Abstract
We describe the characterization of a family 4 UDG1 (uracil DNA glycosylase) from the crenarchaeote Sulfolobus solfataricus. UDG1 is found to have a marked preference for substrates containing a G:U base pair over either A:U or single-stranded uracil-containing DNA substrates. UDG1 is found to interact with the sliding clamp PCNA (proliferating cell nuclear antigen), and does so by a conserved motif in the C-terminus of the protein. S. solfataricus has a heterotrimeric PCNA, and only one of the subunits, PCNA3, interacts with UDG1. We have been unable to detect any stimulation of UDG activity by PCNA, in contrast with the observed effects of PCNA on a number of DNA metabolic enzymes. However, analysis of the effects of Sulfolobus chromatin proteins on UDG1 leads us to propose a mechanistic basis for coupling UDG1 to the replication fork.
Keywords: DNA repair, DNA replication, proliferating cell nuclear antigen (PCNA), Sulfolobus, uracil, uracil DNA glycosylase (UDG)
Abbreviations: DNA polB1, DNA polymerase B1; Fen1, flap endonuclease-1; GST, glutathione S-transferase; hUNG2, human uracil N-glycosylase 2; MCM complex, minichromosome maintenance complex; PCNA, proliferating cell nuclear antigen; PIP, PCNA-interacting protein; RFC, replication factor C; UDG, uracil DNA glycosylase; XPF, xeroderma pigmentosum complementation group F
INTRODUCTION
PCNA (proliferating cell nuclear antigen) is a toroidal molecule that acts as a sliding clamp, tethering a number of DNA replication and repair factors to DNA [1,2]. Homologues of PCNA are found throughout the archaeal and eukaryotic domains of life. The majority of eukaryotes [2] and the euryarchaeal kingdom of archaea possess a single PCNA homologue (e.g. see [3]) that acts as a homotrimer. Intriguingly, organisms belonging to a second kingdom of archaea, the Crenarchaea, encode multiple PCNA homologues. Pyrobaculum aerophilum has two PCNA homologues [4] and Aeropyrum pernix has three [5]. The PCNA subunits of A. pernix have been demonstrated to have the ability to both homo- and hetero-multimerize [5]. Although all three homotrimers could stimulate the activities of A. pernix DNA polymerases, they did so to varying degrees [5]. An even more extreme situation was discovered in the hyperthermophilic crenarchaeote, Sulfolobus solfataricus P2. Like A. pernix, this species encodes three PCNA homologues [6]. Remarkably, unlike A. pernix, the individual subunits do not form stable homotrimers: rather the S. solfataricus PCNA was shown to be a heterotrimer with a defined order of assembly [7]. A dimer of PCNA1 and PCNA2 forms that then recruits PCNA3 to form a heterotrimer [7].
PCNA molecules from a wide variety of species have been shown to act as a tether for a range of DNA replication and repair factors. More specifically, eukaryotic PCNA has been shown to bind, among others, DNA polymerases, DNA ligase 1, Fen1 (flap endonuclease-1), the clamp loader RFC (replication factor C), CAF1 (chromatin assembly factor) and DNA repair factors such as MutS, XPG (xeroderma pigmentosum complementation group G) and UDG (uracil DNA glycosylase) and a number of cell-cycle-regulatory proteins such as p21 (for a review see [2]). Clearly, therefore, PCNA can act as a central nexus for a broad range of DNA metabolic processes and has the potential to integrate and modulate both replicative and post-replicative events.
Within the archaeal domain of life, evidence has been presented for physical and functional interactions between PCNA and DNA polymerases, DNA ligase, Fen1 and the crenarchaeal homologue of the eukaryotic nucleotide excision repair factor, XPF (xeroderma pigmentosum complementation group F) [3,5,7–10]. In addition, physical interactions have been detected between P. aerophilum PCNA subunits and UDGa [11], and between A. fulgidus PCNA and RNaseHII [12].
