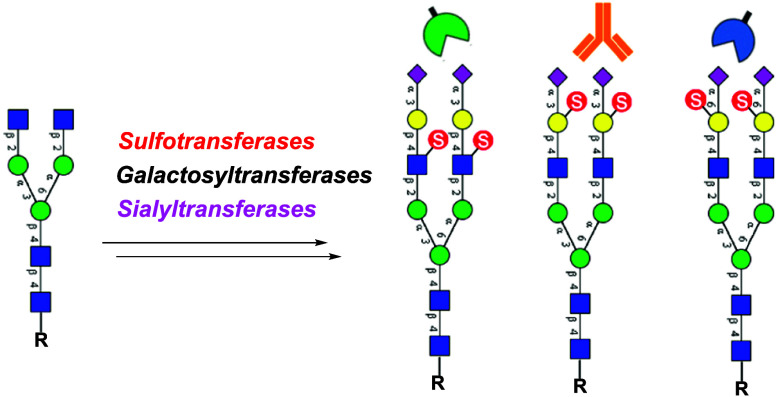
Abstract
Sulfated N-glycans are present in many glycoproteins, which are implicated in playing important roles in biological recognition processes. Here, we report the systematic chemoenzymatic synthesis of a library of sulfated and sialylated biantennary N-glycans and assess their binding to Siglecs and glycan-specific antibodies that recognize them as glycan ligands. The combined use of three human sulfotransferases, GlcNAc-6-O-sulfotransferase (CHST2), Gal-3-O-sulfotransferase (Gal3ST1), and keratan sulfate Gal-6-O-sulfotransferase (CHST1), resulted in asymmetric and symmetric branch-selective sulfation of the GlcNAc and/or Gal moieties of N-glycans. The extension of the sugar chain using α-2,3- and α-2,6-sialyltransferases afforded the sulfated and sialylated N-glycans. These synthetic glycans with different patterns of sulfation and sialylation were evaluated for binding to selected Siglecs and sulfoglycan-specific antibodies using glycan microarrays. The results confirm previously documented glycan-recognizing properties and further reveal novel specificities for these glycan-binding proteins, demonstrating the utility of the library for assessing the specificity of glycan-binding proteins recognizing sulfated and sialylated glycans.
Keywords: sulfated N-glycan, sulfotransferase, sulfation, chemoenzymatic synthesis, Siglecs, glycan array
Introduction
Sulfation is an important carbohydrate modification of the glycans on glycoproteins and glycolipids, which play key roles in biology through the recognition by glycan-binding proteins that recognize them as ligands.1 Sulfation of N-glycans in glycoproteins is elaborated by the action of sulfotransferases that are localized in the Golgi apparatus.2 Sulfotransferases comprise a family of enzymes that catalyze the transfer of a sulfate group from donor substrate 3′-phosphoadenosine-5′-phosphosulfate (PAPS) to acceptor glycoproteins. For complex-type N-glycans, the sulfate groups are usually attached to terminal Galβ1–4GlcNAc (LacNAc) sequences at the C-3 and C-6 positions of galactose and/or the C-6 position of N-acetylglucosamine.3,4 Sulfated N-glycans have been described in a variety of human glycoproteins including several pituitary hormones,5 leutropin,6 thyrotropin,7 Tamm–Horsfall glycoprotein,8,9 and urokinase.10 The roles of sulfated glycans often depend on their biological context and recognition by glycan-binding proteins. To date, they have been documented to play a role in the circulatory half-life of pituitary hormones,5 and to be recognized as ligands of glycan-specific antibodies,11−13 the leukocyte trafficking receptor l-selectin,14,15 and the sialic acid-binding immunoglobulin-like lectins (Siglecs) that modulate immune cell signaling.16,17
There are 14 human and 9 murine Siglecs that are widely expressed on white blood cells of the immune system.16 Many Siglecs are viewed as inhibitory immune checkpoints that contain immunoreceptor tyrosine-based inhibitory motifs (ITIMs) in their cytoplasmic domain. Several others, including human Siglec-14, -15, and -16, have activation functions, whereas Siglec-1 is an endocytic receptor with no signaling function.16,18 The specificity of Siglecs for synthetic sialoglycans has been investigated using glycan microarray and enzyme-linked immunosorbent assay (ELISA)-based assays.19−22 Several Siglecs exhibit preferential binding to sulfate-substituted sialoglycan ligands.12,20−23 For example, Siglec-8 expressed on eosinophils and mast cells was found to have narrow specificity for a sulfated ligand, Neu5Acα2–3[6S]Galβ1–4GlcNAc.22,24,25 In contrast, human Siglec-2 (CD22) preferentially binds a ligand with sulfate on the 6-position of GlcNAc (Neu5Acα2–6Galβ1–4[6S]GlcNAc).20 Although CD22 binds the same ligand without sulfate, the sulfated ligand exhibits higher avidity and is implicated as the physiologically relevant ligand.11,26 The dependence of some Siglecs on sulfated glycans has been observed using cell lines engineered to express specific sulfotransferases.17 Overexpression of the sulfotransferase that creates the 6S-Gal substitution, CHST1,27 was found to enhance binding of human Siglec-3 (CD33), Siglec-5, Siglec-7, Siglec-8, and Siglec-15, while overexpression of the sulfotransferase that creates the 6S-GlcNAc substitution, CHST2,28−31 enhanced binding of both murine and human CD22 and Siglec-9.17 In a related study involving engineered HEK293T cells deficient in N-linked, O-linked, or glycolipid glycans, knock-in of CHST1 enhanced binding of CD33 (Siglec-3), Siglec-7, Siglec-8, and Siglec-15 in an N-linked glycan-dependent manner.32
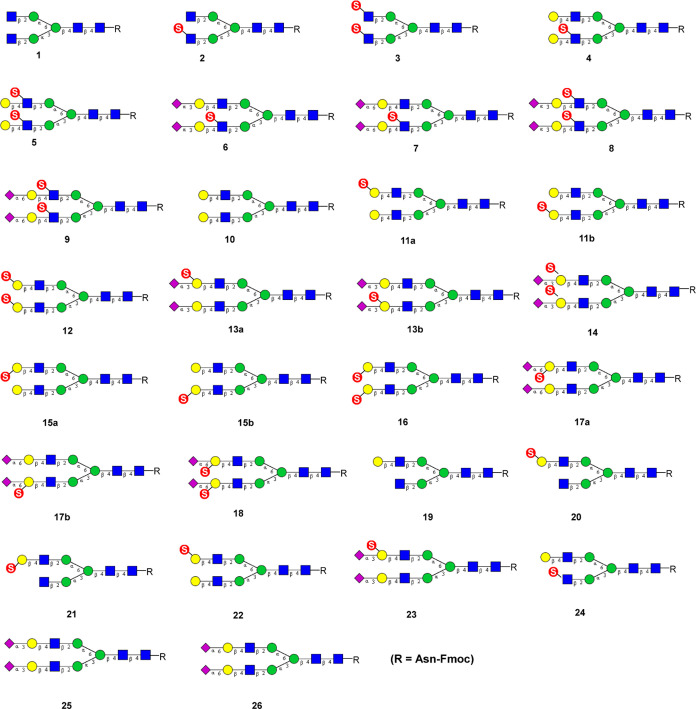
To date, the analysis of the specificity of Siglecs for sulfated-sialosides has used synthetic di- or trisaccharides of more complex natural glycans. Analysis of the specificity of Siglecs for sulfated N-glycans is lacking primarily due to the complexity of the synthesis. Given their biological importance, it would be desirable to evaluate the specificity of Siglecs and other sialic acid-binding proteins for sulfated and sialylated N-glycans. We recently reported the use of human N-acetylglucosamine-6-O-sulfotransferase (CHST2) for in vitro chemoenzymatic synthesis of several sulfated N-glycans, which demonstrated novel branch selectivity in the sulfation of N-glycans.30 Boons and co-workers have recently applied sulfotransferases CHST1 and CHST2 for the synthesis of an array of keratan sulfate oligosaccharides.33 Here, we describe a systematic chemoenzymatic synthesis of site-selectively sulfated and sialylated N-glycans using several sulfotransferases, galactosyl transferases, and sialyltransferases as the enzymes. We found that the combined use of the three human sulfotransferases, including GlcNAc-6-O-sulfotransferase (CHST2), Gal-3-O-sulfotransferase (Gal3ST1), and keratan sulfate Gal-6-O-sulfotransferase (CHST1), enabled the construction of a library of selectively sulfated and sialylated N-glycans (Figure 1). In addition, we used this library augmented with N-glycans of related structure without sulfate to create a glycan microarray to assess the specificity of Siglecs and sulfated-glycan-dependent antibodies. The results reveal novel aspects of their specificity for sulfated and sialylated biantennary N-glycans.
Figure 1.
Structures of the synthetic sulfated N-glycans and related reference compounds.
Results and Discussion
For the systematic synthesis of sulfated and sialylated N-glycans, we focused on the sulfotransferases that attach 6-O-sulfate to GlcNAc (CHST2), 6-O-sulfate to Gal (CHST1), or 3-O-sulfate to Gal (hGal3ST2). These were used in combination with sialyltransferases to elaborate the glycan library illustrated in Figure 1. The strategy for the synthesis of glycans employing each of the sulfotransferases is briefly described below. Most of the enzymatic sulfation reactions were carried out on several milligrams or tens of milligram scales.
Enzymatic Synthesis of GlcNAc-6-O-Sulfated Biantennary N-Glycans
The fluorenylmethyloxycarbonyl (Fmoc) group-tagged asparagine (Asn)-linked N-glycan core (1) was used as the key starting material for the synthesis of the sulfated biantennary N-glycans, which was prepared from the sialylated glycopeptide (SGP) as previously described.30 Introduction of the Fmoc group in the starting N-glycan facilitates the detection and purification of products by reversed-phase high-performance liquid chromatography (HPLC) in the respective enzymatic transformations. For the biosynthesis of sulfated biantennary N-glycans where the sulfate group was attached on the C-6 of the N-acetylglucosamine moiety, human N-acetylglucosamine-6-O-sulfotransferase 1 (CHST2) was used to catalyze sulfation. We have previously shown that CHST2 exhibited a high regioselectivity on the GlcNAc moiety at the Manα1–3Man arm.30 Thus, treatment of 1 with CHST2 and the donor substrate (PAPS) gave the monosulfated N-glycan derivative (2) in over 80% yield. Nevertheless, we found that disulfated N-glycan derivative (3) could be obtained when an excess amount of donor substrate PAPS and a prolonged incubation time were applied, albeit in a relatively low yield (31%) (Scheme 1). For the galactosylation, we expressed and used the bacterial β1,4-galactosyltransferase from Helicobacter pylori (HpGalT), as previous work suggested that HpGalT was able to transfer galactose from UDP-Gal to both 6-O-sulfated terminal GlcNAc and nonsulfated terminal GlcNAc moiety.34 As expected, the treatment of 2 with HpGalT in the presence of donor substrate UDP-Gal gave the fully galactosylated derivative (4) in which only the GlcNAc moiety on the Manα1–3Man branch (lower arm) was sulfated. We also found that the galactosylation of N-glycan carrying two terminal sulfated GlcNAc residues (3) by HpGalT was equally efficient, giving the corresponding galactosylated and disulfated N-glycan (5) in 90% yield. On the other hand, we have previously demonstrated that bacterial β1,4-galactosyltransferase from Neisseria meningitidis could selectively galactosylate the free terminal GlcNAc rather than a sulfated GlcNAc in 2, to give the monosulfated and monogalactosylated N-glycan derivative (24).30
Scheme 1. Enzymatic Synthesis of GlcNAc-6-O-Sulfated Biantennary N-Glycans (R = Asn-Fmoc).
