Abstract
Two mutations (G8363A and A8296G) in the mtDNA (mitochondrial DNA) tRNALys gene have been associated with severe mitochondrial diseases in a number of reports. Their functional significance, however, remains unknown. We have already shown that homoplasmic cybrids harbouring the A8296G mutation display normal oxidative phosphorylation, although the possibility of a subtle change in mitochondrial respiratory capacity remains an open issue. We have now investigated the pathogenic mechanism of another mutation in the tRNALys gene (G8363A) by repopulating an mtDNA-less human osteosarcoma cell line with mitochondria harbouring either this genetic variant alone or an unusual combination of the two mutations (A8296G+G8363A). Cybrids homoplasmic for the single G8363A or the A8296G+G8363A mutations have defective respiratory-chain enzyme activities and low oxygen consumption, indicating a severe impairment of the oxidative phosphorylation system. Generation of G8363A cybrids within a wild-type or the A8296G mtDNA genetic backgrounds resulted in an important alteration in the conformation of the tRNALys, not affecting tRNA steady-state levels. Moreover, mutant cybrids have an important decrease in the proportion of amino-acylated tRNALys and, consequently, mitochondrial protein synthesis is greatly decreased. Our results demonstrate that the pathogenicity of the G8363A mutation is due to a change in the conformation of the tRNA that severely impairs aminoacylation in the absence of changes in tRNA stability. The only effect detected in the A8296G mutation is a moderate decrease in the aminoacylation capacity, which does not affect mitochondrial protein biosynthesis.
Keywords: aminoacylation, mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes (MELAS), myoclonic epilepsy with ragged-red fibres (MERRF), mitochondrial DNA (mtDNA) mutation, mitochondrial tRNALys gene
Abbreviations: FBS, fetal bovine serum; DMEM, Dulbecco's modified Eagle's medium; MELAS, mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes; MERRF, myoclonic epilepsy with ragged-red fibres; mtDNA, mitochondrial DNA; OXPHOS, oxidative phosphorylation
INTRODUCTION
Mitochondria generate most of the cellular ATP by OXPHOS (oxidative phosphorylation). The OXPHOS system is composed of five different complexes embedded in the inner mitochondrial membrane (complexes I–V) and two small electron carriers, namely ubiquinone and cytochrome c [1,2]. Mammalian mitochondria are endowed with their own semi-autonomous genetic system [mtDNA (mitochondrial DNA)] that encodes a limited number of essential genes for OXPHOS biogenesis: 13 polypeptides of complex I [ND1–ND6 and ND4L (subunits 1–6 and 4L of NADH:ubiquinone oxidoreductase], complex III (cytochrome b), complex IV (cytochrome c oxidase subunits I–III) and complex V (ATPases 6–8), as well as the RNA components of the translational apparatus, two rRNAs (12 and 16 S) and 22 tRNAs. The rest of the structural subunits of the OXPHOS system and all the factors involved in OXPHOS assembly and regulation, mtDNA expression and mtDNA maintenance are encoded in the nucleus, translated into cytoplasmic ribosomes and imported to their final mitochondrial location [3]. Therefore OXPHOS defects can be produced by mutations in mitochondrially encoded genes, nuclear genes encoding OXPHOS subunits or in nuclear genes encoding factors directly or indirectly involved in OXPHOS regulation [4–6].
To date, more than 200 mtDNA mutations have been implicated in the pathogenesis of mitochondrial diseases with defective OXPHOS, including large mtDNA rearrangements and point mutations in tRNA, rRNA and protein-coding genes [7]. Mitochondrial diseases are usually multisystemic disorders and produce devastating encephalomyopathies since they affect predominantly high energy-demanding tissues such as the nervous system and skeletal and cardiac muscles [8]. Mutations in protein-coding genes affect a single complex of the OXPHOS system. In contrast, mutations in tRNA genes impair the mitochondrial translation system and therefore affect four of the five OXPHOS complexes, producing combined enzyme deficits of the respiratory chain [9]. Two point mutations, A3243G in the tRNALeu(UUR) gene and A8344G in the tRNALys gene, associated with two well-defined clinical syndromes, MELAS (mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes) and MERRF (myoclonic epilepsy with ragged-red fibres) respectively, are relatively frequent and have been extensively characterized [10–13]. However, the relationship between genotype (mutation) and phenotype (clinical symptoms) has not been understood so far [8]. Heterogeneity is paramount: the same mutation can be associated with diverse clinical manifestations and different mutations can produce the same symptoms. Dosage and distribution of the altered tRNAs in the different tissues of the organism cannot simply account for these phenomena. In addition, the pathogenic mechanisms of the different mtDNA mutations are not fully understood and have not been studied in detail for most of the mutations described so far.
