Abstract
The ORPs (oxysterol-binding-protein-related proteins) constitute an enigmatic family of intracellular lipid receptors that are related through a shared lipid binding domain. Emerging evidence suggests that ORPs relate lipid metabolism to membrane transport. Current data imply that the yeast ORP Kes1p is a negative regulator of Golgi-derived vesicular transport mediated by the essential phosphatidylinositol/phosphatidylcholine transfer protein Sec14p. Inactivation of Kes1p function allows restoration of growth and vesicular transport in cells lacking Sec14p function, and Kes1p function in this regard can be complemented by human ORP1S (ORP1 short). Recent studies have determined that Kes1p and ORP1S both bind phospholipids as ligands. To explore the function of distinct linear segments of ORP1S in phospholipid binding and vesicular transport regulation, we generated a series of 15 open reading frames coding for diagnostic regions within ORP1S. Purified versions of these ORP1S deletion proteins were characterized in vitro, and allowed the identification of a nominal phospholipid binding region. The in vitro analysis was interpreted in the context of in vivo growth and vesicle transport assays for members of the ORP1S deletion set. The results determined that the phospholipid binding domain per se was insufficient for inhibition of vesicular transport by ORP1S, and that transport of carboxypeptidase Y and invertase from the Golgi may be regulated differentially by specific regions of ORP1S/Kes1p.
Keywords: Golgi, oxysterol binding protein, oxysterol-binding-protein related-protein 1 short (ORP1S), Saccharomyces cerevisiae, vesicular transport
Abbreviations: CPY, carboxypeptidase Y; GST, glutathione S-transferase; ORP, oxysterol-binding-protein-related protein; ORP1L, ORP1 long; ORP1S, ORP1 short; OSBP, oxysterol binding protein; PC, phosphatidylcholine; PH, pleckstrin homology; PI, phosphatidylinositol; PI4P, PI 4-phosphate (etc.); PI3,4P2, PI 3,4-bisphosphate (etc.); PI3,4,5P3, PI 3,4,5-trisphosphate (etc.)
INTRODUCTION
The OSBPs (oxysterol binding proteins) and ORPs (OSBP-related proteins) constitute an enigmatic protein family that is united by a signature motif that binds either oxysterols or phospholipids in an OSBP/ORP-isoform-specific manner [1–10]. In humans there are 12 genes that, through differential pre-mRNA splicing, code for at least 16 predicted OSBPs/ORPs [1,3,9–12]. The transcript of the human ORP1 gene is differentially spliced, resulting in the production of two mRNAs encoding ORP1L (long) and ORP1S (short). ORP1L and ORP1S transcripts were found to be expressed ubiquitously, with ORP1L predominating in lung and macrophages, and ORP1S in heart and muscle [3,10]. The ORP1L transcript encodes a 950-amino-acid protein whose N-terminus contains ankyrin repeats, a PH (pleckstrin homology) domain, a FFAT (two phenylalanine residues in an acidic tract) motif for endoplasmic reticulum targeting, and a diagnostic ORP motif of unknown function, while the C-terminus is composed of the signature ORP lipid binding domain. The ORP1S transcript encodes a protein comprising the C-terminal 437 amino acids of ORP1L, and contains only the diagnostic ORP family motif and the lipid binding domain [3,10,11].
The genome of the yeast Saccharomyces cerevisiae encodes seven ORP homologues (Osh1p–Osh7p), of which four are similar to human ORP1S in that they are composed of only the diagnostic ORP family motif and the lipid binding domain [2,5,13–17]. Genetic inactivation of individual yeast ORPs does not result in any obvious phenotype, and indeed all seven members of the yeast ORP family had to be inactivated before lethality was observed [14]. Disruption of the function of any six yeast ORPs in combination was tolerated, implying shared overlapping essential functions within the ORP family, with specialized but non-essential subfunctions for each family member.
Insights into the functional and structural roles of yeast ORPs have mainly been driven by phenotypes associated with loss of function of yeast Osh4p/Kes1p. Genetic and cell biological studies have determined that Kes1p is a negative regulator of Golgi-derived vesicle transport mediated by the essential PI (phosphatidylinositol)/PC (phosphatidylcholine) transfer protein Sec14p [2,5,10,18]. This function is specific to Kes1p, as genetic inactivation of the other yeast ORP family members was unable to bypass the essential function of Sec14p [2]. The combined data indicate that Sec14p functions as a diffusible phospholipid sensor that is required to regulate PC and phosphoinositide homoeostasis in the Golgi.
Previously we demonstrated that human ORP1S, but not the very similar human ORP2, was able to phenocopy Kes1p function with respect to acting as a negative regulator of Sec14p-derived vesicular transport [10]. Purified ORP1S protein was observed to bind phosphatidic acid and phosphoinositides, and it was suggested that ORP1S/Kes1p might regulate vesicle transport through their ability to bind one or more of these lipids. Subsequent studies on Kes1p have determined that purified Kes1p bound several acidic phospholipids, with phosphoinositides bound with the highest affinity [5]. Subsequent genetic and cell biological analyses implied that PI4P (PI 4-phosphate) was the primary candidate to be the Kes1p ligand in vivo.
To define more precisely the phospholipid binding site within the ORP lipid binding domain and to assess the functional role of lipid binding in the regulation of vesicular transport by this family of proteins, we generated a series of ORP1S deletion mutants and analysed the purified proteins for their phospholipid binding ability and specificity in vitro. This analysis was compared with the capacity of a diagnostic set of ORP1S mutants to phenocopy yeast Kes1p in the regulation of Sec14p-mediated vesicular transport in vivo. The results indicate that phospholipid binding in itself is not sufficient for ORP1S function in vivo.