Many of these proteins have been demonstrated to interact with PCNA via a conserved PCNA interaction motif, termed the PIP (PCNA-interacting protein) motif [1]. The crystal structure of human PCNA bound to a PIP motif peptide from p21 has shown that each of the three PCNA subunits can bind a peptide [13]. The presence of three binding sites per PCNA ring led to speculation that multiple partner proteins could bind to a given PCNA molecule, in the so-called ‘tool-belt’ model. Our recent studies of the heterotrimeric PCNA of S. solfataricus provided evidence that supports this theory [7]. We found that the three PCNA subunits, PCNA1, 2 and 3, interacted preferentially with Fen1, DNA polB1 (DNA polymerase B1) and DNA ligase respectively. Furthermore, pull-down assays demonstrated that the PCNA heterotrimer could bridge between Fen1 and DNA ligase and polB1.
Given the precedent from other species, it is likely, however, that S. solfataricus PCNA will interact with additional DNA replication and repair factors. In order to identify additional PCNA-interaction partners in S. solfataricus, we have performed a yeast two-hybrid screen of a S. solfataricus genomic DNA library. In the present paper, we describe the identification and characterization of a specific interaction between PCNA3 and a UDG.
EXPERIMENTAL
Construction of the yeast two-hybrid library
S. solfataricus genomic DNA was partially digested with DNaseI in a buffer containing 50 mM Tris/HCl, pH 8, 10 mM MnCl2 and 50 μg/ml BSA to give a range of fragments from 200 to 2000 bp. EcoRI adaptors (Stratagene) were ligated to the sheared DNA that was first polished with Vent DNA polymerase (NEB). Unligated adaptors were removed by using a SizeSep 400 column (Amersham Biosciences). The adapted sheared genomic DNA was cloned into the EcoRI site of pGADT7 vector (Clontech). Escherichia coli XL10-Gold ultracompetent cells were transformed with the ligation product. We obtained approx. 300000 transformants.
Plasmid constructs
UDG1 was amplified by PCR using various primers containing restriction sites. Oligonucleotide sequences are available from S.D.B. on request. pET33-UDG1 was generated by cloning UDG1 into NcoI and XhoI sites of pET33b (Novagen); the GST (glutathione S-transferase) fusion protein plasmid pGEX-UDG1 was generated by cloning UDG1 into EcoRI and XhoI sites of pGEX-4T3 (Amersham Biosciences). To generate the yeast two-hybrid plasmids, UDG1 was cloned into EcoRI and SalI sites of pGBKT7 (Clontech) and into EcoRI and XhoI sites of pGADT7 (Clontech).
Yeast two-hybrid analysis
The yeast strain AH109 was transformed with the appropriate plasmids (see Figure legends) according to the manufacturer's instructions (Clontech Matchmaker manual). The cells were streaked on either Yc−Trp−Leu or Yc−Trp−Leu−His medium.
Protein purification
Expression plasmids were transformed into E. coli strain Rosetta (DE3) pLysS (Novagen). For the untagged UDG1, the cell pellet was resuspended in Buffer A (20 mM Tris/HCl, pH 8, 150 mM NaCl and 14 mM 2-mercaptoethanol). The cells were lysed by sonication, and the lysate was clarified by centrifugation at 35000 g for 30 min. The supernatant was incubated at 75 °C for 25 min to denature and precipitate the E. coli proteins and then centrifuged at 35000 g for 30 min. The supernatant was loaded on to a 5 ml HiTrap Heparin column (Amersham Biosciences) preequilibrated with buffer A. The protein was eluted using a 50 ml linear gradient of 150–1000 mM NaCl. The UDG1 protein was eluted at 600 mM NaCl.
The GST–UDG protein was immobilized on glutathione–Sepharose beads according to the manufacturer's instructions (Amersham Biosciences). The PCNA subunits were purified as described previously [7]. Alba and Sso7d were purified as described previously [14,15]. The Sso7d overexpression construct was a gift from Professor Malcolm White (Centre for Biomolecular Science, St. Andrews University, St. Andrews, Scotland, U.K.).
