Abstract
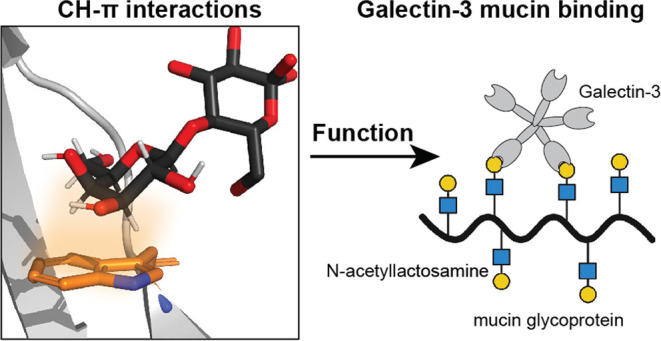
Glycan-binding proteins, or lectins, recognize distinct structural elements of polysaccharides, to mediate myriad biological functions. Targeting glycan-binding proteins involved in human disease has been challenging due to an incomplete understanding of the molecular mechanisms that govern protein–glycan interactions. Bioinformatics and structural studies of glycan-binding proteins indicate that aromatic residues with the potential for CH−π interactions are prevalent in glycan-binding sites. However, the contributions of these CH−π interactions to glycan binding and their relevance in downstream function remain unclear. An emblematic lectin, human galectin-3, recognizes lactose and N-acetyllactosamine-containing glycans by positioning the electropositive face of a galactose residue over the tryptophan 181 (W181) indole forming a CH−π interaction. We generated a suite of galectin-3 W181 variants to assess the importance of these CH−π interactions to glycan binding and function. As determined experimentally and further validated with computational modeling, variants with smaller or less electron-rich aromatic side chains (W181Y, W181F, W181H) or sterically similar but nonaromatic residues (W181M, W181R) showed poor or undetectable binding to lactose and attenuated ability to bind mucins or agglutinate red blood cells. The latter functions depend on multivalent binding, highlighting that weakened CH−π interactions cannot be overcome by avidity. Two galectin-3 variants with disrupted hydrogen bonding interactions (H158A and E184A) showed similarly impaired lactose binding. Molecular simulations demonstrate that all variants have decreased binding orientation stability relative to native galectin-3. Thus, W181 collaborates with the endogenous hydrogen bonding network to enhance binding affinity for lactose, and abrogation of these CH−π interactions is as deleterious as eliminating key hydrogen bonding interactions. These findings underscore the critical roles of CH−π interactions in carbohydrate binding and lectin function and will aid the development of novel lectin inhibitors.
Keywords: galectin, lectin, CH−π interactions, glycan binding, mucin, hemagglutination, multivalency
Introduction
The presence of glycans on the surface of cells emphasizes the need to understand how proteins recognize distinct carbohydrates.1−3 Lectins are nonantibody glycan-binding proteins found in all kingdoms of life that mediate cell–cell interactions, communicate vital information across cell types, and function in host defenses against pathogens.4−7 Galectins are a class of soluble lectins that act in these different capacities through binding β-galactosides. There are at least 12 known human galectins, which share a conserved protein fold in their respective carbohydrate recognition domains (CRDs).8 Galectins are categorized into three classes based on structure: prototype, chimeric, and tandem repeat galectins. Galectin-3, the only known chimeric galectin, is ubiquitously expressed throughout the body. Galectin-3 oligomerizes via its N-terminal domain and plays numerous roles both intracellularly and extracellularly in mediating immune responses, cell adhesion and differentiation, and pathogen clearance.9,10 Dysregulation of galectin-3 function has been implicated in pulmonary fibrosis, cardiovascular disease, and oncogenesis.11−14 The development of potent and selective glycan-mimetic inhibitors of galectin-3 function therefore presents a promising strategy to address a range of human diseases.15,16 To realize this promise, an enhanced understanding of galectin-3 glycan recognition is required.
Protein–ligand interactions are typically driven by the hydrophobic effect, in which a nonpolar ligand’s transition from water, a highly polar environment, into the less polar ligand binding site is enthalpically favored.17 Hydroxyl-rich glycans are far more hydrophilic than most other small molecule ligands. Thus, the glycan desolvation is less favorable enthalpically relative to most protein–small molecule interactions, and the increased entropy from water displacement from a binding pocket is not the major driver of glycan binding.
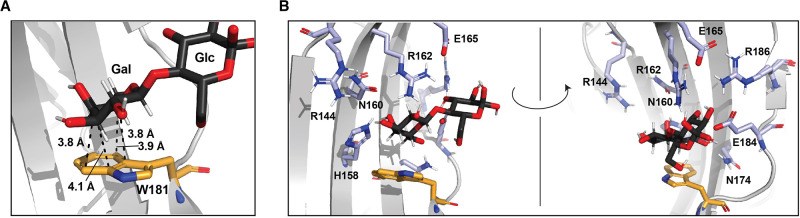
Lectins rely on at least three types of molecular interactions to drive ligand binding: hydrogen bonding, metal ion-mediated glycan hydroxyl group coordination, and CH−π interactions.18 The strength and relative contributions of both hydrogen bonding and metal ion–glycan interactions to lectin–glycan binding have been quantified in protein active sites.19 Increasing evidence suggests that CH−π interactions, which involve donation of electron density from an aromatic system into the σ* orbital of a C–H bond, are key contributors to glycan binding.20−23 Previous bioinformatic studies found that aromatic residues, primarily tryptophan, are enriched in the binding sites of glycan-binding proteins.24 Galectin-3 bears a single tryptophan, W181, in its carbohydrate recognition domain (CRD) that forms CH−π interactions with terminal galactosides and poly-N-acetyllactosamine (LacNAc) motifs (Figure 1A).25 All known mammalian galectins possess a tryptophan residue in an analogous position, further suggesting that this residue plays a role in binding glycans.26
Figure 1.
Structure of the galectin-3 binding site. (A) The structure of the human galectin-3-lactose complex determined by X-ray crystallography to 0.89 Å resolution (PDB code 3ZSJ). The lactose ligand (black) and W181 (light orange) are aligned to engage in a CH−π interaction. The C–H bonds at positions 3, 4, 5, and 6 of the galactose residue are oriented over the indole ring with a favorable arrangement for the formation of three CH−π interactions. (B) Close-up views of key galectin-3 residues that can form hydrogen bonding interactions with lactose.
The galectin-3 CRD is composed of two antiparallel β-sheets, creating an extended binding site divided into subsites A, B, C, and D that can accommodate as many as four monosaccharide units.27 These subsites may also interact with the side chains of glycosylated proteins, including mucins or mucin fragments, as suggested by the tighter binding of glycosylated proteins or peptides.28 With regard to saccharide binding, however, subsite C, which governs binding to β-galactoside-containing polysaccharides, is composed of a conserved series of amino acids, including H158, N160, R162, N174, and E184, which are positioned in close enough proximity to form hydrogen bonds with LacNAc (Figure 1B). The galectin-3 CRD additionally contains W181 located centrally in the binding pocket. As indicated by a high-resolution structure solved using X-ray crystallography, the indole ring of W181 is aligned with the electropositive face of galactose.27 The C3, C4, C5, and C6 C–H bonds of galactose are located approximately 4 Å away from the galectin-3 W181 residue, placing these aliphatic protons in van der Waals contact with the indole ring in a face-on orientation. This protein–ligand alignment positions the key W181 indole ring in an ideal orientation to form CH−π interactions, suggesting that these interactions play a role in galectin-mediated carbohydrate binding. A single amino acid mutation, W181R, has been identified in gastric cancer, suggesting that this CH−π interaction may be required for galectin-3 function.29 Conversely, human galectin-related protein (GRP) shares high sequence homology with other human galectins but does not bind carbohydrates. GRP has an arginine located in a corresponding position to W181, suggesting that hydrogen bonding alone is not sufficient for galactose binding.30 Thus, with the goal of further understanding the molecular mechanisms of lectin function, we sought to determine how these CH−π interactions collaborate with the hydrogen bonding to mediate galectin-3 glycan binding and function.
