Abstract
The major surface glycoprotein (MSG) of Pneumocystis carinii f. sp. carinii is a family of proteins encoded by a family of heterogeneous genes. Messenger RNAs encoding different MSGs each begin with the same 365-bp sequence, called the Upstream Conserved Sequence (UCS), which is in frame with the contiguous MSG sequence. The UCS contains several potential start sites for translation. To determine if translation of MSG mRNAs begins in the UCS, polyclonal antiserum was raised against the 123-amino-acid peptide encoded by the UCS. The anti-UCS serum reacted with a P. carinii protein that migrated at 170 kDa; however, it did not react with the mature MSG protein, which migrates at 116 kDa. A 170-kDa protein was immunoprecipitated with anti-UCS serum and shown to react with a monoclonal antibody against a conserved MSG epitope. To explore the functional role of the UCS in the trafficking of MSG, the nucleotide sequence encoding the UCS peptide was ligated to the 5′ end of an MSG gene and incorporated into a recombinant baculovirus. Insect cells infected with the UCS-MSG hybrid gene expressed a 160-kDa protein which was N-glycosylated. By contrast, insect cells infected with a baculovirus carrying an MSG gene lacking the UCS expressed a nonglycosylated 130-kDa protein. These data suggest that in P. carinii, translation begins in the UCS to produce a pre-MSG protein, which is subsequently directed to the endoplasmic reticulum and processed to the mature form by proteolytic cleavage.
Pneumocystis carinii is a fungus that can cause pneumonia in immunocompromised humans and other mammals (29, 57). Maintenance of P. carinii populations in culture has not yet been achieved. Different genetic varieties of P. carinii (called special forms) are found in different host species (3–5, 15, 33–35, 40, 42, 43, 58). P. carinii f. sp. carinii, the subject of this study, is one of two special forms that have been found in laboratory rats (2).
The predominant protein found on the surface of P. carinii f. sp. carinii is called the major surface glycoprotein (MSG) (1, 12, 17, 19, 23, 32, 37, 51, 52, 56). Other special forms of P. carinii have a similar surface antigen, which is known as either MSG (25, 56) or gpA (10, 11). MSG is thought to play a crucial role in host-pathogen interactions because it is recognized by serum antibodies and T cells from exposed hosts (8, 9, 11, 12, 17, 19, 26, 30, 41) and binds to several host proteins, including fibronectin, surfactant protein A, and surfactant protein D (7, 28, 31, 36, 60).
MSG is actually a family of proteins encoded by a family of heterogeneous genes (9, 13, 16, 18, 44, 45, 59). P. carinii f. sp. carinii contains approximately 100 different MSG genes, which are organized in clusters located at the ends of each chromosome (45, 47, 49, 52–54). It is probable that only one MSG gene is expressed in an individual P. carinii organism at any given time, because only one locus in the genome (known as the MSG expression site) produces mRNA encoding an MSG isoform (6, 47, 48, 54, 55). Different MSG genes can occupy the MSG expression site in different organisms within a population, suggesting that recombination installs 1 of the 100 MSG genes at this unique locus. Such a recombination system would endow P. carinii with the capacity to vary its surface at high frequency.
The expression site locus contains a unique 365-bp sequence (called the Upstream Conserved Sequence, or UCS), which is found at the beginning of each mRNA encoding MSG (55). Examination of the UCS and adjacent MSG-encoding sequences suggests that translation of an MSG peptide might initiate at the first AUG codon, which lies in the UCS, between 17 and 37 nucleotides from the 5′ end of a typical MSG-encoding mRNA molecule (55) (Fig. 1). The first AUG codon of the UCS begins an open reading frame (ORF) that continues through the downstream MSG-encoding sequence in every case examined so far (6, 55). In addition, the UCS portion of this ORF encodes a hydrophobic domain that could function as a signal sequence for translocation of the MSG into the endoplasmic reticulum (6, 54, 55). Such a candidate signal sequence was absent from the conceptual MSG peptide first proposed, which did not include the UCS (18). To test the hypothesis that the primary translation product of an MSG mRNA begins with a peptide encoded by the UCS, antisera were raised against the UCS peptide. The α-UCS sera identified a P. carinii f. sp. carinii protein that has the properties expected of an MSG precursor. The α-UCS sera also indicated that the UCS is not present on the 116-kDa MSG found on the surface of P. carinii f. sp. carinii. Expression studies done with insect cells showed that the UCS can direct an MSG protein to the endoplasmic reticulum, suggesting that one role of the UCS is to direct the MSG to the cell surface. These data suggest that in P. carinii, MSG translation begins in the UCS and that the UCS-MSG protein enters the secretory pathway but that the UCS is ultimately removed to produce the MSG found on the organism’s surface.
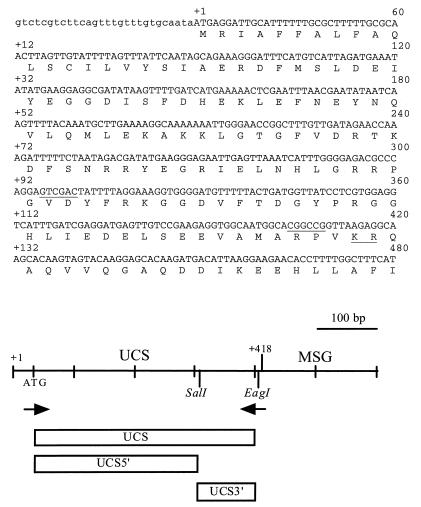
FIG. 1.
Sequences and map of the UCS. In the nucleotide sequence, the uppercase letters show the putative coding sequences, and the lowercase letters indicate the 5′ untranslated region of the UCS-MSG cDNA clone 1 (47). The numerals in the right margin above the nucleotide sequence starting at 60 are aligned with the corresponding nucleotide. Numbers on the left starting with +1 and ending with +132 are encoded amino acid residues, which are shown in single-letter code below the nucleotide sequence, with the amino acid aligned with the second nucleotide of each codon. The SalI and EagI sites are underlined. The underlined KR is a potential site for proteolytic cleavage. The drawing below the sequences is a map of the UCS-MSG locus. Crosshatches mark 100-bp increments. The UCS region extends from +1 to +418. The MSG region begins at +419. The location and orientation of the two primers utilized in the initial PCR are indicated by arrows. The regions expressed as fusion proteins are represented by rectangles labeled UCS, UCS5′, and UCS3′.
