Abstract
Epsilon class GSTs (glutathione transferases) are expressed at higher levels in Anopheles gambiae mosquitoes that are resistant to DDT [1,1,1-trichloro-2,2-bis-(p-chlorophenyl)ethane] than in insecticide-susceptible individuals. At least one of the eight Epsilon GSTs in this species, GSTe2, efficiently metabolizes DDT to DDE [1,1-dichloro-2,2-bis-(p-chlorophenyl)ethane]. In the present study, we investigated the factors regulating expression of this class of GSTs. The activity of the promoter regions of GSTe2 and GSTe3 were compared between resistant and susceptible strains by transfecting recombinant reporter constructs into an A. gambiae cell line. The GSTe2 promoter from the resistant strain exhibited 2.8-fold higher activity than that of the susceptible strain. Six polymorphic sites were identified in the 352 bp sequence immediately upstream of GSTe2. Among these, a 2 bp adenosine indel (insertion/deletion) was found to have the greatest effect on determining promoter activity. The activity of the GSTe3 promoter was elevated to a lesser degree in the DDT-resistant strain (1.3-fold). The role of putative transcription-factor-binding sites in controlling promoter activity was investigated by sequentially deleting the promoter constructs. Several putative transcription-factor-binding sites that are responsive to oxidative stress were present within the core promoters of these GSTs, hence the effect of H2O2 exposure on the transcription of the Epsilon GSTs was investigated. In the DDT-resistant strain, expression of GSTe1, GSTe2 and GSTe3 was significantly increased by a 1-h exposure to H2O2, whereas, in the susceptible strain, only GSTe3 expression responded to this treatment.
Keywords: Anopheles gambiae; glutathione transferase (GST); insecticide resistance; promoter; 1,1,1-trichloro-2,2-bis-(p-chlorophenyl)ethane (DDT)
Abbreviations: AhR, aryl hydrocarbon receptor; AP-1, activator protein 1; CREB, cAMP-response-element-binding protein; DDT, 1,1,1-trichloro-2,2-bis-(p-chlorophenyl)ethane; δEF1, δ elongation factor 1; EST, expressed sequence tag; FOXL1, forkhead box L1; GST, glutathione transferase; indel, insertion/deletion; Inr, initiator; MEF-2, myocyte-specific enhancer-binding factor 2; NFAT, nuclear factor of activated T-cells; NF-κB, nuclear factor κB; ROS, reactive oxygen species; TSS, transcriptional start site; UTR, untranslated region
INTRODUCTION
The GSTs (glutathione transferases) are a superfamily of detoxification proteins that are found in most organisms. They are primarily associated with the detoxification of endogenous and xenobiotic lipophilic compounds by the conjugation of reduced glutathione to their electrophilic centres. However, GSTs are also involved in many other intracellular processes, including protecting against oxidative stress [1,2], transporting intracellular compounds [3], catalysing essential steps in biosynthetic pathways [4] and acting as signalling molecules [5]. Insect GSTs have been classified into at least six classes: Delta, Epsilon, Omega, Sigma, Theta and Zeta [6]. The largest of these classes, the Delta and Epsilon classes, are unique to insects and are probably the primary classes involved in the detoxification of xenobiotics such as insecticides.
Resistance to insecticides is frequently attributed to increased levels of GST activity. GSTs can protect the insect either by increasing the rate of detoxification of insecticide into non-toxic product e.g. the dehydrochlorination of the organochlorine, DDT [1,1,1-trichloro-2,2-bis-(p-chlorophenyl)ethane] [7], and the O-dealkylation or O-dearylation of organophosphate insecticides [8], or by relieving the damage from oxidative stress induced by insecticide exposure [9]. Elevated GST activity is the primary cause of metabolic resistance to DDT in Anopheles mosquitoes. Resistance to this insecticide class contributed to the failure of the malaria-eradication campaign in the 1960s, and is of major concern for sustainable malaria-control programmes today [10].
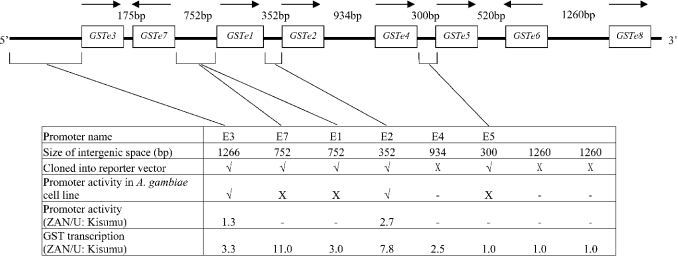
Our previous research on the African malaria vector Anopheles gambiae, identified an Epsilon class GST, GSTe2, with extremely high activity against DDT [11]. The gene encoding this enzyme is one of a cluster of eight Epsilon GST genes, arranged sequentially within 10.5 kb of DNA on division 33B of chromosome 3R (Figure 1). The transcription of GSTe2 and four additional Epsilon GSTs is elevated between 2- and 11-fold in DDT-resistant adult A. gambiae compared with susceptible individuals [12]. Genetic mapping studies showed that the boundaries of a major locus conferring resistance to DDT flanked the Epsilon GST gene cluster [13], and we therefore hypothesized that resistance to DDT in A. gambiae is due to a mutation in a cis-acting factor that controls the expression of one or more Epsilon class GSTs.
Figure 1. Organization of the Epsilon GST gene cluster in A. gambiae and details of the promoter constructs used in the present study.
The direction of transcription is indicated by an arrow. The size of the intergenic space is from the stop codon of the preceding gene to the AUG initiation codon of the following gene (or between AUG initiation codons for genes arranged in head to head orientation) and thus includes UTRs of flanking genes. Experiments not attempted are indicated by a dash, those which gave no activity are indicated by X. The ratio of transcript levels between the ZAN/U and Kisumu strains are the results of previous experiments carried out on adult mosquitoes [12].
Distances between the coding regions of the A. gambiae Epsilon GST genes [referred to as the intergenic space, but in this instance including the UTR (untranslated region) of neighbouring GST genes] range from 175 bp to 1260 bp. The tight clustering of these genes may facilitate the control of expression of multiple members of this class by a common regulatory unit upstream of the Epsilon GST cluster. This phenomenon is very common in prokaryotes. Polycistronic transcription has also been reported in several eukaryotic species, including insects (reviewed in [14]); however, in A. gambiae, the divergent orientation of two of the internal Epsilon GST genes within this cluster (Figure 1) and the variable basal transcription levels of this gene family [12] suggest that these genes are regulated independently. The first aim of the present study was to distinguish between these two alternative hypotheses by determining whether the intergenic DNA within the Epsilon gene cluster could drive gene expression. Having verified that the 352 bp upstream region of GSTe2 is sufficient to drive expression of a reporter gene in vitro, the activity of the GSTe2 and GSTe3 promoters was studied in detail. We identified several variations in these promoters from DDT-resistant and -susceptible strains that account, in part, for the increased transcription of these genes observed in insecticide-resistant strains. In addition, we report that expression of Epsilon GSTs increases in response to oxidative stress.