Previous studies used techniques, including nuclear magnetic resonance (NMR) spectroscopy, X-ray crystallography, and molecular modeling, to quantify the strength of CH−π interactions between glycans using peptides or small molecule systems.31−35 NMR-based studies have shown that phenol, benzene, and indole, present in the aromatic side chains of tyrosine, phenylalanine, and tryptophan respectively, engage in CH−π interactions with β-galactose in water.36 These aromatic π systems preferentially interact with C–H bonds of the alpha face of β-galactose even in the absence of a structured protein active site. Importantly, the indole π system of the tryptophan side chain forms stronger CH−π interactions relative to the less electron-rich aromatic systems in the phenylalanine or tyrosine side chains.20,37 Using a tryptophan-containing β-hairpin model system to eliminate alternative cooperative noncovalent interactions, the magnitude of CH−π interactions between the indole π system and glucose or galactose derivatives was calculated to be small (−0.5 to −0.8 kcal/mol).31,32 In contrast, NMR and computational studies suggest that CH−π interactions between fucose and benzene can contribute 3.0 kcal/mol to binding.36
The previous studies establish that CH−π interactions can contribute to the molecular recognition of carbohydrates by synthetic receptors and to glycan binding in model systems. The relative contributions and potential cooperativity of CH−π interactions with other noncovalent interactions, such as hydrogen bonding, that can occur in protein–glycan interactions are poorly characterized. Thus, our understanding of the contribution of CH−π interactions to glycan binding in the context of a full protein active site and their role in the function of glycan-binding proteins is limited.20
To this end, we investigated the role of CH−π interactions in galectin-3 ligand binding and downstream function. Specifically, we designed a series of galectin-3 W181 variants with aromatic and aliphatic amino acids in place of the native tryptophan to evaluate the contributions of this key interaction. We found that CH−π interactions are required for galectin-3 function. Using isothermal titration calorimetry (ITC), biolayer interferometry (BLI), and further validation with computational modeling using molecular dynamics, we show that disruption of either CH−π interactions or hydrogen bonding interactions via alteration of a single amino acid can abrogate glycan binding. These findings indicate that CH−π and hydrogen bonding interactions functionally cooperate to drive glycan binding by galectin-3. We also show that CH−π interactions are required for downstream functions involving multivalent interactions: hemagglutination and mucin binding. These results highlight the essentiality of CH−π interactions in galectin glycan binding and function, providing further insights into the molecular recognition of glycan ligands by lectins.
Results
Single Amino Acid Variants of W181 Galectin-3C Are Stably Folded
The functional role of CH−π interactions in galectin-3 was assessed using a toolkit of galectin-3 W181 variants (Figure S1). We tested the importance of CH−π interactions on galectin-3 function by generating W181 variants with aromatic systems that vary in size and electron density, as well as variants with nonaromatic side chains. To this end, we examined tyrosine (W181Y), phenylalanine (W181F), and histidine (W181H) variants. Our previous analysis of the Protein Data Bank found that tryptophan is the most enriched, but tyrosine is also overrepresented in carbohydrate binding sites whereas phenylalanine is not.20 Thus, we further aimed to learn whether these bioinformatic data reflect differences in carbohydrate binding capacity. We also designed a variant with histidine (W181H), which retains an aromatic ring but is much less electron-rich relative to tryptophan and typically serves as a hydrogen bond donor rather than a CH−π acceptor. We also replaced W181 with methionine (W181M) to examine the effects of a nonaromatic yet still hydrophobic amino acid on galectin-3 glycan binding. Finally, we tested W181R, as this galectin-3 variant occurs in gastric cancer but the mechanism of pathophysiology is not known.29
To evaluate the glycan-binding capacity of these proteins and eliminate downstream function as a potential confounding variable, we recombinantly produced only the C-terminal carbohydrate recognition domain (galectin-3C) for wild type (WT) and all variants. Following expression and purification, we compared the stabilities of the galectin-3C (Gal3C) variants and WT by differential scanning fluorimetry (DSF) using Sypro Orange, a hydrophobic dye that increases in fluorescence emission upon increasing interaction with hydrophobic residues exposed during protein thermal denaturation.38 We found that WT Gal3C, W181Y Gal3C, W181F Gal3C, W181M Gal3C, and W181H Gal3C were stable at room temperature and thus could be further evaluated for glycan binding (Figure 2A, Figure S2). W181R Gal3C showed no meaningful change in fluorescence emission over the course of the experiment, suggesting that this variant is unable to fold. Thus, we did not use this protein in functional assays. Representative melting curves demonstrate a clear difference in signal between proteins that are stably folded (WT Gal3C, W181Y) as compared to W181R (Figure 2B). WT Gal3C was more stable than all W181 variants. The Tm of W181M Gal3C was slightly less than WT, and variantsW181Y, W181F, and W181H all had a decrease in Tm of at least 10 °C. Pilot studies indicated that an alanine variant (W181A) does not fold, so we did not use it in further studies. Notably, among the three folded aromatic W181 variants, the tryptophan derivative had the highest stability.
Figure 2.
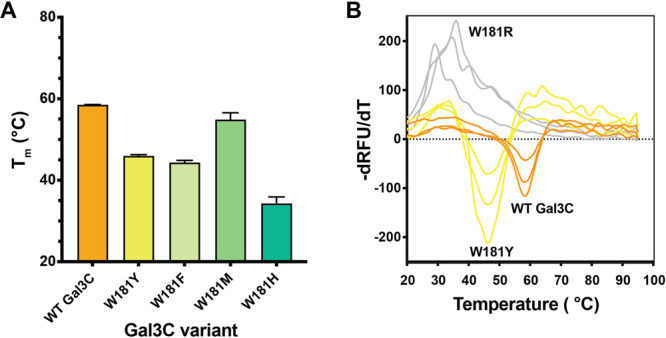
Stability of Gal3C variants. (A) The melting temperature (Tm) of wild-type galectin-3C (WT Gal3C) and W181 variants as measured by differential scanning fluorimetry (DSF) using Sypro Orange dye. All variants tested, with the exception of W181R. displayed a Tm of at least 10 °C greater than 25 °C, the temperature used for downstream glycan binding and functional assays. Bars and whiskers indicate mean ± SEM from n = 3 independent biological replicates. (B) Representative DSF melting curves for wild-type, W181Y, and W181R Gal3C. Melting temperatures were determined from the first derivative of fluorescence with respect to temperature.
