Abstract
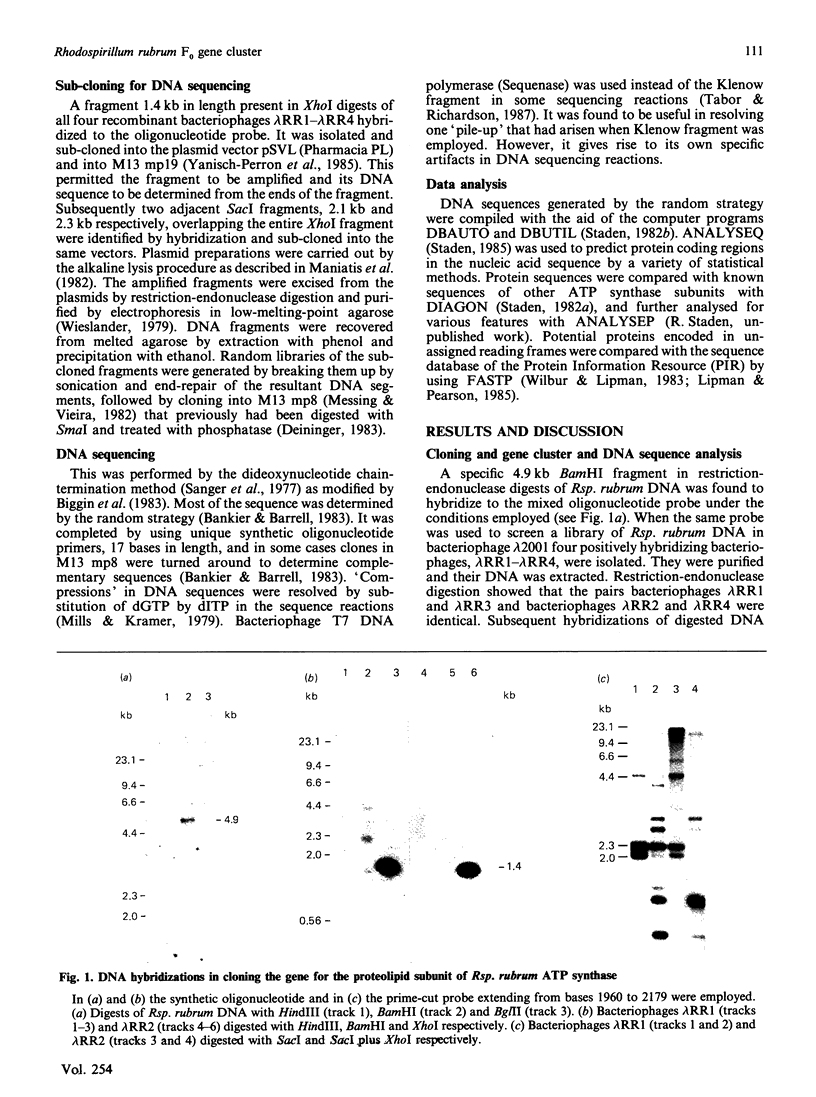
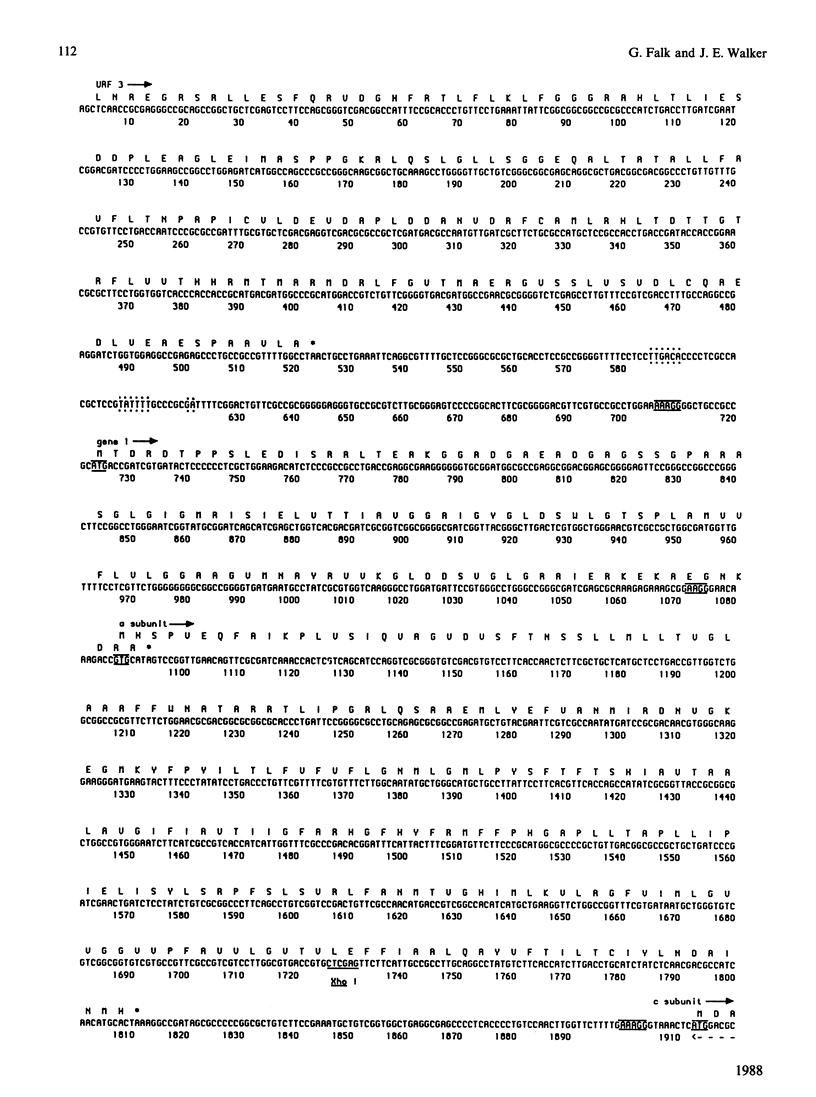
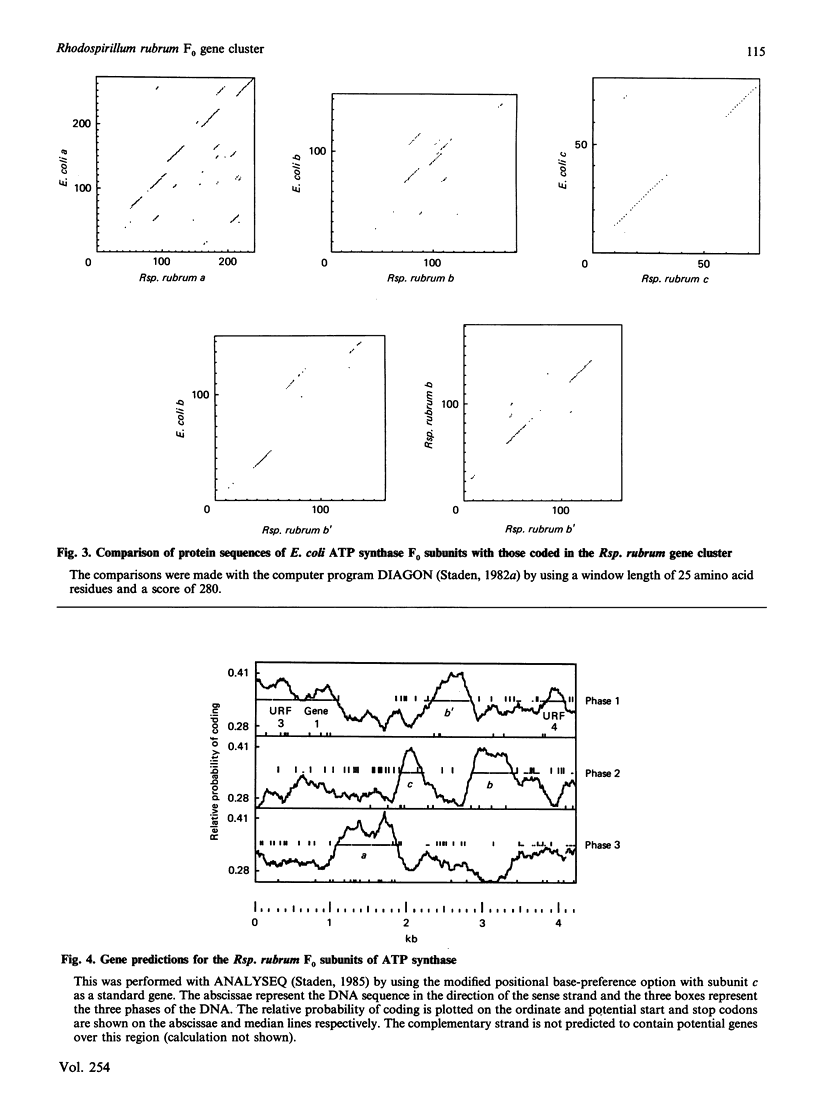
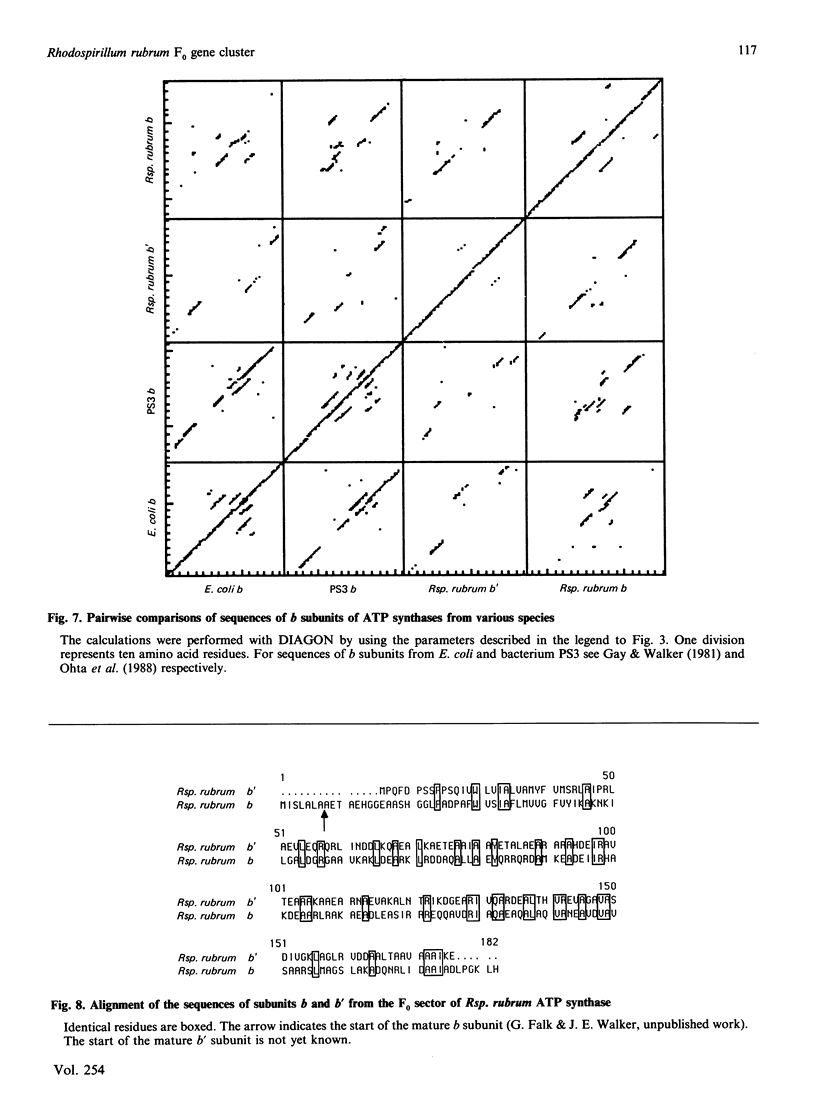
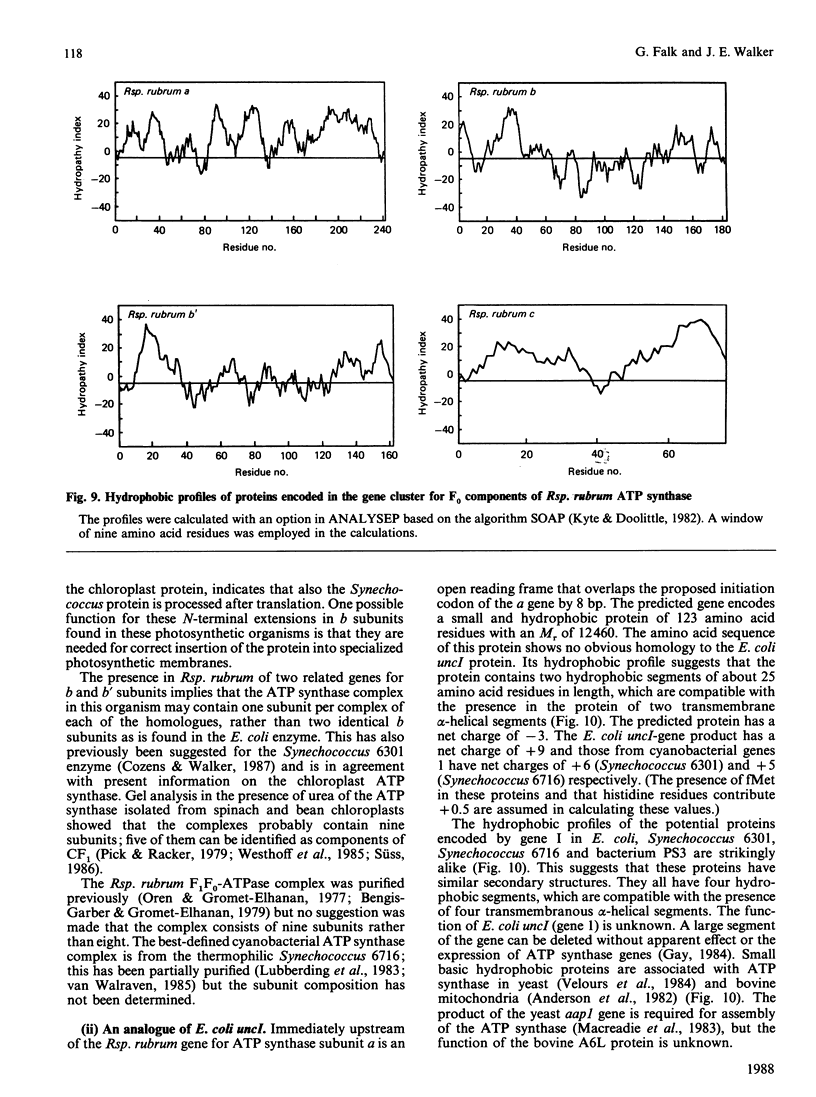
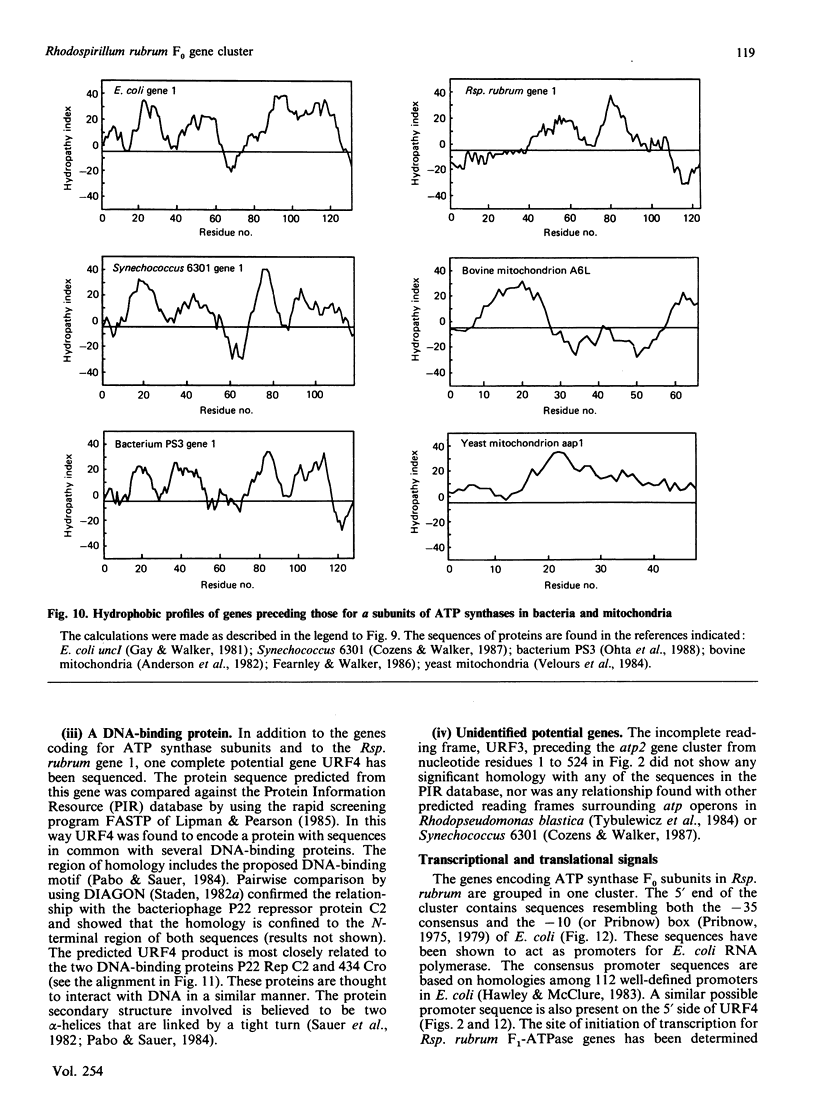
A region was cloned from the genome of the purple non-sulphur photobacterium Rhodospirillum rubrum that contains genes coding for the membrane protein subunits of the F0 sector of ATP synthase. The clone was identified by hybridization with a synthetic oligonucleotide designed on the basis of the known protein sequence of the dicyclohexylcarbodi-imide-reactive proteolipid, or subunit c. The complete nucleotide sequence of 4240 bp of this region was determined. It is separate from an operon described previously that encodes the five subunits of the extrinsic membrane sector of the enzyme, F1-ATPase. It contains a cluster of structural genes encoding homologues of all three membrane subunits a, b and c of the Escherichia coli ATP synthase. The order of the genes in Rsp. rubrum is a-c-b'-b where b and b' are homologues. A similar gene arrangement for F0 subunits has been found in two cyanobacteria, Synechococcus 6301 and Synechococcus 6716. This suggests that the ATP synthase complexes of all these photosynthetic bacteria contain nine different polypeptides rather than eight found in the E. coli enzyme; the chloroplast ATP synthase complex is probably similar to the photosynthetic bacterial enzymes in this respect. The Rsp. rubrum b subunit is modified after translation. As shown by N-terminal sequencing of the protein, the first seven amino acid residues are removed before or during assembly of the ATP synthase complex. The subunit-a gene is preceded by a gene coding for a small hydrophobic protein, as