S. & Alawada, A. Understanding the binding specificities of mRNA targets by the mammalian Quaking protein. Nucl. Acids Res.47, 10564–10579 (2019). 10.1093/nar/gkz877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Healy, F. M., Prior, I. A. & MacEwan, D. J. The importance of Ras in drug resistance in cancer. British J. Pharmacol.179, 2844–2867 (2022). 10.1111/bph.15420 [DOI] [PubMed] [Google Scholar]
- 55.Galiè, M. RAS as supporting actor in breast cancer. Front. Oncol.9, 1199 (2019). 10.3389/fonc.2019.01199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, J. et al. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol. Cancer18, 26 (2019). 10.1186/s12943-019-0954-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi, X. et al. Research progress on the PI3K/AKT signaling pathway in gynecological cancer. Mol. Med. Rep.19, 4529–4535 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chi, D. et al. Estrogen receptor signaling is reprogrammed during breast tumorigenesis. Proc. Natl. Acad. Sci.116, 11437–11443 (2019). 10.1073/pnas.1819155116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang, W., Wang, X., Zhang, C., Xue, L. & Yang, L. Expression and clinical significance of MAPK and EGFR in triple-negative breast cancer. Oncol. Lett.19, 1842–1848 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor, S. et al. Evaluating the evidence for targeting FOXO3a in breast cancer: a systematic review. Cancer Cell Int.15, 1. 10.1186/s12935-015-0156-6 (2015). 10.1186/s12935-015-0156-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dansen, T. B. & Burgering, B. M. Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Boil.18, 421–429 (2008). 10.1016/j.tcb.2008.07.004 [DOI] [PubMed] [Google Scholar]
- 62.Van De Waterbeemd, H. & Gifford, E. ADMET in silico modelling: towards prediction paradise?. Nat. Rev. Drug Discov.2, 192–204 (2003). 10.1038/nrd1032 [DOI] [PubMed] [Google Scholar]
- 63.Gurbani, D. et al. Structure and characterization of a Covalent inhibitor of Src kinase. Front. Mol. Biosci.7, 81. 10.3389/fmolb.2020.00081 (2020). 10.3389/fmolb.2020.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saeed, M. et al. Cytotoxicity of the sesquiterpene lactones neoambrosin and damsin from Ambrosia maritima against multidrug-resistant cancer cells. Front. Pharmacol.6, 267 (2015). 10.3389/fphar.2015.00267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Getlik, M. et al. Hybrid compound design to overcome the gatekeeper T338M mutation in cSrc. J. Med. Chem.52, 3915–3926 (2009). 10.1021/jm9002928 [DOI] [PubMed] [Google Scholar]
- 66.Sarker, D. K., Ray, P., Rouf, R., Shilpi, J. A. & Uddin, S. J. In Silico Molecular docking and dynamic investigations of bioactive phytoconstituents from fenugreek seeds as a potent drug against DPP-IV Enzyme. ACS Food Sci. Technol.3, 1423–1439 (2023). 10.1021/acsfoodscitech.3c00102 [DOI] [Google Scholar]
- 67.Lobanov, M. Y., Bogatyreva, N. & Galzitskaya, O. Radius of gyration as an indicator of protein structure compactness. Mol. Biol.42, 623–628 (2008). 10.1134/S0026893308040195 [DOI] [PubMed] [Google Scholar]
- 68.Bai, F. et al. Free energy landscape for the binding process of Huperzine A to acetylcholinesterase. Proc. Natl. Acad. Sci.110, 4273–4278 (2013). 10.1073/pnas.1301814110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kenny, P. W. Hydrogen-bond donors in drug design. J. Med. Chem.65, 14261–14275 (2022). 10.1021/acs.jmedchem.2c01147 [DOI] [PubMed] [Google Scholar]
