Abstract
Cytokine-induced expression of SOCS (suppressor of cytokine signalling) molecules is important for the negative regulatory control of STAT (signal transduction and activators of transcription)-dependent cytokine signalling, e.g. for the signal transduction of IL-6 (interleukin-6)-type cytokines through the JAK (Janus kinase)/STAT cascade. STAT activation itself represents an important step in the transcriptional activation of SOCS3 gene expression. However, downstream of the STAT-responsive element, the SOCS3 gene contains a GC-rich element in its 5′-upstream region. The aim of the present study was to investigate the implications of this GC-rich element in the transcriptional control of SOCS3 gene expression. In the present study, we show that mutation of this GC-rich element abolishes IL-6-dependent transcriptional activation of the SOCS3 promoter and that Sp3 (specificity protein 3), a ubiquitously expressed transcription factor, but not Sp1 binds to this GC-rich motif, suggesting that Sp3 is involved in the regulation of SOCS3 expression. The results suggest that Sp3 is important for IL-6-induced transcriptional activation of the SOCS3 (gene) promoter and acts as an enhancer of basal as well as induced transcriptional activity, resulting in enhanced SOCS3 mRNA and protein expression. Mutation of Lys-483, a potential target for Sp3 acetylation, inhibited Sp3-mediated enhancement of SOCS3 mRNA expression and SOCS3 promoter activation, indicating that the acetylation of this lysine residue of Sp3 is important for the enhancing effect of Sp3 on SOCS3 expression.
Keywords: cytokine, gene regulation, signal transduction, SOCS3 gene, Sp3, transcription factor
Abbreviations: DMEM, Dulbecco's modified Eagle's medium; EMSA, electrophoretic mobility-shift assay; Epo, erythropoietin; GADPH, glyceraldehyde-3-phosphate dehydrogenase; gp, glycosylated protein; IL, interleukin; HA, haemagglutinin; JAK, Janus kinase; LIF, leukaemia inhibitory factor; MAPK, mitogen-activated protein kinase; RT, reverse transcriptase; SOCS, suppressor of cytokine signalling; Sp, specificity protein; STAT, signal transduction and activators of transcription; SRE, STAT3-responsive element
INTRODUCTION
IL-6 (interleukin-6) belongs to a family of cytokines comprising factors such as IL-11, oncostatin M, LIF (leukaemia inhibitory factor), cardiotrophin-1 and ciliary neurotrophic factor. Depending on the co-acting factors, members of this cytokine family display pro- as well as anti-inflammatory activities. IL-6-type cytokines are key players in the regulation of many physiological and pathophysiological processes such as haematopoiesis, activation of the acute-phase response and regulation of lymphocyte differentiation. They control the expression of many target genes involved in the regulation of cell survival, apoptosis, cell proliferation and differentiation. Thus effective mechanisms controlling the termination of their cellular signal transduction are as important as the initiation of their signalling pathways.
IL-6 exerts its effects through a receptor complex that is initiated on ligand binding to the IL-6-binding subunit gp80 (glycosylated protein 80) and accomplished by the recruitment of the signal-transducing subunit gp130. Subsequently, merging two of these complexes forms the active IL-6 receptor complex. This is supposed to be a critical step, leading to the autophosphorylation and, therefore, activation of JAK-1 (Janus kinase 1), JAK-2 and Tyk-2, which are constitutively associated with gp130. gp130 subsequently tyrosine-phosphorylated at its cytoplasmic tail then recruits transcription factors of the family of STAT1 (signal transduction and activators of transcription 1) and STAT3 through specific phosphotyrosine–SH2 domain interactions. JAK-1 has been shown to be crucial for the tyrosine phosphorylation of the STAT factors that in turn homo- or heterodimerize and translocate to the nucleus, where they bind to enhancer elements of IL-6-inducible genes [1,2].
Different mechanisms contribute to the negative regulation of the JAK/STAT pathway. Kim and Maniatis [3] demonstrated a proteasome-dependent loss of the activated STAT1 in the nucleus. These findings disagree with those of Haspel et al. [4], who proposed a nuclear phosphatase dephosphorylating activated STAT1, which in turn becomes recycled to the cytoplasm, implicating a shuttling of STAT1 between the nucleus and cytoplasm. Moreover, PIAS (protein inhibitors of activated STATs) represent a family of inhibitory proteins that interact with activated STAT proteins by a highly specific mechanism that has not been completely elucidated so far. The mechanism leads to a loss of DNA-binding activity of the activated STAT proteins [5,6].
Another factor negatively regulating IL-6-induced STAT activation is the tyrosine phosphatase SHP2 (SH2-domain-containing protein tyrosine phosphatase), which, on activation by IL-6, also becomes tyrosine-phosphorylated in a Jak1-dependent manner and is recruited to the cytoplasmic tail of gp130 through specific phosphotyrosine–SH2 domain interactions [7]. In this context, Tyr-759 of the cytoplasmic tail of gp130 has been demonstrated to be critical for SHP2 recruitment and negative regulation of gp130-mediated STAT activation [8–10]. Schmitz et al. [11] demonstrated that another molecule, SOCS3 (suppressor of cytokine signalling 3), but not SOCS1, similarly becomes recruited to the gp130 subunit and mediates its inhibitory action by the Tyr-759 recruitment site of the cytoplasmic tail of gp130 [11]. SOCS1 and SOCS3 are members of a protein family of inhibitors of cytokine signalling [12] that are also referred to as JAK-binding proteins [13] or STAT-induced STAT inhibitors [14]. Members of this protein family contain a central SH2 domain as well as a C-terminal domain called the SOCS box. Depending on the cell type examined, SOCS1 and SOCS3 expressions were found to be rapidly induced by IL-6 [15,16]. Since SOCS1 and SOCS3 inhibit the IL-6-induced phosphorylation of JAKs, gp130 and STAT factors, they are often designated as feedback inhibitors of IL-6 signalling. With respect to recent findings demonstrating that, in the absence of SOCS3, IL-6 strongly suppresses liposaccharide-induced release of IL-12 and TNFα (tumour necrosis factor α) in murine SOCS3-deficient macrophages [17], this rather restricted view of the role of particularly SOCS3 in IL-6 signal transduction should be extended to the function of a modulator of IL-6 signalling. Thus, with respect to IL-6 signal transduction, understanding the mechanisms controlling the expression of SOCS3 is of crucial importance.
The transcriptional regulation of cellular SOCS3 expression is not understood completely. Several studies indicate that members of the STAT family of transcription factors are involved in the regulation of SOCS3 expression by factors such as LIF or growth hormone [14,18,19] and the respective STAT3-binding elements of the SOCS3 promoter have been identified so far [18].
However, downstream of the STAT-responsive elements, the SOCS3 gene contains in its 5′-upstream region a GC-rich element. The aim of the present study was to investigate the role of this element in the regulation of SOCS3 expression and to identify transcription factors binding to this element. The results indicate that this GC-rich element is involved in basal and induced transcriptional activation of the SOCS3 promoter and that Sp3 (specificity protein 3), a ubiquitously expressed transcription factor, but not Sp1 binds to this GC-rich motif.
