Abstract
ApoE (apolipoprotein E) is an anti-atherogenic lipid transport protein that plays an integral role in lipoprotein metabolism and cholesterol homoeostasis. Lipid association educes critical functional features of apoE, mediating reduction in plasma and cellular cholesterol levels. The 10-kDa CT (C-terminal) domain of apoE facilitates helix–helix interactions in lipid-free state to promote apoE self-association and helix–lipid interactions during binding with lipoproteins, although the mode of lipid-binding interaction is not well understood. We investigated the mode of lipid-binding interaction and orientation of apoE CT domain on reconstituted lipoproteins. Isolated recombinant human apoE CT domain (residues 201–299) possesses a strong ability to interact with phospholipid vesicles, yielding lipoprotein particles with an apparent molecular mass of ∼600 kDa, while retaining the overall α-helical content. Electron microscopy and non-denaturing PAGE analysis of DMPC (dimyristoylphosphatidylcholine)–apoE CT domain lipoprotein complexes revealed discoidal complexes with a diameter of approx. 17 nm. Cross-linking apoE CT domain on discoidal particles yielded dimeric species as the major product. Attenuated total reflectance Fourier transform IR spectroscopy of phospholipid–apoE CT domain complexes reveals that the helical axis is oriented perpendicular to fatty acyl chains of the phospholipid. Fluorescence quenching analysis of DMPC–apoE CT domain discoidal complexes by spin-labelled stearic acid indicated a relatively superficial location of the native tryptophan residues with respect to the plane of the phospholipid bilayer. Taken together, we propose that apoE CT domain interacts with phospholipid vesicles, forming a long extended helix that circumscribes the discoidal bilayer lipoprotein complex.
Keywords: apolipoprotein E, cross-linking, electron microscopy, IR spectroscopy, lipid-bound conformation, lipoprotein-binding surface
Abbreviations: apoA-I, apolipoprotein A-I; apoE, apolipoprotein E; ATR-FTIR, attenuated total reflectance Fourier transform IR; CT, C-terminal; DMPC, dimyristoylphosphatidylcholine; DMPG, dimyristoylphosphatidylglycerol; 5-DSA, 5-DOXYL stearic acid; 12-DSA, 12-DOXYL stearic acid; DSS, disuccinimidyl suberate; HDL, high-density lipoprotein; LDL, low-density lipoprotein; NT, N-terminal
INTRODUCTION
ApoE (apolipoprotein E) is a 299-residue exchangeable apolipo-protein that is a component of triacylglycerol-rich lipoproteins and a subclass of HDL (high-density lipoprotein). Transgenic studies [1,2], and clinical and biochemical analyses of apoE-deficient subjects [3], reveal the direct relevance of apoE in lipoprotein metabolism, cholesterol homoeostasis and as a modulator of atherogenesis. The role of apoE in causing a reduction of plasma and cellular cholesterol levels is documented by its ability to: (i) act as a ligand for the LDL (low-density lipoprotein) receptor family [4], that leads to cellular uptake and clearance of remnant lipoprotein particles; and (ii) promote cholesterol efflux from peripheral tissues to nascent HDL particles in atherosclerotic lesions [5,6]. In the brain, apoE is involved in cholesterol transport from astrocytes to neurons by means of lipoproteins [7] and in neuronal regeneration following nerve injury [8,9]. In all of these instances, lipid association of apoE is a key factor in determining its functionality.
Lipid association of apoE is mediated by two independently folded domains: the NT (N-terminal) domain (residues 1–191) that bears low-affinity lipoprotein-binding ability and the CT (C-terminal) domain (residues 210–299) that accommodates high-affinity lipid-binding sites [10–12]. The prominent feature of the NT domain is its ability to interact with the LDL receptor via binding sites localized to helix 4 of the four-helix bundle architecture [13], an ability conferred by lipid association. In turn, lipid-binding interaction of apoE NT domain induces dramatic conformational alterations involving opening of the four-helix bundle at putative hinge regions, as determined from surface monolayer [14] and FRET (fluorescence resonance energy transfer) studies [15,16]. The structural re-organization triggered by lipid binding is an a priori requirement for receptor-binding competency of apoE NT domain, which probably bears significant physiological relevance [17,18]. Thus apoE has been proposed to exist in two different lipid-bound conformations: one where the NT domain is in a lipid-free helix-bundle state that is receptor-incompetent, and a second where it is in a lipid-bound, receptor-competent state following helix-bundle opening. Interestingly, both conformers involve ‘anchoring’ of apoE to the lipid particle via the CT domain [17,18]. However, the structural organization of the CT domain in the lipid-bound state and the mode of the lipid-binding interaction of this domain are poorly understood at present.
C-terminal truncation analyses demonstrate that deletion of residues 267–299 impaired 25% of the lipoprotein binding activity, while deleting residues 244–299 and shorter deletion variants abolished entirely binding to VLDL (very-low-density lipoprotein) and resulted in poor binding (∼15–20%) to HDL [19]. Similar impaired lipoprotein association was identified in a naturally occurring apoE truncated variant, apoE-(1–209), with a premature stop codon at position 210 [20]. Other studies confirm that deleting CT residues 192–299 resulted in decreased lipid-binding affinity compared with intact apoE, with a decreased ability to promote cellular phospholipid and cholesterol efflux [21]. In addition to lipoprotein binding, apoE CT domain also mediates protein tetramerization in aqueous solutions via residues 267–299 [19,22,23], probably mediated by intermolecular coiled-coil interaction [24], a motif frequently encountered in protein oligomerization and assembly [25]. However, in the lipid-associated state, apoE has been proposed to exist as a monomer [26], although 5–7 apoE molecules appear to be required to saturate lipoprotein particles with a diameter of ∼25 nm [27].
In the present study, we report the mode of lipid binding of apoE CT domain and the organization of its helices in a lipid-associated state. We evaluate the structural organization of apoE CT domain following reconstitution into nascent lipoprotein particles by CD, ATR-FTIR (attenuated total reflectance Fourier transform IR) and fluorescence spectroscopy.
EXPERIMENTAL
Expression and purification of recombinant apoE CT domain
A pET22b vector encoding apoE (201–299) and bearing a His6 tag at the NT end was used for expression [24]. Recombinant wild-type apoE CT domain was overexpressed in Escherichia coli and purified by affinity chromatography on a Ni-affinity matrix (HiTrap™ chelating column, Amersham Biosciences).