EXPERIMENTAL
Mosquito strains
The ZAN/U strain of A. gambiae was colonized from a DDT-resistant field population from Zanzibar, Tanzania in 1982. Adults of this strain have been maintained under regular DDT selection pressure [15]. The Kisumu strain is a laboratory insecticide-susceptible strain originally colonized from Kisumu, Western Kenya.
Extraction of nucleic acids
Genomic DNA was extracted from approx. 1 g of 1-day-old adult mosquitoes from the ZAN/U or Kisumu strain as described previously [16]. RNA was extracted using the TRI Reagent (Sigma) according to the manufacturer's instructions. Residual genomic DNA was digested with DNase I (Promega), and the RNA was reverse-transcribed into cDNA using Superscript III (Invitrogen) primed with an oligo(dT)18 primer.
Computer-based sequence analysis
In order to determine the maximum extent of the upstream regulatory region of the first gene in the Epsilon gene cluster (GSTe3), the 5.2 kb of DNA preceding the start codon of this gene was retrieved from the A. gambiae genome sequence database (http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?taxid=7165) and searched, using BLASTX (http://www.ncbi.nlm.nih.gov/BLAST), for putative coding regions. A putative transcript (ENSANGG00000014372) was located 5127 bp upstream of the translational start site of GSTe3, and this set the boundary for the analysis of the Epsilon regulatory regions.
The intergenic spaces of the Epsilon GST cluster and the 5.2 kb region upstream of GSTe3 were analysed with the MatInspector program (http://www.genomatrix.de) to identify putative promoter elements and potential transcription-factor-binding sites. The following settings were used: the matrix library was set as vertebrate library, the core similarity at 1.0, and the matrix similarity at 0.95 and 0.9 for the GSTe3 and GSTe2 promoter respectively.
Identification of TSSs (transcriptional start sites)
Putative TSSs for each of the genes were identified by analysis of the Tentative Consensus sequences for A. gambiae ESTs (expressed sequence tags), retrieved from The Institute of Genomic Research (TIGR) (http://www.tigr.org/tdb/tgi/). Primers were designed that either preceded or encompassed the putative TSS of GSTe2 and GSTe3 (Table 1). Reverse primers within the second or third exon of each of these genes were designed and used in PCRs in combination with each of the forward primers. Genomic DNA and cDNA were used as templates, and the PCR conditions were as follows: 0.4 μM each primer, 2 mM MgCl2, 0.2 mM each dNTP, 0.25 units of Taq DNA polymerase (Promega), 40 cycles of 95 °C for 30 s, 55 °C for 40 s and 72 °C for 60 s. The PCR products were resolved on 2% agarose gels and were visualized by ethidium bromide staining.
Table 1. Primers used to map the TSSs.
| Primer name | Sequence |
|---|---|
| 2TSSF1 | 5′-GGCGTTTGCTATGCTTGGGGTCGATGC-3′ |
| 2TSSF2 | 5′-GAACGTGTACGGTGTGTGTC-3′ |
| 2TSSR | 5′-CAACGCGATCCTCGGGGATGTC-3′ |
| 3TSSF1 | 5′-CGAATGGGGATAGCTTCAAATAC-3′ |
| 3TSSF2 | 5′-CGAAAGGGAGAAACGCGCCGTTG-3′ |
| 3TSSR | 5′-CCTGCTTCACTAGATCCTTCGC-3′ |
Preparation of promoter constructs and determination of promoter activity
Primers were designed (Table 2) to amplify the intergenic spaces [plus the 3′ and/or 5′ putative UTRs of the flanking genes] between GSTe7 and GSTe1, GSTe1 and GSTe2, GSTe4 and GSTe5, and 1.3 kb upstream of GSTe3, from genomic DNA extracted from the ZAN/U or Kisumu strains of A. gambiae. Either KpnI or NheI sites were incorporated into the 5′ sequence of the primer to facilitate subsequent cloning into the pGL3-Basic reporter vector (Promega). PCR conditions were as above, except that the DNA polymerase used was Pfu, a proofreading enzyme from Stratagene. The resultant PCR products were incubated with Taq DNA polymerase and dATP, and were cloned into pGEM T-easy vector (Promega). The plasmids were transformed into Escherichia coli XL1-Blue (Stratagene), and positive colonies were identified by PCR using vector-specific primers. For each primer pair, both strands of at least three independent colonies from each mosquito strain were sequenced using a Beckman CEQ 8000 sequencer. Contigs were constructed using Lasergene (DNASTAR), and the consensus sequences for the two strains were aligned using EMBOSS (http://www.ebi.ac.uk/emboss/align/).
Table 2. Primers used to clone promoter constructs.
The KpnI sites in the forward primers and the NheI sites in the reverse primers are underlined. The position of the primers is in relation to the AUG initiation codon in the ZAN/U strain.
| Primer name | Sequence (5′→3′) | Position | Construct name |
|---|---|---|---|
| PG3F1 | GGACAGGTACCGGTTGGATGGGATTTTTCTTCGCG | −1303/−1269 | PGZa3-1; PGKi3-1 |
| PG3F2 | CAAGGGTACCATTTCACAGCGATCGTG | −1215/−1189 | PGZa3-2; PGKi3-2 |
| PG3F3 | CGGACGGTACCCATATTTACCTCAATTCACC | −1050/−1021 | PGZa3-3; PGKi3-3 |
| PG3F4 | GCTCAGGTACCTAAACAATCGATAATTAAACTGC | −1007/−974 | PGZa3-4; PGKi3-4 |
| PG3F5 | GCTCGGTACCTGCGTAAGACTTTGTAGC | −671/−644 | PGZa3-5; PGKi3-5 |
| PG3F6 | CAATGGGTACCGAGCGATCGACTCTGAACAC | −339/−309 | PGZa3-6; PGKi3-6 |
| PG3R | GTGCCATGCTAGCTCGTTGTTGTTGTTGTTGATG | −27/+7 | |
| PG12F1 | GTAACAGGTACCTGCTATTCGCGGTTGTTAAC | −356/−325 | PGZa12-1; PGKi12-1 |
| PG12F2 | GTGTAGGTACCAAAGCACCTAGTAACG | −312/−286 | PGZa12-2; PGKi12-2 |
| PGZa12F6 | GTGTAGGTACCAAAGCACCTAGTAACGTTTTTCTTGTGCATAAAAAACAGGAATTCG | −312/−256 | PGKiMu |
| PGki12F6 | GTGTAGGTACCAAAGCACCTAGTAACGTTTTTCTTGTGCATAAAAAAAACAGGAATTCG | −312/−256 | PGZaMu |
| PG12F3 | CAGGGGTACCCTTCTGCTTTTATGTTGCAGTACG | −265/−232 | PGZa12-3; PGKi12-3 |
| PG12F4 | GAACCGGTACCAGTATGAAATAAATTCGCCG | −229/−199 | PGZa12-4; PGZa12-4 |
| PG12R2 | CAAGAGCTAGCCCCCGAACTGATGATTGGCTGTG | −79/−46 | PGZa12-5,12-6; PGKi12-5,12-6 |
| PG12R | GGACATGCTAGCAGCGAACTAAAAACTGGATGG | −27/+6 | |
| PG71F | CTGCTGGGTACCATCGCTCAGCCTGAAACGCG | −766/−735 | |
| PG71R | GGCATGCTAGCGACTGGGGGTTTCGTCTACG | −26/+5 | |
| PG17F | CTGCTGGCTAGCATCGCTCAGCCTGAAACGCG | −766/−735 | |
| PG17R | GGCATGGTACCGACTGGGGGTTTCGTCTACG | −26/+5 | |
| PG45F | GTGAACGGTACCTGGTTGAGTGAACGCTTCAAGC | −304/−271 | |
| PG45R | CGTTGCGCTAGCTCCGTCGTGATGAGCGGTTCAAG | −26/+9 |
The inserts were cut from the pGEM T-easy vector by digestion with the restriction enzymes KpnI and NheI, gel-purified using Qiagen Qiaquick and ligated into pGL3-Basic luciferase reporter vector pre-digested with KpnI and NheI. After transformation into XL1-Blue, positive colonies were identified by restriction digests, and plasmids were prepared using the Miniprep plasmid-purification kit (Qiagen). The concentration of each plasmid was adjusted to 200 ng/μl using a NanoDrop spectrophotometer (NanoDrop Technologies). The Renilla luciferase reporter vector was used as a control plasmid for the promoter activity assays at a concentration of 1 ng/μl.