EXPERIMENTAL
Materials
High prime labelling kit, Taq polymerase and recombinant Epo (erythropoietin) were purchased from Roche Molecular Biochemicals (Mannheim, Germany); restriction enzymes were obtained from New England Biolabs (Frankfurt, Germany); oligonucleotides were obtained from MWG-Biotech (Ebersberg, Germany); DMEM (Dulbecco's modified Eagle's medium), DMEM nutritional mix F12 and Opti-MEM were from Invitrogen (Karlsruhe, Germany); recombinant human IL-6 was a gift from Dr P. C. Heinrich (Institute of Biochemistry, University of Aachen, Aachen, Germany); murine IL-6 was from PeproTech; and fetal calf serum was from Perbio Science. The One-Step RT (reverse transcriptase)–PCR kit was from Qiagen (Hilden, Germany). The following antibodies were used: rabbit polyclonal antibody specifically raised against Sp1, Sp3 or STAT3 (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.). Monoclonal antibody against GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was purchased from Biodesign (Saco, ME, U.S.A.). Monoclonal antibody against the HA (haemagglutinin) tag was purchased from Babco (Richmond, CA, U.S.A.). Polyclonal antibody specifically raised against acetylated lysine residues was from Abcam (Cambridge, U.K.). Polyclonal SOCS3 antibody was purchased from IBL (Hamburg, Germany).
Cultivation and stimulation of cells
RAW 264.7 cells were cultivated in DMEM, containing 1000 mg glucose/l with glutamax supplemented with 10% (v/v) heat-inactivated fetal calf serum, streptomycin (100 mg/l) and penicillin (60 mg/l). NIH3T3 cells were grown in DMEM containing 4500 mg glucose/l with glutamax supplemented with 10% heat-inactivated fetal calf serum, streptomycin (100 mg/l) and penicillin (60 mg/l).
The human hepatoma cells HepG2 were grown in DMEM/nutritional mix F12 supplemented with 10% fetal calf serum, streptomycin (100 mg/l) and penicillin (60 mg/l). The medium was changed 24 h before experiments were performed.
Nuclear extracts were prepared as described by Andrews and Faller [20]. Protein concentration was determined with a Bio-Rad (Bio-Rad, Munich, Germany) protein assay.
RT–PCR analysis for SOCS3 expression
Transfected NIH3T3 or HepG2 cells were rested in their respective culture medium for approx. 2 days and then stimulated with 1 unit/ml Epo (Roche) for the times indicated. Total RNA was isolated using the RNeasy Mini kit from Qiagen according to the manufacturer's instructions. RT–PCR was performed using 1 μg of total RNA using the One-Step RT–PCR kit from Qiagen according to the manufacturer's instructions. PCR amplification was performed using primer pairs specific for murine and human SOCS3 (upstream primer, 5′-ATGGTCACCCACAGCAAGTT-3′; downstream primer, 5′-TTGTCGGAAGACTGTCAACG-3′) and murine GAPDH (upstream primer, 5′-ACTCCACTCACGGCAAATTC-3′; downstream primer, 5′-ACACATTGGGGGTAGGAACA-3′). The predicted size of the PCR products for SOCS3 and GAPDH were 553 and 573 bp respectively. The optimal number of cycles for RNA of endogenous genes or transcripts from transfected plasmids was determined by Real-time PCR. The PCR products were separated on a 1.5% agarose gel and visualized by ethidium bromide staining.
Plasmids
Standard cloning procedures were performed as outlined by Sambrook and Russel [21]. pGL3-SOCS3-2757Luc containing the promoter region −2757 to +929 of the murine SOCS3 gene fused to the luciferase encoding sequence was kindly provided by Dr S. Melmed (Sinai Research Center, Los Angeles, CA, U.S.A.) and described previously [18]. pGL3-SOCS3-511Luc, -159Luc, -107Luc, -79Luc, -68Luc and -49Luc were generated by PCR and contain the promoter region −511 to +929 and its shortenings of the murine SOCS3 gene fused to the luciferase encoding sequence. Mutations in the SOCS3 promoter-reporter construct and in the Sp3 gene expression plasmid were generated by PCR technique using appropriate oligonucleotides. The sequences of all constructs were controlled by sequencing (MWG-Biotech). The different point mutations introduced into the SOCS3 gene promoter fragment and in the coding sequence of the Sp3 gene are summarized in Table 1. The Epo/gp130 receptor construct was a gift from Dr F. Schaper (Institute of Biochemistry, University of Aachen).
Table 1. Mutations introduced within the SOCS3 promoter or the Sp3 gene.
The wild-type sequences and the mutations introduced (underlined and in boldface) are shown: the mutations introduced within the GC-rich motif and the distal or proximal STAT3-responsive element of the SOCS3 promoter and those mutations introduced into the Sp3 gene to achieve the different lysine mutations.
| Site, region or motif | Sequence |
|---|---|
| Sites | |
| Wild-type distal STAT | 5′-ttacaagaa-3′ |
| Mutated distal STAT | 5′-tcccaggaa-3′ |
| Wild-type proximal STAT | 5′-ttccaggaa-3′ |
| Mutated proximal STAT | 5′-tcccaggaa-3′ |
| Regions | |
| Wild-type GC-rich | 5′-cggggggcggggcg-3′ |
| Mutated GC-rich | 5′-cgggggtagggtcg-3′ |
| Motifs | |
| Wild-type Sp3 IKEE | 5′-atcaaggaagaa-3′ |
| Mutated Sp3 IAEE (K483A) | 5′-atcgcggaagaa-3′ |
| Mutated Sp3 IDEE (K483D) | 5′-atcgatgaagaa-3′ |
Transfection procedure and reporter gene assay
For the reporter gene assay, RAW 264.7 cells were transfected using LIPOFECTAMINE™ 2000 (Invitrogen). Briefly, cells were grown in DMEM containing glucose (1000 mg/l) and supplemented with 10% fetal calf serum on 6-well plates towards 80–95% confluency. Cells were preincubated with 1 ml of Opti-MEM, 1 h before transfection. LIPOFECTAMINE™ 2000 (6 μl) and 3.2 μg of total DNA were prediluted in 250 and 50 μl of Opti-MEM. Diluted reagent and DNA were mixed, incubated for 20 min at room temperature (22–24 °C) and then added to the cells. After 6 h, the medium was changed and incubation was continued in DMEM containing glucose (1000 mg/l), and supplemented with 10% fetal calf serum. Cell lysis and luciferase assays were performed using the dual-luciferase kit (Promega, Madison, WI, U.S.A.) according to the manufacturer's instructions.
Luciferase activity values were normalized to transfection efficiency monitored by the co-transfected Renilla expression vector (Promega). All expression experiments were performed at least in triplicate. Results are presented as percentage of the respective control experiment. Error bars are S.D. Results were analysed using the Student's t test: P<0.05 was considered as statistically significant.
For higher transfection efficiency, cells were transfected using LIPOFECTAMINE™ 2000 and a modified transfection procedure. In brief, 5 μl of LIPOFECTAMINE™ 2000 was diluted in 250 μl of Opti-MEM and 4 μg of DNA was diluted in 50 μl of Opti-MEM. Diluted reagent and DNA were mixed and incubated for 20 min at room temperature. Meanwhile, a confluent flask (75 cm2) of NIH3T3 or HepG2 cells was trypsinized and, after centrifugation, resuspended in their respective culture medium supplemented with 10% fetal calf serum but without antibiotics; subsequently, 300 μl of resuspended cells was added to the transfection mixture. Thereafter, 1 ml of culture medium was added and cells were seeded on a 60 mm culture dish. After 16 h incubation at 37 °C, the medium was replaced by DMEM containing glucose (4500 mg/l) and supplemented with 10% fetal calf serum and cell culture was continued for another 24 h. Thereafter, experiments were performed as outlined in the Results section.