Cross-linking analysis
Cross-linking experiments were carried out using DSS (disuccinimidyl suberate) (Pierce Biotechnology, Rockford, IL, U.S.A.), which is a homobifunctional, primary amine-reactive cross-linker with spacer arm length of ∼12 Å (1 Å=0.1 nm), as described previously [24]. Reaction mixtures contained 0.05 mg of lipid-free apoE CT domain [or DMPC (dimyristoylphos-phatidylcholine)- or DMPG (dimyristoylphosphatidylglycerol)-bound, as described below)] and various concentrations of cross-linker in 25 mM potassium phosphate, pH 7.4. Incubations were carried out for 30 min at room temperature (24 °C). The reaction was quenched by the addition of 1 M Tris/HCl, pH 7.4, and/or non-reducing SDS/PAGE sample treatment buffer for 15 min, followed by electrophoresis on a 4–20% gradient gel to confirm cross-linking. DMSO was used as the solvent for DSS at concentrations not exceeding 2% of total reaction mixtures (v/v).
Lipid binding assay
The ability of apoE CT domain to transform phospholipid vesicles to discoidal bilayer particles was studied by monitoring the changes in right-angle light scattering in a PerkinElmer LS50B spectrofluorimeter [28]. Unilamellar vesicles of DMPG were prepared by extruding through a 200 nm membrane (Avanti Polar Lipids, Alabaster, AL, U.S.A.). Increasing concentrations of apoE CT domain (4–200 μg of protein) were added to 0.2 mg of DMPG vesicles in a thermostatically controlled cuvette containing 20 mM Tris/HCl, pH 7.2, and 150 mM NaCl at 32 °C. The changes in light-scattering intensity were monitored at 560 nm (excitation and emission wavelengths) with a slit width of 3 nm.
Preparation and characterization of DMPC–or DMPG–apoE CT domain complex
DMPC–or DMPG–apoE CT domain discoidal bilayer complexes were prepared as described previously [29]. Briefly, sonicated or extruded phospholipid vesicles and apoE CT domain (2.5:1, w/w) were incubated in 50 mM Tris/HCl, pH 7.5, for 18 h at 24 °C, followed by KBr density gradient ultracentrifugation to separate unbound protein and protein-free lipid vesicles from lipid-bound protein. Fractions containing both protein and lipid were pooled and concentrated. Protein assay was carried out using the bicinchoninic acid method (Pierce Biotechnology), and phospholipids were estimated using the phospholipid assay kit (Wako Chemicals GmbH, Neuss, Germany). Non-denaturing PAGE of the isolated lipoprotein complexes was carried out to evaluate the molecular mass and size of the particles on a 4–20% gradient gel for 20 h at 150 V and stained with Amido Black.
CD spectroscopy
Secondary-structural characterization was carried out on an Applied Photophysics CD spectrometer equipped with a thermostatically controlled cuvette holder. The secondary-structure content of the samples was estimated using CDPRO software [30]. CD spectra were recorded between 250 and 190 nm at 0.5 nm intervals, and averaged over 20000 points.
IR spectroscopy
ATR-FTIR spectra were recorded on a Bruker IFS 55 IR spectrophotometer equipped with a reflectance accessory and a polarizer mount assembly with an aluminium wire grid on a KRS-5 element. The internal reflection element was a germanium ATR plate (50 mm×20 mm×2 mm) with an aperture angle of 45° yielding 25 internal reflections. The samples were dialysed against 5 mM Tris/HCl, pH 7.5, before FTIR analysis. Oriented multilayers were formed by slow evaporation of ∼30 μl of DMPC–apoE CT domain (∼0.5 mg/ml) on one side of the ATR plate under a gentle stream of nitrogen, yielding a semi-dry film bearing residual water molecules. The ATR plate was then sealed in a universal sample holder. Spectra were recorded at a 2 cm−1 nominal resolution. A total of 256 accumulations were performed to improve the signal/noise ratio. The spectrometer was constantly purged with dry air. All measurements were made at 25 °C. Before any analysis, the side-chain contributions to the spectra were subtracted [31].
Secondary-structure analysis by FTIR
Secondary-structure measurements were carried out on samples following deuteration for 1 h as described previously [32]. Briefly, Fourier self-deconvolution was applied to increase the resolution of the spectra in the amide I region. Least squares iterative curve fitting was performed to fit different components of the amide I band revealed by the self-deconvolution to the non-deconvolved spectrum between 1700 and 1600 cm−1. The proportion of various secondary-structural elements was computed as reported in [32].
Orientation of the secondary structures on discoidal bilayer complexes
In an α-helix, the main transition dipole moment lies roughly parallel to the helical axis. It is therefore possible to determine the mean orientation of the α-helix structure from the orientation of the peptide bond C=O group [33]. To obtain this information, spectra of DMPC–apoE CT domain complexes were recorded with parallel and perpendicular polarized incident light with respect to a normal to the ATR plate. Polarization was expressed as the dichroic ratio Ratr=A///A⊥. The mean angle between the helix axis and a normal to the ATR plate surface was then calculated from Ratr. In these calculations, a 38° angle between the long axis of the α-helix and the C=O dipole moment was considered [34]. The γw(CH2) transition at 1202 cm−1, whose dipole lies parallel to the all-trans hydrocarbon chains, was used to characterize the lipid acyl chain orientation [35]. An estimation of the sample film thickness was obtained using the isodichroic ratio [36], based on which the angle for both lipids and protein α-helices was estimated.
Fluorescence quenching
We took advantage of the intrinsic fluorescence properties of apoE CT domain, attributed to the presence of three tryptophan residues, Trp210, Trp264 and Trp276, to assess the depth of location of the fluorophores with respect to the phospholipid bilayer. Quenching of fluorescence emission of DMPC–apoE CT domain lipoprotein complexes was carried out as described previously [37,38] with 5-DSA (5-DOXYL stearic acid) or 12-DSA (12-DOXYL stearic acid), where the DOXYL group (quenching moiety) is located at different depths along the fatty acyl chain. Aliquots of 5-DSA or 12-DSA (1.3 mM stock in ethanol) were added directly to DMPC–apoE CT domain complexes (keeping the final concentration of ethanol ≤1%, v/v), and fluorescence intensities were measured at 340 nm. Effective quenching constants were calculated employing the Stern–Volmer equation, F0/F=1+KSV·[Q], where F0 and F are fluorescence intensities in the absence and the presence of various quencher concentrations [Q] respectively, and KSV is the Stern–Volmer quenching constant [39].
Electron microscopy
DMPC–apoE CT domain complexes were dialysed against ammonium acetate buffer and negatively stained with 2% sodium phosphotungstate, before examination by electron microscopy (Zeiss 10, 80 kV) as described previously [40].
