Abstract
The cuticle constitutes the outermost defensive barrier of most land plants. It comprises a polymeric matrix—cutin, surrounded by soluble waxes. Moreover, the cuticle constitutes the first line of defense against pathogen invasion, while also protecting the plant from many abiotic stresses. Aliphatic monomers in cutin have been suggested to act as immune elicitors in plants. This study analyses the potential of cutin oligomers to activate rapid signaling outputs reminiscent of pattern-triggered immunity in the model plant Arabidopsis. Cutin oligomeric mixtures led to Ca2+ influx and mitogen-activated protein kinase activation. Comparable responses were measured for cutin, which was also able to induce a reactive oxygen species burst. Furthermore, cutin oligomer treatment resulted in a unique transcriptional reprogramming profile, having many archetypal features of pattern-triggered immunity. Targeted spectroscopic and spectrometric analyses of the cutin oligomers suggest that the elicitor compounds consist mostly of two up to three 10,16-dihydroxyhexadecanoic acid monomers linked together through ester bonds. This study demonstrates that cutin breakdown products can act as inducers of early plant immune responses. Further investigation is needed to understand how cutin breakdowns are perceived and to explore their potential use in agriculture.
Keywords: Arabidopsis thaliana, calcium influx, cuticle, cutin oligomers, MAPK activation, Nicotiana benthamiana, pattern-triggered immunity (PTI), RNA-seq, Solanum lycopersicum
Cutin deconstruction releases small molecules consisting mostly of esterified 10,16-dihydroxyhexadecanoic acid units. These molecules elicit early immune responses in plants, strengthening cutin’s defensive role against damage/infection in plants.
Introduction
Plants occupied land environments ~450 million years ago (Delwiche and Cooper, 2015). The transition from water to land habitats exposed plants to numerous challenges imposed by an extremely desiccating environment (Waters, 2003). To control water loss, protect against UV radiation and pathogens, and reinforce the epidermal cell layer, plants developed a hydrophobic barrier—the cuticle (Martin and Rose, 2014; Fich et al., 2016). The cuticle is composed of a polymeric matrix of cutin associated with organic solvent-soluble lipids (waxes) (Yeats and Rose, 2013). In addition, cutin interaction with the polysaccharides that build up the epidermal cell walls has been proposed (Philippe et al., 2020; Xin and Fry, 2021), but the nature of such anchoring remains uncertain.
During infection of the aerial organs of plants, fungal spores release cutin-degrading enzymes—cutinases, having esterase activity (Longhi and Cambillau, 1999) that can disrupt the polymeric matrix and release cutin-derived molecules. Perception of these molecules by the fungus increases the production of cutinases that breach the cuticle barrier, thus allowing the fungus to invade the plant organ (Kolattukudy et al., 1995).
To fend off pathogen invasion, plants have developed a highly specialized mechanism to sense biotic threats by using cell surface pattern recognition receptors (PRRs) (Zipfel, 2014). These receptors perceive pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs), derived from the invading pathogens or from the breakdown of plant tissues, respectively (Zipfel, 2014). Cutin aliphatic monomers (i.e. the major basic elements making up the cutin polymer) have been proposed as DAMPs due to their ability to induce some elements of a canonical immune response, namely the production of reactive oxygen species (ROS) in cucumber, rice, and Arabidopsis thaliana (hereafter Arabidopsis), and the up-regulation of defense-related genes in rice and Arabidopsis (Kauss et al., 1999; Kim et al., 2008; Park et al., 2008). Exogenous application of monomers obtained from plants having augmented cuticular permeability (SlSHN3-OE) increased the resistance of Micro-Tom tomato plants against the fungal pathogen Botrytis cinerea, and activated defense-responsive genes (Buxdorf et al., 2014); however, the nature of the elicitor(s) remains unresolved. Cutin aliphatic monomers were also reported to induce the production of antimicrobial compounds (Serrano et al., 2014). Although cutin monomers have been proposed as DAMPs, their capabilities to elicit other important hallmark early immune responses, for example intracellular calcium influx and activation of mitogen-activated protein kinases (MAPKs), have never been observed (Serrano et al., 2014; Hou et al., 2019). Also, it is unclear if the tested cutin aliphatic monomers are the most potent class of cutin-derived immune elicitors. Esterase-based degradation of cutin progresses through ester cleavage, probably releasing cutin oligomers and not only monomers (Beneloujaephajri et al., 2013). This raises the hypothesis that cutin oligomers can activate signaling outputs reminiscent of pattern-triggered immunity (PTI), similar to that proposed for cutin monomers.
To investigate the hypothesis that cutin oligomers (COMs) can activate PTI responses, the cutin polymer was first isolated from tomato peel (Moreira et al., 2020; Bento et al., 2021), and subsequently broken down through a mild chemical hydrolysis to generate COMs. As such, the ensuing COMs constitute an adequate model of cutins comprising 10,16-dihydroxyhexadecanoic acid (dihydroxy-C16 acid) units. The ability of the produced COMs to activate calcium influx, MAPK activation, and transcriptional reprograming in Arabidopsis was investigated. The results clearly indicate that COMs act as elicitors of rapid immune responses. Spectroscopy and spectrometry analyses suggested that the elicitors are dimers and/or trimers consisting mostly of esterified dihydroxy-C16 acid units, one of which is possibly methylated. The hypothesis that cutin disruption releases oligomers able to act as elicitors of plant immunity, and hence potentially constituting a new class of DAMPs, is discussed.
Materials and methods
Plant growth conditions
Arabidopsis thaliana Col-0 seeds were germinated on soil, and plants were grown for 4 weeks in an Aralab Fitoclima climate chamber with 150 μmol m–2 s–1 light intensity, following a 10 h/14 h day/night cycle, under constant temperature of 20 °C and 60% humidity. Plants were watered automatically for 10 min three times per week. Seeds of A. thaliana Col-0 and the mutants bak1-5 bkk1, cerk1-2AEQ, and bbc (Ranf et al., 2011; Roux et al., 2011; Xin et al., 2016), all in the Col-0 background, were germinated on plates with 0.5× Murashige and Skoog (MS) basal salt mixture supplemented with 1% (w/v) sucrose and 0.9% (w/v) phytoagar. After 4 d, seedlings were transferred to 24-well sterile culture plates containing 0.5× MS mixture supplemented with 1% (w/v) sucrose and grown in sterile conditions in an Aralab Fitoclima climate chamber with 120 μmol m–2 s–1 light intensity, following a 16 h/8 h day/night cycle, under temperatures of 20 °C during the day and 18 °C at night. The growth period was 8, 11, or 14 d depending on the subsequent assays.
Nicotiana benthamiana plants seeds were germinated on soil, and plants were grown for 4 weeks in a greenhouse room with 150 μmol m–2 s–1 light intensity, following 16 h/8 h day/night cycle, under constant temperature of 24 °C. These plants were watered automatically daily for 20 min.
Cutin extraction
Cutin was extracted from tomato pomace as previously described (Moreira et al., 2020). The tomato pomace was obtained from Sumol + Compal, SA., and dried at 60 °C for 1 week to constant weight. Dry pomace was then milled using a Retsch ZM200 electric grinder (granulometry 0.5 mm; 10 000 rpm) and stored at room temperature until further use. In brief, tomato pomace and cholinium hexanoate were mixed (1:10) and incubated for 2 h at 100 °C. The reaction was stopped by the addition of 80 ml of DMSO per gram of tomato pomace. The polyester was recovered by filtration using a nylon membrane filter (0.45 µm). Purification was obtained by washing with an excess of deionized water to remove all traces of DMSO. Purified cutin was then freeze-dried and stored at room temperature for further use. Suspensions of the purified cutin powder (insoluble) of 2 mg ml–1 were prepared in MilliQ water for testing purposes, since they allow reproducible results.
Cutin hydrolysis
To obtain a COM, a sodium methoxide-catalyzed methanolysis was performed by mixing 0.5 g of cutin with 20 ml of a solution of sodium methoxide (0.1 M) in anhydrous methanol, at 40 °C for 2 h without stirring. At the end of the reaction, the mixture was cooled to room temperature and centrifuged (4 °C, 30 min, 4000 g) to recover the non-hydrolyzed cutin fraction. The supernatant (hydrolyzed fraction) was acidified to pH 3–3.5 by addition of 37% HCl and subsequently centrifuged (4 °C, 30 min, 4000 g). The resulting precipitate was recovered, and the supernatant extracted three times by dichloromethane/water partition to release the hydrolysates; and anhydrous sodium sulfate was added to remove traces of water. The solution was concentrated under a constant nitrogen flux at 40 °C. To obtain cutin or COM hydrolysates, a sodium hydroxide alkaline hydrolysis was performed by mixing a solution of 0.5 M NaOH in methanol/water (1:1, v/v) at 95 °C with the cutin/COM powder for 4 h. At the end of the reaction, the mixture was cooled to room temperature, then acidified to pH 3/3.5 with 1 M HCl, and subsequently extracted by dichloromethane/water partition to release the hydrolyzable constituents. The solution was concentrated under a constant nitrogen flux at 40 °C. Stock solutions of the COMs were prepared in heated DMSO. Precise aliquots were dissolved in MilliQ water to reach a final concentration of 2 mg ml–1 or 3 mg ml–1 for testing purposes (see below).