MATERIALS AND METHODS
Antibodies.
RB-E3, RA-E7, RA-C1, RA-C6, RA-C7, RB-C8, and RA-C11 are monoclonal antibodies (MAbs) raised against MSG (21). Polyclonal antibodies to P. carinii f. sp. carinii and to MSG were prepared by immunizing rabbits with purified P. carinii f. sp. carinii organisms and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)-purified MSG.
Generation of plasmids and fusion proteins.
A 365-bp UCS DNA fragment was amplified from the UCS-MSG cDNA1 clone (47) by PCR with primers 1 (5′-GAGGCCTCATTGTGTGCAATAATGAGGATTGCA-3′) and 2 (5′-GGAATTCGGATCCTACATTGCCACCTCTTCGG-3′) (Fig. 1). This PCR product was gel purified, inserted into the EcoRV site of Bluescript SK− (Stratagene, LaJolla, Calif.) and sequenced (38). To produce a plasmid that would express the UCS peptide fused to glutathione S-transferase (GST), the UCS was released from the Bluescript plasmid by digestion with StuI and EcoRI, the sites for which had been incorporated onto the 5′ ends of primers 1 and 2, respectively. The StuI/EcoRI UCS fragment was inserted between the SmaI and EcoRI sites of pGEX-3X (Pharmacia Biotech, Inc., Piscataway, N.J.). The GST-UCS junction was sequenced to confirm that the UCS was in frame with GST. Production of the GST-UCS protein was induced according to the manufacturer’s protocol (Pharmacia Biotech, Inc.) and monitored by SDS-PAGE. The GST-UCS fusion protein was gel purified from the insoluble cell lysate. To produce a plasmid that would express the UCS peptide fused to the gene 9 protein (G9) from bacteriophage T7, the UCS was removed from the Bluescript SK− plasmid as a StuI/EcoRI fragment and inserted between the StuI and EcoRI sites of a vector called pHX9-KS1 (Protein Express, Cincinnati, Ohio) (14) to generate pUCS/pHX9-KS1. Two additional gene 9 fusion plasmids (pG9/UCS5′ and pG9/UCS3′) were made from pUCS and pHX9-KS1. To produce a plasmid that expressed the first 92 amino acids of the UCS peptide (pG9/UCS5′), the 3′ end of the UCS from the UCS/pHX9-KS1 plasmid was deleted by cutting with SalI and EcoRI and filling the single-stranded ends, followed by ligation (38). To produce a plasmid that expressed the last 31 amino acids of the UCS peptide (pG9/UCS-3′), pUCS/pHX9-KS1 was cut with SalI and the ends were filled. Then the DNA was cut with EcoRI, which released the 3′ end of the UCS. The 3′ end of the UCS was ligated into pHX9-KS1 that had been treated in an identical manner (SalI digestion, filling in, and EcoRI digestion) (38).
Production of the G9-UCS, G9-UCS5′, and G9-UCS3′ fusion proteins was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and monitored by SDS-PAGE. The fusion proteins were purified over a nickel column from Qiagen, Inc. (Chatsworth, Calif.).
α-UCS sera.
Polyclonal antibodies were raised against the gel-purified GST-UCS fusion protein by inoculating two New Zealand White rabbits with 100 μg of the fusion protein, followed by additional injections of 50 μg 14, 21, and 49 days later. The serum collected on day 56 was used in this study.
Protein electrophoresis and immunoblot assays.
P. carinii f. sp. carinii was obtained from immunosuppressed rats as previously described (2). Whole-organism homogenates were obtained, solubilized with an equal volume of 2× treatment buffer (0.125 M Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 10% 2-mercaptoethanol) and boiled for 3 min before use. The samples were analyzed by SDS-PAGE on 8 to 16% Novex (San Diego, Calif.) precast gels that were run at 150 V for approximately 1.5 h. Proteins were transferred electrophoretically (100 V for 1 h) from the SDS-PAGE gel onto a nitrocellulose membrane (Micron Separations, Inc., Westborough, Mass.) which was stained with Ponceau red to confirm the protein transfer. The membranes were blocked in TBS (0.02 M Tris-HCl, 0.5 M NaCl [pH 7.5]) with 1% skim milk at room temperature for 45 min. Each blot was incubated overnight at 4°C with antiserum or MAbs diluted 1:1,000 in TBS. The blots were then washed twice for 10 min each time with TBS-0.05% Tween 20 (Bio-Rad, Hercules, Calif.) and incubated for 1.5 h at room temperature with the appropriate phosphatase-labeled conjugate (either goat anti-rabbit immunoglobulin G [IgG] [H plus L] or goat anti-mouse IgG [H plus L] [KPL, Inc., Gaithersburg, Md.]) diluted 1:1,000 in TBS. The blots were washed as described above and then developed with nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP) (Pierce, Rockford, Ill.).
Immunoprecipitation.
P. carinii f. sp. carinii organisms were purified by standard procedures (2). For each immunoprecipitation reaction, a pellet containing 108 organisms was resuspended in 100 μl of solubilization buffer (0.190 M NaCl, 0.006 M EDTA, 0.060 M Tris-HCl [pH 7.4], 4% SDS). The lysate was sonicated for 30 s at 4°C, heated immediately at 100°C for 4 min, and then placed on ice. Next, 100 μl of H2O followed by 800 μl of dilution buffer (0.190 M NaCl, 0.006 M EDTA, 0.050 M Tris [pH 7.4], 2.5% Triton X-100) were added. Centrifugation at 12,000 × g for 30 s was performed, and the supernatant was transferred to a separate tube. Ten microliters of the prebleed serum from rabbit 2 was added to each supernatant and mixed for 1 h at 4°C, followed by centrifugation at 12,000 × g for 2 min. The supernatant was transferred to a separate tube, and 30 μl of protein G-Sepharose (Sigma, St. Louis, Mo.) was added and mixed at room temperature for 2 h. Following centrifugation at 12,000 × g for 30 s, the supernatant was immunoprecipitated with one of the following: 20 μl of α-UCS2 serum or 30 μl of MAb RA-E7 (2 mg/ml) for 1 h, with mixing at 4°C. Thirty microliters of protein G-Sepharose was then added and mixed for 2 h at room temperature. Immunoprecipitations were collected by centrifugation for 30 s at 12,000 × g and washed four times in 0.150 M NaCl-0.050 M Tris (pH 7.4)-0.005 M EDTA-0.1% Triton X-100-0.02% SDS and twice in 0.150 M NaCl-0.050 M Tris (pH 7.5)-0.050 M EDTA. The pellet was then resuspended in 40 μl of 2× treatment buffer, boiled for 3 min, and subjected to centrifugation for 30 s at 12,000 × g. Proteins immunoprecipitated with MAb RA-E7 were separated on a 6% Novex gel, blotted, and reacted to the α-UCS2 serum as described above. Proteins immunoprecipitated with the α-UCS2 serum were separated on an 8 to 16% Novex gel, blotted, and reacted to MAb RA-E7 as described above.