The A. gambiae cell line, Sua 4.0, derived from neonatal larvae of the Suakoko mosquito strain [17], was kindly provided by Dr Hans Michael Müller (Heidelberg, Germany). The cells were maintained in Schneider's insect medium (Sigma) supplemented with 10% (v/v) foetal bovine serum (Sigma), 0.5% (w/v) amphotericin B (Sigma) and 1% (w/v) penicillin/streptomycin mixture (Sigma) at 27 °C.
The Dual-Luciferase™ Reporter Assay (Promega) was used to measure promoter activity. The reporter constructs, containing the A. gambiae promoter sequences inserted upstream of the luciferase gene from the firefly Photinus pyralis, were co-transfected with the internal control, containing the sea pansy Renilla reniformis luciferase under the control of the actin 5C promoter from Drosophila melanogaster, into Sua 4.0 cells using FuGENE 6.0 (Roche), according to a modification of the method of Zhao and Eggleston [18]. Briefly, approx. 5×105 cells per well were plated in 24-well plates 1 day before transfection. Confluence of 50–80% was achieved after 1 day's incubation, which is suitable to obtain high transfection efficiency. For each well of cells to be transfected, 200 ng of the GST promoter construct and 1 ng of Renilla reporter vector were mixed with 0.6 μl of FuGENE 6 in unsupplemented Schneider's insect medium (Sigma) for 20 min at room temperature. The mixture of DNA plasmids and FuGENE reagent was then applied to the appropriate well in triplicate. The transfected cells were incubated at 27 °C for 48 h, and harvested in 100 μl of 1×passive lysis buffer (Promega) per well. The activities of firefly and Renilla luciferase were determined on a luminometer (EG&G Berthold) using the Dual-Luciferase™ Reporter Assay kit. The luciferase activity of each construct was normalized to Renilla activity. For each experiment (repeated twice), triplicate transfections were performed for each construct.
In subsequent experiments, regions of the promoters under study were deleted by re-designing the forward and/or reverse primers (Table 2) and using these to amplify partial fragments of the promoters from the original pGL3-Basic GST promoter plasmids. These fragments were then subcloned into the promoterless pGL3-Basic plasmids, and the inserts were sequenced to verify their integrity. Transfection experiments with these deletion constructs were performed as described above.
Site-directed mutagenesis
Site-directed mutagenesis was used to alter the number of adenosine residues found between bp −266 and −271 in the GSTe2 promoter. Two additional adenosine residues were introduced into the GSTe2 promoter from the ZAN/U strain by PCR using the primer pair pGKi12F6/pG12R (Table 2) and ZAN/U genomic DNA as template. In addition, two adenosine residues were deleted from the Kisumu GSTe2 promoter by PCR using the primer pair pGZa12F6/pG12R (Table 2) and Kisumu genomic DNA as a template. PCR products were subjected to sequencing to confirm the site-directed mutagenesis, and were subcloned into the promoterless reporter vector pGL-3 basic. The constructs containing the mutated fragments of ZAN/U and Kisumu strains were named pGZa-Mu and pGKi-Mu respectively. The mutant constructs, together with pGZa12-2, pGKi12-2 and empty pGL-3 basic vector, were used to transfect Sua 4.0 cells as described above. Each plasmid was transfected in triplicate in two independent experiments.
Induction of Epsilon GST gene expression by treatment with H2O2
To determine the effect of oxidative stress on the expression of Epsilon GST genes, pools of 15 fourth instar larvae from Kisumu and ZAN/U were immersed in 3 mM H2O2 for 1 h, rinsed in distilled water, then left for a further 1 h after treatment and snap-frozen. Three independent RNA extractions were performed on samples of 15 larvae from each larval pool as described above. After DNase digestion and cDNA synthesis, the copy number of three of the Epsilon GST genes (GSTe1, GSTe2 and GSTe3) was determined by real-time PCR as described previously [12]. Expression of the S7 ribosomal protein gene [19] was used to normalize the samples. The copy number of the genes in each of the three cDNA samples was quantified in two independent experiments. The copy number of the Epsilon GSTs was normalized with the copy number of S7 in the same cDNA sample as described previously [12].
Statistical methods
Luciferase activities of constructs and normalized copy numbers of Epsilon GST transcripts were calculated as means±S.D. Each promoter construct was assayed in triplicate in each of two independent experiments. The statistical significance of promoter activities and induction of Epsilon GST gene expression were analysed using Student's t test.