RESULTS
Lipid-binding activity of apoE CT domain
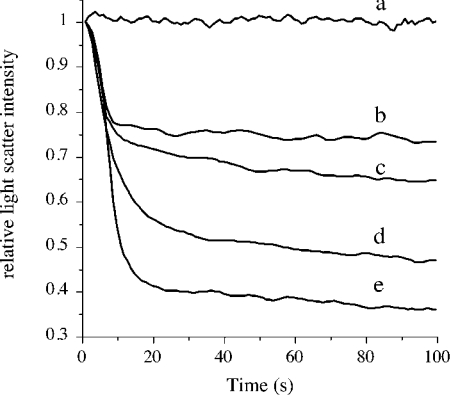
A characteristic feature of apolipoproteins is their ability to interact with vesicular phospholipid bilayer structures (diameter ∼200 nm) and convert them into small lipoprotein particles. This transformation or clearance of phospholipid vesicles has been monitored routinely as changes in intensity of right-angle light scattering [41–44]. We employed DMPG, a negatively charged phospholipid, to study the lipid-binding ability [28] of increasing concentration of apoE CT domain corresponding to lipid/protein mass ratios varying from 50:1 to 1:1. ApoE CT domain displays a concentration-dependent effect in its ability to cause vesicle clearance, as indicated by the decrease in right-angle light scattering intensity following addition of the protein. Figure 1 shows data for 10, 20, 50 and 100 μg of apoE CT domain added to 200 μg of DMPG, corresponding to lipid/protein mass ratios of 20:1, 10:1, 4:1 and 2:1 respectively. This indicates that isolated recombinant apoE CT domain retains lipid-binding ability, allowing us to examine the mechanism of lipid-binding interaction further.
Figure 1. Lipid-binding ability of apoE CT domain.
DMPG vesicles (200 μg) were pre-incubated in 20 mM Tris/HCl, pH 7.2, and 150 mM NaCl at 32 °C followed by addition of varying concentrations of apoE CT domain: a, no added protein; b–e, 10, 20, 50 and 100 μg of apoE CT domain respectively. Transformation of vesicular structures to discoidal particles was followed by right-angle light scattering in a spectrofluorimeter, with the excitation and emission wavelengths set at 560 nm, and the slit width at 3 nm.
Estimation of reconstituted lipoprotein particle diameter
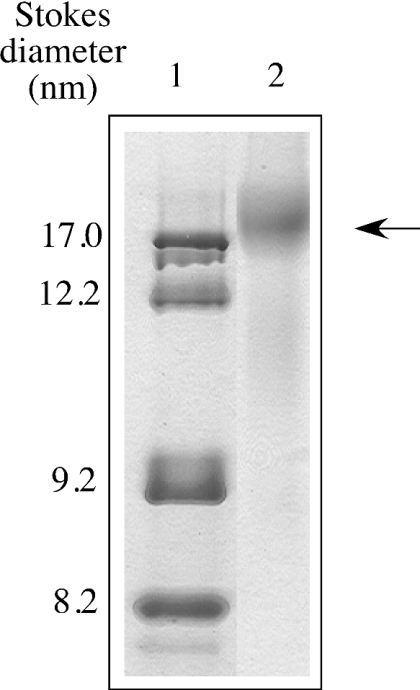
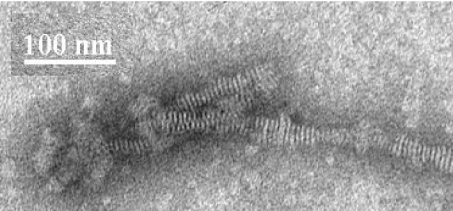
Non-denaturing gradient PAGE analysis of DMPC–apoE CT domain demonstrated lipoprotein particles of 17±2 nm Stokes diameter as the major species, corresponding to an apparent molecular mass of ∼600 kDa (Figure 2). Particles 12 nm in diameter were noted to a minor extent (<10%). Electron microscopic analysis of DMPC–apoE CT domain complexes by negative staining (Figure 3) reveals discoidal structures with an average diameter of 16.6±2.7 nm (n=122), consistent with estimates obtained from native PAGE analysis. Similar discoidal complexes were noted earlier with other apolipoproteins, such as insect apolipophorin III [42,45]. Compositional analysis DMPC/apoE CT domain particles yielded lipid/protein molar ratio of ∼200:1. In the case of DMPG–apoE CT, particles with smaller diameter (∼8 nm) were formed.
Figure 2. Native PAGE analysis of DMPC–apoE CT domain complexes.
Native PAGE analysis was carried out on a 4–20% polyacrylamide continuous gradient gel. Electrophoresis was performed in Tris-glycine buffer, pH 8.4, for 20 h at 150 V, and stained with Amido Black. Lane 1, protein standards; lane 2, DMPC–apoE CT domain complexes (arrow). The particle sizes were calculated from a calibration curve using the following standards and their corresponding Stokes diameters: thyroglobulin, 17 nm; ferritin, 12.2 nm; catalase, 9.2 nm; lactate dehydrogenase, 8.2 nm.
Figure 3. Electron micrographs of DMPC–apoE CT domain complexes.
DMPC–apoE CT domain complexes prepared and isolated as described in the Experimental section. The reconstituted lipoprotein particles were stained with 2% phosphotungstate for visualization. Scale bar, 100 nm.
Secondary-structure characterization of lipid-bound apoE CT domain
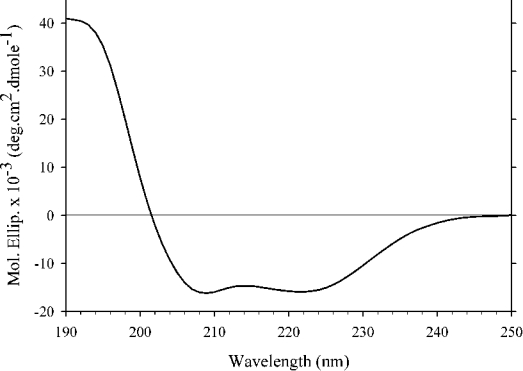
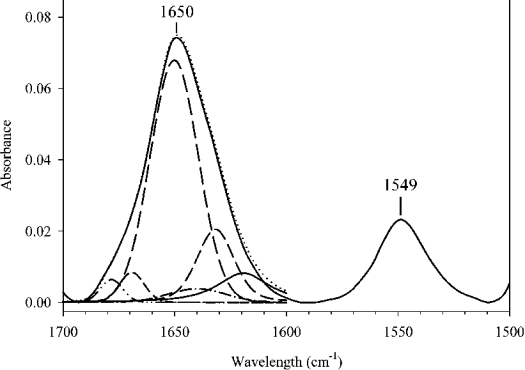
The secondary-structure content of DMPC–apoE CT domain discoidal complexes was analysed by CD (Figure 4) and IR spectroscopy. Far-UV CD spectra of DMPC–apoE CT domain reveal features characteristic of α-helical proteins with troughs at 222 and 208 nm. The α-helical content of DMPC–apoE CT domain complexes (∼58%) and the ratio of molar ellipticities at 222 and 208 nm (∼1.0) were similar to those obtained for lipid-free apoE CT domain [10,24]. A similar observation was made for DMPG–apoE CT domain complexes (results not shown). The IR spectrum of DMPC–apoE CT domain discoidal complex (Figure 5) also displays characteristics features of α-helical structures [46]. The amide I band (1700–1600 cm−1) reveals a sharp peak centred at 1650 cm−1. The maximum intensity of the amide II peak (1600–1500 cm−1) is located at 1549 cm−1. Components of the amide I peak revealed by Fourier self-deconvolution were quantified by least squares iterative curve fitting, as described in [32]. The α-helical content of lipid-bound apoE CT domain was calculated to be ∼62% by FTIR, consistent with CD analysis.