has been observed previously in the atp operons in E. coli, bacterium PS3 and cyanobacteria. A number of features suggest that the Rsp. rubrum cluster of F0 genes is an operon. On its 5' side are found sequences resembling the -10 (Pribnow) and -35 boxes of E. coli promoters, and the gene cluster is followed by a sequence potentially able to form a stable stem-loop structure, suggesting that it acts as a rho-independent transcription terminator. These features and the small intergenic non-coding sequences suggest that the genes are cotranscribed, and so the name atp2 is proposed for this second operon coding for ATP synthase subunits in Rsp. rubrum. The finding that genes for the F0 and F1 sectors of the enzyme are in separate clusters supports the view that these represent evolutionary modules.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., de Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., Young I. G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- Bengis-Garber C., Gromet-Elhanan Z. Purification of the energy-transducing adenosine triphosphatase complex from Rhodospirillum rubrum. Biochemistry. 1979 Aug 7;18(16):3577–3581. doi: 10.1021/bi00583a022. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird C. R., Koller B., Auffret A. D., Huttly A. K., Howe C. J., Dyer T. A., Gray J. C. The wheat chloroplast gene for CF(0) subunit I of ATP synthase contains a large intron. EMBO J. 1985 Jun;4(6):1381–1388. doi: 10.1002/j.1460-2075.1985.tb03790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusilow W. S., Porter A. C., Simoni R. D. Cloning and expression of uncI, the first gene of the unc operon of Escherichia coli. J Bacteriol. 1983 Sep;155(3):1265–1270. doi: 10.1128/jb.155.3.1265-1270.