RESULTS
Prediction of potential core promoter regions
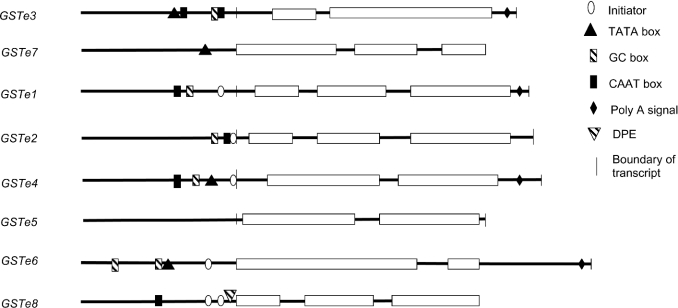
The upstream regions of the Epsilon GST genes were searched for consensus sequences matching core promoter elements using the computer program MatInspector [20]. To identify UTRs of the genes the consensus ESTs, retrieved from the TIGR website (http://www.tigr.org/tdb/tgi/aggi/), were aligned with the genomic DNA sequence of the Epsilon GST gene cluster. A summary of the findings is shown in Figure 2. Four of the eight Epsilon GST genes contain a putative TATA box and one, GSTe8, contains a downstream promoter element [21]. Putative arthropod initiators (Inr) [22] were identified for five of the genes, and, in two of these, GSTe2 and GSTe4, the location of these Inr coincided with the predicted TSS. However, the Inr was found 136 bp upstream of the predicted TSS of GSTe1, suggesting that the sequences in the EST database may not contain the full-length 5′-UTR for this gene. No attempt was made to verify the 5′-UTR of GSTe1 by PCR. The length of the 5′-UTRs, where known, varied from 93 bp in GSTe4 to 16 bp in GSTe5. GSTe5 also had the shortest 3′-UTR (16 bp) contrasting with a 320 bp 3′-UTR in GSTe6.
Figure 2. Scheme of putative core promoter regions of members of the Epsilon GST family.
Exons are shown by boxes, non-coding regions are shown by a horizontal line. Vertical lines denote the boundaries of the UTRs of the genes, where known. Polyadenylation (Poly A) signals were determined from the tentative consensus sequences for A. gambiae ESTs retrieved from the TIGR. Core promoter elements were identified by the MatInspector program. DPE, downstream promotor element.
The computer predictions described above suggested that basal expression of many, if not all, of the Epsilon GSTs may be controlled by core promoters located immediately upstream of the genes. To test this hypothesis, we selected the three shortest intergenic spaces within the Epsilon cluster, plus 1.3 kb of DNA immediately upstream of GSTe3, the first gene in the cluster, for cloning into a reporter construct.
The upstream putative promoter regions of GSTe3, GSTe2 and GSTe5 from the ZAN/U and Kisumu strains were cloned successfully into the reporter vector. In addition, the putative bidirectional promoter between GSTe7 and GSTe1 was inserted in both orientations (see Figure 1 for details of the constructs). For each of the intergenic sequences, the fragment cloned included the UTRs of the flanking genes. These five constructs were transiently transfected into the A. gambiae Sua 4.0 cell line, and their ability to drive heterologous expression of the firefly luciferase was determined.
Only two of these five constructs exhibited promoter activity. The GSTe1, GSTe5 and GSTe7 promoter constructs from both ZAN/U and Kisumu strains were inactive when transfected into the A. gambiae cell line. All eight of the Epsilon GSTs were expressed at varying levels in the adult stage of the mosquito [12], and at least four of the genes were also expressed in the larval stages (results not shown). It therefore seems likely that the absence of activity from the GSTe1, GSTe5 and GSTe7 promoter constructs may be related to their isolation from the endogenous chromosomal environment.
Confirmation of the TSS of GSTe2 and GSTe3
The two promoter regions that were able to drive transcription of the luciferase reporter gene comprised approx. 1300 bp immediately upstream of GSTe3 (the first gene in the Epsilon GST cluster) and the 352 bp fragment between the stop codon of GSTe1 and the translational start AUG codon of GSTe2. A GC box and CAAT box occur 72 and 54 bp upstream of the putative TSS of GSTe3, but no match was found with the consensus arthropod Inr site at the putative TSS. The sequence upstream of GSTe2 does contain an arthropod Inr consensus sequence (TCAGT) [22] 15 bp upstream of the predicted TSS. As we were interested in investigating the importance of 5′-UTRs in controlling expression of their genes, we confirmed the position of the putative TSS of GSTe2 and GSTe3 by PCR, using both cDNA and gDNA as templates. These experiments localized the TSS of GSTe3 to between 66 and 135 bp upstream of the start site of the translation of the gene, in agreement with TIGR alignments predicting a 92 bp 5′-UTR. Similarly, the TIGR prediction of a 48 bp 5′-UTR for GSTe2 matched the UTR range of 46–111 bp determined by our PCR results. Neither of the 5′-UTRs were interrupted by introns (see Figure S1 at http://www.BiochemJ.org/bj/387/bj3870879add.htm for further details).
Comparison of promoter sequences from DDT-resistant and -susceptible mosquitoes
Transcription of both GSTe3 and GSTe2 is significantly higher in the DDT-resistant ZAN/U strain than in the insecticide-susceptible Kisumu strain [12]. Therefore the sequences of these putative promoter regions in the two strains were compared to determine whether mutations in these regions could account for the variation in transcription rates.
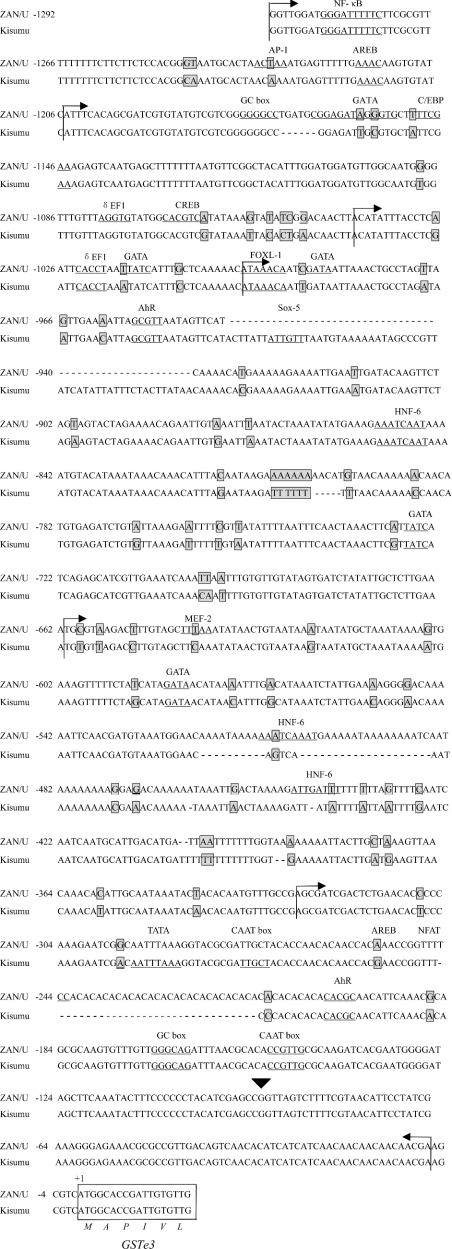
Comparison of the 1266 bp putative promoter region of GSTe3 identified 73 substitutions and ten indels (insertion/deletion) between the strains (Figure 3). These genetic variations include sites of polymorphism within each of the strains. In addition to the core promoter elements described above, numerous putative transcription-factor-binding sites were identified within the upstream regions of GSTe3 (Figure 3). These included putative binding sites for NF-κB (nuclear factor κB), AP-1 (activator protein-1), FOXL1 (Forkhead box L1), AhR (aryl hydrocarbon receptor), GATA box, CREB (cAMP-response-element-binding protein), δEF1 (δ elongation factor 1), AREB (Atp1a1-regulatory-element-binding factor), Sox-5 (Sry-type high-mobility group box protein), MEF-2 (myocyte-specific enhancer-binding factor), NFAT (nuclear factor of activated T-cells) and HNF-6 (hepatocyte nuclear factor 6). Several of these putative binding sites were only present in one of the sequenced strains [for example, binding sites for CREB, NFAT and MEF-2 were identified in the ZAN/U strain, but were absent from Kisumu (Figure 3)].