Figure 4. Far-UV CD spectrum of DMPC–apoE CT domain complex.
Far-UV CD spectrum of DMPC–apoE CT domain complex was recorded in 25 mM sodium phosphate, pH 7.0, from 250 to 190 nm at 24 °C at 0.5 nm intervals, and averaged over 20000 points.
Figure 5. IR spectrum of DMPC–apoE CT domain discoidal complexes in the 1700–1500 cm−1 region (solid curve).
Secondary structure of DMPC–apoE CT domain complex was obtained by analysis of IR spectrum between 1700 and 1600 cm−1 by curve fitting. The different curves used to fit the amide I peak are displayed as broken lines.
Fluorescence quenching analysis
Lipid-based quenchers were employed to estimate the average depth of location of the three tryptophan residues in apoE CT domain with respect to the phospholipid bilayer in the lipoprotein particle. Increasing concentrations of DSA with the DOXYL group located at C-5 (5-DSA) or C-12 (12-DSA) was added directly to DMPC–apoE CT domain complexes in sodium phosphate, pH 7.0, to quench the intrinsic fluorescence emission of tryptophan residues in lipid-bound apoE CT domain. The higher KSV observed for 5-DSA (0.14±0.03 M−1) compared with that of 12-DSA (0.06±0.02 M−1) (Table 1) is indicative of a superficial location of the tryptophan residues proximal to the lipid/water interface.
Table 1. Quenching of intrinsic fluorescence of DMPC–apoE CT domain complexes by 5-DSA and 12-DSA.
Values are means±S.D. from three independent experiments. Tryptophan fluorescence emission intensity was monitored following excitation at 280 nm.
| Quencher | KSV (M−1)* |
|---|---|
| 5-DSA | 0.14±0.03 |
| 12-DSA | 0.06±0.02 |
* KSV is obtained from a plot of F0/F against [Q].
Determination of helix orientation in DMPC–apoE CT domain
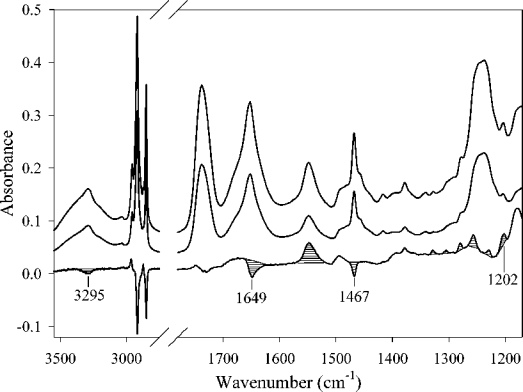
FTIR spectra of DMPC–apoE CT domain complex were recorded with parallel and perpendicular polarized light (Figure 6). The dichroic spectrum was obtained by subtracting the spectrum recorded with the perpendicular polarized light from that recorded with parallel polarized light using the lipid ν(C=O) band as a reference [36]. The respective deviations of absorption bands associated with lipid moieties indicate that the phospholipid hydrocarbon chains in the discs are preferentially oriented perpendicular to the germanium surface. The δ(CH2) band at 1467 cm−1 associated with a dipole oriented perpendicular to the hydrocarbon chain [35,47,48] has a negative deviation in the dichroic spectrum. Conversely, bands of the γw(CH2) progression observed for saturated hydrocarbon chains with all-trans configuration at 1329, 1305, 1279, 1257, 1229 and 1202 cm−1 (Figure 6) and corresponding to dipoles oriented parallel to the hydrocarbon chain have a positive deviation. We used the peak at 1202 cm−1 to quantify the lipid chain orientation. The measured dichroic ratio for this band is 3.1 with an isotropic dichroic ratio of 1.57, i.e. the dichroic ratio observed for an isotropically oriented sample. The lipid ν(C=O) band around 1740 cm−1 provides a good estimate for the isotropic dichroic ratio [36]. On the basis of these dichroic ratio measurements, we calculated the tilt between the acyl chains and a normal to the germanium surface to be 30°. The symmetric stretching mode of vibration of the acyl chain methyl group located at 2872 cm−1 is also oriented parallel to the acyl chain axes [35]. For this band, the calculated dichroic ratio is 3.6, which indicates an orientation of the phospholipid acyl chains that is even more perpendicular to the germanium plate than that indicated by the 1202 cm−1 band. As any additional source of disorder would reduce this tilt, the 30° value is therefore a maximum tilt with respect to a normal to the germanium plate.
Figure 6. IR spectra of DMPC–apoE CT domain complexes.
The top spectrum was obtained with parallel polarized light and the middle spectrum with perpendicular polarized light. The bottom spectrum is the dichroic spectrum obtained by subtracting the middle spectrum (perpendicular polarization) from the top spectrum (parallel polarization). The optical density amplitude of the bottom spectrum has been increased three times with respect to the other spectra. Striped peaks are the most important for the orientation determination (see text).
In the case of the protein component of the complexes, the orientation was determined using the amide I band attributed to apoE CT domain helices. On the dichroic spectrum (Figure 6), a strong negative deviation at 1649 cm−1 was observed, indicating a parallel orientation of the associated dipole to the surface of the germanium plate. From the secondary structure of apoE CT domain complexed with DMPC (see above), and the 1649 cm−1 frequency observed for the deviation, we conclude that the dipole responsible for this deviation is associated with the α-helices of the protein. This amide I dipole, corresponding to the amide ν(C=O) mode of vibration, is roughly oriented in the direction of the helical axes ([34,46], see also the Experimental section). The observed negative deviation indicates that the helices are primarily oriented parallel to the germanium plane and are therefore perpendicular to the hydrocarbon chains of the lipids. This helix orientation is confirmed by the observed negative deviation of the amide A band (at 3292 cm−1), for which the dipole is oriented as in the case of the amide I, as well as by the positive deviation of the amide II band (at 1548 cm−1), which is oriented in the opposite perpendicular direction with respect to the helical axes [34,46]. The dichroic ratio associated with the helical structure in the protein is equal to 1.32, while the mean tilt between the helix axes and a normal to the germanium surface is between 70° and 90°.