EXPERIMENTAL
Reagents
Anti-T7 antibody was purchased from Novagen, and anti-GST (glutathione S-transferase) antibody was purchased from Amersham. Anti-c-Myc antibody and goat anti-mouse conjugated antibodies were purchased from Cell Signaling Technology. Monoclonal antibody against yeast CPY (carboxypeptidase Y) was purchased from Molecular Probes. Phosphoinositides were purchased from Echelon Biosciences Inc., and all other lipids were products of Avanti Polar Lipids.
Construction of ORP1S deletion mutant expression vectors and purification of recombinant ORP1S proteins
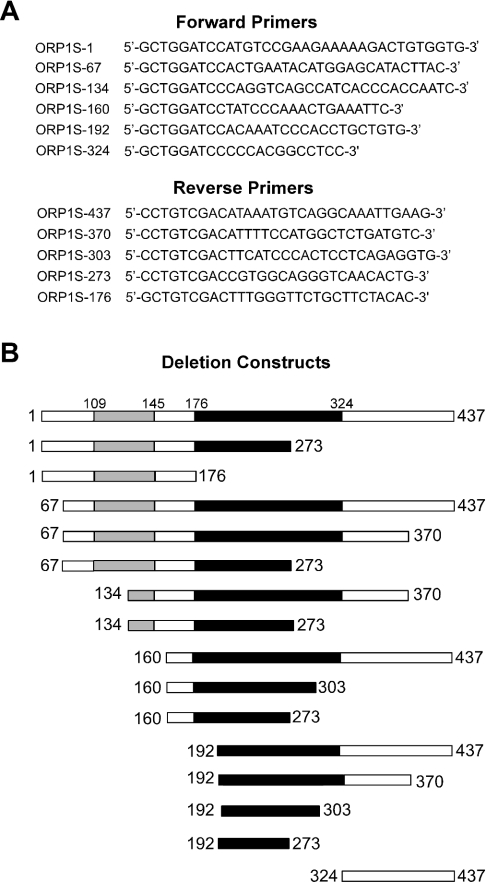
The full-length open reading frame of ORP1S was used as a template to generate ORP1S deletion constructs by PCR using HiFi Platinum Taq polymerase (Invitrogen) with the required primer pairs to generate the desired truncation mutants (Figure 1) and subcloned into pCR2.1-Topo (Invitrogen) for open reading frame confirmation. The full-length and truncated ORP1S open reading frames were subsequently subcloned into the Escherichia coli pET23b (Novagen) expression vector in-frame with the coding sequence for an N-terminal T7 epitope tag and a C-terminal His6 tag. Similarly, full-length open reading frames of ORP1S, the PH domain of mammalian OSBP and yeast KES1 were subcloned into the E. coli pGEX-4T1 (Amersham) expression vector in-frame with the coding sequence to generate a GST fusion protein. Plasmids were transformed into E. coli BL21 (DE3) and protein expression was induced by the addition of 0.5 mM IPTG to exponential-phase cells. After 3 h at 37 °C, cells were lysed with B-PER reagent (Pierce) and purified using either Talon (Clontech) resin and imidazole buffers for His6-tagged proteins or GST–Sepharose (Amersham Pharmacia Biotech) using glutathione buffers for GST fusions proteins. Purified proteins were dialysed overnight against Tris-buffered saline at 4 °C. SDS/PAGE analysis and Coomassie Blue staining of the purified protein preparations gave an estimated purity of >95% for full-length ORP1S and of at least 50% for each ORP1S deletion construct. Western blot analysis using horseradish peroxidase-conjugated anti-T7 antibody detected only parental and ORP1S deletion proteins.
Figure 1. ORP1S deletion constructs used in the present study.
(A) Primer pairs used to generate the ORP1S deletion mutants. (B) Bioinformatics Blast searches and research to date has identified two domains present in ORP1S: the black bar represents the ORP lipid ligand binding region, and the grey bar represents the OSBP domain, which has been identified as being highly conserved in all ORPs from all species.
Fat Western lipid overlay assay
Standard phospholipid binding assays were performed essentially as described [10,19,20] by immobilizing 1 nmol of pure phospholipid on Hybond-C nitrocellulose membranes. Blots were blocked with Tris-buffered saline containing 3% (w/v) fatty acid-free BSA. Purified ORPs were incubated at 4 °C for 12 h with immobilized phospholipids in Tris-buffered saline containing 3% BSA. Blots were washed and incubated with anti-T7 primary antibody (1:5000) or anti-GST primary antibody (1:5000) coupled to horseradish peroxidase for 1 h at room temperature and washed, and phospholipid binding was determined by ECL® (Amersham Pharmacia Biotech).
Cell growth and heterologous ORP1S protein expression in yeast
The yeast strain CMY136 (a ura3 his3 trp1 leu2 sec14-1ts kes1::HIS3) used in the present study has been described previously [10]. The full-length and truncated ORP1S open reading frames were subcloned from pCR2.1-Topo into the yeast expression vector pESC-URA (Invitrogen). In pESC-URA, ORP1S and its deletion mutants are in-frame with an N-terminal c-Myc epitope tag and under the control of the galactose-inducible yeast GAL1 promoter. A patch of cells from a single yeast colony was grown overnight in defined glucose medium (represses GAL1-controlled transcription) containing the appropriate nutrients to ensure plasmid maintenance. Yeast cell concentration was estimated by measuring absorbance at 600 nm, and an identical number of cells were washed and a series of serial dilutions were spotted on to minimal medium agar plates containing the appropriate nutrients required for strain growth and plasmid maintenance with either 2% (w/v) glucose or 2% (w/v) galactose as carbon source.
