Abstract
The ATP-dependent molecular chaperone Hsp90 (heat-shock protein 90) is essential for the maturation of hormone receptors and protein kinases. During the process of client protein activation, Hsp90 co-operates with cofactors/co-chaperones of unique sequence, e.g. Aha1 (activator of Hsp90 ATPase 1), p23 or p50, and with cofactors containing TPR (tetratricopeptide repeat) domains, e.g. Hop, immunophilins or cyclophilins. Although the binding sites for these different types of cofactors are distributed along the three domains of Hsp90, sterical overlap and competition for binding sites restrict the combinations of cofactors that can bind to Hsp90 at the same time. The recently discovered cofactor Aha1 associates with the middle domain of Hsp90, but its relationship to other cofactors of the molecular chaperone is poorly understood. Therefore we analysed whether complexes of Aha1, p23, p50, Hop and a cyclophilin with Hsp90 are disrupted by the other four cofactors by gel permeation chromatography using purified proteins. It turned out that Aha1 competes with the early cofactors Hop and p50, but can bind to Hsp90 in the presence of cyclophilins, suggesting that Aha1 acts as a late cofactor of Hsp90. In contrast with p50, which can bind to Hop, Aha1 does not interact directly with any of the other four cofactors. In vivo studies in yeast and in mammalian cells revealed that Aha1 is not specific for kinase activation, but also contributes to maturation of hormone receptors, proposing a general role for this cofactor in the activation of Hsp90-dependent client proteins.
Keywords: activator of Hsp90 ATPase (Aha1), cofactor, heat-shock protein, heat-shock protein 90 (Hsp90), molecular chaperone, protein folding
Abbreviations: Aha1, activator of Hsp90 ATPase; DOC, deoxycorticosterone; GR, glucocorticoid receptor; Hsp, heat-shock protein; siRNA, small interfering RNA; TPR, tetratricopeptide repeat
INTRODUCTION
The molecular chaperone Hsp90 (heat-shock protein) is a highly conserved, essential, homodimeric molecular chaperone of the eukaryotic cytosol. Many natural substrates of Hsp90 are medically relevant signal-transduction molecules, including the nuclear receptors for steroid hormones and several kinases [1–3]. During the process of substrate protein activation, Hsp90 hydrolyses ATP [4,5] and co-operates with different cofactors/co-chaperones such as Hop (Sti1 in yeast), immunophilins and cyclophilins (e.g. Cpr6 and Cpr7 in yeast), p50 (Cdc37 in yeast), p23 (Sba1 in yeast), Aha1 (activator of Hsp90 ATPase), Tpr2 and CHIP (C-terminus of Hsc70-interacting protein) [6–12] and acts as a part of the multichaperone machine together with Hsp70 and its cofactor Hsp40 [2].
For most of the hormone receptors, it has been shown that a minimum system of Hsp90, Hop, p23 and Hsp70, Hsp40 is sufficient to promote heterocomplex assembly in vitro [2,13–15], but other cofactors such as immunophilins, cyclophilins or the Hsp70-specific cofactor Hip may increase the efficiency of this process, whereas p50 is absent from Hsp90–hormone receptor complexes. On the other hand, p50 plays an essential role in Hsp90-dependent kinase activation [2]. Aha1 has been shown to have a function in kinase activation in vivo [11,12], but it is not known whether this cofactor is also involved in hormone receptor maturation.
Several Hsp90-associated cofactors, e.g. Hop, the immunophilins or cyclophilins, use TPR (tetratricopeptide repeat) motifs to bind to the molecular chaperone [16,17]. Other cofactors, e.g. p23, Aha1 and p50, lack TPR repeats and use unique sequences to associate with Hsp90. Complexes of Hsp90 with one or more of these cofactors have been analysed by different experimental methods such as yeast genetics [18], co-immunoprecipitation [19], CD spectroscopy [12,20], isothermal titration calorimetry [21], gel-filtration chromatography [17] and surface plasmon resonance [21]. Furthermore, structures of different Hsp90 domains in complex with various cofactor fragments have been resolved by X-ray crystallography [16,22–24]. The TPR clamp domain has been identified to bind to the EEVD motif present at the very C-terminal end of the molecular chaperones Hsp90 and Hsp70 [16]. Recently, the structure of Hsp90's middle domain in complex with the N-terminal part of Aha1 and that of the N-terminal domain of Hsp90 in complex with the C-terminal domain of p50 have been presented in [23,24].
To yield well-diffracting protein crystals, truncated versions of Hsp90 and the cofactors Hop, Aha1 and p50 were used for structure analysis [16,23,24]. Moreover, fragments were also used to characterize Hsp90–cofactor complexes by CD spectroscopy, isothermal titration calorimetry and surface plasmon resonance [12,16,21]. Although the importance of these results is highly appreciated, the use of fragments led, at least in some cases, to misinterpretation or neglect of relevant protein interactions. This problem has been addressed for complexes of Hsp90 and Hsp70 with TPR cofactors, such as the immunophilins and Hop [25,26].
Therefore, to investigate the formation of complexes of Hsp90 with p23, p50, Aha1, Hop and Cpr7, we analysed the molecular chaperone and combination in pairs of the five cofactors for competitive binding. To yield a comprehensive view of Hsp90–cofactor interactions and to avoid problems that may arise from the use of protein fragments, these experiments were performed only with full-length versions of the respective proteins by gelpermeation chromatography. The main focus was on characterizing the relationship between Aha1 and the other four cofactors. Moreover, our in vivo studies reveal that Aha1 is involved not only in the activation of kinases as reported recently [11,12] but also in the activation of hormone receptors.
EXPERIMENTAL
Construction of expression plasmids
Expression plasmids for yeast Hsp90, Aha1 and Hop have been reported previously [11]. Yeast p23 and the cyclophilins Cpr6 and Cpr7 were amplified from wild-type yeast DNA and p50 was from a human brain cDNA library (Clontech). PCR products were inserted into pProExHTa expression vector to generate an N-terminal His6 sequence followed by a tobacco etch virus protease cleavage site.