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozens A. L., Walker J. E. The organization and sequence of the genes for ATP synthase subunits in the cyanobacterium Synechococcus 6301. Support for an endosymbiotic origin of chloroplasts. J Mol Biol. 1987 Apr 5;194(3):359–383. doi: 10.1016/0022-2836(87)90667-x. [DOI] [PubMed] [Google Scholar]
- Deininger P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem. 1983 Feb 15;129(1):216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
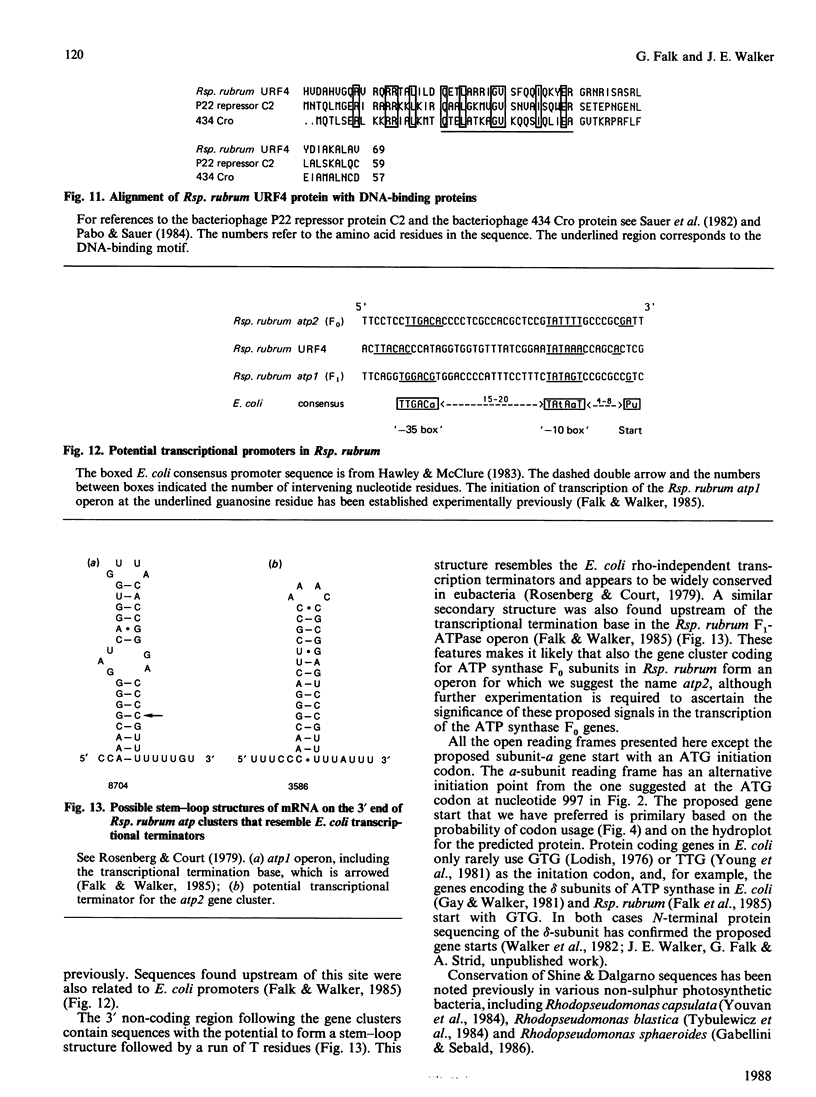
- Falk G., Hampe A., Walker J. E. Nucleotide sequence of the Rhodospirillum rubrum atp operon. Biochem J. 1985 Jun 1;228(2):391–407. doi: 10.1042/bj2280391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk G., Walker J. E. Transcription of Rhodospirillum rubrum atp operon. Biochem J. 1985 Aug 1;229(3):663–668. doi: 10.1042/bj2290663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell P. J., Deininger P. L., Bankier A., Barrell B. Homologous upstream sequences near Epstein-Barr virus promoters. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1565–1569. doi: 10.1073/pnas.80.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnley I. M., Walker J. E. Two overlapping genes in bovine mitochondrial DNA encode membrane components of ATP synthase. EMBO J. 1986 Aug;5(8):2003–2008. doi: 10.1002/j.1460-2075.1986.tb04456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster D. L., Fillingame R. H. Stoichiometry of subunits in the H+-ATPase complex of Escherichia coli. J Biol Chem. 1982 Feb 25;257(4):2009–2015. [PubMed] [Google Scholar]