Invertase secretion
Strain CMY136 transformed with pDB31 (pTPI-SUC2; encodes invertase under the control of the constitutive TPI1 promoter) [21] and the indicated ORP1S fragment under the control of the GAL1 promoter were grown to mid-exponential phase in minimal medium containing the appropriate nutrients required for strain growth and plasmid maintenance with 2% (w/v) galactose as carbon source at 25 °C. Cells were centrifuged and washed with 1 vol. of 2% galactose medium, and resuspended in 0.2 vol. of 2% galactose medium. The culture was shifted to 37 °C for 2 h. The samples were washed twice with 1 ml of cold 10 mM NaN3 and resuspended in 500 μl of the same solution. Each sample was subsequently split into two halves. For each, the non-permeabilized sample was adjusted to 500 μl with 250 μl of 10 mM NaN3. The samples to be permeabilized were similarly adjusted with 10 mM NaN3 containing 0.2% Triton X-100 and subjected to one cycle of freeze–thawing. These samples were used to measure external and total invertase activities respectively. The absorbances of the non-permeabilized samples were measured at 600 nm with a spectrophotometer and served to normalize the data. The secretion index is the ratio of external invertase activity to total invertase activity.
CPY transport
The assay for CPY secretion was performed essentially as described [22]. The c-Myc-tagged ORP1S constructs expressed from the GAL1 promoter were grown to mid-exponential phase in minimal selective medium with galactose as the carbon source. Cells were harvested, washed and resuspended in fresh medium at equivalent cell densities (A600=0.5). After growth for 1 and 3 h, 1.0 ml of each culture was centrifuged at 14000 g for 5 min and 0.8 ml of supernatant was collected. Proteins contained in the extracellular medium were precipitated by adding 0.2 ml of 50% (v/v) trichloroacetic acid along with 1 μg of BSA as carrier protein and incubated on ice for 30 min. The precipitated proteins were collected by centrifugation at 14000 g for 15 min at 4 °C, washed with 100% (v/v) acetone, dried at room temperature, and resuspended in 40 μl of SDS/PAGE sample buffer supplemented with 50 mM Tris/HCl, pH 8.8. Equal volumes were resolved by SDS/PAGE and processed for immunoblots.
Fluorescence microscopy
Cells containing ORP1S constructs in pESC-URA were grown in minimal medium with galactose to exponential phase. Cultures were fixed with 3.7% (v/v) formaldehyde and protein was detected with a mouse anti-c-Myc primary antibody and a goat anti-mouse Texas Red-conjugated secondary antibody as described [23]. Fluorescence microscopy was performed using a Zeiss Axiophot microscope fitted with a plan-neofluor 100× oil immersion objective lens. Images were captured using a Zeiss Axio Cam HR using Axiovision 3.1 software.
RESULTS
In vitro phospholipid binding
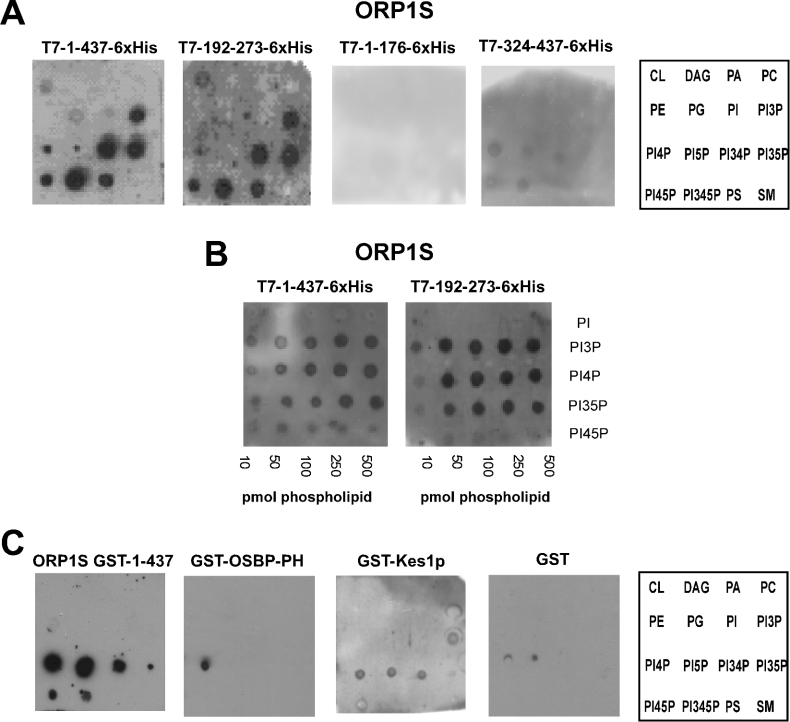
Full-length ORP1S and ORP1S deletion constructs (Figure 1A) were expressed in E. coli in-frame with an N-terminal T7 epitope tag and a C-terminal His6 tag and purified using metal affinity chromatography. Lipid binding ability was determined using the fat Western lipid overlay assay [10,19,20] (Figure 2). The region comprising the first 176 amino acid residues of ORP1S was unable to bind lipids, while very weak affinity for some phosphoinositides was observed for the region comprising amino acid residue 324 through to the end of ORP1S. This weak binding was unexpected, as amino acid residues 324–437 of ORP1S were not believed to bind phospholipids. All other constructs bound a similar set of phospholipids; in vitro ligand binding was highest for various phosphoinositides, with PI3,4,5P3 (PI 3,4,5-trisphosphate), PI3,4P2 (PI 3,4-bisphosphate) and PI3,5P2 serving as the best ligands, followed by PI3P and PI4,5P2, and then by phosphatidylserine, PI4P and PI5P. Weak binding to phosphatidic acid, phosphatidylglycerol and cardiolipin could be observed at higher phospholipid or protein concentrations (results not shown). The smallest region identified as being capable of binding phospholipids with a similar specificity to the wild type comprised amino acids 192–273. We compared the affinity of full-length ORP1S with that of ORP1S-(192–273), and observed that both proteins bound phosphoinositides with similar affinities.