Cross-linking of apoE CT on discoidal particles
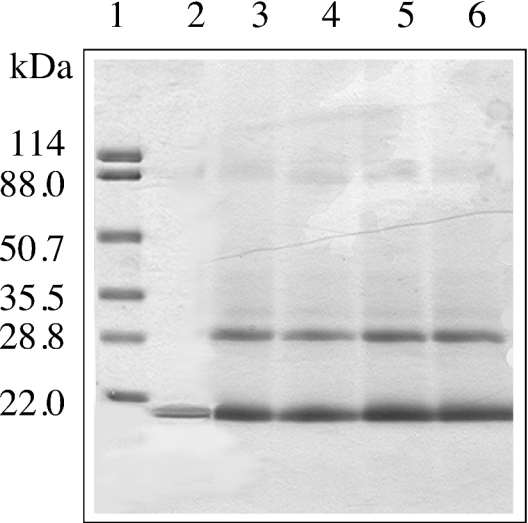
Finally, cross-linking of apoE CT domain was performed subsequent to formation of DMPC discoidal particles. Increasing concentrations of DSS, a cross-linker that bears the ability to access lysine residues from an aqueous and lipid environment, was employed, followed by SDS/PAGE analysis (Figure 7). At all concentrations of DSS employed, a band corresponding to dimeric apoE CT domain (apparent molecular mass of 32±1 kDa) was the major cross-linked species present. Another minor band appeared at ∼37 kDa. Densitometric analysis of the gel showed that the dimeric band (32 kDa) represented 30% of the proteins under all the conditions, while the upper band (37 kDa) contained <2% of the proteins. A similar pattern was noted in the case of DMPG–apoE CT domain (results not shown).
Figure 7. SDS/PAGE of DMPC–apoE CT domain discoidal complexes cross-linked with DSS.
DMPC–apoE CT domain complexes (0.05 mg of protein) were incubated with increasing concentration of DSS for 30 min at room temperature. The reaction was stopped by addition of 1 M Tris/HCl, pH 7.4, and SDS sample treatment buffer, followed by electrophoresis in a 4–20% acrylamide gradient gel. Lane 1, molecular mass markers with the indicated masses in kDa; lane 2, DMPC–apoE CT domain; lanes 3–6, DMPC–apoE CT domain treated with 1-, 10-, 20- and 50-fold molar excess DSS respectively.
DISCUSSION
The lipid-binding interaction is an essential prerequisite for apoE to elicit its physiological role as a ligand for the LDL receptor family of proteins and to promote cholesterol efflux in atherosclerotic lesions. Interestingly, model lipid-bound complexes comprising phospholipids and apoE recapitulate the functional features of apoE on native spherical lipoprotein particles or nascent HDL. Furthermore, lipid binding of apoE is believed to be initiated by the CT domain, which bears a higher lipid-binding affinity than the NT domain, a concept supported by the lower free energy of stabilization for the former and the tendency of apoE to self-associate in a lipid-free state via the CT domain [10,11]. The two domains of apoE are independently folded structural and functional units, and isolated apoE CT domain retains the lipid-binding capability of the intact protein [49,50]. Whereas peptide fragments encompassing discrete segments of apoE (residues 263–286 and 267–286) [51,52] display lipid-binding characteristics, we find it essential to examine the lipid-bound configuration of the entire functional unit encompassed in isolated apoE CT domain.
Sequence-based secondary-structure predictions indicate that apoE CT domain is composed of amphipathic α-helices (class A, residues 203–266, and class G*, residues 268–285) [53], with residues 218–266 bearing a high propensity to form a coiled-coil helix [24]. CD and sedimentation equilibrium analysis of isolated lipid-free apoE CT domain revealed an intermolecular two-stranded coiled-coil helix formation that promotes apoE dimerization and subsequent tetramerization. The ratio of molar ellipticities at 222 and 208 nm in the CD spectrum of α-helical proteins has been employed as a criterion for coiled-coil helix formation [54]. The absorbance at 208 and 222 nm are attributed to the amide π-π* and n-π* transition respectively, with the 208 absorbance (which polarizes parallel with the helical axis) sensitive to whether the helix is unassociated or interacts with a neighbouring helix to form a coiled coil. In the case of lipid-free apoE CT domain, the ratio was calculated to be 1.0, indicative of coiled-coil helix formation [24]. Interestingly, lipid association did not induce any further increase in α-helicity or alteration in the 222/208 nm ratio. We suggest that the overall curvature bestowed on apoE CT domain helices as they circumscribe the discoidal bilayer contributes to the altered absorbance at 208 nm in lipid-associated apoE CT domain, with a resultant maintenance of the ratio at ∼1.0.
In conjunction with fluorescence studies, FTIR analysis provides valuable information regarding the spatial disposition of the helices on the discoidal particles with regard to the lipid bilayer. Using parallax analysis of fluorescence quenching experiments, tryptophan residues 210, 264 and 276 in apoE CT domain are indicated to be located at the same depth in the bilayer, suggesting a perpendicular orientation of the helices. The fluorescence quenching approach provides a limited view of the spatial disposition of selected sites. On the other hand, ATR-FTIR analysis provides a discriminatory approach to determine not only the orientation of a secondary structure (using linear dichroism), but also the nature of this secondary structure (using the wavelength at which the dichroism appears, see above). Therefore, in the present study, we provide the first direct evidence that the entire helical segment of apoE CT domain is oriented perpendicular with respect to the phospholipid fatty acyl chains. This indicates that two helices (∼15 Å diameter each) may be aligned adjacent to each other-shielding the hydrophobic part of the lipid bilayer (34 Å), circumscribing the perimeter of the lipid discoidal particles (the ‘belt model’). This conformation is in agreement with the cross, linking results of the present study that indicate apoE CT probably adopts a dimeric conformation on these particles. A similar orientation has been described for the receptor-binding domain of apoE [47,55]. Since the two domains of apoE are oriented similarly, it is likely that intact apoE also adopts such an extended belt conformation at the periphery of reconstituted discoidal lipoproteins. The perpendicular alignment of helices appears to be the trend for exchangeable apolipoproteins, with apoA-I (apolipoprotein A-I) [56–58] and insect apolipophorin III [46] displaying similar tendencies, while the parallel alignment [49] (the ‘picket-fence model’) appears to be of lesser relevance.
Taking the apparent molecular mass of the lipoprotein particle (∼600 kDa), discoidal particle diameter of approx. 17 nm, in conjunction with the compositional analysis (lipid/protein molar ratio, 200:1) into consideration, an estimated 4–6 apoE CT domain molecules per discoidal particle in a fully extended α-helical conformation are envisaged in a majority of the particles. Furthermore, cross-linking analysis on pre-formed discoidal particles yielded dimers as the predominant species, suggesting that apoE CT domain helices orient with a parallel arrangement. This is indeed the simplest way to envision spatial proximity of the lysine residues involved in cross-linking. Nevertheless, the experiments of the present study cannot rule out the possibility that the helices may adopt an antiparallel arrangement with a variable registry, as also noted for lipid-associated apoA-I [59]. Further studies are in progress to address the issue of apoE helix alignment on lipoprotein particles.