DNA substrates
70-mer, 5′-GTTTAAAGCATTTGAGGGGGATTCAATGAATATTUATGACGATTCCGCAGTATTGGACGCTATCCAGTCT-3′ was 5′-end-labelled with 32P, and was annealed to either 5′-AGACTGGATAGCGTCCAATACTGCGGAATCGTCATAAATATTCATTGAATCCCCCTCAAATGCTTTAAAC-3′ to give a double-stranded substrate with an A:U base pair or to 5′-AGACTGGATAGCGTCCAATACTGCGGAATCGTCATGAATATTCATTGAATCCCCCTCAAATGCTTTAAAC-3′ to give a double-stranded substrate with a G:U base pair. The corresponding substrates containing A:T and G:T base pairs were also prepared.
UDG assay
The assays were performed in 20 μl of buffer containing 50 mM Tris/HCl, pH 8, 1 mM dithiothreitol, 1 mM EDTA, 10 mM KCl, 100 μg/ml BSA, 0.2 pmol of labelled DNA and 0.02 pmol of UDG1, and were incubated for 15–30 min at 65 °C (see Figure legends). The reactions were subjected to hot alkali treatment by addition of 100 mM NaOH and incubation for 10 min at 99 °C. Then HCl was added to 100 mM to neutralize the reaction. The reaction products were separated on a 12% denaturing gel.
RESULTS
Sulfolobus PCNA3 interacts with UDG
We have constructed a yeast two-hybrid library from randomly cleaved S. solfataricus P2 genomic DNA in the yeast/E. coli shuttle vector pGADT7. This library contains 3×105 individual clones with insert size between 200 and 2000 nucleotides. We have screened this library using the S. solfataricus PCNA3 gene (SSO0405 in genome annotation) as bait. Arising from this screen (see the Experimental section for details), we identified a number of interaction partners (Figure 1A).
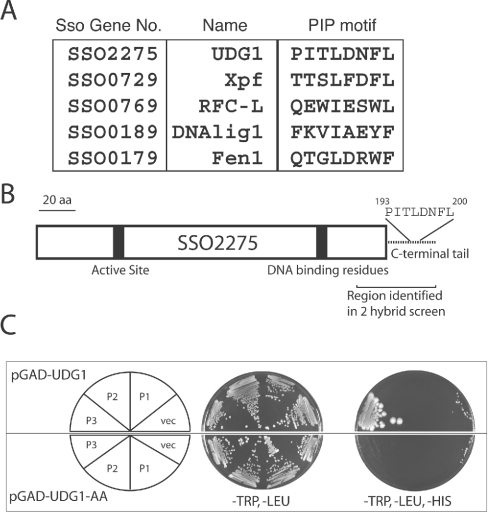
Figure 1. UDG1 interacts specifically with PCNA3 in a PIP motif-dependent manner.
(A) PIP motifs of interaction partners for PCNA3. The PIP motif and position in amino acid residues is given for S. solfataricus UDG1; beneath, the candidate PIP motifs of other PCNA-interaction partners, as well as the derived eukaryotic PIP motif consensus are also shown. DNAlig1, DNA ligase 1. (B) Cartoon of the organization of the predicted protein product of the UDG1 open reading frame. (C) Yeast two-hybrid analysis of the specificity of the UDG1–PCNA3 interaction and its dependence on the PIP motif. The upper panel contains wild-type UDG1 fused to the GAL4 activation domain and in the presence of PCNA1, 2 or 3 fused to the GAL4 DNA-binding domain (or vector alone control, vec). The lower panel contains UDG1 with a mutated PIP motif. The left-hand plate shows growth on medium selecting for plasmids alone (lacking leucine and tryptophan) and the right-hand plate shows growth on medium selecting for interaction of introduced gene products (lacking leucine, tryptophan, histidine).
Among these was a clone containing an insert corresponding to the C-terminal 132 nucleotides of open reading frame SSO2275, annotated as ‘DNA polymerase phage SPO1 N-terminal domain homologue’ [6]. The predicted translation product of the full open reading frame was used in PsiBlast searching of GenBank®. This analysis revealed that the polypeptide encoded by SSO2275 is a member of the family 4 group of UDGs. Hereafter we shall refer to this protein as UDG1 (Figure 1B).