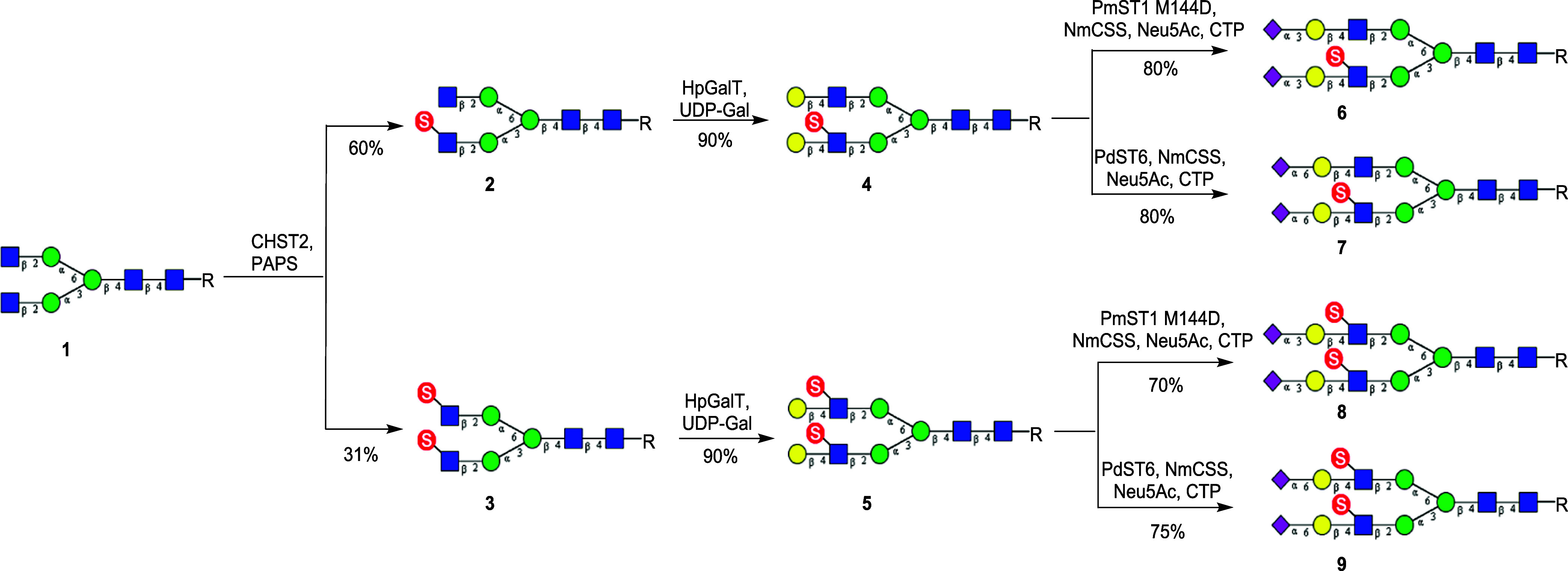
For terminal sialylation, the bacterial α2,3-sialyltransferase mutant (PmST1M144D) from Pasteurella multocida and the bacterial α2,6-sialyltransferase (PdST6) from Photobacterium damsela were employed for α2,3- and α2,6-sialylation, respectively. To decrease the expense of donor substrate CMP-Neu5Ac, CMP-sialic acid synthetase from N. meningitidis (NmCSS) was utilized for in situ generation of CMP-Neu5Ac in the presence of Neu5Ac and CTP. The sialylation process was closely monitored to avoid hydrolysis of the sialylation product, as the bacterial sialyltransferases also demonstrated product hydrolysis activity.35−38 Thus, sialylation of 4 with the PmST1M144D afforded the α2,3-sialylated product (6) in good yield, while sialylation of 4 with the sialyltransferase (PdST6) gave the corresponding α2,6-sialylated N-glycan (7) in high yield (Scheme 1). It was found that in both cases the internal GlcNAc sulfation did not affect the efficiency of the terminal sialyation. Chen and co-workers have previously shown that 6-O-sulfation of the GlcNAc moiety in LacNAc did not influence the efficacy for α2,3-sialylation by PmST1M144D in the synthesis of sulfated sialyl Lewis X oligosaccharide.39 Similarly, the α2,3-sialylated N-glycan (8) and the α2,6-sialylated N-glycan (9) were synthesized through the enzymatic sialylation of 5 using sialyltransferases PmST1M144D and PdST6, respectively (Scheme 1). The resulting sialylated and sulfated N-glycans were isolated by reversed-phase HPLC analysis and characterized by high-resolution mass spectrometry (HRMS) and nuclear magnet resonance (NMR) analysis.
Enzymatic Synthesis of Gal-6-O-Sulfated Biantennary N-Glycans
The keratan sulfate galactose-6-O-sulfotransferase (CHST1) is the key enzyme for the biosynthesis of keratan sulfate, which catalyzes the 6-O-sulfation of the galactose moiety in the context of the keratan sulfate backbone. It has been previously demonstrated that CHST1 can efficiently transfer a sulfate group to the C-6 of the internal galactose of sialylated LacNAc.27 Interestingly, we found that CHST1 also recognized the terminal galactose moieties in N-glycan (10), although the efficiency was relatively low in comparison with that of α2,3-sialylated LacNAc. Thus, treatment of the galactosylated N-glycan (10) with CHST1 in the presence of donor substrate PAPS gave a mixture of three sulfated N-glycans: The two monosulfated N-glycans (11) (50%) which were partially overlapping under HPLC and the disulfated N-glycan (12) (24%) that was readily separated by HPLC. In contrast to CHST2 which preferred the lower arm (the Manα1–3Man branch) GlcNAc moiety for 6-O-sulfation, we found that CHST1 preferred to add a sulfate group to the C-6 of the upper arm (the Manα1–6Man branch) galactose moiety in the N-glycan (10) (Scheme 2). The site selectivity in the sulfation of the two galactose moieties was confirmed by enzymatic transformation based on the substrate selectivity of three glycosidases, coupled with liquid chromatography–mass spectrometry (LC–MS) analysis (Supporting Information, Figures S2–S4). Briefly, the sulfated glycan (10) was treated with the BgaA β1,4-galactosidase, which only removed the nonsulfated galactose moiety. Subsequent treatment of the intermediate with β-N-acetylglucosaminidase S would remove only the exposed terminal GlcNAc moiety. Finally, treatment of the intermediate with the α1,2/3-mannosidase would tell if the Manα1–6Man branch or the Manα1–3Man branch would be exposed, as the α1,2/3-mannosidase would only remove the terminal mannose from a free Manα1–3Man branch. The results indicated that most species in the mixture lost a mannose moiety, suggesting that the CHST1 preferred to add a sulfate group to the C-6 of the upper arm (the Manα1–6Man branch) galactose moiety in the N-glycan (10), while a majority of the galactose moiety in the Manα1–3Man branch was free allowing the sequential trimming by three glycosidases (Figures S2–S4).
Scheme 2. Enzymatic Synthesis of Gal-6-O-Sulfated Biantennary N-Glycans (R = Asn-Fmoc).