- Friedl P., Hoppe J., Gunsalus R. P., Michelsen O., von Meyenburg K., Schairer H. U. Membrane integration and function of the three F0 subunits of the ATP synthase of Escherichia coli K12. EMBO J. 1983;2(1):99–103. doi: 10.1002/j.1460-2075.1983.tb01388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai M., Kanazawa H. Structure and function of proton-translocating adenosine triphosphatase (F0F1): biochemical and molecular biological approaches. Microbiol Rev. 1983 Sep;47(3):285–312. doi: 10.1128/mr.47.3.285-312.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini N., Sebald W. Nucleotide sequence and transcription of the fbc operon from Rhodopseudomonas sphaeroides. Evaluation of the deduced amino acid sequences of the FeS protein, cytochrome b and cytochrome c1. Eur J Biochem. 1986 Feb 3;154(3):569–579. doi: 10.1111/j.1432-1033.1986.tb09437.x. [DOI] [PubMed] [Google Scholar]
- Gay N. J. Construction and characterization of an Escherichia coli strain with a uncI mutation. J Bacteriol. 1984 Jun;158(3):820–825. doi: 10.1128/jb.158.3.820-825.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay N. J., Walker J. E. The atp operon: nucleotide sequence of the promoter and the genes for the membrane proteins, and the delta subunit of Escherichia coli ATP-synthase. Nucleic Acids Res. 1981 Aug 25;9(16):3919–3926. doi: 10.1093/nar/9.16.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe J., Friedl P., Schairer H. U., Sebald W., von Meyenburg K., Jørgensen B. B. The topology of the proton translocating F0 component of the ATP synthase from E. coli K12: studies with proteases. EMBO J. 1983;2(1):105–110. doi: 10.1002/j.1460-2075.1983.tb01389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe J., Montecucco C., Friedl P. Labeling of subunit b of the ATP synthase from Escherichia coli with a photoreactive phospholipid analogue. J Biol Chem. 1983 Mar 10;258(5):2882–2885. [PubMed] [Google Scholar]
