Abstract
XGH (xylogalacturonan hydrolase; GH 28) is an enzyme that is capable of degrading XGA (xylogalacturonan), which is a polymer of α-D-galacturonic acid, highly substituted with β-D-xylose. XGA is present in cell walls of various plants and exudates, such as gum tragacanth. XGA oligosaccharides were derived from an XGH digestion of gum tragacanth, then fractionated, and analysed for their sugar composition and structure by matrix-assisted laser-desorption ionization–time-of-flight MS and nanospray MS. Several oligosaccharides from XGA were identified with different galacturonic acid/xylose ratios including five oligosaccharide isomers. Although XGH can act as an endo-enzyme, product-progression profiling showed that the disaccharide GalAXyl was predominantly produced from XGA by XGH, which indicated also an exolytic action. The latter was further supported by degradation studies of purified oligosaccharide GalA4Xyl3. It was shown that XGH acted from the non-reducing end towards the reducing end of this oligosaccharide, and showed the processive character of XGH. The results from this study further show that although XGH prefers to act between two xylosidated GalA units, it tolerates unsubstituted GalA units in its −1 and +1 subsites.
Keywords: endo-xylogalacturonan hydrolase, gum tragacanth, oligosaccharide, pectin, product-progression profile, xylogalacturonan
Abbreviations: HPAEC, high-performance anion-exchange chromatography; MALDI–TOF-MS, matrix-assisted laser-desorption ionization–time-of-flight mass spectrometry; PG, polygalacturonase; PSD, post-source decay; XGA, xylogalacturonan; XGH, XGA hydrolase
INTRODUCTION
Pectins are complex and highly heterogeneous polysaccharides found in primary cell walls and intercellular regions of higher plants. These biopolymers contain α-(1→4)-linked D-galacturonic acid chains, called the smooth regions of pectins. Besides these linear chains, pectin further consists of the branched polysaccharides rhamnogalacturonan I, rhamonogalacturonan II and XGA (xylogalacturonan), which together are referred to as the hairy regions [1].
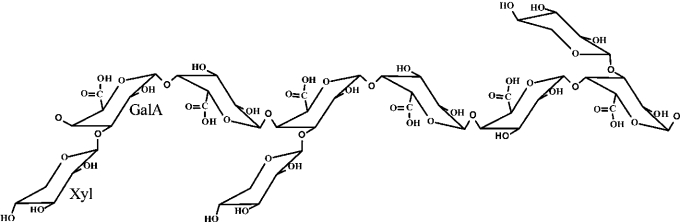
XGA is an α-(1→4)-linked D-galacturonic acid chain, which is highly substituted with β-D-xylose at the C-3 position (Figure 1) [2]. This high-molecular mass polymer, XGA, is present in various plant cell walls, for instance from peas, soya beans, apples, onions, cotton seeds and watermelons, and in exudates such as gum tragacanth from the Astralagus species [3].
Figure 1. Schematic diagram showing the structure of XGA.
Pectins play an important role in the food industry due to their excellent gelling, thickening and stabilizing properties. They are further believed to lower blood cholesterol levels, to protect the gastrointestinal tract and to stimulate the immune system [3,4].
In some industrial food processes, such as the enzymatic clarification of fruit juices, it is crucial that pectins are completely degraded. For this process, several pectinases are available and many of these enzymes have been characterized from a wide variety of microorganisms. Among the most commonly used pectinases are those produced by black aspergilli-like Aspergillus niger. This organism is capable of secreting high levels of enzymes and its products are generally regarded as safe, allowing them to be used in food applications [5,6].
Several pectinases have been isolated from A. niger such as endo-PGs (polygalacturonases), endo-pectate lyases, endo-pectin lyases, pectin methyl esterases, which act on the smooth region of pectin, as well as rhamnogalacturonan acetylesterase, rhamnogalacturonases, arabinases and galactanases that act on the hairy regions of pectin. Most of these enzymes have been thoroughly characterized with respect to their mode of action on their corresponding substrates [7–10].