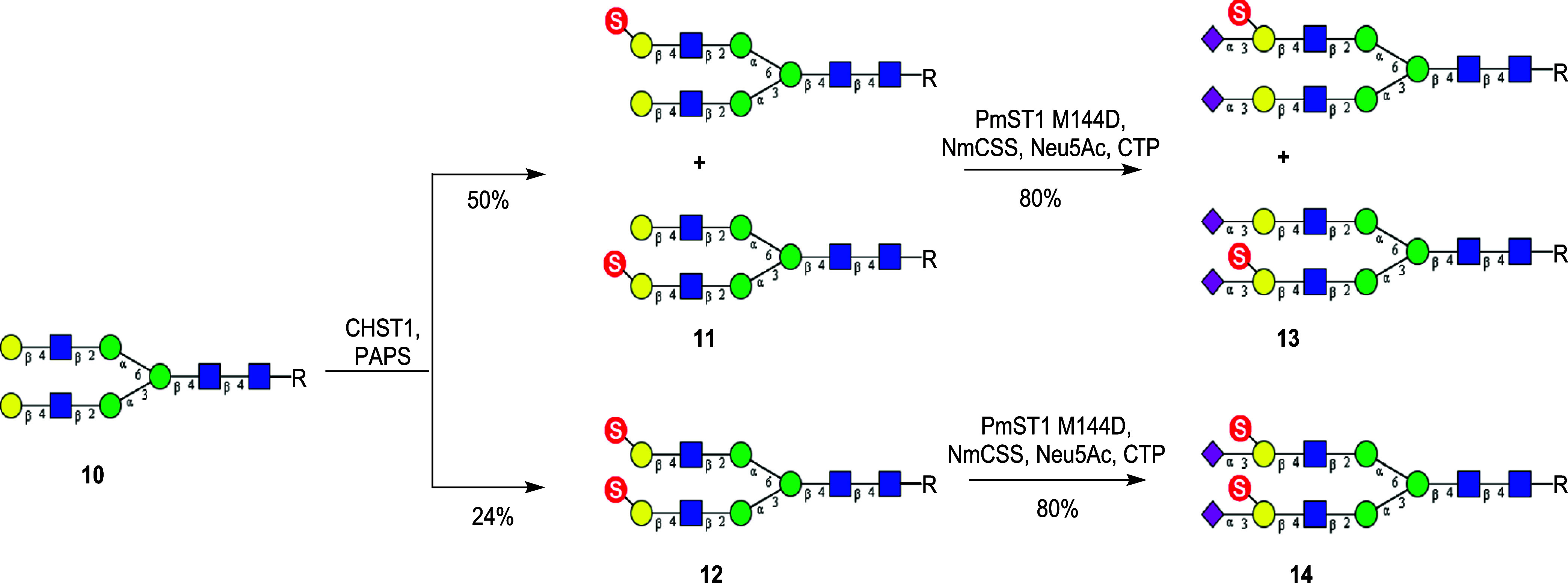
With compounds 11 and 12 in hand, a one-pot two-enzyme system containing PmST1M144D and NmCSS was leveraged to perform the terminal sialylation. It was found that PmST1M144D could efficiently sialylate both sulfated and nonsulfated terminal galactose moieties, affording the α2,3-sialylated N-glycans, 13 and 14, respectively. The substrate promiscuous property of this sialyltransferase was consistent with the previously reported observation.39
Enzymatic Synthesis of Gal-3-O-Sulfated Biantennary N-Glycans
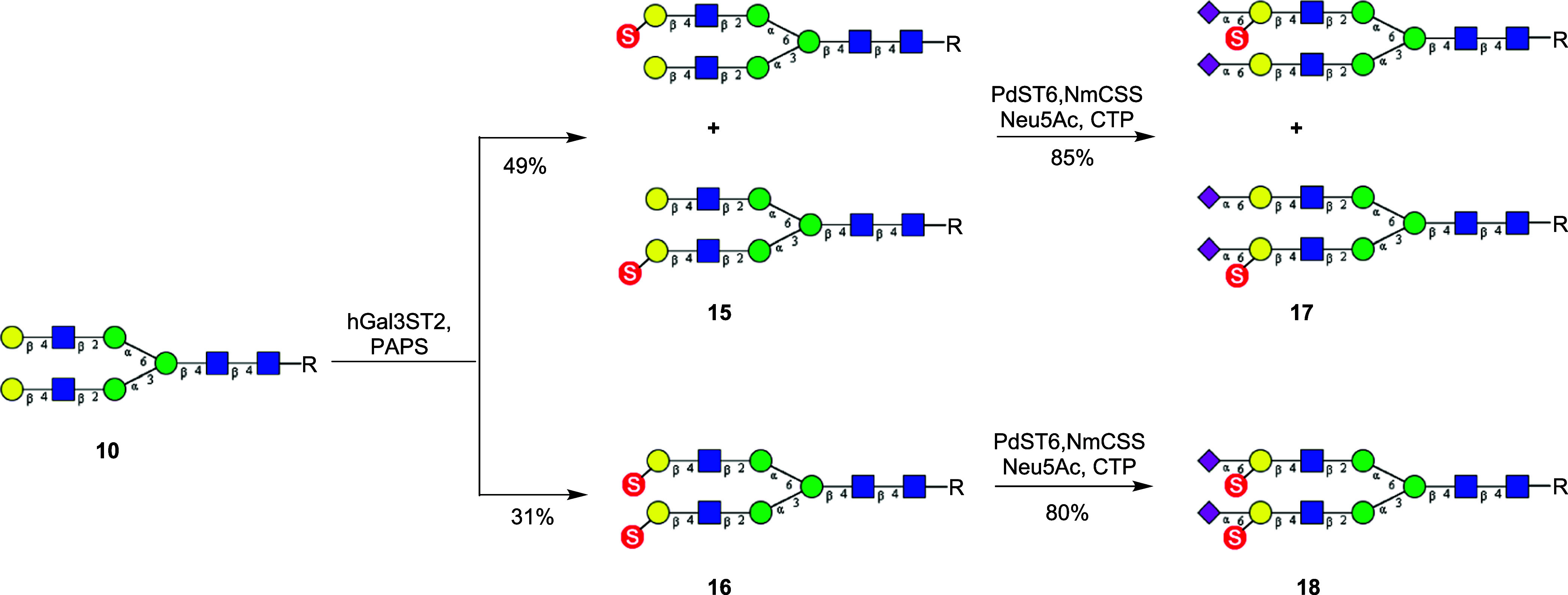
For the synthesis of Gal-3-O-sulfated biantennary N-glycans, we explored human galactose-3-O-sulfotransferase 2 (hGal3ST2) to catalyze the sulfation on C-3 of the terminal galactose moieties in the N-glycan (10). At the beginning, we tried to obtain the purified enzyme after expression in HEK293F cell lines. However, we found that the secretion level was extremely low.40 Therefore, we decided to use cell lysates as the source of the enzyme to perform the sulfation reactions. For the two terminal galactose moieties in N-glycan 10, we found that hGal3ST2 also had preference for the upper arm (the Manα1–6Man branch) galactose moiety for 3-O-sulfation, similar to the selectivity of CHST1. Thus, treatment of 10 with hGal3ST2 gave a mixture of two monosulfated (15) and the disulfated N-glycan derivative (16). The mono- and disulfated products could be easily separated by HPLC. However, an attempt to separate the two monosulfated isomers by HPLC failed. As a result, we used the mixtures of the two monosulfated isomers (15) for further sugar chain extension and subsequent binding studies. The ratio of the two isomers (upper arm/lower arm) was estimated to be 5:2, based on the sulfation selectivity analysis of the hGal3ST2-catalyzed reaction (Supporting Information, Figures S5–S7). Sialylation of 15 by sialyltransferase PdST6 provided the α2,6-sialylated N-glycan (17). Similarly, sialylation of 16 with sialyltransferase PdST6 gave the corresponding α2,6-sialylated N-glycan (18) in excellent yield, in which both internal galactose moieties were sulfated at the C-3 position (Scheme 3). It should be mentioned that this was the first report revealing that the PdST6 sialyltransferase could tolerate 3-O-sulfation of the galactose moiety for efficient α2,6-sialylation.
Scheme 3. Enzymatic Synthesis of Gal-3-O-Sulfated Biantennary N-Glycans (R = Asn-Fmoc).
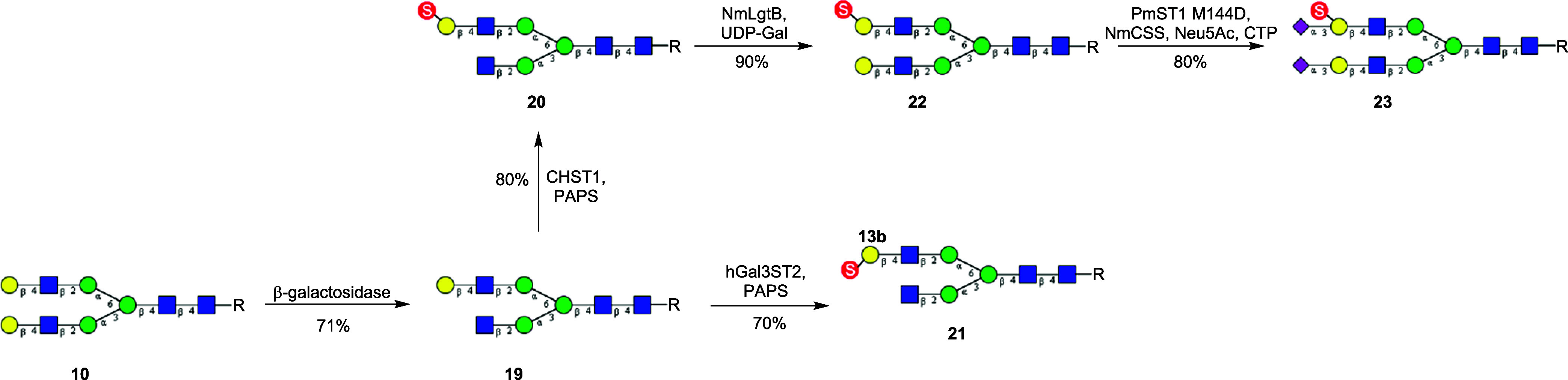
Enzymatic Synthesis of a Homogeneous Mono-Gal-6-O-Sulfated N-Glycans
Since the two isomeric mono-Gal-6-O-sulfated N-glycans (11) were difficult to separate by HPLC, we attempted to synthesize an isomer by an alternative route. First, we took the known selectivity of Escherichia coli β1,4-galactosidase to preferentially remove the lower arm terminal galactose in N-glycan 10 to obtain the monogalactosylated derivative (19). Next, 19 was sulfated by CHST1 and hGal3ST2 to give the Gal-6-O-sulfated and Gal-3-O-sulfated N-glycans, 20 and 21, respectively (Scheme 4). Then, bacterial β1,4-galactosyltransferase from N. meningitidis (NmLgtB) was used to add galactose back to the lower arm terminal GlcNAc to obtain the monosulfated N-glycan derivative (22). Finally, the α2,3-sialylation of 22 was achieved using PmST1M144D as the catalyst, affording the sialylated and monosulfated N-glycan (23) in excellent yield (Scheme 4).
Scheme 4. Enzymatic Synthesis of a Homogeneous Monosulfated Biantennary N-Glycan (R = Asn-Fmoc).