Gal3C Variants Have Reduced Binding Affinity for Lactose
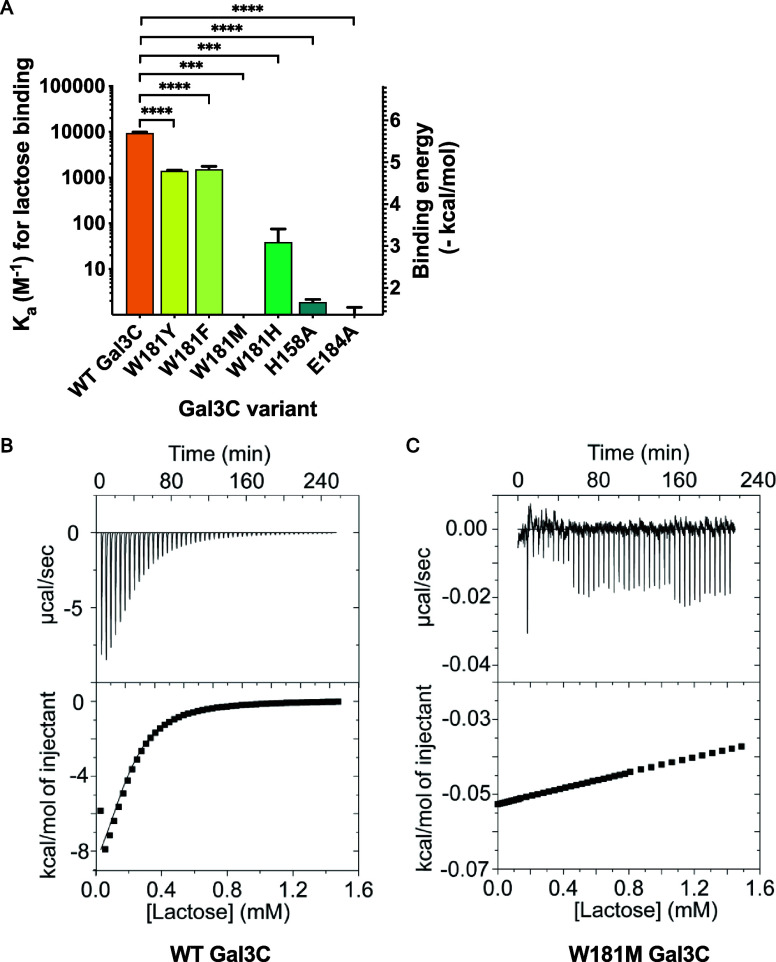
We hypothesized that disruption of CH−π interactions between W181 and lactose, a canonical galectin-3 ligand, would decrease binding. With a suite of stable W181 Gal3C variants in hand, we used isothermal titration calorimetry (ITC) to quantify their lactose-binding capacity. Because the affinities of lectin–glycan complexes are relatively weak as compared to many other types of protein–ligand interactions, we specifically designed our ITC experiments to enhance the precision of our measurements. First, when warranted, we used three injection sizes (2, 5, and 10 μL) to afford a more complete coverage of the binding curve at low lactose concentrations. Second, we analyzed experiments using the dissociation constant (Kd) as our primary determination of lactose binding as opposed to ΔH because the dissociation constant is less sensitive to errors caused by weak binding affinity. Third, we set the number of sites (N) equal to 1 to determine the association constant.39
Using this experimental protocol, we found that the Kd of WT Gal3C for lactose is 110 μM, corresponding to a binding energy of 5.4 kcal/mol (Figure 3A,B, Table S1). Substitution of W181 with any other residue weakens the lactose interaction (Figure S3). The phenylalanine and tyrosine variants, which are smaller and less electron-rich than the native tryptophan, had approximately 10-fold lower affinities for lactose, corresponding to a loss in binding energy of 1.1 kcal/mol. Substitution of tryptophan with histidine, an electron-poor aromatic system, decreases affinity of Gal3C for lactose by 200-fold, corresponding to a 3.3 kcal/mol loss in binding energy. Substitution of tryptophan with methionine abrogated any detectable binding of Gal3C to lactose (Figure 3C). Collectively, these results underscore the importance of CH−π interactions formed by the conserved tryptophan in glycan binding.
Figure 3.
Binding of Gal3C variants to lactose measured by isothermal titration calorimetry (ITC). (A) Affinity of galectin-3C variants. Experiments were conducted at 25 °C in galectin-3 assay buffer. Bars and whiskers indicate mean ± SEM from at least n = 3 independent biological replicates. (B) Representative ITC trace for wild-type Gal3C binding to lactose (Kd = 110 μM). (C) Representative ITC trace for W181M galectin-3C titration with lactose (no detectable binding).
To compare the contributions of the putative CH−π interactions to those of the endogenous hydrogen bonding network in Gal3C, we also made two variants with disrupted hydrogen bonding interactions. These include alanine substitutions at active site residues H158 and E184 to abrogate hydrogen bonding interactions with glycan ligands. The H158A and E184A variants were stably folded at room temperature (Figures S2B,C and S4A). We evaluated lactose binding H158A and E184A using our previously described ITC protocol and found no detectable interaction (Figure 3A, Figure S4C,D). Thus, substitution of W181 with a nonaromatic amino acid (W181M) or substitution of H158 or E184 with alanine completely abolished binding of galectin-3 to lactose. These findings indicate that both hydrogen bonding and CH−π interactions are required for lactose binding. Although hydrogen bonding is known to be critical for glycan binding, our studies indicate that CH−π interactions are at least as important.
Gal3C Variants Do Not Bind LacNAc as Assessed by Biolayer Interferometry
Because of the difficulties of measuring relatively weak protein–ligand binding by ITC, we sought complementary evidence of variant decreases in ligand binding. We therefore used biolayer interferometry (BLI). Specifically, we loaded streptavidin-coated biosensors with biotinylated LacNAc and incubated them with varying concentrations of the four proteins (Figure S5). As a control for specific LacNAc binding, streptavidin-coated biosensors were exposed to biotin prior to incubation with the Gal3C variants. Whereas WT Gal3C showed robust binding to LacNAc above 0.6 μM, W181M Gal3C, H158A Gal3C, and E184A Gal3C did not bind. In combination with the ITC data, these results indicate critical roles in glycan binding not only for hydrogen bonding but also for CH−π interactions.
Gal3C Variants Demonstrate Differential Binding Orientations by Molecular Dynamics Analysis
Gal3C variants W181M, H158A, and E184A had a binding affinity for lactose below detectable levels as determined by ITC and BLI. As such, we utilized a complementary means, molecular dynamics (MD) simulations, to evaluate the glycan-binding properties of these proteins (see “Computational analysis of galectin-3 and variants” in the Supporting Information for methods). Atomic coordinates for WT Gal3C with bound lactose (PDB: 3ZSJ) were used to design W181M, H158A, and E184A variants, which we employed to assess the effects of the deleterious mutations on binding conformation, ligand orientation, and overall energetics (Tables S2 and S3).27
Over the course of our MD simulations, WT Gal3C maintained a lactose-binding orientation consistent with the X-ray-determined structure, with limited dynamic variability, enabling the glucose subunit to form favorable hydrogen bonds with adjacent charged arginine and glutamate residues (Figure 4A,C and Figure S6). All variants had higher dynamic variability than the native protein and occupied at least one alternate orientation in which the lactose ligand rotates to adopt a different orientation (Figure S6). This latter configuration caused galactose to form hydrogen bonds with partners not observed with the native protein. Although some CH−π interactions with W181 were maintained, the E184A and H158A variant proteins often forced the glucose residue out of the binding pocket (Figures S6–S8). In the E184A variant, the lactose adopts an orientation that differs by 180° (Figure S6C). The consequences are that the glucose residue forms hydrogen bonds with H158, R162, N174, and CH−π interactions with W181, whereas the galactose residue engages in hydrogen bonds with W181 and K176 (Figures S6 and S7). During the W181M MD simulations, we also observed complete dissociation of lactose from the binding site (Figure 4B,D). These findings are consistent with the lack of detectable binding to the Gal3C W181, H158A, and E184 variants.
Figure 4.