Addressing the question of accumulation of apoE molecules on lipoprotein particles, it was proposed that apoE transitions from a tetrameric to a monomeric state upon association with LDL-sized spherical microemulsions (∼25 nm diameter) composed of egg phosphatidylcholine and triolein [27]. The model for apoE–phospholipid discoidal complexes proposed in the present study does not prevent the possibility that apoE may exist in a monomeric state on a spherical lipoprotein as proposed previously [26]. It is envisaged that conversion of discoidal nascent HDL structure into spherical lipoprotein particles is likely to be accompanied by repositioning of intermolecular helical contacts of neighbouring apoE molecules depicted on the circumference of the phospholipid bilayer. In other studies, spatial proximity between apoE molecules on discoidal and spherical complexes has been suggested, with evidence of intimate protein–protein contact on the surface of lipoprotein particles [60].
In conclusion, we show that apoE CT domain causes transformation of vesicular bilayer structures. In a previous study, we demonstrated the relevance of intermolecular coiled-coil intermediates with distinct tertiary environment in the unfolding pathway of apoE CT domain [24], indicative of a potential role for dimers during lipid-induced conformational alterations. On nascent discoidal lipoproteins, apoE CT adopts an extended belt-like conformation favouring dimeric interactions. Such a mode of interaction gives us an indication of the structural aspects of recruitment of apoE to cholesterol-poor or nascent HDL particles at sites of atherosclerotic lesions, thereby increasing the capacity to acquire cholesterol [61,62]. Interestingly, it has been shown that the lipoprotein-binding surface of apoE CT domain offers a potential site for interaction with β-amyloid peptide, which is involved in the pathogenesis of Alzheimer's disease [63]. With the emerging role of lipids in Alzheimer's disease, understanding the mode of lipid-binding interaction of apoE is an important first step towards further studies addressing its potential role in amyloid formation.
Acknowledgments
This work was supported by a Pfizer International HDL Research Award, an Alzheimer's Association and a Parkinson's Disease Foundation grant (to V. N.). We thank Dr Cyril M. Kay and Robert Luty for CD measurements. V. R. and E. G. are, respectively, research associate and research director of the National Fund for Scientific Research (Belgium).
References
- 1.Shimano H., Yamada N., Katsuki M., Yamamoto K., Gotoda T., Harada K., Shimada M., Yazaki T. Plasma lipoprotein metabolism in transgenic mice overexpressing apolipoprotein E: accelerated clearance of lipoproteins containing apolipoprotein B. J. Clin. Invest. 1992;90:2084–2091. doi: 10.1172/JCI116091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang S. H., Reddick R. L., Piedrahita J. A., Maeda N. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science. 1992;258:468–471. doi: 10.1126/science.1411543. [DOI] [PubMed] [Google Scholar]
- 3.Schaefer E. J., Gregg R. E., Ghiselli G., Forte T. M., Ordoyas J. M., Zech L. A., Brewer N. B., Jr Familial apolipoprotein E deficiency. J. Clin. Invest. 1986;78:1206–1219. doi: 10.1172/JCI112704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown M. S., Goldstein J. L. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 5.Fielding C. J., Fielding P. E. Molecular physiology of reverse cholesterol transport. J. Lipid Res. 1995;36:211–228. [PubMed] [Google Scholar]
- 6.Von Eckardstein A., Nofer J.-R., Assmann G. High density lipoproteins and arteriosclerosis: role of cholesterol efflux and reverse cholesterol transport. Arterioscler. Thromb. Vasc. Biol. 2001;21:13–27. doi: 10.1161/01.atv.21.1.13. [DOI] [PubMed] [Google Scholar]
- 7.Mauch D. H., Nägler K., Schumacher S., Göritz C., Müller E.-C., Otto A., Pfrieger F. W. CNS synaptogenesis promoted by glia-derived cholesterol. Science. 2001;294:1354–1357. doi: 10.1126/science.294.5545.1354. [DOI] [PubMed] [Google Scholar]
- 8.Mahley R. W., Nathan B. P., Pitas R. E. Apolipoprotein E: structure, function, and possible roles in Alzheimer's disease. Ann. N.Y. Acad. Sci. 1996;777:139–145. doi: 10.1111/j.1749-6632.1996.tb34412.x. [DOI] [PubMed] [Google Scholar]
- 9.Mahley R. W. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 10.Aggerbeck L. P., Wetterau J. R., Weisgraber K. H., Wu C.-S. C., Lindgren F. T. Human apolipoprotein E3 in aqueous solution. II. Properties of the amino- and carboxyl-terminal domains. J. Biol. Chem. 1988;263:6249–6258. [PubMed] [Google Scholar]
- 11.Wetterau J. R., Aggerbeck L. P., Rall S. C., Jr, Weisgraber K. H. Human apolipoprotein E3 in aqueous solution. I. Evidence for two structural domains. J. Biol. Chem. 1988;263:6240–6248. [PubMed] [Google Scholar]
- 12.Weisgraber K. H. Apolipoprotein E: structure–function relationships. Adv. Protein Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 13.Wilson C., Wardell M. R., Weisgraber K. H., Mahley R. W., Agard D. A. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 14.Weisgraber K. H., Lund-Katz S., Phillips M. C. Apolipoprotein E: structure–function correlations. In: Miller N. E., Tall A. R., editors. High Density Lipoproteins and Atherosclerosis III. Amsterdam: Elsevier; 1992. pp. 175–181. [Google Scholar]
- 15.Fisher C. A., Ryan R. O. Lipid binding-induced conformational changes in the N-terminal domain of human apolipoprotein E. J. Lipid Res. 1999;40:93–99. [PubMed] [Google Scholar]
- 16.Fisher C. A., Narayanaswami V., Ryan R. O. The lipid-associated conformation of the low density lipoprotein receptor binding domain of human apolipoprotein E. J. Biol. Chem. 2000;275:33601–33606. doi: 10.1074/jbc.M002643200. [DOI] [PubMed] [Google Scholar]
- 17.Narayanaswami V., Ryan R. O. Molecular basis of exchangeable apolipoprotein function. Biochim. Biophys. Acta. 2000;1483:15–36. doi: 10.1016/s1388-1981(99)00176-6. [DOI] [PubMed] [Google Scholar]