Construction of Sialylated and Sulfated N-Glycan Arrays
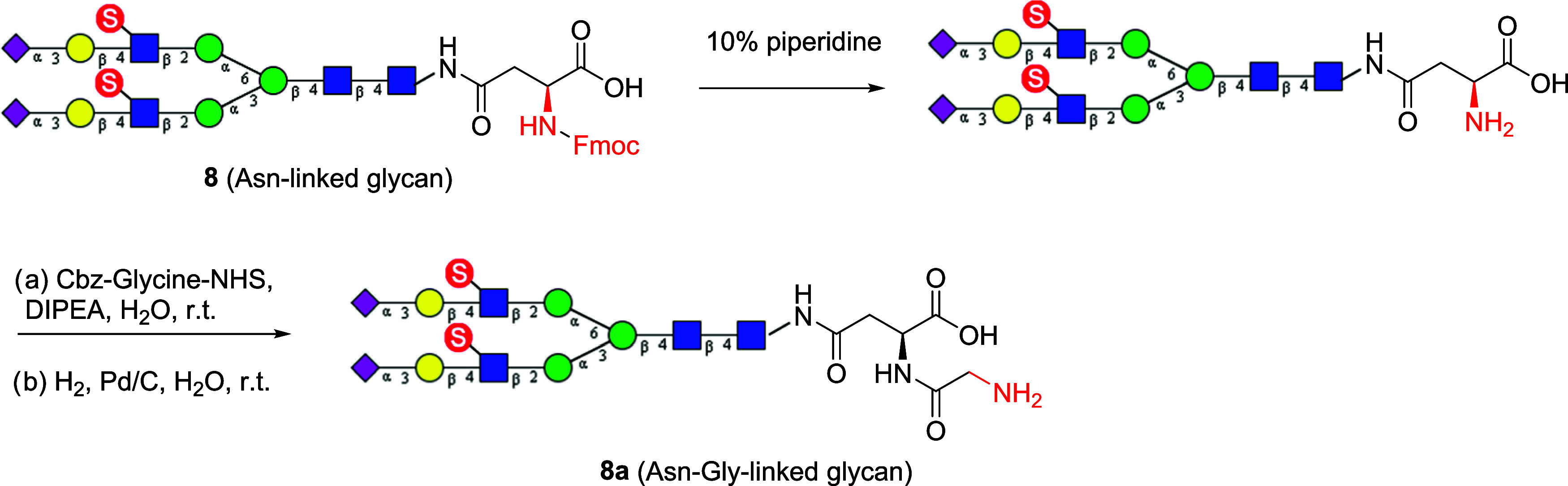
The library of sulfated and sialylated N-glycans in Figure 1 were synthesized in the Fmoc-protected form to facilitate the detection and separation of the enzymatic transformation products. For printing on NHS-activated slides to generate a glycan array, an amine functionality is required. The Fmoc was therefore removed by treatment with 10% piperidine in water. We have previously shown that Asn-terminated glycans printed inefficiently on NHS-activated glass slides even with a free amine moiety,41 probably due to steric hindrance. Thus, we decided to further functionalize the Asn moiety with a glycine moiety to generate a more flexible primary amine. Accordingly, the Asn linker was extended by coupling it with Cbz-protected glycine, followed by Pd-catalyzed hydrogenation, to provide the corresponding N-glycans with an Asn-Gly linker. The procedures were demonstrated using the sialylated and sulfated N-glycan (8) as an example (Scheme 5). We found that all of the Fmoc-protected N-glycans could be efficiently converted to the corresponding N-glycans carrying the Asn-Gly linker. Finally, these free primary amine-containing N-glycans were printed on N-hydroxysuccinimide (NHS) activated glass slides to generate the glycan microarray. We printed glycans with amine linkers at a concentration of 100 μM onto commercial NHS-functionalized glass slides. Concentration is a function of the commercial array and can vary somewhat from batch to batch. The details of the glycan array preparation and property were described in the supplementary glycan microarray document based on MIRAGE (Minimum Information Required for A Glycomics Experiment) guidelines42 (Supporting Information, Table S2).
Scheme 5. Installation of a Free Glycine at the Asn Moiety of the Sulfated N-Glycans for the Construction of Glycan Microarray.
For printing a glycan array, the N-glycan library was augmented with 55 synthetic glycans comprising linear glycan fragments with or without sulfate, and nonsulfated biantennary N-glycans.20,43 The latter included nonsulfated symmetrical biantennary glycans containing one, two, or three LacNAc (Galβ1–4GlcNAc) extensions terminated with Neu5Ac or Neu5Gc. The majority of these glycans were previously reported,20,43 with N-glycans terminated with Neu5Gc sialylated using CMP-Neu5Gc instead of CMP-Neu5Ac.43 This expanded library of 81 glycans (Table S1) was used for constructing the glycan array by printing on NHS-activated slides.43 Since N-glycans in Figure 1 have different numbers on the glycan array, both numbers are shown in Supporting Information, Table S1, and we will refer to the glycan array numbers in normal type (e.g., #28) and by adding the corresponding N-glycan library numbers in bold type as defined in the schemes (e.g., #28/25).
Analysis of the Specificity of Siglecs and Other Glycan-Binding Proteins for Sulfated N-Glycans
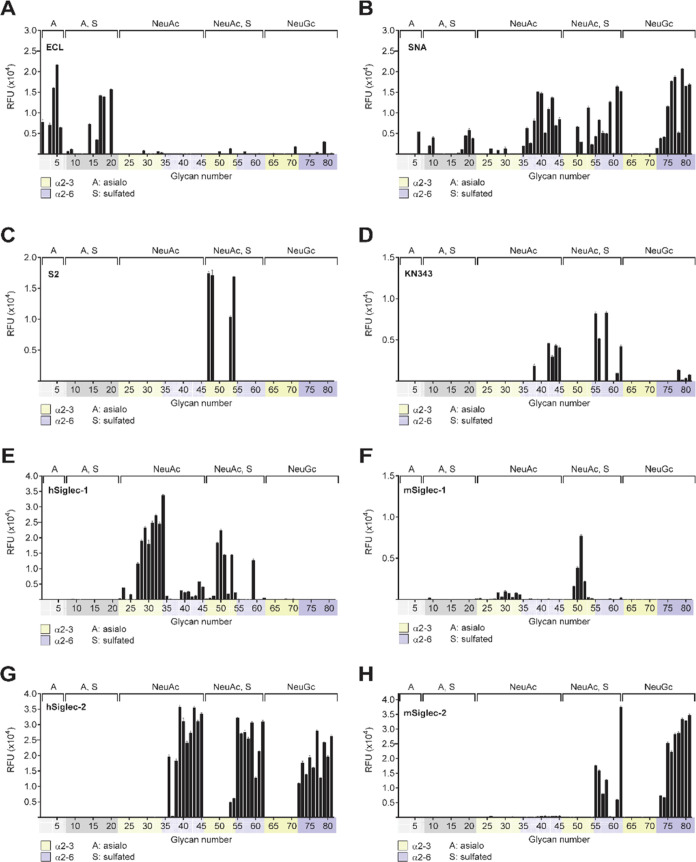
Glycan array analysis was performed on a variety of plant lectins, antiglycan antibodies, and selected members of the Siglec family (Figure 2). The glycan array MIRAGE data for Figure 2 are given in the Supporting Information (Table S3). Several plant lectins of known specificity were tested to validate the array. The Erythrina cristagalli lectin (ECL) is known to specifically bind the terminal sequence Galβ1–4GlcNAc,44 and that binding is blocked by sulfate or sialic acid substitutions on the terminal Gal.45 Indeed, on the glycan array (Figure 2A), ECL binds only to asialo-glycans with one or more terminal Galβ1–4GlcNAc sequences. Binding is blocked by 3S or 6S substitutions on the Gal as expected; however, we find that the 6S substitution on the GlcNAc is tolerated as evidenced by N-glycan #18(8). Notably, only a few of the sialylated glycans (#22–81) exhibit detectable binding by ECL, validating the high degree of sialylation on these glycans that block ECL binding.
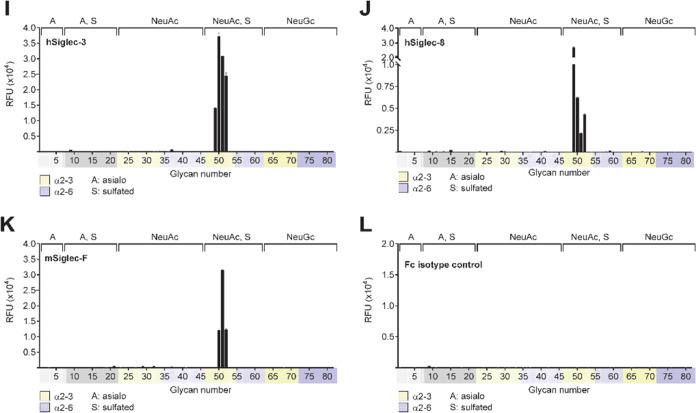
Figure 2.
Glycan array analysis of the specificity of several lectins, monoclonal antibodies, and Siglecs for sulfated, sialylated N-glycans. Analysis of the specificity of selected glycan-binding proteins was conducted with a glycan array of 81 glycans, as described in Experimental Procedures. Briefly, glycan-binding proteins and corresponding detection reagents were overlaid on printed glycan microarrays for 50 min at 22 °C in a humidified chamber. Biotinylated lectins ECL (A) and SNA (B) were overlaid at 10 and 1 μg/mL, respectively, in PBST and detected with AF488-conjugated streptavidin. Monoclonal antibodies S2 (C) and KN343 (D) were run at 10 and 1 μg/mL, respectively, in phosphate-buffered saline (PBS) and were detected with Phycoerythrin (PE)-conjugated αIgM secondary (5 and 0.5 μg/mL, respectively). Human Siglec-1 (E), murine Siglec-1 (F), human CD22 (G), murine CD22 (H), human CD33 (I), Siglec-8 (J), and Siglec-F (K) were analyzed at 50 μg/mL in PBS and detected with AF488-conjugated αIgG secondary (25 μg/mL). The human IgG1 isotype control (L) was also detected with AF488-conjugated αIgG secondary. Graphs depict the mean ± SEM of the relative fluorescence signal (RFU). Glycan structures corresponding to glycan numbers can be found in Supporting Information, Table S1. Controls for sialic acid dependence were assessed with corresponding conserved Arg mutants of each Siglec in Supporting Information, Figure S1. (The glycan array MIRAGE data for Figure S1 are given in the Supporting Information, Table S4).
Sambucus nigra agglutinin (SNA) is widely used to detect the presence of the terminal sequence Neu5Acα2–6Galβ1–4GlcNAc.46,47 Results in Figure 2B strongly underscore this specificity with SNA showing strong binding to nonsulfated glycans with the terminal Neu5Acα2–6Gal linkage and little or no binding to glycans containing sialic acid in the Neu5Acα2–3Gal linkage (#22–34; #63–71). But there are a couple of surprises. SNA binds to sulfated asialo-glycans with the [6S]Galβ1–4GlcNAc sequence (#9, #10, #19/20, #20/11ab, #21/12), and to glycans with both sialic acid and sulfate in the Neu5Acα2–3[6S]Gal sequence (#50/13ab, #51/23) or Neu5Acα2–3Galβ1–4[6S]GlcNAc sequence (#53/6, #54/8). There have been glycan array data sets showing that SNA binds to sulfated nonsialylated oligosaccharides as described in the Consortium of Functional Glycomics (CFG) link (SNA Vector CFG, https://carbogrove.org/showReport.php?resultID=1204). Thus, sulfate substitutions expand the specificity of SNA to selected asialo- and α2–3-sialylated glycans. We noted that #6, a nonsulfated, asialo, biantennary N-linked glycan, showed a blip in the glycan array. We believe this was an anomalous result, since two other structures identical except for the lack of a core fucose (#4, #5/10) showed no binding, as expected.