EMSA (electrophoretic mobility-shift assay)
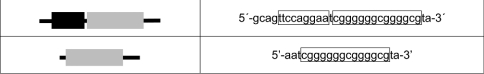
EMSAs were performed as described previously [22]. The protein–DNA complexes were separated on a 4.5% (w/v) polyacrylamide gel containing 7.5% (w/v) glycerol in 0.25× TBE (20 mM Tris base, 20 mM boric acid and 0.5 mM EDTA, pH 8.0) at 20 V/cm for 4 h. Gels were fixed in 10% (v/v) methanol, 10% (v/v) acetic acid and 80% water for 1 h, dried and autoradiographed. Double-stranded 32P-labelled oligonucleotides from the murine SOCS3 promoter were used for EMSA. The sequences used for the oligonucleotides are shown in Figure 1.
Figure 1. Oligonucleotides used as labelled probes for the EMSA.
The STAT-responsive element is depicted as a black box, whereas the GC-rich motif is represented as a grey box. The sequences corresponding to the respective binding sites are marked by a frame.
Immunoprecipitation
For immunoprecipitation, cells grown in a 100 mm dish were stimulated with the respective cytokine at the concentrations indicated. Cells were washed twice with PBS supplemented with 0.1 mM Na3VO4 and solubilized in 1 ml of lysis buffer [1% Triton X-100, 20 mM Tris/HCl (pH 7.4), 136 mM NaCl, 2 mM EDTA, 50 mM β-glycerophosphate, 20 mM sodium pyrophosphate, 1 mM Na3VO4, 4 mM benzamidine, 0.2 mM Pefabloc, 5 μg/ml aprotinin, 5 μg/ml leupeptin and 10% glycerol] at 4 °C. The insoluble material was removed by centrifugation, and the cell lysate was incubated for 3–4 h with specific antibodies and Protein A–agarose (8 mg/ml in lysis buffer) at 4 °C. After centrifugation, the agarose beads were washed three times with wash buffer [0.1% Triton X-100, 20 mM Tris/HCl (pH 7.4), 136 mM NaCl, 2 mM EDTA, 50 mM β-glycerophosphate, 20 mM sodium pyrophosphate, 1 mM Na3VO4, 4 mM benzamidine, 0.2 mM Pefabloc, 5 μg/ml aprotinin, 5 μg/ml leupeptin and 10% glycerol]. The samples were boiled in gel electrophoresis sample buffer and the precipitated proteins were separated by SDS/PAGE (6% gel).
Immunoblotting and immunodetection
For immunoblot analysis, cells were grown in a 60 mm dish and stimulated with the respective cytokine at the concentrations indicated. Cells were washed twice with PBS supplemented with 0.1 mM Na3VO4 and solubilized in 1 ml of the lysis buffer at 4 °C. Protein (50 μg) was subjected to SDS/PAGE (8% gel). The electrophoretically separated proteins were transferred on to PVDF membranes by the semi-dry Western blotting method. Non-specific binding was blocked with 5% (w/v) non-fat dry milk powder in TBS-T [20 mM Tris/HCl (pH 7.4), 137 mM NaCl and 0.1% Tween 20] for 30 min. The blots were incubated overnight at 4 °C or for 2 h at room temperature with primary antibodies at the dilution indicated in TBS-T. After extensive rinsing with TBS-T, blots were incubated with secondary antibodies, goat anti-rabbit IgG or goat anti-mouse IgG conjugated with horseradish peroxidase for 1 h. After further rinsing in TBS-T, the immunoblots were developed with the enhanced chemiluminescence system (ECL®; Amersham Biosciences) following the manufacturer's instructions.
RESULTS
Functional characterization of the elements involved in transcriptional regulation of the SOCS3 gene
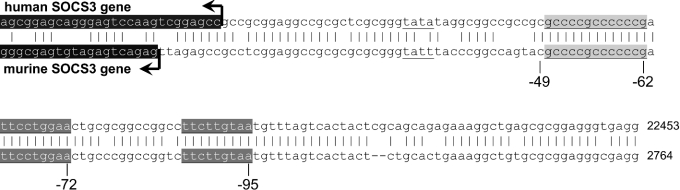
The major evidence for the involvement of STATs in the regulation of SOCS3 expression derives from experiments predominantly performed with LIF, showing that STAT3 binds to a STAT-responsive element of the SOCS3 promoter and is necessary for LIF-induced expression of SOCS3 [18]. As shown in Figure 2, two putative STAT-responsive elements have been identified within the 5′-upstream region of the murine SOCS3 gene mapped to the regions −95 to −87 and −72 to −65 [18].
Figure 2. Comparison of the putative 5′-regulatory region of the human SOCS3 gene with the corresponding sequence of the murine promoter.
Homology screening was performed by BLAST search in the PubMed database using the sequence of the human SOCS3 mRNA (NCBI Entrez Nucleotides database accession no. BC060858) and the 5′-regulatory region of the murine SOCS3 gene (NCBI Entrez Nucleotides database accession no. AF117732). The assumed 5′-upstream regulatory region of the human SOCS3 gene was localized on chromosome 17 (NCBI Entrez Nucleotides database accession no. AC061992). The starting region of the respective human (upper sequence) and murine (lower sequence) transcripts is depicted as black boxes with white letters. The two STAT-binding sites (distal and proximal STAT-responsive elements) are depicted as dark grey boxes with white characters, whereas the GC-rich motif is represented as bright grey boxes with black characters.
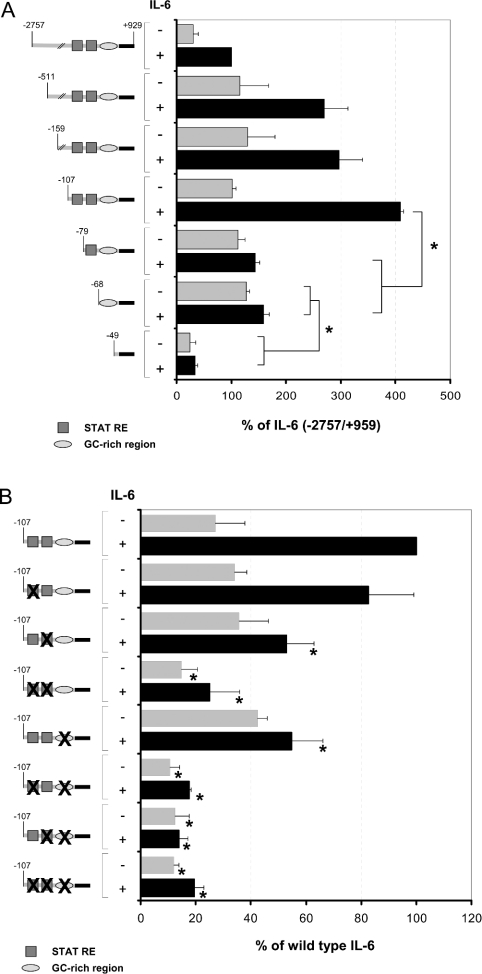
A more detailed analysis of this region, particularly with respect to the GC-rich element located between nt −62 and −49, was performed in the present study. Truncation of the 5′-upstream region of the SOCS3 gene shows that a promoter mutant comprising −107 nt 5′-upstream of the TATA box of the SOCS3 gene is still responsive to IL-6, whereas truncation towards position −79 leads to a promoter mutant that lacks its responsiveness to IL-6 when transfected in RAW 264.7 macrophages (Figure 3A). This indicates that the proximal putative SRE (STAT3-responsive element) located in-between positions −72 and −65 alone is not sufficient to mediate the transcriptional activation induced by IL-6. Furthermore, it can be concluded that the IL-6-responsive region is located downstream of position −107. Further truncation of the promoter to position −68 had no influence on promoter response towards IL-6 stimulation. However, the promoter-reporter gene construct becomes almost completely inactive (Figure 3A) if truncation includes the GC-rich region in-between positions −62 and −49. This suggests that the factors binding to this region are at least responsible for the regulation of the basal transcriptional activity of the SOCS3 promoter controlling the basal expression of the SOCS3 gene.