Immune assays
Leaf discs (collected using a 4 mm biopsy punch) or seedlings were transferred to white 96-well plates (one leaf disc or seedling per well) and equilibrated overnight in sterile ultrapure water (ROS measurements) or coelenterazine solution (calcium measurements). The following day, the equilibration solution was removed, and replaced with a solution containing 100 µg ml–1 up to 2 mg ml–1 COM, cutin hydrolysate, hydroxy palmitic acid (HPA), or cutin (calcium measurements), and mixed with 1 mM luminol, and 10 µg ml–1 horseradish peroxidase (HRP) in the case of ROS measurements. COM at 2 mg ml–1 was tested since this concentration was sufficient to observe a reproducible calcium influx. Positive controls were also prepared with 100 nM (flg22 and Pep1) in MilliQ water, as well as blanks with 0.5% (v/v) DMSO in MilliQ water or MilliQ water alone. Luminescence was detected and measured for 25–45 min using a Tecan Spark microplate reader (seedlings). The plate was scanned each minute, using a 250 ms integration time per well, at a 24 °C constant temperature. Calcium influx responses to COM treatment of Col-0AEQ seedlings and mutants (bak1-4AEQ and cerk1AEQ ) were done by normalization of the aequorin discharge values to allow comparison of distinct genotypes. The aequorin discharge was performed by the addition of a solution [2 M CaCl2, 20% ethanol] to a final concentration of 1 M/10% in each well. To determine the total remaining aequorin luminescence (Lmax), discharge kinetics of each well were measured for 45 s. Calcium concentrations were calculated by the ratio of the elicitor-induced aequorin luminescence counts per s (L) and the total remaining aequorin (Lmax) as previously described (Ranf et al., 2012). All experiments were executed in conditions that ensure minimal disturbance and no physical damage in the leaf discs/seedlings (compounds/mixtures were pipetted to avoid any abrasion of the plant tissue).
MAPK activation
For MAPK activation assays, 14-day-old seedlings were used. The growth medium was removed by inverting the plate on clean paper towels. Seedlings were treated for 30 min with 1 ml of 3 mg ml–1 COM, 100 nM flg22 (positive control), or the corresponding mock solutions (solvent control). The COM concentration was chosen due to its ability to cause a strong reproducible effect. Two seedlings per treatment were dried on clean paper towels, subsequently transferred to 1.5 ml tubes, and instantly frozen in liquid nitrogen. All treated seedlings were stored at –80 °C until further use. Frozen seedlings were pulverized using a nitrogen-cooled plastic micro pestle, then mixed with 150 µl of extraction buffer containing 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 2 mM EDTA, 10% (v/v) glycerol, 2 mM DTT, 1% (v/v) Igepal, and supplemented with protease and phosphatase inhibitors (equivalent to Sigma-Aldrich plant protease inhibitor cocktail and phosphatase inhibitor cocktails #2 and #3). The tissue was then ground at 1800 rpm using an automatic stirrer fitted with a plastic micro pestle. The tubes were centrifuged at 15 000 g for 20 min at 4 °C in a refrigerated microcentrifuge. After centrifugation, 50 μl of extract were transferred to a fresh 1.5 ml Eppendorf tube. Samples were prepared for SDS–PAGE by heating at 80 °C for 10 min in the presence of 6× SDS loading buffer and 100 mM DTT.
Proteins were loaded to a 12% (v/v) polyacrylamide gel, separated at 120 V for ~120 min, and subsequently transferred to a polyvinylidene fluoride (PVDF) membrane at 100 V for 90 min at 4 °C. Membranes were then blocked for 2 h at room temperature or overnight at 4 °C in 5% (w/v) milk in Tris-buffered saline (TBS; 50 mM Tris–HCl pH 7.4, 150 mM NaCl) containing 0.1% (v/v) Tween-20 (TBS-T). Blots were probed in a 1:4000 dilution of the NEB anti-p42/p44-erk primary antibody in 5% BSA in TBS-T for 2 h, followed by washing four times for 10 min each in TBS-T. Blots were then probed with a 1:10 000 dilution of anti-rabbit secondary antibody in 5% milk in TBS-T for 1 h, followed by washing three times for 5 min each in TBS-T. Finally, blots were washed for 5 min in TBS and treated with either standard ECL substrate or SuperSignal West Femto high sensitivity substrate (ThermoFisher Scientific). Blots were imaged using a Bio-Rad ChemiDoc Imaging System (Bio-Rad Laboratories).
RNA extraction and sequencing
For RNA-seq experiments, 14-day-old seedlings were grown as described above. After 9 d of growth in liquid MS medium supplemented with 1% sucrose, the medium was removed from the wells and replaced with 600 µl of fresh liquid MS per well. The following day, 400 µl of 3 mg ml–1 COM in 0.5% DMSO in MilliQ water or the corresponding mock solution were added to each well. Seedlings were treated for 30 min and then two were collected and instantly frozen in liquid nitrogen. In total, four biological replicates were generated for each treatment (COM and mock) and stored at –80 °C for further processing.
Frozen seedlings were pulverized while frozen using a Spex SamplePrep Geno Grinder 2010 at 1500 rpm for 90 s. Total RNA was extracted at 4 °C from two ground seedlings as previously described (Shi and Bressan, 2006) by addition of 900 µl of TRI reagent (Ambion) and 200 µl of chloroform, recovery of 400 µl from the aqueous phase, precipitation with 500 µl of isopropanol, and washing with 70% ethanol. All samples were then solubilized in 30 µl of RNase-free water. Samples were subsequently subjected to DNase treatment using a TURBO DNA-free Kit (Ambion) according to the manufacturer’s instructions. The reaction mix was incubated at 37 °C for 30 min, after which the inactivation reagent was added and incubated for 5 min at room temperature. After centrifugation, the supernatant was transferred to a new tube. Quantification and quality assessment of all RNA samples were evaluated on a TapeStation (Agilent), and the RNA sequencing was performed by the Beijing Genomics Institute (BGI).
RNA-seq data processing
For paired-end RNA-seq, libraries were generated at BGI according to the DNBSEQ stranded mRNA library system. Eight samples were indexed and sequenced using the DNBseq™ sequencing platform (20 million reads per sample). Generated FastQ files were analyzed with FastQC, and any low-quality reads were trimmed with Trimmomatic (Bolger et al., 2014).
All libraries were aligned to the A. thaliana genome assembly TAIR10 with gene annotations from Ensembl Plants v.49 using the HISAT2 v.2.1.0 pipeline (Kim et al., 2015) followed by read counts with HTSeq v. 0.13.5 (Anders et al., 2015). All RNA-seq experiments were carried out with four biological replicates. Differential expression analysis, and quality control principal component analysis (PCA) and MA plots were generated using the DESeq2 v.1.30.0 R package (Love et al., 2014). The genes that showed |log2| >1-fold changes in expression with an adjusted P-value <0.05 are defined as significantly differentially expressed genes (DEGs) in this analysis. Transcript abundance was defined as transcripts per kilobase million (TPM). Gene Ontology (GO) enrichment of the DEGs was performed with the topGO v.2.42.0 R package, using the Fisher exact test to attain significantly enriched categories.
Comparative analysis of transcriptome modification upon elicitor treatment
DEG lists in response to seven elicitors [hydroxy-fatty acid (3-OH-FA), chitooctamer (CO8), elf18, flg22, nlp20, oligogalacturonides (OGs), and Pep1] upon treatment under similar conditions to COM were obtained from Bjornson et al. (2021). This study followed a time course from 5 min to 3 h post-elicitation: a gene list was obtained for each elicitor with genes significantly induced at any time. Abiotic stress treatment analysis for seven abiotic stresses (heat, cold, drought, salt, high osmolarity, UV-B light, and wounding) was also obtained from Bjornson et al. (2021), based on ATH1 microarray experiments presented in Kilian et al. (2007). This study followed a time course from 5 min to 12 h post-elicitation: a gene list was obtained for each elicitor with genes significantly induced at any time up to 3 h. Comparisons and visualizations among DEGs were performed in R using the tools of the tidyverse v.1.3.1 package (Wickham et al., 2019). Spearman correlation among log2 (fold changes) for treatments was calculated using the Hmisc package in R v.4.5-0 and visualized using the corrplot package v.0.89. Annotation data for genes induced specifically by COM were obtained from The Arabidopsis Information Resource (TAIR) via Bioconductor package org.At.tair.db v.3.10.0. The raw data and the processed file are deposited in the BioStudies EMBL-EBI platform with the accession number E-MTAB-12956, and are also accessible through the SRA NCBI public database with the same accession number.
Quantitative analyses of total carbohydrate content
To evaluate the polysaccharide content, each COM sample was subjected to an acid hydrolysis (1 M H2SO4 in methanol) for 4 h at 90 °C. The hydrolyzable sugars were recovered in the supernatant through centrifugation (18 514 g, 4 °C, 20 min) and the pH was neutralized using 5 M NaOH in water. All samples were dried under a flux of nitrogen at room temperature. Quantification of carbohydrates in the dried hydrolysates was performed using the total carbohydrate assay kit from Sigma-Aldrich according to the manufacturer’s instructions. The samples were analyzed in triplicates.
NMR characterization of cutin oligomeric mixtures
NMR spectra of COMs were recorded using an Avance III 800 CRYO (Bruker Biospin, Rheinstetten, Germany). All NMR spectra (1H, 1H-1H COSY, 1H-13C HSQC, and 1H-13C HMBC) were acquired in DMSO-d6 using 5 mm diameter NMR tubes, at 60 °C as follows: 15 mg of COMs in 400 μl of DMSO-d6 (in triplicate) or for validation 40 mg of COMs in 400 μl of DMSO-d6. For quantification purposes, 1.25 mg of benzene (internal standard) was added to each sample. MestReNova, Version 11.04-18998 (Mestrelab Research, S.L.) was used to process the raw data acquired in the Bruker spectrometers.