Protein purification
Expression constructs in pProExHTa were transformed into Escherichia coli BL21(DE3)pLysS cells. Bacteria were grown at 18 °C in Luria–Bertani medium, supplemented with 100 mg/l ampicillin and 34 mg/l chloramphenicol to absorbance A600∼1 and the protein expression was induced for 5 h with 0.25 mM isopropyl β-D-thiogalactoside. After harvesting, proteins were enriched from cell pellets by Ni2+-nitrilotriacetate chromatography at pH 8.0, essentially as described in [11]. Proteins were further purified by ion-exchange chromatography on MonoQ and by gel filtration on Superose 12 (Amersham Biosciences) in 40 mM Hepes/KOH (pH 7.4), 50 mM KCl and 2 mM MgCl2. When desired, His tags were cleaved off by treatment with tobacco etch virus protease.
Protein interaction assays
For gel-filtration analysis, 500 μl samples containing 5 μM or multiples of the indicated combinations of proteins were incubated for 10 min at room temperature (25 °C) and 10 min on ice in 40 mM Hepes/KOH (pH 7.4), 50 mM KCl and 2 mM MgCl2. When p23 was analysed for protein interactions, 2 mM ATP[S] was added and the reaction mixture was incubated for 1 h at 30 °C followed by 10 min on ice. Protein samples were separated on a Superose 12 HR10/30 column equilibrated in 40 mM Hepes/KOH (pH 7.4), 50 mM KCl and 2 mM MgCl2 using an ÄKTA™ chromatography system (Amersham Biosciences). Fractions (500 μl) were collected starting from 6 ml elution volume and analysed by SDS/PAGE (12% gel).
Determination of GR (glucocorticoid receptor) activity in yeast cells
Wild-type yeast W303 and the double knockout strain W303 ΔAHA1/ΔHCH1 have been described recently [11]. Standard methods for growth, transformation and manipulation were used. Cells were grown either on YPD [1% (w/v) yeast extract/2% (w/v) peptone/2% (w/v) glucose] or on synthetic dropout medium, SRaf- or SGal-selective minimal medium (0.67% yeast nitrogen base, supplemented with 2% glucose, raffinose or galactose respectively and with nucleotides and amino acids depending on auxotrophy).
For the analysis of GR activity, cells were transformed with p2HGal/GR/CYC and the reporter plasmid pSX26.1 expressing β-galactosidase under the control of GR response elements [3]. Cultures were grown on SRaff/-His,-Ura medium and GR synthesis was induced by the addition of 2% (w/v) galactose. Receptors were activated at A600∼0.3 by the addition of 10 μM DOC (deoxycorticosterone) for 1 h, cells were collected by centrifugation and β-galactosidase activity was measured using the Galacto-Star kit (Tropix, Bedford, MA, U.S.A.) and normalized to protein concentration of cell lysate.
Cell-culture experiments
DNA sequences encoding human Aha1 [11] were inserted into pcDNA3.1 (Invitrogen) in frame with the C-terminal mycHis tag. GR activity was monitored by a GR-dependent luciferase expression vector (pGRE-luc; Clontech), and a vector expressing β-galactosidase (pSV-β-Gal; Promega) was used as a control to normalize for transfection efficiency as described in [7]. HEK-293 cells in 3 cm dishes were transfected with 0.5 μg of pGRE-luc, 0.5 μg of pSV-β-Gal and 1 μg of pcDNA-Aha1 or empty vector. Transfections using LIPOFECTAMINE™ PLUS (Invitrogen) typically yielded 10–20%. After 24 h, cells were treated with 100 nM of the hormone dexamethasone or an equal volume of ethanol solvent control for another 24 h and then harvested and lysed. The supernatants were tested with the β-galactosidase and luciferase enzyme assay systems (Promega).
For siRNA (small interfering RNA) experiments, a double-stranded synthetic RNA oligomer (Ambion, Austin, TX, U.S.A.) against the sequence 5′-GGTTCAAAATGAAGAAGGC, deduced from the coding DNA region of human Aha1, was used. As a control, the mutated sequence 5′-GGTACAAAATCAAGATG-GC (mutations underlined) or the Silencer negative control #1 siRNA (Ambion) were used. Cells were grown in 6-well plates and transfected with 200 nM siRNA oligomers with Oligofectamine™ (Invitrogen) in reactions separate from the plasmid transfections. Hormone stimulation and enzymic activity assays were performed as above.
To monitor the levels of human Aha1, equal amounts of cell lysate proteins were analysed by immunoblotting with an affinity-purified rabbit polyclonal antibody raised against the peptide sequence NNWHWTERDASNWS from human Aha1. The mouse monoclonal antibodies clone 41 (BD Biosciences) and clone 1D4 (StressGen Biotechnologies) were used to detect GR and glyceraldehyde-3-phosphate dehydrogenase respectively.
Miscellaneous procedures
The Bio-Rad protein assay kit was used to determine protein concentrations.
RESULTS
Direct interactions between Hsp90 cofactors
Hsp90 has the potential to bind various cofactors by means of its three domains. Some of these accessory proteins might not only bind to the Hsp90 chaperone core but associate with each other directly. For example, interactions of p50/Cdc37 with the TPR cofactors Hop and cyclophilin Cpr7 were reported in cell lysates [19] and yeast [27], although this finding has not been verified in a pure biochemical system. Moreover, the identification of the novel Hsp90 cofactor Aha1, which binds to the middle domain of the chaperone [11,23], motivated us to analyse its relationship with other cofactors of the molecular chaperone. From the multitude of Hsp90 cofactors, we chose p23, p50, Aha1, Hop and a cyclophilin, i.e. three unique and two TPR-containing cofactors that are present from yeast to humans and are well characterized regarding their interaction with Hsp90. Recombinant proteins Aha1, p23, Hop and Cpr7 (or Cpr6) were deduced from the respective yeast sequences and purified from E. coli lysates. For p50, the human sequence was used, since complexes of Hsp90 with human p50 exhibit remarkably higher stability compared with those with yeast Cdc37 and have therefore been characterized previously by CD spectroscopy and structure analysis [20,24].