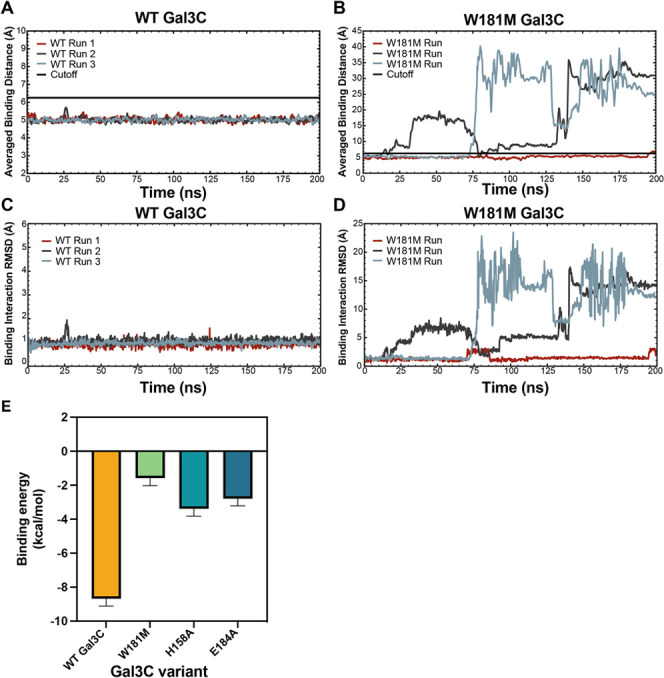
Molecular dynamics simulations of WT vs W181M Gal3C and binding free energy of WT Gal3C and variants. Averaged binding distances over time between WT Gal3C (A) or W181M (B) and the lactose ligand using a 1 ns window. Binding distance is defined as the distance between the center of mass of the lactose ligand and the center of mass of the binding pocket residues H158, R162, W181 (or M181), and E184 in Å. Each 200 ns run is shown in brick, dark grey, and slate blue. The cutoff value of 6.25 Å that is used to determine if the ligand is bound to galectin-3 is shown in black. (C, D) Total RMSD in Å of all atoms in the ligand and the interacting WT (C) or W181M (D) protein residues, H158, R162, W181 (or M181), and E184, for all three simulations. (E) Binding free energy of WT Gal3C, W181M, H158A, and E184A computed with MMGBSA. Bars and whiskers indicate mean ± SEM.
Nearly all binding orientations observed during MD simulations for WT Gal3C and Gal3C variants involve putative CH−π interactions between C–H groups on lactose and Gal3C residues. Even the W181M variant, which cannot form CH−π interactions with position 181, occupies a binding orientation in which H158 rotates to afford compensatory CH−π interactions with the galactose residue (Figure S6). Although CH−π interactions are consistently present in all MD simulations, the participating hydrogen atoms vary and can involve three to four C–H moieties on galactose (all proteins), two to three C–H moieties on glucose (E184A), or a combination of both (H158A and E184A) (Figures S6–S8). Within the binding orientations that contain CH−π interactions between galactose and W181 (e.g., WT Gal3C, W181F, W181Y, and W181H), the interacting hydrogen atoms on the galactose residue vary yet always include at least two hydrogen atoms that reside on carbon atom, 1, 3, 4, 5, or 6 (Figure 4 and Figures S6–S8).
Modeled Binding Affinities for the Gal3C Variants Are Reduced
To evaluate how the conformational differences above relate to binding affinity and determine the contributions from each amino acid, we performed energy decomposition analysis using molecular mechanics-generalized Born and surface area solvation (MMGBSA) to compute the binding free energies (i.e., binding affinities) from the simulations as described in the methods.40 The WT Gal3C binding affinity computed by MMGBSA is −8.7 kcal/mol (Figure 4E and Table S4), similar to the binding affinity of 5.4 kcal/mol determined by ITC. The variant protein with the next strongest calculated binding affinity is −3.4 kcal/mol of H158A, with a binding affinity (Figure 4E and Table S4). This variant also had the least variability in observed binding orientations (Figure S8). These observations suggest some binding interaction between the H158A variant and lactose. Neither ITC nor BLI could detect such affinity, suggesting that the interaction is not strong enough to be physiologically relevant (Figure 3A). The W181M and E184A variants have calculated affinities of −2.8 and −1.4 kcal/mol, respectively (Figure 4E and Table S4). These are weaker calculated affinities than H158A, suggesting that the interactions are weak and not physiologically relevant.
MMGBSA calculations permit decomposition of the interaction strength into contributions from each residue. For WT GalC, this analysis shows that R162, which participates in hydrogen bonding with up to three oxygens on lactose, contributes approximately −7 kcal/mol, the largest favorable binding enthalpic contribution for all variants (Figure S9 and Table S5). W181 has the next highest binding enthalpy contribution of approximately −3 kcal/mol for all simulated proteins excluding W181M (Figure S9 and Table S5). The loss of the Trp residue, as explored in the W181M variant, results in a substantial decrease in overall binding affinity, emphasizing the importance of these CH−π interactions (Figure 4E). H158 and E184 have considerably smaller binding enthalpy contributions as they form longer and less consistently maintained hydrogen bonds. In the case of E184, they may be less favorable than those formed by water (Figure S9 and Table S5). Whereas the H158A variant binding affinity is minimally affected as expected, the E184A variant binding affinity is substantially perturbed because the loss of this residue results in broken salt bridges with two nearby arginine residues R183 and R186, shifting the binding environment substantially (Figure 4E).
Gal3C Variants Minimally Bind Porcine Mucin Glycoproteins
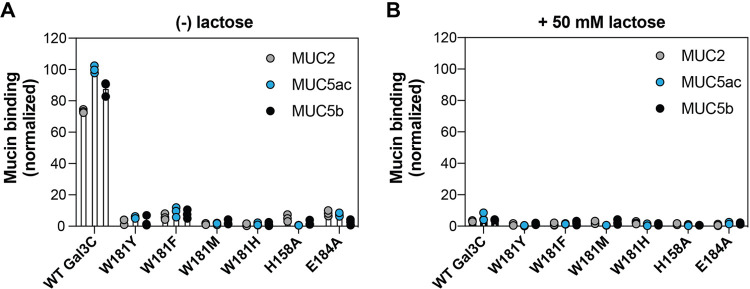
We next evaluated the consequences of disrupting either galectin-3 CH−π interactions or the hydrogen bonding network on the protein’s function. Galectin-3 is abundant at mucosal barriers, where mucins, highly glycosylated proteins that form major structural component of mucus, are highly expressed.41 Mucin glycans help reduce microbial virulence, trap microbial pathogens, and serve as a nutrition source for commensal bacteria.42−44 Both cell surface and secreted mucins bear N-acetyl-lactosamine repeats, and galectin-3 association with surface mucins MUC1 and MUC16 contributes to maintaining epithelial barriers.45−47 We therefore evaluated the capacity of WT Gal3C and all W181, H158, and E184 variants to bind three distinct secreted mammalian mucins found at epithelial surfaces. This assay involves multivalent interactions and thereby provides a distinct test of the role of the importance of different side chain contributions to binding. Specifically, we tested binding of these variants to MUC2, which is primarily expressed in the gastrointestinal tract, and MUC5ac and MUC5b, which are primarily expressed in the respiratory tract.48 We evaluated binding via a dot blot assay, where purified porcine mucins were initially immobilized onto PVDF membranes. Following a blocking step, membranes were incubated with either lectin alone or the lectin in the presence of 50 mM lactose as a competitive inhibitor.
WT Gal3C bound strongly to MUC2, MUC5ac, and MUC5b (Figure 5A). This binding was inhibited with the addition of lactose, suggesting that Gal3C's mucin binding is mediated through glycan recognition (Figure 5B). W181F Gal3C and W181Y Gal3C, variants that retain aromatic amino acid side chains and weak CH−π interactions, showed slight yet detectable binding to all three tested mucins. Again, lactose inhibited these interactions. Similarly, the E184A and H158A Gal3C variants also retained some mucin binding. Conversely, the binding of W181M Gal3C and W181H Gal3C was undetectable. Collectively, these results indicate that changes in a single amino acid involved in hydrogen bonding or CH−π interactions in Gal3C significantly decrease the capacity of the protein to bind physiological ligands. This further highlights that these two types of interactions are each critical for recognition and demonstrates that CH−π interactions with W181 are required for Gal3C’s mucin binding function.
Figure 5.