Expression of MSG in insect cells.
The MSG B gene (49) was amplified by PCR with primer 3 (5′-ACTGATCAATTGATGGCACGGCCGGTTAAGAGG-3′) and primer 4 (5′-ACTGTACAATTGTCATCCATTTTCAAATCGTCTTTCAATG-3′), which contained MunI sites at their 5′ ends. The 3,801-bp MSG B PCR product was gel purified, digested with MunI, and inserted into the EcoRI site of PVL1392 (Invitrogen, Carlsbad, Calif.). A 365-bp UCS fragment was excised from the Bluescript SK−/UCS plasmid with EagI and ligated into the EagI-digested PVL 1392/MSG B plasmid to yield plasmid PVL1392/UCS-MSG. Orientation was confirmed by sequencing. This strategy placed the UCS in frame with the MSG B coding sequence. The PVL 1392/MSG and PVL 1392/UCS-MSG constructs were each transfected into Spodoptera frugiperda (Sf-9) cells (Invitrogen) along with a modified wild-type baculovirus (Baculogold; Pharmingen) from Autographa californica (27, 46). Recombinant viruses containing the UCS-MSG and MSG genes were generated by in situ homologous recombination, plaque purified, and amplified to high titer. Proteins were expressed in Sf-9 cells by infection with the purified recombinant viruses at a multiplicity of infection of 25 to 100 PFU/cell for 1 h. In some experiments, cells were treated with 5 μg of tunicamycin per ml after infection to prevent N-linked glycosylation. Cells were harvested 24 h after infection by scraping and resuspension in phosphate-buffered saline (PBS) containing protease inhibitors. Cell lysates (105 cells/lane) were size fractionated on 6% Novex SDS-PAGE gels and transferred to a nitrocellulose membrane. The membrane was blocked in PBS containing 3% skim milk and 1% Triton-X. The blot was incubated for 1.5 h with MAb RA-E7 diluted 1:250 and then washed with blocking buffer and incubated for 1.5 h with peroxidase-conjugated goat anti-mouse IgG (Bio-Rad) diluted 1:1,000. The blots were washed with PBS containing 1% Triton-X and then developed with o-phenylenediamine.
RESULTS
Identification of a P. carinii f. sp. carinii protein reactive with α-UCS sera.
In order to determine whether the UCS was translated, rabbit antibodies were generated against a GST-UCS fusion protein. Sera from two rabbits (α-UCS2 and α-UCS3) that had been injected with the GST-UCS fusion protein were assayed by immunoblotting for reactivity with the GST-UCS fusion protein and with GST alone. Both α-UCS2 and α-UCS3 sera reacted with the GST-UCS fusion protein and with GST (Fig. 2A and B, lanes 1 and 2).
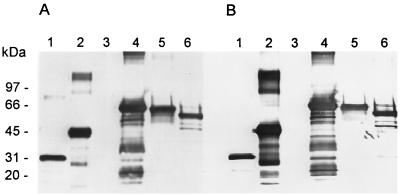
FIG. 2.
Antiserum reactivity with UCS fusion proteins. E. coli lysates containing either GST, G9, or these proteins fused to UCS protein were separated on duplicate 8 to 16% Novex SDS-PAGE gels. Resolved proteins were transferred to a nitrocellulose membrane which was incubated with either α-UCS2 (A) or α-UCS3 (B) serum. Bound rabbit antibodies were detected by reaction with goat anti-rabbit IgG conjugated to alkaline phosphatase. Phosphatase activity was detected by incubation with BCIP-NBT. Lanes: 1, GST; 2, GST-UCS; 3, G9; 4, G9-UCS; 5, G9-UCS-5′; 6, G9-UCS-3′. The positions of Bio-Rad high-molecular-mass markers are indicated to the left of lane 1.
To confirm that the α-UCS sera were recognizing UCS epitopes, three additional fusion proteins were made. These proteins contained part or all of the UCS peptide fused to G9 of bacteriophage T7. In one plasmid, the entire UCS sequence was fused to the G9 gene. Two additional G9 constructs contained either the 5′ end of the UCS (pG9/UCS5′, encoding amino acid residues 1 to 92) or the 3′ end of the UCS (pG9/UCS3′, encoding amino acid residues 94 to 123) fused to G9 (Fig. 1). Lysates from Escherichia coli that had been induced to produce either G9 or one of the three G9-UCS fusion proteins were subjected to SDS-PAGE. The resolved proteins were transferred to a nitrocellulose membrane, which was stained with Ponceau red. In each case, a major band migrated, as expected, from the structure of the plasmid carried by the bacteria (56, 66, 63, and 60 kDA for G9, G9-UCS, G9-5′UCS, and G9-3′UCS, respectively) (data not shown).
The α-UCS sera were tested for ability to recognize G9 and the three UCS-G9 fusion proteins produced in E. coli. Neither α-UCS serum reacted with G9 protein (Fig. 2, lane 3). By contrast, both α-UCS serum samples reacted with all three G9-UCS constructs. Lane 4 contained the G9 fused to the full-length, 123-residue UCS peptide. A strong band is present at 66 kDa, which is the expected mass for this fusion protein. Similarly, lane 5 contained a strongly reactive band at 63 kDa, which is the predicted position of a fusion protein carrying the 92 amino-terminal amino acids of UCS, and lane 6 contained a strongly reactive band at 60 kDa, which is the predicted position of a fusion protein carrying the 31 carboxy-terminal amino acids of UCS. These data showed that the sera from the two rabbits contained antibodies that recognized UCS epitopes and that these epitopes were located in both segments of the UCS peptide tested.