Figure 3. Differential contribution of the GC-rich motif and the proximal or distal STAT-responsive element to basal or IL-6-mediated activation of the murine SOCS3 promoter.
(A) The murine SOCS3 promoter fused to the firefly luciferase cDNA was stepwise truncated and transfected into RAW 264.7 cells. (B) Mutants of the −107/+929 SOCS3 promoter containing either mutations of the GC-rich element, the proximal or the distal STAT-responsive elements alone or combined mutations of the GC-rich element with the proximal and/or the distal STAT-responsive elements as depicted on the left of the Figure were analysed in reporter gene assays for their responses to IL-6. Therefore the wild-type SOCS3 promoter or the respective mutants were transfected into RAW 264.7 cells. An expression vector for β-galactosidase was co-transfected for monitoring transfection efficiency. Cells were stimulated, 2 days after transfection, with IL-6 (300 units/ml) for 16 h as indicated. Luciferase activity in cellular extracts of these cells was determined and normalized to β-galactosidase activity as described in the Experimental section. The two STAT3-binding sites (distal and proximal STAT-responsive elements) are framed and depicted as boxes in dark grey; the GC-rich element is represented as circles in light grey. The SOCS3 promoter fragment cloned 5′ to the luciferase gene of pBL3Luc is depicted as a black bar. Results are expressed as percentage (means±S.D.) of the IL-6-induced activity of full-length SOCS3 promoter construct. Experiments were performed at least in triplicate. *P<0.05, compared with control experiment.
To study the role of the GC-rich element and the two putative STAT-responsive elements within the 5′-region of the SOCS3 gene in more detail, point mutations were introduced into the respective binding sites. As shown in Figure 3(B), introduction of point mutations within the putative distal SRE has little effect when compared with the influence of those introduced in the proximal SRE of the SOCS3 promoter, resulting in almost complete inhibition of IL-6-mediated transcriptional activation of the SOCS3 promoter. Corresponding to the findings described for LIF-induced SOCS3 gene expression, these results suggest that STAT3 mediates its activating effects mainly through the proximal STAT3-responsive region and not through the distal motif. However, considering that removal of the region located in-between positions −107 and −72 by truncation (Figure 3A) almost completely abrogates IL-6 responsiveness of the respective promoter construct; it is quite probable that the proximal SRE is necessary but not sufficient for the transcriptional activation of the SOCS3 gene promoter by IL-6. The observation that mutation of the proposed distal SRE has almost no influence on basal or induced promoter activity might further indicate that an additional not yet identified factor but not STAT3, which requires the region located in-between positions −107 and −72 for DNA binding, is needed for sufficient transcriptional activation of the promoter by IL-6. The view that STAT3 mediates its major effects on transcriptional activation through the proximal SRE is further supported by the fact that the proximal SRE proves to be a strong STAT3-binding motif when analysed for DNA binding (Figure 4), whereas no DNA binding of STAT3 could be detected if the isolated distal responsive element was used for EMSA (results not shown).
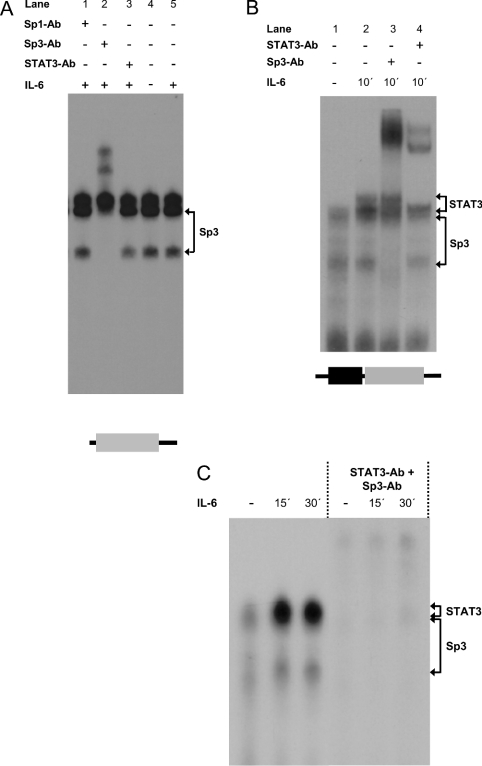
Figure 4. Sp3 binds to the GC-rich region close to the SRE of the SOCS3 promoter.
(A) RAW 264.7 cells were incubated with 300 units/ml IL-6 for the time periods indicated. Nuclear extracts were prepared from these cells and equal amounts of protein were analysed for protein–DNA binding using probes corresponding to the GC-rich element of the SOCS3 promoter. (B, C) RAW 264.7 cells were incubated with 300 units/ml IL-6 for the time period indicated. Nuclear extracts from these cells were analysed for protein–DNA binding using probes comprising both the proximal STAT3 site and the GC-rich element of the 5′-upstream region of the SOCS3 gene. EMSAs were performed as described in the Experimental section. To identify the proteins within the protein–DNA complexes, nuclear extracts were incubated with polyclonal antibodies specific for STAT3α, Sp1 or SP3 as indicated. STAT3–DNA and Sp3–DNA complexes are indicated by arrowheads. STAT3-binding sites are framed and depicted as boxes in dark grey and the GC-rich element is represented by a light grey box.
On the other hand, transcriptional activation of the SOCS3 promoter was similarly impaired on the introduction of a point mutation within the GC-rich element of the 5′-upstream region of the SOCS3 gene. The activity of this construct was only slightly influenced by the introduction of additional point mutations within either the distal or the proximal SRE. These results strongly suggest that STAT3 alone is not sufficient for the transcriptional activation of the SOCS3 gene by IL-6, but at least requires an additional factor mediating its regulatory effects through the functional GC-rich element.
Sp3 binds to the GC-rich region of the SOCS3 promoter and enhances the transcriptional activity of the promoter
GC motifs are known to act as binding motifs for the zinc-finger domain-containing family of transcription factors, Sp1–Sp4. Whereas Sp1 and Sp3 are almost ubiquitously expressed, Sp4 is predominantly neuronal and little is known about the functional implications of Sp2 [23]. With respect to the SOCS3 gene, we focused on Sp1 and Sp3. As shown in Figure 4(A), three different protein–DNA complexes could be identified when nuclear extracts from IL-6-stimulated or unstimulated RAW 264.7 cells were analysed by band-shift assay using an oligonucleotide representing the GC motif of the SOCS3 promoter located in-between positions −66 and −45. Complex formation with this oligonucleotide occurred already in unstimulated RAW 264.7 cells and was not influenced by stimulation of these cells with IL-6. To identify the protein–DNA complexes, supershift analyses with antibodies raised against the Sp1, Sp3 or STAT3 were performed (Figure 4A). Incubation of nuclear extracts from IL-6-stimulated cells with an antiserum specific to Sp3 led to the supershift of the two faster migrating complexes, suggesting that these complexes contain Sp3 bound to the GC motif of the SOCS3 promoter. However, no supershift was observed on incubation with a STAT3- or an Sp1-specific antibody, indicating that neither STAT3 nor Sp1 binds to the GC motif of the SOCS3 promoter. The different mobility of the two Sp3-containing protein–DNA complexes may be due to the fact that three different Sp3 isoforms exist, a 110–115 kDa Sp3 protein and two approx. 60–70 kDa Sp3 species, all of which are recognized by the antibody used for supershift analysis. At present, we have no explanation for the nature of the gel shift band with low mobility.