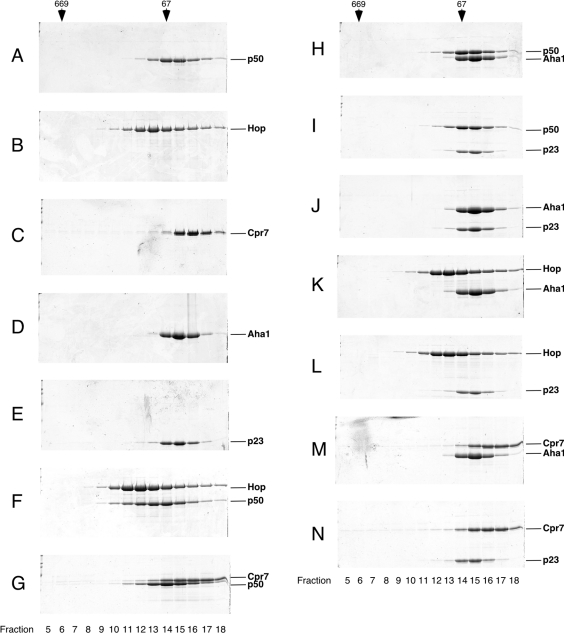
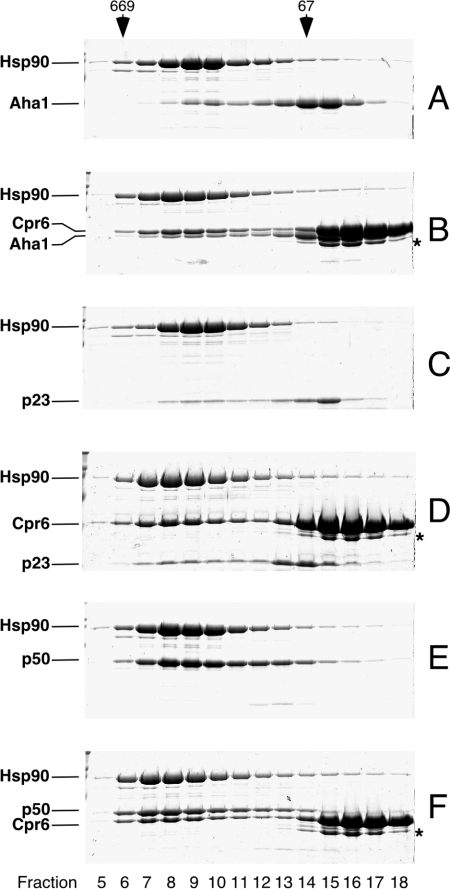
The elution profiles of the individual proteins are represented in Figures 1(A)–1(E). To test for protein interactions, we combined p50 with Hop or Cpr7 and analysed the mixtures by gel-filtration chromatography (Figures 1F and 1G) as described in the Experimental section. A shift in the elution profiles of Hop and p50 to higher molecular mass (Figure 1F, fractions 11–13) compared with the single proteins (Figures 1A, fractions 14–15, and 1B, fractions 12–14) indicates the formation of a stable p50–Hop complex. When p50 was incubated with the other TPR cofactor Cpr7, only a minor shift could be detected (Figure 1G, fractions 11–13), pointing to a more transient stability compared with the p50–Hop complex.
Figure 1. Direct interactions between the cofactors Aha1, p23, p50, Hop and Cpr7.
Purified Aha1, p23, p50, Hop and Cpr7 were incubated as indicated and fractionated by gel-filtration chromatography on a Superose 12 column. Fractions were analysed by SDS/PAGE. Marker proteins are shown on top (thyroglobulin, 669 kDa; BSA, 67 kDa). (A) p50, (B) Hop, (C) Cpr7, (D) Aha1, (E) p23, (F) Hop+p50, (G) Cpr7+p50, (H) Aha1+p50, (I) p50+p23, (J) Aha1+p23, (K) Hop+Aha1, (L) Hop+p23, (M) Cpr7+Aha1 and (N) Cpr7+p23.
In contrast, p50 interacted with neither Aha1 nor p23 (Figures 1H and 1I). Similarly, Aha1 and p23 were unable to form a complex (Figure 1J). Thus the unique cofactors p50, p23 and Aha1 cannot form an interaction network independent of the molecular chaperone Hsp90. We next asked whether Aha1 or p23 is capable of interacting with the TPR cofactors Hop and Cpr7. The elution profiles for mixtures of Hop with Aha1 or p23 (Figures 1K and 1L) or for mixtures of Cpr7 with Aha1 or p23 (Figures 1M and 1N) remained unchanged compared with the proteins alone, consistent with the view that no interaction between these proteins had occurred. Hence, p50 is the only cofactor that can form stable contacts with other Hsp90 cofactors.
Aha1 and p23 are in competition for binding to Hsp90
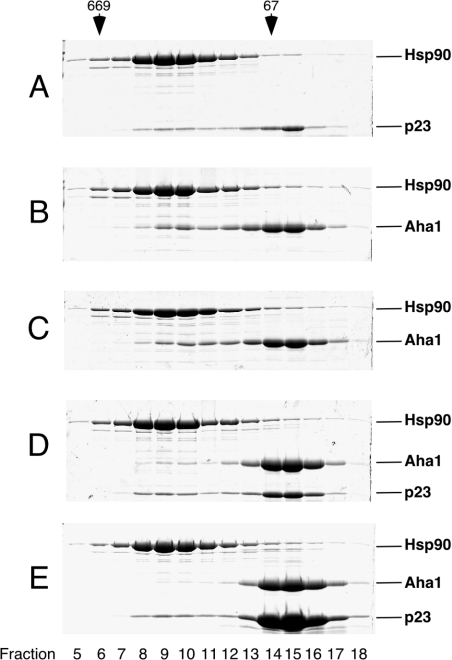
Stable Hsp90–p23 complexes form only at high temperatures in the presence of non-hydrolysable nucleotide analogues such as ATP[S], but dissipate with a half-life of approx. 45 min on removal of the nucleotide [28,29]. Therefore Hsp90 and p23 were incubated at 30 °C for 1 h in the presence of 2 mM ATP[S] after 10 min at 4 °C. Under these conditions, complex formation could be monitored by gel-filtration chromatography in the absence of an expensive nucleotide, although complexes were substoichiometric (Figure 2A). To determine whether these conditions interfere with binding of Aha1 to the molecular chaperone Hsp90, we mixed Aha1 and Hsp90 at 4 °C in the absence of nucleotide as a control (Figure 2B) or at 30 °C with the addition of 2 mM ATP[S] (Figure 2C) and analysed the mixtures for protein interactions. As complex formation was indistinguishable in both cases, these conditions are suitable to analyse competitive binding of Aha1 and p23 to Hsp90. When the molecular chaperone was incubated with Aha1 and increasing concentrations of p23 at 30 °C in the presence of 2 mM ATP[S], a complete disruption of the Hsp90–Aha1 complex was observed (Figures 2D and 2E). This result clearly demonstrates that binding of p23 and Aha1 to Hsp90 is mutually exclusive.