We were the first to report on a family segregating the MERRF syndrome in association with a double mutation, A8296G and G8363A, in the tRNALys gene [14]. The A8296G mutation was practically homoplasmic in all the investigated family members, whereas the proportion of the G8363A mutation correlated with the severity of the phenotype. Both mutations disrupt highly conserved base pairs in the aminoacyl stem of the tRNALys. The A8296G mutation has also been independently described in association with Type II (non-insulin-dependent) diabetes mellitus [15,16], hypertrophic cardiomyopathy [17] and the MELAS syndrome [18]. On the other hand, the G8363A mutation had already been associated with MERRF [19], maternally inherited cardiomyopathy and hearing loss in two pedigrees [20], and with spinocerebellar ataxia and multiple symmetric lipomatosis [21]. More recently, we reported that transmitochondrial hybrids homoplasmic for the A8296G mutation had a normal OXPHOS function [22], a finding which challenges a clear-cut pathogenic role. In the present study, we have generated and extensively characterized transmitochondrial hybrids homoplasmic for the single G8363A mutation and the A8296G/G8363A double mutation. We have found that the A8296G mutation decreases moderately the aminoacylation capacity of the tRNALys, although it does not have consequences for mitochondrial protein biosynthesis. In contrast, biochemical and molecular results demonstrate that the G8363A mutation impairs mitochondrial protein synthesis and compromises severely the function of OXPHOS by altering the conformation of the tRNALys and decreasing drastically its aminoacylation capacity. This phenotype is not modified by the presence of the A8296G mutation.
MATERIALS AND METHODS
Cell lines and media
Two fibroblast cell lines were established from diagnostic skin biopsies obtained from the mother of an MERRF patient harbouring the A8296G-G8363A double mutation [14] and from a patient suffering from cerebellar ataxia and lipomatosis harbouring approx. 80% of the G8363A mutation in the mtDNA tRNAlys gene [21]. Cells were grown in DMEM (Dulbecco's modified Eagle's medium), supplemented with 10% FBS (fetal bovine serum) and 50 μg/ml uridine. The human osteosarcoma cell line 143 B (TK−) was grown in DMEM supplemented with 10% FBS, 100 μg/ml bromodeoxyuridine and 50 μg/ml uridine. Its mtDNA-less derived 206 cell line (ρ0) was grown in DMEM supplemented with 5% FBS and 50 μg/ml uridine.
Establishment of transmitochondrial hybrids
The patients’ derived enucleated fibroblasts (0.5×106 cells) were fused to 106 ρ0-206 cells as described previously [23]. Transmitochondrial hybrid clones (cybrids) were grown in DMEM supplemented with 10% dialysed FBS and 100 μg/ml bromodeoxyuridine. Individual clones were isolated and the mtDNA was analysed for both A8296G and G8363A transitions by the AciI/Asp700 RFLP (restriction-fragment-length polymorphism) method as described in [14].
Respiratory function assays
The rate of oxygen consumption was measured as described previously [22] at 37 °C using a Clark-type platinum polarographic electrode using 3–5×106 exponentially intact cells growing in 2 ml of complete DMEM without glucose. To assess respiratory chain enzymes in cells, approx. 5×106 cells were harvested by trypsinization, washed twice with phosphate buffer and resuspended in 2 ml of Mops/sucrose buffer. Digitonin (200 μg) was added and incubation was performed on ice for 5 min. After centrifugation at 5000 g for 3 min, the pellet was resuspended in 0.5 ml of 10 mM potassium phosphate buffer (pH 7.4) and freeze–thawed twice just before evaluating the enzyme complexes. The activities of rotenone-sensitive NADH-coenzyme Q1 reductase (complex I), succinate dehydrogenase (part of complex II), antimycin-sensitive ubiquinol cytochrome c reductase (complex III), cytochrome c oxidase (complex IV) and citrate synthase were measured in duplicate in each assay, and expressed in terms of nmol of substrate·min−1·(mg of protein)−1. We did not normalize the respiratory chain activities for the activity of citrate synthase since this matrix enzyme is also expressed at high levels in the ρ0 mitochondria of the host cells. Protein concentration was measured by the method of Lowry as described in [22].
Quantification of mitochondrial DNA
To extract total DNA, cells were pooled, washed with PBS, resuspended in 1 ml of lysis buffer (100 mM NaCl, 0.5% SDS, 10 mM Tris and 25 mM EDTA, pH 8.0) and incubated in the presence of 0.2 mg/ml proteinase K at 42 °C for 24 h. DNA was extracted by standard methods [24]. Quantification of mtDNA in fibroblasts, cybrid cell lines and ρ0 cells was performed by slot-blot and Southern-blot analyses. As a probe, we used an [α-32P]dCTP-labelled long mtDNA PCR-amplified fragment (using an oligonucleotide from nucleotide positions 13420–13443 as forward primer and an oligonucleotide from nt 8200–8225 as reverse primer). As loading control, the same membranes were incubated with an [α-32P]dCTP-labelled 18 S rRNA probe. Quantitative analysis was performed by phosphoimager detection.
Quantification of mitochondrial tRNAs
Total RNA (5 μg) extracted from cybrid cell lines was incubated at 90 °C for 5 min, electrophoresed through a 12% polyacrylamide–7 M urea gel in 1×TBE (Tris/borate/EDTA) buffer and electroblotted on to a Zeta-probe membrane at 300 mA for 30 min in 1×TAE (Tris/acetate/EDTA) buffer. Hybridization was performed at 42 °C for 24 h in 6×SSC (1×SSC=0.15 M NaCl/0.015 M sodium citrate), 1×Denhardt's solution, 0.5% SDS, 0.05% sodium pyrophosphate and 100 μg/ml denatured salmon sperm DNA.