- 18.Saito H., Dhanasekaran P., Baldwin F., Weisgraber K. H., Lund-Katz S., Phillips M. C. Lipid binding-induced conformational change in human apolipoprotein E: evidence for two lipid-bound states on spherical particles. J. Biol. Chem. 2001;276:40949–40954. doi: 10.1074/jbc.M106337200. [DOI] [PubMed] [Google Scholar]
- 19.Westerlund J. A., Weisgraber K. H. Discrete carboxyl-terminal segments of apolipoprotein E mediate lipoprotein association and protein oligomerization. J. Biol. Chem. 1993;268:15745–15750. [PubMed] [Google Scholar]
- 20.Lohse P., Brewer H. B., 3rd, Meng M. S., Skarlatos S. I., LaRosa J. C., Brewer H. B., Jr Familial apolipoprotein E deficiency and type III hyperlipoproteinemia due to a premature stop codon in the apolipoprotein E gene. J. Lipid Res. 1992;33:1583–1590. [PubMed] [Google Scholar]
- 21.Gillotte K. L., Zaiou M., Lund-Katz S., Anantharamaiah G. M., Holvoet P., Dhoest A., Palgunachari M. N., Segrest J. P., Weisgraber K. H., Rothblat G. H., Phillips M. C. Apolipoprotein-mediated plasma membrane microsolubilization: role of lipid affinity and membrane penetration in the efflux of cellular cholesterol and phospholipid. J. Biol. Chem. 1999;274:2021–2028. doi: 10.1074/jbc.274.4.2021. [DOI] [PubMed] [Google Scholar]
- 22.Yokoyama S., Kawai Y., Tajima S., Yamamoto A. Behavior of human apolipoprotein E in aqueous solutions and at interfaces. J. Biol. Chem. 1985;260:16375–16382. [PubMed] [Google Scholar]
- 23.Perugini M. A., Schuck P., Howlett G. J. Self-association of human apolipoprotein E3 and E4 in the presence and absence of phospholipid. J. Biol. Chem. 2000;275:36758–36765. doi: 10.1074/jbc.M005565200. [DOI] [PubMed] [Google Scholar]
- 24.Choy N., Raussens V., Narayanaswami V. Inter-molecular coiled-coil formation in human apolipoprotein E C-terminal domain. J. Mol. Biol. 2003;334:527–539. doi: 10.1016/j.jmb.2003.09.059. [DOI] [PubMed] [Google Scholar]
- 25.Kohn W. D., Mant C. T., Hodges R. S. α-Helical protein assembly motifs. J. Biol. Chem. 1997;272:2583–2586. doi: 10.1074/jbc.272.5.2583. [DOI] [PubMed] [Google Scholar]
- 26.Yokoyama S. Self-associated tetramer of human apolipoprotein E does not lead to its accumulation on a lipid particle. Biochim. Biophys. Acta. 1990;1047:99–101. doi: 10.1016/0005-2760(90)90266-z. [DOI] [PubMed] [Google Scholar]
- 27.Funahashi T., Yokoyama S., Yamamoto A. Association of apolipoprotein E with the low density lipoprotein receptor: demonstration of its co-operativity on lipid microemulsion particles. J. Biochem. 1989;105:582–587. doi: 10.1093/oxfordjournals.jbchem.a122708. [DOI] [PubMed] [Google Scholar]
- 28.Weers P. M. M., Van der Horst D., Ryan R. O. Interaction of locust apolipophorin III with lipoproteins and phospholipid vesicles: effect of glycosylation. J. Lipid Res. 2000;41:416–423. [PubMed] [Google Scholar]
- 29.Weintzek M., Kay C. M., Oikawa K., Ryan R. O. Binding of insect apolipophorin III to dimyristoylphosphatidylcholine vesicles: evidence for a conformational change. J. Biol. Chem. 1994;269:4605–4612. [PubMed] [Google Scholar]
- 30.Sreerama N., Woody R. W. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 31.Goormaghtigh E., de Jongh H. H. J., Ruysschaert J.-M. Relevance of protein thin films prepared for attenuated total reflection Fourier transform infrared spectroscopy: significance of the pH. Appl. Spectrosc. 1996;50:1519–1527. [Google Scholar]
- 32.Goormaghtigh E., Cabiaux V., Ruysschaert J.-M. Secondary structure and dosage of soluble and membrane proteins by attenuated total reflection Fourier-transform infrared spectroscopy on hydrated films. Eur. J. Biochem. 1990;193:409–420. doi: 10.1111/j.1432-1033.1990.tb19354.x. [DOI] [PubMed] [Google Scholar]
- 33.Goormaghtigh E., Ruysschaert J.-M. Polarised attenuated total reflection infrared spectroscopy as a tool to investigate the conformation and orientation of membrane components. In: Brasseur R., editor. Molecular Description of Biological Membrane Components by Computer Aided Conformational Analysis. Boca Raton: CRC Press; 1990. pp. 285–329. [Google Scholar]
- 34.Marsh D., Müller M., Schmitt F.-J. Orientation of the infrared transition moments for an α-helix. Biophys. J. 2000;78:2499–2510. doi: 10.1016/S0006-3495(00)76795-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fringeli U. P., Gunthard H. H. Infrared membrane spectroscopy. Mol. Biol. Biochem. Biophys. 1981;31:270–332. doi: 10.1007/978-3-642-81537-9_6. [DOI] [PubMed] [Google Scholar]
- 36.Bechinger B., Ruysschaert J.-M., Goormaghtigh E. Membrane helix orientation from linear dichroism of infrared attenuated total reflection spectra. Biophys. J. 1999;76:552–563. doi: 10.1016/S0006-3495(99)77223-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahoo D., Narayanaswami V., Kay C. M., Ryan R. O. Fluorescence studies of exchangeable apolipoprotein–lipid interactions: superficial association of apolipophorin III with lipoprotein surfaces. J. Biol. Chem. 1998;273:1403–1408. doi: 10.1074/jbc.273.3.1403. [DOI] [PubMed] [Google Scholar]
- 38.Sahoo D., Weers P. M., Ryan R. O., Narayanaswami V. Lipid-triggered conformational switch of apolipophorin III helix bundle to an extended helix organization. J. Mol. Biol. 2002;321:201–214. doi: 10.1016/s0022-2836(02)00618-6. [DOI] [PubMed] [Google Scholar]
- 39.Eftink M. R., Ghiron C. A. Exposure of tryptophanyl residues in proteins: quantitative determination by fluorescence quenching studies. Biochemistry. 1976;15:672–680. doi: 10.1021/bi00648a035. [DOI] [PubMed] [Google Scholar]
- 40.Forte T. M., Nordhausen R. W. Electron microscopy of negatively stained lipoproteins. Methods Enzymol. 1986;128:442–457. doi: 10.1016/0076-6879(86)28086-6. [DOI] [PubMed] [Google Scholar]
- 41.Chetty P. S., Arrese E. L., Rodriguez V., Soulages J. L. Role of helices and loops in the ability of apolipophorin-III to interact with native lipoproteins and form discoidal lipoprotein complexes. Biochemistry. 2003;42:15061–15067. doi: 10.1021/bi035456i. [DOI] [PubMed] [Google Scholar]