Figure 2. Phospholipid binding by linear regions of ORP1S.
(A, B) Purification of His6-tagged ORP1S proteins from their open reading frames in E. coli was by metal chelate resin. Purified ORPs were dialysed overnight against Tris-buffered saline at 4 °C, and fat Western lipid overlays were performed as described in the Experimental section. Full-length ORP1S, ORP1S-(192–273) (the nominal ORP1S lipid binding domain identified in the present study), ORP-(1–176) and ORP-(324–437) are illustrated. Similar results were observed for each ORP1S deletion mutant. (C) Purification of ORP1S GST fusion proteins expressed in E. coli was by glutathione affinity resin. Lipid binding was determined as in (A) and (B) using the GST fusion proteins. GST and the GST–OSBP–PH were used as negative and positive controls respectively, while GST–Kes1p was included for comparison. Results are representative of at least three separate experiments. Abbreviations: CL, cardiolipin; DAG, diacylglycerol; PA, phosphatidic acid; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PS, phosphatidylserine; SM, sphingomyelin.
To further facilitate our analysis of phospholipid binding by ORP1S, we also purified ORP1S as a GST fusion protein and compared its phospholipid binding specificity with that of the PH domain of mammalian OSBP and with that of yeast Kes1p. The PH domain of mammalian OSBP showed a high degree of specificity for PI4P, while yeast Kes1p bound PI3P, PI4P, PI5P and PI3,4P2. Similar to T7–ORP1S–His6, GST–ORP1S bound phosphoinositides, however, the range of phosphoinositides differed, with GST–ORP1S binding PI4P and PI5P very well, followed by PI3,4P2, PI4,5P2=PI3,4,5P3 and then PI3,5P2.
The results establish that ORP1S indeed binds phosphoinositides and that the structure of the lipid binding domain of the ORP1S deletion mutants was intact, as the in vitro binding specificity and affinity were unchanged for the deletion mutants compared with full-length ORP1S protein.
Expression and characterization of ORP1S deletion mutants in vivo
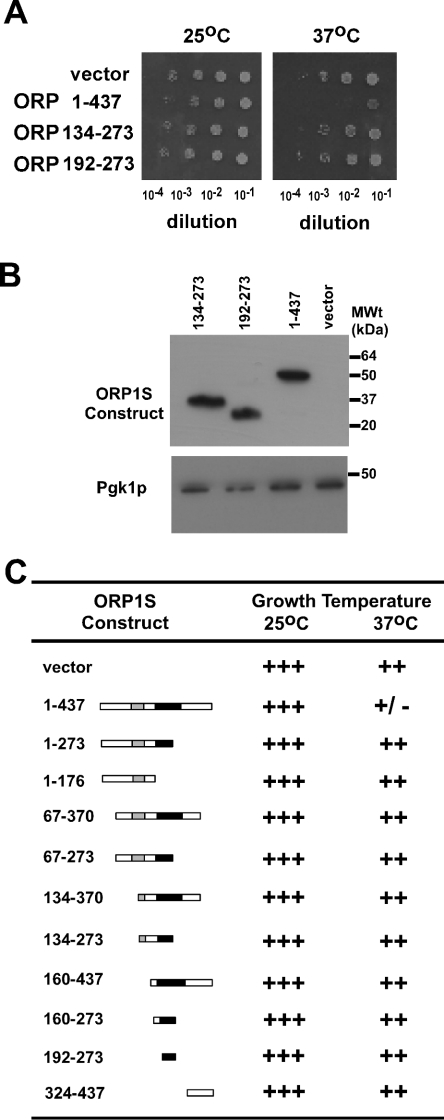
As each of the ORP1S deletion mutants tested was able to bind the same set of phospholipids in vitro, this allowed for the determination of the role of each region of ORP1S, and of lipid binding specifically, in ORP1S in vivo function as a negative regulator of Golgi-derived vesicular transport. It was determined previously that human ORP1S could phenocopy yeast Kes1p and inhibit the Sec14p-mediated regulation of vesicular transport from the Golgi [10]. We assessed the role of a subset of diagnostic ORP1S deletion mutants for their ability to prevent cell growth of a sec14ts kes1− yeast strain at the permissive (25 °C) and non-permissive (37 °C) temperatures for the sec14ts allele (Figure 3A). As observed previously, full-length ORP1S was able to phenocopy the function of Kes1p and prevent growth of the sec14ts kes1− yeast strain at the non-permissive temperature for the sec14ts allele. The ORP1S deletion mutant [ORP1S-(192–273)] that consisted of the nominal phospholipid binding region was unable to phenocopy Kes1p and resulted in growth at the non-permissive temperature. This indicates that the phospholipid binding region of ORP1S in itself is not sufficient for function in vivo.