Despite the rich collection of pectinases in technical enzyme preparations, obtained from A. niger, parts of the hairy regions of pectin remain resistant to degradation and cause membrane fouling in the ultrafiltration process for fruit juice clarification [10]. Being part of the pectic hairy regions, fouling is largely influenced by XGA. Only a little is known about pectinases that can degrade XGA, with the exception of exo-PGs from Aspergillus sp. [9,11,12]. Recently, the enzyme XGH (XGA hydrolase) was discovered in Aspergillus tubingensis, which acts specifically on XGA by cleaving the galacturonic acid backbone in an endo-manner [5,13]. This enzyme belongs to the pectin degrading glycoside hydrolase family 28 (GH 28), which includes PGs and exo-PGs, based on its amino acid sequence similarity to this family [14] [see also Coutinho, P. M. and Henrissat, B. (1999) Carbohydrate-Active Enzymes server at URL: http://afmb.cnrs-mrs.fr/CAZY/]. In addition, the amino acid sequence of XGH contains functionally important residues equivalent to the four conserved active-site segments of PGs [15,16]. All family (GH 28) members, including XGH, act with an inverting mechanism [17].
XGH has been characterized with respect to its kinetic parameters, temperature and pH effects and degradation of differently substituted (xylo-)galacturonans [11].
In the present study, we report on the mode of action of XGH on its substrate XGA. This was realized by the degradation of XGA by XGH, followed by fractionation and quantification and analysis of the oligosaccharides obtained for their sugar abundance, composition and structure. Furthermore, the degradation of a defined-XGA oligosaccharide was studied in a time course to determine the direction of action of XGH.
EXPERIMENTAL
Substrate
Two XGAs derived from acid-modified and alkali-saponified gum tragacanth were used. These XGAs have a Xyl/GalA ratio of 0.29 (XGA-29) and 0.47 (XGA-47) respectively [11].
Enzyme
The enzyme endo-XGH from A. tubingensis was cloned [5] and expressed in the A. niger ‘PlugBug’ of DSM Food Specialities (Delft, the Netherlands). This crude enzyme preparation was purified [11]. The crude and purified enzyme preparation had a specific activity of 77 and 150 units/mg respectively.
Enzyme incubations
XGH was used to digest XGA-29 (final concentration 1 mg/ml) for 1 h at 30 °C. The final enzyme concentration was 3.17 μg/ml. The substrate was dissolved in water in a total volume of 10 ml and had a final pH of 3.6 without additional buffering. The enzyme was inactivated by heating the reaction mixture for 10 min at 100 °C. The digested XGA was concentrated to 5 mg/ml, by freeze-drying and redissolving in water, and was analysed by high-performance size-exclusion chromatography as described previously [18]. Subsequently, the digest was analysed by HPAEC (high-performance anion-exchange chromatography) at pH 12 using pulsed amperometric detection as described previously [19]. The elution of the XGA oligosaccharides by HPAEC was adapted as follows: a combination of two linear gradients was used starting with 0–600 mM sodium acetate in 100 mM NaOH for 50 min, followed by 600–1000 mM sodium acetate in 100 mM NaOH for 5 min.
Preparation of XGA oligosaccharides
For the preparation of XGA oligosaccharides, 300 mg of XGA-29 was digested, as described above, and concentrated to 25 mg/ml before fractionation by HPAEC. All fractions were desalted by H+-Dowex AG 50W X8 (BioRad, Hercules, CA, U.S.A.) treatment. Subsequently, the desalted fractions were analysed by HPAEC for their purity and compared with the complete XGA digest for assignment. A GalA3 standard (5 mg/ml) was used for quantification.
MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight)-MS
The fractionated XGA oligosaccharides were analysed by MALDI–TOF as described previously [18]. Two types of MS spectrometers were used: Voyager-DE RP Biospectrometry work-station (PerSeptive Biosystems, Framingham, MA, U.S.A.), and Ultraflex TOF MS (Bruker Daltonics, Hamburg, Germany). The concentration of the XGA oligosaccharides ranged from 0.25 to 1 mg/ml. The samples were mixed with a matrix solution (1 μl of sample in 1 μl of matrix) on a silver plate. The matrix solution was prepared by dissolving 10 mg of 2,5-dihydroxybenzoic acid in 1 ml mixture of acetonitrile/water (500:500 μl).
XGA oligosaccharides were identified based on their theoretical m/z values. These values were based on the GalA/Xyl ratios, assuming total protonation of GalA and addition of one Na+ or K+.
Labelling
Labelling of the individual XGA oligosaccharides with 18O at their reducing ends was performed as described in [20]. The XGA samples were incubated at 40 °C for 12 days. The progress of labelling was followed by MALDI–TOF-MS and the labelled XGA oligosaccharides were further structurally characterized by PSD (post-source decay) using the Ultraflex-TOF (Bruker Daltonics).