- Hoppe J., Sebald W. The proton conducting F0-part of bacterial ATP synthases. Biochim Biophys Acta. 1984 Apr 9;768(1):1–27. doi: 10.1016/0304-4173(84)90005-3. [DOI] [PubMed] [Google Scholar]
- Johansson B. C., Baltscheffsky M., Baltscheffsky H. Purification and properties of coupling factor (Ca2+-dependent adenosine triphosphatase) from Rhodospirillum rubrum. Eur J Biochem. 1973 Dec 3;40(1):109–117. doi: 10.1111/j.1432-1033.1973.tb03174.x. [DOI] [PubMed] [Google Scholar]
- Johansson B. C., Baltscheffsky M. On the subunit composition of the coupling factor (ATPase) from Rhodospirillum rubrum. FEBS Lett. 1975 May 1;53(2):221–224. doi: 10.1016/0014-5793(75)80024-x. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Lodish H. F. Translational control of protein synthesis. Annu Rev Biochem. 1976;45:39–72. doi: 10.1146/annurev.bi.45.070176.000351. [DOI] [PubMed] [Google Scholar]
- Lubberding H. J., Zimmer G., van Walraven H. S., Schrickx J., Kraayenhof R. Isolation, purification and characterization of the ATPase complex from the thermophilic cyanobacterium Synechococcus 6716. Eur J Biochem. 1983 Dec 1;137(1-2):95–99. doi: 10.1111/j.1432-1033.1983.tb07800.x. [DOI] [PubMed] [Google Scholar]
- Lücke F. K., Klemme J. H. Coupling factor adenosine-5'-triphosphatase from Rhodospirillum rubrum: a simple and rapid procedure for its purification. Z Naturforsch C. 1976 May-Jun;31(5-6):272–279. [PubMed] [Google Scholar]
- MITCHELL P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961 Jul 8;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- Macreadie I. G., Novitski C. E., Maxwell R. J., John U., Ooi B. G., McMullen G. L., Lukins H. B., Linnane A. W., Nagley P. Biogenesis of mitochondria: the mitochondrial gene (aap1) coding for mitochondrial ATPase subunit 8 in Saccharomyces cerevisiae. Nucleic Acids Res. 1983 Jul 11;11(13):4435–4451. doi: 10.1093/nar/11.13.