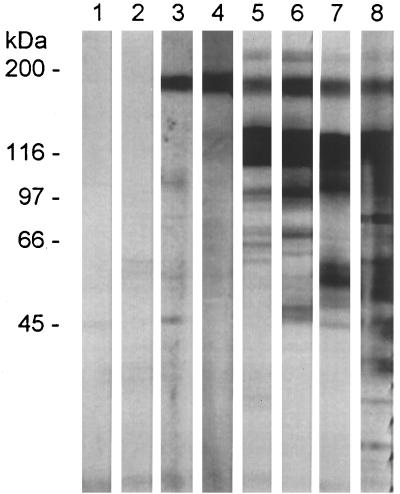
Next, the reactivity of the α-UCS sera with P. carinii f. sp. carinii proteins was assessed by immunoblotting. P. carinii f. sp. carinii organisms were solubilized, and proteins were separated by SDS-PAGE. The proteins were transferred electrophoretically to a nitrocellulose membrane, which was cut into strips. Strips were incubated with α-UCS sera, prebleed sera, and a variety of anti-MSG antibodies. The results are shown in Fig. 3. Lanes 1 and 2 show that the prebleed sera did not react. Lanes 3 and 4 demonstrate that the α-UCS2 and α-UCS3 sera reacted with a single band that migrated at approximately 170 kDa. The same band appeared on strips incubated with two MAbs against MSG (RB-E3 and RA-E7) (21, 22) (lanes 5 and 6), a polyclonal serum against MSG (lane 7), and a polyclonal serum against P. carinii f. sp. carinii (lane 8). As can be seen in lanes 5, 6, and 7, the anti-MSG antibodies recognized a prominent band at 116 kDa, where MSG is normally found. By contrast, the two α-UCS serum samples did not recognize MSG. Similar results were seen when the α-UCS2 serum and MAb RA-E7 were reacted to proteins from eight additional P. carinii f. sp. carinii populations (data not shown).
FIG. 3.
α-UCS sera recognize a 170-kDa protein in P. carinii f. sp. carinii. Proteins in a lysate from P. carinii f. sp. carinii organisms were separated by electrophoresis through an 8 to 16% Novex SDS-PAGE gel. Each lane contained lysate from approximately 107 organisms. After electrophoresis, proteins were transferred to a nitrocellulose sheet, strips of which were incubated with the following: lane 1, prebleed serum 2; lane 2, prebleed serum 3; lane 3, α-UCS2 serum; lane 4, α-UCS3 serum; lane 5, MAb RB-E3; lane 6, MAb RA-E7; lane 7, anti-MSG serum; lane 8, anti-P. carinii serum. Strips 1 to 4, 7, and 8 were reacted with goat anti-rabbit IgG phosphatase-labeled conjugate. Strips 5 and 6 were reacted with goat anti-mouse IgG phosphatase-labeled conjugate. All strips were developed in BCIP-NBT. The positions of the Bio-Rad high-molecular-mass markers are indicated to the left of lane 1.
The data described above identified a 170-kDa band that contained at least two MSG epitopes, as determined by the reactivity of this protein with MAbs RA-E7 and RB-E3. To determine if this band also contained additional epitopes found on MSG, eight P. carinii f. sp. carinii populations (each from a different rat) were assayed for reactivity to five different MAbs (21, 22), each of which recognizes distinct MSG epitopes (22, 50). The data obtained from the analysis of one P. carinii f. sp. carinii population are shown in Fig. 4. Each MAb reacted with a band at 170 and 116 kDa, suggesting that the 170-kDa band contains many MSG epitopes. Similar results were found in the other P. carinii f. sp. carinii populations examined for reactivity to the same MAbs (data not shown).
FIG. 4.
Reactivity of MSG-specific MAbs with P. carinii f. sp. carinii proteins. P. carinii f. sp. carinii proteins were separated by electrophoresis through an 8 to 16% Novex SDS-PAGE gel. Each lane contained lysate from approximately 107 organisms. Proteins were transferred to a nitrocellulose sheet, strips of which were incubated with the following mouse MAbs: lane 1, RA-E7; lane 2, RA-C1; lane 3, RA-C6; lane 4, RA-C7; lane 5, RB-C8; lane 6, RA-C11. Bound antibodies were detected as described in the legend to Fig. 3. Positions of molecular mass markers are indicated to the left of lane 1.
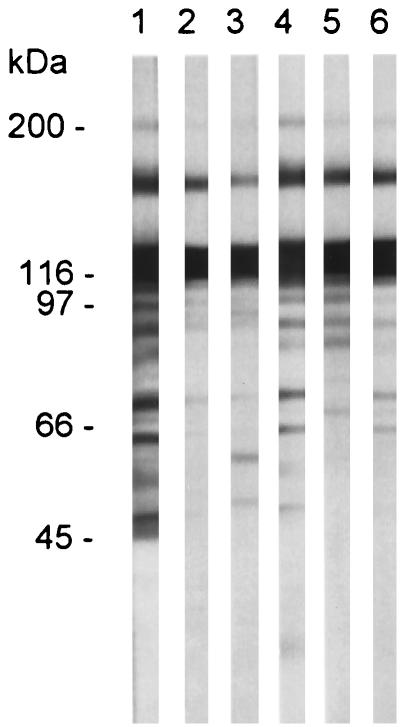
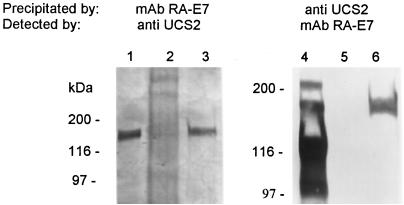
These immunoblot data suggested the presence of a protein at 170 kDa containing both MSG and UCS epitopes. To test this interpretation, P. carinii f. sp. carinii proteins were fractionated by immunoprecipitation with either the α-UCS2 serum or MAb RA-E7. Figure 5 shows that the α-UCS serum precipitated a 170-kDa protein that reacted with MAb RA-E7 (lane 6) and that MAb RA-E7 precipitated a 170-kDa protein that reacted with the α-UCS serum (lane 3). Immunoprecipitation with α-UCS2 serum eliminated the other bands that were reactive with MAb RA-E7, showing that this procedure was effective in purifying the 170-kDa material.
FIG. 5.