Figure 2. Aha1 and p23 are in competition for binding to Hsp90.
Purified Hsp90, Aha1 and p23 were incubated as indicated and fractionated by gel-filtration chromatography on a Superose 12 column. Protein concentrations were 5 μM or multiples thereof, as indicated. Fractions were analysed by SDS/PAGE. Marker proteins are shown on top (thyroglobulin, 669 kDa; BSA, 67 kDa). (A) Hsp90+p23 at 30 °C, 2 mM ATP[S]. (B) Hsp90+Aha1. (C) Hsp90+Aha1 at 30 °C, 2 mM ATP[S]. (D) Hsp90+Aha1+p23 at 30 °C, 2 mM ATP[S]. (E) Hsp90+Aha1+25 μM p23 at 30 °C, 2 mM ATP[S].
The cofactor p50 competes with Aha1 but not with p23 for binding to Hsp90
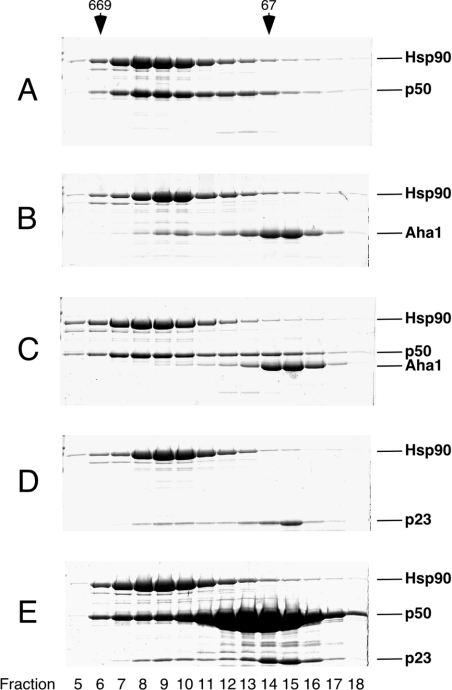
The three cofactors p50, p23 and Aha1 do not interact with each other (Figure 1) and have different requirements to associate with Hsp90. Recently, it has been established by structure analysis that a C-terminal fragment of p50 binds to the N-terminal domain of Hsp90 [24]; accordingly, complex formation between full-length Hsp90 and p50 is shown in Figure 3(A). We next sought to determine whether p50 can bind to the molecular chaperone Hsp90 in the presence of two other unique cofactors Aha1 and p23. When p50, together with Aha1, was probed for interaction with Hsp90, p50 apparently disrupted the Hsp90–Aha1 complex (compare Figure 3C with Figure 3B), indicating a clear competition between the two cofactors, which use adjacent domains for interaction with Hsp90. In contrast, when the Hsp90–p23 complex was probed with a 10-fold excess of p50, p23 as well as p50 remained bound to the molecular chaperone (compare Figure 3E with Figure 3D). Although p23 and p50 interact with the N-terminal domain of Hsp90, their binding sites seem to be non-overlapping.
Figure 3. The cofactor p50 competes with Aha1 but not with p23 for binding to Hsp90.
Purified Hsp90, Aha1, p50 and p23 were incubated as indicated and fractionated by gel-filtration chromatography on a Superose 12 column. Protein concentrations were 5 μM or multiples thereof, as indicated. Fractions were analysed by SDS/PAGE. Marker proteins are shown on top (thyroglobulin, 669 kDa; BSA, 67 kDa). (A) Hsp90+p50, (B) Hsp90+Aha1 (same as Figure 2B), (C) Hsp90+p50+Aha1, (D) Hsp90+p23 at 30 °C, 2 mM ATP[S] (same as Figure 2A), (E) Hsp90+p23+50 μM p50 at 30 °C, 2 mM ATP[S].
The early TPR cofactor Hop competes with Aha1, p23 and p50 for binding to Hsp90
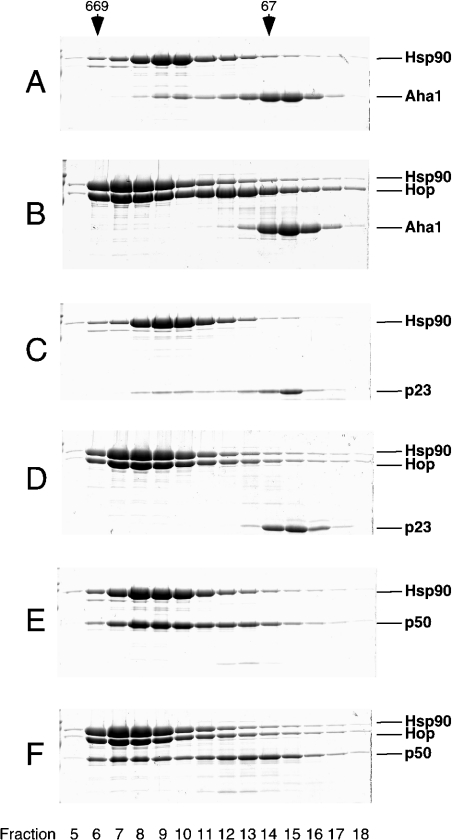
Aha1 and p23 interact with Hsp90 at sites different from the TPR adapter site for Hop. When the Hsp90–Aha1 complex was incubated with a 2-fold excess of Hop, the complex was entirely disrupted [11] (compare Figure 4B with Figure 4A). Similarly, Hop also competed the binding of p23 to Hsp90 (compare Figure 4D with Figure 4C), consistent with the view that p23 is a late cofactor and Hop is an early cofactor of Hsp90 [30,31].