A more complex picture emerges from the analysis of unstimulated or IL-6-stimulated nuclear extracts with an oligonucleotide (GC/SRE) comprising both the GC motif and the proximal SRE located in-between positions −76 and −45 of the SOCS3 promoter. According to the findings for the isolated GC-rich element, complex formation of two different protein–DNA complexes could be observed when nuclear extracts isolated from either unstimulated or IL-6-stimulated RAW 264.7 cells were analysed using this GC/SRE oligonucleotide (Figure 4B). As suggested by supershift analysis, these two protein–DNA complexes were found to contain Sp3 (Figure 4B, lane 4) when nuclear extracts isolated from cells stimulated with IL-6 were analysed using antibodies raised against Sp3. However, stimulation of RAW 264.7 (Figure 4) with IL-6 induces the formation of additional protein–DNA complexes of lower mobility when nuclear extracts from IL-6-stimulated RAW 264.7 cells (Figure 4B) were analysed in an EMSA using the GC/SRE oligonucleotide. Supershift analysis reveals that these protein–DNA complexes contain STAT3, indicating that IL-6 induces STAT3 binding to this element. Differences in mobility of these complexes is quite probably due to the formation of STAT3 homo- or heterodimers, with additional proteins, e.g. STAT1, recruited to the transcriptional complex on stimulation with IL-6. Analysis of IL-6-stimulated and unstimulated nuclear extracts using both STAT3- and Sp3-specific antibodies in combination led to a shift of almost all the protein–DNA complexes formed with the GC/SRE oligonucleotide (Figure 4C). This suggests that STAT3 and Sp3 are the major transcription factors binding to the GC/SRE oligonucleotide.
In summary, these results support the view that binding of STAT factors towards the responsive sites within the SOCS3 promoter is under the direct control of IL-6 and therefore a major event in transcriptional control of the SOCS3 promoter by IL-6. On the other hand, binding of Sp3 but not Sp1 to the GC-rich element of the SOCS3 promoter does not seem to be due to the direct regulatory control of cytokine signalling, at least for IL-6, since the DNA-binding intensity of Sp3 is not regulated by IL-6. However, as suggested by the promoter analysis, Sp3 binding is a prerequisite for promoter activation by activated STAT-3.
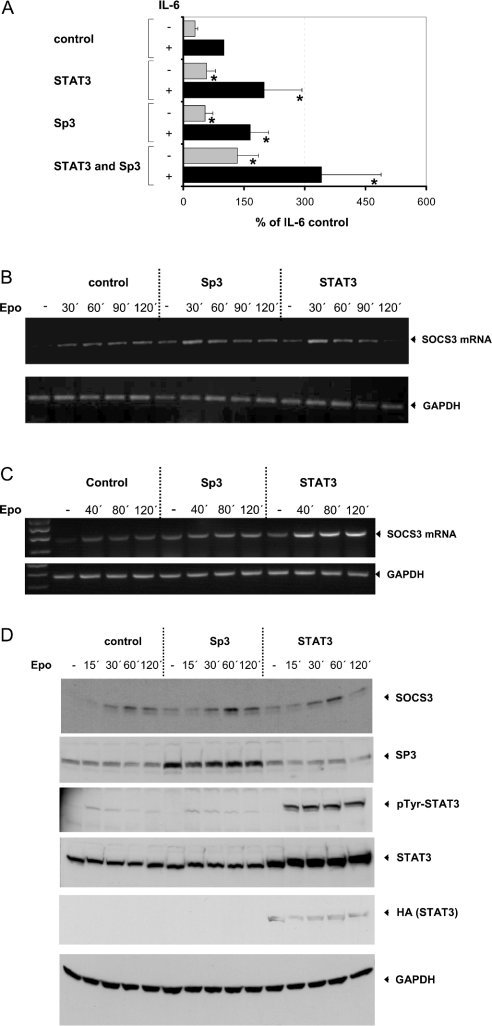
To test whether Sp3 has an influence on transcriptional activation of the SOCS3 promoter, the effect of co-transfected Sp3 on basal and IL-6-induced transcriptional activation of an SOCS3 promoter gene construct was analysed. RAW 264.7 cells were co-transfected with Sp3 or STAT3 alone or in combination with an SOCS3 promoter-reporter construct. Basal and IL-6-dependent induction of the reporter gene was analysed. As shown in Figure 5(A), co-transfection of Sp3 enhances basal as well as IL-6-induced transcriptional activation of the SOCS3 promoter, suggesting that Sp3 has a positive regulatory role in the transcriptional activation of SOCS3 gene expression. This enhancement was almost as effective as the transcriptional enhancement of the SOCS3 promoter achieved by co-transfection of STAT3. Moreover, Sp3 and STAT3 had additive effects on basal and IL-6-mediated transcriptional activation of the SOCS3 promoter as shown by co-transfection of Sp3 together with STAT3.
Figure 5. Sp3 as well as STAT3 induces basal and IL-6-mediated transcriptional activation of an SOCS3 promoter construct and leads to enhanced expression of SOCS3 protein.
(A) RAW 264.7 cells were co-transfected with a reporter gene construct containing the SOCS3 promoter fused to the firefly luciferase gene and a control plasmid or expression vector for Sp3- or HA-tagged STAT3 as indicated. An expression vector for β-galactosidase was co-transfected for monitoring the transfection efficiency. Cells were stimulated, 2 days after transfection, either with or without IL-6 (300 units/ml) for 16 h as shown. Luciferase activity in cellular extracts of these cells was determined and normalized to β-galactosidase activity as described in the Experimental section. NIH3T3 cells (B, D) and HepG2 cells (C) were co-transfected with an Epo/gp130 receptor chimaera including the extracellular part of the Epo receptor and the transmembrane and cytoplasmic domains of the gp130 signal-transducing receptor subunit of the IL-6 receptor complex and a control plasmid or expression vector for Sp3- or HA-tagged STAT3 as depicted. (B, C) Cells were stimulated with 1 unit/ml Epo for the time periods indicated and total RNA was isolated as described in the Experimental section. Subsequently, total RNA was analysed by RT–PCR as described in the Experimental section using primers specific for SOCS3 (upper panel). For loading control, RT–PCR was repeated using primer pairs specific for GAPDH (lower panel). The optimal number of cycles for each primer pair was determined by real-time PCR. The experiments shown are representative of at least three independent experiments. (D) Cellular extracts were prepared and equal amounts of the protein were separated by SDS/PAGE and analysed by Western blotting with a polyclonal antibody from rabbit specifically raised against SOCS3 (topmost panel). For loading and transfection controls, the membrane was reprobed with an Sp3-specific polyclonal antibody (second panel), an antibody specifically recognizing STAT3 tyrosine-phosphorylated at Tyr-759 (third panel) or a STAT3-specific polyclonal antibody (fourth panel) and an HA- (fifth panel) or GAPDH-specific (bottommost panel) monoclonal antibody. *P<0.05, compared with control experiment.
In support of these results, basal and gp130-mediated SOCS3 RNA (Figure 5B) and protein (Figure 5D) expressions were enhanced when NIH3T3 cells were co-transfected with Sp3 or STAT3. In this experiment, Epo/gp130 chimaeric receptors containing the extracellular domain of the Epo receptor and the transmembrane and cytoplasmic domain of gp130 were co-transfected to induce selectively gp130-mediated SOCS3 expression in transfected cells. Basal as well as Epo-induced peak expression level of endogenous SOCS3 RNA (Figure 5B) and protein (Figure 5D) were enhanced to a similar extent when compared with the respective control experiment by co-transfection of either Sp3 or STAT3. The influence of Sp3 on SOCS3 expression was not due to increased STAT3 activity, as substantiated by detection of Epo-dependent STAT3 tyrosine phosphorylation using an antibody specifically recognizing STAT3 when tyrosine-phosphorylated at Tyr-759.