GC-MS characterization of cutin oligomeric mixtures
To release the hydrolyzable constituents, the COMs were treated with a solution of 0.5 M NaOH in methanol:water [1:1 (v/v)] at 95 °C for 4 h. The mixture was cooled to room temperature and acidified to pH 3–3.5 with 1 M HCl, spiked with a known concentration of hexadecane (internal standard), and extracted three times with dichloromethane. Anhydrous sodium sulfate was added to the organic phase to remove water and concentrated under a nitrogen flow. The non-hydrolyzable fraction was recovered by filtration (cellulose nitrate filter) and subsequently washed, dried, and weighed (recalcitrance). The COM samples were also analyzed directly (i.e. not subjected to alkaline hydrolysis). The dried samples were derivatized in N,O-bis(trimethylsilyl)trifluoroacetamide containing 1% (v/v) trimethylchlorosilane in pyridine (5:1), for 30 min at 90 °C. The derivatives were then analyzed by GC-MS (Agilent: 7820A GC and 5977B quadrupole MS; HP-5MS column) as follows: ramp temperature 80 °C, then 2 °C min–1 to 310 °C for 15 min. The MS scan mode, with source at 230 °C and electron impact ionization (EI+, 70 eV), was used for all samples. Data acquisition was accomplished by MSD ChemStation (Agilent Technologies); compounds were identified based on the equipment spectral library (Wiley-National Institute of Standards and Technology) and quantified using external standards of the major classes of the aliphatic monomers (heptadecanoic acid, hexadecanedioic acid, and pentadecanol). All samples were analyzed in triplicates, each with technical duplicates.
LC-MS/MS characterization of COMs
The LC-MS/MS protocol was adapted from Bhunia et al. (2018). The experiments were performed in a Q Exactive Focus™ Hybrid Quadrupole-Orbitrap™ Mass Spectrometer coupled to a Dionex Ultimate 3000 UHPLC. HPA was used as a standard and prepared in isopropanol:methanol:acetonitrile (1:1:1) at a concentration of 200 ng µl–1. The samples were prepared in the same way at 1 µg µl–1. Separation was achieved in a Waters XBridge column C18 (2.1 × 150 mm, 3.5 µm particle size, P/N 186003023), using a gradient of an increasing percentage of 20 mM ammonium formate in isopropanol (IPA):acetonitrile (ACN) (75:25) (B) and a decreasing percentage of ACN:water (60:40) with 20 mM ammonium formate (A). The total method time was 77 min, the flow rate was 0.4 ml min–1, and the column was kept at 37 °C. The data were acquired using the Xcalibur software v.4.0.27.19 (Thermo Scientific). The method consisted of several cycles of full MS scans (R=70 000; scan range=100–1500 m/z) followed by three ddMS2 scans (R=17 500; NCE 30 V) in positive and negative mode. External calibration was performed using LTQ ESI Positive/Negative Ion Calibration Solution (Thermo Scientific). Generated mass spectra were processed using Compound Discoverer 3.2 (Thermo) for small molecule identification. The search was performed against the mass list with provided molecular formulas (dimers, trimers), as well as the mzCloud MS2, KEGG, and ChEBI MS1 databases. A 3 ppm mass tolerance was used. The minimum peak intensity (MS1) for detection was 106. A manual validation of the assignments for the identified oligomers was performed by inspection of the MS2 fragmentation profiles against the theoretical fragmentation generated on Mass Frontier 8.0 (Thermo). Theoretical chemical structures for the identified oligomers were generated in ChemDraw 21.0.0.
LDI-TOF and MALDI-TOF analyses of COMs and corresponding hydrolysates
The samples were analyzed by laser desorption/ionization (LDI)-time-of-flight (TOF) MS and by matrix-assisted laser desorption/ionization (MALDI)-time-of-flight (TOF) MS. As control for monomers, total alkaline hydrolysate of cutin was used. As oligomer control, a batch of oligomers [degree of polymerization (DP) DP1–DP8] were produced from purified cutin monomers as previously described (Marc et al., 2021) with slight modification. The polymerization time was reduced to 8 h and the oligomers were extracted from the polymer by hot (70 °C) ethanol extraction.
For the LDI-TOF analyses, the samples were deposited on a polished steel MALDI target plate and analyzed without any preparation. For the MALDI-TOF analyses, samples were mixed with the matrix solution composed of DHB (2,5-dihydroxybenzoic acid) 3 mg ml–1 in 75% methanol, with 2.5 mM LiCl, in a 1:3 ratio (v/v). The mixture (1 μl) was deposited on a polished steel MALDI target plate. Measurements were performed on a rapifleX MALDI‐TOF spectrometer (Bruker Daltonics, Bremen, Germany) equipped with a Smartbeam 3D laser (355 nm, 10 000 Hz) and controlled using the Flex Control 4.0 software package. The mass spectrometer was operated in reflectron mode with positive polarity for MALDI-TOF analyses and in negative polarity for LDI-TOF analyses. Spectra were acquired in the range of 180–5000 m/z. Neither the MALDI-TOF nor the LDI-TOF used in these experiments can observe the signal of the free p-coumaric acid.
Results and discussion
Cutin polymer activates a ROS burst
We hypothesized that the degradation of the plant polyester cutin is coordinated with the release of polymeric/oligomeric variants capable of eliciting hallmark early plant immune responses. We first tested the ability of a cutin polymeric variant to induce a ROS burst in Arabidopsis. The ROS burst was measured through a well-established luminol-based assay (Zhu et al., 2016).
The composition of COMs is highly conserved among species, regardless of distinct C16:C18 ratios (Moore et al., 2016). Although Arabidopsis has an unusual dicarboxylic acid-rich cutin, dihydroxy-C16 acid units are systematically found in all cutinized tissues (Bonaventure et al., 2004; Molina et al., 2006; Li-Beisson et al., 2009). Accordingly, as cutin source, we purified cutin from tomato pomace—enriched in dihydroxy-C16 acid units—since its high availability as an agroindustry residue (European Commission, 2021) enables the production of large amounts of polymeric structures. Specifically, to obtain the cutin, an ionic liquid extractant was applied to isolate a highly pure cutin polymer (hereafter simply referred as cutin), showing minor ester cleavage (Moreira et al., 2020). This method ensures faster and simpler purification of cutin compared with the conventional enzyme-based isolation (Moreira et al., 2020).
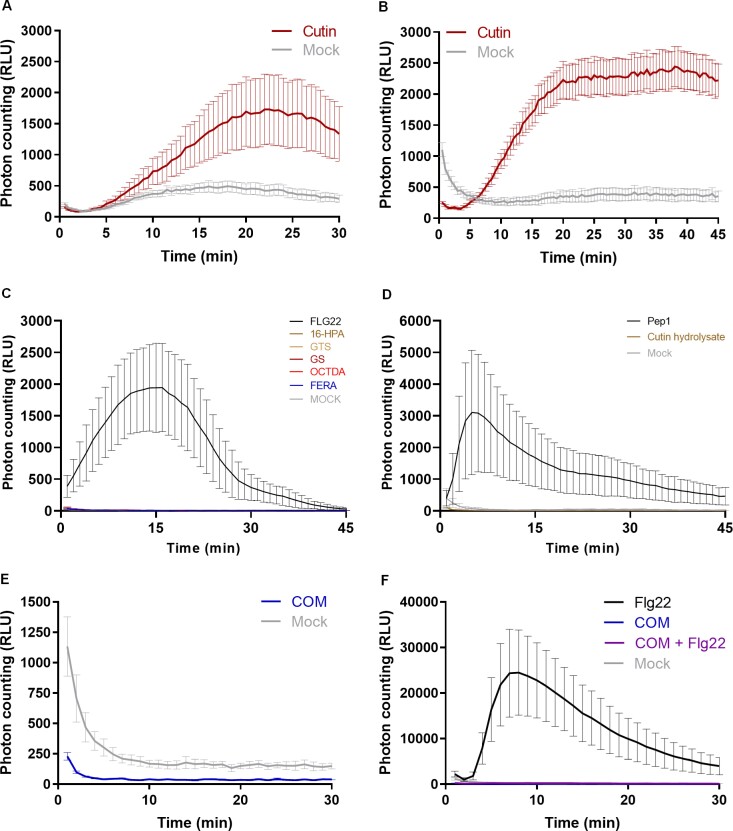
Exposure of Arabidopsis seedlings to cutin (suspension in MilliQ water) resulted in a clear ROS burst (Fig. 1A, B; Supplementary Fig. S1). Flg22, a 22 amino acid peptide derived from bacterial flagellin, is a well-established strong inducer of plant immunity (Felix et al., 1999) (used here as positive control). The effect was reproducible, and the response was not depleted at 45 min post-treatment (Fig. 1B). In contrast, pure compounds (commercially available), which are representative of cutin constituents, long chain fatty acids, hydroxycinnamic acids, or fatty acid monoglycerides having variable side chains, did not induce a ROS burst under the tested conditions (Fig. 1C; Supplementary Fig. S2). Cutin hydrolysates, which are obtained by an extensive hydrolysis of the cutin, consist almost exclusively of aliphatic monomers with a few aromatic monomers (Escórcio et al., 2022). These hydrolysates also did not elicit a ROS burst (Fig. 1D). Collectively, the results suggest that once the polymeric backbone of the plant polyester is deconstructed to its constituent monomeric pieces, its capability to elicit a ROS burst is lost; hence some preservation of the polymeric backbone might be required for elicitation of a ROS burst.
Fig. 1.