The 170-kDa protein immunoprecipitates with UCS and MSG antibodies. The UCS-MSG protein was immunoprecipitated with the α-UCS2 serum and MAb RA-E7. The resulting precipitate was subjected to electrophoresis in either a 6% Novex gel for the MAb RA-E7 precipitate (lanes 1 to 3) or an 8 to 16% Novex gel for the α-UCS2 precipitate (lanes 4 to 6). Each gel was blotted to nitrocellulose and reacted to either the α-UCS2 serum (lanes 1 to 3) or MAb RA-E7 (lanes 4 to 6). The blots were developed as described in the legend to Fig. 3. Lanes 1 and 4 contain an aliquot of the immunoprecipitation-starting material. Lanes 2 and 5 contain the precipitates produced in the preclearing step from the MAb RA-E7 and α-UCS2 immunoprecipitations, respectively. Lanes 3 and 6 contain the immunoprecipitates obtained with MAb RA-E7 and α-UCS2 serum, respectively. The positions of the molecular mass markers are indicated to the left of lanes 1 and 4.
Expression of a UCS-MSG protein in a heterologous system.
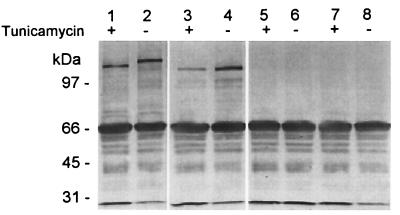
The previous results indicated that the UCS is translated. The predicted amino acid sequence of the UCS contains a putative signal sequence (6, 55). To determine if the UCS can provide a functional signal sequence and direct the UCS-MSG precursor protein to the endoplasmic reticulum, a UCS-MSG protein was expressed in insect cells with recombinant baculoviruses. The results are shown in Fig. 6. The UCS-MSG protein produced in Sf-9 cells migrated with a mass of approximately 160 kDa (lane 2). Incubation of the UCS-MSG virus-infected cells with tunicamycin resulted in more rapid migration of the protein, indicating that N-glycosylation of the protein was inhibited (compare lanes 1 and 2). By contrast, a baculovirus carrying an MSG gene lacking the UCS produced a 130-kDa protein species (lane 4). Incubation of the infected insect cells with tunicamycin had no effect on the migration of this protein (compare lanes 3 and 4). These immunoreactive bands were not present in the control lanes containing lysates of Sf-9 cells infected with an unrelated recombinant baculovirus carrying a rat cDNA encoding surfactant protein A (SP-A) (lanes 5 and 6) or uninfected cells (lanes 7 and 8). These data demonstrate that the UCS is required for N-glycosylation of MSG in insect cells and suggest that the UCS directs nascent MSG to the endoplasmic reticulum.
FIG. 6.
The UCS is required for N-glycosylation of MSG in insect cells. Insect cells were infected with recombinant baculoviruses containing genes encoding UCS-MSG (lanes 1 and 2), MSG (lanes 3 and 4), or SP-A, an unrelated protein (lanes 5 and 6). Lysates from uninfected cells are in lanes 7 and 8. The cells were incubated in the absence (−) or presence (+) of 5 μg of tunicamycin per ml for 24 h and harvested by scraping. Cell lysates were fractionated on a 6% Novex gel, transferred to nitrocellulose, and reacted with MAb RA-E7. The blot was developed by incubation with antimouse IgG horseradish peroxidase-labeled conjugate and horseradish peroxidase-dependent oxidation of o-phenylenediamine. The positions of Bio-Rad low-molecular-mass markers are indicated to the left of lane 1.
DISCUSSION
The α-UCS sera identified a P. carinii f. sp. carinii protein that has the properties expected of an MSG precursor. Such a precursor was anticipated based on the structure of mRNA encoding MSG isoforms (6, 54, 55). If the first AUG in the UCS was used to initiate translation, the primary translation product of an MSG mRNA would be a 134-kDa peptide beginning with a peptide encoded by the UCS and ending with an MSG isoform. Immunoblotting of proteins from P. carinii f. sp. carinii detected a band larger than MSG that reacted with anti-UCS antibodies as well as with several MAbs known to recognize different epitopes on MSG, suggesting that the band detected by immunoblotting was a protein that contained both UCS and MSG determinants. This protein was subsequently isolated by immunoprecipitation.
The putative MSG precursor migrated at an apparent molecular mass of 170 kDa, which is greater than the 134 kDa predicted from the sequence of MSG mRNA. It is not clear why the UCS-MSG protein did not migrate as far as expected on the basis of the predicted amino acid sequence. One factor to consider is that the 170-kDa value of the mass of the UCS-MSG protein is an approximation of an apparent mass. The actual mass could be closer to that predicted from the conceptual amino acid sequence. Aberrant migration during SDS-PAGE is a common phenomenon and could contribute to the high apparent mass. A second possibility is that the UCS-MSG peptide is actually longer than predicted. However, this possibility seems unlikely because two groups have reported that the 5′ end of mRNA encoding MSG extends no more than 56 nucleotides upstream of the translation initiation site that begins the ORF encoding a 134-kDa UCS-MSG peptide (6, 55). Therefore, the amino terminus of the UCS-MSG peptide can contain no more than 18 additional amino acids, even in the unlikely event that translation starts upstream of the first AUG codon. Similarly, the carboxyl terminus of MSG is clearly defined from numerous mRNA sequences (6, 16, 18, 20, 53). Another possible cause of the slow migration of the UCS-MSG band is posttranslational modification. Glycosylation of the UCS-MSG protein would be expected to occur because MSG on the surface of P. carinii is glycosylated (19, 25, 32, 37, 52). About 10 kDa of the apparent mass of MSG is removable by treatment with N-glycosidase (25, 37, 52). Thus, N-linked glycosylation would be expected to increase the mass of UCS-MSG protein to at least 144 kDa. Some or all of the additional apparent mass might be due to other sugars. In this regard, it is important to appreciate that the sugar content of MSG itself is not entirely clear. While N-linked sugars account for a modest fraction of this mass, chemical deglycosylation by treatment with trifluoromethanesulfonic acid reduced the apparent size of MSG to 68 kDa (37). Whether or not this is the mass of the core protein of MSG is not known, because the amino and carboxyl termini of MSG have not been sequenced and other data on the core protein have not been reported. In any event, the sugar content of the UCS-MSG precursor would not necessarily be the same as that of mature MSG, and only direct analysis of the UCS-MSG protein can determine its structure.