Nanospray MS
Dynamic nanospray MS was performed on an LCQ ion-trap (Finnigan MAT 95, San Jose, CA, U.S.A.) as described previously [18]. For MS analysis of XGA oligosaccharides, settings were adapted as follows: the flow rate was set at 5 μl/min, the spray voltage at 4.5 kV, and a 20–30% of relative collision energy for MS2 and higher was applied.
Product-progression profiling of XGH
An XGA-29 solution (1 mg/ml) was incubated with purified XGH (final concentration 0.35 μg/ml) at 30 °C. The digest had a total volume of 20 ml. At different time intervals (0–48 h), samples were taken and incubated for 10 min at 100 °C to terminate the reaction. An extra enzyme dose (final concentration 0.70 μg/ml) was added to the digest after 24 h.
By freeze-drying and redissolving in water, samples were concentrated five times more and were subsequently analysed by HPAEC along with identified XGA oligosaccharides and GalA3 (5 mg/ml).
Preparation and degradation of oligosaccharide GalA4Xyl3
Another batch of XGA from gum tragacanth batch, similar to XGA-47, as described by Beldman et al. [11], was degraded by XGH under the same conditions as described above. This digest was fractionated by HPAEC as described before and was isolated.
GalA4Xyl3 was labelled and structurally characterized by MALDI–TOF PSD. Subsequently, GalA4Xyl3 was degraded with purified XGH under the conditions described above in the Enzyme incubations subsection. At different intervals (0–24 h), a sample was taken and incubated for 10 min at 100 °C to terminate the reaction. An extra enzyme dose (final concentration 0.70 μg/ml) was added to the digest after 8 h. The progress of degradation in all samples was analysed as described in [18] by MALDI–TOF-MS (Ultraflex TOF MS; Bruker Daltonics).
RESULTS AND DISCUSSION
HPLC analysis of XGA digest
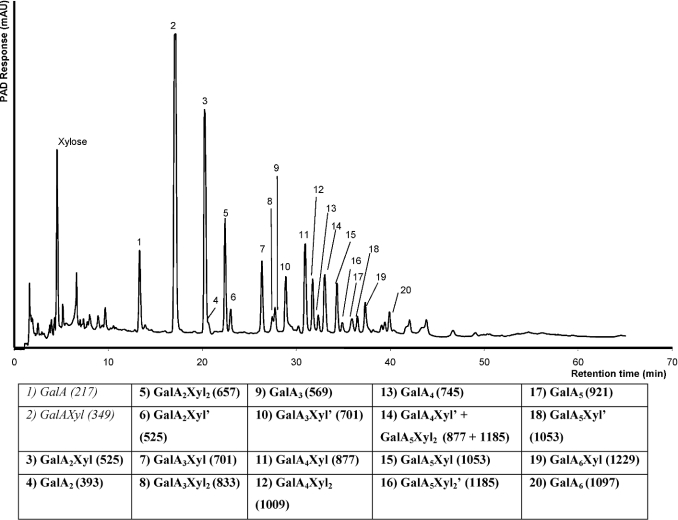
XGA-29 was incubated with endo-XGH for 24 h and subsequently analysed by high-performance size-exclusion chromatography. An endo-type of degradation of XGA was observed as shown previously [11], although analysis by HPAEC (Figure 2) revealed the presence of more XGA oligosaccharides when compared with the earlier reports. This is explained by the application of higher concentrations of the degraded substrate on to the column. In addition to the previously observed oligosaccharides, high-molecular mass XGA products (shown as peaks 5–20) were noticeable after 1 h of enzymatic degradation of XGA.
Figure 2. HPAEC elution profile of an XGA-29 digest.
The included table summarizes the newly identified XGA oligosaccharides (bold; see text for details). The m/z values of the XGA oligosaccharides (including the addition of an H+ and Na+) are shown in parentheses. The accentuated XGA oligosaccharides (′) are isomers with different distribution of xylose over the GalA backbone.
Identification of XGA oligosaccharides
An XGA-29 digest was preparatively fractionated by HPAEC to isolate the individual oligomeric products. All fractions were subjected to HPAEC again to check their purity. By MALDI–TOF-MS these purified XGA oligosaccharides were analysed for their sugar composition. Figure 2 shows the HPAEC profile of the XGA digest from which eighteen XGA oligosaccharides were identified. The previously observed products Xyl and GalA, as well as the dimer GalAXyl were also observed.