Cutin polymer, but not pure cutin constituents, activates a ROS burst. Luminescence-based detection of apoplastic ROS in Arabidopsis Col-0 leaf discs upon treatment with 2 mg ml–1 cutin in MilliQ water for 30 min (A) and 45 min (B); (C) 1 mM of commercially available pure monomers [16-hydroxypalmitic acid (16-HPA), octanedioic acid (OCTDA), and ferulic acid (FERA)] and oligomers [glyceryl stearate (GS) and glyceryl tristearate (GTS)], in 10% ethanol in MilliQ water for 45 min; (D) 1 mg ml–1 of cutin hydrolysate obtained after alkaline hydrolysis of cutin; (E) 2 mg ml–1 of COM obtained through the methanolysis of cutin in 0.5% DMSO in MilliQ water; and (F) co-treatment with 2 mg ml–1 COM and 100 nM Flg22 in 0.5% DMSO in MilliQ water. In all the assays, mock consists of the solvent. The positive controls were Flg22 (100 nM) or Pep1 (100 nM).
To test if small chains of monomers linked together through ester linkages [i.e. oligomers (<7)] could act as elicitors, we prepared COMs. To produce these, cutin was depolymerized through a mild chemical hydrolysis and the released molecules were collected (see the Materials and methods). The produced COMs were unable to trigger a ROS burst (Fig. 1E). However, no effect was detected when the seedings were co-treated with flg22 and COM, although flg22 alone clearly induced a ROS burst (Fig. 1F). This result suggests that constituents of the COM preparation interfered with the reporter of luminescence. In fact, the COM contains phenolic compounds (Supplementary Table S1), and phenol oxidation has been reported to inactivate HRP activity in a concentration-dependent manner (i.e. enzyme:inhibitor ratio) (Mao et al., 2013). Cutin hydrolysates (mixture of all hydrolyzable cutin monomers) also contain low levels of phenolic compounds (Escórcio et al., 2022). There are alternative methods for ROS measurement; for example, DAB (3,3'-diaminobenzidine) staining has been used to detect the accumulation of intracellular ROS in response to treatment with cutin aliphatic monomers, specifically HPA (Kim et al., 2008; Park et al., 2008). Thus, while we observed that cutin aliphatic monomers did not induce an apoplastic ROS burst using a luminol-based assay, we cannot disregard the possibility that accumulation of intracellular ROS might occur at extended post-treatment periods.
The results show that treatment with cutin induced a ROS burst (Fig. 1A) but none of the tested pure cutin constituents that are commercially available did so (Fig. 1C). Due to the aforementioned technical limitation, to further test the activity of COMs as potential elicitors of plant immunity, we converged towards the calcium response—another hallmark early immune response.
Cutin and COMs, but not cutin hydrolysates, activate a calcium influx
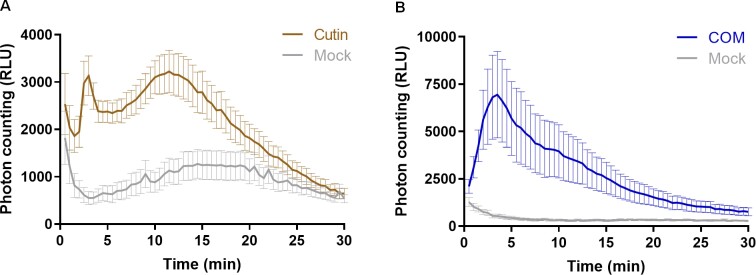
Both cutin and COMs showed a clear and reproducible induction of calcium influx in Arabidopsis plants expressing aequorin (Fig. 2A, B), a widely used calcium-activated reporter of immune responses in plants (Mithöfer and Mazars, 2002). The response patterns were however different: cutin response was bimodal with maximum values at 3 min and 12 min (Fig. 2A), whereas COM response was monomodal with a maximum between 3 min and 5 min (Fig. 2B). This result suggests that cutin may comprise several classes of chemical elicitors, one of which is prevalent in the COM fraction. Nicotiana benthamiana plants expressing aequorin (Segonzac et al., 2011) when exposed to COMs also showed a calcium influx having a monomodal response type (Supplementary Fig. S3A). Since the eliciting molecules were similarly recognized by both tested plants, the elicitors are probably not species specific, consistent with the conservation of dihydroxy-C16 acid units in cutinized tissues.
Fig. 2.
Cutin and COMs, but not cutin hydrolysates, activate a calcium influx. Luminescence-based detection of calcium influx in Arabidopsis seedlings expressing the calcium reporter aequorin, upon treatment with: (A) 1 mg ml–1 cutin in MilliQ water; (B) 2 mg ml–1 COM in 0.5% DMSO in MilliQ water. In all the assays, mock consists of the solvent.
Finally, no induction of a calcium influx was observed upon treatment of seedlings with increasing concentrations of either HPA or cutin hydrolysates (Supplementary Fig. S3B, C). Collectively, these data validate the opening hypothesis that cutin small oligomers may act as elicitors of PTI. Mild deconstruction of the polymer potentiates its capability to induce a calcium influx, but its complete depolymerization abolished this eliciting effect.
Cutin oligomers trigger MAPK activation
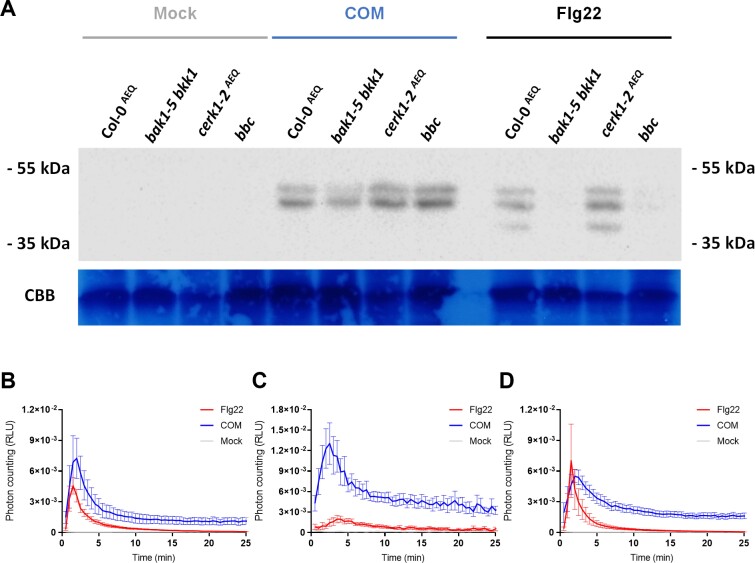
PTI signaling events occurring downstream to elicitor perception involve the activation of MAPKs (Yu et al., 2017; DeFalco and Zipfel, 2021). Accordingly, the capabilities of COM to activate MAPKs in Arabidopsis Col-0 seedlings were evaluated. The immunoblot-based assay allows the detection of the phosphorylated (active) forms of MAPK 3, 4, 6, and 11 during PTI signaling (Willmann et al., 2014). Short (10 min) exposure of seedlings to COM activated hallmark MAPK activation, similar to that observed when the plants were exposed to flg22 (Fig. 3A). In addition to the wild-type plants, three mutants were also tested: single, cerk1-2 (Ranf et al., 2011); double, bak1-5 bkk1 (Roux et al., 2011); and triple, bak1-5 bbk1 cerk1 (bbc) (Xin et al., 2016). CHITIN ELICITOR RECEPTOR KINASE 1 (CERK1) is a common co-receptor for LysM-type PRRs (Macho and Zipfel, 2014) while BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED KINASE 1 (BAK1) and BAK1-LIKE KINASE 1 (BKK1) are common co-receptors for leucine-rich repeat-type PRRs (Tang et al., 2017). Thus, differences in the response pattern of the selected mutants may reveal potential families of PRR(s) that recognize the elicitor(s) within COM. The results showed that COM induced a clear activation of MAPKs in all the mutants tested, similar to that observed in the wild-type plants (Fig. 3A). Similar results were observed for the calcium influx response upon COM treatment in the bak1-4AEQ and cerk1AEQ mutants (Fig. 3B–D). The observation that MAPK activation and calcium influx were similar in all mutants is suggestive of a perception mechanism independent of the families of PRRs known to associate with CERK1 and SERKs. Ultimately, these results suggest that COMs triggered a MAPK-mediated signaling cascade, opening up the hypothesis that COM exposure also involves transcriptional reprogramming.
Fig. 3.
Cutin oligomers trigger MAPK activation. (A) Western blot evaluation of MAPK activation in 14-day-old Arabidopsis seedlings from wild-type Col-0AEQ plants and the bak1-5 bkk1, cerk1-2AEQ, and bbc mutants, upon treatment with 3 mg ml–1 COM in 0.5% DMSO in MilliQ water. (B–D) Luminescence-based detection of calcium influx in A. thaliana Col-0 seedlings (B) and deletion mutants bak1-4 (C) and cerk1 (D), expressing the calcium reporter aequorin, upon treatment with 2 mg ml–1 COM in 0.5% DMSO in MilliQ water. Values are normalized by the aequorin discharge measurements. In all the assays, mock consists of the solvent and the positive control is Flg22 (100 nM).