Figure 4. The early TPR cofactor Hop competes with Aha1, p23 and p50 for binding to Hsp90.
Purified Hsp90, Aha1, Hop, Hch1, p23 and p50 were incubated as indicated and fractionated by gel-filtration chromatography on a Superose 12 column. Fractions were analysed by SDS/PAGE. Marker proteins are shown on top (thyroglobulin, 669 kDa; BSA, 67 kDa). (A) Hsp90+Aha1 (same as Figure 2B), (B) Hsp90+Hop+Aha1, (C) Hsp90+p23 at 30 °C, 2 mM ATP[S] (same as Figure 2A), (D) Hsp90+Hop+p23 at 30 °C, 2 mM ATP[S], (E) Hsp90+p50 (same as Figure 3B), (F) Hsp90+Hop+p50.
It has been reported that binding of p50 and Hop to Hsp90 is mutually exclusive [32], although both proteins are now known to use separate binding sites to interact with Hsp90 [24]. When Hop was added to the Hsp90–p50 complex, the p50 peak (Figure 4E, fractions 8–10) was split into two peaks at lower and higher molecular masses (Figure 4F, fractions 7–8 and 13–15). The appearance of a backward shift (fractions 13–15) is consistent with the finding that Hop competes with p50 for binding to Hsp90 [32], most probably due to sterical restrictions. However, the shift to higher molecular mass (fractions 7–8), which persisted even in the presence of severalfold excess of Hop (results not shown), suggests the formation of a complex consisting of the three proteins, Hsp90, Hop and p50, of which p50 or Hop acts as the central component to which Hsp90 and the other cofactor are bound. This view is consistent with the formation of a Hop–p50 complex (Figure 1) and is supported by the observation of direct Hop–p50 interactions in yeast [27].
The late TPR cofactors Cpr6 and Cpr7 do not interfere with the binding of p50, p23 or Aha1 to Hsp90
During the process of hormone receptor activation, the early cofactor Hop is replaced by late TPR cofactors occupying the TPR adapter site on the molecular chaperone [1,2,9]. We asked whether binding of late TPR cofactors, such as the cyclophilins Cpr6 or Cpr7, would be compatible with association of the unique cofactors Aha1, p23 and p50 to Hsp90. When a mixture of Hsp90 and Aha1 (Figure 5A) was incubated with excess of Cpr6, a complex consisting of the three components Hsp90, Aha1 and Cpr6 was formed (Figure 5B). Similarly, when Hsp90–p23 (Figure 5C) or Hsp90–p50 (Figure 5E) complexes were challenged with excess of Cpr6, complexes comprising Hsp90, one of the unique cofactors p23 or p50 and the TPR cofactor Cpr6 (Figures 5D and 5F) were formed. Similar results were obtained when Cpr7 was used instead of Cpr6 (results not shown).
Figure 5. The late TPR cofactor Cpr6 does not interfere with binding of Aha1, p23 or p50 to Hsp90.
Purified Hsp90, Aha1, Cpr6, p23 and p50 were incubated as indicated and fractionated by gel-filtration chromatography on a Superose 12 column. Protein concentrations were 5 μM or manifolds thereof, as indicated. Fractions were analysed by SDS/PAGE. Marker proteins are shown on top (thyroglobulin, 669 kDa; BSA, 67 kDa). (A) Hsp90+Aha1 (same as Figure 2B), (B) Hsp90+25 μM Cpr6+Aha1, (C) Hsp90+p23 at 30 °C, 2 mM ATP[S] (same as Figure 2A), (D) Hsp90+25 μM Cpr6+p23 at 30 °C, 2 mM ATP[S], (E) Hsp90+p50 and (F) Hsp90+p50+25 μM Cpr6; *, a contaminating degradation product of the Cpr6 preparation.
Aha1 assists Hsp90 in kinase and hormone receptor activation
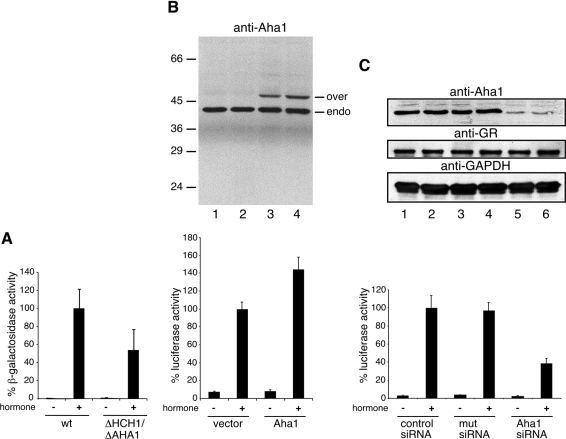
It has been shown previously that activity of the kinase v-Src is dependent on Aha1 [11,12]. We therefore asked whether this cofactor is specific for kinase maturation or whether Aha1 also assists the activation of other Hsp90 client proteins. For this reason, a GR expression plasmid together with a reporter plasmid to monitor GR activity was co-transformed into wild-type yeast W303 and into W303 ΔAHA1/ΔHCH1, a double knockout strain in which Aha1 and its yeast-specific homologue Hch1 have been deleted [11]. Expression of GR was induced by the addition of 2% galactose and hormone receptors were activated by DOC as described in the Experimental section. As shown recently, the activity of v-Src was decreased in ΔAHA1/ΔHCH cells [11,12]. Similarly, when we assayed GR activity in W303 ΔAHA1/ΔHCH1 cells as described in the Experimental section, a decrease to approx. 50% of the wild-type cells was observed (Figure 6A), similar to the previously reported decrease of v-Src activity in this deletion strain [11].
Figure 6. Aha1 assists Hsp90 in kinase and hormone receptor activation.