Binding of Gal3C and variants to isolated porcine mucins. (A) Determination of strep-tagged WT Gal3C or variant binding to porcine mucins MUC2, MUC5ac, and MUC5b using a dot blot assay. Purified mucins were spotted onto PVDF membranes and incubated with 10 μM of WT Gal3C or variant bearing a C-terminal His6 tag. Protein binding was visualized with an anti-His6 HRP-conjugated antibody. (B) Binding of WT Gal3C or variant with the addition of 50 mM lactose as a competitive glycan ligand. Mucin binding was quantified with the Fiji image suite and normalized to the highest intensity signal observed. Bars and whiskers indicate mean ± SEM from n = 3 independent biological replicates.
Galectin-3 Variants Have Decreased Hemagglutination Capacity
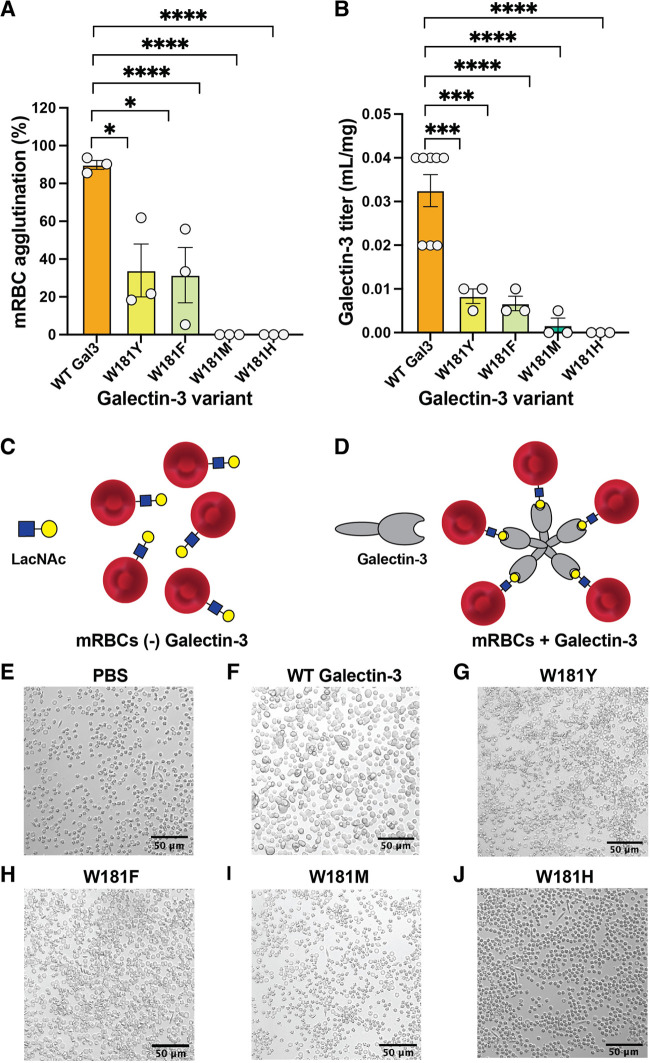
To further investigate the importance of the W181 CH−π interactions, we asked whether disrupting these interactions impairs hemagglutination. Galectin-3 can agglutinate murine and human erythrocytes, a process that depends on multivalent binding.49,50 This activity is driven by binding of the C-terminal CRD to extended lactosamine-containing N-glycans on erythrocyte membrane proteins and further oligomerization of the protein mediated by the N-terminal domain.51,52 We therefore tested whether W181 variants could agglutinate murine erythrocytes to a similar level as WT. Because agglutination requires the N-terminal domain in addition to the CRD, we cloned and expressed full-length galectin-3 W181 variants. Native galectin-3 (200 nM) showed robust murine erythrocyte (mRBC) agglutination (Figure 6A). Consistent with previous calorimetry and mucin binding data, the W181F and W181Y variants agglutinated mRBCs at 6.25 μM but not at lower concentrations. Thus, these variants are approximately 30-fold less potent. No hemagglutination was observed with W181M or W181H variants. We obtained similar results using a plate-based hemagglutination assay, where galectin-3 W181 variants with a abrogated CH−π interactions could not agglutinate mRBCs (Figure 6B). Thus, disrupting the W181 CH−π interactions has a dramatic effect on hemagglutination activity and cell clustering, highlighting the critical role of these interactions in galectin-3 function.
Figure 6.
Agglutination of mouse red blood cells (mRBCs) by galectin-3 and variants. (A) Percentage agglutination of mouse red blood cells by full-length galectin-3 and W181 variants following 30 min of treatment. mRBC exposure to the W181H or W181M galectin-3 variants did not result in hemagglutination at any concentration, nor did treatment with PBS as a negative control. (B) Agglutination of a 0.5% suspension of mouse red blood cells in 96-well plates using dilutions of galectin-3 and W181 variants in PBS. Titer is shown in inverse concentration units. Plots depict at least n = 3 independent biological replicates. (C) mRBCs with LacNAc in the absence of galectin-3. (D) Proposed model of galectin-3-mediated mRBC agglutination by binding multiple blood cells per pentamer in a galectin lattice. Representative images of mRBCs treated with PBS (E), 200 μg/mL WT galectin-3 (F), W181Y (G), W181F (H), W181M (I), or W181H (J). Bars and whiskers indicate mean ± SEM. *P < 0.05; ***P < 0.001; ****P < 0.0001 by analysis of variance (ANOVA) with Tukey’s multiple comparison test.
Discussion
Lectin–glycan interactions play critical roles in biology. Galectin-3 is a soluble lectin that binds to β-galactoside-containing residues, and galectin-3-mediated inflammatory signaling is associated with human diseases including pulmonary fibrosis, cardiovascular disease, oncogenesis, coronary artery disease, and Alzheimer’s disease.11−14,53,54 A highly conserved tryptophan residue, W181, forms CH−π interactions with galectin-3 glycan ligands. However, the relative contribution of these CH−π interactions to galectin-3-mediated glycan binding and function has not been evaluated.
We thus designed and expressed a series of Gal3C W181 variants where we replaced the native tryptophan with amino acids containing smaller aromatic systems (W181F, W181Y), an electron-poor aromatic ring (W181H), or a hydrophobic, nonaromatic side chain (W181M). As evaluated by ITC, relative to WT Gal3C, the W181F, W181Y, and W181H variants all exhibit reduced lactose binding. Notably, the W181H variant, which possesses the most electron-poor aromatic system, suffered the greatest loss of binding affinity. Moreover, the W181M variant, which has a similar stability as native Gal3C, exhibited no detectable binding to lactose, indicating that the proximity of a nonpolar surface to the C–H bonds of a β-galactose residue fails to promote binding. These results are inconsistent with a model in which the main role of the aromatic residue is to engage solely in hydrophobic interactions. They indicate that an electron-rich aromatic residue facilitates recognition of the electropositive face of the galactose residue by Gal3C, highlighting the importance of the W181 CH−π interaction. For comparison, we generated Gal3C H158A and E184A variants, in which key hydrogen bonding interactions were disrupted. As expected, abrogating hydrogen bonding interactions disrupted lactose or LacNAc binding. These findings indicate that hydrogen bonding and CH−π interactions functionally cooperate to drive galectin-3-glycan binding.
To evaluate the relevance of these CH−π interactions on galectin-3 function, we assessed the variants’ capacity to interact with mucins and agglutinate mammalian red blood cells. These processes involve multivalent binding, so we hypothesized that they would serve as sensitive reporters of the contributions of different side-chain interactions. The H158A and E184A variants were weak binders, as expected from the importance of hydrogen bonding in glycan binding. Whereas WT Gal3C strongly bound porcine MUC2, MUC5ac, and MUC5b, interactions with the W181F, W181Y, W181H, and W181M variants were minimal. Similarly, the H158A and E184A variants minimally bound these mucins. Thus, disrupting the key W181 CH−π interactions was as deleterious as abrogating hydrogen bonding interactions. Indeed, full-length galectin-3 agglutinated mRBCs, but W181 variants bearing disrupted CH−π interactions did not. Thus, the W181 CH−π interactions are required for galectin-3 mucin and hemagglutination functions, emphasizing the physiological importance of this contribution.