Figure 3. Regulation of cell growth by ORP1S deletion mutants.
(A) CMY136 yeast (a ura3 his3 trp1 leu2 sec14-1ts kes1::HIS3) were transformed with the pESC-URA vector containing ORP1S or the indicated ORP1S deletion mutants in-frame with an N-terminal Myc epitope tag and under the control of the galactose-inducible GAL1 promoter. Identical numbers of exponential-phase cells were harvested and washed, and serial dilutions were spotted on to minimal medium agar plates containing the appropriate nutrients required for strain growth and plasmid maintenance, with galactose as carbon source, and grown at 25 °C or 37 °C for 4 days. (B) Western blot analysis of the expression levels of Myc-tagged ORP1S mutants, with Pgk1p as a loading control. (C) Growth phenotypes for all ORP1S deletion constructs that were expressed in yeast strain CMY136. Results are representative of at least three separate experiments.
Bioinformatics analysis has uncovered several conserved regions in the ORP family, with the most notable being termed the OSBP domain (Figure 1) [5,11]. This region of amino acids is conserved across species, and in human ORP1S it encompasses amino acid residues 109–145. Within the OSBP region are two stretches of amino acid residues that are diagnostic, encompassing K110PFNPLLGET119 and E134QVSHHPP141 in ORP1S. Within the deletion set of ORP1S proteins that were constructed, some had this motif split in half, while others lacked the N- or C-terminal region of ORP1S. Determination of the ability of the various proteins to effect substantial growth inhibition of the sec14ts kes1− yeast strain at the non-permissive temperature for the sec14ts allele revealed that only full-length ORP1S was capable of phenocopying Kes1p (Figures 3A and 3C). To ensure that the growth phenotypes observed were not due to trivial reasons such as protein expression amount or subcellular location, Western blots and immunofluorescence experiments were performed for ORP1S, ORP1S-(192–273) and ORP1S-(134–273). All three proteins were expressed at comparable levels, as assessed by Western blot, and were localized to the cytoplasm, as determined by immunofluorescence (Figure 3B and results not shown). This distribution of ORP1S and its deletion constructs was similar to that reported for endogenous Kes1p in yeast [5,24] and for ORP1S in Chinese hamster ovary cells [3,10].
Secretory competence of ORP1S deletion mutants
Growth phenotypes observed upon inactivation of Sec14p function normally correlate with the most apparent phenotypes associated with its inactivation, which are cessation of vesicular transport from the Golgi to the plasma membrane, as assessed by invertase secretion, and a substantially reduced rate of CPY transport from the Golgi to the vacuole [2,5,10,18]. To test if the alterations in growth observed upon expression of the ORP1S mutant proteins in the sec14ts kes1− yeast strain at the non-permissive temperature for the sec14ts allele were due to alterations in Sec14p-mediated vesicle transport, the effects of these proteins on both invertase secretion and CPY transport were determined.
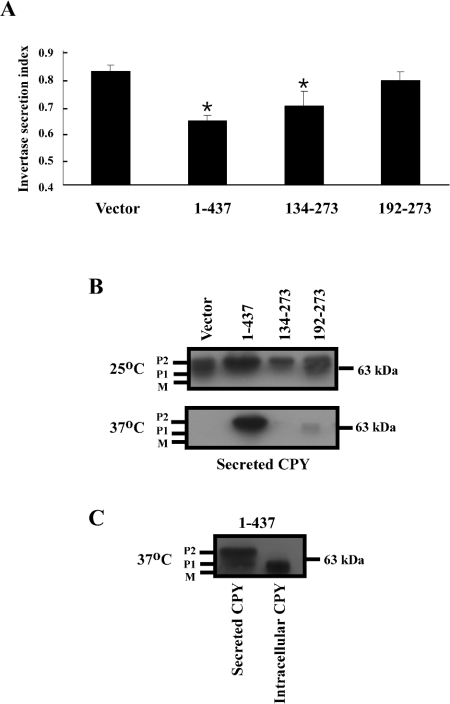
Invertase transcription is induced upon a shift from a high-glucose to a low-glucose medium. However, ORP1S and the deletion mutants derived from ORP1S were expressed from the GAL1 promoter, and thus glucose could not be used as a carbon source for growth, as ORP1S and its derivatives would not be expressed in glucose-containing medium. To circumvent this difficulty, invertase expression was driven by the constitutive TPI1 promoter, and thus expression of invertase was now independent of the carbon source. The constitutive expression of invertase results in a higher level of measured invertase secretion in cells with secretion blocks, due to pre-existing invertase being present in the periplasmic space before the secretion block is imposed; however, secretion blocks are still accurately reflected by a decrease in measurable secreted invertase [21]. As has been observed previously, sec14ts kes1− cells secreted most of their invertase, indicating that vesicular transport from the Golgi to the plasma membrane is intact (Figure 4A). Expression of full-length ORP1S, which prevented the growth of sec14ts kes1− cells as ORP1S can phenocopy Kes1p function, led to a decrease in invertase secretion even in the presence of pre-existing secreted invertase due to the promoter systems used. Expression of ORP1S-(134–273) or ORP1S-(192–273) resulted in intermediate levels of invertase secretion (Figure 4).
Figure 4. Regulation of vesicular transport by diagnostic linear regions of ORP1S.