Figure 3. Sequence of the GSTe3 promoter.
Alignment of the sequence upstream from GSTe3 in the ZAN/U and Kisumu strains of A. gambiae obtained using the EMBOSS pairwise alignment programme (http://www.ebi.ac.uk/emboss/align/). Gaps inserted to maintain sequence alignment are shown by a horizontal dash. Potential transcription factors and the consensus sequences of high homology (100% core similarity and 95% matrix similarity) identified by MatInspector are underlined. Allelic variations between the two strains are boxed and shaded grey. The putative TSS of GSTe3 is indicated by a triangle. The start of the coding sequence of GSTe3 is boxed, and the amino acid translation is shown below the sequence alignment. Arrows indicate the boundaries of the constructs used for promoter analysis. AREB, Atp1a1-regulatory-element-binding factor; C/EBP, CCAAT/enhancer-binding protein; Sox-5, Sry-type high-mobility group box protein; HNF-6, hepatocyte nuclear factor 6.
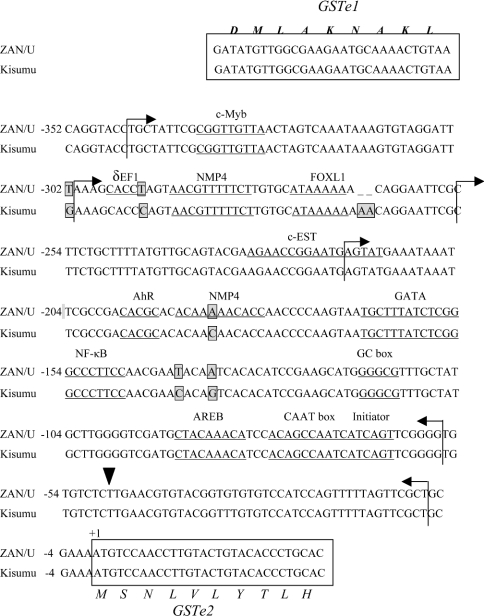
Five substitutions and a 2-adenosine indel were identified in the 352 bp GSTe2 promoter sequence from ZAN/U and Kisumu (Figure 4). The genetic variations were all conserved within each of the two strains. Again, multiple putative transcription-factor-binding sites were located within this fragment some of which were only present in one of the two strains (Figure 4).
Figure 4. Sequence of GSTe2 promoter.
Details are as for GSTe3 in Figure 3. NMP4, nuclear matrix protein 4.
Activity of promoters from DDT-resistant and -susceptible mosquitoes
To compare the activity of the Epsilon GST promoter elements from the DTT-susceptible and resistant strains of A. gambiae, separate constructs were prepared containing the upstream sequence of GSTe2 from ZAN/U and Kisumu (pZA2-1 and pKi2-1) and the upstream sequence of GSTe3 from both strains (pZA3-1 and pKi3-1) cloned into the pGL3-Basic luciferase reporter vector. The activities of the orthologous promoter elements from both strains were compared by transiently transfecting the A. gambiae cell line with each construct and comparing the normalized luciferase activity. All four constructs resulted in increases in luciferase activity (ranging from 31× to 95×) when compared with the promoterless pGL3-Basic vector (Figures 5 and 6). Both of the promoter elements derived from the ZAN/U strain were significantly more active than their Kisumu counterparts [GSTe2 Z/K ratio 2.7 (P<0.01); GSTe3 Z/K ratio 1.3 (P<0.05), where Z is ZAN/U and K is Kisumu].
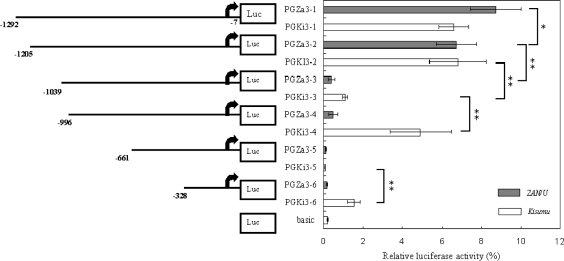
Figure 5. Normalized firefly luciferase expression following transfection into A. gambiae Sua 4.0 cells of a panel of progressive deletion constructs from the upstream sequence of GSTe3.
The arrows represent the putative TSS of GSTe3. Distances are from the A of the AUG initiation codon of GSTe3. Values shown on the panel on the right are the mean (±S.D.) luciferase activities (normalized to control Renilla activity) for two separate experiments performed in triplicate. The promoterless pGL3-Basic vector is designated basic. The promoter activity of the various constructs was analysed by pairwise Student's t tests. Statistically significant differences are shown by square brackets whose ends designate the sequences being compared (*P<0.05, **P<0.01).
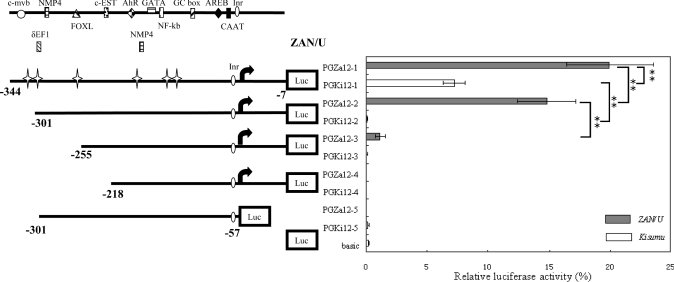
Figure 6. Normalized firefly luciferase expression following transfection into A. gambiae Sua 4.0 cells of a panel of progressive deletion constructs from the intergenic spacer region between GSTe1 and GSTe2.
The arrows represent the putative TSS of GSTe2. Distances are from the A of the AUG initiation codon of GSTe2. Values shown on the panel on the right are the mean (±S.D.) luciferase activities (normalized to control Renilla activity) for two separate experiments performed in triplicate. Potential transcription-factor-binding sites are shown above the constructs. Cross stars denote allelic variations in the promoter region between the two strains. The promoterless pGL3-Basic vector is designated basic. Promoter activity of constructs was analysed by pairwise Student's t test. Statistically significant differences are shown by square brackets whose ends designate the sequences being compared (**P<0.01). AREB, Atp1a1-regulatory-element-binding factor; NMP4, nuclear matrix protein 4.