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F. H., Castro M. M., Aboul-ela F., Tinoco I., Jr Base pairing involving deoxyinosine: implications for probe design. Nucleic Acids Res. 1985 Dec 20;13(24):8927–8938. doi: 10.1093/nar/13.24.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J. E. Expression of the unc genes in Escherichia coli. J Bioenerg Biomembr. 1988 Feb;20(1):19–39. doi: 10.1007/BF00762136. [DOI] [PubMed] [Google Scholar]
- McCarthy J. E., Schairer H. U., Sebald W. Translational initiation frequency of atp genes from Escherichia coli: identification of an intercistronic sequence that enhances translation. EMBO J. 1985 Feb;4(2):519–526. doi: 10.1002/j.1460-2075.1985.tb03659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Mills D. R., Kramer F. R. Structure-independent nucleotide sequence analysis. Proc Natl Acad Sci U S A. 1979 May;76(5):2232–2235. doi: 10.1073/pnas.76.5.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. A chemiosmotic molecular mechanism for proton-translocating adenosine triphosphatases. FEBS Lett. 1974 Jul 15;43(2):189–194. doi: 10.1016/0014-5793(74)80997-x. [DOI] [PubMed] [Google Scholar]
- Ohta S., Yohda M., Ishizuka M., Hirata H., Hamamoto T., Otawara-Hamamoto Y., Matsuda K., Kagawa Y. Sequence and over-expression of subunits of adenosine triphosphate synthase in thermophilic bacterium PS3. Biochim Biophys Acta. 1988 Mar 30;933(1):141–155. doi: 10.1016/0005-2728(88)90064-3. [DOI] [PubMed] [Google Scholar]
- Ohtsuka E., Matsuki S., Ikehara M., Takahashi Y., Matsubara K. An alternative approach to deoxyoligonucleotides as hybridization probes by insertion of deoxyinosine at ambiguous codon positions. J Biol Chem. 1985 Mar 10;260(5):2605–2608. [PubMed] [Google Scholar]