The identified XGA oligosaccharides have various GalA/Xyl ratios. In addition, five pairs of oligosaccharide isomers were found to have a similar sugar composition but a different elution time on HPAEC analysis. This can be explained by a different distribution of Xyl over the GalA backbone as the xylose side chain may be linked to a different GalA residue or to a different–OH group (C-2 or C-3) on the same GalA residue. These pairs of oligosaccharide isomers are shown as peaks 3 and 6, 7 and 10, 11 and 14, 14 and 16, and 15 and 18. To avoid confusion, the isomers are identified as shown in the table of Figure 2.
A measurable amount of xylose was present in the digest. This indicates that some xylose was released by enzymatic or chemical hydrolysis from XGA and/or the oligosaccharides during the digestion. The total amount of free xylose however was estimated to be only approx. 2% of the total amount present in the substrate.
Structural characterization of XGA oligosaccharides
Oligosaccharides GalA2Xyl, GalA2Xyl′, GalA3Xyl, GalA3Xyl2 and GalA3Xyl′ as shown in Figure 2 were further structurally analysed. The oligosaccharide GalA4Xyl3 (derived from XGA-47) was also structurally analysed and used for a detailed degradation analysis by XGH (see below).
For structural characterization, these oligosaccharides were 18O-labelled at their reducing end by acid-catalysed exchange in H218O, which increases their size in m/z 2-fold. All labelled XGA oligosaccharides and their corresponding fragments that contain the label are indicated by an asterix.
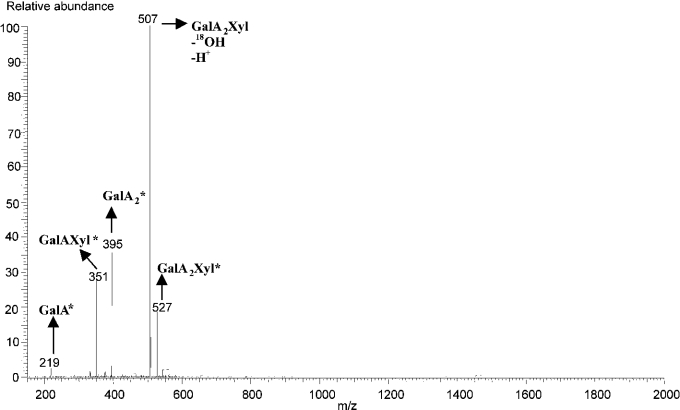
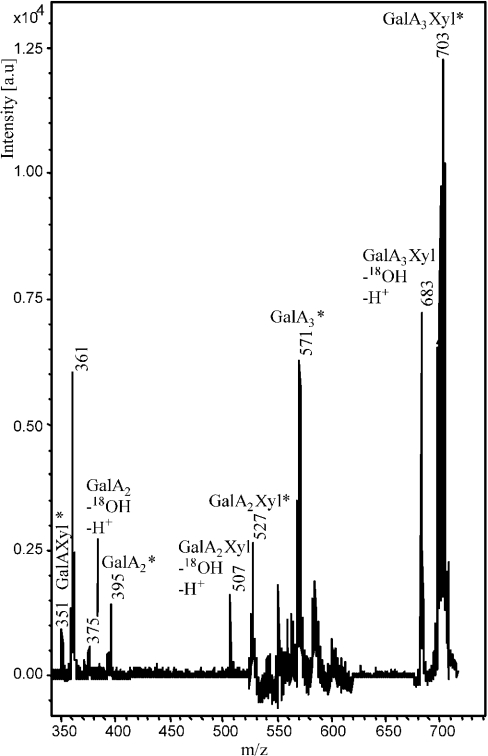
The labelled oligosaccharides GalA2Xyl* and GalA2Xyl′* were subjected to nanospray MS. Figure 3 shows an MS2 spectrum of the fragmented parent ion at m/z 527, corresponding to the 18O-labelled oligosaccharide GalA2Xyl. The produced fragments were [GalA*] (m/z 219), [GalAXyl*] (m/z 315), [GalA2*] (m/z 395) and [GalA2Xyl–OH–H+] (m/z 507). The presence of fragment GalAXyl* showed that xylose was substituted at the reducing GalA unit.
Figure 3. Nanospray MS/MS analysis of GalA2Xyl* (m/z 527).