Delineation of the GSTe3 promoter
Six different promoter constructs were prepared from the putative GSTe3 promoter element from both A. gambiae strains. For the ZAN/U strain, promoter activity generally decreased with successive deletions, although no significant difference was observed between promoter constructs pGZa3-3 and pGZa3-4 or between pGZa3-5 and pGZa3-6. The most dramatic decrease in luciferase expression occurred when the 166 bp between −1205 and −1039 were deleted (Figure 5). Exclusion of this region, which contains several putative transcription-factor-binding sites, including GC box, GATA, δEF1 and CREB, reduced expression of the ZAN/U promoter by 94%.
The activity of the Kisumu GSTe3 promoter also decreased as deletions were made from the 5′ end, again with a dramatic reduction in activity (84%) when the 160 bp (a 6 bp indel between the strains is present in this region) downstream from −1205 were removed. Further deletion of the Kisumu promoter resulted, however, in an increase in luciferase expression levels. Promoter activity significantly increased when the Kisumu promoter element was deleted by an additional 43 bp, suggesting that the deleted region from −1039 to −996 might encompass a potential negative regulatory element (a repressor) in the Kisumu strain, although none was identified by computational analysis of this region. A second, smaller, increase in promoter activity was noted when 300 bp was removed from the Kisumu construct containing 661 bp upstream of the initiation codon. Again, we speculate that a repressor element may be located between −296 and −596 in the Kisumu strain, but not in the corresponding region from the ZAN/U strain. Interestingly, both of the regions that contain putative repressors are characterized by large indels between the strains.
Delineation of the GSTe2 promoter
Deleting 43 bp from the 5′ end of the GSTe2 promoter element decreased the activity of the ZAN/U promoter by 25.8% and eliminated the activity of the Kisumu promoter element (Figure 6). This 43 bp fragment contained the 3′-UTR of the preceding gene, GSTe1. Deletion of an additional 46 bp from the ZAN/U construct decreased luciferase expression to only 5-fold above the promoterless control plasmid. The deleted fragment contains a putative binding site for the FOXL1 transcription factor and, in the ZAN/U strain only, a putative binding site for the transcription factor δEF1. A final deletion of an additional 37 bp from the 5′ end obliterated a potential c-EST binding site and destroyed all promoter activity (Figure 6).
In addition to deleting the 5′ end of the promoter element, we also investigated the effect of removing the 5′-UTR of GSTe2. Removing the 70 bp immediately preceding the AUG initiation codon destroyed the activity of the promoter (Figure 6). Thus the results from these deletion experiments show that the essential core promoter of GSTe2 includes the 5′-UTR of GSTe2 and the 3′-UTR of the preceding gene, GSTe1.
Site-directed mutagenesis
The dramatic difference between the promoter activity of plasmid pGZa12-2 and pGKi12-2 (activity is 122-fold higher in the ZAN/U plasmid) indicated that one or more sequence polymorphisms within the intergenic space between GSTe1 and GSTe2 (excluding the 3′-UTR of GSTe1, which is absent from this plasmid, and the 5′-UTR, which is identical in sequence in both strains) is critical for conferring the elevated activity seen in the ZAN/U GSTe2 promoter. As the activity of the ZAN/U promoter decreases dramatically when a further 46 bp are deleted, we speculated that sequence polymorphisms between −301 and −255 bp were important in determining transcription rates. Two sequence variations were noted within this region. A T/C substitution at −293 creates a possible δEF1-binding site in ZAN/U and a 2 bp indel is found immediately downstream of a putative FOXL1-binding site (Figure 4). The role of each of these polymorphisms in detecting the activity of the GSTe2 promoter was studied in turn.
The putative δEF1-binding site was introduced into the Kisumu construct pGKi12-2 by mutating C into T using the primer pG12F2 (Table 2). The lack of promoter activity of this construct suggests that the T/C nucleotide at −293 is not key to controlling transcription of GSTe2.
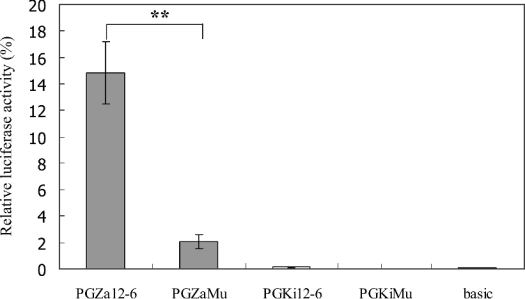
To study the role of the AA indel at −265 bp, two additional adenosine residues were introduced into the ZAN/U plasmid at −265 by site-directed mutagenesis. The ability of the mutated plasmid to drive luciferase expression was reduced dramatically (approx. 7-fold) compared with the wild-type ZAN/U plasmid (Figure 7). The reciprocal experiment was also performed by deleting two adenosine residues at −265 in the Kisumu plasmid. In this case, no significant difference in promoter activity was observed between the wild-type and mutated Kisumu plasmids (Figure 7).
Figure 7. Effects of altering the number of consecutive adenosine residues in the GSTe2 promoter on luciferase activity.
The number of adenosine residues at −265 in the GSTe2 promoter was altered by site-directed mutagenesis. The resultant constructs were transfected into A. gambiae Sua 4.0 cells and the luciferase activity was measured. The mean (±S.D.) luciferase activities (normalized to control Renilla activity) for two separate experiments performed in triplicate are shown. Pairwise Student's t tests were used for statistical analysis (**P<0.01).
H2O2-inducible expression of Epsilon GSTs
Several putative binding sites for transcription factors responsive to oxidative stress were identified in the GSTe2 promoter, including FOXL1, NF-κB and AhR. As expression of some GSTs from other organisms increases in response to oxidative stress, we measured the effect of H2O2 treatment on expression of three A. gambiae Epsilon GSTs. Quantitative PCR results showed expression of GSTe1, GSTe2 and GSTe3 in larvae of the ZAN/U strain was induced by a 1-h exposure to H2O2. In the Kisumu strain, however, only expression of GSTe3 was induced by H2O2 exposure (Figure 8).
Figure 8. Induction of GST expression by H2O2 exposure.
To test whether GSTe3, GSTe1 and GSTe2 in A. gambiae are inducible by oxidative stress, larvae of Kisumu and ZAN/U strains were exposed to 3 mM H2O2 for 1 h. mRNA was extracted and reversed-transcribed into cDNA. The copy number of the Epsilon GST, GSTe3, GSTe1 and GSTe2 (inset) was determined by real-time PCR. The data are from duplicates of three biological replicates. The transcript copy number in control (no H2O2 exposure) was compared with the H2O2-exposed samples using the pairwise Student's t test (**P<0.01; N.S., not significant).
Expression of GSTe3 was induced 3.3-fold in ZAN/U and 2.3-fold in Kisumu by H2O2 exposure (Figure 8). Expression of GSTe1 and GSTe2 (Figure 8) increased 4-fold and 1.8-fold respectively after 1 h of H2O2 treatment of ZAN/U larvae. Basal expression of GSTe2 in larvae was higher than that of GSTe1 and GSTe3 in both strains. Expression of GSTe2 in the absence of H2O2 was approx. 3.6-fold higher than in the susceptible strain (Figure 8).