- 70.Wade, R. C. & Goodford, P. J. The role of hydrogen-bonds in drug binding. Progr. Clin. Biol. Res.289, 433–444 (1989). [PubMed] [Google Scholar]
- 71.Wang, C., Greene, D. A., Xiao, L., Qi, R. & Luo, R. Recent developments and applications of the MMPBSA method. Front. Mol. Biosci.4, 87 (2018). 10.3389/fmolb.2017.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jha, V., Macchia, M., Tuccinardi, T. & Poli, G. Three-dimensional interactions analysis of the anticancer target c-Src kinase with its inhibitors. Cancers12, 2327 (2020). 10.3390/cancers12082327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khan, M. A., Jain, V. K., Rizwanullah, M., Ahmad, J. & Jain, K. PI3K/AKT/mTOR pathway inhibitors in triple-negative breast cancer: a review on drug discovery and future challenges. Drug Discov. Today24, 2181–2191 (2019). 10.1016/j.drudis.2019.09.001 [DOI] [PubMed] [Google Scholar]
- 74.Kulkoyluoglu-Cotul, E., Arca, A. & Madak-Erdogan, Z. Crosstalk between estrogen signaling and breast cancer metabolism. Trends Endocrinol. Metabolism30, 25–38 (2019). 10.1016/j.tem.2018.10.006 [DOI] [PubMed] [Google Scholar]
- 75.Wright, K. L. et al. Ras signaling is a key determinant for metastatic dissemination and poor survival of luminal breast cancer patients. Cancer Res.75, 4960–4972 (2015). 10.1158/0008-5472.CAN-14-2992 [DOI] [PubMed] [Google Scholar]
- 76.Bullock, M. FOXO factors and breast cancer: outfoxing endocrine resistance. Endocrine-Related Cancer23, R113–R130 (2016). 10.1530/ERC-15-0461 [DOI] [PubMed] [Google Scholar]
- 77.Rivera-Torres, J. & San José, E. Src tyrosine kinase inhibitors: new perspectives on their immune, antiviral, and senotherapeutic potential. Front. Pharmacol.10, 460745 (2019). 10.3389/fphar.2019.01011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song, M., Bode, A. M., Dong, Z. & Lee, M.-H. AKT as a therapeutic target for cancer. Cancer Res.79, 1019–1031 (2019). 10.1158/0008-5472.CAN-18-2738 [DOI] [PubMed] [Google Scholar]
- 79.De Vivo, M., Masetti, M., Bottegoni, G. & Cavalli, A. Role of molecular dynamics and related methods in drug discovery. J. Med. Chem.59, 4035–4061 (2016). 10.1021/acs.jmedchem.5b01684 [DOI] [PubMed] [Google Scholar]
- 80.Baby, K. et al. Computational drug repurposing of Akt-1 allosteric inhibitors for non-small cell lung cancer. Sci. Rep.13, 7947 (2023). 10.1038/s41598-023-35122-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen, J., Elfiky, A., Han, M., Chen, C. & Saif, M. W. The role of Src in colon cancer and its therapeutic implications. Clin. Colorectal Cancer13, 5–13 (2014). 10.1016/j.clcc.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 82.Zhong, W. et al. SHC4 orchestrates β-catenin pathway-mediated metastasis in triple-negative breast cancer by promoting Src kinase autophosphorylation. Cancer Lett.582, 216516 (2024). 10.1016/j.canlet.2023.216516 [DOI] [PubMed] [Google Scholar]
- 83.Zou, H. et al. Tyrosine phosphorylation-mediated YAP1-TFAP2A interactions coordinate transcription and trastuzumab resistance in HER2+ breast cancer. Drug Resist. Updates73, 101051 (2024). 10.1016/j.drup.2024.101051 [DOI] [PubMed] [Google Scholar]
- 84.Riggio, M. et al. AKT1 and AKT2 isoforms play distinct roles during breast cancer progression through the regulation of specific downstream proteins. Sci. Rep.7, 44244 (2017). 10.1038/srep44244 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All software is freely available, and the manuscript and supporting information file contain the data used to reach the conclusion.