The tRNALys was detected using a synthetic 20-mer oligonucleotide (spanning mtDNA nucleotide positions 8361–8342), which had been labelled with [γ-32P]ATP at its 5′-end. After hybridization, the blot was washed twice for 30 min with 6×SSC, 0.1% SDS and 0.05% sodium pyrophosphate at 42 °C. The membrane was stripped out and rehybridized under the same conditions with a [γ-32P] 5′-end-labelled 21-mer oligonucleotide corresponding to mtDNA positions 597–577 of the tRNAPhe gene. The ratio of tRNALys to tRNAPhe was calculated by quantification of the respective signals either by densitometry or phosphoimager analysis.
Single-strand conformational analysis of mitochondrial tRNAs
Total RNA (8 μg) extracted from cybrid cell lines was electrophoresed through a 6% non-denaturing polyacrylamide (60:1) gel in 1×TBE buffer as described previously [25]. To detect the tRNALys and tRNAPhe, we used the same labelled probes and hybridization conditions described above.
Analysis of the aminoacylation of mitochondrial tRNAs
Total RNA from cells was extracted under acidic conditions to preserve aminoacylated tRNAs using a previously described method [26] with minor modifications. Total RNA (3 μg) was electrophoresed through an 8% non-denaturing polyacrylamide (29:1) gel in 0.1 M sodium acetate (pH 5.0) buffer at 100–200 V for 18–20 h, with the buffer being continuously recirculated at 4 °C. To deacylate tRNAs, an aliquot of total RNA was boiled for 10 min (pH 8.0) before electrophoresis. Then, the gel was electroblotted on to a Zeta-probe membrane and hybridized with specific tRNALys and tRNAPhe probes.
Mitochondrial protein synthesis
To analyse the translation products of mitochondrial genes in the different cybrid cell lines, subconfluent cultures were labelled with [35S]methionine in the presence of 100 μg of emetine·(ml of medium)−1 by the method of Chomyn [27] with minor modifications. Cells were labelled with 125 μCi of [35S]methionine for 2 h, washed with phosphate buffer and harvested. After centrifugation at 400 g, cells were resuspended in lysis buffer [20 mM Tris/HCl (pH 7.5), 0.1% SDS, 5 mM MgCl2 and 1 mM PMSF]. Total protein (100 μg) was electrophoresed and detected by autoradiography.
RESULTS
Generation of transmitochondrial cybrids
In a previous study, we generated transmitochondrial cybrids by fusing the human mtDNA-less ρ0-206 cell line with fibroblasts obtained from an MERRF patient containing >95% of the A8296G combined with 50% G8363A mutations in mtDNA, as described in the Materials and methods section. More than 60 clones were isolated, most of them being homoplasmic for the A8296G mutation and the remainder containing different proportions of the A8296G-G8363A double mutation. Several clones were selected for further biochemical and molecular characterization, essentially homoplasmic (>99%) for the single A8296G mutation [22] or homoplasmic (>99%) for both A8296G and G8363A mutations (the present study). Since the A8296G mutation is virtually homoplasmic in the patient's derived skin fibroblasts, it was impractical to continue the isolation of clones to obtain cybrids homoplasmic for the G8363A mutation alone. To achieve this, we generated additional transmitochondrial cybrids fusing the human mtDNA-less ρ0-206 cell line with fibroblasts obtained from a patient suffering from spinocerebellar ataxia and multiple lipomas, containing approx. 80% of the G8363A mutation. We isolated several independent clones homoplasmic for the G8363A mutation. We used as control a cybrid cell line obtained by the fusion of the ρ0-206 cell line with fibroblasts homoplasmic for wild-type mtDNA. The genetic characterization of the different cybrid cell lines was performed by PCR–RFLP as described previously [14] (results not shown). All the clones contained similar amounts of mtDNA as clones containing wild-type mtDNA, suggesting that there is no negative selection against the mutation.
The G8363A mutation produces severe biochemical defects of the respiratory chain in cybrid cell lines
We investigated the respiratory capacity of at least two independent clones harbouring either wild-type mtDNA or the specific kind of mutations by measuring the specific activities of the mitochondrial respiratory chain complexes I–IV and the rate of oxygen consumption in intact cells. We have reported previously that homoplasmic single A8296G mutant clones showed normal values in the respiratory chain activities and oxygen consumption, comparable with those of homoplasmic wild-type mtDNA clones [22]. In contrast, cybrid clones homoplasmic for the G8363A single mutant or A8296G-G8363A double mutant showed a clear defect of the mitochondrial respiratory chain complexes I, III and IV, with a mean residual activity of 36% (complex I) and 10% (complexes III and IV) in mutant clones when compared with wild-type lines or homoplasmic >95% A8296G clones (Table 1). Oxygen consumption was severely impaired in these clones (Table 1; mean decrease was 95%, P<0.001).
Table 1. Respiratory function assays in cybrid cell lines.