The screen that identified the udg1 gene used PCNA3 as bait. Previously, we have found that the three S. solfataricus PCNAs have distinct preferred interaction partners [7]. We wished to determine whether UDG1 might also show selectivity in its choice of partner protein. Accordingly, we cloned the full-length open reading frame of udg1 into pGADT7 and tested for its ability to interact with PCNA1, 2 and 3, as well as control constructs. As can be seen in the upper panel of Figure 1(C), interaction was detectable with PCNA3 only. Next we sought to verify the specificity of the protein–protein interaction.
As discussed above, many proteins have been observed to interact with PCNA, allowing the determination of a consensus interaction motif, or PIP element. This corresponds to the general consensus of QXX(L/M/I)XX(F,Y,H)(F,Y) where X is any amino acid [1]. The initial clone identified in the two-hybrid screen contained the C-terminal 43 amino acids. Examination of the predicted polypeptide sequence of this region of UDG1 revealed the presence of a motif of sequence PITLDNFL from residues 193 to 200 of the protein. Although not showing exact correspondence to the consensus PIP motif, this nevertheless is similar to the PIP motif of the known S. solfataricus PCNA3 interactors XPF and Fen1 (Figure 1A) [7,9]. Using site-directed mutagenesis, we altered the codons for the phenylalanine and leucine to encode alanine, and we termed the resultant mutant protein UDG1-AA. We then employed UDG1-AA in yeast two-hybrid assays. We first verified that the wild-type and UDG1-AA fusion proteins were expressed to similar levels in yeast (results not shown). Importantly, when UDG1-AA was tested in the two-hybrid assay, we were unable to detect any interaction with PCNA3 (Figure 1C, lower panel), indicating the importance of the candidate PIP motif in mediating the interaction.
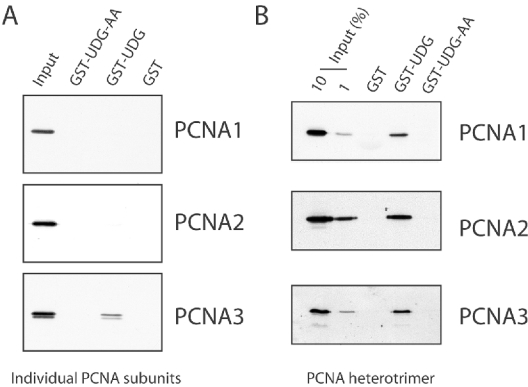
To test whether the interactions detected by two-hybrid analysis were direct, we next expressed wild-type UDG1 and UDG1-AA as fusion proteins with GST. We then employed these proteins in pull-down assays with purified recombinant PCNA1, PCNA2 or PCNA3. In agreement with the two-hybrid results, we could only detect an interaction between GST fused to wild-type UDG1 and PCNA3. Importantly, the interaction was not detected with GST–UDG1-AA, nor did PCNA1 or PCNA2 bind detectably to either wild-type UDG1 or UDG1-AA (Figure 2A). However, when pulldown assays were performed with the assembled heterotrimeric PCNA, instead of isolated subunits, it was now possible to detect PCNA1 and PCNA2 being pulled down as well as PCNA3, by GST–UDG1 (Figure 2B).
Figure 2. UDG interacts with the PCNA complex.
(A) A 3 μg amount of the indicated GST fusion protein was immobilized on GST–Sepharose and incubated with 1 μg of purified recombinant PCNA1, PCNA2 or PCNA3. Following extensive washing, beads were boiled in SDS/PAGE loading buffer, and proteins were separated by SDS/PAGE. The bound proteins were detected by Western blotting with specific antisera as indicated. (B) As in (A), except that the GST fusion protein was incubated with 1 μg of the PCNA heterotrimer.