(A) Yeast cells W303 wild-type (wt) and W303 ΔAHA1/ΔHCH1 were co-transformed with GR and the reporter plasmid pSX26.1 containing β-galactosidase under the control of GR response elements. GR was activated by the addition of 10 μM DOC as described in the Experimental section. GR-dependent β-galactosidase activity was measured using the GalactoStar kit (Tropix) and normalized to protein concentration of the lysate. Activities are expressed as the averages of at least three independent experiments, and error bars are indicated. (B) HEK-293 cells were transfected with a plasmid encoding a luciferase reporter gene downstream of GR response elements and a plasmid encoding β-galactosidase serving as a control for transfection efficiency. Empty vector (lanes 1–2) or a vector encoding mycHis-tagged human Aha1 (lanes 3–4) were co-transfected, together with the reporter and control plasmids. Cells were treated for 24 h with 100 nM dexamethasone where indicated, and harvested. Cell lysates were tested for luciferase activity and normalized against β-galactosidase activity and protein concentration. Error bars show the S.D. for at least three independent experiments. Top: immunoblotting with an antibody raised against Aha1; transfected overexpressed mycHis-tagged Aha1 (over) is visible as a band above the endogenous species (endo). Bottom: GR-activated normalized luciferase activity. Activities are expressed as the averages of at least three independent experiments, and error bars are indicated. (C) A double-stranded siRNA oligomer against the human Aha1 RNA was transfected into cells (lanes 5–6) and control experiments included a negative control oligonucleotide (lanes 1–2) or a mutated sequence (lanes 3–4) (see the Experimental section for details). Cells were co-transfected, 24 h after transfection, with the luciferase and β-galactosidase plasmids, and 48 h after transfection with siRNA oligomers cells were treated for another 24 h with 100 nM dexamethasone where indicated and harvested. Cell lysates were tested for luciferase activity and normalized against β-galactosidase activity and protein concentration. Top: total cell lysates were immunoblotted for endogenous Aha1, GR, and for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control. Bottom: normalized GR-dependent luciferase activity under conditions indicated. Activities are expressed as the averages for at least three independent experiments, and error bars are indicated.
To corroborate further the dependence of hormone receptor activation on Aha1, we analysed the activation of endogenous GR in mammalian cells. To measure this activity, we transfected HEK-293 cells with a plasmid encoding a luciferase reporter gene downstream of GR response elements, as well as a control plasmid encoding constitutively expressed β-galactosidase [7]. Dexa-methasone treatment caused a strong activation of endogenous GR relative to untreated cells, as measured by luciferase expression normalized to β-galactosidase levels (Figure 6B, lanes 1–2). Cotransfection of mycHis-tagged Aha1 led to overexpression of Aha1 at a level similar to Tpr2, another cofactor of Hsp90 in mammalian cells [7], and increased hormone-dependent GR activation to approx. 150% of the control cells lacking exogenous Aha1 (Figure 6B, lanes 3–4). This outcome demonstrates the contribution of Aha1 to the activation of the hormone receptor GR similar to its effect on v-Src activity.
Next, we tested the effect of decreasing the expression of Aha1 on GR activity by the siRNA approach described in the Experimental section. The double-stranded siRNA oligomer decreased the Aha1 levels drastically (Figure 6C, lanes 5–6) compared with control cells transfected with either a negative control siRNA (Figure 6C, lanes 1–2) or a point-mutated siRNA oligomer (Figure 6C, lanes 3–4). However, siRNA treatment did not decrease GR expression levels. Interestingly, Aha1 down-regulation by the siRNA technique also decreased the hormone-dependent activation of GR to approx. 40% (Figure 6C, lanes 5–6) when compared with the control.
These results obtained by deletion of Aha1 and Hch1 in yeast and by overexpression and silencing of Aha1 in human cells clearly demonstrate that Aha1 stimulates the Hsp90 chaperone system in vivo and contributes to activation of the hormone receptor GR, similar to its effect on v-Src activity that has been reported recently [11,12]. However, since deletion of Aha1 and Hch1 in yeast and significant silencing of Aha1 in mammalian cells (Figure 6C) does not abolish but rather decreases client protein activation, Aha1 seems to improve the efficiency of the Hsp90 chaperone machine, but is not essential for its overall function.
DISCUSSION
Hsp90 is the core component of a molecular machine working together with several cofactors that regulate the chaperone's activity. In general, during the process of hormone receptor activation, Hop, p23 and an immunophilin or cyclophilin bind to Hsp90 in a defined order. On the other hand, kinases are processed by Hsp90 in association with p50 [2]. The recent discovery of Aha1 [11,12] increases the number of possible Hsp90–cofactor complexes and gives additional complexity to this established scheme. In the present study, we have shown that Aha1 contributes not only to the activation of the kinase v-Src but also to the activation of the hormone receptor GR. This observation defines further the relationship between Aha1 and other key cofactors involved in client protein activation by the Hsp90 chaperone system.
Initial reports contained contradictory results about the coexistence of other cofactors in Hsp90–Aha1 complexes, most probably due to the use of protein fragments [11,12]. Using only full-length versions of cofactor proteins, we found that Aha1 competes with the early cofactors Hop and p50, which inhibit ATPase activity of the molecular chaperone [20,31], but does not compete with the late cofactor Cpr6 (or Cpr7) for binding to Hsp90. Accordingly, Aha1 is absent from early Hsp90 cofactor complexes and can thus be considered a late Hsp90 cofactor. In the process of kinase maturation, this suggests that a p50-regulated step of ATPase inhibition is followed by a distinct stage of Aha1-stimulated Hsp90 ATPase activity. Similarly, Aha1 is proposed to act on hormone receptor activation after Hop has been replaced by a late TPR cofactor such as Cpr6 (or Cpr7) [11]. Moreover, we found that association of Aha1 and p23 with Hsp90 is mutually exclusive, but the chronological order of their contribution to hormone receptor maturation has not yet been established. However, a test system that can discriminate between Aha1 and p23 action will have to be set up for this purpose in the future.
The variety of Hsp90–cofactor complexes is limited by sterical overlap and competition for binding sites, and the client protein itself seems to select on the composition of Hsp90–cofactor complexes [2]. In general, p50 (for most of the kinases) and a set of Hop, p23 and a cyclophilin/immunophilin (for most of the hormone receptors) act as specific Hsp90 cofactors respectively. Our results (Figures 1 and 4) and evidence from experiments in yeast [27] clearly demonstrate a stable interaction between Hop and p50. This observation points to the existence of Hop–p50–Hsp90 and/or p50–Hop–Hsp90 complexes, in which one cofactor acts as the central component. Such a complex would integrate the findings of Silverstein et al. [32], who reported competition of Hop and p50 for the TPR adapter site on Hsp90, and the observations of Hartson et al. [19], who found that Hsp90 co-adsorbs with p50 and Hop. Moreover, the presence of a complex consisting of Hsp90, Hop and p50 is in support of the stabilizing effects observed for p50 and Hop on Hsp90–kinase complexes in vivo [33].