Results are expressed as means±S.D.; each individual clone was analysed twice. *P<0.001 and **P<0.01 indicate statistical significance with respect to wild-type (wt) and 100% A8296G cell lines respectively, and ***P<0.05 indicates statistical significance with respect to 100% A8296G cell lines, by the two-tailed, Mann–Whitney U test for unpaired samples. Oxygen consumption is expressed in terms of fmol·min−1·cell−1 and mean specific enzyme activity is expressed in terms of (nmol of substrate)·min−1·(mg of protein)−1.
| Mean specific enzyme activity | ||||||
|---|---|---|---|---|---|---|
| Cell line | Oxygen consumption | Complex I | Complex II | Complex III | Complex IV | Citrate synthase |
| Wt (n=6) | 4.6±0.9 | 14.3±1.2 | 10.8±3.5 | 104.0±25 | 54±8 | 200±26 |
| 8363 (n=2) | 0 | 5.6±0.8*,*** | 10.2±1.6 | 15.0±4.5** | 5.1±2.8* | 188±22 |
| 8296 (n=9) | 4.5±0.8 | 11.8±3.4 | 9.1±4.0 | 93.1±24 | 57±3 | 183±22 |
| 8296/8363 (n=6) | 0.1±0.1* | 3.9±1.0* | 11.1±3.0 | 5.5±4.5* | 6.4±2.6* | 188±22 |
| 143B (n=3) | 4.4±0.5 | 18.1±3.5 | 14.1±3.7 | 102±23 | 70±20 | 189±30 |
| 143B/206 (ρ0) (n=3) | 0 | 1.5±0.4 | 11.1±0.8 | 2±1.8 | 1±0.8 | 167±15 |
Mitochondrial tRNALys steady-state levels and conformation analysis
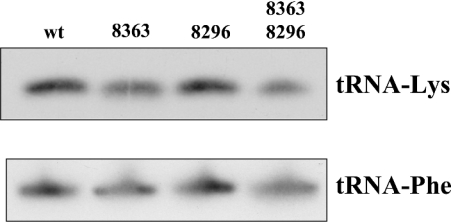
The potential effect of the G8363A mutation on the steady-state level of the tRNALys was investigated by Northern-blot analysis. Total RNA was extracted from mutants and wild-type cybrids, electrophoresed on agarose–formaldehyde gels and probed with a DNA fragment containing the tRNALys gene labelled with [α-32P]-dCTP. As control, we have quantified the steady-state level of the tRNAPhe using a specific tRNAPhe probe (Figure 1). The ratios between the signals obtained with a tRNALys gene probe and a tRNAPhe gene probe used as control were similar in all cases, indicating that neither the G8363A mutation alone nor the A8296G-G8363A double mutations significantly decrease the stability of the tRNALys. To find out whether the mutations introduce changes in the conformation of the tRNA, we performed non-denaturing single-strand analysis as described in the Materials and methods section. In contrast with the lack of effect observed in tRNA stability, the G8363A mutation produces a drastic shift in the mobility of the tRNALys, and practically the entire tRNA shows a delayed migration (Figure 2). Interestingly, the tRNALys harbouring the A8296G-G8363A double mutations showed a slightly increased delay in the electrophoretic migration compared with the tRNALys harbouring the single G8363A mutation. This result clearly demonstrated that the conformation of the tRNALys is not changed by the A8296G mutation and its alteration is due to the G8363A mutation. Furthermore, the A8296G mutation has some synergistic role in the change induced by the G8363A mutation.
Figure 1. Steady-state levels of tRNALys in transmitochondrial wild-type and mutant cybrid cell lines.
Total RNA extracted from various cybrid cell lines was analysed by Northern blotting using specific tRNALys and tRNAPhe probes. Equal amounts of RNA (5 μg) were fractionated on 12% polyacrylamide–7 M urea gels, transferred on to a nylon membrane, hybridized with the tRNALys and tRNAPhe radiolabelled probes and autoradiographed. Signals from at least four independent experiments from each cybrid cell line were quantified by densitometric and/or phosphoimager analyses. No statistically significant differences were detected (the Kruskal–Wallis method).
Figure 2. tRNA conformational analysis.
Total RNA (8 μg) extracted from the various cybrid cell lines was electrophoresed on a 6% polyacrylamide gel under non-denaturing conditions, transferred on to a nylon membrane, hybridized with the tRNALys radiolabelled probe and autoradiographed. The same filter was rehybridized with the tRNAPhe probe and analysed as described above.
Synthesis of mitochondrial proteins
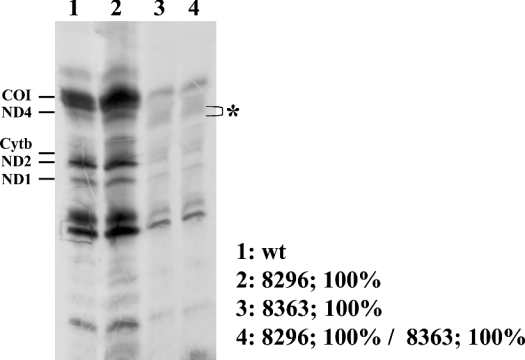
Figure 3 shows a representative experiment to investigate the effect of the mutations on the mitochondrial translation capacity. In contrast with homoplasmic A8296G cybrids [22], homoplasmic G8363A and double mutant A8296G-G8363A cybrids showed a marked decrease in the incorporation of [35S]methionine into the mitochondrially encoded polypeptides. A landmark of previously characterized tRNALys mutations is the presence of abnormal polypeptides generated by premature termination of translation. In particular, one of them, designated pMERRF, is derived from premature termination of COI (subunit 1 of cytochrome c oxidase) and migrates below the ND4 polypeptide [10]. Although we have not detected a prominent truncated product in mutant cells, in the mitochondrial translation pattern of G8363A and A8296G-G8363A cybrids there are relative increases in the radioactive signal in the region that migrates below the ND4 polypeptide (indicated by an asterisk in Figure 3), suggesting that, in the mutant cybrids, there are some accumulations of truncated polypeptides. These results confirm that the behaviour of the mtDNA harbouring the A8296G mutation is similar to wild-type and indicate that the G8363A mutation has a profound effect on mitochondrial protein synthesis.