As shown in Figure 5(C), co-transfection of either Sp3 or STAT3 in the human hepatoma cell line HepG2 enhanced basal as well as Epo-induced peak expression level of endogenous SOCS3 mRNA to a similar extent when compared with the respective experiment in murine cell lines. Although the human SOCS3 promoter has not been cloned so far, these results indicate that, beside STAT3, Sp3 is also involved in the regulation of the human SOCS3 gene. This hypothesis is further supported by the fact that the putative STAT-responsive region of the human SOCS3 gene identified by a homology screening by BLAST search on chromosome 17 (NCBI Entrez Nucleotides database accession no. AC061992) using the sequences for the human SOCS3 mRNA (NCBI Entrez Nucleotides database accession no. BC060858) and the published sequence of the murine SOCS3 promoter (NCBI Entrez Nucleotides database accession no. AF117732) features a high homology to the respective sequence of the murine SOCS3 gene (Figure 2).
Mutation of Lys-483 of Sp3 down-regulates enhancement of SOCS3 expression by co-transfected Sp3
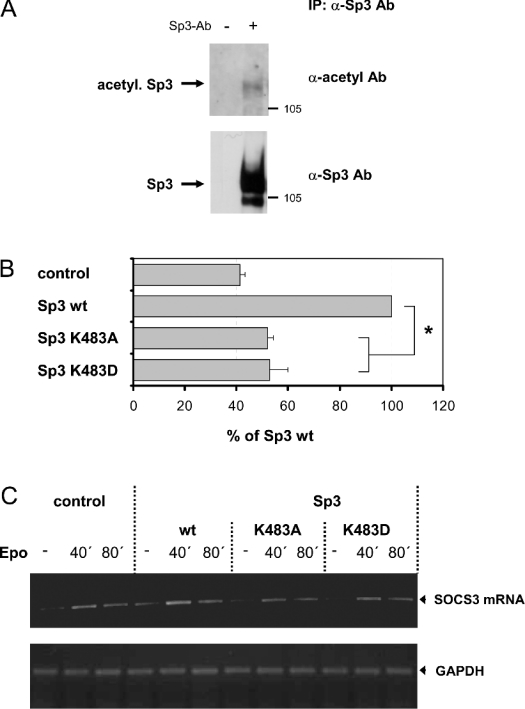
Recent work demonstrated that Sp3 is highly acetylated in vivo [24] and that acetylated Sp3 might act as a transcriptional activator [25]. Thus SP3 acetylation might be relevant for the transcriptional regulation of the expression of SOCS3. Immunoprecipitation followed by analysis of the precipitated Sp3 for its acetylation using antibodies specific for acetylated lysine residues showed that Sp3 is acetylated in RAW 264.7 cells (Figure 6A).
Figure 6. Sp3 is acetylated at a lysine residue in NIH3T3 as well as in a macrophage cell line.
(A) Nuclear extracts from RAW 264.7 were prepared and incubated with antibodies against Sp3 (αSp3 Ab) for immunoprecipitation (IP: Sp3). Protein–antibody complexes were separated by SDS/PAGE and analysed by Western blotting with a polyclonal antibody specifically raised against acetylated lysine residues as described in the Experimental section. Thereafter, blots were stripped and reprobed with a polyclonal antibody specific for Sp3. The experiments shown here are representative of at least three independent experiments. (B) RAW 264.7 cells were co-transfected with a reporter gene construct containing the SOCS3 promoter fused to the firefly luciferase gene and a control plasmid or expression vector for Sp3 or mutants of Sp3, where lysine was mutated to alanine or aspartic acid respectively. An expression vector for Renilla luciferase was co-transfected for monitoring transfection efficiency. Luciferase activity was determined as described in Figure 3. (C) NIH3T3 cells were co-transfected with the Epo/gp130 receptor chimaera and a control plasmid or expression vector for Sp3 or mutants of Sp3, where lysine was mutated to alanine or aspartic acid respectively. Expression of SOCS3 mRNA was determined by RT–PCR as described in Figure 5(B). For loading control, RT–PCR was repeated using primer pairs specific for GAPDH (lower panel). The results shown here are representative of at least three independent experiments.
As shown recently, wild-type Sp3 is acetylated to a significantly higher degree than an Sp3 mutant lacking Lys-483, suggesting that this lysine residue is an important target for post-translational modification of Sp3 by acetylation [24]. Therefore point mutations within the KEE motif of the inhibitory domain of Sp3 were introduced, mutating Lys-483 to alanine (K483A, Lys483→Ala) or aspartic acid (K483D). When compared with wild-type Sp3, mutation of Sp3 at Lys-483 significantly decreases its ability to enhance the transcriptional activation of an SOCS3 promoter-reporter gene construct co-transfected in RAW 264.7 cells (Figure 6B). According to this, an enhancement of Epo-induced SOCS3 mRNA expression in NIH3T3 cells achieved by co-transfection of Sp3 wild-type was decreased when Lys-483 of Sp3 was mutated to alanine (K483A) or aspartic acid (K483D), as shown in Figure 6(C). This suggests that, with respect to the regulation of SOCS3 expression, Lys-483 mediates, most probably by its covalent modification through acetylation, enhancing effects on the transcriptional activity of Sp3. However, one should consider that mutation of this lysine residue probably does not engender a dominant-negative Sp3 mutant, antagonizing endogenous Sp3 action. Thus it is quite conceivable that the transcriptional inactivation of Sp3 resulting from mutation of Lys-483 is much stronger than that suggested by the experiments.
DISCUSSION
Control of SOCS3 gene expression occurs at least partially at the transcriptional level. However, the mechanisms involved in the transcriptional regulation of SOCS3 expression are incompletely understood. It has been shown that members of the STAT family of transcription factors are crucial. The SOCS3 promoter contains STAT1/STAT3-binding elements, which are necessary and sufficient for LIF-dependent activation of the SOCS3 promoter; consequently, co-transfection of a dominant-negative mutant of STAT3 down-regulates LIF-induced induction of the SOCS3 gene [18]. Further evidence exists to suggest that STAT5β participates in the regulation of SOCS3, since up-regulation of SOCS3 in the liver in response to stimulation with growth hormone is inhibited in mice deficient of STAT5β, and STAT5β activated by stimulation with insulin was demonstrated to bind to the STAT1/SRE within the SOCS3 promoter [19,26]. Moreover, several lines of evidence suggest that activation of members of the MAPK (mitogen-activated protein kinase) family such as the ERK-type MAPKs (where ERK stands for extracellular-signal-regulated kinase) or the MKK6/p38MAPK cascade (where MKK6 stands for MAPK kinase 6) are involved in the regulation of SOCS3 expression in response to IL-6 [27], TNFα [28], liposaccharide [28], CpG-DNA [29], PMA [30], fibroblast growth factor-2 [30] or infection with Listeria monocytogenes [31].