- Oren R., Gromet-Elhanan Z. Coupling factor adenosine triphosphatase-complex of Rhodospirillum rubrum. Isolation of an oligomycin-sensitive Ca2+, Mg2+--ATPase. FEBS Lett. 1977 Jul 1;79(1):147–150. doi: 10.1016/0014-5793(77)80371-2. [DOI] [PubMed] [Google Scholar]
- Oren R., Weiss S., Garty H., Caplan S. R., Gromet-Elhanan Z. ATP synthesis catalyzed by the ATPase complex from Rhodospirillum rubrum reconstituted into phospholipid vesicles together with bacteriorhodopsin. Arch Biochem Biophys. 1980 Dec;205(2):503–509. doi: 10.1016/0003-9861(80)90133-2. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Pick U., Racker E. Purification and reconstitution of the N,N'-dicyclohexylcarbodiimide-sensitive ATPase complex from spinach chloroplasts. J Biol Chem. 1979 Apr 25;254(8):2793–2799. [PubMed] [Google Scholar]
- Pribnow D. Bacteriophage T7 early promoters: nucleotide sequences of two RNA polymerase binding sites. J Mol Biol. 1975 Dec 15;99(3):419–443. doi: 10.1016/s0022-2836(75)80136-7. [DOI] [PubMed] [Google Scholar]
- Robinson A., Austen B. The role of topogenic sequences in the movement of proteins through membranes. Biochem J. 1987 Sep 1;246(2):249–261. doi: 10.1042/bj2460249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982 Jul 29;298(5873):447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- Schneider E., Altendorf K. All three subunits are required for the reconstitution of an active proton channel (F0) of Escherichia coli ATP synthase (F1F0). EMBO J. 1985 Feb;4(2):515–518. doi: 10.1002/j.1460-2075.1985.tb03658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R., McLachlan A. D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982 Jan 11;10(1):141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tybulewicz V. L., Falk G., Walker J. E. Rhodopseudomonas blastica atp operon. Nucleotide sequence and transcription. J Mol Biol. 1984 Oct 25;179(2):185–214. doi: 10.1016/0022-2836(84)90465-0. [DOI] [PubMed] [Google Scholar]