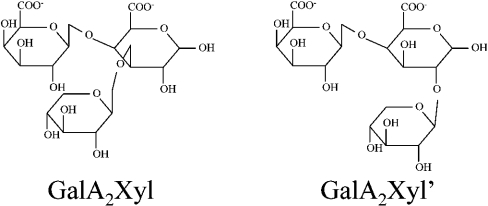
In the MS2 spectrum of GalA2Xyl′* also, the fragment GalAXyl* (m/z 351) was found, which showed that in this oligosaccharide the xylose was also present at the reducing GalA. However, the MS2 spectra showed a different ratio of fragments from GalA2Xyl* and GalA2Xyl′*. This difference in fragmentation might be explained by different xylose linkages at the reducing GalA unit for these two oligosaccharides. Although xylose is known to be attached at the C-3 position of GalA in XGA, the presence of other types of linkages could occur, yet in small proportions [2]. Therefore we suggest that the major product GalA2Xyl, shown as peak 3 in Figure 2, carries xylose at the C-3 position of the reducing GalA unit, whereas the minor product GalA2Xyl′, shown as peak 10, carries xylose at the C-2 position. The proposed structures of GalA2Xyl and GalA2Xyl′ are presented in Figure 4.
Figure 4. Proposed structures of XGA oligosaccharides GalA2Xyl and GalA2Xyl′.
It is quite probable that the difference in substitution with xylose at the reducing GalA of GalA2Xyl and GalA2Xyl′ causes a different elution on HPAEC. A similar phenomenon has been reported before for xylan oligosaccharides substituted with arabinose at either C-2 or C-3 of the xylose residue [21].
The other labelled oligosaccharides were analysed by MALDI–TOF using PSD. Figure 5 shows a PSD spectrum of the (fragmented) parent ion at m/z 703, corresponding to GalA3Xyl*. In Table 1 the results are summarized for fragments that were observed with PSD of GalA3Xyl* as well as of GalA3Xyl′*, GalA3Xyl2 and GalA4Xyl3*.
Figure 5. MALDI–TOF PSD analysis of GalA3Xyl* (m/z 703).
Table 1. Observed fragments derived from MALDI-TOF PSD spectra of labelled oligosaccharides GalA3Xyl and GalA3Xyl′, GalA4Xyl3 and unlabelled oligosaccharide GalA3Xyl2 along with the drawn structures.
●, GalA; X, xylose; ○, reducing GalA; *, the label at the reducing end of the oligosaccharide. Numbers in parentheses represent m/z values.
The derived structures were rationalized as follows. GalA3Xyl*: as shown in Figure 5 and Table 1, the presence of the GalAXyl* fragment indicates xylose substitution at the reducing GalA unit of GalA3Xyl*. GalA3Xyl′*: PSD analysis of this oligosaccharide resulted in the appearance of fragment GalAXyl, but did not reveal fragments GalAXyl* and GalA2Xyl*. The absence of the latter two fragments indicates that no xylose substitution occurs at the reducing and internal GalA units. Therefore possibilities other than xylose substitution at the non-reducing end of GalA3Xyl′* can be excluded. GalA3Xyl2: labelling of this oligosaccharide failed for unknown reasons; however, PSD analysis could resolve the structure of this unlabelled oligosaccharide. The absence of the GalA2Xyl2 fragment demonstrated that the xyloses are not substituted to two adjacent GalA units. Therefore the only possibility was the substitution of the reducing and non-reducing ends of GalA3Xyl2. GalA4Xyl3*: PSD analysis on GalA4Xyl3* showed the absence of GalAXyl*, which suggests a free-reducing GalA unit. Therefore each of the other three GalA units must be substituted with a xylose.
Product-progression profiling of XGH
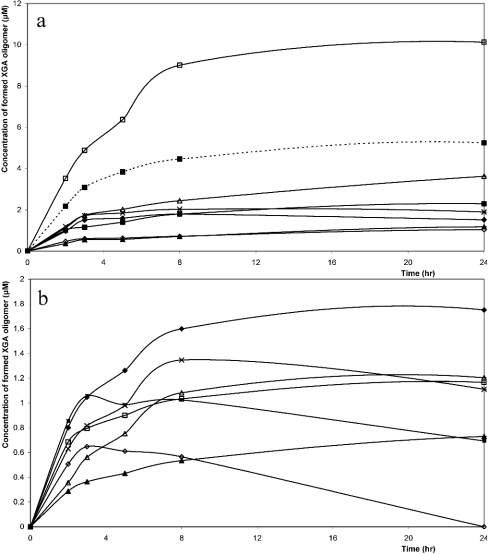
The appearance of products during the degradation of XGA by XGH was studied. This was performed by degrading XGA-29 with purified XGH for different time intervals followed by HPAEC analysis and quantification (Figure 6). The co-eluting XGA oligosaccharides were plotted together in the product-progression profiles.