- 42.Weers P. M. M., Kay C. M., Ryan R. O. Conformational changes of an exchangeable apolipoprotein, apolipophorin III from Locusta migratoria, at low pH: correlation with lipid binding. Biochemistry. 2001;40:7754–7760. doi: 10.1021/bi010410f. [DOI] [PubMed] [Google Scholar]
- 43.Weers P. M., Narayanaswami V., Choy N., Luty R., Hicks L., Kay C. M., Ryan R. O. Lipid binding ability of human apolipoprotein E N-terminal domain isoforms: correlation with protein stability? Biophys. Chem. 2003;100:481–492. doi: 10.1016/s0301-4622(02)00300-9. [DOI] [PubMed] [Google Scholar]
- 44.Segall M. L., Dhanasekaran P., Baldwin F., Anantharamaiah G. M., Weisgraber K. H., Phillips M. C., Lund-Katz S. Influence of apoE domain structure and polymorphism on the kinetics of phospholipid vesicle solubilization. J. Lipid Res. 2002;43:1688–1700. doi: 10.1194/jlr.m200157-jlr200. [DOI] [PubMed] [Google Scholar]
- 45.Dettloff M., Weers P. M. M., Niete M., Kay C. M., Ryan R. O., Wiesner A. An N-terminal three-helix fragment of the exchangeable insect apolipoprotein apolipophorin III conserves the lipid binding properties of wild-type protein. Biochemistry. 2001;40:3150–3157. doi: 10.1021/bi0013804. [DOI] [PubMed] [Google Scholar]
- 46.Goormaghtigh E., Cabiaux V., Ruysschaert J.-M. Determination of soluble and membrane protein structure by Fourier transform infrared spectroscopy. III. Secondary structures. Subcell. Biochem. 1994;23:405–450. doi: 10.1007/978-1-4615-1863-1_10. [DOI] [PubMed] [Google Scholar]
- 47.Raussens V., Narayanaswami V., Goormaghtigh E., Ryan R. O., Ruysschaert J.-M. Alignment of the apolipophorin-III α-helices in complex with dimyristoylphosphatidylcholine: a unique spatial orientation. J. Biol. Chem. 1995;270:12542–12547. doi: 10.1074/jbc.270.21.12542. [DOI] [PubMed] [Google Scholar]
- 48.Raussens V., Fisher C. A., Goormaghtigh E., Ryan R. O., Ruysschaert J.-M. The low density lipoprotein receptor active conformation of apolipoprotein E: helix organization in N-terminal domain-phospholipid disc particles. J. Biol. Chem. 1998;273:25825–25830. doi: 10.1074/jbc.273.40.25825. [DOI] [PubMed] [Google Scholar]
- 49.De Pauw M., Vanloo B., Dergunov A. D., Devreese A.-M., Baert J., Brasseur R., Rosseneu M. Composition and structural and functional properties of discoidal and spherical phospholipid–apoE3 complexes. Biochemistry (Moscow) 1997;62:251–263. [PubMed] [Google Scholar]
- 50.Morrow J. A., Segall M. L., Lund-Katz S., Phillips M. C., Knapp M., Rupp B., Weisgraber K. H. Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry. 2000;39:11657–11666. doi: 10.1021/bi000099m. [DOI] [PubMed] [Google Scholar]
- 51.Sparrow J. T., Sparrow D. A., Fernando G., Culwell A. R., Kovar M., Gotto A. M., Jr Apolipoprotein E: phospholipid binding studies with synthetic peptides from the carboxyl terminus. Biochemistry. 1992;31:1065–1068. doi: 10.1021/bi00119a015. [DOI] [PubMed] [Google Scholar]
- 52.Wang G., Pierens G. K., Treleaven W. D., Sparrow J. T., Cushley R. J. Conformations of human apolipoprotein E(263–286) and E(267–289) in aqueous solutions of sodium dodecyl sulfate by CD and 1H NMR. Biochemistry. 1996;35:10358–10366. doi: 10.1021/bi960934t. [DOI] [PubMed] [Google Scholar]
- 53.Segrest J. P., Jones M. K., DeLoof H., Brouillette C. G., Venkatachalapathi M., Anantharamaiah G. M. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 1992;33:141–166. [PubMed] [Google Scholar]
- 54.Zhou N. E., Kay C. M., Hodges R. S. Synthetic model proteins: positional effects of interchain hydrophobic interactions on stability of two-stranded α-helical coiled-coils. J. Biol. Chem. 1992;267:2664–2670. [PubMed] [Google Scholar]
- 55.Narayanaswami V., Maiorano J. N., Dhanasekaran P., Ryan R. O., Phillips M. C., Lund-Katz S., Davidson W. S. Helix orientation of the functional domains in apolipoprotein E in discoidal high density lipoprotein particles. J. Biol. Chem. 2004;279:14273–14279. doi: 10.1074/jbc.M313318200. [DOI] [PubMed] [Google Scholar]
- 56.Borhani D. W., Rogers D. P., Engler J. A., Brouillette C. G. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12291–12296. doi: 10.1073/pnas.94.23.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koppaka V., Silvestro L., Engler J. A., Brouillette C. G., Axelsen P. H. The structure of human lipoprotein A-I: evidence for the “belt” model. J. Biol. Chem. 1999;274:14541–14544. doi: 10.1074/jbc.274.21.14541. [DOI] [PubMed] [Google Scholar]
- 58.Li H., Lyles D. S., Thomas M. J., Pan W., Sorci-Thomas M. G. Structural determination of lipid-bound ApoA-I using fluorescence resonance energy transfer. J. Biol. Chem. 2000;275:37048–37054. doi: 10.1074/jbc.M005336200. [DOI] [PubMed] [Google Scholar]
- 59.Li H., Lyles D. S., Pan W., Alexander E., Thomas M. J., Sorci-Thomas M. G. ApoA-I structure on discs and spheres: variable helix registry and conformational states. J. Biol. Chem. 2002;277:39093–39101. doi: 10.1074/jbc.M206770200. [DOI] [PubMed] [Google Scholar]
- 60.Gordon V., Innerarity T. L., Mahley R. W. Formation of cholesterol- and apoprotein E-enriched high density lipoproteins in vitro. J. Biol. Chem. 1983;258:6202–6212. [PubMed] [Google Scholar]
- 61.Koo C., Innerarity T. L., Mahley R. W. Obligatory role of cholesterol and apolipoprotein E in the formation of large cholesterol-enriched and receptor-active high density lipoproteins. J. Biol. Chem. 1985;260:11934–11943. [PubMed] [Google Scholar]
- 62.Dergunov A. D., Vorotnikova Y. Y., Visvikis S., Siest G. Homo- and hetero-complexes of exchangeable apolipoproteins in solution and in lipid-bound form. Spectrochim. Acta Part A. 2003;59:1127–1137. doi: 10.1016/s1386-1425(02)00298-6. [DOI] [PubMed] [Google Scholar]
- 63.Mahley R. W., Huang Y. Apolipoprotein E: from atherosclerosis to Alzheimer's disease and beyond. Curr. Opin. Lipidol. 1999;10:207–217. doi: 10.1097/00041433-199906000-00003. [DOI] [PubMed] [Google Scholar]