- Velours J., Esparza M., Hoppe J., Sebald W., Guerin B. Amino acid sequence of a new mitochondrially synthesized proteolipid of the ATP synthase of Saccharomyces cerevisiae. EMBO J. 1984 Jan;3(1):207–212. doi: 10.1002/j.1460-2075.1984.tb01785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Gay N. J. E. coli F1-ATPase interacts with a membrane protein component of a proton channel. Nature. 1982 Aug 26;298(5877):867–869. doi: 10.1038/298867a0. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Gay N. J. The unc operon. Nucleotide sequence, regulation and structure of ATP-synthase. Biochim Biophys Acta. 1984 Sep 6;768(2):164–200. doi: 10.1016/0304-4173(84)90003-x. [DOI] [PubMed] [Google Scholar]
- Wieslander L. A simple method to recover intact high molecular weight RNA and DNA after electrophoretic separation in low gelling temperature agarose gels. Anal Biochem. 1979 Oct 1;98(2):305–309. doi: 10.1016/0003-2697(79)90145-3. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Young I. G., Rogers B. L., Campbell H. D., Jaworowski A., Shaw D. C. Nucleotide sequence coding for the respiratory NADH dehydrogenase of Escherichia coli. UUG initiation codon. Eur J Biochem. 1981 May;116(1):165–170. doi: 10.1111/j.1432-1033.1981.tb05314.x. [DOI] [PubMed] [Google Scholar]
- Youvan D. C., Alberti M., Begusch H., Bylina E. J., Hearst J. E. Reaction center and light-harvesting I genes from Rhodopseudomonas capsulata. Proc Natl Acad Sci U S A. 1984 Jan;81(1):189–192. doi: 10.1073/pnas.81.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]