Figure 6. Product-progression profiles of XGH on the XGA analogue.
(a) GalA (△), GalAXyl (□), GalA2Xyl (---■---), GalA2 (◇), GalA2Xyl2 (×), GalA2Xyl′ (▲), GalA3Xyl (■), GalA3Xyl2+GalA3 (◆); (b) GalA3Xyl′ (△), GalA4Xyl (◆), GalA4Xyl2 (■), GalA4 (▲), GalA4Xyl′+GalA5Xyl2 (×), GalA5Xyl (□), GalA5Xyl2′ (◇). Note the different scaling of (a) and (b).
It was assumed that the response factors of the individual oligosaccharides under the conditions of HPAEC were not substantially different from each other. This was based on the HPAEC analysis of GalA and GalA oligosaccharides with a degree of polymerization of 2–5 in equal molar concentrations for which no substantial differences in response factors were found (results not shown). In addition, from the literature it is known that only a small difference exists in the response factor for GalA and GalAXyl [9]. This demonstrates that xylose substitution has no remarkable effect on the response factor of XGA oligosaccharides.
The product-progression profiles show a predominant production of GalAXyl, with a minor production of linear oligosaccharides like GalA2, GalA3 and GalA4, indicating that XGH prefers to act between two xylosylated GalA units. This implies that subsites −1 and +1 of XGH have a high affinity for xylosylated GalA units. Linear oligosaccharides were not degraded after prolonged digestion, which shows the dependence of XGH for xylose substitution as well as the absence of other galacturonosidases (i.e. exo-PG). To our knowledge, only exo-PGs from Aspergillus sp. are known to accept xylose-substituted galacturonic acid in its subsite −1, because they produce the dimer GalAXyl from XGA [12,22]. Although exo-PG has a pronounced sequence similarity to XGH, this latter enzyme contains only a part of the conserved regions characteristic of exo-PGs. This indicates the uniqueness of XGH [16].
XGH can also act between two GalA units of which one is xylosylated. This is supported by the production of oligosaccharides, such as GalA3Xyl and GalA3Xyl′ (peaks 7 and 10 in Figure 2 respectively), that have no xylose substitution at the non-reducing or at the reducing end respectively (Table 1).
As shown in Figure 6, GalA4Xyl2 was further degraded, which demonstrates that XGH prefers to act on XGA oligosaccharides with a backbone of at least four GalA units. In addition, GalA5Xyl2′ was degraded more efficiently than GalA4Xyl2, which demonstrates that an increase in the GalA backbone by one GalA unit significantly increases the XGH activity. Furthermore, the rapid degradation of GalA5Xyl2′ is in line with the decrease in the GalA4Xyl′–GalA5Xyl2 pair in the progression profiles. GalA4Xyl and GalA5Xyl were not further degraded, which demonstrates that increased xylose substitution enhances XGH activity.
Degradation of GalA4Xyl3
To study the direction of action of XGH, a highly substituted oligosaccharide with at least four GalA units in the backbone that can potentially be hydrolysed more than once by XGH is required. Furthermore, this oligosaccharide should have an asymmetrical structure with respect to xylose substitution for the recognition of the reducing end. As none of the oligosaccharides that were produced from XGA-29 met this requirement, XGA-47 with a higher degree of substitution with xylose was used to prepare the desired oligosaccharide. Partial degradation of this substrate resulted in the production of significant amounts of oligosaccharide GalA4Xyl3, which appeared suitable for further degradation studies. This oligosaccharide was purified to near homogeneity (>95%) from the XGA digest by preparative HPAEC and structurally characterized by MALDI–TOF PSD as described in the previous subsection.