Activity of UDG1
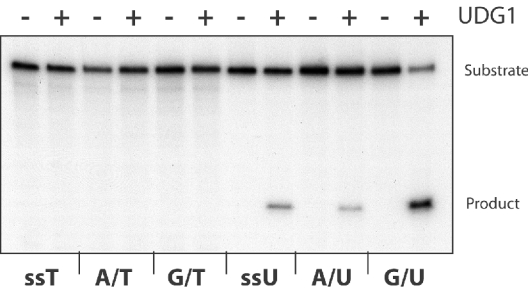
We next sought to determine the activity of the UDG1 protein. To this end, we purified a recombinant version of the full-length protein using a combination of heat clarification of E. coli extract, followed by heparin–Sepharose chromatography. First, we examined the activity of the protein on a number of synthetic oligonucleotide substrates. The results of this analysis revealed that the protein had no activity detectable on A:T base pairs or G:T mismatches. However, substrates with either A:U or G:U base pairs, or single-stranded DNA containing uracil, were cleaved by the enzyme. The preferred substrate appears to be the G:U-containing double-stranded oligonucleotide (Figure 3).
Figure 3. Substrate specificity of UDG1.
A 0.2 pmol amount of single-stranded 70-mer substrate (ssT or ssU) or double-stranded 70-mer substrate (A/T, G/T, A/U or G/U) was incubated with or without 0.02 pmol of UDG1 at 65 °C for 15 min. Reaction products were treated as described in the Experimental section, and resolved on a 12% denaturing polyacrylamide gel before drying and detection of products by autoradiography.
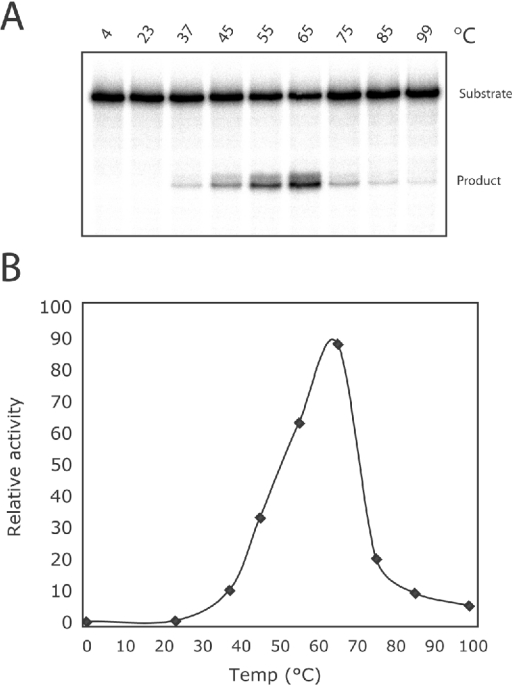
S. solfataricus is a hyperthermophile, growing optimally at 80 °C. We next tested the optimal temperature for the assay. As can be seen from Figure 4, the enzyme has peak activity at 65 °C, and, while product is still detectable at 75, 85 and 99 °C, it is at a markedly reduced level at these elevated temperatures (Figure 4).
Figure 4. Temperature dependence of UDG1 activity.
(A) A 0.2 pmol amount of double-stranded 70-mer substrate (G:U) was incubated with 0.02 pmol of UDG1 at the indicated temperature for 30 min. (B) Quantification of the activity of UDG1 at different temperatures as shown in (A). The dried gel was exposed to a phosphor storage screen and the amount of the product was quantified using the ImageQuant software.
Given that the interaction of many enzymes with PCNA results in a stimulation of their activity, we wished to test whether this was the case for UDG1. However, despite testing a wide range of temperatures and ionic conditions, and varying the relative amounts, ratios and order of addition of UDG1, PCNA and DNA, we were unable to detect any specific PCNA-dependent modulation of the activity of UDG1 either by individual PCNA subunits or the assembled PCNA heterotrimer (Figure 5 and results not shown).
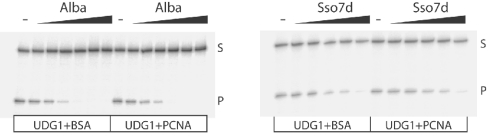
Figure 5. Effect of chromatin proteins on the UDG1 activity.
A 0.2 pmol amount of double-stranded 70-mer substrate (G:U) was incubated with 0, 1.17, 2.34, 4.68, 9.4, 18.8 or 37.5 pmol of Alba (left-hand panel) or 0, 12.5, 25,100, 200 or 400 pmol of Sso7d (right-hand panel) for 10 min at 60 °C, and 1.5 pmol of BSA or heterotrimeric PCNA was added and incubated for a further 10 min before the addition of 0.01 pmol of UDG1 and incubation for a further 15 min. S, substrate; P, product.