Cutin oligomer treatment induced a transcriptional reprogramming consistent with activation of PTI
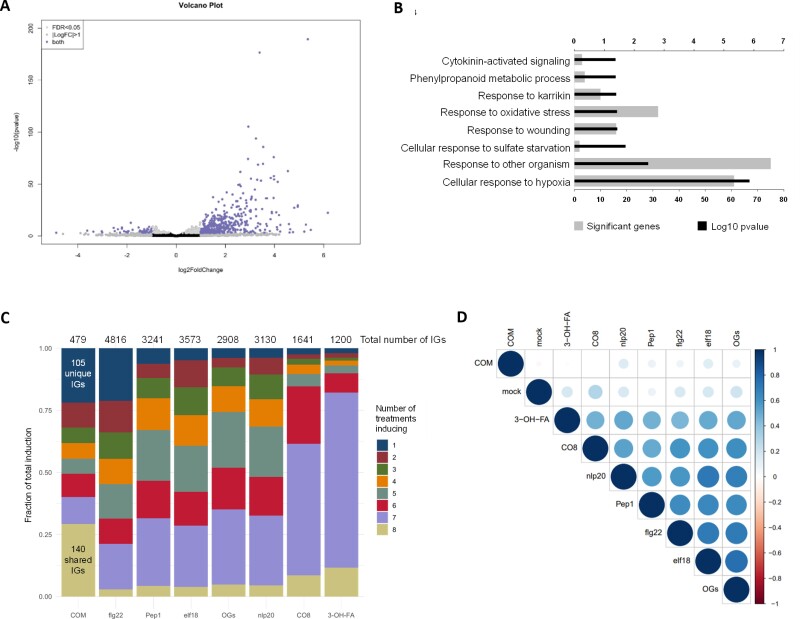
We evaluated the transcriptional reprogramming in Arabidopsis seedlings upon a 30 min treatment with COM compared with mock control (RNA-seq). Previous studies covering distinct PTI elicitors showed significant responses at 30 min post-treatment (Bjornson et al., 2021). PCA demonstrates that their transcriptomic profiles are clearly separating from each other (Supplementary Fig. S4). A total of 528 DEGs resulted from the COM treatment, of which 479 genes were up-regulated, while only 49 were down-regulated compared with the mock treatment (Fig. 4A). Enriched GO categories were only obtained for the subset of up-regulated genes due to the small number of down-regulated genes. An enrichment for terms related to activation of plant immunity, particularly ‘response to wounding’ and ‘response to other organism’, was noticed (Fig. 4B).
Fig. 4.
COM treatment induces a transcriptional reprogramming consistent with activation of PTI. (A) Volcano plot representing the statistically significant (adjusted P-value <0.05) differentially expressed genes (log2 fold change ≤ –1 or ≥1), in Arabidopsis Col-0 plants upon treatment with COM for 30 min (479 up-regulated genes and 49 down-regulated genes). (B) GO term enrichment analysis of the genes that showed up-regulation upon treatment with COM for 30 min. (C) Comparison of genes induced by treatment of COM with those induced by seven other elicitors of PTI. The total number of induced genes (IGs) for each elicitor (over all time points in Bjornson et al. 2021) is presented and the numbers of genes induced by all treatments or solely by COM are highlighted. (D) Correlation plot depicting changes in gene expression between all the elicitors evaluated in the comparative analysis: all transcriptomes compared at 30 min post-treatment.
The observed transcriptional reprogramming induced by the COM treatment was compared with that induced (i.e. up-regulated) by seven other well-characterized elicitors of plant immunity, recently reported by Bjornson et al. (2021). The COM effect presents similarity with that of the other elicitors: 140 induced genes (~30%) responded to COM and the other PTI elicitors (Fig. 4C). The transcriptional reprogramming induced by COM has, however, some uniqueness since 105 induced genes (~20%) were not induced by any of the other tested elicitors (Fig. 4C). In fact, such a level of specificity in transcriptional reprogramming was previously only observed for flg22 (Bjornson et al., 2021) (Fig. 4C). The lower number of genes induced by COM treatment could be related to the single time point used, different from the other elicitors of PTI where multiple time points were used. The genes induced only by COM (and not by the other elicitors) were compared with genes found to be up-regulated under abiotic stress (seven types of stresses were considered, see the Materials and methods). We observed that among these, 32 genes were induced solely by COM and not by any of the abiotic stresses (Supplementary Tables S2, S3), further suggestive of a certain degree of uniqueness of the effect of COMs.
The uniqueness of the COM treatment was further demonstrated through a correlation analysis of all elicitor transcriptomic datasets at the 30 min time point (Fig. 4D). At this time point, the effect of COMs is not well correlated with any of the other tested elicitors, for example bacterial 3-OH-FA and fungal CO8. It also showed no correlation with the effect of OGs originating from plant cell wall pectin degradation. Cutin anchoring to the cell wall is a long-standing debate, but the involvement of polysaccharide-based moieties has been suggested (Philippe et al., 2020). Polysaccharides can be found in very low amounts in cutin isolated using ionic liquid extractant (Bento et al., 2021), but it remains an unresolved question if the detected polysaccharide moieties are covalently linked to cutin. No glycoside-type linkages were detected in the NMR spectral fingerprint of a highly concentrated COM sample: 40 mg to allow the detection of low-intensity signals (Supplementary Fig. S5A). Several molecules derived from cell wall polysaccharides can act as elicitors, for example OGs (Ferrari et al., 2013), cellobiose (de Azevedo Souza et al., 2017), arabinoxylan oligosaccharides (Mélida et al., 2020), and mixed-linked β-1,3/1,4-glucans (Rebaque et al., 2021). However, the reported amounts for their eliciting effects (de Azevedo Souza et al., 2017) usually range from µg ml–1 to mg ml–1. These levels are higher than those detected in the COM preparations that were observed to contain only picograms of hydrolyzable sugars per mg of COM (Supplementary Fig. S5B). The acquired data thus indicate that the molecules within the COM preparation acting as elicitors have a lipidic nature.
Guiding principles on the chemistry of cutin oligomers able to elicit a rapid immune response
COM preparations have been shown to consist of oligomers and monomers (Escórcio et al., 2022). During infection, pathogens can secrete enzymes able to hydrolyze ester-type linkages present in cutin (Serrano et al., 2014), breaking the structural integrity of the cutin barrier to allow pathogen invasion of the infected plant tissue. To mimic such progressive attack of cutin, after obtaining a COM preparation, the non-hydrolyzed cutin fraction was recovered. The recovered cutin was subjected to a second round of mild hydrolysis and subsequently processed to obtain a COM II fraction. The signal intensity of the COM II-induced calcium influx was >2-fold higher than that observed after COM treatment (Supplementary Fig. S6). This observation suggests that COM II might be more enriched in active elicitors compared with COM.
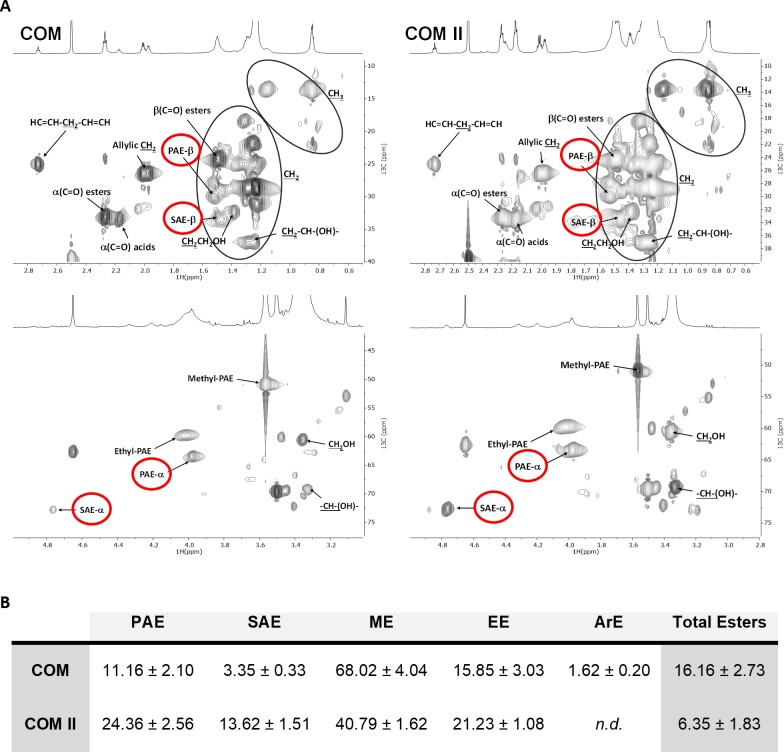
The free monomers were detected (and quantified) by GC-MS analysis, which also differentiates the methylated derivatives formed during the cleavage of esters through the methanolysis reaction (Supplementary Table S1). The presence of oligomers was directly inferred from the detection of both primary (PAEs) and secondary aliphatic esters (SAEs) through NMR analyses, specifically in the HSQC spectral fingerprints of either COM preparation (Fig. 5A). The integration of their corresponding 1H-NMR signals, relative to an internal standard, was used to infer the relative amount of PAEs (i.e. linear) and SAEs (i.e. branched) (Fig. 5B). Both types of esterification have been reported before in the spectral fingerprint of several cutin variants (Moreira et al., 2020; Bento et al., 2021; Escórcio et al., 2022). The estimated relative abundances of methyl esters in COM range from 40% to 70% of the total esters, consistent with the GC-MS data (Supplementary Table S1). To depolymerize all the oligomers present, the COM preparation was subjected to hydrolysis and reanalyzed by GC-MS. Comparison of the resultant monomeric profiles of the COM and the resulting hydrolysate revealed monomers increasing in abundance after hydrolysis (Supplementary Table S1). The major aliphatic monomer of cutin, dihydroxy-C16 acid, is probably the major building block of the oligomers, distantly followed by 9,10-epoxy-18-hydroxyoctadecanoic acid, nonanedioic acid, and hexadecanedioic acid.