Prior experimental analyses have suggested that CH−π interactions contribute relatively weak stabilization that ranges from 0.6 to 2 kcal/mol per monosaccharide–aromatic interaction depending on the structure of the glycan and aromatic partner.21 These and other studies have suggested that CH−π interactions are less favorable than hydrogen bonding interactions. Our MMGBSA calculations on snapshots sampled from extensive (i.e, 200 ns) molecular dynamics trajectories indicate that the W181 residue in native galectin-3 contributes 3.2 kcal/mol stabilization to lactose binding, which is a greater energetic contribution than that of H158 or E184, two key hydrogen bonding partners. Although MMGBSA may overestimate the binding contributions, it provides qualitative support for the importance of W181 interactions. These findings indicate that CH−π interactions can be more favorable than hydrogen bonding interactions and may contribute more to glycan binding than previously thought.
Our results build on previous studies and demonstrate the essentiality of the CH−π interactions in galectin-3 function. Our findings are consistent with those obtained by analyzing linear free energy relationships of the interaction of β-methyl galactose with substituted indoles in water.20 A larger, polarizable aromatic π system of higher electron density correlates with stronger Gal3C CH−π interactions with lactose. Histidine is an especially weak π donor system because of its electronics. Finally the lack of binding to the methionine variant highlights the previously observed scarcity of aliphatic residues in glycan binding sites.
Patients with biliary tract cancer and gastric adenocarcinoma were identified to possess somatic mutations corresponding to galectin-3 R162H and W181R variants, respectively.29 Our MMGBSA calculations found that R162 and W181 are the galectin-3 residues with the largest binding enthalpy contributions to lactose binding. Further, we found that recombinant W181R galectin-3 was not stably folded, suggesting that this mutation is deleterious to galectin-3 function. Thus, identification of a mutation leading to variation in a key hydrogen bonding residue (R162) or the residue essential for the galectin-3 CH−π interactions (W181) in cancer tissue suggests no functional galectin-3 would be produced. These findings provide additional evidence that these interactions are both required for glycan binding and emphasizes the essentiality of the W181 CH−π interactions for galectin-3 function. As functional galectin-3 is implicated in cancer, additional studies examining the consequences of these mutations on the production and maintenance of tumors are of great interest.
Glycomimetic inhibitors of galectin-3 function can potentially address numerous human diseases.55 Unlike hydrogen bonding interactions, CH−π interactions are not available to the carbohydrate ligand in solution. Thus, the entirety of the CH−π interaction energy contributes to the binding energy of the protein–carbohydrate interaction. Collectively, these observations suggest the potential utility of designing galectin-3 inhibitors by developing competitive ligands that form a stronger W181–glycan interactions relative to W181−β-galactose. Binding of galectin-3 to mucin glycoprotein MUC1 on epithelial cells increases epidermal growth factor receptor (EGFR) activation and signaling, thereby contributing to cancer progression.56 Further, galectin-3 leads to an upregulation of MUC2 in human colon cancer cells, suggesting a role in colon cancer metastasis.57 Our results reveal that the disruption of the galectin-3 CH−π interactions can inhibit galectin-3 mucin binding. This loss of function may prevent galectin-mediated tumorigenesis and metastasis in some cancers.
β-Galactose has the capacity to engage in especially strong CH−π interactions because H5 is strongly electropositive due to the inductive effect of the proximal ring oxygen and overlap of the σC5–H-5 bond with the σC4–O4* antibond.20 Further, CH−π interactions are enhanced by the β-galactose axial 4-hydroxyl group, which draws electron density away from the aliphatic protons on C3 and C5, increasing their acidity and enhancing the interaction with an aromatic system.20,24 Augmenting the CH−π interactions by further polarizing the C–H bond or engaging in edge-on stacking with the tryptophan π-system could enhance the ability of a glycomimetic ligand to bind a lectin that employs CH−π interactions to bind its glycan partner. From a protein engineering perspective, our results suggest that CH−π interactions can be exploited to engineer glycan affinity. This strategy is conducive for engineering binding to saccharide residues with strongly polarized CH bonds, including β-galactose, β-galactosamine, β-mannose, and β-mannosamine.
Conclusions
Our results indicate that CH−π interactions with galectin-3 W181 are required for glycan binding, mucin binding, and hemagglutination function. Abrogating CH−π interactions has a similarly deleterious effect as abolishing hydrogen bonding interactions, emphasizing the essentiality of these interactions in driving galectin-3 binding to lactose. Decreasing the strength of CH−π interactions dramatically impacts galectin-3′s ability to participate in mucin binding and hemagglutination, functions that depend on multivalent binding. Together, our findings may be harnessed to develop novel inhibitors of glycan-binding proteins including galectin-3 and engineer carbohydrate-binding proteins with differential glycan selectivity for a range of basic science and therapeutic applications.
Acknowledgments
This research was supported by the National Institute of Allergy and Infectious Diseases under grant R01 AI055258 (L.L.K) and the National Institute of Biomedical Imaging and Bioengineering R01 EB017755 (K.R.). This work was also supported by the U.S. Department of Energy, Office of Science, Office of Advanced Scientific Computing, Office of Basic Energy Sciences, via the Scientific Discovery through Advanced Computing (SciDAC) program (H.J.K.). ITC experiments were performed at the University of Wisconsin (UW)-Madison Biophysics Instrumentation Facility, which is supported by UW-Madison, NSF grant BIR-9512577, NIH grant S10 RR13790, the Massachusetts Institute of Technology Biophysical Instrumentation Facility, and the Center for Development of Therapeutics at the Broad Institute of MIT and Harvard. Microscopy experiments were performed at the Microscopy Core of the Koch Institute for Integrated Cancer Research at MIT, supported by National Cancer Institute grant P30-CA14051. We thank Prof. Barbara Imperiali (MIT) for helpful discussions and generous access to equipment and Elizabeth M. Ward for experimental assistance. We thank Dr. Amanda Peiffer for assistance with structural analysis using PyMol. R.C.D. thanks the Molecular Biosciences Training Grant (T32GM007215) at UW-Madison for support.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.4c00357.