Effect of chromatin proteins on the activity of UDG1
The UDG1 assays described above were performed on naked DNA substrates. In the context of the archaeal cell, DNA is compacted by association with small basic proteins. In S. solfataricus, the best characterized of these proteins are members of the Sulfolobus-specific Sul7d family (Sso7d in S. solfataricus) and the highly conserved Alba [16]. We titrated purified recombinant Alba and Sso7d individually into UDG1 assays. Significantly, we found that both Alba and Sso7d repressed the activity of UDG1. The effect of Sso7d was markedly less than that of Alba. Thus it appears that the formation of archaeal chromatin is highly repressive to UDG1 activity (Figure 5). No specific direct interactions could be detected between either chromatin protein and UDG1 (results not shown). It should be noted that the addition of PCNA to these reactions had no significant effect.
DISCUSSION
One of the commonest pro-mutagenic events encountered by cells is the spontaneous deamination of cytosine followed by tautomeric shift to create uracil. This leads to generation of a G:U base pair, which, if uncorrected before DNA replication, can eventually lead to an A:T base pair substituting for the correct G:C base pair. As well as occurring in situ on DNA, uracil can be formed in the context of deoxycytidine triphosphate precursor and lead to incorporation of deoxyuridine monophosphate by DNA polymerase during replication. Deamination of cytosine to uracil is promoted at elevated temperatures. Thus hyperthermophilic organisms might be anticipated to possess highly efficient machineries for detecting and removing uracil bases in DNA. A number of UDG activities have been detected in hyperthermophilic archaea and bacteria [17]. The family 4 enzymes are found in a range of species and, although in a distinct family from hUNG2 (human uracil N-glycosylase 2), have similar substrate preference to the human protein [11,18,19]. Interestingly, hUNG2 has been shown to interact with PCNA, and has been proposed to have a primary role in post-replicative removal of uracil from DNA [19].
Given the precedent for PCNA-mediated stimulation of a range of DNA replication and repair associated enzymes [1,2], we were initially somewhat surprised to be unable to detect any enhancement of the UDG1 activity by PCNA.
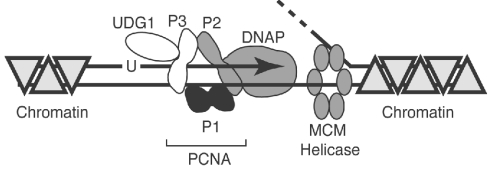
However, in the light of our observation of the repressive effect of archaeal chromatin proteins on UDG1 activity, we propose the model illustrated in Figure 6. We found that the archaeal chromatin proteins strongly repress the activity of UDG1. Replication of cellular DNA takes place in the context of chromatin, and the replication machinery must displace DNA-bound proteins, including chromatin proteins, which would otherwise act as a block to the passage of replication forks. This task is likely to be performed by the replicative helicase; in archaea, this is widely presumed to be the MCM (minichromosome maintenance) complex. Indeed, recent work has revealed that an archaeal MCM complex (from Methanothermobacter thermoautotrophicum) can even disrupt a biotin–streptavidin complex on artificial DNA substrates [20]. Thus it is likely that, as the replication fork proceeds, the MCM complex will remove chromatin ahead of the fork. It is also likely that archaeal chromatin will re-form on the newly synthesized daughter strands behind the DNA polymerase. Therefore tethering UDG1 to the fork via PCNA provides a simple mechanism to ensure that dUMP incorporated by the polymerase is rapidly detected and also ensures that the UDG1 is targeted to regions of naked DNA, i.e. the replication fork. Furthermore, as UDG1 interacts with PCNA3 and DNA polymerase interacts with PCNA2 [7], it is likely that UDG activity can be directly coupled to progression of the replication fork.
Figure 6. Model for the action of UDG at the replication fork.