(A) Strain CMY136 transformed with pDB31 (pTPI-SUC2; encodes invertase under the control of the constitutive TPI1 promoter) and the indicated ORP1S fragment under the control of the GAL1 promoter were grown at 25 °C to mid-exponential phase in minimal medium with 2% (w/v) galactose as carbon source. Cells were washed, shifted to 37 °C for 2 h, and invertase secretion was measured as described in the Experimental section. The results are means±S.D. of three separate experiments performed in triplicate. Student's two-tailed t test was used to determine significant differences from the vector control (*P<0.05). (B) ORP1S constructs expressed from the GAL1 promoter were grown as in (A). Cells were pelleted by centrifugation and proteins contained in the supernatant were precipitated by adding trichloroacetic acid, collected by centrifugation, washed with 100% acetone, dried, and resuspended in SDS/PAGE sample buffer. Samples were resolved by SDS/PAGE and processed for CPY immunoblots. (C) Pelleted cells from (B) were processed for CPY immunoblots and compared with secreted CPY levels to compare the processing fate of internal compared with secreted CPY. Results in (B) and (C) are representative of three separate experiments.
Normally, newly synthesized CPY is modified by N-glycosylation in the endoplasmic reticulum (P1 form), then transported to the Golgi, where it is modified further by core glycosylation (P2 form), and subsequently transported to the vacuole, where it undergoes regulated proteolysis to produce the mature (M) form of the enzyme. A delay or block in Golgi-to-vacuole transport often results in re-routing of the P1 and P2 forms of CPY into the plasma membrane secretion pathway [13,22]. Cells lacking Sec14p function possess defects in both the Golgi-to-vacuole and Golgi-to-plasma-membrane trafficking pathways, as defects in the kinetics of CPY processing to the vacuole from the Golgi and in invertase secretion from the cell are both associated with inactivation of Sec14p function [2,10,18]. It was determined previously that the rescue of the CPY Golgi-to-vacuole transport delay caused by inactivation of KES1 in cells lacking Sec14p function could be re-instated by expression of human ORP1S in sec14ts kes1− yeast [10]. In the present study we addressed the relative delays in Golgi-to-vacuole compared with Golgi-to-plasma-membrane transport in sec14ts kes1− cells grown at the non-permissive temperature for sec14ts by determining CPY secretion from these cells. In sec14ts kes1− cells grown at the permissive temperature, cells expressing full-length ORP1S, ORP1S-(134–173), ORP1S-(192–273) or empty vector all secreted P1 and P2 versions of CPY (Figure 4B). Growth at the non-permissive temperature resulted in unchanged levels of CPY secretion in sec14ts kes1− cells expressing full-length ORP1S, while cells expressing ORP1S-(134–273), ORP1S-(192–273) or empty vector displayed substantially reduced CPY secretion compared with that observed for the same cells grown at the permissive temperature. These results also imply that the Golgi-to-vacuole pathway is not as active as the Golgi-to-plasma-membrane vesicular transport route in sec14ts kes1− cells grown at the permissive temperature for sec14ts, resulting in some secretion of CPY regardless of the presence or absence of ORP1S. CPY was only secreted at the non-permissive temperature for sec14ts function in sec14ts kes1− cells expressing full-length ORP1S, indicating that Golgi-to-plasma-membrane transport of CPY is intact in cells that lack Sec14p function, but that CPY transport is prevented if both Sec14p and Kes1p function are lacking. Intracellular CPY in sec14ts kes1− cells expressing ORP1S was present in the mature form (Figure 4C), indicating that it is only the endoplasmic reticulum and Golgi forms of CPY that are being secreted.
DISCUSSION
The ORPs are a family of intracellular lipid binding receptors that appear to relate alterations in lipid metabolism to membrane transport within cells through as yet poorly defined mechanisms [1–8,10,11,14,16,25,26].
Kes1p and ORP1S are negative regulators of Sec14p function, as ablation of Kes1p function results in bypass of the normally essential requirement for Sec14p, and Kes1p function can be completely complemented by human ORP1S in this regard [2,5,10]. In the present work we determined that, in sec14ts kes1− yeast expressing ORP1S, the secretion of CPY was contrary to the phenotype observed for invertase secretion when compared with vector control cells, implying that (i) transport of CPY and invertase from the Golgi may be regulated differentially by Sec14p and ORP1S/Kes1p, or (ii) invertase and CPY are transported from the Golgi to the plasma membrane by different routes.
Our in vitro characterization of the phospholipid binding ability and specificity of purified proteins comprising ORP1S linear regions revealed that both specificity and binding could be localized to amino acid residues 192–273. Expression of the ORP1S deletion set in the sec14ts kes1− yeast strain allowed determination of the in vivo function of the phospholipid binding domain and other ORP1S analytical regions. The ORP1S phospholipid binding domain alone was unable to inhibit Golgi-derived vesicular transport, implying either that phospholipid binding is dispensable for the regulation of vesicular transport or that phospholipid binding must act in concert with another domain within ORP1S to facilitate inhibition of vesicular transport [11]. Our analysis of the entire ORP1S deletion set revealed that essentially the entire ORP1S protein is essential for inhibition of membrane transport, implying that all regions of ORP1S play a required role in the regulation of vesicular transport. Further studies of potential ORP1S/Kes1p-interacting proteins will aid in facilitating precise roles for each region.