Fig. 5.
NMR shows that cutin oligomers comprise both primary and secondary esters. (A) NMR characterization of cutin-derived COMs. Magnification of the HSQC spectral regions corresponding to aliphatics for each sample. Some correlations (unlabeled) are uncertain or unidentified. (B) NMR quantification of the relative abundances of ester types present in each COM calculated through the integration of signals in the corresponding 1H-NMR spectra. Ester types detected on this analysis include PAE (primary aliphatic esters), SAE (secondary aliphatic esters), ME (methyl esters), EE (ethyl esters), and ArE (aromatic esters). Esters that were not detected on a sample are labeled as n.d.
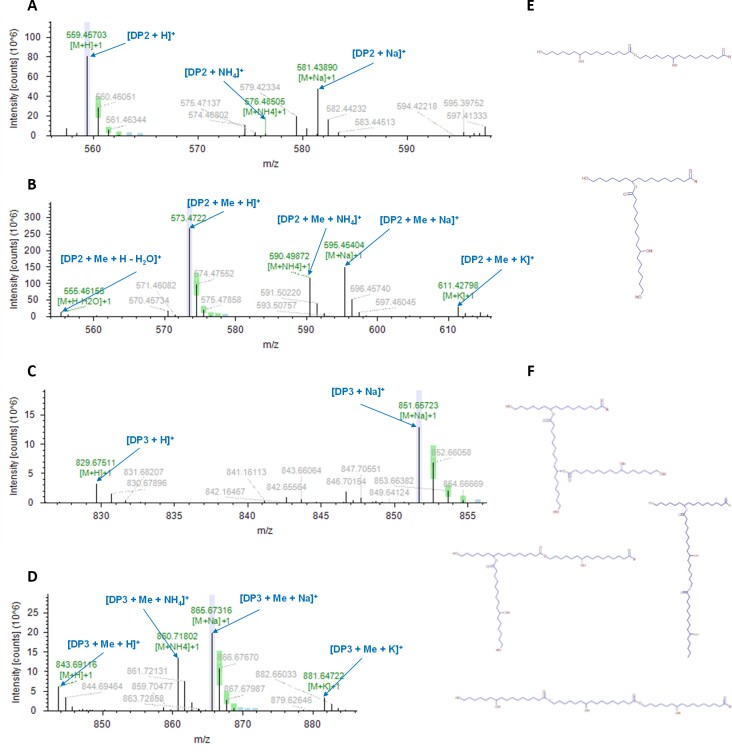
A preliminary LC-MS/MS analysis was performed targeting the exact masses of dihydroxy-C16 acid dimers and trimers with or without one methylation (Supplementary Table S4A). Pure HPA was used to set up the method (see the Materials and methods). The outputs (Compound Discovery 3.2) were unsupervised since the software automatically computes the most likely ions/adducts to be generated in negative/positive modes for each given mass (Supplementary Table S4B). In both COMs, dimers were putatively identified, namely two dihydroxy-C16 acid molecules esterified, methylated, or not—DP2 (Fig. 6A, B; Supplementary Fig. S7A, B). A trimer of dihydroxy-C16 acid molecules, with or without one methylation, was putatively identified only in COM II—DP3 (Fig. 6C, D; Supplementary Fig. S7C, D). These molecules can be a linear chain, yet one side branch is possible (Fig. 6E, F). The NMR quantification data suggest that linear esters are on average 2- to 3-fold more abundant than branched esters (Fig. 5B), accordingly the linear DP3 chain is more likely to exist.
Fig. 6.
LC-MS/MS identifies the COMs as dimers and trimers of ester-linked 10,16-dihydroxyhexadecanoic acid molecules. LC-MS/MS characterization of COM II (representative spectra for both COMs) in positive mode. MS1 spectra of the detected dimer composed of ester-linked molecules of 10,16-dihydroxyhexadecanoic acid (DP2) in the non-methylated (A) and methylated (B) forms. MS1 spectra of the detected trimer composed of ester-linked molecules of 10,16-dihydroxyhexadecanoic acid (DP3) in the non-methylated (C) and methylated (D) forms. Possible chemical configurations of the putatively identified oligomers (E, F), where R corresponds to an OH or CH3 group for the non-methylated and methylated forms, respectively. Lavender color indicates when the labeled centroid matches the monoisotopic mass of the expected compound ion; green color indicates when the labeled centroid matches the delta mass and the relative intensity of the theoretical isotope pattern within the specified tolerances; blue color indicates when the expected centroid for this m/z value might be missing because its theoretical intensity is at the level of the baseline noise. The corresponding MS2 spectra are shown in Supplementary Fig. S7.
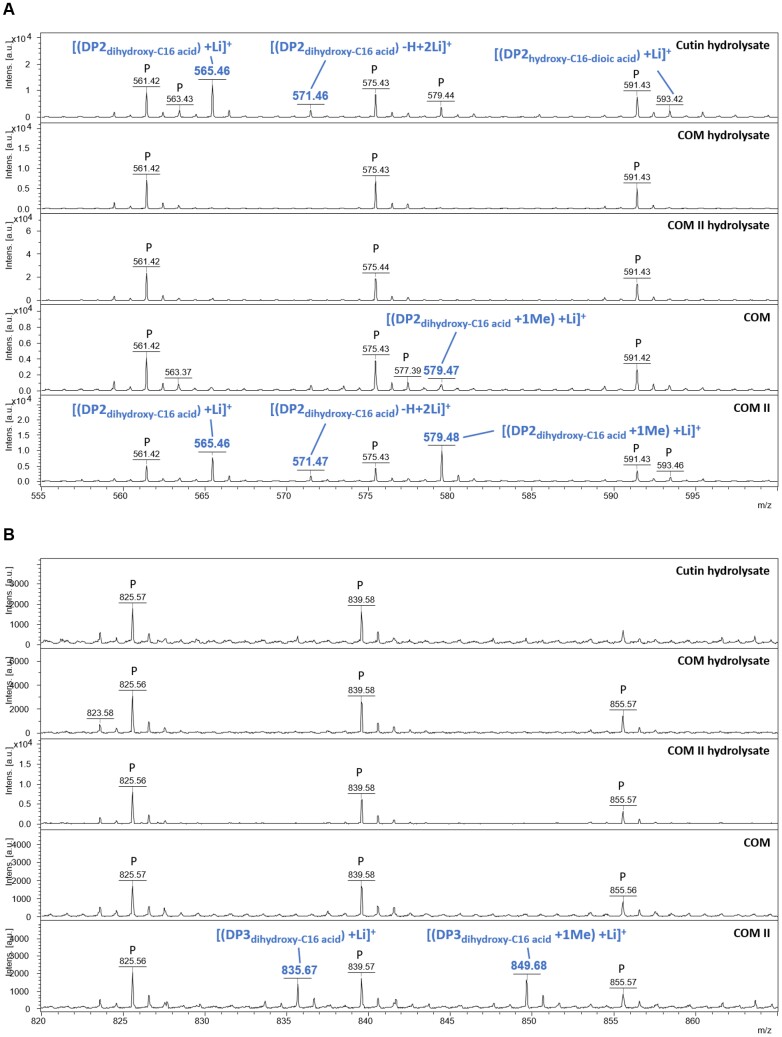
A MALDI-TOF method was developed to screen for the putative presence of oligomers up to octamers (Supplementary Fig. S8). The MALDI-TOF analyses showed the presence of the most abundant free monomers in COM and COM II, some of which were in the methylated form (Supplementary Table S5). The controls—cutin and COM hydrolysates—also contain the same non-methylated monomers (Supplementary Table S5). In the COMs, dimers were identified, namely DP2, both methylated and not methylated (Fig. 7A), consistent with the LC-MS/MS. To selectively observe species capable of self-ionization such as aromatics, samples analyzed by MALDI-TOF were also analyzed without addition of the ionization matrix (LDI-TOF). On the LDI-TOF spectra, other dimers detected consist of a dihydroxy-C16 acid esterified to coumaric acid without, or with one or two methylations—DP2c (Supplementary Fig. S9). The DP2c methylated molecules were only detected in the COM. The cutin hydrolysate (control) showed the presence of the non-methylated forms of DP2 in MALDI-TOF and DP2c in LDI-TOF (Supplementary Table S5). NMR analyses of 40 mg of either COM showed the presence of esterified aromatics only in COM (Supplementary Fig. S10). Finally, the methylated-DP3 and its non-methylated form were identified in COM II (Fig. 7B), regardless of being undetected in COM, possibly due to lower relative abundance. Collectively the data on the COMs (and cutin hydrolysates) suggest that amongst the identified oligomeric species, the best PTI elicitor candidates are linear dimers or trimers composed of dihydroxy-C16 acid units, one of which is possibly methylated.
Fig. 7.
MALDI-TOF confirms that COMs comprise dimers and trimers of ester-linked 10,16-dihydroxyhexadecanoic acid molecules. MALDI-TOF (+) spectra of cutin hydrolysate, COM hydrolysates, and COMs. The range 555–600 Da corresponds to expected masses for DP2s (A). The range 820–865 Da corresponds to expected masses for DP3s (B). Annotations were deduced from exact mass measurements. ‘P’ indicates an ion from polyethylene glycol contamination.
Are the cutin oligomers capable of activating signaling outputs reminiscent of PTI a new class of DAMPs?