The α-UCS sera indicated that the UCS is not present on the 116-kDa MSG found on the surface of P. carinii. The 116-kDa MSG failed to react with anti-UCS antibodies, which were shown to recognize determinants residing within the last 25% of the UCS peptide. This suggests that more than 75% of the UCS is ultimately removed from the 170-kDa protein. An alternative explanation for the lack of reactivity between the α-UCS sera and the 116-kDa MSG is that the determinants recognized by the sera were masked. This possibility seems less likely than proteolytic processing for four reasons. First, multiple determinants were recognized by the α-UCS sera. Second, proteolytic processing would also explain the smaller apparent mass of the MSG found on the cell surface. Third, retention of the UCS on surface MSG would seem disadvantageous because it could increase vulnerability of the pathogen to attack by the host immune system. A final reason to propose that the UCS is removed from the putative MSG precursor is that a family of proteases that could serve to remove the UCS has been recently described (24). These proteases are highly related to subtilisin-like proteases, which are enzymes that cleave peptide chains after paired basic amino acid residues (39). Interestingly, the predicted amino acid sequences of all known MSG molecules contain a lysine-arginine pair at the junction between the UCS and the remainder of the MSG (Fig. 1) (6, 16, 18, 47, 54). Cleavage of the 170-kDa precursor after this lysine-arginine would remove all of the UCS peptide from the mature MSG and explain the lack of reactivity of these molecules with the α-UCS sera.
ACKNOWLEDGMENTS
We thank Protein Express for the generation of the UCS constructs and the UCS antiserum.
This work was supported by a Public Health Service grant (ROI A1 36701) from the National Institutes of Health (J.R.S.), a Career Investigator grant from the American Lung Association (F.X.M.), and the Medical Research Service Department of Veterans Affairs and grant AI 36701 from the National Institutes of Health (P.D.W.).
REFERENCES
- 1.Angus C W, Tu A, Vogel P, Qin M, Kovacs J A. Expression of variants of the major surface glycoprotein of Pneumocystis carinii. J Exp Med. 1996;183:1229–1234. doi: 10.1084/jem.183.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cushion M T, Kaselis M, Stringer S L, Stringer J R. Genetic stability and diversity of Pneumocystis carinii infecting rat colonies. Infect Immun. 1993;61:4801–4813. doi: 10.1128/iai.61.11.4801-4813.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dei-Cas E, Mazars E, Ferragut C O, Durand I, Aliouat E-M, Dridba M, Palluault F, Cailliez J-C, Seguy N, Tibayrenc M, Mullet C, Creusy C, Camus D. Ultrastructural, genomic, isoenzymatic and biological features make it possible to distinguish rabbit Pneumocystis from other mammal Pneumocystis strains. J Eukaryot Microbiol. 1994;41:84S. [PubMed] [Google Scholar]
- 4.De Luca A, Ortona E, Margutti P, Visconti E, Tamburrini E, Siracusano A. Different amplification efficiency and nucleotide sequence variation in various Pneumocystis isolates from humans and rats. J Eukaryot Microbiol. 1994;41:85S. [PubMed] [Google Scholar]
- 5.Edlind T D, Bartlett M S, Weinberg G A, Prah G N, Smith J W. The beta-tubulin gene from rat and human isolates of Pneumocystis carinii. Mol Microbiol. 1992;6:3365–3373. doi: 10.1111/j.1365-2958.1992.tb02204.x. [DOI] [PubMed] [Google Scholar]
- 6.Edman J C, Hatton T W, Nam M, Turner R, Mei Q, Angus C W, Kovacs J A. A single expression site with a conserved leader sequence regulates variation of expression of the Pneumocystis carinii family of major surface glycoprotein genes. DNA Cell Biol. 1996;15:989–999. doi: 10.1089/dna.1996.15.989. [DOI] [PubMed] [Google Scholar]
- 7.Ezekowitz R A, Williams D J, Koziel H, Armstrong M Y, Warner A, Richards F F, Rose R M. Uptake of Pneumocystis carinii mediated by the macrophage mannose receptor. Nature. 1991;351:155–158. doi: 10.1038/351155a0. [DOI] [PubMed] [Google Scholar]
- 8.Fisher D J, Gigliotti F, Zauderer M, Harmsen A G. Specific T-cell response to a Pneumocystis carinii surface glycoprotein (gp120) after immunization and natural infection. Infect Immun. 1991;59:3372–3376. doi: 10.1128/iai.59.10.3372-3376.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garbe T R, Stringer J R. Molecular characterization of clustered variants of genes encoding major surface antigens of human Pneumocystis carinii. Infect Immun. 1994;62:3092–3101. doi: 10.1128/iai.62.8.3092-3101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gigliotti F. Host species-specific antigenic variation of a mannosylated surface glycoprotein of Pneumocystis carinii. J Infect Dis. 1992;165:329–336. doi: 10.1093/infdis/165.2.329. [DOI] [PubMed] [Google Scholar]
- 11.Gigliotti F, Ballou L R, Hughes W T, Mosley B D. Purification and initial characterization of a ferret Pneumocystis carinii surface antigen. J Infect Dis. 1988;158:848–854. doi: 10.1093/infdis/158.4.848. [DOI] [PubMed] [Google Scholar]
- 12.Graves D C, McNabb S J N, Worley M A, Downs T D, Ivey M H. Analyses of rat Pneumocystis carinii antigens recognized by human and rat antibodies by using Western immunoblotting. Infect Immun. 1986;54:96–103. doi: 10.1128/iai.54.1.96-103.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haidaris P J, Wright T W, Gigliotti F, Haidaris C G. Expression and characterization of a cDNA clone encoding an immunodominant surface glycoprotein of Pneumocystis carinii. J Infect Dis. 1992;166:1113–1123. doi: 10.1093/infdis/166.5.1113. [DOI] [PubMed] [Google Scholar]