Figures S1–S9, Tables S1–S5, materials and methods, supplementary DSF and ITC data, MD simulations, and MMGBSA energy decomposition for key galectin-3 residues (PDF)
Author Contributions
○ R.C.D. and R.S.C. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Pilobello K. T.; Mahal L. K. Deciphering the glycocode: the complexity and analytical challenge of glycomics. Curr. Opin Chem. Biol. 2007, 11, 300–305. 10.1016/j.cbpa.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Reily C.; Stewart T. J.; Renfrow M. B.; Novak J. Glycosylation in health and disease. Nat. Rev. Nephrol 2019, 15, 346–366. 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo K.; Marth J. D. Glycosylation in cellular mechanisms of health and disease. Cell 2006, 126, 855–867. 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Lis H.; Sharon N. Lectins: Carbohydrate-Specific Proteins That Mediate Cellular Recognition. Chem. Rev. 1998, 98, 637–674. 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- Tsaneva M.; Van Damme E. J. M. 130 years of Plant Lectin Research. Glycoconj J. 2020, 37, 533–551. 10.1007/s10719-020-09942-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon N.; Lis H. History of lectins: from hemagglutinins to biological recognition molecules. Glycobiology 2004, 14, 53R–62R. 10.1093/glycob/cwh122. [DOI] [PubMed] [Google Scholar]
- Wesener D. A.; Dugan A.; Kiessling L. L. Recognition of microbial glycans by soluble human lectins. Curr. Opin Struct Biol. 2017, 44, 168–178. 10.1016/j.sbi.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerke H.; Dias-Baruffi M.; Cummings R. D.; Arthur C. M.; Stowell S. R. Galectins: An Ancient Family of Carbohydrate Binding Proteins with Modern Functions. Methods Mol. Biol. 2022, 2442, 1–40. 10.1007/978-1-0716-2055-7_1. [DOI] [PubMed] [Google Scholar]
- Park A. M.; Hagiwara S.; Hsu D. K.; Liu F. T.; Yoshie O. Galectin-3 Plays an Important Role in Innate Immunity to Gastric Infection by Helicobacter pylori. Infect. Immun. 2016, 84, 1184–1193. 10.1128/IAI.01299-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.; Xu X.; Cheng H.; Miller M. C.; He Z.; Gu H.; Zhang Z.; Raz A.; Mayo K. H.; Tai G.; Zhou Y. Galectin-3 N-terminal tail prolines modulate cell activity and glycan-mediated oligomerization/phase separation. Proc. Natl. Acad. Sci. U. S. A. 2021, 118, 1–11. 10.1073/pnas.2021074118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciacchitano S.; Lavra L.; Morgante A.; Ulivieri A.; Magi F.; De Francesco G. P.; Bellotti C.; Salehi L. B.; Ricci A. Galectin-3: One Molecule for an Alphabet of Diseases, from A to Z. Int. J. Mol. Sci. 2018, 19, 379. 10.3390/ijms19020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouffette S.; Botez I.; De Ceuninck F. Targeting galectin-3 in inflammatory and fibrotic diseases. Trends Pharmacol. Sci. 2023, 44, 519–531. 10.1016/j.tips.2023.06.001. [DOI] [PubMed] [Google Scholar]
- Papaspyridonos M.; McNeill E.; de Bono J. P.; Smith A.; Burnand K. G.; Channon K. M.; Greaves D. R. Galectin-3 is an amplifier of inflammation in atherosclerotic plaque progression through macrophage activation and monocyte chemoattraction. Arterioscler Thromb Vasc Biol. 2008, 28, 433–440. 10.1161/ATVBAHA.107.159160. [DOI] [PubMed] [Google Scholar]
- Liu F. T.; Rabinovich G. A. Galectins as modulators of tumour progression. Nat. Rev. Cancer 2005, 5, 29–41. 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- Slack R. J.; Mills R.; Mackinnon A. C. The therapeutic potential of galectin-3 inhibition in fibrotic disease. Int. J. Biochem. Cell Biol. 2021, 130, 105881 10.1016/j.biocel.2020.105881. [DOI] [PubMed] [Google Scholar]
- Marino K. V.; Cagnoni A. J.; Croci D. O.; Rabinovich G. A. Targeting galectin-driven regulatory circuits in cancer and fibrosis. Nat. Rev. Drug Discov 2023, 22, 295–316. 10.1038/s41573-023-00636-2. [DOI] [PubMed] [Google Scholar]
- Biela A.; Nasief N. N.; Betz M.; Heine A.; Hangauer D.; Klebe G. Dissecting the hydrophobic effect on the molecular level: the role of water, enthalpy, and entropy in ligand binding to thermolysin. Angew. Chem., Int. Ed. Engl. 2013, 52, 1822–1828. 10.1002/anie.201208561. [DOI] [PubMed] [Google Scholar]
- Drickamer K. Making a fitting choice: common aspects of sugar-binding sites in plant and animal lectins. Structure 1997, 5, 465–468. 10.1016/S0969-2126(97)00202-5. [DOI] [PubMed] [Google Scholar]
- Weis W. I.; Drickamer K. Structural basis of lectin-carbohydrate recognition. Annu. Rev. Biochem. 1996, 65, 441–473. 10.1146/annurev.bi.65.070196.002301. [DOI] [PubMed] [Google Scholar]
- Hudson K. L.; Bartlett G. J.; Diehl R. C.; Agirre J.; Gallagher T.; Kiessling L. L.; Woolfson D. N. Carbohydrate-Aromatic Interactions in Proteins. J. Am. Chem. Soc. 2015, 137, 15152–15160. 10.1021/jacs.5b08424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asensio J. L.; Arda A.; Canada F. J.; Jimenez-Barbero J. Carbohydrate-aromatic interactions. Acc. Chem. Res. 2013, 46, 946–954. 10.1021/ar300024d. [DOI] [PubMed] [Google Scholar]
- Hsu C. H.; Park S.; Mortenson D. E.; Foley B. L.; Wang X.; Woods R. J.; Case D. A.; Powers E. T.; Wong C. H.; Dyson H. J.; Kelly J. W. The Dependence of Carbohydrate-Aromatic Strengths on the Structure of the Carbohydrate. J. Am. Chem. Soc. 2016, 138, 7636–7648. 10.1021/jacs.6b02879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser J.; Kozmon S.; Mishra D.; Hammerova Z.; Wimmerova M.; Koca J. The CH-pi Interaction in Protein-Carbohydrate Binding: Bioinformatics and In Vitro Quantification. Chemistry 2020, 26, 10769–10780. 10.1002/chem.202000593. [DOI] [PubMed] [Google Scholar]
- Kiessling L. L.; Diehl R. C. CH-pi Interactions in Glycan Recognition. ACS Chem. Biol. 2021, 16, 1884–1893. 10.1021/acschembio.1c00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. C.; Lin H. Y.; Tu Z.; Kuo Y. H.; Hsu S.-T. D.; Lin C. H. Dissecting the Structure-Activity Relationship of Galectin-Ligand Interactions. Int. J. Mol. Sci. 2018, 19, 392. 10.3390/ijms19020392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meynier C.; Guerlesquin F.; Roche P. Computational studies of human galectin-1: role of conserved tryptophan residue in stacking interaction with carbohydrate ligands. J. Biomol Struct Dyn 2009, 27, 49–58. 10.1080/07391102.2009.10507295. [DOI] [PubMed] [Google Scholar]
- Saraboji K.; Hakansson M.; Genheden S.; Diehl C.; Qvist J.; Weininger U.; Nilsson U. J.; Leffler H.; Ryde U.; Akke M.; Logan D. T. The carbohydrate-binding site in galectin-3 is preorganized to recognize a sugarlike framework of oxygens: ultra-high-resolution structures and water dynamics. Biochemistry 2012, 51, 296–306. 10.1021/bi201459p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M. C.; Yegorova S.; Pitteloud J. P.; Chavaroche A. E.; Andre S.; Arda A.; Minond D.; Jimenez-Barbero J.; Gabius H. J.; Cudic M. Thermodynamic Switch in Binding of Adhesion/Growth Regulatory Human Galectin-3 to Tumor-Associated TF Antigen (CD176) and MUC1 Glycopeptides. Biochemistry 2015, 54, 4462–4474. 10.1021/acs.biochem.5b00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. G.; Bamford S.; Jubb H. C.; Sondka Z.; Beare D. M.; Bindal N.; Boutselakis H.; Cole C. G.; Creatore C.; Dawson E.; et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D.; Ge H.; Sun J.; Gao Y.; Teng M.; Niu L. Crystal structure of the C-terminal conserved domain of human GRP, a galectin-related protein, reveals a function mode different from those of galectins. Proteins 2008, 71, 1582–1588. 10.1002/prot.22003. [DOI] [PubMed] [Google Scholar]