Transcriptional regulation of genes is commonly the result of the concerted action of activating and inhibiting proteins binding to distinct promoter elements. Since SOCS3 deficiency causes early embryonic lethality and SOCS3 is known to play a pivotal role in the regulation of cytokine action, detailed understanding of the mechanisms involved in the regulation of this factor is of general importance. The aim of the present study was to investigate the mechanisms involved in the transcriptional regulation of SOCS3. It is shown that Sp3 acts as a transcriptional activator of the SOCS3 promoter, modulating the basal and induced transcriptional activation of the SOCS3 gene (Figures 4 and 5). Furthermore, the Sp3-responsive element, mediating the regulatory effects of Sp3 on the SOCS3 promoter, has been identified as a GC-rich motif close to the TATA box of the SOCS3 promoter (Figures 3 and 4). As suggested by previous studies, the SREs located in-between positions −64 and −72 is especially essential for IL-6-dependent transcriptional activation of the SOCS3 gene (Figures 3 and 4). However, IL-6-induced transcriptional activation of the SOCS3 gene was also disrupted by the introduction of point mutations within the GC-rich element. These findings suggest that a functional SRE on its own is not sufficient for transcriptional activation of the SOCS3 gene by IL-6, but rather requires a functional GC-rich element. With respect to this, it is interesting to note that a G-rich element, but not the two putative STAT-binding sites, was identified as the major motif for IL-6-, growth hormone- and glucocorticoid-dependent regulation of the rat SOCS3 promoter [32]. These results are in agreement with the present findings and support the view that the GC-rich motif is important for the regulation of SOCS3 expression. The fact that transcriptional activation of the SOCS3 promoter by IL-6 requires both a functional SRE and the intact GC-rich motif might point towards a possible protein–protein interaction of Sp3 and STAT3 essential for transcriptional activation. However, in extensive co-precipitation studies using endogenous as well as overexpressed proteins, basal or IL-6-induced physical interaction of these two proteins could not be detected (results not shown).
Reports on the transcriptional properties of Sp3 appear contradictory. In co-transfection experiments in SL2 insect cells, Sp3 has been shown to act as a transcriptional activator [33–35], whereas in other studies Sp3 remained inactive or showed only weak transcriptional activity [36–38]. Particularly, transcriptionally inactive Sp3 competes with Sp1 for DNA binding, thereby inhibiting Sp1-dependent transcriptional activation [36,39]. The function of SP3 appears to depend, at least in part, on the structure and arrangement of the respective recognition sites. Sp3 is often only a weak activator of promoters containing multiple binding elements, whereas promoters with only one recognition element, such as the SOCS3 promoter, can be activated in response to Sp3 [23,40,41].
At present, little is known about the modulation of transcriptional properties of Sp3 by covalent modifications. Recently, it has been demonstrated that Sp3 is highly acetylated in vivo [24] and that acetylated Sp3 might act as a transcriptional activator [25]. With respect to this, Lys-483 has been suggested to be an important target for acetylation [24]. In the present study, it is shown that mutation of the lysine residue impairs Sp3-dependent transcriptional activation and expression of the SOCS3 gene, suggesting that acetylation of Sp3 is important for the enhancing effect of Sp3 on transcriptional control of SOCS3 gene expression. However, using the Sp3-free environment of Drosophila Schneider SL2 cells, Sp3 mutated at Lys-483 enhances rather than inhibits the transcriptional activation of a reporter gene construct containing two adjacent GC boxes, whereas wild-type Sp3 has only a small effect on the transcriptional activation of the reporter construct [24]. On the other hand, the results of an in vitro transcription assay, using GC-box-binding protein-depleted nuclear extracts from HeLa cells reconstituted with either recombinant wild-type Sp3 or mutated Sp3 (SP3SD: lacking 13 amino acids including Lys-483), suggest that Sp3 is a potent transcriptional activator, whereas the acetylation-deficient mutant seems to be less potent, particularly when used at low concentrations [24]. Thus the cellular context seems to be essential for the actions exerted by Sp3 and the influence of acetylation on its transcriptional properties. From the results presented here, it can be concluded that, with respect to the regulation of the SOCS3 gene in mammalian cells, Sp3 acetylation seems to be relevant for the enhanced transcriptional activity exerted by Sp3 to support SOCS3 gene expression. However, it is not clear to what extent acetylation of Sp3 is subject to cellular control mechanisms. Another interesting but open question is whether cell-type-dependent variations of the basal acetylation rate of Sp3 is relevant for cell-type-specific basal expression levels of SOCS3 molecules.
Acknowledgments
We thank J. Matthes for the excellent technical assistance. We are grateful to Dr F. Schaper and Dr P. C. Heinrich for providing us the expression vectors for the Epo/gp130 chimaera and recombinant human IL-6. We thank Dr G. Suske (Marburg, Germany) for providing us the expression vectors for Sp3 and Dr S. Melmed for providing the full-length SOCS3 promoter-reporter gene construct. This work was supported by the Deutsche Forschungsgemeinschaft (Bonn, Germany), through the Sonderforschungsbereich 575 ‘Experimentelle Hepatologie’, and by the Bennigsen-Foerder Preis.
References
- 1.Heinrich P. C., Behrmann I., Haan S., Hermanns H. M., Muller-Newen G., Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heinrich P. C., Behrmann I., Muller-Newen G., Schaper F., Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J. 1998;334:297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim T. K., Maniatis T. Regulation of interferon-gamma-activated STAT1 by the ubiquitin-proteasome pathway. Science. 1996;273:1717–1719. doi: 10.1126/science.273.5282.1717. [DOI] [PubMed] [Google Scholar]
- 4.Haspel R. L., Salditt-Georgieff M., Darnell J. J. E. The rapid inactivation of nuclear tyrosine phosphorylated Stat1 depends upon a protein tyrosine phosphatase. EMBO J. 1996;15:6262–6268. [PMC free article] [PubMed] [Google Scholar]
- 5.Chung C. D., Liao J. Y., Liu B., Rao X. P., Jay P., Berta P., Shuai K. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 6.Liu B., Liao J. Y., Rao X. P., Kushner S. A., Chung C. D., Chang D. D., Shuai K. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. U.S.A. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stahl N., Farruggella T. J., Boulton T. G., Zhong Z., Darnell J. E., Jr, Yancopoulos G. D. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science. 1995;267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 8.Schaper F., Gendo C., Eck M., Schmitz J., Grimm C., Anhuf D., Kerr I. M., Heinrich P. C. Activation of the protein tyrosine phosphatase SHP2 via the interleukin-6 signal transducing receptor protein gp130 requires tyrosine kinase Jak1 and limits acute-phase protein expression. Biochem. J. 1998;335:557–565. doi: 10.1042/bj3350557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim H., Hawley T. S., Hawley R. G., Baumann H. Protein tyrosine phosphatase 2 (SHP-2) moderates signaling by gp130 but is not required for the induction of acute-phase plasma protein genes in hepatic cells. Mol. Cell. Biol. 1998;18:1525–1533. doi: 10.1128/mcb.18.3.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Symes A., Stahl N., Reeves S. A., Farruggella T., Servidei T., Gearan T., Yancopoulos G., Fink J. S. The protein tyrosine phosphatase SHP-2 negatively regulates ciliary neurotrophic factor induction of gene expression. Curr. Biol. 1997;7:697–700. doi: 10.1016/s0960-9822(06)00298-3. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz J., Weissenbach M., Haan S., Heinrich P. C., Schaper F. SOCS3 exerts its inhibitory function on interleukin-6 signal transduction through the SHP2 recruitment site of gp130. J. Biol. Chem. 2000;275:12848–12856. doi: 10.1074/jbc.275.17.12848. [DOI] [PubMed] [Google Scholar]