Interpretation of the in vitro lipid binding results for ORP1S must be made with caution. Previous results have indicated that in vitro lipid binding specificity of ORP family members may not reflect in vivo ligands [5,27]. Using a liposome-based assay for lipid binding, the in vitro specificity of yeast Kes1p was highest for PI4,5P2; however, this did not correlate with the identity of its apparent in vivo ligand as PI4P [5]. Our overlay to assess potential Kes1p lipid ligands observed that Kes1p bound PI4P, PI3P, PI5P and PI3,4P2. As in vitro assays use purified protein and equimolar lipid amounts to assess specificity and affinity, and in vivo ligand binding is affected by the amount and location of each lipid within the cell and by competition for lipid binding by other cellular proteins whose amounts can in turn vary, in vitro results rarely allow one to draw conclusions with respect to in vivo lipid ligands. Indeed, previous in vitro analysis of ORP1S phospholipid binding specificity determined that PI3P and phosphatidic acid were optimal in vitro ligands [10]; however, in that study PBS was used as the buffer against which the purified ORPs were dialysed, while in the present work Tris-buffered saline was used, illustrating how minor alterations in in vitro buffer composition can alter lipid binding specificity and affinity.
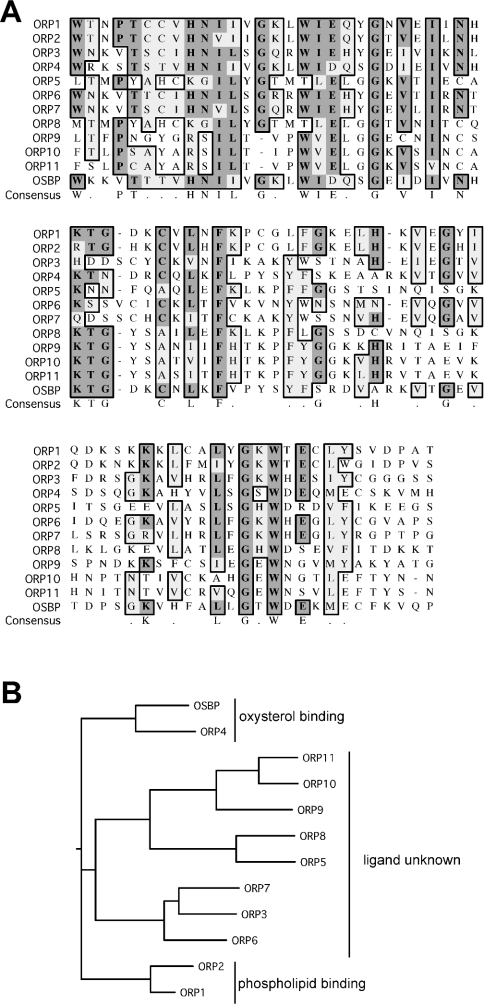
Regardless of lipid specificity, our results definitively identify a nominal phospholipid binding region for ORP1S. Alignment of this region of ORP1S with the analogous regions of all other human ORP family members, and dendogram analysis based on this region, identifies several interesting groupings (Figure 5). OSBP and ORP4 form one discrete group, and these two ORP family members have been demonstrated to bind sterols [1,6,9]. ORP1 and ORP2 have been found to bind phospholipids [10], and they form a separate discrete group that possesses the least degree of similarity to the lipid binding region of OSBP and ORP4. The ligand binding similarities of the remaining ORP family members are represented by a third arm of the dendogram that lies between these two discrete groups. The lipid binding specificities of these ORP family members have yet to be determined, and will require direct experimentation, as they cannot be predicted based on similarities within the nominal lipid binding region identified herein to ORPs of known ligand binding specificity. Interpreted in the light of the identification of the nominal phospholipid binding region of ORP1S, it can be suggested either (i) that the same region of OSBP/ORP4 binds sterols instead of phospholipids, or (ii) that OSBP/ORP4 bind sterols at a site different from the phospholipid binding region of ORP1 and ORP2, and that this region of OSBP/ORP4 may not bind lipids at all. Continued structural and functional analysis of ORPs, coupled with in-depth genetic analyses, should further our understanding of the shared biochemical roles of particular domains, along with disparate roles for each ORP family member in the regulation of lipid homoeostasis within cells and cellular organelles.
Figure 5. Alignment of the nominal phospholipid binding region of ORP family members.
(A) Sequence alignment of the nominal lipid binding region of human ORP1 with the same region of all other human ORP family members. Identical amino acid residues are boxed in dark grey, whereas similar residues are boxed in light grey. (B) Dendogram analysis of the nominal lipid binding region of human ORPs using CLUSTALW in SLOW pairwise mode using the BLOSUM30 matrix including hydrophobic and residue specific penalties.
Acknowledgments
We thank members of the Atlantic Research Centre for helpful discussions, and Randy Scheckman and Neale Ridgway for the TPI1-SUC2 and pGEX-OSBP-PH plasmids respectively. This work was supported by an operating grant from the National Sciences and Engineering Research Council of Canada, a Canada Research Chair (to C.R.M.), and an IWK Health Centre Graduate Studentship (to G.D.F).