DISCUSSION
We have shown previously that DDT resistance in A. gambiae is conferred by overexpression of an Epsilon GST, GSTe2, that is mediated, at least in part, by mutations in a cis-acting regulatory factor(s) [23]. A central aim of the present study was to identify the mechanism responsible for the up-regulation of this gene, with the eventual goal of developing a robust assay to detect this mutation in field populations.
The active GSTe2 promoter is one of the smallest characterized in insects. The distance between the stop codon of the preceding gene, GSTe1, and the start codon of GSTe2 is only 352 bp. The UTRs of both genes are important for maximal GSTe2 promoter activity. Deleting the 5′-UTR of GSTe2 obliterates promoter activity, and, surprisingly, deletion of the 3′-UTR of GSTe1 decreases activity by 26%. The activity of the full-length GSTe2 promoter construct is 2.7-fold higher in the DDT-resistant strain than in the -susceptible strain, and this correlates well with the elevation in basal transcript copy number observed in the larvae of the resistant strain (3.6-fold, Figure 8). The 1.3 kb sequence upstream of GSTe3 in the ZAN/U strain had slightly higher activity than the corresponding region from the Kisumu strain (1.3-fold elevation), although there was no significant difference in transcript copy number between the strains for this gene (Figure 8). In adult ZAN/U mosquitoes, GSTe2 expression was elevated 8-fold and GSTe3 expression was elevated 3-fold [12], suggesting that different regulatory elements may control expression in different life stages.
Several sequence polymorphisms were observed in the promoter region of GSTe2. To determine which of these are responsible for variation in promoter activity, the 352 bp promoter was progressively deleted, and the activity of each new construct was compared in the two strains. A 46 bp region between −301 and −255 was implicated. Two polymorphic sites are found within this section, one of which lies within a putative recognition site for the transcription factor δEF1 and the second introduces an AA indel. Mutating the first of these to introduce a δEF1 site in the Kisumu strain did not increase promoter activity. It is unlikely that δEF1 is important in regulating expression of GSTe2, since, in all cases where binding sites for this transcription factor have been identified, two recognition sites separated by 20–50 bp are required [24]. A single δEF1 site is found in the GSTe2 promoter. Interestingly, however, two δEF1 sites separated by 51 bp are found in the promoter of GSTe3. Deletion of one of these virtually eliminates GSTe3 promoter activity.
The second candidate polymorphic site in the GSTe2 promoter is the 2 bp AA indel located adjacent to a putative FOXL1 motif. Introduction of two adenosine residues into the ZAN/U promoter, while retaining the three variable sites downstream in the ZAN/U promoter sequences, decreased the activity of this construct 7-fold. The reverse experiment, in which two adenosine residues were deleted from the Kisumu construct, did not restore promoter activity to this strain. Thus, presumably, in addition to the number of adenosine residues present, one or more of the three polymorphic sites located downstream from this indel is important in determining GSTe2 promoter activity. An analysis of this sequence from other DDT-resistant and -susceptible strains or field-caught individuals is needed to confirm the role of these polymorphisms in the GSTe2 promoter in conferring DDT resistance.
The promoters of both GSTe2 and GSTe3 contain putative binding sites for several transcription factors that mediate response to oxidative stress in other species [25–28]. These include L1, AP-1, NF-κB and AhR. GSTs can protect against oxidative stress either by direct peroxidase activity or by detoxifying secondary products that are generated by ROS (reactive oxygen species) [29]. Furthermore, expression of GSTs is induced by exposure to oxidative stress in several species [30–33]. We therefore investigated the effect of treatment with H2O2 on the expression of the A. gambiae Epsilon GSTs. Increased expression of GSTe3 following H2O2 exposure has been reported previously in A. gambiae [34], and, in agreement with this, we observed a significant increase in transcript copy number in both DDT-susceptible and -resistant strains. Interestingly, for the two additional Epsilon GSTs examined, a response was only detectable in the DDT-resistant, ZAN/U, larvae. The reason for the differing response of the strains is unclear, and could be due to changes in regulatory proteins, rather than the promoters themselves. However, it is intriguing that the DDT-resistant strain is more responsive to oxidative stress. Exposure to insecticides induces the production of ROS, and insecticide resistance has been linked to the elevated expression of a Delta GST with high peroxidase activity [9]. Perhaps the role of GSTs in the ZAN/U strain in conferring resistance to insecticides extends beyond their ability to metabolize the insecticides.
Online data
Acknowledgments
This work was supported by a project grant from the Wellcome Trust (to H. R. and J. H.) and a Leverhulme Trust studentship to Y. D.
References
- 1.Singh S. P., Coronella J. A., Benes H., Cochrane B. J., Zimniak P. Catalytic function of Drosophila melanogaster glutathione S-transferase DmGSTS1-1 (GST-2) in conjugation of lipid peroxidation end products. Eur. J. Biochem. 2001;268:2912–2923. doi: 10.1046/j.1432-1327.2001.02179.x. [DOI] [PubMed] [Google Scholar]
- 2.Sawicki R., Singh S. P., Mondal A. K., Benes H., Zimniak P. Cloning, expression and biochemical characterization of one Epsilon-class (GST-3) and ten Delta-class (GST-1) glutathione S-transferases from Drosophila melanogaster, and identification of additional nine members of the Epsilon class. Biochem. J. 2003;370:661–669. doi: 10.1042/BJ20021287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Listowsky I., Abramovitz M., Homma H., Niitsu Y. Intracellular binding and transport of hormones and xenobiotics by glutathione-S-transferases. Drug Metab. Rev. 1988;19:305–318. doi: 10.3109/03602538808994138. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn A. C., Woollatt E., Sutherland G. R., Board P. G. Characterization and chromosome location of the gene GSTZ1 encoding the human Zeta class glutathione transferase and maleylacetoacetate isomerase. Cytogenet. Cell Genet. 1998;83:109–114. doi: 10.1159/000015145. [DOI] [PubMed] [Google Scholar]
- 5.Cho S. G., Lee Y. H., Park H. S., Ryoo K., Kang K. W., Park J., Eom S. J., Kim M. J., Chang T. S., Choi S. Y., et al. Glutathione S-transferase mu modulates the stress-activated signals by suppressing apoptosis signal-regulating kinase 1. J. Biol. Chem. 2001;276:12749–12755. doi: 10.1074/jbc.M005561200. [DOI] [PubMed] [Google Scholar]