- 14.Howell M L, Blumenthal K M. Cloning and expression of a synthetic gene for Cerebratulus lacteus neurotoxin B-IV. J Biol Chem. 1989;264:15268–15273. [PubMed] [Google Scholar]
- 15.Keely S, Pai H J, Baughman R, Sidman C, Sunkin S M, Stringer J R, Stringer S L. Pneumocystis species inferred from analysis of multiple genes. J Eukaryot Microbiol. 1994;41:94S. [PubMed] [Google Scholar]
- 16.Kitada K, Wada M, Nakamura Y. Multi-gene family of major surface glycoproteins of Pneumocystis carinii: full-size cDNA cloning and expression. DNA Res. 1994;1:57–66. doi: 10.1093/dnares/1.2.57. [DOI] [PubMed] [Google Scholar]
- 17.Kovacs J A, Halpern J L, Swan J C, Moss J, Parrillo J E, Masur H. Identification of antigens and antibodies specific for Pneumocystis carinii. J Immunol. 1988;140:2023–2031. [PubMed] [Google Scholar]
- 18.Kovacs J A, Powell F, Edman J C, Lundgren B, Martinez A, Drew B, Angus C W. Multiple genes encode the major surface glycoprotein of Pneumocystis carinii. J Biol Chem. 1993;268:6034–6040. [PubMed] [Google Scholar]
- 19.Linke M J, Cushion M T, Walzer P D. Properties of the major antigens of rat and human Pneumocystis carinii. Infect Immun. 1989;57:1547–1555. doi: 10.1128/iai.57.5.1547-1555.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linke M J, Smulian A G, Stringer J R, Walzer P D. Characterization of multiple unique cDNAs encoding the major surface glycoprotein of rat-derived Pneumocystis carinii. Parasitol Res. 1994;80:478–486. doi: 10.1007/BF00932694. [DOI] [PubMed] [Google Scholar]
- 21.Linke M J, Smulian A G, Yoshihara P, Walzer P D. Production and characterization of monoclonal antibodies specific for the major surface glycoprotein of Pneumocystis carinii. J Eukaryot Microbiol. 1994;41:99S–100S. [PubMed] [Google Scholar]
- 22.Linke, M. J., S. M. Sunkin, R. P. Andrews, J. R. Stringer, and P. D. Walzer. Expression, structure, and location of epitopes of the major surface glycoprotein of Pneumocystis carinii f. sp. carinii. Clin. Diagn. Lab. Immunol., in press. [DOI] [PMC free article] [PubMed]
- 23.Linke M J, Walzer P D. Analysis of a surface antigen of Pneumocystis carinii. J Protozool. 1989;36:60S–61S. doi: 10.1111/j.1550-7408.1989.tb02701.x. [DOI] [PubMed] [Google Scholar]
- 24.Lugli E B, Allen A G, Wakefield A E. A Pneumocystis carinii multi-gene family with homology to subtilisin-like serine proteases. Microbiology. 1997;143:2223–2236. doi: 10.1099/00221287-143-7-2223. [DOI] [PubMed] [Google Scholar]
- 25.Lundgren B, Lipschik G Y, Kovacs J A. Purification and characterization of a major human Pneumocystis carinii surface antigen. J Clin Invest. 1991;87:163–170. doi: 10.1172/JCI114966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundgren B, Lundgren J D, Nielsen T, Mathiesen L, Nielsen J O, Kovacs J A. Antibody responses to a major Pneumocystis carinii antigen in human immunodeficiency virus-infected patients with and without P. carinii pneumonia. J Infect Dis. 1992;165:1151–1155. doi: 10.1093/infdis/165.6.1151. [DOI] [PubMed] [Google Scholar]
- 27.McCormack F X, Calvert H M, Watson P W, Smith D L, Mason R J, Voelker D R. The structure and function of surfactant protein A, hydroxyproline and carbohydrate deficient proteins. J Biol Chem. 1994;269:5833–5841. [PubMed] [Google Scholar]
- 28.McCormack F X, Festa A L, Andrews R P, Linke M J, Walzer P D. The carbohydrate recognition domain of surfactant protein A mediates binding to the major surface glycoprotein of Pneumocystis carinii. Biochemistry. 1997;36:8092–8099. doi: 10.1021/bi970313f. [DOI] [PubMed] [Google Scholar]
- 29.Mills J. Pneumocystis carinii and Toxoplasma gondii infections in patients with AIDS. Rev Infect Dis. 1986;8:1001–1011. doi: 10.1093/clinids/8.6.1001. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura Y, Tanabe K, Egawa K. Structure of major surface determinants and DNA diagnosis of Pneumocystis carinii. J Protozool. 1989;36:58S–60S. doi: 10.1111/j.1550-7408.1989.tb02699.x. [DOI] [PubMed] [Google Scholar]
- 31.O’Riordan D M, Standing J E, Kwon K-Y, Chang D, Crouch E C, Limper A H. Surfactant protein D interacts with Pneumocystis carinii and mediates organism adherence to alveolar macrophages. J Clin Invest. 1995;95:2699–2710. doi: 10.1172/JCI117972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pesanti E L, Shanley J D. Glycoproteins of Pneumocystis carinii: characterization by electrophoresis and microscopy. J Infect Dis. 1988;158:1353–1359. doi: 10.1093/infdis/158.6.1353. [DOI] [PubMed] [Google Scholar]
- 33.Peters S E, English K, Laakkonen J, Gurnell J. DNA analysis of Pneumocystis carinii infecting Finnish and English shrews. J Eukaryot Microbiol. 1994;41:108S. [PubMed] [Google Scholar]
- 34.Peters S E, Wakefield A E, Whitwell K E, Hopkin J M. Pneumocystis carinii pneumonia in thoroughbred foals: identification of a genetically distinct organism by DNA amplification. J Clin Microbiol. 1994;32:213–216. doi: 10.1128/jcm.32.1.213-216.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pneumocystis Workshop. Revised nomenclature for Pneumocystis carinii. J Eukaryot Microbiol. 1994;41:121S–122S. [PubMed] [Google Scholar]