Figure 3. Mitochondrial protein synthesis in transmitochondrial wild-type and mutant cybrid cell lines.
Cells were labelled with [35S]methionine in the presence of emetine as described in the Materials and methods section. A fluorogram of the mitochondrial translation products after fractionation on an SDS 15–20% polyacrylamide gel is shown. The position of different mitochondrial polypeptides is indicated; *, presence of potential abnormal translation products in mutant cybrid cells.
tRNALys aminoacylation
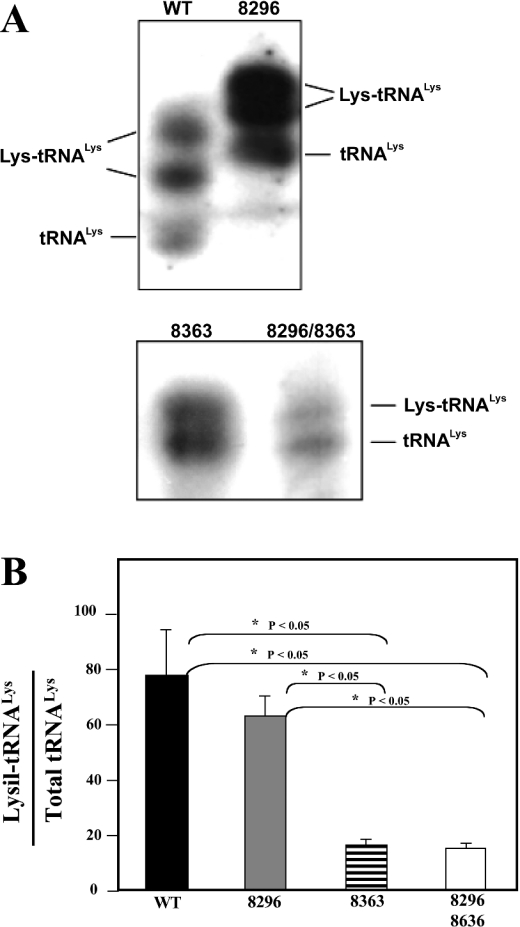
To determine whether the G8363A mutation impairs the aminoacylation capacity of the tRNALys, we isolated, from purified mitochondria, total RNA under conditions that preserved the aminoacylated tRNAs. Total RNAs were electrophoresed on 6.5% polyacrylamide–8 M urea gels, electroblotted on to nylon membranes and hybridized with specific tRNALys and tRNAPhe probes as described in the Materials and methods section. Aminoacylated and non-aminoacylated tRNALys can clearly be observed under these conditions in both wild-type and mutant cybrids, although the mobility of the wild-type tRNALys and Lys-tRNALys is somewhat slower compared with the mutant ones (Figure 4A). When the RNA samples were heated and electrophoresed under the same conditions, a total conversion of the aminoacylated-tRNALys in its deacylated form was observed (results not shown). As a control of the loading of the gel, we rehybridized the filters with a tRNAPhe-specific probe. Under these conditions, the acylated and deacylated tRNAPhe forms were not resolved (results not shown).
Figure 4. Aminoacylation of tRNALys in transmitochondrial wild-type and mutant cybrid cell lines.
(A) Total mitochondrial RNA was extracted under acidic conditions and electrophoresed on an 8% non-denaturing polyacrylamide gel as described in the Materials and methods section. RNAs were transferred on to a nylon membrane, hybridized with a specific tRNALys-radiolabelled probe and autoradiographed. (B) Signals corresponding to the aminoacylated and deacylated forms of the tRNALys obtained in at least three independent experiments were densitometrically quantified. Results are expressed as a percentage of the aminoacylated form±S.D.; *P<0.05, protected least significant difference test.
Quantitative densitometric analysis of the autoradiograms showed a moderate but significant decrease in the percentage of the aminoacylated form of the tRNALys harbouring the A8296G mutation (61%; Figure 4B) when compared with tRNALys wild-type (76%). The tRNALys harbouring the G8363A mutation had a drastic reduction in the amount of its aminoacylated form (18%), indicating that a change in the conformation of the tRNA produced by the G8363A mutation impairs drastically its aminoacylation capacity. The presence of the A8296G mutation did not act synergistically, since the same percentage of aminoacylated forms was present in the tRNALys containing the two mutations (A8296G+G8363A).