The plant cell wall barrier is an important interface during plant–microbe interactions, where cutin is the outermost polymeric component. Plants are able to recognize damage caused by pathogens, and elicit immune responses, for example upon recognition of cell wall-derived fragments acting as DAMPs. As such, the cell wall barrier orchestrates key responses of the plant interaction with the surrounding environment. This rationale has defined the major hypothesis of our study, namely that cutin oligomers are able to activate responses suggestive of PTI. In Arabidopsis, COMs, obtained through methanolysis of a purified model cutin comprising dihydroxy-C16 acid units, elicited several hallmark immune responses, including calcium influx (Fig. 2) and MAPK activation (Fig. 3A), and a transcriptional response comprising features similar to those activated by well-characterized elicitors (Fig. 4). The perception mechanism of the COMs, which was observed to be independent of BAK1/BKK1 and CERK1 co-receptors (Fig. 3A–D), remains unresolved. Chemical analyses identified that the COMs contain trimers and dimers (Figs 6, 7). Further, none of the analytical methods used (NMR, GC-MS, LC-MS, and MALDI-TOF) could detect any other compound different from cutin constituents in the COMs used, thus indicating that the observed eliciting activity is most probably not due to contaminants. The strongest elicitor candidates are the dihydroxy-C16 acid dimers (DP2) or trimers (DP3) carrying a methylation. DP2, whether methylated or not, were detected in both COMs. In contrast, DP3 and DP2c (dihydroxy-C16 acid esterified to coumaric acid), with or without methylation, were only detected in COM II and COM, respectively. The non-methylated DP2 and DP2c were present in cutin hydrolysates unable to elicit a calcium burst, though the threshold for PTI activation remains unknown. The methylation increases the oligomer’s lipophilicity, possibly favoring its diffusion; a hypothesis that requires focused analysis. Methyl esters are, for example, present in seeds (Annarao et al., 2008) and vegetable oil (Di Pietro et al., 2020). However, the isolated cutin polymer, which is deprived of methyl esters, elicited a rapid ROS burst (Fig. 1A, B) and calcium influx (Fig. 2A). This observation questions the requirement of methylation for immune activation. Fungal lipases can generate methyl esters, for example from vegetable oil (Li et al., 2007). In plants, the modification of cutin degradation products by microbial methyltransferases remains unknown in the context of PTI. However, methylation to potentiate the eliciting effect of cutin oligomers may inspire alternative biotechnological valorization paths for fruit pomaces.
Both the cutin polymer (with a minor degree of structural damage, tested as a suspension in water due to its insolubility) and the generated oligomers (tested in stable water/DMSO preparations) activated responses suggestive of a role as PTI elicitors. Previous work by others showed that some cutin monomers also activated some aspects of plant immunity (Kauss et al., 1999; Kim et al., 2008; Park et al., 2008). A step-by-step activation of specific elements of plant immunity by cutin having distinct degrees of structural damage constitutes an appealing concept that deserves further investigation. The release of oligomeric elicitors during plant infection requires validation to attain a mechanistic understanding of cutin’s multiple functions in plant–pathogen interactions. The identity of the precise COM elicitors remains putative, and efficient syntheses are needed to obtain pure compounds and advance towards precise analysis of their response pathways. The COM concentrations used are high, but this is simply because COMs comprise both active and non-active cutin constituents; that is, the concentration of the active dimers/trimers is likely to be low. However, COMs clearly activated multiple rapid responses indicative of PTI. These results open up the hypothesis that COMs constitute a new class of DAMPs, reliant on future experimental evidence that they can be released during infection, potentially via the action of fungal cutinases. Regardless, their production from agro-industrial residues constitutes a promising value chain and may support development of sustainable agricultural bio-based treatments to increase disease resistance.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. ROS measurements in A. thaliana Col-0 seedlings treated with cutin.
Fig. S2. ROS measurements in A. thaliana Col-0 seedlings treated with commercially available model compounds.
Fig. S3. Calcium measurements in N. benthamiana leaf discs treated with COM and A. thaliana seedlings treated with cutin monomers.
Fig. S4. Principal component analysis (PCA) performed with all samples used for RNA sequencing.
Fig. S5. NRM characterization (HSQC) of COM and quantification of the total carbohydrates present in COM samples.
Fig. S6. Calcium measurements in A. thaliana seedlings treated with COM II.
Fig. S7. LC-MS/MS characterization of COM in positive mode for detection of methylated and non-methylated forms of DP2 and DP3 oligomers.
Fig. S8. MALDI-TOF (+) spectrum of a reference sample containing DPs of 10,16-dihydroxyhexadecanoic acid.
Fig. S9. LDI-TOF (–) spectra of cutin hydrolysate and COMs.
Fig. S10. NMR characterization (HSQC) of the presence of aromatic esters in COM and COM II samples.
Table S1. GC-MS characterization of COM before and after alkaline hydrolysis.
Table S2. List of genes uniquely induced by COM treatment.
Table S3. List of genes induced by both COM treatment and abiotic stress conditions.
Table S4A. Lists of putative oligomeric targets for LC-MS/MS analysis and of putatively identified oligomers (DP2 and DP3) by LC-MS/MS in COM.
Table S5. List of all putatively identified monomers and oligomers in COM by MALDI-TOF (+) and LDI-TOF (–).
Acknowledgements
The authors are grateful to Pedro Lamosa (ITQB NOVA) for support with the NMR analyses. MS data were generated by the Mass Spectrometry Unit (UniMS), ITQB/iBET, Oeiras, Portugal. We thank Stefanie Ranf for the gift of cerk1-2AEQ seeds. All members of the Silva Pereira and Zipfel labs are also thanked for useful discussions.
Contributor Information
Carlos J S Moreira, Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Oeiras, Portugal; Institute of Plant and Microbial Biology, Zurich-Basel Plant Science Center, University of Zurich, Zurich, Switzerland.
Rita Escórcio, Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Oeiras, Portugal.
Artur Bento, Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Oeiras, Portugal.
Marta Bjornson, Institute of Plant and Microbial Biology, Zurich-Basel Plant Science Center, University of Zurich, Zurich, Switzerland.
Laura Herold, Institute of Plant and Microbial Biology, Zurich-Basel Plant Science Center, University of Zurich, Zurich, Switzerland.
Ana S Tomé, Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Oeiras, Portugal.
Celso Martins, Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Oeiras, Portugal.
Mathieu Fanuel, PROBE research infrastructure, BIBS Facility, INRAE, Nantes, France; Research Unit Biopolymers Interaction Assemblies, INRAE, Nantes, France.
Isabel Martins, Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Oeiras, Portugal.
Bénédicte Bakan, Research Unit Biopolymers Interaction Assemblies, INRAE, Nantes, France.
Cyril Zipfel, Institute of Plant and Microbial Biology, Zurich-Basel Plant Science Center, University of Zurich, Zurich, Switzerland; The Sainsbury Laboratory, University of East Anglia, Norwich Research Park, Norwich, UK.
Cristina Silva Pereira, Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Oeiras, Portugal.
Daolong Dou, Nanjing Agricultural University, China.
Author contributions
CSP and CZ: supervision and data interpretation; CSP: preparing the final version of the manuscript. All authors have made substantial contributions to the acquisition, analysis, and interpretation of data, and contributed to the drafting of the manuscript: CJSM, AB, and RE (cutin and COM preparation); CJSM and LH (plant experiments); CJSM, MB, and CM (RNA seq); AST, CJSM, and RE (GC-MS); AB and RE (NMR); IM and CJSM (LC-MS/MS); BB and MF (MALDI-TOF); CJSM (preparation of the initial draft of the manuscript). All authors read and approved the final version of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interests.
Funding
We acknowledge funding from the European Research Council through grant ERC 2014-CoG-647928 and from Fundação para a Ciência e Tecnologia (FCT) by Project MOSTMICRO ITQB (UIDB/04612/2020 and UIDP/04612/2020), LS4FUTURE Associated Laboratory (LA/P/0087/2020), from the PESSOA program (Prpc no. 441.00) to CSP, and from the University of Zurich to CZ. The NMR data were acquired at CERMAX, ITQB-NOVA, Oeiras, Portugal with equipment funded by FCT. CJSM is grateful to Aralab (Portugal) for the PhD contract 06/PlantsLife/2017 and to EMBO for the short-term fellowship (#8003). RE and IM are grateful for FCT funding for the PhD scholarship BD/06435/2021 and for the working contract financed by national funds under norma transitória D.L. no. 57/2016, respectively.
Data availability
All relevant data are available in the manuscript. Supplementary figures and tables are provided to support the results presented. The transcriptomic data have been deposited in the BioStudies EMBL-EBI platform with the accession number E-MTAB-12956, and are also accessible through the SRA NCBI public database with the same accession number.