Two antiglycan antibodies reported to bind sialylated and sulfated glycans were analyzed. The monoclonal antibody S2 has been reported to be specific for the sequence Neu5Acα2–3Galβ1–4[6S]GlcNAc with or without Fucα1–3-linked to the GlcNAc.48 This strict specificity is clearly seen in Figure 2C with all four glycans containing this sulfated sequence, including the two synthetic N-glycans (#53/6, #54/8). The other monoclonal KN343 has been shown to have remarkable specificity for glycans containing the sequence Neu5Acα2–6Galβ1–4[6S]GlcNAc and has been used to implicate this sequence as the preferred ligand of CD22 on human B cells.12 Results in Figure 2D show that KN343 does indeed bind this sequence when expressed on N-glycans (#61/7, #62/9) and does not bind to the corresponding N-glycans missing either sialic acid (#17/4, #18/5) or sulfate (#39–41). However, KN343 surprisingly bound to nonsulfated N-glycans if the terminal Neu5Acα2–6Galβ1–4GlcNAc sequence was extended with one or two additional Galβ1–4GlcNAc repeats (#42–45), and bound weakly to the same glycans with Neu5Gc instead of Neu5Ac (#78–81). Thus, while the 6S substitution of GlcNAc enhances the affinity of KN343, there is not an absolute dependence on the sulfate for binding, and extended glycans with the Neu5Acα2–6Gal sequence can serve as ligands for this antibody.
Importantly, we assessed the impact of sulfate substitutions on the recognition of N-glycan ligands of several members of the Siglec family. Human and murine Siglec-1 exhibit strong homology and are expressed predominately in macrophages of both species. Both have been reported to bind preferentially to sialosides with the terminal Neu5Acα2–3Gal linkage.19,49 As seen in Figure 2E,F, both bind preferentially to N-glycans terminated with the Neu5Acα2–3Galβ1–4GlcNAc sequence (#28–30). However, they differ in their impact of sulfation. For hSiglec-1, the 6S-Gal and 6S-GlcNAc substitutions reduce avidity (compare #28/25 with #52/14 and #54/8, respectively) and for mSiglec-1, the 6S-Gal substitution increases avidity (compare #28/25 with #52/14).
Human CD22 (hCD22, Siglec-2) is known for its strong preference for ligands terminating in the Neu5Acα2–6Gal or Neu5Gc-α2–6Gal linkage with minimal binding to the Neu5Acα2–3Gal or Neu5Gcα2–3Gal linkages,16,18,26 and that a 6S-GlcNAc substitution somewhat enhances avidity.26 As shown in Figure 2G, hCD22 binds to N-glycans with the Neu5Acα2–6Galβ1–4[6S]GlcNAc sequence on one antenna (#61/7) or both antennae (#62/9) equivalently to the nonsulfated N-glycans (#39–45), in keeping with results obtained with trisaccharide fragments.26 In addition, however, we found that hCD22 could accommodate the 3S-Gal substitution on one (#59/17ab) or both antennae (#60/18). Murine CD22 (mCD22; Figure 2H) shows strong binding to glycans with the Neu5Gcα2–6Gal linkage (#72–81) and no binding to N-glycans with the Neu5Acα2–6Galβ1–4GlcNAc sequence (#39–45), in keeping with its known preference for Neu5Gc over Neu5Ac.16,18 Although there were no sulfated Neu5Gc containing N-glycans in the library, it is notable that 6S-GlcNAc substitution of N-glycans terminated with Neu5Acα2–6Galβ1–4GlcNAc showed significant binding (#55–58, #61/7, #62/9), in keeping with the reported increased avidity of mCD22 for 6S-GlcNAc substituted for glycan fragments containing this sequence.
Three additional Siglecs were tested on the sulfated N-glycan array. Human CD33 (Siglec-3; Figure 2I), human Siglec-8 (Figure 2J), and murine Siglec-F (Figure 2K) exhibited high specificity for N-glycans containing the terminal sequence Neu5Acα2–3[6S]Galβ1–4GlcNAc (#50/13ab, #51/23, #52/14). Remarkably, murine Siglec-F binds only N-glycans containing this sequence, while human Siglec-3 and Siglec-8 also bind the corresponding Neu5Acα2–3[6S]Galβ1–4GlcNAc trisaccharide (#49), and none of the three binds to the corresponding nonsulfated glycans (#23–34). Thus, particularly in the context of N-glycans, the 6S-Gal substitution strongly enhances the avidity for these three Siglecs. The strong enhancement of avidity by the 6S-Gal substitution has been previously documented for Siglec-8 and Siglec-F using glycan microarrays with the corresponding trisaccharide fragments,21,23,25,50 or glycolipids.22,50 The impact of the 6S-Gal substitution has also been observed by the enhancement of Siglec binding to cells overexpressing CHST1 that makes this substitution.17,32 It has been further demonstrated that suppressing the expression of complex-type N-glycans, as shown with MGAT-KO cells, results in a decrease in the binding of Siglec-3 and -8 to the cells, suggesting the involvement of [6S]Galβ1-4GlcNAc epitope on N-glycans in Siglec recognition.32 Our synthetic sulfated N-glycan array data presented here are consistent with the observation from the cell-based glycan array studies.
Conclusions
A systematic chemoenzymatic synthesis of site-selectively sulfated and sialylated N-glycans is achieved with the combined use of selected sulfotransferases, galactosyl transferases, and sialyltransferases as the enzymes. This study reveals novel glycan antenna selectivity of the GlcNAc-6-O-sulfotransferase (CHST2), the Gal-3-O-sulfotransferase (Gal3ST1), and the keratan sulfate Gal-6-O-sulfotransferase (CHST1), which enables site-selective sulfation of N-glycans. Glycan microarray analysis of the synthetic sulfated and sialylated N-glycans with a panel of Siglecs and sulfated-glycan-dependent antibodies reveals novel aspects of their specificity for sulfated and sialylated biantennary N-glycans, demonstrating the utility of the library of sulfated, sialylated N-glycans for analysis of the specificity of glycan-binding proteins. Future work should be directed to the synthesis of a broader library of structurally well-defined and selectively modified N-glycans for further investigation of the effects of sulfation on molecular recognition of Siglecs and related glycan-binding proteins.
Methods
Materials
Uridine 5′-diphospho-galactose (UDP-Gal), N-acetylneuraminic acid (Neu5Ac), and cytidine 5′-triphosphate disodium salt (CTP) were purchased from BIOSYNTH-Carbosynth. N-acetylglucosaminidase S and α1,2/3-mannosidase were purchased from New England Biolabs. E. coli β-galactosidase (G5635), Fmoc-Cl, Pronase, trifluoroacetic acid, and formic acid were purchased from Millipore-Sigma. Cbz-Gly-Osu was purchased from Sigma-Aldrich. All chemicals were used without further purification unless otherwise stated. CMP-sialic acid synthetase from N. meningitidis (NmCSS),51 α2,3-sialyltransferase mutant M144D from P. multocida (PmST1M144D),35,36 α2,6-sialyltransferase from P. damsel (PdST6),37 sialidase from Micromonospara Viridifaciens,52 ATP sulfurylase from Kluyveromyces lactis (KAST),53 APS kinase from Penicillium chrysogrenum (APSK),53 inorganic pyrophosphatase from E. coliK12(PPA),53 β1,4-galactosyltransferase from N. meningitidis (NmLgtB),34 and H. pylori (HpGalT)34 were expressed and purified following the respective, previously described procedures. PAPS was either kindly provided by Prof. Jian Liu or synthesized and purified as described previously.53
HPLC
Analytical reversed-phase HPLC was executed on a Thermo Scientific instrument with a YMC C18 column (5 μL, 4.6 × 250 mm2). The column was eluted with a linear gradient of 10–40% acetonitrile with 0.1% trifluoroacetic acid (v/v) for 30 min at a flow rate of 1 mL per min under monitoring by a UV detector at 266 nm. Preparative reversed-phase HPLC was carried out on a Waters 600 HPLC instrument with a YMC-Actus Triart C18 column (5 μm, 20 × 250 mm2) and SymmetryPrep C18 column (7 μm, 19 × 300 mm2). The YMC-Actus Triart C18 column was eluted with a linear gradient of 0–10% acetonitrile with 0.1% ammonium hydroxide (v/v) for 30 min at a flow rate of 10 mL/min under monitoring by a UV detector at 266 nm. The SymmetryPrep C18 column was eluted with a linear gradient of 10–50% acetonitrile with 0.1% formic acid (v/v) for 30 min at a flow rate of 10 mL per min under the monitoring by UV detector at 266 nm.
HRMS
High-resolution mass spectrometry was performed on an Exactive Plus Orbitrap Mass Spectrometer (Thermo Scientific).
NMR
1H, 13C, and H–C HSQC NMR spectra were recorded on either a 400 or 600 MHz spectrometer (Bruker, Tokyo, Japan) with deuterium oxide (D2O) as the solvent.
Protein Expression and Purification of Human Sulfotransferases
The coding region of human keratan sulfate galactose-6-O-sulfotransferase (CHST1), N-acetylglucosamine 6-O-sulfotransferase 1 (CHST2), and galactose-3-O-sulfotransferase 2 (hGal3ST2) was inserted into pGEn2 vector, which was used for transient expression of HEK293F cells as previously described.40,54 The protein purification of CHST1 and CHST2 was executed as described previously.30,40,54 For hGal3ST2, after transient expression, the culture media was spun down at 4000 rpm 4 °C for 10 min. The cell pellets were resuspended into 1× PBS buffer and sonicated for 10 min (10 s on, 10 s off). The cell lysate was utilized as the enzyme source.
Sulfation Position Determination
A 5 μL reaction mixture containing monosulfated G2-Asn-Fmoc (50 μg), 1× PBS buffer pH 7.4, and β1,4-galactosidase (13 μg, BgaA) was incubated at 37 °C for 1 h. The reaction was monitored by MALDI-TOF MS. When terminal galactose was completely hydrolyzed, β-N-acetylglucosaminidase S (4U) and 1× glycobuffer 1 (CaCl2 5 mM, sodium acetate 50 mM) were added into the reaction. The reaction was monitored by MALDI-TOF MS. When the N-acetylglucosamine was fully removed, 1× BSA and 32 U α-1,2/3-mannosidase were added into the reaction mixture. The product was analyzed by MALDI-TOF MS.