For simplicity, only the leading strand assembly is shown. The lagging strand template is indicated by a dotted line at the top of the Figure. The three PCNA subunits, PCNA1 (P1), PCNA2 (P2) and PCNA3 (P3), are shown in black, grey and white respectively. PCNA2 interacts with DNA polymerase (DNAP). As we have revealed in the present study, PCNA3 interacts with UDG1 (white oval). The hexameric presumptive replicative helicase, the MCM complex, is shown as a hexagonal arrangement of subunits. The direction of extension of newly synthesized DNA is shown by the arrowhead, and a uracil is indicated. The archaeal chromatin proteins (Alba and Sso7d) are indicated by triangles.
Our data therefore support a model of the UDG1 performing an immediately post-replicative function. As this UDG1 cannot detect uracil in the context of archaeal chromatin, it is possible that alternative uracil DNA glycosylases exist that perform a genomemonitoring function in bulk non-replicating DNA. In this light, a number of additional UDG1 activities have been identified in other archaea [17], and it is possible that one of these additional molecules performs this function. Additionally, it has been observed that archaeal B-type DNA polymerases possess a read-ahead uracil-sensing function [21,22].
Finally, is the assembly of UDG1 on PCNA regulated? Our previous work has revealed that the heterotrimeric Sulfolobus PCNA can simultaneously bind Fen1, DNA polB1 and DNA ligase 1 via interactions with PCNA1, 2 or 3 respectively [7], providing a model for co-ordinated action at the lagging strand. However, in addition, PCNA1 and 3 have been shown to be capable of interaction with XPF [9]. Therefore PCNA3 has the capacity to interact with at least three distinct factors, DNA ligase, XPF and UDG1, as well as the clamp loader, RFC. As all these factors interact via similar mechanisms with PCNA3, this poses the question of how the assembly of these various factors on PCNA is co-ordinated and regulated. Do PCNA trimers on the lagging and leading strands have discrete assemblies or is the generation of complexes simply stochastic? It is apparent that goals for ongoing research include investigation of the stoichiometry of PCNA at the replication fork in vivo, determination of the presence (if any) of covalent modifications of either PCNA or associated factors and the establishment of a defined in vitro replication system to allow these complex questions to be addressed.
Acknowledgments
This work was supported by the Medical Research Council.
References
- 1.Warbrick E. The puzzle of PCNA's many partners. BioEssays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 2.Vivona J. B., Kelman Z. The diverse spectrum of sliding clamp interacting proteins. FEBS Lett. 2003;546:167–172. doi: 10.1016/s0014-5793(03)00622-7. [DOI] [PubMed] [Google Scholar]
- 3.Cann I. K. O., Ishino S., Hayashi I., Komori K., Toh H., Morikawa K., Ishino Y. Functional interactions of a homolog of proliferating cell nuclear antigen with DNA polymerases in Archaea. J. Bacteriol. 1999;181:6591–6599. doi: 10.1128/jb.181.21.6591-6599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fitz-Gibbon S. T., Ladner H., Kim U. J., Stetter K. O., Simon M. I., Miller J. H. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. U.S.A. 2002;99:984–989. doi: 10.1073/pnas.241636498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daimon K., Kawarabayasi Y., Kikuchi H., Sako Y., Ishino Y. Three proliferating cell nuclear antigen-like proteins found in the hyperthermophilic archaeon Aeropyrum pernix: interactions with the two DNA polymerases. J. Bacteriol. 2002;184:687–694. doi: 10.1128/JB.184.3.687-694.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.She Q., Singh R. K., Confalonieri F., Zivanovic Y., Allard G., Awayez M. J., Chan-Weiher C. C. Y., Clausen I. G., Curtis B. A., De Moors A., et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dionne I., Nookala R. K., Jackson S. P., Doherty A. J., Bell S. D. A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Cell. 2003;11:275–282. doi: 10.1016/s1097-2765(02)00824-9. [DOI] [PubMed] [Google Scholar]