DISCUSSION
To date, 12 different mutations in the mtDNA tRNALys gene have been described in association with an array of mitochondrial diseases [7]. We first documented a A8296G-G8363A double mutant in the mtDNA tRNALys gene in a family with MERRF [14]. Both mutations have been reported in isolation in independent pedigrees [15–21]. Interestingly, we have recently reported that homoplasmic A8296G cybrid clones have normal respiratory chain function, as shown by oxygen consumption, lactate production and respiratory chain enzyme activities [22]. However, the possibility that the mutation might cause subtle defects of oxidative phosphorylation or a defect depending on the nuclear background was not fully excluded.
To investigate the pathogenic role of the G8363A mutation, we have generated and extensively characterized cybrid clones harbouring homoplasmic levels of mutant mtDNAs in two different mtDNA tRNALys genetic backgrounds, i.e. wild-type and A8296G. Homoplasmic G8363A as well as G8363A+A8296G mutants caused a drastic decrease in the activities of the respiratory-chain complexes I, III and IV, which contain mtDNA-encoded subunits, whereas the activity of complex II (containing only nuclear DNA-encoded subunits) remained normal. Consistently, we observed a marked decrease in oxygen consumption and in mitochondrial protein synthesis in clones harbouring homoplasmic G8363A and A8296G-G8363A mutations, whereas oxygen consumption and the mitochondrial synthetic capacity in clones homoplasmic for the single A8296G mutation were similar to wild-type. The G8363A mutation produced a drastic conformational change in the tRNALys that is slightly increased by the presence of the A8296G mutation. Furthermore, there was evidence of a significant decrease (57–59%) in the aminoacylation capacity of the tRNALys harbouring the G8363A mutation, of the same range as detected in the tRNALys harbouring the A8296G-G8363A double mutant. In contrast, the presence of the A8296G mutation decreases only moderately (15–17%) the aminoacylation capacity of the tRNALys.
These results indicate that a decrease in the aminoacylation capacity is, at least in part, the pathogenic molecular mechanism causing the collapse in mitochondrial protein synthesis and OXPHOS defects of the G8363A mutation. This pathogenic mechanism is not significantly modified by the presence of the A8296G mutation. Aminoacylation defects have been found in other tRNALys point mutations analysed in transmitochondrial cybrids. The well-established pathogenic A8344G mutation in the tRNALys gene produces a mild decrease in tRNA stability and tRNA aminoacylation, decreasing tRNALys lysine charging capacity in mitochondria [10]. In addition, the G8313A mutation in the tRNALys produces a strong decrease in the tRNA stability and decreases its aminoacylation capacity [28]. In contrast, neither the G8363A mutation nor the A8296G-G8363A mutation alters the steady-state level of the tRNALys, indicating that the stability of the tRNALys is not altered by the presence of the mutations. In addition, the steady-state levels of the flanking mRNAs (COII and ATPases 6–8) are similar in wild-type and mutant cybrids (results not shown), suggesting that the processing of the heavy-strand pre-mRNA is correct at the tRNALys site.
If our results demonstrate unequivocally that the G8363A mutation is pathogenic, the potential pathogenicity of the A8296G mutation is still puzzling. The high degree of conservation of this specific nucleotide in the tRNALys gene during evolution, together with the fact that several pedigrees are found in which the mutation segregates with the affected phenotype and the absence of the mutation in a large number of ethnically matched normal controls contrast with the apparent lack of a disease-causative role in a commonly used experimental cell system. The modest decrease in the aminoacylation capacity of the tRNALys that we observed does not suffice to affect the respiratory function. It is possible that the mutation achieves its pathogenic effects when present within a specific nuclear background or, alternatively, that it impairs mitochondrial physiology by a mechanism not related to the OXPHOS function. In this regard, it has been observed that cells with the A3243G mutation, a well-known pathogenic mutation in the mitochondrial tRNALeu(UUR) gene, produced an impaired mitochondrial translation machinery that depends heavily on the specific nuclear background (reviewed in [29]). The A3243G mutation is related to a profound decrease in the steady-state level and aminoacylation capacity of the tRNALeu(UUR) in some cases [11,30], whereas it decreases the association of mRNA with mitoribosomes in other cases [31]. However, aminoacylation is practically unaffected in some other cell lines and impaired mitochondrial protein synthesis occurs because of misreading of the phenylalanine codon due to a decrease in the modification of the U present in the wobble position of the anti-codon [32]. Moreover, it has been described in a different cell system that a defect in processing of the pre-mRNA at the tRNALeu(UUR) site leads to the accumulation of an intermediary transcript that might contribute to the impairment of mitochondrial protein synthesis [13,33]. Finally, and most interestingly, an RNA circularization technique has demonstrated that the decrease in total and/or aminoacylated tRNALeu(UUR levels also varies considerably in vivo when muscle biopsied samples from MELAS patients are used [34]. This does not apply to biopsied material from MERRF patients harbouring mutations in the tRNALys gene [34]. These results suggest that trans-acting factors and/or compensatory mechanisms that depend on the nuclear background modulate the expression of the mtDNA mutations. In this regard, established cybrid cell lines with defined molecular pathogenic mechanisms may be critical for identifying these factors in the near future [29].