General Procedures for Enzymatic Reactions
CHST1-Catalyzed Sulfation at C6 Position on Galactose
Reaction mixtures containing G2-Asn-Fmoc (1 equiv), PAPS (2.5 equiv), Tris buffer (50 mM, pH 7), MgCl2 (5 mM), NaF (10 mM), ATP (0.1 mM), and CHST1 (2 wt %/wt relative to G2-Asn-Fmoc) were incubated at 37 °C overnight. The reactions were monitored by analytical HPLC and MALDI-ToF. More enzyme and donor substrate were added into the mixture for longer incubation until the majority of substrate was converted. The reaction was quenched by heating at 95 °C for 5 min. The products were purified by preparative HPLC. The fractions containing products were pooled and lyophilized for compound characterization and next step synthesis.
hGal3ST2-Catalyzed Sulfation at C3 Position on Galactose
Reaction mixtures containing G2-Asn-Fmoc (1 equiv), PAPS (2.5 equiv), Tris buffer (100 mM, pH 7.5), MgCl2 (10 mM), NaF (10 mM), ATP (1 mM) and appropriate amount of cell lysate of hGal3ST2 were incubated at 37 °C overnight. The reactions were monitored by analytical HPLC and MALDI-TOF. The reaction was quenched by heating at 95 °C for 5 min. The products were purified by preparative HPLC. The fractions containing products were pooled and lyophilized for compound characterization and next step synthesis.
CHST2-Catalyzed Sulfation at C6 Position on N-Acetylglucosamine
Reaction mixtures containing G0-Asn-Fmoc (1 equiv), PAPS (2.5 equiv), MES buffer (100 mM, pH 6), MgCl2 (5 mM), NaF (10 mM), ATP (1 mM) and CHST2 (2 wt %/wt relative to G0-Asn-Fmoc) were incubated at 37 °C overnight. The reactions were monitored by analytical HPLC and MALDI-TOF. The reaction was quenched by heating at 95 °C for 5 min. The products were purified by preparative HPLC. The fractions containing products were pooled and lyophilized for compound characterization and next step synthesis.
NmLgtB-Catalyzed β1,4-Galactosylation
Reaction mixtures containing acceptor substrates (1 equiv), UDP-Gal (1.5 equiv), Tris buffer (100 mM, pH 7.5), MnCl2 (10 mM), and NmLgtB (2 wt %/wt relative to acceptor substrate) were incubated at 37 °C. The reaction was monitored by analytical HPLC and MALDI-TOF. When the substrate was completely converted to the product, the reaction was quenched by heating to 95 °C for 5 min. The products were purified by preparative HPLC. The fractions containing products were pooled and lyophilized for compound characterization and next step synthesis.
HpGalT-Catalyzed β1,4-Galactosylation
Reaction mixtures containing acceptor substrates (1 equiv), UDP-Gal (2.5 equiv), Tris buffer (100 mM, pH 7.5), MgCl2 (20 mM), and HpGalT (5 wt %/wt relative to acceptor substrate) were incubated at 37 °C. The reactions were monitored by analytical HPLC and MALDI-TOF. When the substrate was completely converted to the product, the reactions were quenched by heating at 95 °C for 5 min. The products were purified by preparative HPLC. The fractions containing products were pooled and lyophilized for compound characterization and next step synthesis.
PmST1M144D-Catalyzed α2,3-Sialylation
Reaction mixtures containing acceptor substrates (1 equiv), Neu5Ac (3 equiv), CTP (3 equiv), Tris buffer (100 mM, pH 7.5), MgCl2 (20 mM), NmCSS (3% w/w relative to acceptor substrate), and PmST1M144D (3% w/w relative to acceptor substrate) were incubated at 37 °C. The reactions were closely monitored by analytical HPLC and MALDI-TOF. When most of the substrate was converted, the reaction was quenched by heating at 95 °C for 5 min. The products were purified by preparative HPLC. The fractions containing products were pooled and lyophilized for compound characterization.
PdST6-Catalyzed α2,6-Sialylation
Reaction mixtures containing acceptor substrates (1 equiv), Neu5Ac (3 equiv), CTP (3 equiv), Tris buffer (100 mM, pH 7.5), MgCl2 (20 mM), NmCSS (10% w/w relative to acceptor substrate), and PdST6 (10% w/w relative to acceptor substrate) were incubated at 37 °C. The reactions were closely monitored by analytical HPLC and MALDI-TOF. When the majority of starting material was converted, the reaction was quenched by heating at 95 °C for 5 min. The product was purified by preparative HPLC. The fractions containing products were pooled and lyophilized for compound characterization.
β-Galactosidase-Catalyzed Hydrolysis of Galactose
A reaction mixture containing G2-Asn-Fmoc, MgCl2 (5 mM), Tris buffer (100 mM, pH 7.3), and E. coli β-galactosidase (1 U per 2.5 μmol substrate) was incubated at 37 °C overnight. The reaction was monitored by analytical HPLC and MALDI-TOF. The reaction was quenched by heating at 95 °C for 5 min. The product was purified by preparative HPLC. The fractions containing products were pooled and lyophilized for compound characterization and next step synthesis.
General Procedure for Removing the Fmoc Group from the Sulfated N-Glycans and Coupling with Cbz-Gly-NHS to Provide the Asn-Gly-Linked Glycans for Glycan Arrays
To a solution of the respective Fmoc-protected N-Glycan (1–2 mg) in 90 μL of Milli-Q water was added 10 μL of piperidine, and the reaction mixture was incubated at room temperature and monitored by MALDI-TOF. When the reaction was completed, the solution was diluted with 600 μL of Milli-Q water before lyophilization. The residue was redissolved in 300 μL of Milli-Q water with adding sodium bicarbonate (2 equiv/acid group). The mixture was extracted with 2 × 300 μL chloroform to remove the released Fmoc. The water layer was dried with a SpeedVac vacuum concentrator and the resulting Asn-linked glycan was dissolved in distilled water (500 μL) containing Cbz-Gly-NHS (4 mol. equiv.) and DIPEA (4 mol. equiv.). The mixture was incubated at r.t. for 2 h, then the mixture was loaded on a C18 SPE cartridge and the column was eluted with 5–20% aqueous MeCN. The fractions containing the N-glycans were pooled and dried. The residue was dissolved in water (500 μL) containing palladium on charcoal (5 mg), which was subjected to hydrogenation to remove the Cbz protecting group under a hydrogen atmosphere (supplied with a hydrogen balloon) for 5 h. The mixture was filtered, and the filtrate was dried with a SpeedVac vacuum concentrator. The identity of the resulting Asn-Gly-linked glycan was confirmed by high-resolution ESI-TOF MS analysis. The respective Asn-Gly-linked glycan was used for printing glycan arrays without further purification.
Glycan Array Analysis
The N-glycan library comprising 81 compounds (Table S1) was printed on NHS-activated slides (Schott Minifab), in replicates of 6 as previously described.20,43 Analysis conducted for the binding of lectins, antibodies, and Siglec-Fc chimeras is briefly described below. Biotinylated lectins ECL and SNA (Vector Laboratories) were precomplexed in a 2:1 ratio with AF488-Streptavidin (Biolegend) at 10 and 1 μg/mL, respectively, in 200 μL of phosphate-buffered saline (PBS) for 15 min at 4 °C before being applied to the array. Monoclonal antibodies S2 (purified from hybridoma kindly provided by Romain Ballet from Stanford University, with permission from Hiroto Kawashima from Chiba University) and KN343 (kindly provided by Noriyuki Yuasa and Yuji Matsuzaki from Tokyo Chemical Industry) were prepared at 10 and 1 μg/mL, respectively, in 50 μL of PBS with 0.05% Tween-20 (PBST). The slides were washed in PBST prior to application of 5 μg/mL PE-conjugated anti-IgM (Jackson ImmunoResearch) in 300 μL of PBST. Siglec-Fc chimeras produced as previously described55 were run at 50 μg/mL precomplexed with 25 μg/mL AF488-conjugated anti-hIgG (Jackson Immuno Research Laboratories) in 50 μL PBS for 15 min at 4 °C prior to application to the slide. Coverslips were applied to arrays overlaid with 50 μL of protein to conserve material but were not required for reagents overlaid in 300 μL. Each protein was incubated on the array surface in a humidified chamber for 55 min at 22°C before sequential washing with PBST, PBS, and water prior to imaging. Fluorescent signal was detected and measured using an Innoscan 1100AL confocal microarray scanner and Mapix software (Innopsys). Each glycan was printed with 6 replicates. The mean and standard error of the mean (SEM) were calculated for four of the replicates after omitting the other two with the highest and lowest signals.
Acknowledgments
The authors acknowledge Prof. Jian Liu (The University of North Carolina) for providing the 3′-phosphoadenosine-5′-phosphosulfate (PAPS) and other members of the Wang Lab for technical support and helpful discussions. They wish to acknowledge the help and encouragement of Landon Edgar, Chika Kikuchi, Andrew Thompson, and Brett Garabedian. This work was supported in part by the National Institutes of Health (NIH grants R01GM080374 and R01AI155716 to L.-X.W.; R01AI114730 to J.C.P.; and R01GM130915 and R21HD110982 to K.W.M.).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.4c00307.
Glycan array compound structure and ID (Table S1); supplementary glycan microarray document based on MIRAGE guidelines (Table S2); glycan array MIRAGE data for Figure 2 (Table S3); glycan array MIRAGE data for Figure S1 (Table S4); glycan array analysis with the Siglec lectin conserved Arg mutants (Figure S1); characterization of the antennary selectivity of the terminal GlcNAc moieties in the sulfation catalyzed by CHST1 (Figures S2–S4); characterization of the antennary selectivity of the terminal galactose moieties in the sulfation catalyzed by hGal3ST2 (Figures S5–S7); NMR and HRMS data for all key sulfated N-glycans and sialylated/sulfated N-glycans; and HRMS analysis of the corresponding Asn-Gly-linked glycans (PDF)
Author Contributions
# K.H. and E.E.B. contributed equally to this work. CRediT: Kun Huang investigation, methodology, writing-original draft; Eleanor E. Bashian investigation, methodology, writing-review & editing; Guanghui Zong investigation, writing-review & editing; Corwin M. Nycholat investigation, methodology, writing-review & editing; Ryan McBride investigation, resources, writing-review & editing; Margaryta Gomozkova investigation, writing-review & editing; Shengyang C Wang investigation; Chin Huang resources, writing-review & editing; Digantkumar G. Chapla resources, writing-review & editing; Edward N. Schmidt investigation, resources; Matthew S. Macauley funding acquisition, resources, supervision, writing-review & editing; Kelley W Moremen funding acquisition, resources, supervision, writing-review & editing; James C Paulson conceptualization, funding acquisition, supervision, writing-review & editing; Lai-Xi Wang conceptualization, funding acquisition, supervision, writing-review & editing.
The authors declare no competing financial interest.