References
- Anders S, Pyl PT, Huber W.. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annarao S, Sidhu OP, Roy R, Tuli R, Khetrapal CL.. 2008. Lipid profiling of developing Jatropha curcas L. seeds using 1H NMR spectroscopy. Bioresource Technology 99, 9032–9035. [DOI] [PubMed] [Google Scholar]
- Beneloujaephajri E, Costa A, L’Haridon F, Métraux J-P, Binda M.. 2013. Production of reactive oxygen species and wound-induced resistance in Arabidopsis thaliana against Botrytis cinerea preceded and depend on a burst of calcium. BMC Plant Biology 13, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento A, Moreira CJS, Correia VG, et al. 2021. Quantification of structure–property relationships for plant polyesters reveals suberin and cutin idiosyncrasies. ACS Sustainable Chemistry and Engineering 9, 15780–15792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhunia RK, Showman LJ, Jose A, Nikolau BJ.. 2018. Combined use of cutinase and high-resolution mass-spectrometry to query the molecular architecture of cutin. Plant Methods 14, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson M, Pimprikar P, Nürnberger T, Zipfel C.. 2021. The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity. Nature Plants 7, 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventure G, Beisson F, Ohlrogge J, Pollard M.. 2004. Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. The Plant Journal 40, 920–930. [DOI] [PubMed] [Google Scholar]
- Buxdorf K, Rubinsky G, Barda O, Burdman S, Aharoni A, Levy M.. 2014. The transcription factor SlSHINE3 modulates defense responses in tomato plants. Plant Molecular Biology 84, 37–47. [DOI] [PubMed] [Google Scholar]
- de Azevedo Souza C, Li S, Lin AZ, Boutrot F, Grossmann G, Zipfel C, Somerville SC.. 2017. Cellulose-derived oligomers act as damage-associated molecular patterns and trigger defense-like responses. Plant Physiology 173, 2383–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco TA, Zipfel C.. 2021. Molecular mechanisms of early plant pattern-triggered immune signaling. Molecular Cell 81, 3449–3467. [DOI] [PubMed] [Google Scholar]
- Delwiche CF, Cooper ED.. 2015. The evolutionary origin of a terrestrial flora. Current Biology 25, R899–R910. [DOI] [PubMed] [Google Scholar]
- Di Pietro ME, Mannu A, Mele A.. 2020. NMR determination of free fatty acids in vegetable oils. Processes 8, 410–418. [Google Scholar]
- Escórcio R, Bento A, Tomé AS, Correia VG, Rodrigues R, Moreira CJS, Marion D, Bakan B, Silva Pereira C.. 2022. Finding a needle in a haystack: producing antimicrobial cutin-derived oligomers from tomato pomace. ACS Sustainable Chemistry and Engineering 10, 11415–11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Commission. 2021. The tomato market in the EU: Vol. 1: production and area statistics. Brussels: European Commission - DG. [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T.. 1999. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. The Plant Journal 18, 265–276. [DOI] [PubMed] [Google Scholar]
- Ferrari S, Savatin DV, Sicilia F, Gramegna G, Cervone F, De Lorenzo G.. 2013. Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Frontiers in Plant Science 4, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fich EA, Segerson NA, Rose JKC.. 2016. The plant polyester cutin: biosynthesis, structure, and biological roles. Annual Review of Plant Biology 67, 207–233. [DOI] [PubMed] [Google Scholar]
- Hou S, Liu Z, Shen H, Wu D.. 2019. Damage-associated molecular pattern-triggered immunity in plants. Frontiers in Plant Science 10, 646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauss H, Fauth M, Merten A, Jeblick W.. 1999. Cucumber hypocotyls respond to cutin monomers via both an inducible and a constitutive H2O2-generating system. Plant Physiology 120, 1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K.. 2007. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal 50, 347–363. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. 2015. 2015. HISAT: a fast spliced aligner with low memory requirements. Nature Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Park JH, Kim MC, Cho SH.. 2008. Cutin monomer induces expression of the rice OsLTP5 lipid transfer protein gene. Journal of Plant Physiology 165, 345–349. [DOI] [PubMed] [Google Scholar]
- Kolattukudy PE, Rogers LM, Li D, Hwang CS, Flaishman MA.. 1995. Surface signaling in pathogenesis. Proceedings of the National Academy of Sciences, USA 92, 4080–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Du W, Liu D.. 2007. Rhizopus oryzae IFO 4697 whole cell-catalyzed methanolysis of crude and acidified rapeseed oils for biodiesel production in tert-butanol system. Process Biochemistry 42, 1481–1485. [Google Scholar]
- Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F.. 2009. Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proceedings of the National Academy of Sciences, USA 106, 22008–22013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhi S, Cambillau C.. 1999. Structure–activity of cutinase, a small lipolytic enzyme. Biochimica et Biophysica Acta 1441, 185–196. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zipfel C.. 2014. Plant PRRs and the activation of innate immune signaling. Molecular Cell 54, 263–272. [DOI] [PubMed] [Google Scholar]
- Mao L, Luo S, Huang Q, Lu J.. 2013. Horseradish peroxidase inactivation: heme destruction and influence of polyethylene glycol. Scientific Reports 3, 3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc M, Risani R, Desnoes E, et al. 2021. Bioinspired co-polyesters of hydroxy-fatty acids extracted from tomato peel agro-wastes and glycerol with tunable mechanical, thermal and barrier properties. Industrial Crops and Products 170, 113718. [Google Scholar]
- Martin LBB, Rose JKC.. 2014. There’s more than one way to skin a fruit: formation and functions of fruit cuticles. Journal of Experimental Botany 65, 4639–4651. [DOI] [PubMed] [Google Scholar]
- Mélida H, Bacete L, Ruprecht C, Rebaque D, del Hierro I, López G, Brunner F, Pfrengle F, Molina A.. 2020. Arabinoxylan-oligosaccharides act as damage associated molecular patterns in plants regulating disease resistance. Frontiers in Plant Science 11, 1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithöfer A, Mazars C.. 2002. Aequorin-based measurements of intracellular Ca2+-signatures in plant cells. Biological Procedures Online 4, 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina I, Bonaventure G, Ohlrogge J, Pollard M.. 2006. The lipid polyester composition of Arabidopsis thaliana and Brassica napus seeds. Phytochemistry 67, 2597–2610. [DOI] [PubMed] [Google Scholar]
- Moore I, Gao C, Wang H, Fernández V, Guzmán-Delgado P, Graça J, Santos S, Gil L.. 2016. Cuticle structure in relation to chemical composition: re-assessing the prevailing model. Frontiers in Plant Science 7, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira CJS, Bento A, Pais J, et al. 2020. An ionic liquid extraction that preserves the molecular structure of cutin shown by nuclear magnetic resonance. Plant Physiology 184, 592–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Suh MC, Kim TH, Kim MC, Cho SH.. 2008. Expression of glycine-rich protein genes, AtGRP5 and AtGRP23, induced by the cutin monomer 16-hydroxypalmitic acid in Arabidopsis thaliana. Plant Physiology and Biochemistry 46, 1015–1018. [DOI] [PubMed] [Google Scholar]
- Philippe G, Geneix N, Petit J, Guillon F, Sandt C, Rothan C, Lahaye M, Marion D, Bakan B.. 2020. Assembly of tomato fruit cuticles: a cross-talk between the cutin polyester and cell wall polysaccharides. New Phytologist 226, 809–822. [DOI] [PubMed] [Google Scholar]
- Ranf S, Eschen-Lippold L, Pecher P, Lee J, Scheel D.. 2011. Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. The Plant Journal 68, 100–113. [DOI] [PubMed] [Google Scholar]
- Ranf S, Grimmer J, Pöschl Y, Pecher P, Chinchilla D, Scheel D, Lee J.. 2012. Defense-related calcium signaling mutants uncovered via a quantitative high-throughput screen in Arabidopsis thaliana. Molecular Plant 5, 115–130. [DOI] [PubMed] [Google Scholar]
- Rebaque D, Hierro I, López G, et al. 2021. Cell wall-derived mixed-linked β-1,3/1,4-glucans trigger immune responses and disease resistance in plants. The Plant Journal 106, 601–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, Vries S de, Zipfel C.. 2011. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. The Plant Cell 23, 2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C, Feike D, Gimenez-Ibanez S, Hann DR, Zipfel C, Rathjen JP.. 2011. Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiology 156, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Coluccia F, Torres M, L’Haridon F, Métraux J-P.. 2014. The cuticle and plant defense to pathogens. Frontiers in Plant Science 0, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Bressan R.. 2006. RNA extraction. Methods in Molecular Biology 323, 345–348. [DOI] [PubMed] [Google Scholar]
- Tang D, Wang G, Zhou J-M.. 2017. Receptor kinases in plant–pathogen interactions: more than pattern recognition. The Plant Cell 29, 618–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters ER. 2003. Molecular adaptation and the origin of land plants. Molecular Phylogenetics and Evolution 29, 456–463. [DOI] [PubMed] [Google Scholar]
- Wickham H, Averick M, Bryan J, et al. 2019. Welcome to the Tidyverse. Journal of Open Source Software 4, 1686. [Google Scholar]
- Willmann R, Haischer DJ, Gust AA.. 2014. Analysis of MAPK activities using MAPK-specific antibodies. Methods in Molecular Biology 1171, 27–37. [DOI] [PubMed] [Google Scholar]
- Xin A, Fry SC.. 2021. Cutin:xyloglucan transacylase (CXT) activity covalently links cutin to a plant cell-wall polysaccharide. Journal of Plant Physiology 262, 153446. [DOI] [PubMed] [Google Scholar]
- Xin X-F, Nomura K, Aung K, Velásquez AC, Yao J, Boutrot F, Chang JH, Zipfel C, He SY.. 2016. Bacteria establish an aqueous living space in plants crucial for virulence. Nature 539, 524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JKC.. 2013. The formation and function of plant cuticles. Plant Physiology 163, 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Feng B, He P, Shan L.. 2017. From chaos to harmony: responses and signaling upon microbial pattern recognition. Annual Review of Phytopathology 55, 109–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Jia Z, Trush MA, Li YR.. 2016. A highly sensitive chemiluminometric assay for real-time detection of biological hydrogen peroxide formation. Reactive Oxygen Species 1, 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. 2014. Plant pattern-recognition receptors. Trends in Immunology 35, 345–351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available in the manuscript. Supplementary figures and tables are provided to support the results presented. The transcriptomic data have been deposited in the BioStudies EMBL-EBI platform with the accession number E-MTAB-12956, and are also accessible through the SRA NCBI public database with the same accession number.