Acknowledgments
This work was supported by grants from Ministerio de Educación y Ciencia of Spain, Comunidad de Madrid and Instituto de Salud Carlos III, Redes de centros RCMN (C03/08) and Temáticas (G03/011). The work in F. M. S.'s laboratory was partially supported by the Italian Ministry of Health. We thank M. Zeviani for his kind collaboration during the first stage of this work.
References
- 1.Scheffler I. E. Mitochondria. New York: Wiley-Liss; 1999. [Google Scholar]
- 2.Garesse R., Vallejo C. G. Animal mitochondrial biogenesis and function: a regulatory cross-talk between two genomes. Gene. 2001;263:1–16. doi: 10.1016/s0378-1119(00)00582-5. [DOI] [PubMed] [Google Scholar]
- 3.Attardi G., Schatz G. Biogenesis of mitochondria. Annu. Rev. Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Moreno M. A., Bornstein B., Campos Y., Arenas J., Garesse R. The pathogenic role of point mutations affecting the translational initiation codon of mitochondrial genes. Mol. Genet. Metab. 2000;70:238–240. doi: 10.1006/mgme.2000.3005. [DOI] [PubMed] [Google Scholar]
- 5.Smeitink J. A. Mitochondrial disorders: clinical presentation and diagnostic dilemmas. J. Inherit. Metab. Dis. 2003;26:199–207. doi: 10.1023/a:1024489218004. [DOI] [PubMed] [Google Scholar]
- 6.Zeviani M., Spinazzola A. Mitochondrial disorders. Curr. Neurol. Neurosci. Rep. 2003;3:423–432. doi: 10.1007/s11910-003-0026-9. [DOI] [PubMed] [Google Scholar]
- 7.Servidei S. Mitochondrial encephalomyopathies:gene mutation. Neuromuscul. Disord. 2003;13:848–853. doi: 10.1016/s0960-8966(03)00209-8. [DOI] [PubMed] [Google Scholar]
- 8.DiMauro S., Schon E. A. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 9.Leonard J. V., Schapira A. H. Mitochondrial respiratory chain disorders I: mitochondrial DNA defects. Lancet. 2000;355:299–304. doi: 10.1016/s0140-6736(99)05225-3. [DOI] [PubMed] [Google Scholar]
- 10.Enriquez J. A., Chomyn A., Attardi G. MtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNA(Lys) and premature translation termination. Nat. Genet. 1995;10:47–55. doi: 10.1038/ng0595-47. [DOI] [PubMed] [Google Scholar]
- 11.El Meziane A., Lehtinen S. K., Holt I. J., Jacobs H. T. Mitochondrial tRNALeu isoforms in lung carcinoma cybrid cells containing the np 3243 mtDNA mutation. Hum. Mol. Genet. 1998;7:2141–2147. doi: 10.1093/hmg/7.13.2141. [DOI] [PubMed] [Google Scholar]
- 12.Helm M., Florentz C., Chomyn A., Attardi G. Search for differences in post-transcriptional modification patterns of mitochondrial DNA-encoded wild-type and mutant human tRNALys and tRNALeu(UUR) Nucleic Acids Res. 1999;27:756–763. doi: 10.1093/nar/27.3.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaufmann P., Koga Y., Shanske S., Hirano M., DiMauro S., King M. P., Schon E. A. Mitochondrial DNA and RNA processing in MELAS. Ann. Neurol. 1996;40:172–180. doi: 10.1002/ana.410400208. [DOI] [PubMed] [Google Scholar]
- 14.Arenas J., Campos Y., Bornstein B., Ribacoba R., Martin M. A., Rubio J. C., Santorelli F. M., Zeviani M., DiMauro S., Garesse R. A double mutation (A8296G and G8363A) in the mitochondrial DNA tRNA (Lys) gene associated with myoclonus epilepsy with ragged-red fibers. Neurology. 1999;52:377–382. doi: 10.1212/wnl.52.2.377. [DOI] [PubMed] [Google Scholar]
- 15.Kameoka K., Isotani H., Tanaka K., Kitaoka H., Ohsawa N. Impaired insulin secretion in Japanese diabetic subjects with an A-to-G mutation at nucleotide 8296 of the mitochondrial DNA in tRNA(Lys) Diabetes Care. 1998;21:2034–2035. doi: 10.2337/diacare.21.11.2034. [DOI] [PubMed] [Google Scholar]
- 16.Kameoka K., Isotani H., Tanaka K., Azukari K., Fujimura Y., Shiota Y., Sasaki E., Majima M., Furukawa K., Haginomori S., et al. Novel mitochondrial DNA mutation in tRNA(Lys) (8296A→G) associated with diabetes. Biochem. Biophys. Res. Commun. 1998;245:523–527. doi: 10.1006/bbrc.1998.8437. [DOI] [PubMed] [Google Scholar]
- 17.Akita Y., Koga Y., Iwanaga R., Wada N., Tsubone J., Fukuda S., Nakamura Y., Kato H. Fatal hypertrophic cardiomyopathy associated with an A8296G mutation in the mitochondrial tRNA(Lys) gene. Hum. Mutat. 2000;15:382. doi: 10.1002/(SICI)1098-1004(200004)15:4<382::AID-HUMU15>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 18.Sakuta R., Honzawa S., Murakami N., Goto Y., Nagai T. Atypical MELAS associated with mitochondrial tRNA(Lys) gene A8296G mutation. Pediatr. Neurol. 2002;27:397–400. doi: 10.1016/s0887-8994(02)00456-3. [DOI] [PubMed] [Google Scholar]