Supplementary Material
References
- Honke K.; Taniguchi N. Sulfotransferases and sulfated oligosaccharides. Med. Res. Rev. 2002, 22 (6), 637–654. 10.1002/med.10020. [DOI] [PubMed] [Google Scholar]
- Klaassen C. D.; Boles J. W. Sulfation and sulfotransferases 5: the importance of 3′-phosphoadenosine 5′-phosphosulfate (PAPS) in the regulation of sulfation. FASEB J. 1997, 11 (6), 404–418. 10.1096/fasebj.11.6.9194521. [DOI] [PubMed] [Google Scholar]
- Merkle R. K.; Elbein A. D.; Heifetz A. The effect of swainsonine and castanospermine on the sulfation of the oligosaccharide chains of N-linked glycoproteins. J. Biol. Chem. 1985, 260 (2), 1083–1089. 10.1016/S0021-9258(20)71210-X. [DOI] [PubMed] [Google Scholar]
- Barboza M.; Duschak V. G.; Fukuyama Y.; Nonami H.; Erra-Balsells R.; Cazzulo J. J.; Couto A. S. Structural analysis of the N-glycans of the major cysteine proteinase of Trypanosoma cruzi. Identification of sulfated high-mannose type oligosaccharides. FEBS J. 2005, 272 (15), 3803–3815. 10.1111/j.1742-4658.2005.04787.x. [DOI] [PubMed] [Google Scholar]
- Baenziger J. U.; Green E. D. Pituitary glycoprotein hormone oligosaccharides: structure, synthesis and function of the asparagine-linked oligosaccharides on lutropin, follitropin and thyrotropin. Biochim. Biophys. Acta, Rev. Biomembr. 1988, 947 (2), 287–306. 10.1016/0304-4157(88)90012-3. [DOI] [PubMed] [Google Scholar]
- Weisshaar G.; Hiyama J.; Renwick A. G. Site-specific N-glycosylation of human chorionic gonadotrophin--structural analysis of glycopeptides by one- and two-dimensional 1H NMR spectroscopy. Glycobiology 1991, 1 (4), 393–404. 10.1093/glycob/1.4.393. [DOI] [PubMed] [Google Scholar]
- Hiyama J.; Weisshaar G.; Renwick A. G. The asparagine-linked oligosaccharides at individual glycosylation sites in human thyrotrophin. Glycobiology 1992, 2 (5), 401–409. 10.1093/glycob/2.5.401. [DOI] [PubMed] [Google Scholar]
- van Rooijen J. J. M.; Kamerling J. P.; Vliegenthart J. F. Sulfated di-, tri- and tetraantennary N-glycans in human Tamm-Horsfall glycoprotein. Eur. J. Biochem. 1998, 256 (2), 471–487. 10.1046/j.1432-1327.1998.2560471.x. [DOI] [PubMed] [Google Scholar]
- Toma G.; Bates J. M. Jr.; Kumar S. Uromodulin (Tamm-Horsfall protein) is a leukocyte adhesion molecule. Biochem. Biophys. Res. Commun. 1994, 200 (1), 275–282. 10.1006/bbrc.1994.1445. [DOI] [PubMed] [Google Scholar]
- Bergwerff A. A.; Van Oostrum J.; Kamerling J. P.; Vliegenthart J. F. The major N-linked carbohydrate chains from human urokinase. The occurrence of 4-O-sulfated, (alpha 2–6)-sialylated or (alpha 1–3)-fucosylated N-acetylgalactosamine(beta 1–4)-N-acetylglucosamine elements. Eur. J. Biochem. 1995, 228 (3), 1009–1019. 10.1111/j.1432-1033.1995.tb20351.x. [DOI] [PubMed] [Google Scholar]
- Kannagi R.; Ohmori K.; Kimura N. Anti-oligosaccharide antibodies as tools for studying sulfated sialoglycoconjugate ligands for siglecs and selectins. Glycoconjugate J. 2009, 26 (8), 923–928. 10.1007/s10719-008-9122-z. [DOI] [PubMed] [Google Scholar]
- Kimura N.; Ohmori K.; Miyazaki K.; Izawa M.; Matsuzaki Y.; Yasuda Y.; Takematsu H.; Kozutsumi Y.; Moriyama A.; Kannagi R. Human B-lymphocytes express alpha2–6-sialylated 6-sulfo-N-acetyllactosamine serving as a preferred ligand for CD22/Siglec-2. J. Biol. Chem. 2007, 282 (44), 32200–32207. 10.1074/jbc.M702341200. [DOI] [PubMed] [Google Scholar]
- Nakashima K.; Sakai Y.; Hoshino H.; Umeda Y.; Kawashima H.; Sekido Y.; Ishizuka T.; Kobayashi M. Sulfated Glycans Recognized by S1Monoclonal Antibody can Serve as a Diagnostic Marker for Malignant Pleural Mesothelioma. Lung 2022, 200 (3), 339–346. 10.1007/s00408-022-00531-4. [DOI] [PubMed] [Google Scholar]
- Mir G. H.; Helin J.; Skarp K. P.; Cummings R. D.; Makitie A.; Renkonen R.; Leppanen A. Glycoforms of human endothelial CD34 that bind L-selectin carry sulfated sialyl Lewis x capped O- and N-glycans. Blood 2009, 114 (3), 733–741. 10.1182/blood-2009-03-210237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppanen A.; Parviainen V.; Ahola-Iivarinen E.; Kalkkinen N.; Cummings R. D. Human L-selectin preferentially binds synthetic glycosulfopeptides modeled after endoglycan and containing tyrosine sulfate residues and sialyl Lewis x in core 2 O-glycans. Glycobiology 2010, 20 (9), 1170–1185. 10.1093/glycob/cwq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S.; Paulson J. C. Siglecs as Immune Cell Checkpoints in Disease. Annu. Rev. Immunol. 2020, 38, 365–395. 10.1146/annurev-immunol-102419-035900. [DOI] [PubMed] [Google Scholar]
- Jung J.; Enterina J. R.; Bui D. T.; Mozaneh F.; Lin P. H.; Nitin; Kuo C. W.; Rodrigues E.; Bhattacherjee A.; Raeisimakiani P.; Daskhan G. C.; St Laurent C. D.; Khoo K. H.; Mahal L. K.; Zandberg W. F.; Huang X.; Klassen J. S.; Macauley M. S. Carbohydrate Sulfation As a Mechanism for Fine-Tuning Siglec Ligands. ACS Chem. Biol. 2021, 16 (11), 2673–2689. 10.1021/acschembio.1c00501. [DOI] [PubMed] [Google Scholar]
- Crocker P. R.; Paulson J. C.; Varki A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007, 7 (4), 255–266. 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- Blixt O.; Collins B. E.; van den Nieuwenhof I. M.; Crocker P. R.; Paulson J. C. Sialoside specificity of the siglec family assessed using novel multivalent probes: identification of potent inhibitors of myelin-associated glycoprotein. J. Biol. Chem. 2003, 278 (33), 31007–31019. 10.1074/jbc.M304331200. [DOI] [PubMed] [Google Scholar]
- Blixt O.; Head S.; Mondala T.; Scanlan C.; Huflejt M. E.; Alvarez R.; Bryan M. C.; Fazio F.; Calarese D.; Stevens J.; Razi N.; Stevens D. J.; Skehel J. J.; van Die I.; Burton D. R.; Wilson I. A.; Cummings R.; Bovin N.; Wong C. H.; Paulson J. C. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. U.S.A. 2004, 101 (49), 17033–17038. 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. S.; Alvarez R. A.; Mehta P.; Bovin N. V.; Blixt O.; White J. R.; Schnaar R. L. Glycan array screening reveals a candidate ligand for Siglec-8. J. Biol. Chem. 2005, 280 (6), 4307–4312. 10.1074/jbc.M412378200. [DOI] [PubMed] [Google Scholar]
- Campanero-Rhodes M. A.; Childs R. A.; Kiso M.; Komba S.; Le Narvor C.; Warren J.; Otto D.; Crocker P. R.; Feizi T. Carbohydrate microarrays reveal sulphation as a modulator of siglec binding. Biochem. Biophys. Res. Commun. 2006, 344 (4), 1141–1146. 10.1016/j.bbrc.2006.03.223. [DOI] [PubMed] [Google Scholar]
- Tateno H.; Crocker P. R.; Paulson J. C. Mouse Siglec-F and human Siglec-8 are functionally convergent paralogs that are selectively expressed on eosinophils and recognize 6′-sulfo-sialyl Lewis X as a preferred glycan ligand. Glycobiology 2005, 15 (11), 1125–1135. 10.1093/glycob/cwi097. [DOI] [PubMed] [Google Scholar]
- Kiwamoto T.; Brummet M. E.; Wu F.; Motari M. G.; Smith D. F.; Schnaar R. L.; Zhu Z.; Bochner B. S. Mice deficient in the St3gal3 gene product alpha2,3 sialyltransferase (ST3Gal-III) exhibit enhanced allergic eosinophilic airway inflammation. J. Allergy Clin. Immunol. 2014, 133 (1), 240–247e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gil A.; Porell R. N.; Fernandes S. M.; Wei Y.; Yu H.; Carroll D. J.; McBride R.; Paulson J. C.; Tiemeyer M.; Aoki K.; Bochner B. S.; Schnaar R. L. Sialylated keratan sulfate proteoglycans are Siglec-8 ligands in human airways. Glycobiology 2018, 28 (10), 786–801. 10.1093/glycob/cwy057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley M. S.; Kawasaki N.; Peng W.; Wang S. H.; He Y.; Arlian B. M.; McBride R.; Kannagi R.; Khoo K. H.; Paulson J. C. Unmasking of CD22 Co-receptor on Germinal Center B-cells Occurs by Alternative Mechanisms in Mouse and Man. J. Biol. Chem. 2015, 290 (50), 30066–30077. 10.1074/jbc.M115.691337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii T.; Fukuta M.; Habuchi O. Sulfation of sialyl N-acetyllactosamine oligosaccharides and fetuin oligosaccharides by keratan sulfate Gal-6-sulfotransferase. Glycobiology 2000, 10 (2), 203–211. 10.1093/glycob/10.2.203. [DOI] [PubMed] [Google Scholar]
- Akama T. O.; Misra A. K.; Hindsgaul O.; Fukuda M. N. Enzymatic synthesis in vitro of the disulfated disaccharide unit of corneal keratan sulfate. J. Biol. Chem. 2002, 277 (45), 42505–42513. 10.1074/jbc.M207412200. [DOI] [PubMed] [Google Scholar]
- Grunwell J. R.; Bertozzi C. R. Carbohydrate sulfotransferases of the GalNAc/Gal/GlcNAc6ST family. Biochemistry 2002, 41 (44), 13117–13126. 10.1021/bi020507h. [DOI] [PubMed] [Google Scholar]