- 12.Starr R., Willson T. A., Viney E. M., Murray L. J., Rayner J. R., Jenkins B. J., Gonda T. J., Alexander W. S., Metcalf D., Nicola N. A., et al. A family of cytokine-inducible inhibitors of signalling. Nature (London) 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 13.Endo T. A., Masuhara M., Yokouchi M., Suzuki R., Sakamoto H., Mitsui K., Matsumoto A., Tanimura S., Ohtsubo M., Misawa H., et al. A new protein containing an SH2 domain that inhibits JAK kinases. Nature (London) 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 14.Naka T., Narazaki M., Hirata M., Matsumoto T., Minamoto S., Aono A., Nishimoto N., Kajita T., Taga T., Yoshizaki K., et al. Structure and function of a new STAT-induced STAT inhibitor. Nature (London) 1997;387:924–929. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 15.Fujimoto M., Naka T. Regulation of cytokine signaling by SOCS family molecules. Trends Immunol. 2003;24:659–666. doi: 10.1016/j.it.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 16.Krebs D. L., Hilton D. J. SOCS proteins: negative regulators of cytokine signaling. Stem Cells. 2001;19:378–387. doi: 10.1634/stemcells.19-5-378. [DOI] [PubMed] [Google Scholar]
- 17.Yasukawa H., Ohishi M., Mori H., Murakami M., Chinen T., Aki D., Hanada T., Takeda K., Akira S., Hoshijima M., et al. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat. Immunol. 2003;4:551–556. doi: 10.1038/ni938. [DOI] [PubMed] [Google Scholar]
- 18.Auernhammer C. J., Bousquet C., Melmed S. Autoregulation of pituitary corticotroph SOCS-3 expression: characterization of the murine SOCS-3 promoter. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6964–6969. doi: 10.1073/pnas.96.12.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davey H. W., McLachlan M. J., Wilkins R. J., Hilton D. J., Adams T. E. STAT5b mediates the GH-induced expression of SOCS-2 and SOCS-3 mRNA in the liver. Mol. Cell. Endocrinol. 1999;158:111–116. doi: 10.1016/s0303-7207(99)00175-6. [DOI] [PubMed] [Google Scholar]
- 20.Andrews N. C., Faller D. V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J., Russel D. Plainview, NY: Cold Spring Harbor Laboratory Press; 2000. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 22.Wegenka U. M., Buschmann J., Lütticken C., Heinrich P. C., Horn F. Acute-phase response factor, a nuclear factor binding to acute-phase response elements, is rapidly activated by interleukin-6 at the posttranslational level. Mol. Cell. Biol. 1993;13:276–288. doi: 10.1128/mcb.13.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 24.Braun H., Koop R., Ertmer A., Nacht S., Suske G. Transcription factor Sp3 is regulated by acetylation. Nucleic Acids Res. 2001;29:4994–5000. doi: 10.1093/nar/29.24.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ammanamanchi S., Freeman J. W., Brattain M. G. Acetylated sp3 is a transcriptional activator. J. Biol. Chem. 2003;278:35775–35780. doi: 10.1074/jbc.M305961200. [DOI] [PubMed] [Google Scholar]
- 26.Emanuelli B., Peraldi P., Filloux C., Sawka-Verhelle D., Hilton D., Van Obberghen E. SOCS-3 is an insulin-induced negative regulator of insulin signaling. J. Biol. Chem. 2000;275:15985–15991. doi: 10.1074/jbc.275.21.15985. [DOI] [PubMed] [Google Scholar]
- 27.Bode J. G., Ludwig S., Freitas C. A., Schaper F., Ruhl M., Melmed S., Heinrich P. C., Haussinger D. The MKK6/p38 mitogen-activated protein kinase pathway is capable of inducing SOCS3 gene expression and inhibits IL-6-induced transcription. Biol. Chem. 2001;382:1447–1453. doi: 10.1515/BC.2001.178. [DOI] [PubMed] [Google Scholar]
- 28.Bode J. G., Nimmesgern A., Schmitz J., Schaper F., Schmitt M., Frisch W., Haussinger D., Heinrich P. C., Graeve L. LPS and TNFalpha induce SOCS3 mRNA and inhibit IL-6-induced activation of STAT3 in macrophages. FEBS Lett. 1999;463:365–370. doi: 10.1016/s0014-5793(99)01662-2. [DOI] [PubMed] [Google Scholar]
- 29.Dalpke A. H., Opper S., Zimmermann S., Heeg K. Suppressors of cytokine signaling (SOCS)-1 and SOCS-3 are induced by CpG-DNA and modulate cytokine responses in APCs. J. Immunol. 2001;166:7082–7089. doi: 10.4049/jimmunol.166.12.7082. [DOI] [PubMed] [Google Scholar]
- 30.Terstegen L., Gatsios P., Bode J. G., Schaper F., Heinrich P. C., Graeve L. The inhibition of interleukin-6-dependent STAT activation by mitogen-activated protein kinases depends on tyrosine 759 in the cytoplasmic tail of glycoprotein 130. J. Biol. Chem. 2000;275:18810–18817. doi: 10.1074/jbc.M904148199. [DOI] [PubMed] [Google Scholar]
- 31.Stoiber D., Stockinger S., Steinlein P., Kovarik J., Decker T. Listeria monocytogenes modulates macrophage cytokine responses through STAT serine phosphorylation and the induction of suppressor of cytokine signaling 3. J. Immunol. 2001;166:466–472. doi: 10.4049/jimmunol.166.1.466. [DOI] [PubMed] [Google Scholar]
- 32.Paul C., Seiliez I., Thissen J. P., Le Cam A. Regulation of expression of the rat SOCS-3 gene in hepatocytes by growth hormone, interleukin-6 and glucocorticoids mRNA analysis and promoter characterization. Eur. J. Biochem. 2000;267:5849–5857. doi: 10.1046/j.1432-1327.2000.01395.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhao L., Chang L. S. The human POLD1 gene. Identification of an upstream activator sequence, activation by Sp1 and Sp3, and cell cycle regulation. J. Biol. Chem. 1997;272:4869–4882. [PubMed] [Google Scholar]
- 34.Udvadia A. J., Templeton D. J., Horowitz J. M. Functional interactions between the retinoblastoma (Rb) protein and Sp-family members: superactivation by Rb requires amino acids necessary for growth suppression. Proc. Natl. Acad. Sci. U.S.A. 1995;92:3953–3957. doi: 10.1073/pnas.92.9.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang Y., Robinson D. F., Dennig J., Suske G., Fahl W. E. Transcriptional regulation of the SIS/PDGF-B gene in human osteosarcoma cells by the Sp family of transcription factors. J. Biol. Chem. 1996;271:11792–11797. doi: 10.1074/jbc.271.20.11792. [DOI] [PubMed] [Google Scholar]
- 36.Hagen G., Muller S., Beato M., Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majello B., De Luca P., Hagen G., Suske G., Lania L. Different members of the Sp1 multigene family exert opposite transcriptional regulation of the long terminal repeat of HIV-1. Nucleic Acids Res. 1994;22:4914–4921. doi: 10.1093/nar/22.23.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dennig J., Hagen G., Beato M., Suske G. Members of the Sp transcription factor family control transcription from the uteroglobin promoter. J. Biol. Chem. 1995;270:12737–12744. doi: 10.1074/jbc.270.21.12737. [DOI] [PubMed] [Google Scholar]
- 39.Ghayor C., Chadjichristos C., Herrouin J. F., Ala-Kokko L., Suske G., Pujol J. P., Galera P. Sp3 represses the Sp1-mediated transactivation of the human COL2A1 gene in primary and de-differentiated chondrocytes. J. Biol. Chem. 2001;276:36881–36895. doi: 10.1074/jbc.M105083200. [DOI] [PubMed] [Google Scholar]
- 40.Dennig J., Beato M., Suske G. An inhibitor domain in Sp3 regulates its glutamine-rich activation domains. EMBO J. 1996;15:5659–5667. [PMC free article] [PubMed] [Google Scholar]
- 41.Birnbaum M. J., van Wijnen A. J., Odgren P. R., Last T. J., Suske G., Stein G. S., Stein J. L. Sp1 trans-activation of cell cycle regulated promoters is selectively repressed by Sp3. Biochemistry. 1995;34:16503–16508. doi: 10.1021/bi00050a034. [DOI] [PubMed] [Google Scholar]