- 36.Pottratz S T, Martin W J. Role of fibronectin in Pneumocystis carinii attachment to cultured lung cells. J Clin Invest. 1990;85:351–356. doi: 10.1172/JCI114445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radding J A, Armstrong M Y, Ullu E, Richards F F. Identification and isolation of a major cell surface glycoprotein of Pneumocystis carinii. Infect Immun. 1989;57:2149–2157. doi: 10.1128/iai.57.7.2149-2157.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Siezen R J, de Vos W M, Leunissen J A M, Dijkstra B W. Homology modelling and protein engineering strategy of subtilases, the family of subtilisin-like serine proteinases. Protein Eng. 1991;4:719–737. doi: 10.1093/protein/4.7.719. [DOI] [PubMed] [Google Scholar]
- 40.Sinclair K, Wakefield A E, Banerji S, Hopkin J M. Pneumocystis carinii organisms derived from rat and human hosts are genetically distinct. Mol Biochem Parasitol. 1991;45:183–184. doi: 10.1016/0166-6851(91)90042-5. [DOI] [PubMed] [Google Scholar]
- 41.Smulian A G, Sullivan D W, Linke M J, Halsey N A, Quinn T C, MacPhail A P, Hernandez Avila M A, Hong S T, Walzer P D. Geographic variation in the humoral response to Pneumocystis carinii. J Infect Dis. 1993;167:1243–1247. doi: 10.1093/infdis/167.5.1243. [DOI] [PubMed] [Google Scholar]
- 42.Stringer J R. The identity of Pneumocystis carinii: not a single protozoan, but a diverse group of exotic fungi. Infect Agents Dis. 1993;2:109–117. [PubMed] [Google Scholar]
- 43.Stringer J R, Stringer S L, Zhang J, Baughman R, Smulian A G, Cushion M T. Molecular genetic distinction of Pneumocystis carinii from rats and humans. J Eukaryot Microbiol. 1993;40:733–741. doi: 10.1111/j.1550-7408.1993.tb04468.x. [DOI] [PubMed] [Google Scholar]
- 44.Stringer S L, Garbe T, Sunkin S M, Stringer J R. Genes encoding antigenic surface glycoproteins in Pneumocystis from humans. J Eukaryot Microbiol. 1993;40:821–826. doi: 10.1111/j.1550-7408.1993.tb04481.x. [DOI] [PubMed] [Google Scholar]
- 45.Stringer S L, Hong S T, Giuntoli D, Stringer J R. Repeated DNA in Pneumocystis carinii. J Clin Microbiol. 1991;29:1194–1201. doi: 10.1128/jcm.29.6.1194-1201.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Summers M D, Smith G E. A manual for baculovirus vectors and insect cell culture procedures. College Station: Texas Agricultural Experiment Station; 1988. [Google Scholar]
- 47.Sunkin S M, Stringer J R. Translocation of surface antigen genes to a unique telomeric expression site in Pneumocystis carinii. Mol Microbiol. 1996;19:283–295. doi: 10.1046/j.1365-2958.1996.375905.x. [DOI] [PubMed] [Google Scholar]
- 48.Sunkin S M, Stringer J R. Residence at the expression site is necessary and sufficient for the transcription of surface antigen genes of Pneumocystis carinii. Mol Microbiol. 1997;25:147–160. doi: 10.1046/j.1365-2958.1997.4461806.x. [DOI] [PubMed] [Google Scholar]
- 49.Sunkin S M, Stringer S L, Stringer J R. A tandem repeat of rat-derived Pneumocystis carinii genes encoding the major surface glycoprotein. J Eukaryot Microbiol. 1994;41:292–300. doi: 10.1111/j.1550-7408.1994.tb01509.x. [DOI] [PubMed] [Google Scholar]
- 50.Sunkin S M. Ph.D. thesis. Cincinnati, Ohio: University of Cincinnati; 1996. [Google Scholar]
- 51.Tanabe K, Takasaki S, Watanabe J, Kobata A, Egawa K, Nakamura Y. Glycoproteins composed of major surface immunodeterminants of Pneumocystis carinii. Infect Immun. 1989;57:1363–1368. doi: 10.1128/iai.57.5.1363-1368.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Underwood A P, Louis E J, Borts R H, Stringer J R, Wakefield A E. Pneumocystis carinii telomere repeats are composed of TTAGGG and the subtelomeric sequence contains a gene encoding the major surface glycoprotein. Mol Microbiol. 1996;19:273–281. doi: 10.1046/j.1365-2958.1996.374904.x. [DOI] [PubMed] [Google Scholar]
- 53.Wada M, Kitada K, Saito M, Egawa K, Nakamura Y. cDNA sequence diversity and genomic clusters of major surface glycoprotein genes of Pneumocystis carinii. J Infect Dis. 1993;168:979–985. doi: 10.1093/infdis/168.4.979. [DOI] [PubMed] [Google Scholar]
- 54.Wada M, Nakamura Y. Unique telomeric expression site of major surface glycoprotein genes of Pneumocystis carinii. DNA Res. 1996;3:55–64. doi: 10.1093/dnares/3.2.55. [DOI] [PubMed] [Google Scholar]
- 55.Wada M, Sunkin S M, Stringer J R, Nakamura Y. Antigenic variation by positional control of major surface glycoprotein gene expression in Pneumocystis carinii. J Infect Dis. 1995;171:1563–1568. doi: 10.1093/infdis/171.6.1563. [DOI] [PubMed] [Google Scholar]
- 56.Walzer P D, Linke M J. A comparison of the antigenic characteristics of rat and human Pneumocystis carinii by immunoblotting. J Immunol. 1987;138:2257–2265. [PubMed] [Google Scholar]
- 57.Walzer P D, Pearl D P, Krogstad D J, Rawson P G, Schultz M G. Pneumocystis carinii pneumonia in the United States: epidemiologic, clinical and diagnostic features. Ann Intern Med. 1974;80:83–93. doi: 10.7326/0003-4819-80-1-83. [DOI] [PubMed] [Google Scholar]
- 58.Weinberg G, Edlind T, Lu J, Lee C, Bauer N, Durant P. Genetic diversity of Pneumocystis carinii from different host species at the beta-tubulin gene locus and at the internal transcribed spacer regions of the rRNA gene cluster. J Eukaryot Microbiol. 1994;41:118S. [PubMed] [Google Scholar]
- 59.Wright T W, Simpson Haidaris P J, Gigliotti F, Harmsen A G, Haidaris C G. Conserved sequence homology of cysteine-rich regions in genes encoding glycoprotein A in Pneumocystis carinii derived from different host species. Infect Immun. 1994;62:1513–1519. doi: 10.1128/iai.62.5.1513-1519.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zimmerman P E, Voelker D R, McCormack F X, Paulsrud J R, Martin W J. 120-kD surface glycoprotein of Pneumocystis carinii is a ligand for surfactant protein A. J Clin Invest. 1992;89:143–149. doi: 10.1172/JCI115554. [DOI] [PMC free article] [PubMed] [Google Scholar]