- Huang K.; Li C.; Zong G.; Prabhu S. K.; Chapla D. G.; Moremen K. W.; Wang L. X. Site-selective sulfation of N-glycans by human GlcNAc-6-O-sulfotransferase 1 (CHST2) and chemoenzymatic synthesis of sulfated antibody glycoforms. Bioorg. Chem. 2022, 128, 106070 10.1016/j.bioorg.2022.106070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S. Y.; Hsiao C. T.; Izawa M.; Yusa A.; Ishida H.; Nakamura S.; Yagi H.; Kannagi R.; Khoo K. H. Distinct substrate specificities of human GlcNAc-6-sulfotransferases revealed by mass spectrometry-based sulfoglycomic analysis. J. Biol. Chem. 2018, 293 (39), 15163–15177. 10.1074/jbc.RA118.001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büll C.; Nason R.; Sun L.; Van Coillie J.; Madriz Sorensen D.; Moons S. J.; Yang Z.; Arbitman S.; Fernandes S. M.; Furukawa S.; McBride R.; Nycholat C. M.; Adema G. J.; Paulson J. C.; Schnaar R. L.; Boltje T. J.; Clausen H.; Narimatsu Y. Probing the binding specificities of human Siglecs by cell-based glycan arrays. Proc. Natl. Acad. Sci. U.S.A. 2021, 118 (17), e2026102118 10.1073/pnas.2026102118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Vos G. M.; Huang C.; Chapla D.; Kimpel A. L. M.; Moremen K. W.; de Vries R. P.; Boons G. J. Exploiting Substrate Specificities of 6-O-Sulfotransferases to Enzymatically Synthesize Keratan Sulfate Oligosaccharides. JACS Au 2023, 3 (11), 3155–3164. 10.1021/jacsau.3c00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau K.; Thon V.; Yu H.; Ding L.; Chen Y.; Muthana M. M.; Wong D.; Huang R.; Chen X. Highly efficient chemoenzymatic synthesis of beta1–4-linked galactosides with promiscuous bacterial beta1–4-galactosyltransferases. Chem. Commun. 2010, 46 (33), 6066–6068. 10.1039/c0cc01381a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiarto G.; Lau K.; Qu J.; Li Y.; Lim S.; Mu S.; Ames J. B.; Fisher A. J.; Chen X. A sialyltransferase mutant with decreased donor hydrolysis and reduced sialidase activities for directly sialylating LewisX. ACS Chem. Biol. 2012, 7 (7), 1232–1240. 10.1021/cb300125k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.; Chokhawala H.; Karpel R.; Yu H.; Wu B.; Zhang J.; Zhang Y.; Jia Q.; Chen X. A multifunctional Pasteurella multocida sialyltransferase: a powerful tool for the synthesis of sialoside libraries. J. Am. Chem. Soc. 2005, 127 (50), 17618–17619. 10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]
- Sun M.; Li Y.; Chokhawala H. A.; Henning R.; Chen X. N-Terminal 112 amino acid residues are not required for the sialyltransferase activity of Photobacterium damsela alpha2,6-sialyltransferase. Biotechnol. Lett. 2008, 30 (4), 671–676. 10.1007/s10529-007-9588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.; Huang S.; Chokhawala H.; Sun M.; Zheng H.; Chen X. Highly efficient chemoenzymatic synthesis of naturally occurring and non-natural alpha-2,6-linked sialosides: a P. damsela alpha-2,6-sialyltransferase with extremely flexible donor-substrate specificity. Angew. Chem., Int. Ed. 2006, 45 (24), 3938–3944. 10.1002/anie.200600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra A.; Yu H.; Tasnima N.; Muthana M. M.; Li Y.; Zeng J.; Kenyond N. J.; Louie A. Y.; Chen X. Systematic Chemoenzymatic Synthesis of O-Sulfated Sialyl Lewis x Antigens. Chem. Sci. 2016, 7 (4), 2827–2831. 10.1039/C5SC04104J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen K. W.; Ramiah A.; Stuart M.; Steel J.; Meng L.; Forouhar F.; Moniz H. A.; Gahlay G.; Gao Z.; Chapla D.; Wang S.; Yang J. Y.; Prabhakar P. K.; Johnson R.; Rosa M. D.; Geisler C.; Nairn A. V.; Seetharaman J.; Wu S. C.; Tong L.; Gilbert H. J.; LaBaer J.; Jarvis D. L. Expression system for structural and functional studies of human glycosylation enzymes. Nat. Chem. Biol. 2018, 14 (2), 156–162. 10.1038/nchembio.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nycholat C. M.; McBride R.; Ekiert D. C.; Xu R.; Rangarajan J.; Peng W.; Razi N.; Gilbert M.; Wakarchuk W.; Wilson I. A.; Paulson J. C. Recognition of sialylated poly-N-acetyllactosamine chains on N- and O-linked glycans by human and avian influenza A virus hemagglutinins. Angew. Chem., Int. Ed. 2012, 51 (20), 4860–4863. 10.1002/anie.201200596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; McBride R.; Stoll M.; Palma A. S.; Silva L.; Agravat S.; Aoki-Kinoshita K. F.; Campbell M. P.; Costello C. E.; Dell A.; Haslam S. M.; Karlsson N. G.; Khoo K. H.; Kolarich D.; Novotny M. V.; Packer N. H.; Ranzinger R.; Rapp E.; Rudd P. M.; Struwe W. B.; Tiemeyer M.; Wells L.; York W. S.; Zaia J.; Kettner C.; Paulson J. C.; Feizi T.; Smith D. F. The minimum information required for a glycomics experiment (MIRAGE) project: improving the standards for reporting glycan microarray-based data. Glycobiology 2017, 27 (4), 280–284. 10.1093/glycob/cww118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W.; de Vries R. P.; Grant O. C.; Thompson A. J.; McBride R.; Tsogtbaatar B.; Lee P. S.; Razi N.; Wilson I. A.; Woods R. J.; Paulson J. C. Recent H3N2 Viruses Have Evolved Specificity for Extended, Branched Human-type Receptors, Conferring Potential for Increased Avidity. Cell Host Microbe 2017, 21 (1), 23–34. 10.1016/j.chom.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura Y.; Nakamura-Tsuruta S.; Kominami J.; Sharon N.; Kasai K.; Hirabayashi J. Systematic comparison of oligosaccharide specificity of Ricinus communis agglutinin I and Erythrina lectins: a search by frontal affinity chromatography. J. Biochem. 2007, 142 (4), 459–469. 10.1093/jb/mvm153. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Yu G.; Han Z.; Yang B.; Hu Y.; Zhao X.; Wu J.; Lv Y.; Chai W. Specificities of Ricinus communis agglutinin 120 interaction with sulfated galactose. FEBS Lett. 2011, 585 (24), 3927–3934. 10.1016/j.febslet.2011.10.035. [DOI] [PubMed] [Google Scholar]
- Shang C.; Van Damme E. J. Comparative analysis of carbohydrate binding properties of Sambucus nigra lectins and ribosome-inactivating proteins. Glycoconjugate J. 2014, 31 (5), 345–354. 10.1007/s10719-014-9527-9. [DOI] [PubMed] [Google Scholar]
- Shibuya N.; Goldstein I. J.; Broekaert W. F.; Nsimba-Lubaki M.; Peeters B.; Peumans W. J. The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(alpha 2–6)Gal/GalNAc sequence. J. Biol. Chem. 1987, 262 (4), 1596–1601. 10.1016/S0021-9258(19)75677-4. [DOI] [PubMed] [Google Scholar]
- Hirakawa J.; Tsuboi K.; Sato K.; Kobayashi M.; Watanabe S.; Takakura A.; Imai Y.; Ito Y.; Fukuda M.; Kawashima H. Novel Anti-carbohydrate Antibodies Reveal the Cooperative Function of Sulfated N- and O-Glycans in Lymphocyte Homing. J. Biol. Chem. 2010, 285 (52), 40864–40878. 10.1074/jbc.M110.167296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman-Van der Linden E. C.; Varki A. New aspects of siglec binding specificities, including the significance of fucosylation and of the sialyl-Tn epitope. Sialic acid-binding immunoglobulin superfamily lectins. J. Biol. Chem. 2000, 275 (12), 8625–8632. 10.1074/jbc.275.12.8625. [DOI] [PubMed] [Google Scholar]
- Yu H.; Gonzalez-Gil A.; Wei Y.; Fernandes S. M.; Porell R. N.; Vajn K.; Paulson J. C.; Nycholat C. M.; Schnaar R. L. Siglec-8 and Siglec-9 binding specificities and endogenous airway ligand distributions and properties. Glycobiology 2017, 27 (7), 657–668. 10.1093/glycob/cwx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.; Yu H.; Karpel R.; Chen X. Chemoenzymatic synthesis of CMP-sialic acid derivatives by a one-pot two-enzyme system: comparison of substrate flexibility of three microbial CMP-sialic acid synthetases. Bioorg. Med. Chem. 2004, 12 (24), 6427–6435. 10.1016/j.bmc.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Watson J. N.; Dookhun V.; Borgford T. J.; Bennet A. J. Mutagenesis of the conserved active-site tyrosine changes a retaining sialidase into an inverting sialidase. Biochemistry 2003, 42 (43), 12682–12690. 10.1021/bi035396g. [DOI] [PubMed] [Google Scholar]
- Zhou X.; Chandarajoti K.; Pham T. Q.; Liu R.; Liu J. Expression of heparan sulfate sulfotransferases in Kluyveromyces lactis and preparation of 3′-phosphoadenosine-5′-phosphosulfate. Glycobiology 2011, 21 (6), 771–780. 10.1093/glycob/cwr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L.; Forouhar F.; Thieker D.; Gao Z.; Ramiah A.; Moniz H.; Xiang Y.; Seetharaman J.; Milaninia S.; Su M.; Bridger R.; Veillon L.; Azadi P.; Kornhaber G.; Wells L.; Montelione G. T.; Woods R. J.; Tong L.; Moremen K. W. Enzymatic basis for N-glycan sialylation: structure of rat alpha2,6-sialyltransferase (ST6GAL1) reveals conserved and unique features for glycan sialylation. J. Biol. Chem. 2013, 288 (48), 34680–34698. 10.1074/jbc.M113.519041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues E.; Jung J.; Park H.; Loo C.; Soukhtehzari S.; Kitova E. N.; Mozaneh F.; Daskhan G.; Schmidt E. N.; Aghanya V.; Sarkar S.; Streith L.; St Laurent C. D.; Nguyen L.; Julien J. P.; West L. J.; Williams K. C.; Klassen J. S.; Macauley M. S. A versatile soluble siglec scaffold for sensitive and quantitative detection of glycan ligands. Nat. Commun. 2020, 11 (1), 5091 10.1038/s41467-020-18907-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.