Interestingly, Hsp90 forms complexes with p50 and p23 at the same time (Figure 3), although both cofactors make contacts with the N-terminal domain of the molecular chaperone for interaction. Moreover, co-association of p50 and p23 with the molecular chaperone has been reported during in vitro assembly of Hsp90 with the protein kinases Fes, HRI or Lck [19,34]. In contrast with p23, p50 is an early cofactor with the ability to arrest Hsp90 ATPase activity and to recruit client proteins [24]. Therefore p50–Hsp90–p23 complexes may represent a kind of transitional stage within the Hsp90 reaction cycle on the way from intermediate towards mature Hsp90 complexes.
Using yeast as a model organism to delete Aha1 and its relative Hch1, and by overexpression and silencing of Aha1 in mammalian cells, we have shown in the present study that Aha1 increases the efficiency of the Hsp90 chaperone system to activate the hormone receptor GR. However, as already demonstrated for the kinase v-Src, the cofactor Aha1 is not essential for this activation process but rather renders the Hsp90 chaperone machine more effective. This capacity might become crucial under conditions of cellular stress [11,12]. Similarly, the cofactors Hop and p23 are not essential in yeast [18,35], although future experiments are necessary to examine the situation in mammalian cells.
The rate of Hsp90 ATP hydrolysis is controlled by dampers and throttles. ATPase activity is inhibited by Hop and p50, which allow the loading of client proteins on to Hsp90 and to a lower extent by p23 [12,20,31]. In the absence of those cofactors, stimulation of Hsp90 by Aha1 confers high ATPase activity to the molecular chaperone, which results in efficient activation of client proteins in vivo. Remarkably, it has been reported recently that Hsp90 isolated from tumour cells or tumour tissue is in a conformation with high affinity for the Hsp90 inhibitor 17-allylaminogeldanamycin and has strikingly higher ATPase activity compared with Hsp90 from healthy sources [36]. This characteristic was proposed to be due to increased presence of tumour Hsp90 in chaperone complexes with cofactors, unlike Hsp90 from normal cells or tissue [36]. Given that Hop, p50 and p23 inhibit ATP hydrolysis of the molecular chaperone [12,20,31], Aha1 might account for the high ATPase activity of tumour Hsp90. Since stimulation of the Hsp90 ATPase rate makes client protein activation more efficient, targeting Hsp90–Aha1 complexes may provide a strategy to decrease the activity of disease-causing signalling molecules that are dependent on this molecular chaperone. Since Aha1 can be silenced quite efficiently (Figure 6C), which results in decreased client protein activity in vivo, targeting the step of ATPase stimulation within the Hsp90 reaction cycle by RNA interference may offer new perspectives for therapeutic intervention in the treatment of diseases like cancer [37].
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 284/project Z3) and by the European Commission (QLK3-CT2000-00720). We thank Dr S. Lindquist (Whitehead Institute, Cambridge, MA, U.S.A.) for generously providing plasmids for the expression of v-Src, GR and pSX26.1 in yeast.
References
- 1.Pratt W. B., Toft D. O. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 2.Pratt W. B., Toft D. O. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 3.Nathan D. F., Lindquist S. Mutational analysis of hsp90 function: interactions with a steroid receptor and a protein kinase. Mol. Cell. Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panaretou B., Prodromou C., Roe S. M., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obermann W. M., Sondermann H., Russo A. A., Pavletich N. P., Hartl F. U. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J. Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchner J. Hsp90 & Co.–a holding for folding. Trends Biochem. Sci. 1999;24:136–141. doi: 10.1016/s0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- 7.Brychzy A., Rein T., Winklhofer K. F., Hartl F. U., Young J. C., Obermann W. M. J. Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J. 2003;22:3613–3623. doi: 10.1093/emboj/cdg362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caplan A. J. Hsp90's secrets unfold: new insights from structural and functional studies. Trends Cell Biol. 1999;9:262–268. doi: 10.1016/s0962-8924(99)01580-9. [DOI] [PubMed] [Google Scholar]
- 9.Young J. C., Moarefi I., Hartl F. U. Hsp90: a specialized but essential protein-folding tool. J. Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Hohfeld J., Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 11.Lotz G. P., Lin H., Harst A., Obermann W. M. J. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J. Biol. Chem. 2003;278:17228–17235. doi: 10.1074/jbc.M212761200. [DOI] [PubMed] [Google Scholar]
- 12.Panaretou B., Siligardi G., Meyer P., Maloney A., Sullivan J. K., Singh S., Millson S. H., Clarke P. A., Naaby-Hansen S., Stein R., et al. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol. Cell. 2002;10:1307–1318. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- 13.Dittmar K. D., Banach M., Galigniana M. D., Pratt W. B. The role of DnaJ-like proteins in glucocorticoid receptor hsp90 heterocomplex assembly by the reconstituted hsp90.p60.hsp70 foldosome complex. J. Biol. Chem. 1998;273:7358–7366. doi: 10.1074/jbc.273.13.7358. [DOI] [PubMed] [Google Scholar]
- 14.Kosano H., Stensgard B., Charlesworth M. C., McMahon N., Toft D. The assembly of progesterone receptor-hsp90 complexes using purified proteins. J. Biol. Chem. 1998;273:32973–32979. doi: 10.1074/jbc.273.49.32973. [DOI] [PubMed] [Google Scholar]