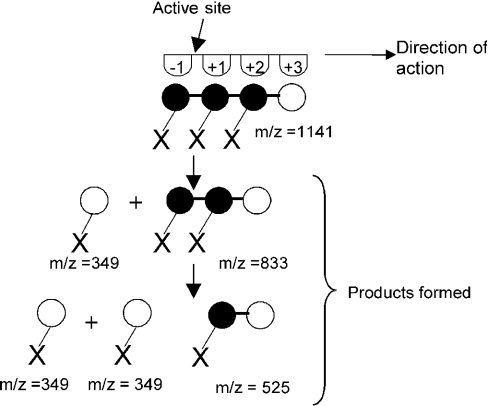
GalA4Xyl3 was degraded with purified XGH for different time intervals followed by MALDI–TOF-MS analysis (results not shown). Degradation of the oligosaccharide GalA4Xyl3 for 5 min resulted in the formation of significant amounts of GalAXyl and GalA3Xyl2. After 24 h of incubation, the main products left were GalA2Xyl and GalAXyl. This shows that XGH acts mainly from the non-reducing end of GalA4Xyl3 towards the reducing end, cleaving off GalAXyl step-by-step. Furthermore, the smallest substrate hydrolysable appeared to be GalA3Xyl2. Since a small amount of GalA2Xyl2 was also produced, it can be concluded that XGH also acts, however less favourably, at the internal glycosidic bond of GalA4Xyl3. This mode of action is depicted schematically in Figure 7. The number of four subsites has been taken arbitrarily.
Figure 7. Proposed model of the action mechanism of XGH towards GalA4Xyl3.
●, GalA; X, xylose; ○, reducing GalA. The number of subsites is arbitrarily taken as four.
Although XGH is recognized as an endo-enzyme, it primarily behaves in an exolytic way during degradation of XGA. Unlike endo-PGs that are known to attack the GalA chain randomly [8,23], XGH acted on GalA4Xyl3 from the non-reducing end towards the reducing end, which is indicative of an exo-acting enzyme [8,9,24].
The exo-character is also in accord with the higher sequence homology of XGH to exo-PGs when compared with endo-PGs [5]. The stepwise release of GalAXyl from GalA4Xyl3 suggests a processive behaviour of XGH.
Acknowledgments
This research was supported by Senter IOP (Innovatief Onderzoeks Programma), The Netherlands (IGE01021).
References
- 1.Vincken J.-P., Schols H. A., Ooomen R. J. F. J., Beldman G., Visser R. G. F., Voragen A. G. J. Pectin-the hairy thing. In: Voragen A. G. J., Schols H., Visser R., editors. Advances in Pectin and Pectinase Research. Dordrecht: Kluwer Academic Publishers; 2003. pp. 47–59. [Google Scholar]
- 2.Aspinall G. O., Baillie J. Gum tragacanth. Part I. Fractionation of the gum and the structure of tragacanthic acid. J. Chem. Soc. 1963;318:1702–1714. [Google Scholar]
- 3.Thibault J.-F., Ralet M.-C. Pectins, their origin, structure and functions. In: McCleary B. V., Prosky L., editors. Advanced Dietary Fibre Technology. Oxford: Blackwell Science; 2001. pp. 369–378. [Google Scholar]
- 4.Yamada H. Contribution of pectins on health care. In: Visser J., Voragen A. G. J., editors. Pectins and Pectinases. Amsterdam: Elsevier; 1996. pp. 173–190. [Google Scholar]
- 5.Van der Vlugt-Bermans C. J. B., Meeuwsen P. J. A., Voragen A. G. J., van Ooyen A. J. J. Endo-xylogalacturonan hydrolase, a novel pectinolytic enzyme. Appl. Environ. Microbiol. 2000;66:36–41. doi: 10.1128/aem.66.1.36-41.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Vries R. P. Regulation of Aspergillus genes encoding plant cell wall polysaccharide-degrading enzymes; relevance for industrial production. Appl. Microbiol. Biotechnol. 2003;61:10–20. doi: 10.1007/s00253-002-1171-9. [DOI] [PubMed] [Google Scholar]
- 7.Searle-van Leeuwen M. J. F., Van den Broek L. A. M., Schols H. A., Beldman G., Voragen A. G. J. Rhamnogalacturonan acetylesterase: a novel enzyme from Aspergillus aculeatus, specific for the deacetylation of hairy (ramified) regions of pectins. Appl. Microbiol. Biotechnol. 1992;38:347–349. [Google Scholar]
- 8.Benen J. A. E., van Alebeek G.-J. W. M., Voragen A. G. J., Visser J. Mode of action analysis and structure-function relationships of aspergillus niger pectinolytic enzymes. In: Voragen A. G. J., Schols H., Visser R., editors. Advances in Pectin and Pectinase Research. Dordrecht: Kluwer Academic Publishers; 2003. pp. 235–256. [Google Scholar]