- Laughrey Z. R.; Kiehna S. E.; Riemen A. J.; Waters M. L. Carbohydrate-pi interactions: what are they worth?. J. Am. Chem. Soc. 2008, 130, 14625–14633. 10.1021/ja803960x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehna S. E.; Laughrey Z. R.; Waters M. L. Evaluation of a carbohydrate-pi interaction in a peptide model system. Chem. Commun. (Camb) 2007, 39, 4026–4028. 10.1039/b711431a. [DOI] [PubMed] [Google Scholar]
- Chen W.; Enck S.; Price J. L.; Powers D. L.; Powers E. T.; Wong C. H.; Dyson H. J.; Kelly J. W. Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins. J. Am. Chem. Soc. 2013, 135, 9877–9884. 10.1021/ja4040472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio M. The CH/pi hydrogen bond in chemistry. Conformation, supramolecules, optical resolution and interactions involving carbohydrates. Phys. Chem. Chem. Phys. 2011, 13, 13873–13900. 10.1039/c1cp20404a. [DOI] [PubMed] [Google Scholar]
- Montalvillo-Jimenez L.; Santana A. G.; Corzana F.; Jimenez-Oses G.; Jimenez-Barbero J.; Gomez A. M.; Asensio J. L. Impact of Aromatic Stacking on Glycoside Reactivity: Balancing CH/pi and Cation/pi Interactions for the Stabilization of Glycosyl-Oxocarbenium Ions. J. Am. Chem. Soc. 2019, 141, 13372–13384. 10.1021/jacs.9b03285. [DOI] [PubMed] [Google Scholar]
- del Carmen Fernandez-Alonso M.; Cañada F. J.; Jiménez-Barbero J.; Cuevas G. Molecular recognition of saccharides by proteins. Insights on the origin of the carbohydrate-aromatic interactions. J. Am. Chem. Soc. 2005, 127, 7379–7386. 10.1021/ja051020+. [DOI] [PubMed] [Google Scholar]
- Vandenbussche S.; Diaz D.; Fernandez-Alonso M. C.; Pan W.; Vincent S. P.; Cuevas G.; Canada F. J.; Jimenez-Barbero J.; Bartik K. Aromatic-carbohydrate interactions: an NMR and computational study of model systems. Chemistry 2008, 14, 7570–7578. 10.1002/chem.200800247. [DOI] [PubMed] [Google Scholar]
- Niesen F. H.; Berglund H.; Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc 2007, 2, 2212–2221. 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- Turnbull W. B.; Daranas A. H. On the value of c: can low affinity systems be studied by isothermal titration calorimetry?. J. Am. Chem. Soc. 2003, 125, 14859–14866. 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- Miller B. R. 3rd; McGee T. D. Jr.; Swails J. M.; Homeyer N.; Gohlke H.; Roitberg A. E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput 2012, 8, 3314–3321. 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- Pelaseyed T.; Bergstrom J. H.; Gustafsson J. K.; Ermund A.; Birchenough G. M.; Schutte A.; van der Post S.; Svensson F.; Rodriguez-Pineiro A. M.; Nystrom E. E.; et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014, 260, 8–20. 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler K. M.; Carcamo-Oyarce G.; Turner B. S.; Dellos-Nolan S.; Co J. Y.; Lehoux S.; Cummings R. D.; Wozniak D. J.; Ribbeck K. Mucin glycans attenuate the virulence of Pseudomonas aeruginosa in infection. Nat. Microbiol 2019, 4, 2146–2154. 10.1038/s41564-019-0581-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J.; Aoki K.; Turner B. S.; Lamont S.; Lehoux S.; Kavanaugh N.; Gulati M.; Valle Arevalo A.; Lawrence T. J.; Kim C. Y.; et al. Mucin O-glycans are natural inhibitors of Candida albicans pathogenicity. Nat. Chem. Biol. 2022, 18, 762–773. 10.1038/s41589-022-01035-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M.; Barua N.; Ip M. Mucin-degrading gut commensals isolated from healthy faecal donor suppress intestinal epithelial inflammation and regulate tight junction barrier function. Front. Immunol. 2022, 13, 1021094. 10.3389/fimmu.2022.1021094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarp M. A.; Clausen H. Mucin-type O-glycosylation and its potential use in drug and vaccine development. Biochim. Biophys. Acta 2008, 1780, 546–563. 10.1016/j.bbagen.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Leclaire C.; Lecointe K.; Gunning P. A.; Tribolo S.; Kavanaugh D. W.; Wittmann A.; Latousakis D.; MacKenzie D. A.; Kawasaki N.; Juge N. Molecular basis for intestinal mucin recognition by galectin-3 and C-type lectins. FASEB J. 2018, 32, 3301–3320. 10.1096/fj.201700619R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argüeso P.; Guzman-Aranguez A.; Mantelli F.; Cao Z.; Ricciuto J.; Panjwani N. Association of cell surface mucins with galectin-3 contributes to the ocular surface epithelial barrier. J. Biol. Chem. 2009, 284, 23037–23045. 10.1074/jbc.M109.033332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C. E.; Krupkin M.; Smith-Dupont K. B.; Wu C. M.; Bustos N. A.; Witten J.; Ribbeck K. Comparison of Physicochemical Properties of Native Mucus and Reconstituted Mucin Gels. Biomacromolecules 2023, 24, 628–639. 10.1021/acs.biomac.2c01016. [DOI] [PubMed] [Google Scholar]
- Liu F. T.; Hsu D. K.; Zuberi R. I.; Hill P. N.; Shenhav A.; Kuwabara I.; Chen S. S. Modulation of functional properties of galectin-3 by monoclonal antibodies binding to the non-lectin domains. Biochemistry 1996, 35, 6073–6079. 10.1021/bi952716q. [DOI] [PubMed] [Google Scholar]
- Zhang T.; Miller M. C.; Zheng Y.; Zhang Z.; Xue H.; Zhao D.; Su J.; Mayo K. H.; Zhou Y.; Tai G. Macromolecular assemblies of complex polysaccharides with galectin-3 and their synergistic effects on function. Biochem. J. 2017, 474, 3849–3868. 10.1042/BCJ20170143. [DOI] [PubMed] [Google Scholar]
- Ochieng J.; Green B.; Evans S.; James O.; Warfield P. Modulation of the biological functions of galectin-3 by matrix metalloproteinases. Biochim. Biophys. Acta 1998, 1379, 97–106. 10.1016/S0304-4165(97)00086-X. [DOI] [PubMed] [Google Scholar]
- Denecke J.; Kranz C.; Nimtz M.; Conradt H. S.; Brune T.; Heimpel H.; Marquardt T. Characterization of the N-glycosylation phenotype of erythrocyte membrane proteins in congenital dyserythropoietic anemia type II (CDA II/HEMPAS). Glycoconj J. 2008, 25, 375–382. 10.1007/s10719-007-9089-1. [DOI] [PubMed] [Google Scholar]
- Liao Y. H.; Teng M. S.; Juang J. J.; Chiang F. T.; Er L. K.; Wu S.; Ko Y. L. Genetic determinants of circulating galectin-3 levels in patients with coronary artery disease. Mol. Genet. Genomic Med. 2020, 8, e1370 10.1002/mgg3.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.; Zheng Y.; Xu D.; Sun Z.; Yang H.; Yin Q. Galectin-3: a key player in microglia-mediated neuroinflammation and Alzheimer’s disease. Cell Biosci. 2021, 11, 78. 10.1186/s13578-021-00592-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst B.; Magnani J. L. From carbohydrate leads to glycomimetic drugs. Nat. Rev. Drug Discov 2009, 8, 661–677. 10.1038/nrd2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piyush T.; Chacko A. R.; Sindrewicz P.; Hilkens J.; Rhodes J. M.; Yu L. G. Interaction of galectin-3 with MUC1 on cell surface promotes EGFR dimerization and activation in human epithelial cancer cells. Cell Death Differ. 2017, 24, 1937–1947. 10.1038/cdd.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas S. P.; Yunker C. K.; Sternberg L. R.; Byrd J. C.; Bresalier R. S. Expression of human intestinal mucin is modulated by the beta-galactoside binding protein galectin-3 in colon cancer. Gastroenterology 2002, 123, 817–826. 10.1053/gast.2002.35395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.