- 15.Morishima Y., Kanelakis K. C., Murphy P. J. M., Lowe E. R., Jenkins G. J., Osawa Y., Sunahara R. K., Pratt W. B. The Hsp90 cochaperone p23 is the limiting component of the multiprotein Hsp90/Hsp70-based chaperone system in vivo where it acts to stabilize the client protein Hsp90 complex. J. Biol. Chem. 2003;278:48754–48763. doi: 10.1074/jbc.M309814200. [DOI] [PubMed] [Google Scholar]
- 16.Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell (Cambridge, Mass.) 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 17.Young J. C., Obermann W. M., Hartl F. U. Specific binding of tetratricopeptide repeat proteins to the C-terminal 12-kDa domain of hsp90. J. Biol. Chem. 1998;273:18007–18010. doi: 10.1074/jbc.273.29.18007. [DOI] [PubMed] [Google Scholar]
- 18.Fang Y. F., Fliss A. E., Rao J., Caplan A. J. Sba1 encodes a yeast Hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol. Cell. Biol. 1998;18:3727–3734. doi: 10.1128/mcb.18.7.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartson S. D., Irwin A. D., Shao J., Scroggins B. T., Volk L., Huang W., Matts R. L. p50(cdc37) is a nonexclusive Hsp90 cohort which participates intimately in Hsp90-mediated folding of immature kinase molecules. Biochemistry. 2000;39:7631–7644. doi: 10.1021/bi000315r. [DOI] [PubMed] [Google Scholar]
- 20.Siligardi G., Panaretou B., Meyer P., Singh S., Woolfson D. N., Piper P. W., Pearl L. H., Prodromou C. Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J. Biol. Chem. 2002;277:20151–20159. doi: 10.1074/jbc.M201287200. [DOI] [PubMed] [Google Scholar]
- 21.Brinker A., Scheufler C., Von Der Mulbe F., Fleckenstein B., Herrmann C., Jung G., Moarefi I., Hartl F. U. Ligand discrimination by TPR domains. Relevance and selectivity of EEVD-recognition in Hsp70×Hop×Hsp90 complexes. J. Biol. Chem. 2002;277:19265–19275. doi: 10.1074/jbc.M109002200. [DOI] [PubMed] [Google Scholar]
- 22.Meyer P., Prodromou C., Hu B., Vaughan C., Roe S. M., Panaretou B., Piper P. W., Pearl L. H. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol. Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 23.Meyer P., Prodromou C., Liao C., Hu B., Roe S. M., Vaughan C. K., Vlasic I., Panaretou B., Piper P. W., Pearl L. H. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 2004;23:511–519. doi: 10.1038/sj.emboj.7600060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roe S. M., Ali M. M., Meyer P., Vaughan C. K., Panaretou B., Piper P. W., Prodromou C., Pearl L. H. The Mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50(cdc37) Cell (Cambridge, Mass.) 2004;116:87–98. doi: 10.1016/s0092-8674(03)01027-4. [DOI] [PubMed] [Google Scholar]
- 25.Carrigan P. E., Nelson G. M., Roberts P. J., Stoffer J. N., Riggs D. L., Smith D. F. Multiple domains of the co-chaperone Hop are important for Hsp70 binding. J. Biol. Chem. 2004;279:16185–16193. doi: 10.1074/jbc.M314130200. [DOI] [PubMed] [Google Scholar]
- 26.Cheung-Flynn J., Roberts P. J., Riggs D. L., Smith D. F. C-terminal sequences outside the tetratricopeptide repeat domain of FKBP51 and FKBP52 cause differential binding to Hsp90. J. Biol. Chem. 2003;278:17388–17394. doi: 10.1074/jbc.M300955200. [DOI] [PubMed] [Google Scholar]
- 27.Abbas-Terki T., Briand P. A., Donze O., Picard D. The Hsp90 co-chaperones Cdc37 and Sti1 interact physically and genetically. Biol. Chem. 2002;383:1335–1342. doi: 10.1515/BC.2002.152. [DOI] [PubMed] [Google Scholar]
- 28.Sullivan W., Stensgard B., Caucutt G., Bartha B., McMahon N., Alnemri E. S., Litwack G., Toft D. Nucleotides and two functional states of hsp90. J. Biol. Chem. 1997;272:8007–8012. doi: 10.1074/jbc.272.12.8007. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan W. P., Owen B. A., Toft D. O. The influence of ATP and p23 on the conformation of hsp90. J. Biol. Chem. 2002;277:45942–45948. doi: 10.1074/jbc.M207754200. [DOI] [PubMed] [Google Scholar]
- 30.Johnson B. D., Schumacher R. J., Ross E. D., Toft D. O. Hop modulates hsp70/hsp90 interactions in protein folding. J. Biol. Chem. 1998;273:3679–3686. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- 31.Prodromou C., Siligardi G., O'Brien R., Woolfson D. N., Regan L., Panaretou B., Ladbury J. E., Piper P. W., Pearl L. H. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 1999;18:754–762. doi: 10.1093/emboj/18.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silverstein A. M., Grammatikakis N., Cochran B. H., Chinkers M., Pratt W. B. p50(cdc37) binds directly to the catalytic domain of Raf as well as to a site on hsp90 that is topologically adjacent to the tetratricopeptide repeat binding site. J. Biol. Chem. 1998;273:20090–20095. doi: 10.1074/jbc.273.32.20090. [DOI] [PubMed] [Google Scholar]
- 33.Lee P., Shabbir A., Cardozo C., Caplan A. J. Sti1 and Cdc37 can stabilize Hsp90 in chaperone complexes with a protein kinase. Mol. Biol. Cell. 2004;15:1785–1792. doi: 10.1091/mbc.E03-07-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nair S. C., Toran E. J., Rimerman R. A., Hjermstad S., Smithgall T. E., Smith D. F. A pathway of multi-chaperone interactions common to diverse regulatory proteins: estrogen receptor, Fes tyrosine kinase, heat shock transcription factor Hsf1, and the aryl hydrocarbon receptor. Cell Stress Chaperones. 1996;1:237–250. doi: 10.1379/1466-1268(1996)001<0237:apomci>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang H. C., Nathan D. F., Lindquist S. In vivo analysis of the Hsp90 cochaperone Sti1 (p60) Mol. Cell. Biol. 1997;17:318–325. doi: 10.1128/mcb.17.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamal A., Thao L., Sensintaffar J., Zhang L., Boehm M. F., Fritz L. C., Burrows F. J. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature (London) 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 37.Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J., et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature (London) 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]