- 8.Kelman Z., Hurwitz J. A unique organization of the protein subunits of the DNA polymerase clamp loader in the archaeon Methanobacterium thermoautotrophicum ΔH. J. Biol. Chem. 2000;275:7327–7336. doi: 10.1074/jbc.275.10.7327. [DOI] [PubMed] [Google Scholar]
- 9.Roberts J. A., Bell S. D., White M. F. An archaeal XPF repair endonuclease dependent on a heterotrimeric PCNA. Mol. Microbiol. 2003;48:361–372. doi: 10.1046/j.1365-2958.2003.03444.x. [DOI] [PubMed] [Google Scholar]
- 10.Seybert A., Scott D. J., Scaife S., Singleton M. R., Wigley D. B. Biochemical characterisation of the clamp/clamp loader proteins from the euryarchaeon Archaeoglobus fulgidus. Nucleic Acids Res. 2002;30:4329–4338. doi: 10.1093/nar/gkf584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H. J., Chiang J. H., Fitz-Gibbon S., Lebel M., Sartori A. A., Jiricny J., Slupska M. M., Miller J. H. Direct interaction between uracil-DNA glycosylase and a proliferating cell nuclear antigen homolog in the crenarchaeon Pyrobaculum aerophilum. J. Biol. Chem. 2002;277:22271–22278. doi: 10.1074/jbc.M201820200. [DOI] [PubMed] [Google Scholar]
- 12.Motz M., Kober I., Girardot C., Loeser E., Bauer U., Albers M., Moeckel G., Minch E., Voss H., Kilger C., Koegl M. Elucidation of an archaeal replication protein network to generate enhanced PCR enzymes. J. Biol. Chem. 2002;277:16179–16188. doi: 10.1074/jbc.M107793200. [DOI] [PubMed] [Google Scholar]
- 13.Gulbis J. M., Kelman Z., Hurwitz J., O'Donnell M., Kuriyan J. Structure of the C-terminal region of p21WAF1/CIP1 complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 14.Bell S. D., Botting C. H., Wardleworth B. N., Jackson S. P., White M. F. The interaction of Alba, a conserved archaeal, chromatin protein, with Sir2 and its regulation by acetylation. Science. 2002;296:148–151. doi: 10.1126/science.1070506. [DOI] [PubMed] [Google Scholar]
- 15.Edmondson S. P., Shriver J. W. DNA-binding proteins Sac7d and Sso7d from Sulfolobus. Methods Enzymol. 2001;334:129–145. doi: 10.1016/s0076-6879(01)34463-4. [DOI] [PubMed] [Google Scholar]
- 16.White M. F., Bell S. D. Holding it together: chromatin in the archaea. Trends Genet. 2003;18:621–626. doi: 10.1016/s0168-9525(02)02808-1. [DOI] [PubMed] [Google Scholar]
- 17.Sartori A. A., Schar P., Fitz-Gibbon S., Miller J. H., Jiricny J. Biochemical characterization of uracil processing activities in the hyperthermophilic archaeon Pyrobaculum aerophilum. J. Biol. Chem. 2001;276:29979–29986. doi: 10.1074/jbc.M102985200. [DOI] [PubMed] [Google Scholar]
- 18.Kavli B., Sundheim O., Akbari M., Otterlei M., Nilsen H., Skorpen F., Aas P. A., Hagen L., Krokan H. E., Slupphaug G. HUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 2002;277:39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 19.Otterlei M., Warbrick E., Nagelhus T. A., Haug T., Slupphaug G., Akbari M., Aas P. A., Steinsbekk K., Bakke O., Krokan H. E. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin J. H., Jiang Y., Grabowski B., Hurwitz J., Kelman Z. Substrate requirements for duplex DNA translocation by the eukaryal and archaeal minichromosome maintenance helicases. J. Biol. Chem. 2003;278:49053–49062. doi: 10.1074/jbc.M308599200. [DOI] [PubMed] [Google Scholar]
- 21.Greagg M. A., Fogg M. J., Panayotou G., Evans S. J., Connolly B. A., Pearl L. H. A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9045–9050. doi: 10.1073/pnas.96.16.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fogg M. J., Pearl L. H., Connolly B. A. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat. Struct. Biol. 2002;9:922–927. doi: 10.1038/nsb867. [DOI] [PubMed] [Google Scholar]