- 19.Ozawa M., Nishino I., Horai S., Nonaka I., Goto Y. I. Myoclonus epilepsy associated with ragged-red fibers: a G-to-A mutation at nucleotide pair 8363 in mitochondrial tRNA(Lys) in two families. Muscle Nerve. 1997;20:271–278. doi: 10.1002/(SICI)1097-4598(199703)20:3<271::AID-MUS2>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 20.Santorelli F. M., Mak S. C., El-Schahawi M., Casali C., Shanske S., Baram T. Z., Madrid R. E., DiMauro S. Maternally inherited cardiomyopathy and hearing loss associated with a novel mutation in the mitochondrial tRNA(Lys) gene (G8363A) Am. J. Hum. Genet. 1996;58:933–939. [PMC free article] [PubMed] [Google Scholar]
- 21.Casali C., Fabrizi G. M., Santorelli F. M., Colazza G., Villanova M., Dotti M. T., Cavallaro T., Cardaioli E., Battisti C., Manneschi L., et al. Mitochondrial G8363A mutation presenting as cerebellar ataxia and lipomas in an Italian family. Neurology. 1999;52:1103–1104. doi: 10.1212/wnl.52.5.1103. [DOI] [PubMed] [Google Scholar]
- 22.Bornstein B., Mas J. A., Fernandez-Moreno M. A., Campos Y., Martin M. A., del Hoyo P., Rubio J. C., Arenas J., Garesse R. The A8296G mtDNA mutation associated with several mitochondrial diseases does not cause mitochondrial dysfunction in cybrid cell lines. Hum. Mutat. 2002;19:234–239. doi: 10.1002/humu.10050. [DOI] [PubMed] [Google Scholar]
- 23.King M. P., Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J., Fritsch E. F., Maniatis T. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 25.Hao H., Moraes C. T. A disease-associated G5703A mutation in human mitochondrial DNA causes a conformational change and a marked decrease in steady-state levels of mitochondrial tRNA(Asn) Mol. Cell. Biol. 1997;17:6831–6837. doi: 10.1128/mcb.17.12.6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enriquez J. A., Attardi G. Analysis of aminoacylation of human mitochondrial tRNAs. Methods Enzymol. 1996;264:183–196. doi: 10.1016/s0076-6879(96)64019-1. [DOI] [PubMed] [Google Scholar]
- 27.Chomyn A. In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 1996;264:197–211. doi: 10.1016/s0076-6879(96)64020-8. [DOI] [PubMed] [Google Scholar]
- 28.Bacman S. R., Atencio D. P., Moraes C. T. Decreased mitochondrial tRNALys steady-state levels and aminoacylation are associated with the pathogenic G8313A mitochondrial DNA mutation. Biochem. J. 2003;374:131–136. doi: 10.1042/BJ20030222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs H. T., Holt I. J. The np 3243 MELAS mutation: damned if you aminoacylate, damned if you don't. Hum. Mol. Genet. 2000;9:463–465. doi: 10.1093/hmg/9.4.463. [DOI] [PubMed] [Google Scholar]
- 30.Janssen G. M., Maassen J. A., van Den Ouweland J. M. The diabetes-associated 3243 mutation in the mitochondrial tRNA(Leu(UUR)) gene causes severe mitochondrial dysfunction without a strong decrease in protein synthesis rate. J. Biol. Chem. 1999;274:29744–29748. doi: 10.1074/jbc.274.42.29744. [DOI] [PubMed] [Google Scholar]
- 31.Chomyn A., Enriquez J. A., Micol V., Fernandez-Silva P., Attardi G. The mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episode syndrome-associated human mitochondrial tRNALeu(UUR) mutation causes aminoacylation deficiency and concomitant reduced association of mRNA with ribosomes. J. Biol. Chem. 2000;275:19198–19209. doi: 10.1074/jbc.M908734199. [DOI] [PubMed] [Google Scholar]
- 32.Yasukawa T., Suzuki T., Ueda T., Ohta S., Watanabe K. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J. Biol. Chem. 2000;275:4251–4257. doi: 10.1074/jbc.275.6.4251. [DOI] [PubMed] [Google Scholar]
- 33.Schon E. A., Koga Y., Davidson M., Moraes C. T., King M. P. The mitochondrial tRNA(Leu)(UUR)) mutation in MELAS: a model for pathogenesis. Biochim. Biophys. Acta. 1992;1101:206–209. [PubMed] [Google Scholar]
- 34.Börner G. V., Zeviani M., Tiranti V., Carrara F., Hoffmann S., Gerbitz K. D., Lochmuller H., Pongratz D., Klopstock T., Melberg A., et al. Decreased aminoacylation of mutant tRNAs in MELAS but not in MERRF patients. Hum. Mol. Genet. 2000;9:467–475. doi: 10.1093/hmg/9.4.467. [DOI] [PubMed] [Google Scholar]