- 6.Ranson H., Claudianos C., Ortelli F., Abgrall C., Hemingway J., Sharakhova M. V., Unger M. F., Collins F. H., Feyereisen R. Evolution of supergene families associated with insecticide resistance. Science. 2002;298:179–181. doi: 10.1126/science.1076781. [DOI] [PubMed] [Google Scholar]
- 7.Clark A. G., Shamaan N. A. Evidence that DDT-dehydrochlorinase from the house fly is a glutathione S-transferase. Pestic. Biochem. Physiol. 1984;22:249–261. [Google Scholar]
- 8.Chiang F. M., Sun C. N. Glutathione transferase isozymes of diamondback moth larvae and their role in the degradation of some organophosphorus insecticides. Pestic. Biochem. Physiol. 1993;45:7–14. [Google Scholar]
- 9.Vontas J. G., Small G. J., Hemingway J. Glutathione S-transferases as antioxidant defence agents confer pyrethroid resistance in Nilaparvata lugens. Biochem. J. 2001;357:65–72. doi: 10.1042/0264-6021:3570065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO. Technical Report No. 818. Geneva: World Heath Organization; 1992. Vector Resistance to Pesticides. Fifteenth report of the WHO Expert Committee on Vector Biology and Control. [PubMed] [Google Scholar]
- 11.Ortelli F., Rossiter L. C., Vontas J., Ranson H., Hemingway J. Heterologous expression of four glutathione transferase genes genetically linked to a major insecticide-resistance locus from the malaria vector Anopheles gambiae. Biochem. J. 2003;373:957–963. doi: 10.1042/BJ20030169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding Y., Ortelli F., Rossiter L. C., Hemingway J., Ranson H. The Anopheles gambiae glutathione transferase supergene family: annotation, phylogeny and expression profiles. BMC Genomics. 2003;4:35. doi: 10.1186/1471-2164-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ranson H., Jensen B., Wang X., Hemingway J., Collins F. H. Genetic mapping of two loci affecting DDT resistance in the malaria vector Anopheles gambiae. Insect Mol. Biol. 2000;9:499–507. doi: 10.1046/j.1365-2583.2000.00214.x. [DOI] [PubMed] [Google Scholar]
- 14.Blumenthal T. Gene clusters and polycistronic transcription in eukaryotes. BioEssays. 1998;20:480–487. doi: 10.1002/(SICI)1521-1878(199806)20:6<480::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 15.Ranson H., Collins F., Hemingway J. The role of alternative mRNA splicing in generating heterogeneity within the Anopheles gambiae class I glutathione S-transferase family. Proc. Natl. Acad. Sci. U.S.A. 1998;95:14284–14289. doi: 10.1073/pnas.95.24.14284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaughan A., Rodriguez M., Hemingway J. The independent gene amplification of electrophoretically indistinguishable B esterases from the insecticide-resistant mosquito Culex quinquefasciatus. Biochem. J. 1995;305:651–658. doi: 10.1042/bj3050651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catteruccia F., Nolan T., Blass C., Muller H. M., Crisanti A., Kafatos F. C., Loukeris T. G. Toward Anopheles transformation: Minos element activity in anopheline cells and embryos. Proc. Natl. Acad. Sci. U.S.A. 2000;97:2157–2162. doi: 10.1073/pnas.040568397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y. G., Eggleston P. Comparative analysis of promoters for transient gene expression in cultured mosquito cells. Insect Mol. Biol. 1999;8:31–38. doi: 10.1046/j.1365-2583.1999.810031.x. [DOI] [PubMed] [Google Scholar]
- 19.Salazar C. E., Mills-Hamm D., Kumar V., Collins F. H. Sequence of a cDNA from the mosquito Anopheles gambiae encoding a homologue of human ribosomal protein S7. Nucleic Acids Res. 1993;21:4147. doi: 10.1093/nar/21.17.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quandt K., Frech K., Karas H., Wingender E., Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burke T. W., Kadonaga J. T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 22.Cherbas L., Cherbas P. The arthropod initiator: the capsite consensus plays an important role in transcription. Insect Biochem. Mol. Biol. 1993;23:81–90. doi: 10.1016/0965-1748(93)90085-7. [DOI] [PubMed] [Google Scholar]
- 23.Ranson H., Rossiter L., Ortelli F., Jensen B., Wang X., Roth C. W., Collins F. H., Hemingway J. Identification of a novel class of insect glutathione S-transferases involved in resistance to DDT in the malaria vector Anopheles gambiae. Biochem. J. 2001;359:295–304. doi: 10.1042/0264-6021:3590295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Remacle J. E., Kraft H., Lerchner W., Wuytens G., Collart C., Verschueren K., Smith J. C., Huylebroeck D. New mode of DNA binding of multi-zinc finger transcription factors: δEF1 family members bind with two hands to two target sites. EMBO J. 1999;18:5073–5084. doi: 10.1093/emboj/18.18.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgering B. M., Kops G. J. Cell cycle and death control: long live Forkheads. Trends Biochem. Sci. 2002;27:352–360. doi: 10.1016/s0968-0004(02)02113-8. [DOI] [PubMed] [Google Scholar]
- 26.Kops G. J., Dansen T. B., Polderman P. E., Saarloos I., Wirtz K. W., Coffer P. J., Huang T. T., Bos J. L., Medema R. H., Burgering B. M. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature (London) 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 27.Pinkus R., Weiner L. M., Daniel V. Role of oxidants and antioxidants in the induction of AP-1, NF-κB, and glutathione S-transferase gene expression. J. Biol. Chem. 1996;271:13422–13429. doi: 10.1074/jbc.271.23.13422. [DOI] [PubMed] [Google Scholar]
- 28.Haddad J. J. Oxygen sensing and oxidant/redox-related pathways. Biochem. Biophys. Res. Commun. 2004;316:969–977. doi: 10.1016/j.bbrc.2004.02.162. [DOI] [PubMed] [Google Scholar]
- 29.Hayes J. D., McLellan L. I. Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Radical Res. 1999;31:273–300. doi: 10.1080/10715769900300851. [DOI] [PubMed] [Google Scholar]
- 30.Chen W., Singh K. B. The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J. 1999;19:667–677. doi: 10.1046/j.1365-313x.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- 31.Singhal S. S., Godley B. F., Chandra A., Pandya U., Jin G. F., Saini M. K., Awasthi S., Awasthi Y. C. Induction of glutathione S-transferase hGST 5.8 is an early response to oxidative stress in RPE cells. Invest. Ophthalmol. Vis. Sci. 1999;40:2652–2659. [PubMed] [Google Scholar]
- 32.Zelck U. E., Von Janowsky B. Antioxidant enzymes in intramolluscan Schistosoma mansoni and ROS-induced changes in expression. Parasitology. 2004;128:493–501. doi: 10.1017/s0031182004004895. [DOI] [PubMed] [Google Scholar]
- 33.Fiander H., Schneider H. Compounds that induce isoforms of glutathione S-transferase with properties of a critical enzyme in defense against oxidative stress. Biochem. Biophys. Res. Commun. 1999;262:591–595. doi: 10.1006/bbrc.1999.1262. [DOI] [PubMed] [Google Scholar]
- 34.Kumar S., Christophides G. K., Cantera R., Charles B., Han Y. S., Meister S., Dimopoulos G., Kafatos F. C., Barillas-Mury C. The role of reactive oxygen species on Plasmodium melanotic encapsulation in Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14139–14144. doi: 10.1073/pnas.2036262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.