- 9.Sakamoto T., Bonnin E., Quemener B., Thibault J.-F. Purification and characterisation of two exo-polygalacturonases from Aspergillus niger able to degrade xylogalacturonan and acetylated homogalacturonan. Biochim. Biophys. Acta. 2002;1572:10–18. doi: 10.1016/s0304-4165(02)00277-5. [DOI] [PubMed] [Google Scholar]
- 10.Vincken J.-P., Voragen A. G. J., Beldman G. Enzymes degrading rhamnogalacturonan and xylogalacturonan. In: Whitaker J. R., Voragen A. G. J., Wong D. W. S., editors. Handbook of Food Enzymology. New York: Marcel Dekker; 2003. pp. 931–941. [Google Scholar]
- 11.Beldman G., Vincken J.-P., Schols H. A., Meeuwsen P. J. A., Herweijer M., Voragen A. G. J. Degradation of differently substituted xylogalacturonans by endoxylangalacturonan hydrolase and endopolygalacturonases. Biocatal. Biotransform. 2003;21:189–198. [Google Scholar]
- 12.Kester H., Benen J., Visser J. The exo-polygalacturonase from Aspergillus tubingensis is also active on xylogalacturonan. Biotechnol. Appl. Biochem. 1999;30:53–57. [PubMed] [Google Scholar]
- 13.Herweijer M. A., Vincken J.-P., Meeuwsen P. J. A., van der Vlught-Bergmans C. J. B., Beldman G., van Ooyen A. J. J., Voragen A. G. J. Endo-xylogalacturonan hydrolase. In: Voragen A. G. J., Schols H., Visser R., editors. Advances in Pectin and Pectinase Research. Dordrecht: Kluwer Academic Publishers; 2003. pp. 257–266. [Google Scholar]
- 14.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coutinho P. M., Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert H. J., Davies G., Henrissat B., Svensson B., editors. Recent Advances in Carbohydrate Bioengineering. Cambridge: The Royal Society of Chemistry; 1999. pp. 3–12. [Google Scholar]
- 16.Markovic O., Janecek S. Pectin degrading glycoside hydrolases of family 28: sequence-structural features, specificities and evolution. Protein Eng. 2001;14:615–631. doi: 10.1093/protein/14.9.615. [DOI] [PubMed] [Google Scholar]
- 17.Henrissat B., Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 18.Kabel M. A., Schols H. A., Voragen A. G. J. Complex xylo-oligosaccharides identified from hydrothermally treated Eucalyptus wood and brewery's spent grain. Carbohydr. Polym. 2002;50:191–200. [Google Scholar]
- 19.Van Alebeek G.-J., Christensen T. M. I. E., Schols H. A., Mikkelsen J. D., Voragen A. G. J. Mode of action of pectin lyase A of Aspergillus niger on differently C6-substituted oligogalacturonides. J. Biol. Chem. 2002;277:25929–25936. doi: 10.1074/jbc.M202250200. [DOI] [PubMed] [Google Scholar]
- 20.Van Alebeek G.-J. W. M., Zabotina O., Beldman G., Schols H. A., Voragen A. G. J. Structural analysis of (methyl-esterified) oligogalacturonides using post-source decay matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Mass Spectrom. 2000;35:831–840. doi: 10.1002/1096-9888(200007)35:7<831::AID-JMS7>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Van Laere K. M. J., Beldman G., Voragen A. G. J. A new arabinofuranohydrolase from Bifidobacterium adolescentis able to remove arabinosyl residues from double-substituted xylose units in arabinoxylan. Appl. Microbiol. Biotechnol. 1997;47:231–235. doi: 10.1007/s002530050918. [DOI] [PubMed] [Google Scholar]
- 22.Beldman G., van den Broek L. A. M., Schols H. A., Searle-van Leeuwen M. J. F., van Laere K. M. J., Voragen A. G. J. An exogalacturonase from Aspergillus aculeatus able to degrade Xylogalacturonan. Biotechnol. Lett. 1996;18:707–712. [Google Scholar]
- 23.Bonnin E., Le G. A., Korner R., Van A. G., Christensen T., Voragen A. G. J., Roepstorff P., Caprari C., Thibault J. F. Study of the mode of action of endopolygalacturonase from Fusarium moniliforme. Biochim. Biophys. Acta. 2001;15:301–309. doi: 10.1016/s0304-4165(01)00141-6. [DOI] [PubMed] [Google Scholar]
- 24.Chellegatti M. A. D., Fonseca M. J. V., Said S. Purification and partial characterization of exopolygalacturonase I from Penicillium frequentans. Microbiol. Res. 2002;157:19–24. doi: 10.1078/0944-5013-00127. [DOI] [PubMed] [Google Scholar]