References
- 1.Dawson P. A., Ridgway N. D., Slaughter C. A., Brown M. S., Goldstein J. L. cDNA cloning and expression of oxysterol-binding protein, an oligomer with a potential leucine zipper. J. Biol. Chem. 1989;264:16798–16803. [PubMed] [Google Scholar]
- 2.Fang M., Kearns B. G., Gedvilaite A., Kagiwada S., Kearns M., Fung M. K., Bankaitis V. A. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 1996;15:6447–6459. [PMC free article] [PubMed] [Google Scholar]
- 3.Johansson M., Bocher V., Lehto M., Chinetti G., Kuismanen E., Ehnholm C., Staels B., Olkkonen V. M. The two variants of oxysterol binding protein-related protein-1 display different tissue expression patterns, have different intracellular localization, and are functionally distinct. Mol. Biol. Cell. 2003;14:903–915. doi: 10.1091/mbc.E02-08-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagace T. A., Byers D. M., Cook H. W., Ridgway N. D. Chinese hamster ovary cells overexpressing the oxysterol binding protein (OSBP) display enhanced synthesis of sphingomyelin in response to 25-hydroxycholesterol. J. Lipid Res. 1999;40:109–116. [PubMed] [Google Scholar]
- 5.Li X., Rivas M. P., Fang M., Marchena J., Mehrotra B., Chaudhary A., Feng L., Prestwich G. D., Bankaitis V. A. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J. Cell Biol. 2002;157:63–77. doi: 10.1083/jcb.200201037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ridgway N. D., Dawson P. A., Ho Y. K., Brown M. S., Goldstein J. L. Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J. Cell Biol. 1992;116:307–319. doi: 10.1083/jcb.116.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridgway N. D., Lagace T. A., Cook H. W., Byers D. M. Differential effects of sphingomyelin hydrolysis and cholesterol transport on oxysterol-binding protein phosphorylation and Golgi localization. J. Biol. Chem. 1998;273:31621–31628. doi: 10.1074/jbc.273.47.31621. [DOI] [PubMed] [Google Scholar]
- 8.Storey M. K., Byers D. M., Cook H. W., Ridgway N. D. Cholesterol regulates oxysterol binding protein (OSBP) phosphorylation and Golgi localization in Chinese hamster ovary cells: correlation with stimulation of sphingomyelin synthesis by 25-hydroxycholesterol. Biochem. J. 1998;336:247–256. doi: 10.1042/bj3360247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C., JeBailey L., Ridgway N. D. Oxysterol-binding-protein (OSBP)-related protein 4 binds 25-hydroxycholesterol and interacts with vimentin intermediate filaments. Biochem. J. 2002;361:461–472. doi: 10.1042/0264-6021:3610461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y., Liu Y., Ridgway N. D., McMaster C. R. Novel members of the human oxysterol-binding protein family bind phospholipids and regulate vesicle transport. J. Biol. Chem. 2001;276:18407–18414. doi: 10.1074/jbc.M101204200. [DOI] [PubMed] [Google Scholar]
- 11.Lehto M., Olkkonen V. M. The OSBP-related proteins: a novel protein family involved in vesicle transport, cellular lipid metabolism, and cell signalling. Biochim. Biophys. Acta. 2003;1631:1–11. doi: 10.1016/s1388-1981(02)00364-5. [DOI] [PubMed] [Google Scholar]
- 12.Olkkonen V. M., Levine T. P. Oxysterol binding proteins: in more than one place at one time? Biochem. Cell Biol. 2004;82:87–98. doi: 10.1139/o03-088. [DOI] [PubMed] [Google Scholar]
- 13.Bankaitis V. A., Morris A. J. Lipids and the exocytotic machinery of eukaryotic cells. Curr. Opin. Cell Biol. 2003;15:389–395. doi: 10.1016/s0955-0674(03)00076-0. [DOI] [PubMed] [Google Scholar]
- 14.Beh C. T., Cool L., Phillips J., Rine J. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics. 2001;157:1117–1140. doi: 10.1093/genetics/157.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine T. P., Munro S. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr. Biol. 1998;8:729–739. doi: 10.1016/s0960-9822(98)70296-9. [DOI] [PubMed] [Google Scholar]
- 16.Levine T. P., Munro S. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol. Biol. Cell. 2001;12:1633–1644. doi: 10.1091/mbc.12.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loewen C. J., Roy A., Levine T. P. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cleves A. E., McGee T. P., Whitters E. A., Champion K. M., Aitken J. R., Dowhan W., Goebl M., Bankaitis V. A. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dowler S., Currie R. A., Campbell D. G., Deak M., Kular G., Downes C. P., Alessi D. R. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dowler S., Kular G., Alessi D. R. Protein lipid overlay assay. Sci. STKE 2002. 2002:PL6. doi: 10.1126/stke.2002.129.pl6. [DOI] [PubMed] [Google Scholar]
- 21.Brada D., Schekman R. Coincident localization of secretory and plasma membrane proteins in organelles of the yeast secretory pathway. J. Bacteriol. 1988;170:2775–2783. doi: 10.1128/jb.170.6.2775-2783.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belden W. J., Barlowe C. Deletion of yeast p24 genes activates the unfolded protein response. Mol. Biol. Cell. 2001;12:957–969. doi: 10.1091/mbc.12.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howe A. G., Zaremberg V., McMaster C. R. Cessation of growth to prevent cell death due to inhibition of phosphatidylcholine synthesis is impaired at 37 °C in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:44100–44107. doi: 10.1074/jbc.M206643200. [DOI] [PubMed] [Google Scholar]
- 24.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'shea E. K. Global analysis of protein localization in budding yeast. Nature (London) 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 25.Wyles J. P., McMaster C. R., Ridgway N. D. Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J. Biol. Chem. 2002;277:29908–29918. doi: 10.1074/jbc.M201191200. [DOI] [PubMed] [Google Scholar]
- 26.Roy A., Levine T. P. Multiple pools of PtdIns 4-phosphate detected using the pleckstrin homology domain of Osh2p. J. Biol. Chem. 2004;279:44683–44689. doi: 10.1074/jbc.M401583200. [DOI] [PubMed] [Google Scholar]
- 27.Levine T. P., Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr. Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]